Abstract
Background
The immune checkpoint, indoleamine 2,3-dioxygenase 1, is under investigation as target of novel immunotherapies for cancers, including head and neck squamous cell carcinomas (HNSCC). The aim of our study was to analyze DNA methylation of the encoding gene (IDO1) in HNSCC.
Methods
Methylation of three CpG sites within the promoter, promoter flank, and gene body was investigated and correlated with mRNA expression, immune cell infiltration, mutational burden, human papillomavirus (HPV)-status, and overall survival in a cohort of N = 528 HNSCC patients obtained from The Cancer Genome Atlas. In addition, IDO1 immunohistochemistry and DNA methylation analysis was performed in an independent cohort of N = 138 HNSCC samples.
Findings
Significant inverse correlations of IDO1 methylation and IDO1 mRNA expression were found in the promoter and promoter flank region (Spearman's ρ = −0.163 and ρ = −0.377, respectively) while a positive correlation was present in the gene body (ρ = 0.502; all P < 0.001). IDO1 DNA methylation significantly correlated with IDO1 protein expressing immune cells as well as tumor cells. IDO1 promoter flank hypermethylation was significantly associated with poor overall survival (P < 0.001). In addition, we discovered significant correlations between IDO1 methylation and expression with RNA signatures of immune cell infiltrates and with HPV-status, mutational load (methylation only), and interferon γ signature.
Interpretation
Our results suggest IDO1 expression levels are epigenetically regulated by DNA methylation. This study provides rationale to test IDO1 methylation as potential biomarker for prediction of response to IDO1 immune checkpoint inhibitors in HNSCC.
Keywords: Indoleamine 2,3-dioxygenase 1; IDO1, Biomarker; DNA methylation; Head and neck squamous cell carcinoma; HPV; Immunotherapy; Prognosis; Prediction
Research in context
Evidence before this study
Indoleamine 2,3-dioxygenase 1 (IDO1) is a promising target for immunotherapy. However, a phase III trial testing the IDO1 inhibitor, epacadostat, together with PD-1 inhbitor, pembrolizumab, has recently missed the first primary endpoint. Reports on the regulation of IDO1 on an epigenetic level are rare. Novel insights into this regulation might provide a rationale for the development of mechanism-driven biomarkers for patient stratification. We analyzed the head and neck squamous cell carcinoma (HNSCC) cohort included in The Cancer Genome Atlas database and searched in the Gene Expression Omnibus (GEO) database for information on IDO1 expression and methylation in cell lines and leukocytes. To validate our findings, we performed protein expression analysis by immunohistochemistry to study immune microenvironment and IDO1 expression in HNSCC.
Added value of this study
Our study provides evidence of epigenetic regulation of IDO1 by DNA methylation in HNSCCs. We identified significant correlations between IDO1 methylation and expression (mRNA and protein), with immune cell infiltrates, mutational load, HPV, interferon γ signature, and patient outcome.
Implications of all the available evidence
Taking all available evidence into account, IDO1 methylation should be considered as potential biomarker for prediction of response to anti-IDO1 immune checkpoint inhibitors in HNSCC. IDO1 methylation testing should be included into biomarker programs of clinical trials that include IDO1 inhibitors.
1. Introduction
65,410 new cases of oral cavity, pharyngeal, and laryngeal tumors are estimated to be diagnosed in 2019 in the United States [1]. Moreover, it is estimated that 358,144 patients worldwide with cancer of the lip, oral cavity, oropharynx, hypopharynx, and larynx will die from the disease in 2018 [2]. The majority of malignant tumors in the head and neck region are of squamous cell origin. Thus, head and neck squamous cell carcinomas (HNSCCs) represent a major health burden worldwide.
HNSCC is associated with certain environmental risk factors like smoking and alcohol abuse as well as infection with high risk human papillomavirus (HPV). Patients with HPV-associated cancers (low-risk tumors) experience significantly longer overall survival than patients with tumors associated with classical risk factors like smoking and alcohol abuse (high-risk tumors) [3,4]. Despite the development of new therapies for HNSCC the prognosis remains dismal once recurrent or metastatic disease occurs. The anti-EGFR antibody, cetuximab, in combination with chemotherapy, is the most common treatment regimen for advanced or metastatic disease [5]. Recently, immunotherapy has emerged as a promising treatment for HNSCC. The immune checkpoint inhibitor, nivolumab, targeting the immune checkpoint programmed cell death 1 (PD-1) receptor has been approved for second line therapy based on the results of the CheckMate 141 trial [6]. This trial demonstrated an overall survival benefit for patients receiving nivolumab, in regardless of HPV-status [7]. In addition, another antibody targeting PD-1, pembrolizumab, and antibodies targeting PD-1 ligand 1 (PD-L1), atezolizumab and durvalumab, have demonstrated significant antitumor activity [8,9]. Pembrolizumab has recently been approved as first-line therapy in recurrent and metastatic HNSCC in combination with platinum therapy and 5-FU [10].
Other immunotherapeutic agents are being developed and progressing to clinical trials such as the indoleamine 2,3-dioxygenase 1 (IDO1) inhibitors, epacadostat and navoximod [11], [12], [13]. IDO1 is the rate-limiting enzyme in the conversion of the essential amino acid tryptophan to kynurenine. IDO1 is highly expressed in many tumor types and has been shown to play a role in immunosuppression, through increased tryptophan metabolism, in the tumor microenvironment (TME) [14,15]. Increased IDO1 expression can lead to suppression of antitumoral T cells, differentiation of CD4+ T cells into immunosuppressive regulatory T cells (Tregs), and polerisation of antigen-presenting cells into a tolerogenic phenotype [16,17]. Overexpression of IDO1 in various tumor tissues is associated with worse overall survival [15,18]. IDO1 inhibitors could thus restore function of anti-tumoral T cells and shift the TME from immunosuppressive to immunogenic [19].
The IDO1 inhibitor navoximod was well tolerated in a phase I trial and stable disease responses were observed in 8 (36%) out of 22 patients [13]. Recent results from the phase I/II ECHO-202/KEYNOTE-037 trial demonstrated encouraging antitumor activity of epacadostat in combination with pembrolizumab [11]. In combination with nivolumab, epacadostat also improved disease control in the HNSCC cohort of the phase I/II ECHO-204 trial. However, epacadostat failed to demonstrate therapeutic benefit in combination with immune checkpoint blockade in a malignant melanoma phase III trial and thus several other trials have been put on hold [20,21]. Nevertheless, researchers offered reasons for the failed trial and recommend a further clinical investigation of IDO inhibitors. Since IDO1 remains a promising immunotherapeutic target, a better understanding of its regulation resulting in the development of companion biomarkers is needed in order to identify subgroups of patients that are likely to benefit from treatment. Predictive biomarkers are best studied in the context of anti-PD-1 immunotherapies. Tumor mutational burden, tumor programmed cell death ligand 1 (PD-L1) expression, the intensity of intratumoral CD8+ T cell infiltrates, and an interferon γ or T cell inflamed profile have each been proposed as distinct features of response to immune checkpoint blockage [22], [23], [24].
DNA methylation is an epigenetic mechanism associated with transcriptional gene activity and involved in fundamental biological processes, i.e. embryogenesis, imprinting, X chromosome inactivation, aging, and differentiation [25,26]. In the context of immunotherapies, DNA methylation plays a role in T cell differentiation and T cell exhaustion [27], [28], [29]. Recent reports suggest a role for DNA methylation in the regulation of immune checkpoint genes, i.e. PD-1, PD-L1, PD-1 ligand 2 (PD-L2), and cytotoxic T-lymphocyte associated protein 4 (CTLA4) in various malignancies, including HNSCC [30], [31], [32], [33], [34], [35], [36], [37], [38]. Furthermore, CTLA4 methylation in tumors from patients with metastastatic melanoma can be used to predict response to anti-PD-1 and anti-CTLA-4 directed immune checkpoint blockage [39]. First reports also suggest an epigenetic regulation of IDO1 via DNA methylation [40,41].
Our present study aims to understand the epigenetic regulation of IDO1 via DNA methylation and its association with tumor immunogenicity in HNSCC. This could help to stratify subgroups of patients who are likely to benefit from IDO1-inhibitor therapy.
2. Materials and methods
2.1. Patients and data acquisition
TCGA cohort: Results are based on data generated by The Cancer Genome Atlas Research Network (TCGA, http://cancergenome.nih.gov/). Informed consent was obtained by the TCGA Research Network from all patients in accordance with the Helsinki Declaration of 1975. Clinico-pathological data were obtained from the TCGA Research Network. Data were available for N = 528 cancer tissue samples and N = 50 matched normal adjacent tissue (NAT) samples. Overall survival was censored after 60 months to account only for tumor-specific death. Molecular data was adopted from studies previously published by the TCGA Research Network [42,43].
UKB cohort: For validation purposes, we included N = 138 formalin-fixed and paraffin-embedded HNSCCs from patients treated at the University Hospital Bonn (UKB). The study protocol was approved by the Institutional Review Board (vote no. 187/16). HPV status was assessed by p16 immunohistochemistry.
2.2. Cell lines and isolated immune cells
We included methylation data from three HPV-positive (UPCI:SCC090, 93VU-147 T, UM:SCC047) and three HPV-negative (UPCI:SCC003, UPCI:SCC036, and PCI-30) HNSCC cell lines previously generated by Lechner et al. (Gene Expression Omnibus (GEO) accession number: GSE38271; National Center for Biotechnology Information (NCBI), Bethesda, MD, USA) and from N = 20 HPV-negative HNSCC cell lines (BHY, BICR22, CAL-27, CAL-33, Detroit562, FADU, HN, JHU-011, JHU-022, SAS, SAT, SCC-15, SCC-25, SCC-4, SCC-9, SKN-3, A253, HSC-3, OSC-19, Ca9-22) provided by Iorio et al. (GSE68379) [44,45]. Furthermore, we included additional 13 HNSCC cell lines (eleven HPV-negative: UM-SCC-14B, UM-SCC-14A, UM-SCC-24A, UM-SCC-24B, UD-SCC-1, UM-SCC-11B, UD-SCC-4, UM-SCC-10BPT, UD-SCC-5, UT-SCC-33, UT-SCC-60B; two HPV-positive: UD-SCC-2, UM-SCC-104) provided by the University Hospital Dusseldorf.
Methylation data from isolated immune cells (CD4+ T cells, CD8+ T cells, regulatory T cells, B cells, and monocytes) were obtained from three previous studies which included isolated immune cells obtained from N = 26 healthy donors from Scotland (GSE87650), N = 6 healthy Israeli women (GSE71244), and N = 72 healthy American individuals (GSE59250) [46], [47], [48].
2.3. Immune cell infiltrates
TCGA cohort: Thorsson et al. developed RNASeq signatures as surrogate marker for immune cell infiltrates which provides quantitative data on infiltrating leukocytes. This includes signatures for naive B cells, memory B cells, naive CD4+ T cells, activated and resting CD4+ memory T cells, T follicular helper cells, Tregs, CD8+ T cells, γδ T cells, activated and resting NK cells, plasma cells, macrophages (including monocytes and M0/M1/M2 macrophages), dendritic cells (including resting and activated dendritic cells), mast cells (including activated and resting mast cells) for N = 468 tumor samples [49].
UKB cohort: Infiltrates of CD45+, CD3+, CD8+, CD4+, and IDO1+ immune cells were quantified via immunohistochemistry as described below.
2.4. Methylation analysis
Gene methylation data (β-values) generated using the Infinium HumanMethylation450 BeadChip (Illumina, Inc., San Diego, CA, USA) technology were downloaded from the UCSC Xena browser (TCGA cohort, www.xena.ucsc.edu) and GEO webpage (HNSCC cell lines and isolated immune cells). In addition, we performed methylation analysis of additional HNSCC cell lines using the Infinium MethylationEPIC BeadChip (Illumina, Inc.) following the manufacturer's instructions.
Methylation analysis of tumor tissues from the UKB cohort was performed after macrodissection of tumor areals from sections mounted on glass slides and subsequent lysis and bisulfite conversion using the innuCONVERT Bisulfite All-In-One Kit (Analytik Jena, Jena, Germany) following the manufacturer's instructions. We applied quantitative real-time PCR methylation analysis developed by Lehmann and Kreipe [50] in order to determine the methylation levels at the CpG site targeted by bead 3 within the gene body (forward primer: agggttttgttttggtttgttttga, reverse primer: ccaaaaaacccattaaactatatctatt, probemethylated: 6-FAM-tgttacgttagtaagtacgtaaagt-BHQ-1, probeunmethylated: HEX-tttgttatgttagtaagtatgtaaagtaaa-BHQ-1). PCR reactions were performed in 20 µl volumes containing 10 ng bisulfite converted DNA (quantified via UV–vis spectrophotometry) and 0.4 µM each primer and each probe. PCR buffer conditions were used as previously described [51]. Real-time PCR was carried out using a 7900HT Fast Real-Time PCR system (Applied Biosystems, Waltham, MA, USA) applying the following temperature profile: 10 min at 95 °C and 45 cycles with 15 s at 95 °C and 45 s at 52 °C. Percentage methylation levels were calculated using cycle treshold (CT) values obtained from probes specifically binding to methylated (CTmethylated) and unmethylated (CTunmethylated) DNA, respectively, using the following formula: Methylation [%] = 100%/(1 + 2CTmethylated–CTunmethylated).
2.5. mRNA expression analysis
mRNA expression data were downloaded from the TCGA Research Network (http://cancergenome.nih.gov/). mRNA expression data were available for N = 21 samples of NATs and N = 521 samples of cancer tissues. Data were generated employing the Illumina HiSeq 2000 RNA Sequencing Version 2 analysis (Illumina, Inc., San Diego, CA, USA) and normalized counts (n.c.) per gene were reported.
2.6. Immunohistochemistry
Tissue sections from formalin-fixed, paraffin-embedded tumor tissues were freshly cut (4 µm) and mounted on Super Frost Plus slides (Thermo Fisher, Waltham, MA, USA). After deparaffinization and rehydration, the sections were washed with 550 mM Tris-buffered saline (TBS).
IDO1 and p16INK4a antigen retrieval for was performed in Target Retrieval Solution (IDO1: pH6, p16INK4a: pH9) for 10 min at 100 °C (#S169984-2, Dako / Agilent Technologies, Inc., Santa Clara, CA, USA). After blocking, the sections were incubated with the primary IDO1 and p16INK4a antibodies (IDO1: dilution 1:1000, rabbit polyclonal antibody anti-IDO1 #HPA023072, Sigma-Aldrich, St. Louis, MO, USA; p16INK4a: dilution 1:100, mouse monoclonal antibody clone JC2, #MSK123-05, Zytomed Systems GmbH, Berlin, Germany) for 60 min at room temperature. After washing with 550 mM TBS, the visualization was performed with the Dako REAL Detection System Alkaline Phosphatase/RED (Dako / Agilent Technologies, #K5005) on the Dako Autostainer and were contrasted with Mayer´s Hemalum solution (Merck Millipore, Billerica, MA, USA, #HX73030749). IDO1 positive cells (tumor or immune cells) were calculated as a percentage of total cells.
Immunohistochemical staining of CD45, CD3, CD8, and CD4 was carried out using monoclonal antibodies; anti-CD45 (1:100, clones 2B11 + PD7/26, #M070101, Dako / Agilent Technologies), anti-CD3 (1:200, clone LN10, #NCL-L-CD3-565, Leica Biosystems, Wetzlar, Germany), anti-CD8 (1:50, clone C8/144B, #M710301, Dako / Agilent Technologies), and anti-CD4 (1:20, clone SP35, #503–3354, Zytomed Systems GmbH), respectively, using the Dako Omnis system (Dako / Agilent Technologies). The Envision FLEX Magenta, High pH kit (#GV900, Dako / Agilent Technologies) was used for visualization. Labeling was performed after heat pretreatment for 20 min at 95 °C in EnVision FLEX TRS Low pH (#GC80511-2, Dako / Agilent Technologies; anti-CD4 and anti-CD45 antibodies) or EnV FLEX TRS, High pH (anti-CD3 and anti-CD8 antibodies). The incubation period for the primary antibodies was 30 min. Protein expression of CD45, CD3, CD4, CD8 was evaluated by V.S., as percentage of all cells.
2.7. Statistics
SPSS (version 23.0; SPSS Inc., Chicago, IL, USA) was employed to perform statistical analyses. Correlations were calculated using Spearman's rank correlation (Spearman's ρ). Mean values were compared using the Wilcoxon–Mann–Whitney U test (two groups). Multiple comparisons between groups were tested with one-way ANOVA and post-hoc Bonferroni test. Cox Proportional Hazards regressions of overall survival were conducted using log2-transfomed methylation and mRNA expression levels. Kaplan–Meier analysis of overall survival was performed based on an optimized cut-off for result dichotomization. P-values refer to log-rank test and Wald test, respectively. Two-sided P-values < 0.05 were considered statistically significant.
3. Results
3.1. IDO1 methylation correlates with IDO1 expression
We used beads cg10262052 (bead 1), cg08465774 (bead 2), and cg24188163 (bead 3) from the Infinium HumanMethylation450 BeadChip to assess methylation levels at CpG sites within the IDO1 gene. These beads target CpG sites in the promoter region, promoter flank, and gene body of the IDO1 gene, respectively (as seen in Fig. 1). DNA methylation levels correlated significantly with the respective mRNA expression both in normal adjacent and tumor tissues (Table 1). We observed a negative correlation between methylation and mRNA expression for the beads targeting the promoter and the promoter flank region (beads 1 and 2). A positive correlation between methylation and mRNA expression, however, was observed at a CpG site within the gene body (bead 3).
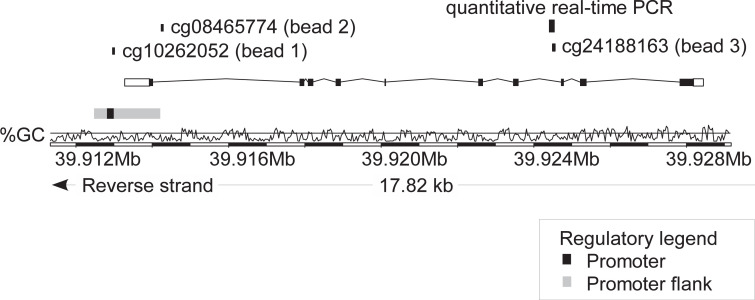
Fig. 1.
Genomic organization of the IDO1 gene. Shown is the IDO1 transcript ENST00000518237.6, CG-density, target sites of HumanMethylation450 BeadChip beads, and the target sequence of the quantitative real-time PCR assay. The modified illustration was exported from www.ensemble.org (Version 89.38) and is based on Genome Reference Consortium Human Build 38 patch release 10 (GRCh38.p10). cg10262052 (bead 1) targets the central promoter site, bead cg08465774 (bead 2) probes the intragenic promoter flank, and bead cg24188163 (bead 3) targets the gene body.
Table 1.
IDO1 methylation (%) and mRNA expression (n.c.) in normal adjacent tissues, HPV-negative and HPV–positive HNSCC tissues. IDO1 methylation was determined at three different loci targeted by HumanMethylation450 BeadChip beads (Fig. 1) in N = 528 HNSCC patients from The Cancer Genome Atlas. mRNA expression data was available for N = 521 patients. Methylation and expression data could be correlated in N = 20 normal adjacent tissues and N = 521 tumor tissues. Significant data are shown in boldface. P-values refer to Wilcoxon–Mann–Whitney U test and Spearman's ρ correlations, respectively.
| Mean methylation [%] / mRNA expression [n.c.]; [95% CI] |
Correlation methylation with mRNA expression |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Analyte | Normal adjacent tissue | All tumors | P-value (normal adjacent tissues vs. tumors) | HPV-negative tumors | HPV-positive tumors | P-value (HPV-positive vs. -negative | Tumor |
Normal adjacent tissue |
||
| Spearman's ρ | P-value | Spearman's ρ | P-value | |||||||
| IDO1 mRNA expression | 393 [122–909] | 1191 [982–1400] | 0.001 | 976 [723–1230] | 2127 [952–3301] | 0.001 | NA | NA | NA | NA |
| IDO1 methylation cg10262052 | 49.1 [44.3–54.0] | 17.5 [16.3–18.7] | <0.001 | 17.6 [16.0–19.3] | 24.4 [17.1–31.7] | 0.34 | −0.163 | <0.001 | −0.463 | 0.040 |
| IDO1 methylation cg08465774 | 88.4 [86.7–90.0] | 70.3 [68.8–71.8] | <0.001 | 73.2 [71.1–75.4] | 58.8 [52.8–64.9] | <0.001 | −0.377 | <0.001 | −0.534 | 0.015 |
| IDO1 methylation cg24188163 | 59.0 [55.6–62.4] | 64.4 [62.7–66.1] | 0.003 | 64.0 [61.4–66.6] | 65.5 [60.4–70.6] | 0.88 | 0.502 | <0.001 | 0.105 | 0.66 |
NA: Not Applicable.
We further investigated the correlation between IDO1 methylation and protein expression in an independent cohort comprised of N = 138 HNSCC tumors (UKB cohort). DNA methylation was assessed using a quantitative real-time PCR assays that targets the CpG site within the gene body probed by bead 3 (Fig. 1). We selected this CpG site because it showed the strongest correlation with IDO1 mRNA expression in the TCGA cohort (Table 1). Mean methylation at this locus in tumor tissue from the UKB cohort was 81.9% [95% CI 79.4–84.5%] compared to 64.4% [95% CI 62.7–66.1%] in the TCGA cohort. Differences in methylation in the TCGA cohort (evaluated by next-generation sequencing) and the independent UKB cohort (evaluated by real-time PCR) might be attributable to the different techniques which are not calibrated. IDO1 protein expression was quantified via immunohistochemistry. Strong expression was found in a subset of infiltrating immune cells as well as in tumor cells (Fig. 2). The percentage of IDO1 expressing cells (tumor and immune cells) positively correlated with IDO1 gene body methylation (Spearman's ρ = 0.181, P = 0.036). Similarly significant positive correlations were observed between IDO1 methylation and the proportion of IDO1 expressing tumor cells (ρ = 0.197, P = 0.022; Table 2) as well as with distinct subsets of immune cells (as described below).
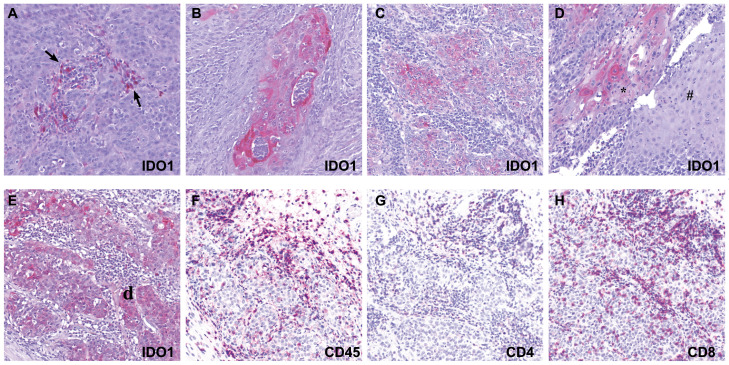
Fig. 2.
Immunohistochemical staining of IDO1, CD45, CD4, and CD8 in HNSCCs. A-D: IDO1 expression in tumor, immune cells, and adjacent tissues. IDO1 expression (red chromagen) in immune cells (arrows); surrounding tumor tissue negative for IDO1 (A). IDO1 expression in tumor tissue; no expression in surrounding immune cells (B, C). IDO1 expression in invasive HNSCC (*) in contrast to negative expression in normal adjacent tissue (#) (D). E-F: Immune microenvironement in HNSCC: IDO1 expression in tumor and immune cells (E) and CD45+ (F), CD4+ (G), and CD8+ (H) expression in tumor-adjacent immune cells (all red chromagen). Original magnification x 10.
Table 2.
Percentage of distinct cell populations in HNSCC tumors and their correlation with IDO1 methylation. Immunohistochemical quantification of percentage of CD45+ leukocytes, T cells (CD3+, CD8+, and CD4+), IDO1+ immune cells, and IDO1+ tumor cells of total cells and their correlation with IDO1 methylation in N = 138 HNSCCs. Shown are numbers of negative (without any expressing cells) and positive (tumors with fractions of expressing cells), mean percentage fraction of expressing cells in positive tumors and the correlations between distinct immune cell subsets and IDO1+ tumor cells with IDO1 methylation. IDO1 methylation was quantified using a quantitative real time PCR assay that targets the CpG sites probed by bead 3 (cg24188163, Fig. 1). Significant data are shown in boldface.
| Correlations (Spearman's ρ, P-value) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cell population | Negative tumors (without expressing cells) | Positive tumors (with expressing cells) | Percentage of total cells (mean [95% CI]; min-max)a | CD45+ leukocytes | CD3+ T cells | CD8+ T cells | CD4+ T cells | IDO1+ immune cells | IDO1+ tumor cells | IDO1 methylation |
| CD45+ leukocytes | 0 (0%) | 138 (100%) | 42 [37–46]; 5–90 | 1 | 0.842; <0.001 | 0.764; <0.001 | 0.777; <0.001 | 0.239; 0.005 | 0.215; 0.012 | 0.125; 0.14 |
| CD3+ T cells | 0 (0%) | 138 (100%) | 15 [13–17]; 1–60 | 0.842; <0.001 | 1 | 0.886; <0.001 | 0.884; <0.001 | 0.213; 0.013 | 0.201; 0.019 | 0.212; 0.013 |
| CD8+ T cells | 0 (0%) | 138 (100%) | 8 [7–9]; 1–45 | 0.764; <0.001 | 0.886; <0.001 | 1 | 0.720; <0.001 | 0.245; 0.004 | 0.219; 0.010 | 0.209; 0.014 |
| CD4+ T cells | 0 (0%) | 138 (100%) | 7 [6–8]; 1–40 | 0.777; <0.001 | 0.884; <0.001 | 0.720; <0.001 | 1 | 0.122; 0.16 | 0.149; 0.083 | 0.230; 0.007 |
| IDO1+ immune cells | 68 (49.3%) | 70 (50.7%) | 2 [1–3]; 1–30 | 0.239; 0.005 | 0.213; 0.013 | 0.245; 0.004 | 0.122; 0.16 | 1 | 0.446; <0.001 | 0.171; 0.048 |
| IDO1+ tumor cells | 104 (75.4%) | 34 (24.6%) | 5 [3–9]; 1–35 | 0.215; 0.012 | 0.201; 0.019 | 0.219; 0.010 | 0.149; 0.083 | 0.446; <0.001 | 1 | 0.197; 0.022 |
positive tumors only.
3.2. Correlation of IDO1 DNA methylation with IFN-γ signature
In the UKB cohort, we detected a significant, positive correlation between the percentage of IDO1-expressing tumor cells and IDO1-expressing immune cells (Spearman's ρ = 0.446, P < 0.001; Table 2). This finding might point to a similar regulation of IDO1 expression in tumor and immune cells, e.g. via cytokines in the TME. IFN-γ is a cytokine that induces IDO1 expression and an IFN-γ signature is proposed to correlate with response to immune checkpoint inhibitors [22,23]. We therefore performed correlative analyses of mRNA expression levels of genes involved in the IFN-γ regulatory pathway (IFNG, STAT1, STAT2, JAK2, and IRF9) with IDO1 methylation and IDO1 mRNA expression in HNSCC tumor tissue from the TCGA cohort. We observed a positive correlation between IDO1 mRNA expression and all IFN-γ signature genes. We found a negative correlation between DNA methylation at the promotor flank region and a positive correlation between DNA methylation at gene body of IDO1 (bead 2 and 3, respectively) with mRNA expression of IFN-γ signature genes. DNA methylation of CpG site located in the promoter of IDO1 (bead 1) only negatively correlated with STAT2 mRNA expression (Table 3).
Table 3.
Correlations of IDO1 methylation and mRNA expression with mRNA expression of interferon γ signature genes. mRNA expression of IFNG, STAT1, STAT2, JAK2, and IRF9 is used as surrogate marker for an interferon γ signature. Matched methylation and mRNA expression data were obtained from N = 521 HNSCCs. Significant data are shown in boldface.
|
IFNG |
STAT1 |
STAT2 |
JAK2 |
IRF9 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Analyte | Spearman's ρ | P-value | Spearman's ρ | P-value | Spearman's ρ | P-value | Spearman's ρ | P-value | Spearman's ρ | P-value |
| IDO1 mRNA | 0.821 | <0.001 | 0.737 | <0.001 | 0.579 | <0.001 | 0.686 | <0.001 | 0.446 | <0.001 |
| IDO1 methylation cg10262052 (bead 1) | −0.039 | 0.38 | −0.050 | 0.26 | −0.094 | 0.033 | 0.036 | 0.41 | −0.078 | 0.076 |
| IDO1 methylation cg08465774 (bead 2) | −0.299 | <0.001 | −0.139 | 0.001 | −0.107 | 0.014 | −0.214 | <0.001 | −0.106 | 0.016 |
| IDO1 methylation cg24188163 (bead 3) | 0.461 | <0.001 | 0.602 | <0.001 | 0.521 | <0.001 | 0.445 | <0.001 | 0.382 | <0.001 |
3.3. IDO1 DNA methylation correlates with leukocyte infiltration
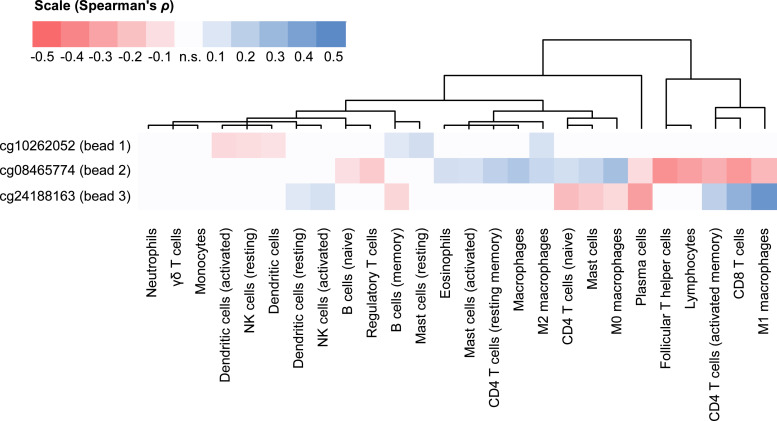
As shown above, we found strong IDO1 protein expression in a small number of immune cells. Consequently, we further investigated the correlations of IDO1 methylation with infiltrates of immune cells. Immune cell infiltration was determined using mRNA expression signatures of distinct subpopulations of immune cells which was developed by Thorsson and colleagues [49]. While we found only weak correlations of IDO1 DNA promoter methylation (bead 1) with six out of the 26 analyzed signatures (three negative [including dendritic cells and resting NK cells] and three positive correlations [memory B cells, resting mast cells, and M2 macrophages]), significant correlations were present for IDO1 promoter flank methylation (bead 2) and 16 immune signatures (Fig. 3). Of note, positive correlations with promoter flank methylation (bead 2) were found for M0/M2 macrophages while M1 macrophages correlated negatively. Additional positive correlations are related to eosinophils, naïve and resting memory CD4+ T cells, and activated mast cells. Most lymphocytes, including naïve B cells, Tregs, follicular T helper cells, activated memory CD4+ T cells, and CD8+ T cells showed negative correlations with promoter flank methylation. In accordance with the oppositional correlations of methylation and mRNA expression at the promoter flank compared to the gene body (bead 3), we detected significant correlations for ten out of 26 immune cell signatures, that were (with plasma cells representing the only exception) oppositely directed, namely memory B cells, naïve CD4+ T cells, mast cells, and M0 macrophages (negative correlations) and resting dendritic cells, activated NK cells, activated memory CD4+ cells, CD8+ T cells, and M1 macrophages (positive correlations).
Fig. 3.
Clustered correlations of IDO1 methylation with signatures of tumor infiltrating immune cells. mRNA expression (RNASeq) signatures described in Thorsson et al. were used to estimate infiltration of distinct immune cell subsets [49]. These signatures were correlated with IDO1 methylation at three CpG sites. Only statistically significant (P < 0.05) Spearman's ρ rank correlation coefficients were used for unsupervised clustering and illustration (n.s.: not significant).
We further corroborated the correlation of IDO1 gene body methylation (assessed via real-time PCR) and leukocyte infiltration in the UKB cohort. Since the results presented above are solely based on RNA signatures of immune cell infitrates, we quantified the proportion of CD45+ leukocytes, CD3+ lymphocytes, CD4+ T cells, and CD8+ T cells via immunohistochemistry in order to validate the results on the level of protein expression. We were able to confirm significant positive correlations between IDO1 gene body methylation and CD3+ lymphocytes, (Spearman's ρ = 0.212, P = 0.013) as well as with CD8+ T cells (ρ = 0.209, P = 0.014) and CD4+ T cells (ρ = 0.230, P = 0.007; Table 2). No significant correlation was found between IDO1 methylation and total leukocyte (CD45+) infiltrates. However, IDO1+ immune cells as well as IDO1+ tumor cells correlated significantly with total leukocyte (CD45+) as well as with total T cell infiltrates (CD3+).
3.4. IDO1 methylation in distinct immune cell populations
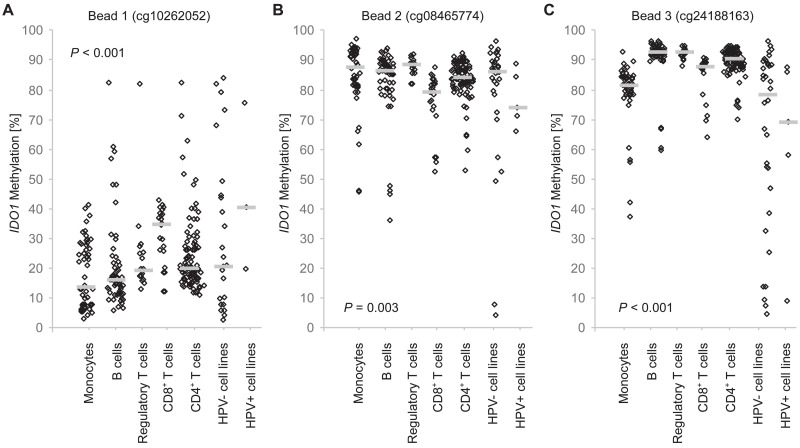
Our results as presented above suggest a significant association of IDO1 methylation with tumor immunity. We therefore investigated IDO1 methylation of purified immune cell subpopulations in peripheral blood from healthy donors. When comparing differential methylation in isolated leukocytes from healthy donors, we found significant differences (Supplemental Table 1, Fig. 4). Significant differences of promoter methylation (bead 1) were found between tumor cells and monocytes and B cells as well as between monocytes and CD8+ T cells and between B cells and CD8+ T cells. Methylation of the CpG site located within the promoter flank, however, showed only significant differential methylation between monocytes and CD8+ T cells and between CD8+ T cells and regulatory T cells. In particular, the CpG site within the gene body targeted by bead 3 showed high and significant methylation differences between tumor cells and immune cells as well as between monocytes and CD4+, CD8+, and regulatory T cells (Supplemental Table 1, Fig. 4)
Fig. 4.
IDO1 methylation in distinct leukocytes, HPV-positive, and HPV-negative cell lines. IDO1 methylation at three sites (A: cg10262052, B: cg08465774, C: cg24188163) in isolated leukocytes (N = 52 monocytes, N = 60 B cells, N = 24 CD8+ T cells, N = 94 CD4+ T cells, N = 18 Tregs) from healthy donors, HPV-positive (cg08465774, cg24188163: N = 5; cg10262052 N = 3), and HPV-negative tumor cell lines (cg08465774, cg24188163: N = 34; cg10262052 N = 23). P-values refer to one-way ANOVA. Bars indicate median methylation levels. Results (P-values) from pairwise Bonferroni post-hoc comparisons are listed in Supplemental Table 1.
3.5. IDO1 DNA methylation and mRNA expression stratified by HPV status
We observed significant differences in IDO1 DNA methylation and its mRNA expression in HPV-negative and HPV-positive tumors (Table 1). Tumors not associated with HPV (HPV-negative) showed higher mean DNA methylation (73.3%, CI: 71.1%−75.4%) at the promoter flank region (bead 2) than HPV-positive tumors (58.8%, [95% CI: 52.8%−64.9%], P < 0.001, Wilcoxon–Mann–Whitney U test). We did not observe significant differences in methylation at the other two investigated loci within the TCGA cohort. In the UKB cohort, however, HPV-positive (p16-positive) tumors showed higher methylation compared to HPV-negative (p16-negative) tumors at the CpG site within the gene body (p16-negative: N = 103, methylation 79.7%, [95% CI: 76.6–82.9%]; p16-positive: N = 35, methylation 88.7%, [95% CI: 86.2–91.1%]; P = 0.010, Wilcoxon–Mann–Whitney U test). Lower IDO1 mRNA expression was observed in HPV-negative tumors (973 n.c., [95% CI: 723–1230]) in comparison with HPV-positive tumors (2127 n.c., [95% CI: 952–3301], P = 0.001) in the TCGA cohort. Concordantly, we found a lower percentage of IDO1-expressing immune cells and tumor cells in HPV-negative vs. HPV-positive tumors in the UKB cohort (IDO1+ immune cells: HPV+: 1.2% [95% CI: 0.77–1.7%]; HPV-: 1.0% [95% CI: 0.35–1.6]; P = 0.007; IDO1+ tumor cells: HPV+: 2.7% [95% CI: 0.28–5.0]; HPV-: 1.0% [95% CI: 0.31–1.0]; P = 0.040, Wilcoxon–Mann–Whitney U test). However, no methylation differences were observed in HPV-negative HNSCC cell lines, when compared with HPV-positive HNSCC cell lines (Fig. 4, Supplemental Table 1) which can not be interpretated due to the small number of investigated cell lines. On the other hand, we found significantely higher infiltrates of CD3+ (P = 0.021) and CD8+ (P = 0.028; Wilcoxon–Mann–Whitney U test) T cells in HPV-positive compared to HPV-negative tumors within the UKB cohort. The difference in the immune infiltration may be the reason for the observed difference of IDO1 methylation between the groups.
3.6. Correlation of IDO1 DNA methylation with overall survival
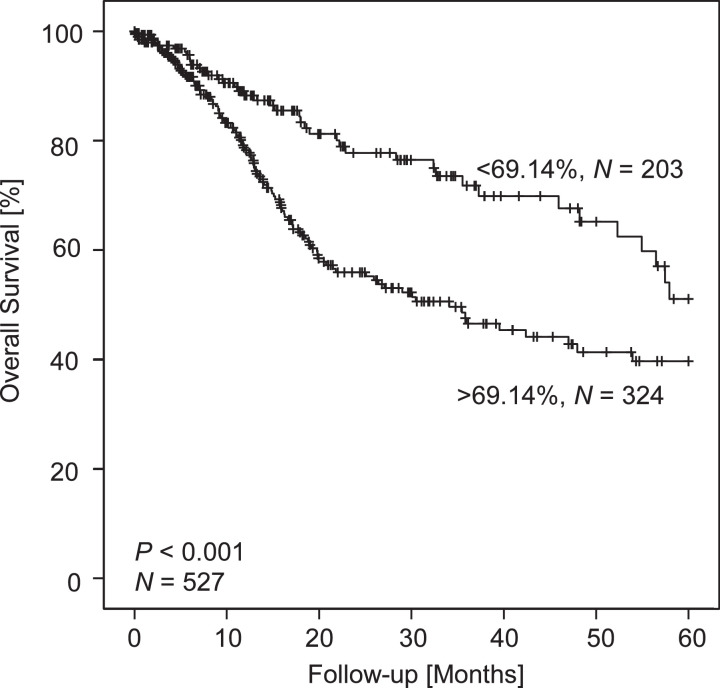
Next, we analyzed the correlation of log2-transformed methylation and mRNA expression levels with patients’ overall survival. A Cox proportional hazards analysis revealed a significant increase in risk of death (HR: 1.53, [95% CI: 1.02–2.30], P = 0.041, Wald test, Table 4) in patients showing higher methylation levels at CpG site within the promoter flank targeted by bead 2. No association with overall survival was observed for CpG sites targeted by bead 1 and 3. We further corroborated our results in a Kaplan-Meier analysis of overall survival of patients stratified according to methylation at the CpG site probed by bead 2. We used an optimized cut-off of 69.14% methylation for the dichotomization of methylation levels. We applied the cut-off that yielded the lowest P-value (P < 0.001, log-rank test) when comparing the overall survival of the hypermethylated and hypomethylated groups (Fig. 5). Patients with tumors habouring IDO1 hypermethylation at CpG site 2 had a median overall survival of 34.1 [95% CI: 22.5–45.7] months whilst the median survival within the patient group with IDO1 hypomethylated tumors was not reached within 5 years of follow-up.
Table 4.
Correlations of IDO1 methylation and mRNA expression with overall survival and mutational load in HNSCC patients. Methylation and mRNA were analyzed as continuous log2-transformed variates. Significant data are shown in boldface. P-values refer to Wald test and Spearman's ρ correlations, respectively.
| Correlation with overall survival (N = 527, methylation; N = 521, mRNA)) |
Correlation with mutational load (N = 256) |
|||
|---|---|---|---|---|
| Analyte | Hazard ratio [95% CI] | P-value | Spearman's ρ | P-value |
| IDO1 mRNA | 0.95 [0.90–1.01] | 0.12 | −0.015 | 0.81 |
| IDO1 methylation cg10262052 | 1.09 [0.92–1.29] | 0.38 | −0.252 | <0.001 |
| IDO1 methylation cg08465774 | 1.53 [1.02–2.30] | 0.041 | −0.161 | 0.010 |
| IDO1 methylation cg24188163 | 1.09 [0.82–1.45] | 0.56 | −0.154 | 0.014 |
Fig. 5.
Kaplan–Meier analysis of overall survival in HNSCC patients stratified according to IDO1 methylation. Patient samples were dichotomized based on an optimized cutoff (69.14% methylation at CpG within the promoter flank targeted by bead cg08465774). Shown are results from N = 527 HNSCC patients from The Cancer Genome Atlas. P-value refers to log-rank test.
3.7. Correlation of IDO1 DNA methylation with tumor mutational load
All CpG sites (targeted by beads 1–3) showed a significant negative correlation between methylation and tumor mutational load (bead 1: Spearman's ρ = −0.252, P < 0.001; bead 2: ρ = −0.161, P = 0.010; bead 3: ρ = −0.154, P = 0.014; Table 4). No significant correlation was observed for IDO1 mRNA expression and mutational load.
3.8. Correlation of IDO1 with PD-1/PD-L1 DNA methylation and mRNA expression
PD-1 promoter methylation as determined using beads cg20805133, cg00795812, cg27051683, cg17322655, and cg03889044 has previously been shown to be a strong prognostic biomarker for overall survival in the same cohort under investigation [32]. In addition, PD-L1 (CD274) methylation at a CpG site within the CD274 promoter (targeted by bead cg19724470) has been reported to be strongly correlated to PD-L1 expression in the TCGA cohort [30]. Since immune checkpoint genes are frequently coexpressed, and PD-1 and IDO inhibitors are being trialed in combination therapy, we further analyzed the correlation of PD-1/PD-L1 methylation and mRNA expression with IDO1. We found a strong PD-1/PD-L1 mRNA coexpression with IDO1 (PD-1: Spearman's ρ = 0.790, P < 0.001; PD-L1: ρ = 0.586, P < 0.001; N = 521). Furthermore, we detected significant positive correlations of IDO1 methylation of CpG site targeted by beads 2 and 3 (N = 528; bead 2: Spearman's ρ = 0.313, P < 0.001; bead 3: ρ = 0.109, P = 0.012) with average PD-1 promoter methylation determined via the five beads mentioned above. No significant correlation between methylation of IDO1 CpG site targeted by bead 1 and PD-1 promoter methylation was present (Spearman's ρ = 0.023, P = 0.60). Of note, we found significant positive correlations between PD-L1 methylation (cg19724470) and IDO1 methylation targeted by bead 1 (Spearman's ρ = 0.156, P < 0.001) and 2 (Spearman's ρ = 0.294, P < 0.001) but a negative correlation for bead 3 (Spearman's ρ = −0.277, P < 0.001).
4. Discussion
IDO1 inhibitors are being tested as promising immunomodulatory agents in patients with advanced cancer, albeit with mixed results thus far [11], [12], [13]. We aimed to investigate the epigenetic regulation by methylation of the IDO1 gene in the tumor immune microenvironment.
DNA methylation is frequently associated with transcriptional gene activity depending on the locus of methylation, e.g. promoter region or gene body [52,53]. In the present study we observed that IDO1 gene expression is negatively associated with promoter and promoter flank methylation which is consistent with the current model of epigenetic regulation whereby methylation in this region often results in decreased gene expression. Our results are concordant with published results from breast and esophageal tumors where IDO1 promoter methylation has been shown to be associated with reduced gene expression [40,41]. In contrast, gene body methylation is well known to correlate positively with mRNA expression [53]. Concordantly, we observed a positive correlation of IDO1 DNA methylation in the gene body with IDO1 mRNA expression as well as with IDO1 protein expression.
HNSCCs are a heterogeneous group of cancers comprising several distinct subgroups that are associated with risik factors, i.e. HPV-infection and smoking [42,54]. A study conducted in patients with HPV-negative oral squamous cell cancers found significant IDO1 overexpression in low-risk tumors that were not attributable to alcohol and tobacco abuse compared to high-risk tumors from smokers and drinkers [55]. In our study, a significant difference in IDO1 methylation status and IDO1 expression (mRNA and protein) was observed between high-risk HPV-negative and low-risk HPV-positive tumors. HPV-associated tumors showed significantly lower DNA methylation in the promoter flank region which correlated with increased mRNA expression. The association of IDO1 expression and methylation with regard to distinct etiologies, e.g. HPV-infection and smoking, needs further investigation. Integration of the viral genome might result in significant changes in the methylome and thus could explain this result [54,56]. However, DNA methylation differences between virus-associated and non-associated tumors could also be explained by different tumor microenvironments. Recently, Thorsson et al. reported IDO1 methylation and expression to be significantly different between immune subtypes across 33 cancer types [49]. This is supported by our observation that distinct immune cell subsets and tumor cells exhibit different IDO1 methylation levels. Concordantly, hypomethylation of the promoter flank region and an increase in IDO1 mRNA expression also correlated positively with infiltration of tumor tissue by CD8+ and CD4+ T cells. This is suggestive of a positive feedback mechanism in which IDO1 expression is upregulated in T cell inflamed tumors. Accordingly, Carrero et al. were able to show, in the same TCGA HNSCC cohort, that HPV-positive tumors harbor a T cell rich microenvironement which might account for the better prognosis in this patient cohort [57]. This finding is in line with our observation that HPV-positive tumors were higher infiltrated with CD3+ and CD8+ lymphocytes. Of note, we also found a significant correlation between IDO1+ immune cells and IDO+ tumor cells in HNSCCs, suggesting that there may be a similar mechanism for IDO1 induction in these cells, e.g. via proinflammatory cytokines in the TME. For example, it has been shown that IFN-γ released by T cells induces IDO1 expression [58] which is in line with the strong correlation between IDO1 expression and an IFN-γ signature as shown in our study.
In conclusion, DNA methylation analysis of IDO1 could serve as a biomarker to identify tumors that show a T cell-inflamed signature (including IDO1 expressing immune cells) with an upregulated IFN-γ response. These conditions have been shown to be necessary but not always sufficient for clinical benefit from PD-1 immune checkpoint inhibition [59]. These patients might benefit from IDO1 inhibitors, either alone or in combination with other checkpoint inhibitors, e.g. PD-1 immune checkpoint inhibitors, since PD-L1 is also upregulated by IFN-γ [60].
Expression of PD-L1 in tumor tissue and/or surrounding immune cells is currently the best predictor for response to immune checkpoint inhibition. However, its clinical performance is limited and more accurate biomarkers or biomarkers that add informative value are needed. We did observe a positive correlation of PD-1/PD-L1 and IDO1 mRNA expression and methylation in our cohort which is in line with a previous report [61]. The strong correlations between IDO1 methylation with the expression of other immune checkpoints, TILs, and an interferon γ suggests IDO1 methylation as a biomarker for general tumor immunogenicity. Again, it would be interesting to investigate in a prospective setting, if IDO1 methylation could act as predictive biomarker for response to immune checkpoint inhibition, including anti-IDO1 and/or anti-PD-1 antibodies. A multitude of PD-L1 tests and scoring systems are currently used and their harmonization is challenging [62]. A simple quantitative DNA methylation test might allow for a highly reproducible testing between laboratories.
One predictor for response to immunotherapy with checkpoint inhibitors is a high tumor mutational load with a high number of nonsynonymous mutations. These likely result in a high number of immunogenic neoantigens [63]. In the present study, IDO1 DNA methylation of all three analyzed CpG sites was negatively correlated with tumor mutational burden. We did not observe an association of IDO1 expression with mutational load on the mRNA level so the relevance of the aforementioned findings is unclear.
A study investigating the microenvironment of malignant melanomas described an interesting feedback mechanism. The authors found that in tumors with high infiltration of CD8+ T cells, immunosuppressive pathways including IDO1, PD-L1, and regulatory T cells are also upregulated [64]. Our study supports this finding in that IDO1 DNA methylation and expression is correlated with T cell infiltration and methylation and mRNA expression of PD-L1 and PD-1. This could provide an additional rationale for targeting immunosuppressive pathways like the ones regulated by IDO1 and PD-L1.
Our results demonstrate the complex relationship between tumor microenvironment and epigenetic regulation of the immune suppressive gene IDO1. While biomarker studies in patient cohorts from IDO1 inhibitor trials are still awaited [65], analysis of DNA methylation status of IDO1 could potentially help select subgroups of patients with high IDO1 expression and possibily a high level of anti-tumoral T cell infiltration, who are likely to benefit from IDO1 inhibitor therapy. Further studies including anti-IDO1 treated HNSCC patient cohorts are needed to test the possibility of using IDO1 methylation as a predictive biomarker for IDO1 inhibitors. While the predictive value of IDO1 methylation testing is purely speculative based on the present data, it is well acknowledged that most likely not only a single parameter is sufficient to reliably predict response to immunotherapy. Hence, IDO1 methylation needs to be tested together with other potentially predictive factors in order to assess potential additive value. If IDO1 methylation indeed shows some predictive value, its implementation into clinical testing might harbor several advantages. DNA methylation testing is inexpensive and can be performed even in small amounts of formalin-fixed and paraffin-embedded clinical specimens [51,66,67]. It needs to be mentioned that a significant number of clinically relevant specimens are too small to allow for an analysis of all parameters. An additional advantage of DNA methylation testing is its biological stability while mRNA and protein expression might highly be effected by the sample processing procedure. Moreover, since a diploid cell contains only two alleles of each gene, the individual contribution of each cell to the overall methylation signal obtained from a heterogeneous sample is the same. In contrast, protein and mRNA quantification might be impaired by small subgroups of cells that highly express the gene and thereby mask the lower expression of the majority of cells.
In conclusion, our results suggest IDO1 expression levels are epigenetically regulated by DNA methylation. IDO1 methylation correlates with features of responsiveness (PD-L1 expression, CD8+ T cell infiltrates, tumor mutational burden, and an IFN-γ signature) to immune checkpoint inhibitors. Our study provides rationale to test IDO1 methylation as potential biomarker for prediction of response to IDO1 immune checkpoint inhibitors.
Funding source
The study was funded by the University Hospital Bonn.
Declaration of Competing Interest
None.
CRediT authorship contribution statement
Verena Sailer: Data curation, Formal analysis, Methodology, Visualization, Writing - original draft. Ulrike Sailer: Data curation, Formal analysis, Methodology, Visualization. Emma Grace Bawden: Writing - review & editing. Romina Zarbl: Data curation, Formal analysis, Methodology, Visualization. Constanze Wiek: Funding acquisition, Resources. Timo J. Vogt: Data curation, Formal analysis, Methodology, Visualization. Joern Dietrich: Data curation, Formal analysis, Methodology, Visualization. Sophia Loick: Data curation, Formal analysis, Methodology, Visualization. Ingela Grünwald: Data curation, Formal analysis, Methodology, Visualization. Marieta Toma: Data curation, Formal analysis, Methodology, Visualization. Carsten Golletz: Data curation, Formal analysis, Methodology, Visualization. Andreas Gerstner: Funding acquisition, Resources. Glen Kristiansen: Funding acquisition, Resources. Friedrich Bootz: Funding acquisition, Resources. Kathrin Scheckenbach: Funding acquisition, Resources. Jennifer Landsberg: Conceptualization, Project administration, Supervision, Validation, Writing - review & editing. Dimo Dietrich: Data curation, Formal analysis, Methodology, Visualization, Conceptualization, Project administration, Supervision, Validation, Writing - review & editing.
Acknowledgment
Dimo Dietrich owns patents and patent applications on biomarker technologies and methylation of immune checkpoint genes as predictive and prognostic biomarkers (DE 10 2016 005 947.8, DE 10 2015 009 187.5, DE 10 2017 125 780.2, PCT/EP2016/001237). The patents are licensed to Qiagen GmbH (Hilden, Germany). Dimo Dietrich is a consultant of Qiagen. The University Hospital Bonn (PI Dimo Dietrich) receives research funding from Qiagen. Dimo Dietrich is a consultant for AJ Innuscreen GmbH (Berlin, Germany), a 100% daughter company of Analytik Jena AG (Jena, Germany), and receives royalties from product sales (innuCONVERT kits).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2019.09.038.
Appendix. Supplementary materials
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Fakhry C., Zhang Q., Nguyen-Tan P.F., Rosenthal D., El-Naggar A., Garden A.S. Human papillomavirus and overall survival after progression of oropharyngeal squamous cell carcinoma. J Clin Oncol. 2014;32:3365–3373. doi: 10.1200/JCO.2014.55.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ang K.K., Harris J., Wheeler R., Weber R., Rosenthal D.I., Nguyen-Tân P.F. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363:24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taberna M., Oliva M., Mesía R. Cetuximab-Containing combinations in locally advanced and recurrent or metastatic head and neck squamous cell carcinoma. Front Oncol. 2019;9:383. doi: 10.3389/fonc.2019.00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferris R.L., Blumenschein G, Jr., Fayette J., Guigay J., Colevas A.D., Licitra L. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N Engl J Med. 2016;375:1856–1867. doi: 10.1056/NEJMoa1602252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferris R.L., Blumenschein G, Jr., Fayette J., Guigay J., Colevas A.D., Licitra L. Nivolumab vs investigator's choice in recurrent or metastatic squamous cell carcinoma of the head and neck: 2-year long-term survival update of Checkmate 141 with analyses by tumor PD-L1 expression. Oral Oncol. 2018;81:45–51. doi: 10.1016/j.oraloncology.2018.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colevas A.D., Bahleda R., Braiteh F., Balmanoukian A., Brana I., Chau N.G. Safety and clinical activity of atezolizumab in head and neck cancer: results from a phase I trial. Ann Oncol. 2018;29:2247–2253. doi: 10.1093/annonc/mdy411. [DOI] [PubMed] [Google Scholar]
- 9.Segal N.H., Ou S.I., Balmanoukian A., Fury M.G., Massarelli E., Brahmer J.R. Safety and efficacy of durvalumab in patients with head and neck squamous cell carcinoma: results from a phase I/II expansion cohort. Eur J Cancer. 2019;109:154–161. doi: 10.1016/j.ejca.2018.12.029. [DOI] [PubMed] [Google Scholar]
- 10.Cohen E.E.W., Bell R.B., Bifulco C.B., Burtness B., Gillison M.L., Harrington K.J. The society for immunotherapy of cancer consensus statement on immunotherapy for the treatment of squamous cell carcinoma of the head and neck (HNSCC) J Immunother Cancer. 2019;7:184. doi: 10.1186/s40425-019-0662-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitchell T.C., Hamid O., Smith D.C., Bauer T.M., Wasser J.S., Olszanski A.J. Epacadostat plus pembrolizumab in patients with advanced solid tumors: phase I results from a multicenter, open-label phase I/II trial (ECHO-202/KEYNOTE-037) J Clin Oncol. 2018 doi: 10.1200/JCO.2018.78.9602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jung K.H., LoRusso P., Burris H., Gordon M., Bang Y.J., Hellmann M.D. Phase I study of the indoleamine 2,3-dioxygenase 1 (IDO1) inhibitor navoximod (GDC-0919) administered with PD-L1 inhibitor (Atezolizumab) in advanced solid tumors. Clin Cancer Res. 2019;25:3220–3228. doi: 10.1158/1078-0432.CCR-18-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nayak-Kapoor A., Hao Z., Sadek R., Dobbins R., Marshall L., Vahanian N.N. Phase I study of the indoleamine 2,3-dioxygenase 1 (IDO1) inhibitor navoximod (GDC-0919) in patients with recurrent advanced solid tumors. J Immunother Cancer. 2018;6:61. doi: 10.1186/s40425-018-0351-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adams S., Braidy N., Bessede A., Brew B.J., Grant R., Teo C. The kynurenine pathway in brain tumor pathogenesis. Cancer Res. 2012;72:5649–5657. doi: 10.1158/0008-5472.CAN-12-0549. [DOI] [PubMed] [Google Scholar]
- 15.Prendergast G.C., Malachowski W.J., Mondal A., Scherle P., Muller A.J. Indoleamine 2,3-dioxygenase and its therapeutic inhibition in cancer. Int Rev Cell Mol Biol. 2018;336:175–203. doi: 10.1016/bs.ircmb.2017.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maliniemi P., Laukkanen K., Väkevä L., Dettmer K., Lipsanen T., Jeskanen L. Biological and clinical significance of tryptophan-catabolizing enzymes in cutaneous T-cell lymphomas. Oncoimmunology. 2017;6 doi: 10.1080/2162402X.2016.1273310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yentz S., Smith D. Indoleamine 2, 3-dioxygenase (IDO) inhibition as a strategy to augment cancer immunotherapy. BioDrugs. 2018;32:311–317. doi: 10.1007/s40259-018-0291-4. [DOI] [PubMed] [Google Scholar]
- 18.Godin-Ethier J., Hanafi L.A., Piccirillo C.A., Lapointe R. Indoleamine 2,3-dioxygenase expression in human cancers: clinical and immunologic perspectives. Clin Cancer Res. 2011;17:6985–6991. doi: 10.1158/1078-0432.CCR-11-1331. [DOI] [PubMed] [Google Scholar]
- 19.Prendergast G.C., Malachowski W.P., DuHadaway J.B., Muller A.J. Discovery of IDO1 inhibitors: from bench to bedside. Cancer Res. 2017;77:6795–6811. doi: 10.1158/0008-5472.CAN-17-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komiya T., Huang C.H. Updates in the clinical development of epacadostat and other indoleamine 2,3-dioxygenase 1 inhibitors (IDO1) for human cancers. Front Oncol. 2018;8:423. doi: 10.3389/fonc.2018.00423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Long G.V., Dummer R., Hamid O., Gajewski T.F., Caglevic C., Dalle S. Epacadostat plus pembrolizumab versus placebo plus pembrolizumab in patients with unresectable or metastatic melanoma (ECHO-301/KEYNOTE-252): a phase 3, randomised, double-blind study. Lancet Oncol. 2019;20:1083–1097. doi: 10.1016/S1470-2045(19)30274-8. [DOI] [PubMed] [Google Scholar]
- 22.Keenan T.E., Burke K.P., Van Allen E.M. Genomic correlates of response to immune checkpoint blockade. Nat Med. 2019;25:389–402. doi: 10.1038/s41591-019-0382-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gibney G.T., Weiner L.M., Atkins M.B. Predictive biomarkers for checkpoint inhibitor-based immunotherapy. Lancet Oncol. 2016;17:e542–e551. doi: 10.1016/S1470-2045(16)30406-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Topalian S.L., Taube J.M., Anders R.A., Pardoll D.M. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer. 2016;16:275–287. doi: 10.1038/nrc.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson A.A., Akman K., Calimport S.R., Wuttke D., Stolzing A., de Magalhães J.P. The role of DNA methylation in aging, rejuvenation, and age-related disease. Rejuvenation Res. 2012;15:483–494. doi: 10.1089/rej.2012.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robertson K.D. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 27.Ghoneim H.E., Fan Y., Moustaki A., Abdelsamed H.A., Dash P., Dogra P. De novo epigenetic programs inhibit PD-1 blockade-mediated t cell rejuvenation. Cell. 2017;170:142–157. doi: 10.1016/j.cell.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scharer C.D., Barwick B.G., Youngblood B.A., Ahmed R., Boss J.M. Global DNA methylation remodeling accompanies CD8 t cell effector function. J Immunol. 2013;191:3419–3429. doi: 10.4049/jimmunol.1301395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Durek P., Nordstrom K., Gasparoni G., Salhab A., Kressler C., de Almeida M. et al. epigenomic profiling of human CD4(+) T cells supports a linear differentiation model and highlights molecular regulators of memory development. Immunity. 2016;45:1148–1161. doi: 10.1016/j.immuni.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 30.Franzen A., Vogt T.J., Muller T., Dietrich J., Schrock A., Golletz C. PD-L1 (CD274) and PD-L2 (PDCD1LG2) promoter methylation is associated with hpv infection and transcriptional repression in head and neck squamous cell carcinomas. Oncotarget. 2018;9:641–650. doi: 10.18632/oncotarget.23080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rover L.K., Gevensleben H., Dietrich J., Bootz F., Landsberg J., Goltz D. PD-1 (PDCD1) promoter methylation is a prognostic factor in patients with diffuse lower-grade gliomas harboring isocitrate dehydrogenase (IDH) mutations. EBioMedicine. 2018;28:97–104. doi: 10.1016/j.ebiom.2018.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goltz D., Gevensleben H., Dietrich J., Schroeck F., de Vos L., Droege F. PDCD1 (PD-1) promoter methylation predicts outcome in head and neck squamous cell carcinoma patients. Oncotarget. 2017;8:41011–41020. doi: 10.18632/oncotarget.17354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goltz D., Gevensleben H., Dietrich J., Dietrich D. PD-L1 (CD274) promoter methylation predicts survival in colorectal cancer patients. Oncoimmunology. 2017;6 doi: 10.1080/2162402X.2016.1257454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goltz D., Gevensleben H., Dietrich J., Ellinger J., Landsberg J., Kristiansen G. Promoter methylation of the immune checkpoint receptor PD-1 (PDCD1) is an independent prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients following radical prostatectomy. Oncoimmunology. 2016;5 doi: 10.1080/2162402X.2016.1221555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gevensleben H., Holmes E.E., Goltz D., Dietrich J., Sailer V., Ellinger J. PD-L1 promoter methylation is a prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients following radical prostatectomy. Oncotarget. 2016;7:79943–79955. doi: 10.18632/oncotarget.13161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Micevic G., Thakral D., McGeary M., Bosenberg M. PD-L1 methylation regulates PD-L1 expression and is associated with melanoma survival. Pigment Cell Melanoma Res. 2018;32:435–440. doi: 10.1111/pcmr.12745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Y.P., Zhang J., Wang Y.Q., Liu N., He Q.M., Yang X.J. The immune molecular landscape of the B7 and TNFR immunoregulatory ligand-receptor families in head and neck cancer: a comprehensive overview and the immunotherapeutic implications. Oncoimmunology. 2017;6 doi: 10.1080/2162402X.2017.1288329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goltz D., Gevensleben H., Grunen S., Dietrich J., Kristiansen G., Landsberg J. PD-L1 (CD274) promoter methylation predicts survival in patients with acute myeloid leukemia. Leukemia. 2017;31:738–743. doi: 10.1038/leu.2016.328. [DOI] [PubMed] [Google Scholar]
- 39.Goltz D., Gevensleben H., Vogt T.J., Dietrich J., Golletz C., Bootz F. CTLA4 methylation predicts response to anti-PD-1 and anti-CTLA-4 immunotherapy in melanoma patients. JCI Insight. 2018;3 doi: 10.1172/jci.insight.96793. pii: 96793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dewi D.L., Mohapatra S.R., Blanco Cabanes S., Adam I., Somarribas Patterson L.F., Berdel B. Suppression of indoleamine-2,3-dioxygenase 1 expression by promoter hypermethylation in ER-positive breast cancer. Oncoimmunology. 2017;6 doi: 10.1080/2162402X.2016.1274477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kiyozumi Y., Baba Y., Okadome K., Yagi T., Ogata Y., Eto K. Indoleamine 2, 3-dioxygenase 1 promoter hypomethylation is associated with poor prognosis in patients with esophageal cancer. Cancer Sci. 2019;110:1863–1871. doi: 10.1111/cas.14028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cancer Genome Atlas Network Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kandoth C., McLellan M.D., Vandin F., Ye K., Niu B., Lu C. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lechner M., Fenton T., West J., Wilson G., Feber A., Henderson S. Identification and functional validation of HPV-mediated hypermethylation in head and neck squamous cell carcinoma. Genome Med. 2013;5(2):15. doi: 10.1186/gm419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M. A landscape of pharmacogenomic interactions in cancer. Cell. 2016;166:740–754. doi: 10.1016/j.cell.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ventham N.T., Kennedy N.A., Adams A.T., Kalla R., Heath S., O'Leary K.R. Integrative epigenome-wide analysis demonstrates that DNA methylation may mediate genetic risk in inflammatory bowel disease. Nat Commun. 2016;7:13507. doi: 10.1038/ncomms13507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mamrut S., Avidan N., Staun-Ram E., Ginzburg E., Truffault F., Berrih-Aknin S. Integrative analysis of methylome and transcriptome in human blood identifies extensive sex- and immune cell-specific differentially methylated regions. Epigenetics. 2015;10:943–957. doi: 10.1080/15592294.2015.1084462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Absher D.M., Li X., Waite L.L., Gibson A., Roberts K., Edberg J. Genome-wide DNA methylation analysis of systemic lupus erythematosus reveals persistent hypomethylation of interferon genes and compositional changes to CD4+ T-cell populations. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thorsson V., Gibbs D.L., Brown S.D., Wolf D., Bortone D.S., Ou Yang T.H. The immune landscape of cancer. Immunity. 2018;48:812–830. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lehmann U., Kreipe H. Real-time PCR-based assay for quantitative determination of methylation status. Methods Mol Biol. 2004;287:207–218. doi: 10.1385/1-59259-828-5:207. [DOI] [PubMed] [Google Scholar]
- 51.Jung M., Uhl B., Kristiansen G., Dietrich D. Bisulfite conversion of dna from tissues, cell lines, buffy coat, ffpe tissues, microdissected cells, swabs, sputum, aspirates, lavages, effusions, plasma, serum, and urine. Methods Mol Biol. 2017;1589:139–159. doi: 10.1007/7651_2015_260. [DOI] [PubMed] [Google Scholar]
- 52.Ehrlich M., Lacey M. DNA methylation and differentiation: silencing, upregulation and modulation of gene expression. Epigenomics. 2013;5:553–568. doi: 10.2217/epi.13.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang X., Han H., De Carvalho D.D., Lay F.D., Jones P.A., Liang G. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014;26:577–590. doi: 10.1016/j.ccr.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brennan K., Koenig J.L., Gentles A.J., Sunwoo J.B., Gevaert O. Identification of an atypical etiological head and neck squamous carcinoma subtype featuring the CPG island methylator phenotype. EBioMedicine. 2017;17:223–236. doi: 10.1016/j.ebiom.2017.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Foy J.P., Bertolus C., Michallet M.C., Deneuve S., Incitti R., Bendriss-Vermare N. The immune microenvironment of HPV-negative oral squamous cell carcinoma from never-smokers and never-drinkers patients suggests higher clinical benefit of IDO1 and PD1/PD-L1 blockade. Ann Oncol. 2017;28:1934–1941. doi: 10.1093/annonc/mdx210. [DOI] [PubMed] [Google Scholar]
- 56.Minarovits J., Demcsak A., Banati F., Niller H.H. Epigenetic dysregulation in virus-associated neoplasms. Adv Exp Med Biol. 2016;879:71–90. doi: 10.1007/978-3-319-24738-0_4. [DOI] [PubMed] [Google Scholar]
- 57.Carrero I., Liu H.C., Sikora A.G., Milosavljevic A. Histoepigenetic analysis of HPV- and tobacco-associated head and neck cancer identifies both subtype-specific and common therapeutic targets despite divergent microenvironments. Oncogene. 2019;38:3551–3568. doi: 10.1038/s41388-018-0659-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yasui H., Takai K., Yoshida R., Hayaishi O. Interferon enhances tryptophan metabolism by inducing pulmonary indoleamine 2,3-dioxygenase: its possible occurrence in cancer patients. PNAS. 1986;83:6622–6626. doi: 10.1073/pnas.83.17.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ayers M., Lunceford J., Nebozhyn M., Murphy E., Loboda A., Kaufman D.R. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Investig. 2017;127:2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Garcia-Diaz A., Shin D.S., Moreno B.H., Saco J., Escuin-Ordinas H., Rodriguez G.A. Interferon receptor signaling pathways regulating PD-L1 and PD-L2 expression. Cell Rep. 2017;19:1189–1201. doi: 10.1016/j.celrep.2017.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krähenbühl L., Goldinger S.M., Mangana J., Kerl K., Chevolet I., Brochez L. A longitudinal analysis of IDO and PDL1 expression during immune- or targeted therapy in advanced melanoma. Neoplasia. 2018;20:218–225. doi: 10.1016/j.neo.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scheel A.H., Dietel M., Heukamp L.C., Johrens K., Kirchner T., Reu S. Harmonized PD-L1 immunohistochemistry for pulmonary squamous-cell and adenocarcinomas. Mod Pathol. 2016;29:1165–1172. doi: 10.1038/modpathol.2016.117. [DOI] [PubMed] [Google Scholar]
- 63.Rizvi N.A., Hellmann M.D., Snyder A., Kvistborg P., Makarov V., Havel J.J. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Spranger S., Spaapen R.M., Zha Y., Williams J., Meng Y., Ha T.T. Up-regulation of PD-L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Sci Transl Med. 2013;5:200ra116. doi: 10.1126/scitranslmed.3006504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Muller A.J., Manfredi M.G., Zakharia Y., Prendergast G.C. Inhibiting IDO pathways to treat cancer: lessons from the ECHO-301 trial and beyond. Semin Immunopathol. 2019;41:41–48. doi: 10.1007/s00281-018-0702-0. [DOI] [PubMed] [Google Scholar]
- 66.Uhl B., Gevensleben H., Tolkach Y., Sailer V., Majores M., Jung M. PITX2 DNA methylation as biomarker for individualized risk assessment of prostate cancer in core biopsies. J Mol Diagn. 2017;19:107–114. doi: 10.1016/j.jmoldx.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 67.Dietrich D., Lesche R., Tetzner R., Krispin M., Dietrich J., Haedicke W. Analysis of DNA methylation of multiple genes in microdissected cells from formalin-fixed and paraffin-embedded tissues. J Histochem Cytochem. 2009;57:477–489. doi: 10.1369/jhc.2009.953026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.