Abstract
Background
Mitochondrial functions are exploited in cancer and provide a validated therapeutic target. However, how this process is regulated has remained mostly elusive and the identification of new pathways that control mitochondrial integrity in cancer is an urgent priority.
Methods
We studied clinically-annotated patient series of primary and metastatic prostate cancer, representative cases of multiple myeloma (MM) and publicly available genetic databases. Gene regulation studies involved chromatin immunoprecipitation, PCR amplification and Western blotting of conditional Myc-expressing cell lines. Transient or stable gene silencing was used to quantify mitochondrial functions in bioenergetics, outer membrane permeability, Ca2+ homeostasis, redox balance and cell death. Tumorigenicity was assessed by cell proliferation, colony formation and xenograft tumour growth.
Findings
We identified Mitochondrial Fission Factor (MFF) as a novel transcriptional target of oncogenic Myc overexpressed in primary and metastatic cancer, compared to normal tissues. Biochemically, MFF isoforms, MFF1 and MFF2 associate with the Voltage-Dependent Anion Channel-1 (VDAC1) at the mitochondrial outer membrane, in vivo. Disruption of this complex by MFF silencing induces general collapse of mitochondrial functions with increased outer membrane permeability, loss of inner membrane potential, Ca2+ unbalance, bioenergetics defects and activation of cell death pathways. In turn, this inhibits tumour cell proliferation, suppresses colony formation and reduces xenograft tumour growth in mice.
Interpretation
An MFF-VDAC1 complex is a novel regulator of mitochondrial integrity and actionable therapeutic target in cancer.
Keywords: Mitochondria, MFF, Cell death, Tumour metabolism, VDAC1, Cancer therapy
Research in context.
Evidence before this study
Mitochondria are hubs of multiple signalling pathways that become invariably subverted in cancer to sustain energy production, aberrant cell survival and buffering of oxidative stress. Despite extensive efforts, the regulators of mitochondrial integrity in cancer have not been clearly identified, and this has hampered the development of new therapies.
Added value of this study
We identified a novel crosstalk between mitochondrial pathways exploited in cancer, including mitochondrial dynamics and the control of mitochondrial cell death. This involves a new molecular association between Mitochondrial Fission Factor (MFF) and the Voltage-Dependent Anion Channel-1 (VDAC1) at the organelle outer membrane, which is required to maintain cancer bioenergetics, prevent cell death and support tumour growth.
Implications of all the available evidence
Disruption of protein-protein interactions at the mitochondrial outer membrane is feasible and has generated successful cell-death modifying drugs currently in clinical practice. Such therapeutic targeting of an MFF-VDAC1 complex may provide a novel strategy to shut off multiple mitochondrial functions exploited for tumour progression.
Alt-text: Unlabelled Box
1. Introduction
As tumours tend to shift their metabolism towards glycolysis, even when oxygen is present [1], the importance of mitochondria in cancer has long remained uncertain [2]. This perception has now changed [3] as reprogramming of mitochondrial functions has emerged as a key driver of primary and metastatic disease [4] and validated therapeutic target [5]. The molecular requirements of this process are beginning to emerge, but mitochondrial-directed cancer therapies are still in infancy [6], there is a paucity of druggable mitochondrial targets [7], and even established drugs designed to (re)activate mitochondrial cell death [8], including Bcl2 antagonists [8] or Smac mimetics [9] have limited disease indications and often short-lived efficacy due to resistance mechanisms [10].
In this context, mitochondria are hubs of multiple cell death pathways [11]. How these processes are reprogrammed in cancer to promote aberrant cell survival is far from understood [12]. However, a sudden increase in the permeability of the mitochondrial outer membrane [13], mainly driven by pro-apoptotic Bcl2 family proteins [14], is a requirement of some cell death mechanisms culminating with the release of apoptogenic proteins in the cytosol [11]. Isoforms of Voltage-Dependent Anion Channel (VDAC) are key effectors of this process [15], maintaining ion and metabolite exchange [16] and overall mitochondrial homeostasis, including in cancer [17]. The mechanisms that connect increased mitochondrial outer membrane permeability to downstream events of inner membrane permeability transition, with loss of transmembrane potential, uncoupling of the TCA cycle and activation of regulated necrosis remain to be delineated [12], and the identity of key regulators in these responses has remained elusive [18].
Another mitochondrial pathway that is commonly exploited in cancer is mitochondrial dynamics [19], an adaptive process that continuously adjusts the size, shape and subcellular position of mitochondria to changes in cellular environment(s) [20]. In particular, mitochondrial fragmentation, or fission initiates with the binding of Dynamin-Related Protein-1 (Drp1) [21] to one of its receptors, Mitochondrial Fission Factor (MFF) on the mitochondrial outer membrane [22,23]. This process has been implicated in tumour progression [24], and there is evidence that mitochondrial fission couples to the machinery of organelle cell death, including apoptosis [25], mitochondrial integrity [26] and proapoptotic Bcl2 proteins [27], but a direct, mechanistic link between effectors of mitochondrial fission and mitochondrial cell death has not been identified.
In this study, we uncovered a novel molecular interface between mitochondrial dynamics and the control of mitochondrial integrity as a potential therapeutic target in disparate tumours.
2. Materials and methods
2.1. Patient samples
A clinically-annotated series of 192 patients with histologically confirmed diagnosis of primary prostate cancer [28] together with 17 prostate cancer metastases to different organs were used in this study (Supplementary Table S1). Archival tissues and clinical records were obtained from Fondazione IRCCS Ca’ Granda Hospital in Milan (Italy) under a protocol approved by the Institutional Review Boards (IRB) of Fondazione IRCCS Ca’ Granda-Ospedale Maggiore Policlinico (code 1381/11). Because of the retrospective nature of this study and the use of data anonymization practices, the need for written informed consent was waived. For patients with primary prostate cancer, epithelial tissue samples for normal prostate, prostatic intraepithelial neoplasia (PIN) and prostatic adenocarcinoma (AdCa) were arranged in tissue microarrays (TMA) blocks for immunohistochemical evaluations. Distant metastases were analysed as full sections. Representative bone marrow-derived samples of monoclonal gammopathy of uncertain significance (MGUS, n = 7) or multiple myeloma (n = 8) were also examined by immunohistochemistry. Patient samples for this study were collected between the years 2004–2006 (primary prostate cancer), 2012–2012 (metastatic prostate cancer) and 2009–2018 (multiple myeloma and MGUS).
2.2. Plasmids and gene silencing
cDNA clones encoding human MFF1, MFF5 or control vector were purchased from GeneCopoeia (Cat. n. EX-Z4766, EX-Z0675). An MFF2 cDNA was obtained from Addgene. Transfection of plasmid DNA (1 μg) was carried out using 2 μl X-Treme gene HP (Roche) for 24 h. For gene knockdown experiments, tumour cells were transfected with control, non-targeting small interfering RNA (siRNA) pool (D-001810, Dharmacon) or specific siRNA pools targeting MFF (Santa Cruz#sc-94736) or individual MFF-directed siRNA sequences (Santa Cruz #Sc-94,36-A and C). An additional siRNA sequence targeting MFF (SantaCruz #SC-94736-A: 5′-GAACAAAGAACGUGCUAAAUUUU-3′) was synthesized by Dharmacon. Cells were transfected with the various siRNA (30–60 nM) in the presence of Lipofectamine RNAiMAX (Invitrogen) at a 1:1 ratio (vol siRNA 20 μM:vol Lipofectamine RNAiMAX) and processed as described [29]. Two independent shRNA sequences were also used for targeting human MFF: TRCN0000167581 and TRCN0000343573 (Sigma Aldrich). An empty pLKO-based lentivirus was used as control. Individual clones of PC3 and DU145 cells stably expressing MFF-directed shRNA sequences were generated by infection with lentiviral particles, followed by a 2-week selection in the presence of puromycin at 2 μg/ml.
2.3. Cells and cell culture
Human prostate adenocarcinoma (LNCaP, C4-2, C4-2B, PC3 and DU145), normal prostatic epithelial (RWPE-1) and human glioblastoma (LN229) cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA), and maintained in culture according to the supplier's specifications. Benign prostatic hyperplasia (BPH-1) cells were a gift from Dr. Simon Hayward (Vanderbilt University, Nashville, TN) and primary human foreskin fibroblasts (HFF) were a gift from Dr. Meenhard Herlyn (The Wistar Institute, Philadelphia, PA). Neuroblastoma SHEP21N, SHEP21-NMycER cells containing a conditionally-regulated N-Myc transgene were as described [30] and used in recent studies of mitochondrial reprogramming in cancer [29]. In these cells, treatment with 50 ng/ml doxycycline (Dox) for 48 h suppresses N-Myc expression, whereas addition of 4-hydroxytamoxifen (4OHT, 0.5 μg/ml) results in strong N-Myc induction. Cell passaging was limited to <40 passages from receipt and cell lines were authenticated by STR profiling with AmpFlSTR Identifiler PCR Amplification Kit (Life Technologies) at the Wistar Institute's Genomics Shared Resource. Mycoplasma free-cultures were confirmed at the beginning of the studies, and every 2 months afterwards, by direct PCR amplification using Bioo Scientific Mycoplasma Primer Sets (cat. #375501) and Hot Start polymerase (QIAGEN).
2.4. Chromatin immunoprecipitation (ChIP)
PC3 cells transfected with siCtrl or Myc-directed siRNA (siMyc) for 72 h were used for ChIP experiments, as described [29] and incubated with non-binding rabbit IgG or a rabbit monoclonal antibody to Myc (Abcam #Ab32072). Real-time PCR amplification of chromatin fragments was carried out using SYBR green master mix (Applied Biosystems) on an ABI7500 sequence detection system.
2.5. Immunofluorescence and confocal microscopy
PC3 or DU145 cells transfected with siCtrl or MFF-directed siRNA (siMFF), or alternatively, expressing control plasmid or MFF cDNA were fixed in formalin/PBS (4% final concentration) and processed for immunofluorescence with primary antibodies against MFF (1:100) or MTCO2 (mitochondrial cytochrome c oxidase subunit II, 1:500) as described [29]. Secondary antibodies conjugated to Alexa488, TRITC or Alexa633 were used. F-actin was stained with phalloidin Alexa (1:200 dilution). After washes, slides were analysed by Z-stack imaging and confocal microscopy (SPF5 II, Leica). At least 70 cells per sample were analysed and mitochondrial morphologies were classified as fragmented (individual round- or rod-shaped organelles, >80% displaying an axial length of <4 μm), intermediate (majority <4 μm), or tubular (often interconnected in branched networks, >80% displaying a length of >4 μm).
2.6. Immunoprecipitation (IP)
Cell extracts prepared in 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA containing 1% CHAPS, EDTA-free Protease Inhibitor Cocktail (Sigma-Aldrich) and Phosphatase Inhibitor Cocktail PhosSTOP (Roche) were immunoprecipitated with anti-Flag-conjugated beads (Sigma-Aldrich) and processed as described [31].
2.7. Mitochondrial outer membrane permeability
PC3 cells (3 × 105) transfected with siCtrl or siMFF were stained with calcein (0.01 μM) and cobalt chloride (0.4 μM) (MitoProbe Transition Pore Assay, Molecular Probes, cat# M34153) for 15 min in HBSS (with calcium and without phenol red), washed in PBS, pH 7.4, and analysed on a FACS Celesta flow cytometer at 488 nm excitation and emission filters. Intact cells were gated in the FSC/SSC plot to exclude small debris.
2.8. Mitochondrial membrane potential
Normal prostatic epithelial RWPE1 or BPH-1 cells or prostate cancer PC3 or DU145 cells were transfected with siCtrl or siMFF and analysed on a FACS Calibur flow cytometer, with the TMRE signal as FL1. Intact cells were gated in the FSC/SSC plot to exclude small debris. The resulting FL1 data were plotted on a histogram.
2.9. Cellular respiration and mitochondrial ROS
PC3 or DU145 cells were transfected with siCtrl or siMFF and analysed after 48 h for Oxygen Consumption Rates (OCR) or Extracellular Acidification Rates (ECAR) using an Extracellular Flux System 24 Instrument (Seahorse Bioscience, Billerica, MD) using 2.5 × 104 cells plated in each well of a Seahorse XF24 cell culture plate (100 μl). After 4 h 150 μl of media was added to each well, and the cells were grown for 24 h at 37 °C in 5% CO2. The media was then exchanged with unbuffered DMEM XF assay media (Seahorse Bioscience) supplemented with 2 mM glutaMAX, 1 mM sodium pyruvate and 5 mM glucose (pH 7.4 at 37 °C) for 30 min at 37 °C and ~0.04% CO2 before the experiment. OCR was monitored in basal condition (before any addition) and after sequential addition of oligomycin (1.25 μM), FCCP (0.4 μM), and antimycin plus rotenone (0.25 μM), all dissolved in DMSO. OCR was quantified after three cycles of mixing (150 s), waiting (120 s), and measuring (210 s). This cycle was repeated following each injection. In some experiments, PC3 cells transfected with siCtrl or siMFF were incubated with 5 μM MitoSOX Red (Life Technology) for 10 min in complete medium, washed three times in warm PBS and counted. Ten thousand stained cells in 100 μl of PBS were analysed on a microplate florescent reader (Ex/Em = 510/580 nm, Molecular Devices). Unlabelled cells were used as basal control.
2.10. Calcium measurements
PC3 cells (3 × 105) transfected with siCtrl or siMFF were stained with calcium-sensitive dyes to determine changes in calcium concentrations in cytosol, mitochondria and endoplasmic reticulum (ER). Calcium levels in cytosol and ER were measured by staining the cells with FLUO3-AM (1 μM) and FLUO-5 N (5 μM), respectively for 1 h, washed in PBS, pH 7.4, and analysed on LSR18 flow cytometer at 506/526 nm excitation and emission filters, respectively. Rhod2-AM (10 μM) was used to measure calcium levels in mitochondria. For these experiments, cells were stained for 1 h and analysed by flow cytometry at 552/581 nm excitation and emission filters respectively. Intact cells were gated in the FSC/SSC plot to exclude small debris.
2.11. Mitophagy assay
Mitophagy was examined using a FACS-based analysis of mitochondrial targeted mKeima-Red fluorescence reporter (Addgene, cat. #56018). PC3 cells stably expressing mKeima were transfected with siCtrl or siMFF for 72 h. Cells were detached by trypsin treatment, washed and suspended in PBS followed by analysis on an LSR 18 flow cytometer at 405 and 561 nm lasers and 610/20 filters. Intact cells were gated in the FSC/SSC plot to exclude small debris.
2.12. Cell death
PC3 or DU145 cells (1 × 106) were transfected with siCtrl or siMFF, labelled for Annexin V and propidium iodide (PI) (BD Biosciences) and analysed by multiparametric flow cytometry. Alternatively, mitochondria-associated changes in cell viability were quantified by an MTT assay or Trypan blue dye exclusion assay and light microscopy. In some experiments, PC3 cells transfected with siCtrl or siMFF were analysed for proteolytic processing of PARP with or without stress stimuli, H2O2, nutrient deprivation or chemotherapeutic drugs, doxorubicin or etoposide, by Western blotting.
2.13. Colony formation assay
Four hundred PC3 or DU145 cells stably expressing pLKO or two independent MFF-targeting shRNA were plated in triplicate in 6-multiwell plates and quantified macroscopically for colony formation as described [31].
2.14. Animal studies
Studies involving vertebrate animals (rodents) were carried out in accordance with the Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011). Protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of The Wistar Institute (protocol #112625 and 112610). Groups of 4–6 weeks-old male athymic nude mice (Crl:NU(NCr)-Foxn1nu, Charles River Laboratory) were injected s.c. with PC3 clones (5 × 106 cells) stably transduced with pLKO or MFF-directed shRNA and tumour volume was quantified with a calliper over a two-week interval.
2.15. Immunohistochemistry
Four μm-thick sections from each tissue block were stained with an antibody to MFF (Protein Tech#17090-1-AP) using diaminobenzidine (DAB) as a chromogen. Immunohistochemistry was performed using Benchmark Ultra Roche Ventana immunostainer (Roche Group, Tucson, AZ). All slides were counterstained with haematoxylin. Two pathologists (V.V. and S.F.) blinded to clinical data evaluated and scored all slides. When discrepancies occurred, the case was further reviewed to reach an agreement score.
2.16. Statistical analysis
Data were analysed using the two-sided unpaired t, chi-square or Kruskal-Wallis (with p value correction for multiple testing) tests using a GraphPad software package (Prism 8.1) for Windows. Data are expressed as mean ± SD of replicates from a representative experiment out of at least two or three independent determinations or as mean ± SD of three individual experiments. A p value of <0.05 was considered as statistically significant.
3. Results
3.1. Differential MFF overexpression in cancer
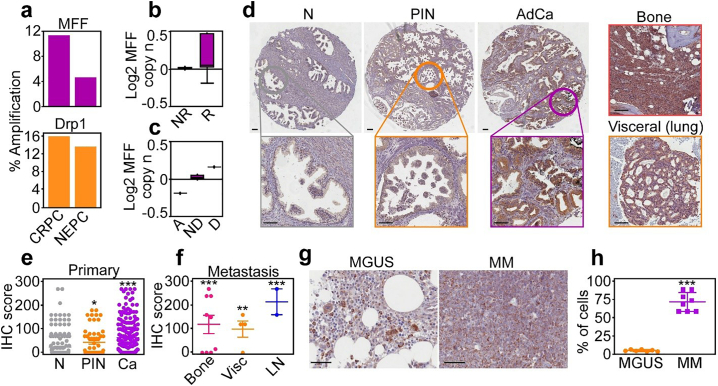
We began this study by examining the expression of mitochondrial fission effectors, Drp1 [24] and its outer mitochondrial membrane receptor, MFF [22] in prostate cancer. Analysis of public databases showed that MFF and Drp1 were amplified in castration-resistant and neuroendocrine prostate cancer (Fig. 1a), correlating with prostate cancer relapse (Fig. 1b) and abbreviated patient survival (Fig. 1c). In a cohort of 192 patients with localized and metastatic prostate cancer (Supplementary Table S1), we found that MFF levels increased from normal prostate to prostatic intraepithelial neoplasia (PIN) and were the highest in localized (Fig. 1d and e) and metastatic prostate cancer to lymph nodes, bones and visceral sites (Fig. 1d and f), by immunohistochemistry. These metastatic sites stained positive for prostate-specific antigen (PSA), confirming their prostatic origin (Supplementary Fig. S1a and b). In this patient series, increased MFF expression correlated with high Gleason grade (Supplementary Fig. S1c), but not tumour size (Supplementary Fig. S1d).
Fig. 1.
MFF overexpression in cancer. (a) Amplification of MFF and Drp1 in prostate cancer (77 patients, 107 samples). CRPC, castration-resistant prostate cancer; NEPC, neuroendocrine prostate cancer. (b) TCGA correlation (n = 380) between MFF expression and prostate cancer progression. NR, no recurrence at 5 years; R, recurrence at 5 years. (c) TCGA correlation (n = 380) of MFF expression (log MFF copy number) and prostate cancer survival. A, alive at 5 years; ND, alive with no evidence of disease at 5 years; D, dead with disease at 5 years. (d) MFF expression by immunohistochemistry in a patient cohort of localized and metastatic prostate cancer (n = 192). N, normal; PIN, prostatic intraepithelial neoplasia; AdCa, adenocarcinoma. Insets, image magnification of selected regions. Right, MFF expression in prostate cancer metastases to bone or lungs. Scale bar, 100 μm. (e and f) Quantification of MFF expression by immunohistochemistry (IHC) in primary (e) or metastatic (f) prostate cancer. *, p = .01; **, p = .003; ***, p < .0001 (by two-sided unpaired t-test; all compared to normal prostate). Vis, visceral; LN, lymph nodes. (g and h) MFF expression in representative patient samples of Monoclonal Gammopathy of Uncertain Significance (MGUS, n = 7) or Multiple Myeloma (MM, n = 8) by immunohistochemistry (g) and quantification of IHC score (h). Scale bar, 50 μm. ***, p < .0001 (by two-sided unpaired t-test).
As an independent approach, we next looked at a genetic model of prostate cancer, i.e. Transgenic Adenocarcinoma of the Mouse Prostate (TRAMP). Prostatic tumours formed in TRAMP mice, including neuroendocrine (NE) prostate cancer, well-differentiated adenocarcinoma (AdCa) and phyllodes-type tumours all expressed high levels of MFF, by immunohistochemistry (Supplementary Fig. S2a). In contrast, Drp1 was present at low levels in tumours from TRAMP mice (Supplementary Fig. S2a). Consistent with these observations, most prostate cancer cell lines expressed MFF, with the highest levels observed in castration-resistant as opposed to androgen receptor (AR)-positive cell types (Supplementary Fig. S2b). Other regulators of mitochondrial fusion (MFN1, MFN2, Opa1) or fission (Drp1) showed more variable expression in prostate cancer cell lines (Supplementary Fig. S2b).
Next, we asked if MFF was overexpressed in other tumour types. We found that MFF was highly expressed in patients with non-small cell lung cancer (NSCLC) including cases of adenocarcinoma (AdCa) and squamous cell carcinoma (SCC), compared to normal bronchus, by immunohistochemistry (our unpublished observations). In addition, MFF was strongly expressed in cases of multiple myeloma (MM), compared to patients with monoclonal gammopathy of uncertain significance (MGUS), by immunohistochemistry (Fig. 1g and h).
3.2. MFF is a novel Myc transcriptional target
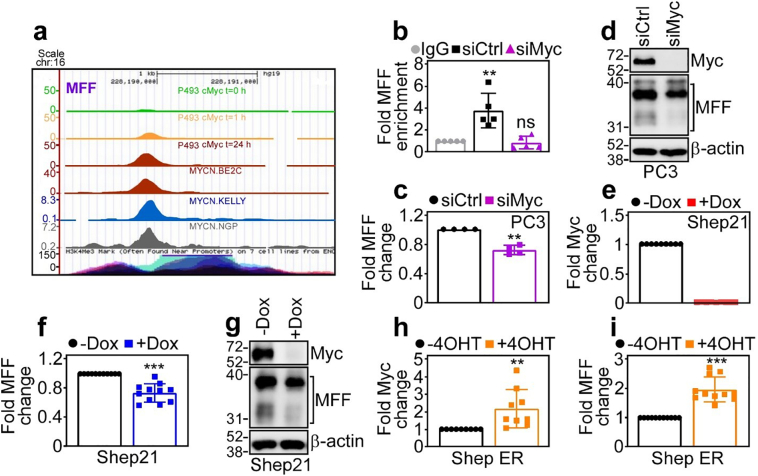
To begin elucidating how MFF becomes overexpressed in cancer, we next focused on the Myc oncogene, which is a key disease driver in prostate cancer and MM. Analysis of ChIP-Seq tracks demonstrated time-dependent accumulation of Myc at the MFF promoter in Burkitt lymphoma P493 cells as well as neuroblastoma BE2C, Kelly and NGP cell lines (Fig. 2a). In chromatin immunoprecipitation (ChIP) experiments, Myc readily bound to the promoter of MFF in PC3 cells, in a reaction abolished by siRNA knockdown of Myc (Fig. 2b). Consistent with these data, Myc silencing by siRNA, but not control siRNA, reduced MFF mRNA levels (Fig. 2c) and protein expression (Fig. 2d) in PC3 cells, by quantitative PCR and Western blotting, respectively.
Fig. 2.
MFF is a novel transcriptional target of oncogenic Myc. (a) ChIP-Seq tracks of time-dependent Myc accumulation at the MFF promoter in Burkitt lymphoma P493 cells at three time points (t = 0, 1 and 24 h) after removal of doxycycline (Dox) or neuroblastoma BE2C, Kelly or NGP cell lines. (b) ChIP of Myc accumulation at the MFF promoter in PC3 cells transfected with control non-targeting siRNA (siCtrl) or Myc-directed siRNA (siMyc). IgG, non-binding IgG. Mean ± SD. **, p = .004; ns, not significant (by two-sided unpaired t-test). (c and d) PC3 cells were transfected with siCtrl or siMyc and analysed for MFF expression by quantitative PCR (c) or Western blotting (d). Mean ± SD. **, p = .002 (by two-sided unpaired t-test). (e) Neuroblastoma Shep21 cells stably transfected with a Dox-regulated conditional Myc-directed shRNA were analysed for changes in Myc mRNA expression in the presence or absence of Dox, by quantitative PCR. Mean ± SD. (f and g) The conditions are as in (e) and Shep21 cells were analysed for MFF expression with or without Dox by quantitative PCR (f) or Western blotting (g). Mean ± SD. ***p < .0001 (by two-sided unpaired t-test). (h and i) Shep ER neuroblastoma cells stably transfected with a 4OHT-inducible N-Myc transgene were analysed for changes in Myc (h) or MFF (i) mRNA expression with or without 4OHT by quantitative PCR. Mean ± SD. ***p < .0001 (by two-sided unpaired t-test).
To independently validate these results, we next used the model of Shep21 neuroblastoma cells engineered with doxycycline (Dox)-regulated conditional ablation of Myc, or, alternatively, 4-hydroxy-tamoxifen (4OHT)-dependent Myc induction (Shep21-ER). Addition of Dox abolished Myc mRNA levels in Shep21 cells (Fig. 2e), and this was associated with reduced MFF mRNA (Fig. 2f) and protein (Fig. 2g) expression, compared to control transfectants. Reciprocally, addition of 4OHT to Shep21-ER cells increased the mRNA levels of Myc (Fig. 2h) as well as MFF (Fig. 2i), compared to cultures in the absence of 4OHT.
3.3. MFF controls mitochondrial fission in cancer
Based on these data, we next examined the function of MFF in cancer. Processing of the human MFF locus is predicted to generate at least five protein isoforms by alternative splicing (Supplementary Fig. S3a). Of these, MFF1 and MFF2 were the most abundantly expressed isoforms in PC3 cells (Supplementary Fig. S3b). In addition, transfection of Flag-MFF1 in these settings produced levels of recombinant protein comparable to endogenous MFF1 (Supplementary Fig. S3b). Using this approach, expression of MFF1 in PC3 cells resulted in extensive mitochondrial fragmentation, i.e. fission, and loss of mitochondrial elongation (Supplementary Fig. S3c and d), consistent with a role of this pathway in mitochondrial dynamics [22].
To carry out reciprocal experiments, we next established two independent siRNA sequences that reduce the expression of all MFF isoforms in PC3 cells (Supplementary Fig. S4a). In parallel, we also generated clones of DU145 and PC3 cells stably transduced with pLKO or MFF-directed shRNA, resulting in loss of endogenous MFF levels, compared to pLKO-transduced cultures (Supplementary Fig. S4b). MFF knockdown in these settings did not significantly affect mitochondrial dynamics, as comparable fractions of elongated or fragmented mitochondria were observed in control transfectants and MFF-silenced cells (Supplementary Fig. S4c and d). Consistent with these data, MFF silencing did not affect mitochondrial mass in DU145 or PC3 cells (Supplementary Fig. S4e). The fact that loss of MFF does not affect mitochondrial dynamics is consistent with a proposed role for other mitochondrial outer membrane receptor(s) mediating Drp1 recruitment [32] and organelle fission [33].
3.4. MFF associates with VDAC1 at the mitochondrial outer membrane
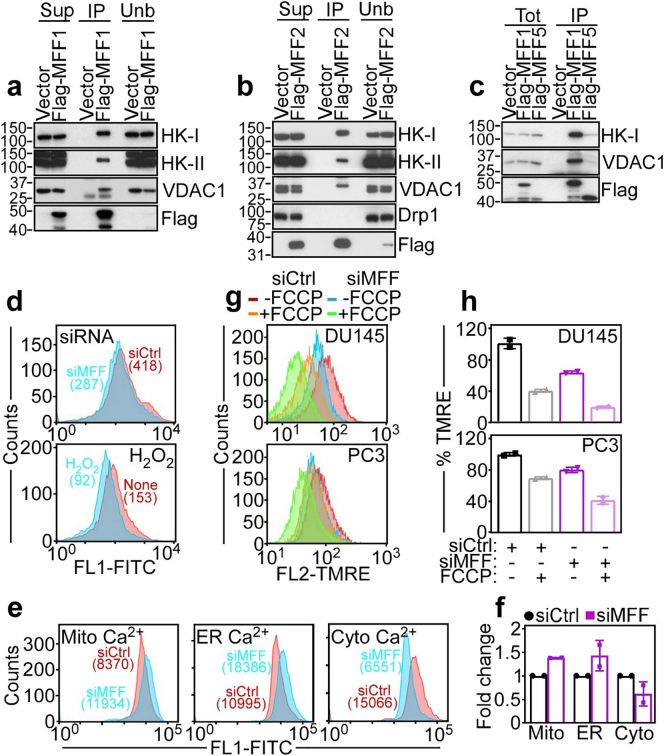
To test whether MFF had functions in cancer other than mitochondrial fission, we next carried out a proteomics screen for MFF-associated molecules in PC3 cells (our unpublished observations). One of the top hits in the screen was VDAC1 (our unpublished observations). Consistent this prediction, MFF1 (Fig. 3a) and MFF2 (Fig. 3b) co-immunoprecipitated with VDAC1 in PC3 cells, in vivo. Other mitochondrial outer membrane proteins that bind VDAC1, including hexokinase-I (HK-I) and -II (HK-II) were also present in the MFF-VDAC1 complex in PC3 cells (Fig. 3a and b). In contrast, the MFF ligand, Drp1 did not co-immunoprecipitate with the MFF-VDAC1 complex (Fig. 3b). Finally, this interaction was selective as another MFF isoform, MFF5 did not associate with VDAC1 or HK-I, by co-immunoprecipitation in PC3 cells (Fig. 3c).
Fig. 3.
MFF regulation of mitochondrial outer membrane permeability. (a–c) PC3 cells were transfected with vector, Flag-MFF1 (a), Flag-MFF2 (b) or Flag-MFF5 (c), immunoprecipitated (IP) with an antibody to Flag and analysed by Western blotting. Sup, supernatant; Unb, unbound. (d) PC3 cells transfected with siCtrl or siMFF (top) or treated with H2O2 (bottom) were labelled with calcein in the presence of CoCl2 and analysed by flow cytometry. Numbers indicate fluorescence units per each condition. None, untreated. (e and f) PC3 cells transfected with siCtrl or siMFF were incubated with Fluoro3-AM and FLUO-5 N or, alternatively Rhod2-AM, and changes in mitochondrial (Mito)-, endoplasmic reticulum (ER)- or cytosolic (Cytosol)-associated Ca 2+ levels were determined by flow cytometry (e, representative experiment) and quantified (f). Numbers correspond to fluorescence units. Mean ± SD. (g) DU145 (top) or PC3 (bottom) cells were transfected with siCtrl or MFF-directed siRNA (siMFF) and analysed for mitochondrial inner membrane potential by TMRE staining and flow cytometry with or without suboptimal concentrations of the uncoupler, FCCP. Representative experiment (n = 3). (h) The conditions are as in (g) and TMRE staining was quantified in siRNA-transfected DU145 and PC3 cells. Mean ± SD.
Based on these data, we next asked if MFF regulated VDAC1 function in cancer [17]. In these experiments, disruption of the MFF-VDAC1 complex by MFF knockdown triggered increased permeability of the mitochondrial outer membrane, by quantification of mitochondrial calcein fluorescence and flow cytometry (Fig. 3d). This response was quantitatively comparable to the increase in mitochondrial outer membrane permeability induced by H2O2 treatment (Fig. 3d) and was not further increased in the combination of H2O2 plus MFF knockdown (Supplementary Fig. S5a). Consistent with increased outer membrane permeability, MFF silencing in PC3 was also associated with increased levels of mitochondrial-associated Ca2+ and concomitant decrease in cytosolic Ca2+ (Fig. 3e and f). Conversely, ER-associated Ca2+ concentrations were not significantly different in control or MFF-targeted cells (Fig. 3e and f). In addition, MFF silencing in DU145 or PC3 cells was associated with loss of mitochondrial inner membrane potential, in a reaction further amplified by suboptimal concentrations of the uncoupler, carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP) (Fig. 3g and h). Conversely, MFF silencing did not depolarize mitochondria in normal prostatic epithelial BPH-1 or RWPE1 cells (Supplementary Fig. S5b), which had basal levels of mitochondrial transmembrane potential comparable to tumour cells (Fig. 3g and Supplementary Fig. S5b).
3.5. MFF control of tumour bioenergetics
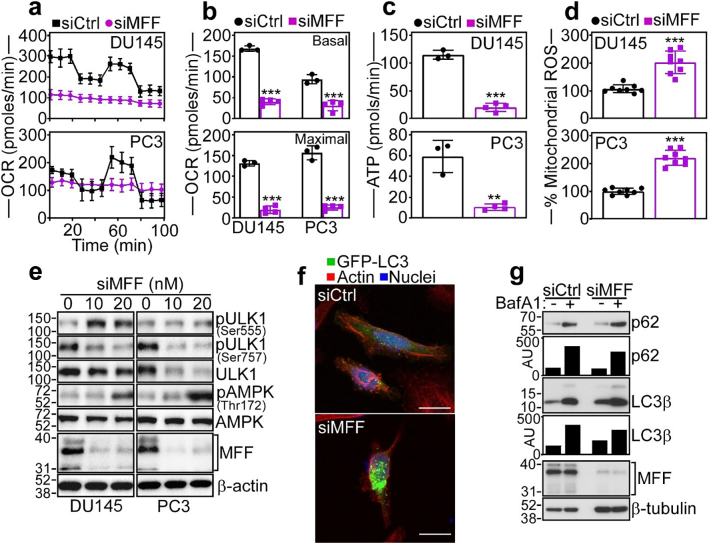
Depolarization of the mitochondrial inner membrane uncouples the TCA cycle, and, accordingly, MFF-targeted cells exhibited extensive bioenergetics defects. This included suppression of Oxygen Consumption Rates (OCR), a marker of oxidative phosphorylation (Fig. 4a), with significant decrease in both basal and maximal respiratory capacity (Fig. 5b). Further, MFF knockdown in DU145 and PC3 cells was accompanied by loss of Extracellular Acidification Rates (ECAR) (Supplementary Fig. S5c) and decreased glycolysis as well as glycolytic capacity, compared to control transfectants (Supplementary Fig. S5d).
Fig. 4.
MFF regulation of mitochondrial oxidative metabolism. (a) DU145 (top) or PC3 (bottom) cells were transfected with siCtrl or siMFF and analysed for oxygen consumption rates (OCR) on a Seahorse XFe96 Bioenergetics Flux Analyzer. Mean ± SD (n = 3). (b) The conditions are as in (a) and siRNA-transfected DU145 or PC3 cells were quantified for basal (top) and maximal (bottom) respiratory capacity. Mean ± SD (n = 3). ***, p = .0009 - <0.0001 (by two-sided unpaired t-test). (c) The conditions are as in (a) and the rate of ATP production was quantified in siRNA-transfected cells. Mean ± SD. ***, p < .0001; **, p = .001 (by two-sided unpaired t-test). (d) siRNA-transfected DU145 (top) or PC3 (bottom) cells as in (a) were analysed for mitochondrial superoxide (mitoSox) production by fluorescence microscopy. Mean ± SD. ***, p < .0001 (by two-sided unpaired t-test). (e) DU145 or PC3 cells transfected with increasing concentrations of siMFF (10–20 nM) were analysed by Western blotting. p, phosphorylated. (f) PC3 cells expressing LC3-GFP were transfected with siCtrl or siMFF and analysed by confocal fluorescence microscopy for analysis of cells with punctate LC3-GFP staining. Scale bar, 20 μm. (g) PC3 cells transfected with siCtrl or siMFF were incubated with or without the autophagy inhibitor Bafilomycin A1 (BafA1) and analysed by Western blotting. Bars, densitometric quantification of p62 and LC3β protein bands. AU, arbitrary units.
Fig. 5.
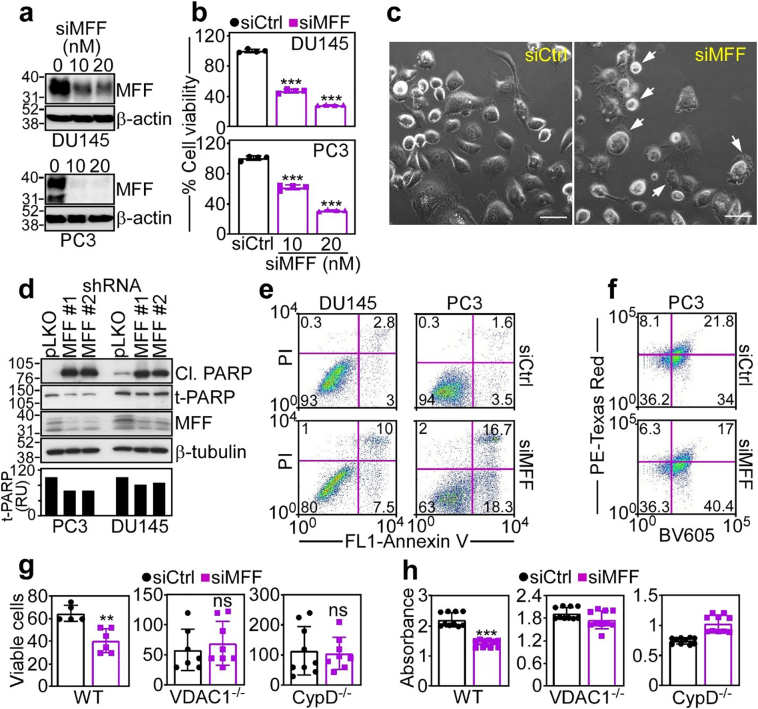
MFF inhibition of mitochondrial cell death. (a and b) DU145 (top) or PC3 (bottom) cells transfected with siCtrl or increasing concentrations of siMFF (100–200 nM) were analysed by Western blotting (a) and mitochondrial-dependent cell viability was determined by an MTT assay (b). Mean ± SD (n = 3). ***, p < .0001 (by two-sided unpaired t-test). (c) PC3 cells were transfected with siCtrl or siMFF and analysed for cellular morphology by light contrast microscopy. Arrows, cells with membrane blebbing and chromatin condensation. Scale bars, 50 μm. (d) Two independent clones of DU145 or PC3 cells stably expressing shMFF (MFF #1 and MFF #2) or pLKO were analysed by Western blotting. Cl., cleaved. Bar graph, densitometric quantification of total PARP (t-PARP) bands. RU, relative units. (e) DU145 (left) or PC3 (right) cells were transfected with siCtrl or siMFF and analysed for Annexin V and propidium iodide (PI) staining by multiparametric flow cytometry. The percentage of cells in each quadrant is indicated. (f) PC3 cells stably expressing a mitochondrial Keima-Red reporter were transfected with siCtrl or siMFF and analysed for pH-sensitive changes in fluorescence emission (PE-Texas Red) indicative of mitophagy. The percentage of cells in each quadrant is indicated. Representative experiment. BV605, Brilliant Violet 605 (g and h). WT, VDAC1−/− or CypD−/− MEF were transfected with siCtrl or siMFF and analysed for cell viability by Trypan blue exclusion and direct cell counting after 48 h (g) or an MTT assay (h). Mean ± SD. ***, p < .0001; **, p = .001; ns, not significant (by two-sided unpaired t-test). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Consistent with impaired bioenergetics, MFF silencing nearly completely abolished ATP production in prostate cancer cells (Fig. 4c). This was associated with oxidative stress and increased production of mitochondrial-derived ROS after MFF knockdown (Fig. 4d). Because of these bioenergetics defects, MFF-targeted DU145 and PC3 cells exhibited hallmarks of nutrient deprivation, with increased phosphorylation (Thr172) of the energy sensor, AMPK (Fig. 4e), differential phosphorylation of the autophagy activator, ULK1 with increased phosphorylation on the AMPK (Ser555) and decreased phosphorylation on the mTOR (Ser757) site (Fig. 4e), and induction of autophagy, with increased punctate LC3 staining, by fluorescence microscopy (Fig. 4f, Supplementary Fig. S5e). Accordingly, loss of MFF resulted in the accumulation of autophagy markers, p62 and LC3β, compared to control transfectants, in a response further augmented by treatment with the autophagy inhibitor, Bafilomycin A1 (Fig. 4g).
3.6. MFF regulates mitochondrial cell death
Consistent with a general collapse of mitochondrial integrity, concentration-dependent depletion of MFF by siRNA (Fig. 5a) was associated with mitochondrial-dependent killing of DU145 and PC3 cells (Fig. 5b). This response had hallmarks of apoptosis, including membrane blebbing and chromatin condensation (Fig. 5c), proteolytic processing of PARP (Fig. 5d) and increased Annexin V labelling (Fig. 5e). When combined with other stress stimuli, including nutrient deprivation, oxidative stress (H2O2) or chemotherapeutic agents, etoposide or doxorubicin, MFF knockdown further augmented PARP cleavage in prostate cancer cells (Supplementary Fig. S6a). In addition to mitochondrial cell death, VDAC1 has been proposed as a regulator of Parkin-dependent mitophagy [34]. Conversely, we found that knockdown of MFF in PC3 cells did not significantly affect pH-sensitive fluorescence emissions of a mitochondrial Keima-Red reporter (Fig. 5f), which quantifies mitophagy (Supplementary Fig. S6b), compared to control transfectants.
To further characterize the cell death response induced by MFF silencing, we next used genetically-modified mouse embryonic fibroblasts (MEF) that express endogenous levels of MFF1 and MFF2 (Supplementary Fig. S6c). Consistent with the data above, MFF silencing induced cell death in wild type (WT) MEF, as determined by Trypan blue exclusion and light microscopy (Fig. 5g). In contrast, MEF knockout for VDAC1 (Supplementary Fig. S6c) were entirely resistant to cell death induced by MFF silencing (Fig. 5g). Similarly, MEF with homozygous deletion of Cyclophilin D (CypD), an essential effector of regulated necrosis, were protected from cell death after MFF loss (Fig. 5g). We next carried out similar studies by quantifying mitochondrial-associated cell death by an MTT assay. Similar to the data above, MFF silencing killed WT MEF, whereas VDAC1 knockout or CypD knockout MEF were not affected (Fig. 5h).
3.7. MFF is required for tumour growth
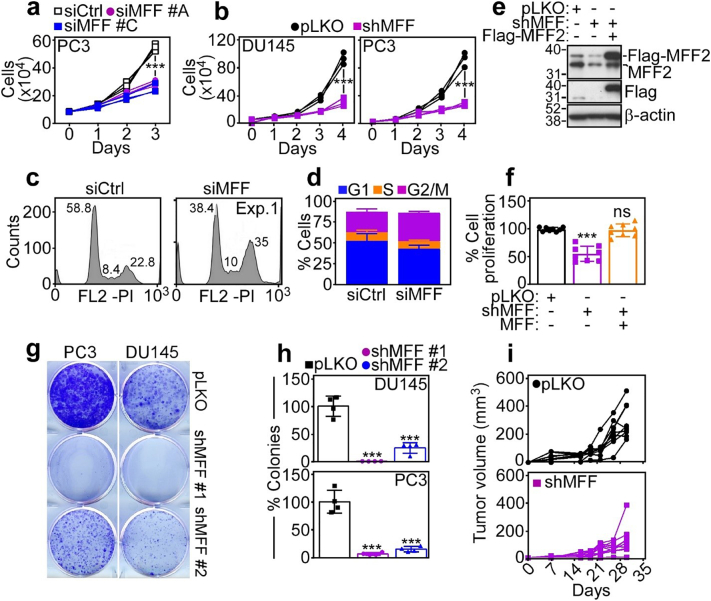
Based on these data, we next looked at the impact of MFF targeting on tumorigenesis. siRNA silencing of MFF inhibited PC3 (Fig. 6a) or DU145 (Supplementary Fig. S6d) cell proliferation, compared to control transfectants. Stable knockdown of MFF by shRNA gave similar results, suppressing DU145 or PC3 cell proliferation, compared to pLKO cultures (Fig. 6b). This response was not limited to prostate cancer, as MFF silencing inhibited glioblastoma LN229 or neuroblastoma SK-N-SH cell proliferation (Supplementary Fig. S6d). Consistent with oxidative stress in these settings, MFF knockdown caused extensive cell cycle defects in tumour cells (Fig. 6c, Supplementary Fig. S6f), characterized by accumulation of cells with G2/M DNA content (Fig. 6d). To confirm the specificity of this response, we next reconstituted MFF-depleted PC3 cells with shRNA-insensitive MFF2 cDNA (Fig. 6e). In these experiments, re-expression of MFF2 restored PC3 cell proliferation to levels of pLKO transfectants (Fig. 6f). When analysed for hallmark of tumorigenicity, shRNA knockdown of MFF potently suppressed colony formation (Fig. 6g and h), and inhibited s.c. xenograft tumour (PC3) growth in immunocompromised mice (Fig. 6i). As control, pLKO transfectants exhibited extensive colony formation (Fig. 6g and h) and gave rise to exponentially growing tumours in mice (Fig. 6i).
Fig. 6.
MFF regulation of tumour cell proliferation. (a) PC3 cells were transfected with siCtrl or two independent MFF-directed siRNA (siMFF #A and siMFF #C) and analysed for cell proliferation by direct cell counting during a 3-day interval. Each tracing corresponds to an individual experiment (n = 3). ***, p = .0002–0.0003 (by two-sided unpaired t-test). (b) DU145 (left) or PC3 (right) cells stably transduced with pLKO or MFF-directed shRNA (shMFF) were analysed for cell proliferation by direct cell counting at the indicated time intervals. Each tracing corresponds to an individual experiment (n = 3). ***, p < .0001 (by two-sided unpaired t-test). (c and d) PC3 cells transfected with siCtrl or siMFF were analysed for DNA content by PI staining and flow cytometry (c, representative experiment; numbers correspond to the percentage of cells in each peak) and the percentage of cells in the indicated cell cycle phase was quantified (d). Mean ± SD (n = 2). (e and f) PC3 cells transduced with pLKO or shMFF were reconstituted with MFF2 cDNA and analysed by Western blotting (e) or cell proliferation by direct cell counting after 72 h (f). The position of endogenous or Flag-MFF2 is indicated. Mean ± SD (n = 8). ***, p < .0001; ns, not significant (by two-sided unpaired t-test). (g and h) PC3 or DU145 cells stably transduced with two independent MFF-directed shRNA (clones #1 and #2) or pLKO were analysed for colony formation after 14 d by crystal violet staining (g, representative experiment) and quantified (h). Mean ± SD (n = 4). ***, p ≤.0001–0.003 (by two-sided unpaired t-test). (i) PC3 cells stably transduced with pLKO (top) or shMFF (bottom) were injected s.c. on the flanks of immunocompromised athymic nude mice and tumour growth was measured at the indicated time intervals with a calliper. Each symbol corresponds to an individual tumour. Tumour measurements (mm3) at day 30 are as follows: pLKO, 292.3 ± 40.1 (n = 9); MFF shRNA, 124.4 ± 35.4 (n = 9). **, p = .006 (by two-sided unpaired t-test). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
In this study, we have shown that a regulator of mitochondrial dynamics, MFF is a direct transcriptional target of oncogenic Myc, and becomes overexpressed in primary and metastatic cancer, compared to normal tissues. Biochemically, MFF isoforms MFF1 or MFF2 associate with VDAC1 at the mitochondrial outer membrane. Disruption of this complex by MFF silencing increases the permeability of the mitochondrial outer membrane followed by general collapse of organelle functions with Ca2+ unbalance, loss of inner membrane potential, extensive bioenergetics defects and oxidative stress. In turn, this activates multiple mitochondrial cell death pathways resulting in preclinical inhibition of tumour cell proliferation, suppression of colony formation and reduced xenograft tumour growth in mice.
Although deregulated mitochondrial fission [19] is commonly observed in cancer and implicated in advanced disease traits [20,24], a contribution of MFF in these responses has not been previously investigated. The role of oncogenic Myc [35] in this pathway, which may be responsible for the overexpression of MFF in primary and metastatic tumours, in vivo, fits well with an expanding role of Myc in mitochondrial reprogramming in cancer. In addition to oxidative phosphorylation gene expression [36] and modulation of multiple bioenergetics pathways [37], Myc-directed transcription has been associated with mitochondrial dynamics, promoting changes in organelle structure that favour therapy resistance [38] or heightened subcellular mitochondrial trafficking to fuel tumour cell invasion and metastasis [29].
Aside from its role in mitochondrial fission, in agreement with previous observations [22,23], we uncovered here a much broader function of MFF in preserving organelle integrity in cancer. Biochemically, this involved a novel complex between MFF and VDAC1 at the mitochondrial outer membrane. Structurally organized as a transmembrane β-barrel with an N-terminal domain arranged as an α-helix [16], VDAC1 acts as a voltage-gated channel controlling the exchange of small ions and metabolites across the mitochondrial outer membrane, sustaining a host of organelle functions [15]. The complex phenotype of MFF-targeted cells, characterized by a general collapse of mitochondrial bioenergetics, Ca2+ influx, oxidative stress potentially responsible for downstream cell cycle defects and induction of multiple cell death pathways is consistent with this view, and suggests that MFF is a novel regulator of VDAC1 functions in cancer. The loss of HK-I [39] and HK-II [40] from an MFF-VDAC1 complex is expected to further exacerbate this response, increasing cell death and preventing compensatory bioenergetics via glycolysis.
How MFF regulates VDAC1 functions, especially with respect to cell death induction, remains to be fully elucidated. We know that VDAC1 contributes to mitochondrial outer membrane permeability in concert with Bcl2 family proteins [11], but the role of channel conductance in this response has been debated [12]. Evidence from electrophysiologic studies and reconstitution experiments in artificial membranes suggests that VDAC1 assembles in higher-order multimers that modulate channel conductance in an “open” or “closed” configuration [15]. It is possible that MFF binding to VDAC1 regulates this process in tumour cells, shutting off channel conductance and opposing cell death without affecting Parkin-dependent mitophagy [34]. Such pro-survival function is consistent with a recent role of MFF in maintaining the cancer stem cell compartment in prostate cancer [41], and may counter the propensity to cell death associated with mitochondrial fission [25], facilitating its exploitation for tumour growth [42,43] and metastasis [44,45].
Disruption of an MFF-VDAC1 complex by MFF silencing was associated with morphologic and biochemical hallmarks of apoptosis [14], including Annexin V reactivity, PARP cleavage and VDAC1 dependence. However, MFF targeting induced features of other cell death pathways as well. For instance, the phenotype of severe nutrient deprivation induced by MFF silencing was associated with autophagy [46], which has been implicated in crosstalk with other cell death mechanisms [47], whereas homozygous deletion of the obligatory effector of regulated necrosis [48], CypD [49] entirely prevented cell death induced by MFF silencing. While the relative contribution of autophagy, apoptosis and regulated necrosis to the inhibition of tumour growth after MFF silencing remains to be delineated, the coexistence of multiple cell death pathways is not uncommon as a consequence of acute mitochondrial dysfunction [50] and reflects mechanistic overlap of cell death responses [51].
Consistent with a general collapse of mitochondrial integrity, MFF targeting delivered preclinical anticancer activity, with inhibition of tumour cell proliferation, suppression of colony formation and impaired tumour growth in mice. The differential overexpression of MFF in cancer, coupled with the insensitivity of normal cells to mitochondrial depolarization after MFF loss suggests that an MFF-VDAC1 complex may provide an actionable therapeutic target in cancer. Because only MFF1 and MFF2 bind VDAC1, in vivo, sequence(s) uniquely present in these isoforms, specifically Val199-Arg271 in MFF1 and the corresponding region Val 148-Arg220 in MFF2, would be predicted to form the VDAC1 binding interface and regulate channel conductance [15]. As it is now feasible to disrupt protein-protein interactions at the mitochondrial outer membrane [8], isoform-specific MFF mimetics targeting these sequences may compete for VDAC1 binding, trigger global organelle collapse and exert anticancer activity, in vivo. Compared to current mitochondrial-directed therapies [14], targeting the MFF-VDAC1 complex may offer broader indications in heterogeneous tumours and its mechanism of action involving multiple cell death pathways may evade drug resistance maintained by antiapoptotic Bcl2 proteins [10].
Funding sources
This work was supported by National Institutes of Health (NIH) grants P01 CA140043 (D.C.A) and R35 CA220446 (D.C.A.), by Fondazione Cariplo (2014-1148 to V.V.), by the Italian Minister of Health-Ricerca Corrente program 2017 (to S.F.), and by the National Research Foundation of Korea funded by the Ministry of Education (2018R1D1A1B07048104, 2018R1A6A1A03025810) and the Ministry of Science and ICT (2014M3A9D8034459). Y.C·C is the recipient of the Research Fund (1.170074.01) of Ulsan National Institute of Science and Technology (UNIST) and the National Research Foundation of Korea (2014M3A9D8034459) funded by the Ministry of Science and ICT. A.M.S. is supported by a fellowship from the Doctorate School in Molecular and Translational Medicine at the University of Milan. Support for Core Facilities utilized in this study was provided by Cancer Center Support Grant (CCSG) CA010815 to The Wistar Institute. The funders had no role in study design, data collection, data analysis, interpretation or writing of the report.
Author contributions
J.H.S.,Y.C.C., E.A. and D.C.A. conceived the project; J.H.S., Y.C.C., Y.G.L. and E.A. performed experiments of MFF-VDAC interaction, mitochondrial outer membrane permeability, modulation of organelle cell death, tumour cell proliferation, metabolic reprogramming and characterization of tumour growth in mice; D.S.G. analysed immunohistochemical staining in TRAMP mice; A.M.S., S.F., G.G. and V.V. analysed MFF expression in primary and metastatic patient cohorts; U.G. provided clinical samples; J.H.S., Y.C.C., E.A. and D.C.A. analysed data; and J.H.S., Y.C.C., E.A. and D.C.A. wrote the paper. All authors read and approved the final version of the paper.
Declaration of Competing Interest
None.
Acknowledgments
We thank Dr. Gyorgy Hajnoczky for VDAC1 knockout MEF.
Footnotes
Funding: National Institutes of Health; Italian Minister of Health; National Research Foundation of Korea.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.09.017.
Contributor Information
Young Chan Chae, Email: ychae@unist.ac.kr.
Dario C. Altieri, Email: daltieri@wistar.org.
Appendix A. Supplementary data
Supplementary material
References
- 1.Ward P.S., Thompson C.B. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21(3):297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gatenby R.A., Gillies R.J. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4(11):891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 3.Anderson R.G., Ghiraldeli L.P., Pardee T.S. Mitochondria in cancer metabolism, an organelle whose time has come? Biochim Biophys Acta Rev Cancer. 2018;1870(1):96–102. doi: 10.1016/j.bbcan.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vyas S., Zaganjor E., Haigis M.C. Mitochondria and Cancer. Cell. 2016;166(3):555–566. doi: 10.1016/j.cell.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Croce C.M., Reed J.C. Finally, an apoptosis-targeting therapeutic for cancer. Cancer Res. 2016;76(20):5914–5920. doi: 10.1158/0008-5472.CAN-16-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fulda S., Galluzzi L., Kroemer G. Targeting mitochondria for cancer therapy. Nat Rev Drug Discov. 2010;9(6):447–464. doi: 10.1038/nrd3137. [DOI] [PubMed] [Google Scholar]
- 7.Caino M.C., Altieri D.C. Molecular pathways: mitochondrial reprogramming in tumor progression and therapy. Clin Cancer Res. 2016;22(3):540–545. doi: 10.1158/1078-0432.CCR-15-0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ashkenazi A., Fairbrother W.J., Leverson J.D., Souers A.J. From basic apoptosis discoveries to advanced selective BCL-2 family inhibitors. Nat Rev Drug Discov. 2017;16(4):273–284. doi: 10.1038/nrd.2016.253. [DOI] [PubMed] [Google Scholar]
- 9.Fulda S. Promises and challenges of smac mimetics as cancer therapeutics. Clin Cancer Res. 2015;21(22):5030–5036. doi: 10.1158/1078-0432.CCR-15-0365. [DOI] [PubMed] [Google Scholar]
- 10.Birkinshaw R.W., Gong J.N., Luo C.S., Lio D., White C.A., Anderson M.A. Structures of BCL-2 in complex with venetoclax reveal the molecular basis of resistance mutations. Nat Commun. 2019;10(1):2385. doi: 10.1038/s41467-019-10363-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galluzzi L., Vitale I., Aaronson S.A., Abrams J.M., Adam D., Agostinis P. Molecular mechanisms of cell death: recommendations of the nomenclature committee on cell death 2018. Cell Death Differ. 2018;25(3):486–541. doi: 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Izzo V., Bravo-San Pedro J.M., Sica V., Kroemer G., Galluzzi L. Mitochondrial permeability transition: new findings and persisting uncertainties. Trends Cell Biol. 2016;26(9):655–667. doi: 10.1016/j.tcb.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 13.Tait S.W., Green D.R. Mitochondria and cell death: outer membrane permeabilization and beyond. Nat Rev Mol Cell Biol. 2010;11(9):621–632. doi: 10.1038/nrm2952. [DOI] [PubMed] [Google Scholar]
- 14.Adams J.M., Cory S. The BCL-2 arbiters of apoptosis and their growing role as cancer targets. Cell Death Differ. 2018;25(1):27–36. doi: 10.1038/cdd.2017.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Magri A., Reina S., De Pinto V. VDAC1 as pharmacological target in cancer and neurodegeneration: focus on its role in apoptosis. Front Chem. 2018;6:108. doi: 10.3389/fchem.2018.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colombini M. VDAC structure, selectivity, and dynamics. Biochim Biophys Acta. 2012;1818(6):1457–1465. doi: 10.1016/j.bbamem.2011.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mazure N.M. VDAC in cancer. Biochim Biophys Acta. 2017;1858(8):665–673. doi: 10.1016/j.bbabio.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 18.Carroll J., He J., Ding S., Fearnley I.M., Walker J.E. Persistence of the permeability transition pore in human mitochondria devoid of an assembled ATP synthase. Proc Natl Acad Sci U S A. 2019;116(26):12816–12821. doi: 10.1073/pnas.1904005116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Youle R.J., van der Bliek A.M. Mitochondrial fission, fusion, and stress. Science. 2012;337(6098):1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eisner V., Picard M., Hajnoczky G. Mitochondrial dynamics in adaptive and maladaptive cellular stress responses. Nat Cell Biol. 2018;20(7):755–765. doi: 10.1038/s41556-018-0133-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trotta A.P., Chipuk J.E. Mitochondrial dynamics as regulators of cancer biology. Cell Mol Life Sci. 2017;74(11):1999–2017. doi: 10.1007/s00018-016-2451-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otera H., Wang C., Cleland M.M., Setoguchi K., Yokota S., Youle R.J. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J Cell Biol. 2010;191(6):1141–1158. doi: 10.1083/jcb.201007152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu R., Chan D.C. The mitochondrial fission receptor Mff selectively recruits oligomerized Drp1. Mol Biol Cell. 2015;26(24):4466–4477. doi: 10.1091/mbc.E15-08-0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Senft D., Ronai Z.A. Regulators of mitochondrial dynamics in cancer. Curr Opin Cell Biol. 2016;39:43–52. doi: 10.1016/j.ceb.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suen D.F., Norris K.L., Youle R.J. Mitochondrial dynamics and apoptosis. Genes Dev. 2008;22(12):1577–1590. doi: 10.1101/gad.1658508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu S., Wang P., Zhang H., Gong G., Gutierrez Cortes N., Zhu W. CaMKII induces permeability transition through Drp1 phosphorylation during chronic beta-AR stimulation. Nat Commun. 2016;7 doi: 10.1038/ncomms13189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Renault T.T., Floros K.V., Elkholi R., Corrigan K.A., Kushnareva Y., Wieder S.Y. Mitochondrial shape governs BAX-induced membrane permeabilization and apoptosis. Mol Cell. 2015;57(1):69–82. doi: 10.1016/j.molcel.2014.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forno I., Ferrero S., Russo M.V., Gazzano G., Giangiobbe S., Montanari E. Deregulation of MiR-34b/Sox2 predicts prostate cancer progression. PLoS One. 2015;10(6) doi: 10.1371/journal.pone.0130060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agarwal E., Altman B.J., Ho Seo J., Bertolini I., Ghosh J.C., Kaur A. Myc regulation of a mitochondrial trafficking network mediates tumor cell invasion and metastasis. Mol Cell Biol. 2019;39(14) doi: 10.1128/MCB.00109-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ushmorov A., Hogarty M.D., Liu X., Knauss H., Debatin K.M., Beltinger C. N-myc augments death and attenuates protective effects of Bcl-2 in trophically stressed neuroblastoma cells. Oncogene. 2008;27(24):3424–3434. doi: 10.1038/sj.onc.1211017. [DOI] [PubMed] [Google Scholar]
- 31.Caino M.C., Seo J.H., Wang Y., Rivadeneira D.B., Gabrilovich D.I., Kim E.T. Syntaphilin controls a mitochondrial rheostat for proliferation-motility decisions in cancer. J Clin Invest. 2017;127(10):3755–3769. doi: 10.1172/JCI93172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osellame L.D., Singh A.P., Stroud D.A., Palmer C.S., Stojanovski D., Ramachandran R. Cooperative and independent roles of the Drp1 adaptors Mff, MiD49 and MiD51 in mitochondrial fission. J Cell Sci. 2016;129(11):2170–2181. doi: 10.1242/jcs.185165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kalia R., Wang R.Y., Yusuf A., Thomas P.V., Agard D.A., Shaw J.M. Structural basis of mitochondrial receptor binding and constriction by DRP1. Nature. 2018;558(7710):401–405. doi: 10.1038/s41586-018-0211-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geisler S., Holmstrom K.M., Skujat D., Fiesel F.C., Rothfuss O.C., Kahle P.J. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12(2):119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 35.Toyoshima M., Howie H.L., Imakura M., Walsh R.M., Annis J.E., Chang A.N. Functional genomics identifies therapeutic targets for MYC-driven cancer. Proc Natl Acad Sci U S A. 2012;109(24):9545–9550. doi: 10.1073/pnas.1121119109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oran A.R., Adams C.M., Zhang X.Y., Gennaro V.J., Pfeiffer H.K., Mellert H.S. Multi-focal control of mitochondrial gene expression by oncogenic MYC provides potential therapeutic targets in cancer. Oncotarget. 2016;7(45):72395–72414. doi: 10.18632/oncotarget.11718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stine Z.E., Walton Z.E., Altman B.J., Hsieh A.L., Dang C.V. MYC, metabolism, and cancer. Cancer Discov. 2015;5(10):1024–1039. doi: 10.1158/2159-8290.CD-15-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Casinelli G., LaRosa J., Sharma M., Cherok E., Banerjee S., Branca M. N-Myc overexpression increases cisplatin resistance in neuroblastoma via deregulation of mitochondrial dynamics. Cell Death Dis. 2016;2 doi: 10.1038/cddiscovery.2016.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Villinger S., Briones R., Giller K., Zachariae U., Lange A., de Groot B.L. Functional dynamics in the voltage-dependent anion channel. Proc Natl Acad Sci U S A. 2010;107(52):22546–22551. doi: 10.1073/pnas.1012310108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pastorino J.G., Shulga N., Hoek J.B. Mitochondrial binding of hexokinase II inhibits Bax-induced cytochrome c release and apoptosis. J Biol Chem. 2002;277(9):7610–7618. doi: 10.1074/jbc.M109950200. [DOI] [PubMed] [Google Scholar]
- 41.Civenni G., Bosotti R., Timpanaro A., Vazquez R., Merulla J., Pandit S. Epigenetic control of mitochondrial fission enables self-renewal of stem-like tumor cells in human prostate cancer. Cell Metab. 2019 Aug 6;30(2):303–313.e6. doi: 10.1016/j.cmet.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 42.Kashatus J.A., Nascimento A., Myers L.J., Sher A., Byrne F.L., Hoehn K.L. Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth. Mol Cell. 2015;57(3):537–551. doi: 10.1016/j.molcel.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Serasinghe M.N., Wieder S.Y., Renault T.T., Elkholi R., Asciolla J.J., Yao J.L. Mitochondrial division is requisite to RAS-induced transformation and targeted by oncogenic MAPK pathway inhibitors. Mol Cell. 2015;57(3):521–536. doi: 10.1016/j.molcel.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caino M.C., Seo J.H., Aguinaldo A., Wait E., Bryant K.G., Kossenkov A.V. A neuronal network of mitochondrial dynamics regulates metastasis. Nat Commun. 2016;7 doi: 10.1038/ncomms13730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao J., Zhang J., Yu M., Xie Y., Huang Y., Wolff D.W. Mitochondrial dynamics regulates migration and invasion of breast cancer cells. Oncogene. 2013;32(40):4814–4824. doi: 10.1038/onc.2012.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levine B., Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176(1–2):11–42. doi: 10.1016/j.cell.2018.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goodall M.L., Fitzwalter B.E., Zahedi S., Wu M., Rodriguez D., Mulcahy-Levy J.M. The autophagy machinery controls cell death switching between apoptosis and necroptosis. Dev Cell. 2016;37(4):337–349. doi: 10.1016/j.devcel.2016.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Conrad M., Angeli J.P., Vandenabeele P., Stockwell B.R. Regulated necrosis: disease relevance and therapeutic opportunities. Nat Rev Drug Discov. 2016;15(5):348–366. doi: 10.1038/nrd.2015.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baines C.P., Kaiser R.A., Purcell N.H., Blair N.S., Osinska H., Hambleton M.A. Loss of cyclophilin D reveals a critical role for mitochondrial permeability transition in cell death. Nature. 2005;434(7033):658–662. doi: 10.1038/nature03434. [DOI] [PubMed] [Google Scholar]
- 50.Declercq W., Takahashi N., Vandenabeele P. Dual face apoptotic machinery: from initiator of apoptosis to guardian of necroptosis. Immunity. 2011;35(4):493–495. doi: 10.1016/j.immuni.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 51.Lemasters J.J. V. Necrapoptosis and the mitochondrial permeability transition: shared pathways to necrosis and apoptosis. Am J Physiol. 1999;276(1):G1–G6. doi: 10.1152/ajpgi.1999.276.1.G1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material