Abstract
Background
Colorectal cancer (CRC) is one of the high incidences tumours and is ranked second in cancer-related mortality. Even though great progress has been made, there are no effective therapeutic strategies for late stage and metastatic CRC patients. Acidity is one characteristic of the tumour microenvironment. However, how cancer cells respond to this acidic environment surrounding them remains largely unknown, especially in colorectal cancer.
Methods
Proton sensor receptor expression was analysed in GEO and TCGA datasets. The expression of GPR4 in CRC specimens was confirmed by western blotting and immunohistochemistry (IHC). The role of GPR4 in CRC progression was analysed both in vitro and in vivo. Pharmacological intervention, immunofluorescence and gene set enrichment analyses were performed to reveal the underlying molecular mechanisms of GPR4.
Findings
We found that GPR4 was upregulated in CRC samples. In addition, its high expression correlated with late stage tumours and poor overall survival in patients. Furthermore, loss-of-function assays proved that GPR4 promoted CRC carcinogenesis and metastatic ability. Mechanistically, GPR4 was activated by extracellular protons in the tumour microenvironment and enhanced RhoA activation and F-actin rearrangement, leading to LATS activity inhibition, YAP1 nuclear translocation and oncogene transcription.
Interpretation
The expression of GPR4 is upregulated in colorectal cancer and is associated with shorter overall survival time in CRC patients. These findings reveal the novel roles of GPR4 in CRC progression and suggest GPR4 might be a new therapeutic target for CRC treatment.
Keywords: Extracellular acidification, GPR4, RhoA, Hippo pathway
Research in context.
Evidence before this study
Acidosis has been defined as a characteristic of the tumour microenvironment. Previous researches showed that proton-sensing G-protein coupled receptors play important roles in responding to the extracellular H+. However, searching for “colorectal cancer”, “acidosis”, and “tumour microenvironment” as keywords in PubMed, we found that how colorectal cancer response to acidic microenvironment largely remained unknown. In addition, the roles of GPR4 in tumour progression were poorly studied by searching for GPR4 as keyword in PubMed. Also, the mechanism by which cancer cells take advantages of GPR4 and acidosis tumour microenvironment largely remained unknown by searching for “GPR4” and “cancer” as keywords in PubMed.
Added value of this study
Our studies revealed that GPR4, whose expression is positively associated with poor prognosis in CRC, responded to extracellular H+ and promoted the growth and migration of CRC cancer cells. Mechanistic studies indicated that activated GPR4 triggers YAP1 nuclear localization and downstream target gene expression via the RhoA-F-actin axis. Furthermore, mouse model experiments showed that GPR4 deletion suppressed CRC tumour progression, indicating that it may be a promising therapeutic target.
Implications of all the available evidence
Our findings showed that GPR4 inhibition suppressed CRC tumour progression both in vitro and in vivo. Recently, the GPR4 selective antagonist NE-52-QQ57 had been developed. Our study established a primary foundation for GPR4 translational research, and future efforts will attempt to elucidate the effect of GPR4 in non-tumour cells and evaluate its potential for translational applications.
Alt-text: Unlabelled Box
1. Introduction
Colorectal cancer (CRC) is one of the most commonly diagnosed cancer and the second leading cause of cancer-related death worldwide [1]. Despite the decreasing trend in deaths and new cases of CRC from 1992 to 2016, the incidence of CRC has been increasing in the United States among individuals younger than 50 years old [2]. In addition, patients with advanced and metastatic cancer, accounting for more than half of CRC cases, have fewer therapy strategies and thus an extremely poor prognosis [3]; these statistics highlight our incomplete understanding of the mechanism of CRC initiation and progression.
The characteristics of the solid tumour microenvironment are multifarious. The combination of weak vascular infiltration, insufficient oxygen, and enhanced lactate efflux from cancer cells, which prefer glycolysis, in solid tumours causes pH values of the tumour environment decrease to 6.5 [4]. Previous studies reported that tumour microenvironment acidification occurs in the early stage of cancer progression [5]. With the progression of hyperplasia, cancer cells gradually separate from the basement tissue and vasculature in space, resulting in cell exposure to a hypoxic environment and metabolic reprogramming to a more glycolytic-dependent status, also called the Warburg effect [6], which promotes proton accumulation in the interstitial fluid due to limited oxygen diffusion and increased acid production. Therefore, understanding the mechanisms by which transformed cells adapt to the acidic surrounding is critical to understanding how cancer progresses to local invasion.
Cells sense the extracellular plasma pH mainly through proton-sensing G-protein coupled receptors, including GPR4, OGR1 (GPR68) TDAG8 (GPR65), and G2A (GPR132) [7,8]. Under normal physiological conditions, GPR4 plays a crucial role in mediating neuronal respiratory sensitivity to CO2 homeostasis [9,10] and renal acid excretion [11]. Additionally, GPR4 was also reported to take part in the immune response by recruiting monocytes and neutrophils [12,13]. At the same time, GPR4 expression was reported to be upregulated in breast cancer, ovarian cancer, and liver cancer [14]. Early primary osteoblast tissue and osteosarcoma cancer cells have strong GPR4 expression at their membranes [7]. After activation under acidic pH conditions, GPR4 was reported to regulate endothelial cell growth, and its deletion reduced angiogenesis, resulting in abrogated tumour growth in GPR4−/−mice [15]. Similar to a number of GPCR superfamily members, GPR4 was reported to activate the canonical downstream signalling pathway. Overexpression of GPR4 constitutively mediates cAMP accumulation as observed in HUVECs [16]. Ectopic GPR4 expression could increase the activity of p115RhoGEF, which is regulated by G12 or G13 proteins [17]. Additionally, Gq/PLC is one of the dominant downstream signalling pathways of GPR4 acted as a pH-sensing receptor [18]. Further structure-dependent studies revealed that the three residues of GPR4 at 79, 165, and 269 are important for triggering multiple intracellular signalling pathways through a conformational change of the receptor for G-protein coupling [18].
In this study, we aimed to explore how cancer cells respond to acidic microenvironments in CRC and how such knowledge might directly influence the progression of CRC. By datasets analysis, we found that upregulated proton-sensor-receptor GPR4 in CRC cells could be activated by extracellular H+, following activation of the RhoA signalling axis to regulate F-actin; finally, the hippo pathway is activated to trigger oncogene expression and CRC progression.
2. Materials and methods
2.1. Cell lines and culture method
Colorectal cancer cell lines COLO302, DLD1, HCT116, LOVO, SW480, SW620 and the murine-derived colorectal cancer cell MC38 were obtained from the ATCC. All cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fatal bovine serum (FBS, Gibco), 1% penicillin and 1% streptomycin (Life Technologies). For the function and mechanism assay, cells were grown in pH 6.8 medium; otherwise, they were grown at pH 7.4 medium. Cells with <20 passages were used in this study. Cells were tested for mycoplasma contamination through a MycoProbe Kit (CUL001B, R&D), and cells with negative test results were used in this study.
2.2. Lentiviral production and cell transduction
GPR4 shRNA and negative control shRNA were constructed into the pLKO.1 plasmid (#10878, Addgene) with the target sequences shRNA1CCCUCUACAUCUUUGUCAU (human), shRNA2 CCAUCACAGUUUGGCGAUU (human), shGPR4GCGUCUACCUGAUGAACUU (mouse) and shNC AUACCUGAAGCGAGAUUG. The abovementioned plasmids were transfected into 293 T cells with REV, RRE and VSVG to package the lentivirus. After 48 h of transfection, the cell culture supernatants were collected, spun for 15 min at 3000 rpm, and then passed through a 0.22-μm filter. Viral supernatants were used to infect target cells supplemented with polybrene for 24 h. The infected cells were then cultured in medium supplied with 2.5 mg/ml puromycin for one week.
2.3. Quantitative PCR and chromatin immunoprecipitation (ChIP) PCR
A PureLink™ RNA Mini Kit was used to isolate RNA according to the standard protocol. Total mRNA was further transcribed to cDNA by using an M-MLV Reverse Transcriptase cDNA synthesis kit (Invitrogen). RNA expression level was quantified by Q-PCR with SYBR-Green PCR kit (Invitrogen) mixed with cDNAs and gene-specific primers; reactions were performed on an ABI qPCR 7500. β-actin was used as mRNA loading control. The 2(-ΔΔCt) method was used to analyse the relative mRNA expression levels of genes. The RT-PCR primers are listed in Supplementary Table 1. As ChIP-PCR, 1% formaldehyde was used to fix the interaction between DNA and protein, following sonicate to shear DNA into fragment. After that, anti-TEAD1 (ab133533, Abcam) was incubated with those prepared samples and immunoprecipitated. Q-PCR was performed to detected the DNA precipitated by anti-TEAD1 antibody. The ChIP-PCR primers are listed in Supplementary Table 2.
2.4. Western blots
Cell lysate for immunoblotting was performed according to the standard protocol. Briefly, cells were lysed with buffer that freshly added protease and phosphatase inhibitor cocktail (B15001, Bimake); then, 4%–20% SDS polyacrylamide gels was used to separate proteins, following transferred onto 0.22-μm NC membranes (Thermo Fisher Scientific), and incubated with primary antibodies at 4 °C overnight. The next day, HRP-conjugated secondary antibodies were incubated at room temperature (RT) for 2 h. β-actin was used for the total protein loading control. The primary antibodies and dilutions as follows: GPR4(1:1000, ab75330, Abcam), Phospho-MST1 (T183) (1:1000, 49,322, Cell signalling), MST1(1:1000, 3682, Cell signalling), Phospho-LATS1/2(T1079 + T1041)(1:1000, ab111344, Abcam), Phospho-YAP1 (S127) (1:1000, 13,088, Cell signalling), YAP1(1:1000, 14,074, Cell signalling), β-actin (1:3000, ab8226, Abcam).
2.5. Cell proliferation assay
Cell Counting Kit-8 was used for cell proliferation assays. Briefly, cells were diluted to 3000 cells/100 μl; then, they were seeded into 96-well plates and incubated for 12 h. Next, the cells were cultured in pH 6.8 DMEM. Then, 10% CCK-8 reagent was mixed with 90% medium and incubated with the cells for 1 h at incubator. OD450 was measured to indicate cell proliferation through PerkinElmer microplate reader.
2.6. Colony formation assay
For the colony formation assay, 3000 cells/well were seeded into 6-well plates and cultured with different medium or different reagent treatments for 2 weeks. Then, 4% paraformaldehyde was used to fix the cells for 2 h at RT; the cells were stained with a 0.25% crystal violet buffer and analysed. The difference of groups was compared by subtracting the extraction buffer value from the previously stained cells and measuring the absorbance at OD 570 nm.
2.7. Wound healing assay
A wound healing assay was performed using Culture-Insert 2 Wells in a μ-Dish (Ibidi). Briefly, the cells were seeded onto the insert, placed in 6-well plates and grown overnight. Then, the inserts were removed, and the cells were washed with PBS. The ratio of wound healing was observed by collecting images at the indicated time points. The experiment was repeated twice.
2.8. In vitro invasion assay
Before the experiment, cells receiving the indicated treatments were cultured in basic DMEM without FBS for 12 h. Next, 1 × MaxGel ECM (E0282, Sigma) was added to the 8-μm hanging cell culture insert (Millipore). Then, 20,000 cells were suspended in 100 μl basic DMEM and seeded into the insert. The insert was then placed in a 24-well plate contained 500 μl DMEM supplemented with 20% FBS and cultured for another 12 h. Then, the cells were fixed and stained with a crystal violet solution. The non-invasive cells on top of the membrane was then removed. Five fields for each membrane were photographed to determine the invasive cells. The difference of groups was counted by using an inverted microscope equipped with either a 10× objective and plotted as the number of invading cells.
2.9. 3D on-top culture
A 3D on-top culture was performed as previously reported [19]. Briefly, a 48-well plate was coated with 100 μl Matrigel and placed at 37 °C for 15 min to allow the gel to harden. HCT116 and HT29 colorectal cancer cells were seeded into the coated plate and incubated at cell incubator for 30 min. Next, DMEM supplemented with 10% Matrigel was added into the culture plate.
2.10. Luciferase promoter activity assay
A Luciferase promoter activity assay was performed according in line with standard method. Briefly, CTGF promoter region was constructed into a pGL3-BL plasmid (Promega). A total of 1 μg CTGF promoter plasmid and 100 ng Renilla reporter plasmid were transfected into colorectal cancer cells, which received the indicated treatment following another 48 h of culture. Then, luciferase activity was detected with a microplate reader. The luciferase activity of Renilla acted as a loading control.
2.11. RhoA activation assay
When RhoA is bound to GTP, its status is activated. The amount of GTP-RhoA was measured with a RhoA activation assay kit (Cytoskeleton, BK036). HCT116 and HT29 cell were lysis and centrifuged at 13,000 g for 10 min at 4 °C. The pellet was discarded, and the supernatants were used for the following assay. Ten percent of the supernatant was mixed directly with loading buffer, and the leftover supernatant was mixed with beads conjugated with the Rho-binding domain of rhotekin for 2 h at 4 °C. Then, the beads were washed twice with wash buffer, eluted with WB loading buffer.
2.12. F-actin and G-actin ratio assay
The ratio of G-actin to F-actin was determined using a G-actin/F-actin In Vivo Assay Biochem Kit (BK037, Cytoskeleton) in line with standard procedure. GPR4 knockdown HCT116 cells and control cells were scraped from culture plates and suspended in LAS2 buffer to prepare the lysates. After that, the lysates were centrifuged at 350 rpm for 10 min. The supernatants were collected and centrifuged at 100,000 g for 1 h. Then, supernatants that containing G-actin and pellets that containing F-actin were mixed with loading buffer and prepared for immunoblotting.
2.13. Immunofluorescence staining
HCT116 WT or GPR4-depleted CRC were plated into 8-well u-Chamber (Ibidi, Germany) and incubated overnight. After that, the cells were fixed and permeabilized with buffer supplied with 0.05%Triton X-100 for 3 min; then, the cells were washed with PBS containing 0.05% Tween-20 twice. Next, the cells were blocked with 5% BSA buffer for 2 h at room temperature. For YAP1 staining, cells were incubated with anti-YAP1 antibody (#14074, 1:100, Cell signalling) overnight at 4 °C. On the second day, cells were washed twice and incubated with goat anti-rabbit antibodies conjugated with Alexa 488 at a 1:300 for 1 h. Cells were incubated with Phalloidin-iFluor 594 (ab176757, Abcam) to stain F-actin. After that, the cells were washed twice and supplied with anti-fade fluorescence medium with DAPI (S2110, Solarbio), and visualized with a confocal microscope.
2.14. Protein immunoprecipitation
First, colorectal cancer cells were rinsed twice with PBS. A fraction of each lysate was kept as a loading control, and the leftover lysates were subsequently incubated with IgG (ab172730, Abcam), anti-YAP1(14,074, Cell signalling) or anti-TEAD1 (ab133533, Abcam) antibodies crosslinked to Protein G Dynabeads (10003D, Invitrogen) for 2 h; then, the samples were rinsed three times with PBS containing 0.05% Tween-20. After that, the precipitates were eluted from the beads, prepared foe WB.
2.15. Histology and immunostaining
Tumours resected from mice were fixed immediately, followed by paraffin embedding according to a standard protocol. For IHC staining, tumour sections were deparaffinized. Sodium citrate was used for antigen retrieval at 95 °C for 10 min. Then, 3% H2O2 was used to deplete endogenous peroxidase activity at 37 °C for 30 min. Next, the tumour tissues were blocked with 5% BSA, followed by incubation with anti-GPR4 (ab188931, Abcam), anti-YAP1 (14,074, Cell signalling), anti-TEAD1 (ab133533, Abcam), anti-Ki67 (#9449, Cell signalling), anti-Myc (ab32072, Abcam) or anti-Active RhoA -GTP (26,904, NewEast Biosciences) antibody. On the second day, the sections were washed three times with PBS and incubated with the corresponding HRP-linked secondary antibodies for 1 h at RT. IHC staining signals were presented with DAB Kit (Cell Signalling).
2.16. Patients and samples
Patients with CRC were drawn from Ren Ji Hospital, School of Medicine, Shanghai Jiao Tong University. Study protocol that strictly in line with International Ethical Guidelines for Biomedical Research Involving Human Subjects was approved by the Research Ethics Committee of Ren Ji Hospital, School of Medicine, Shanghai Jiao Tong University. Written informed consent was provided to all the patients before the enrolment. Patients that had received no anti-tumour therapy and signed written informed consent were enrolled in this study.
2.17. Organoid culture
Colorectal cancer organoids were cultured according to a previously described method [20]. Briefly, primary cancer cells were isolated from tumour tissue and digested by buffer contained 1.5 mg/ml collagenase II and 20 μg/ml hyaluronidase for 30 min at 37 °C. Then, the cells received the shGPR4 virus or indicated treatment and were cultured with organoid culture medium reported by previous study [21].
2.18. Animal studies
Animal studies were performed in line with the rules for the care and use of laboratory animals from the National Academy of Sciences. Animal experiments were approved by Institutional Animal Care and Use Committee of Shanghai jiaotong University (Shanghai, P.R. China). For subcutaneous mouse model, 1 × 106 shNC or shGPR4 HCT116 cells were diluted in 200 μL basic DMEM and injected into the male 8-week-old nude mice. Mouse tumour weights were measured using an analytical balance. Tumour volumes were evaluated with calipers and expressed in mm3 using the formula: V = π /6 x (Dmax2 x Dmin/2) [22]. For the liver metastasis mouse model, 1 × 106 shNC or shGPR4 MC38 cells were injected into the spleens of 8-week-old male C57BL/6 J mice. For the liver metastasis mouse survival studies, the moribund state defined by the IACUC was used as the endpoint. Liver tissue was then resected from the sacrificed mice to analyse the metastasis burden.
2.19. Statistical analysis
The data presented in this study were the mean ± standard deviation (SD). GraphPad Prism 7 was used for statistical analysis. Student's t-tests were applied to determine significant differences of two groups. Normal error distribution analysis was performed before Student's t-tests were applied, and no differences were observed. One-way ANOVA was used to determine differences among more than two groups. A p-value <.05 was considered statistically significant.
3. Results
3.1. GPR4 was overexpressed and predicted a poor prognosis in CRC
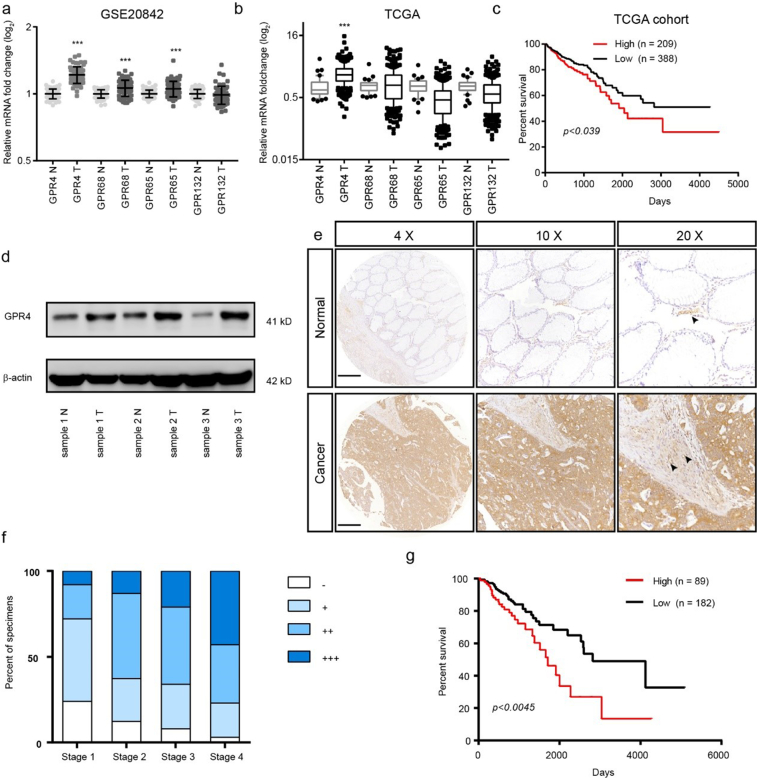
To determine which proton-sensing receptor responds to the extracellular acidic tumour microenvironment, we analysed the paired CRC GEO and CRC TCGA dataset (Fig. 1a–b). The results showed that only GPR4 was upregulated in both datasets. Further analysis of the CRC TCGA dataset suggested that GPR4 expression was positively associated with shorter overall survival time in CRC patients (Fig. 1c). To validate these results, we first detected protein expression level of GPR4 in 3 CRC and adjacent tissue samples. Indeed, the expression of GPR4 was greatly increased in the tumour tissues (Fig. 1d). Next, we investigated the correlation between the expression profile of GPR4 and the overall prognosis in a tissue microarray contained 317 CRC specimens; 291 of these specimens with clinical information were chosen for the prognostic analysis. Consistently, the IHC results showed that GPR4 expression was markedly increased in the CRC samples(Fig. 1e). The sample expression levels of GPR4 were scored as -, +, ++, or +++, considering the area percentage and intensity of staining (Supplementary Fig. 1). Notably, the expression of GPR4 gradually increased with colorectal cancer progression (Fig. 1f). Furthermore, Kaplan-Meier analyses were applied to evaluate the clinical correlation between GPR4 expression and the survival rate of CRC patients (Fig. 1g). Our data reveal that patients with high GPR4 expression presented shorter survival times than those with low GPR4 expression. Together, these data indicated that GPR4 upregulated in CRC and might play a role in CRC progression.
Fig. 1.
GPR4 was overexpressed and predicted a poor prognosis in CRC.
(a) GPR4, GPR68, GPR65 and GPR132 relative mRNA expression levels in colorectal cancer and non-tumour tissues in the GSE20842 dataset. (b) GPR4, GPR68, GPR65 and GPR132 relative mRNA expression levels in colorectal cancer and non-tumour tissues in the TCGA dataset. (c) Kaplan-Meier analyses of the overall survival of the Cancer Genome Atlas (TCGA) cohort patients with high GPR4 expression versus those with low GPR4 expression. (d) GPR4 protein expression in CRC tumour and adjacent non-tumour tissues. (e) Representative examples of CRC tissue array immunostaining for GPR4 in CRC tumour tissues and corresponding non-tumour tissue. Endothelial cells pointed out with black arrow. Scale bars, 500 μm. (f) Statistical analysis of GPR4 staining results for different tumour stages. (g) Kaplan-Meier analyses of the overall survival of patients with colorectal cancer based on GPR4 IHC staining. Differences between the groups in (a, b) were analysed by unpaired Student's t-test (* P < .05; ** P < .01; *** P < .001). Differences between two groups in (c, g) were analysed by log-rank test. (* P < .05; ** P < .01; *** P < .001).
3.2. Genetically inhibiting GPR4 suppressed the growth and migration of CRC cancer cells
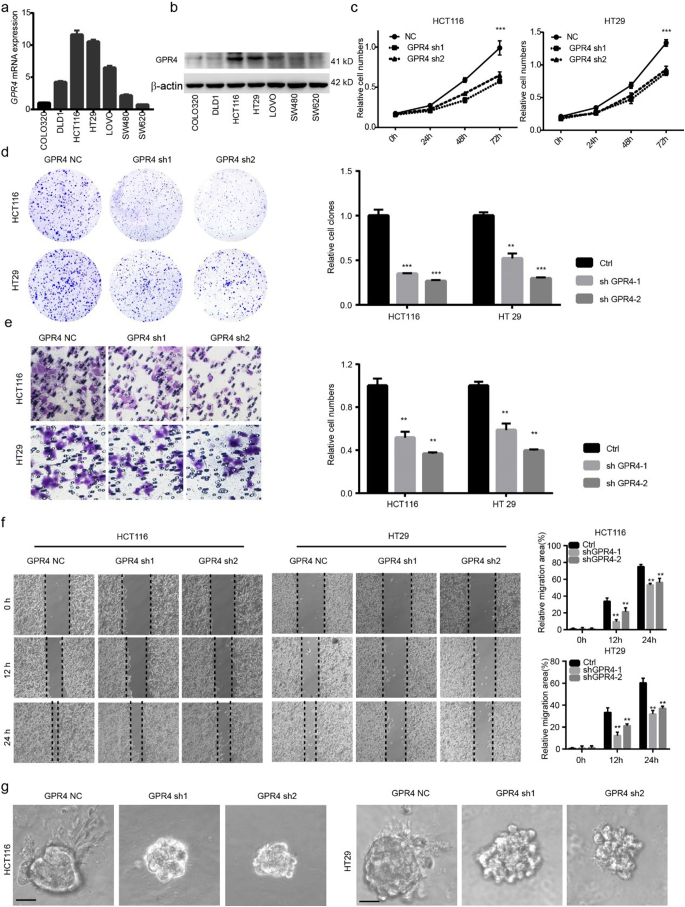
To give insight into whether GPR4 is the major proton sensor receptor that response to acidic microenvironment in CRC progression, firstly, GPR4 and others proton sensor receptor were silenced by siRNA and cultured in pH 6.8 medium. The results showed that GPR4 depletion significantly suppressed the growth of CRC cancer cells, but not other receptors (Supplementary Fig. 2a). Next, GPR4 was depleted via shRNA in HCT116 and HT29 cells (Supplementary Fig. 2b–c), two colorectal cancer cell lines with relatively high GPR4 expression (Fig. 2a–b). The results showed that the growth ability and colony-forming ability were markedly lower in GPR4-silenced CRC cells than in the corresponding control cells (Fig. 2c–d). Furthermore, we aimed to study the role of GPR4 in tumour cell migration. Cell invasion assays were performed using control and GPR4 knockdown cells. The results revealed that GPR4 depletion also significantly inhibited tumour migration (Fig. 2e). Consistent with the cell invasion results, shRNA-mediated depletion of GPR4 in HCT116 and HT29 cells significantly impaired their wound healing ability (Fig. 2f). Considering that the two-dimensional (2D) culture system may be much different than the in vivo environment, a three-dimensional (3D) on-top growth method was used to assess CRC cancer cell tumorigenesis ability. Interestingly, small spheres and few invasive structures were found in the GPR4-depleted cancer cells (Fig. 2g and Supplementary Fig. 2d). To further assess whether the biological effects are acidic pH dependent, we performed all the above experiments under physiological pH 7.4. The results showed that GPR4 depletion did not significantly impair the proliferation, migration and invasion of CRC cancer cells under physiological pH (Supplementary Fig. 3a–f), which indicated that the biological effects of GPR4 are indeed acidic pH-dependent. Taken together, our results indicated that GPR4 depletion inhibited the growth and migration of CRC cancer cells.
Fig. 2.
Genetically inhibiting GPR4 suppressed the proliferation, migration of CRC cancer cells.
(a–b) GPR4 expression in mRNA (a), protein (b) level of different colorectal cancer cell lines. (c) CCK-8 cell proliferation assay of control or GPR4-silenced HCT116 and HT29 cells in pH 6.8 culture medium. (d) Colony formation assay of control or GPR4-silenced HCT116 and HT29 cells in pH 6.8 culture medium. Statistical of colony formation assay were presented in the right panel. (e) Cell invasion assay of control or GPR4-silenced HCT116 and HT29 cells in pH 6.8 culture medium. Statistical of cell invasion assay were presented in the right panel. (f) Wound healing assay of control or GPR4-silenced HCT116 and HT29 cells in pH 6.8 culture medium. Statistical of wound healing assay were presented in the right panel (g) 3D on-top growth assay of control or GPR4-silenced HCT116 and HT29 cells in pH 6.8 culture medium. Scale bars, 100 μm. Differences between the control and other groups in (c) were analysed by one-way ANOVA test (* P < .05; ** P < .01; *** P < .001). Differences between the groups in (d, e and f) were analysed by unpaired Student's t-test (* P < .05; ** P < .01; *** P < .001).
3.3. Activated GPR4 triggers hippo pathway
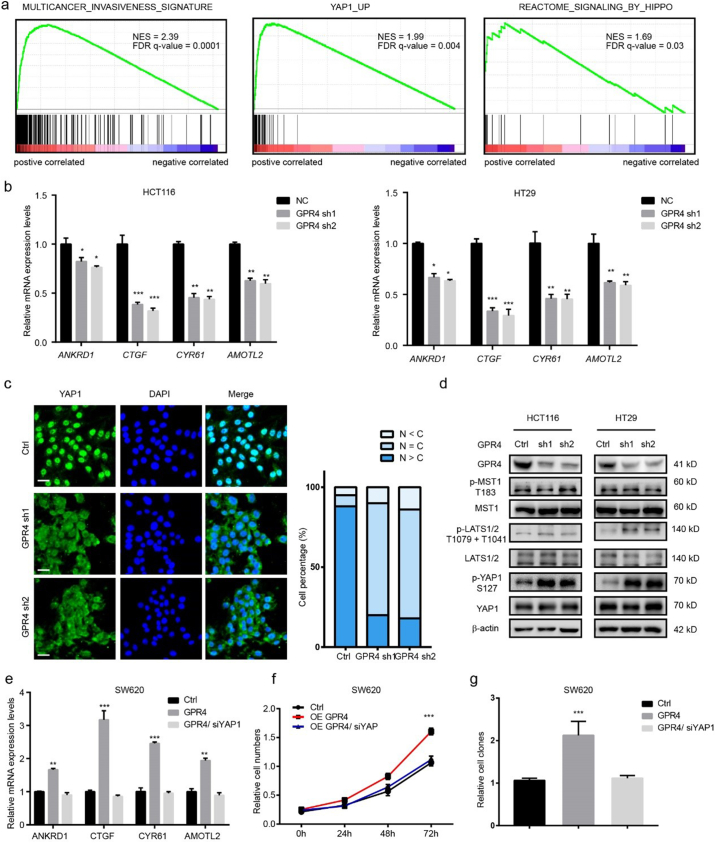
To understand the underlying mechanism through which activated GPR4 promotes CRC progression, Gene Set Enrichment Analysis (GSEA) [23] was performed on TCGA colorectal cancer datasets. The results showed that high GPR4 expression was positively associated with the “invasiveness signature”, “YAP1_UP” and “signalling_BY_HIPPO” (Fig. 3a), indicating that the hippo pathway may be involved in the growth-promoting effect of activated GPR4. To validate this hypothesis, four YAP1 target genes, ANKRD1, CTGF, CYR61 and AMOTL2, were detected in GPR4 stable knockdown HCT116 and HT29 cells. Q-PCR results showed that YAP1-regulated gene expression was greatly decreased (Fig. 3b). Next, an immunofluorescence was performed to analyse the subcellular location of YAP1. Statistical data showed that the nuclear localization of YAP1 was significantly reduced upon shRNA-mediated GPR4 knockdown in HCT116 cells (Fig. 3c). Furthermore, the phosphorylation levels of key signalling members of the hippo pathway were detected in GPR4-depleted CRC cell lines. The immune blot results showed that p-LATS1/2(T1079 + T1041) and p-YAP1 (S127) expression was obviously upregulated; however, GPR4 knockdown barely affected the expression of the canonical LATS1/2 upstream kinase p-MST1 (T183) (Fig. 3d). In line with this, immunoblots results showed that the phosphorylation level of LATS1 and YAP1 significantly increased in pH 7.4 compared to pH 6.8 with unaffected GPR4 expression no matter in low or high cell density (Supplementary Fig. 4a–b). To further determine whether YAP1 is necessary for the oncogenic role of GPR4 in CRC, YAP1 was deleted with siRNA in GPR4-overexpressing SW620 and SW480 cells, CRC cell lines with relatively low GPR4 expression. Consistently, the YAP1 target genes sharply increased after the ectopic expression of GPR4. However, GPR4 could no longer activate these genes after YAP1 knockdown (Fig. 3e and Supplementary Fig. 4a), indicating the growth promotive effect of GPR4 was mediated by YAP1. In line with this finding, the growth-promoting and invasion-enhancing effects of GPR4 were also blocked by siRNA-mediated YAP1 depletion (Fig. 3f–g and Supplementary Fig. 4d–g). Taken together, these findings reveal that activated GPR4 triggers hippo pathway which depends on YAP1 nuclear location.
Fig. 3.
Activated GPR4 triggers hippo pathway.
(a) Gene set enrichment analysis of RNA-seq data for patients with high GPR4 expression versus low GPR4 expression from the TCGA dataset. (b) Q-PCR of the relative mRNA levels of the YAP1 target genes ANKRD1, CTGF, CYR61, and AMOTL2 in GPR4-depleted HCT116 and HT29 cells. (c) Immunofluorescence staining of YAP1 in HCT116 control and GPR4 knockdown cells. Statistical analysis of YAP1 localization is presented in the right panel. Scale bars, 20 μm. (d) Expression of GPR4, p-MST1 (T183), MST1, p-LATS1/2 (T1079+T1041), LATS1/2, p-YAP1 (S127), YAP1 in HCT116 and HT29 GPR4-depleted or control cells by western blotting. (e) Q-PCR analysis of the relative mRNA levels of the indicated YAP1 target genes ANKRD1, CTGF, CYR61, and AMOTL2 in SW620 cells with GPR4 overexpression or GPR4 overexpression plus YAP1 knockdown mediated by siRNA or the corresponding control cells. (f) cell proliferation, colony formation (g) of SW620 cells with GPR4 overexpression or GPR4 overexpression plus YAP1 silencing or the corresponding control cells. Differences between the groups in (b, e and g) were analysed by unpaired Student's t-test (* P < .05; ** P < .01; *** P < .001). Differences between the control and other groups in (f) were analysed by one-way ANOVA test (* P < .05; ** P < .01; *** P < .001).
3.4. YAP1 cooperates with TEAD1 to activate oncogene expression under the regulation of GPR4
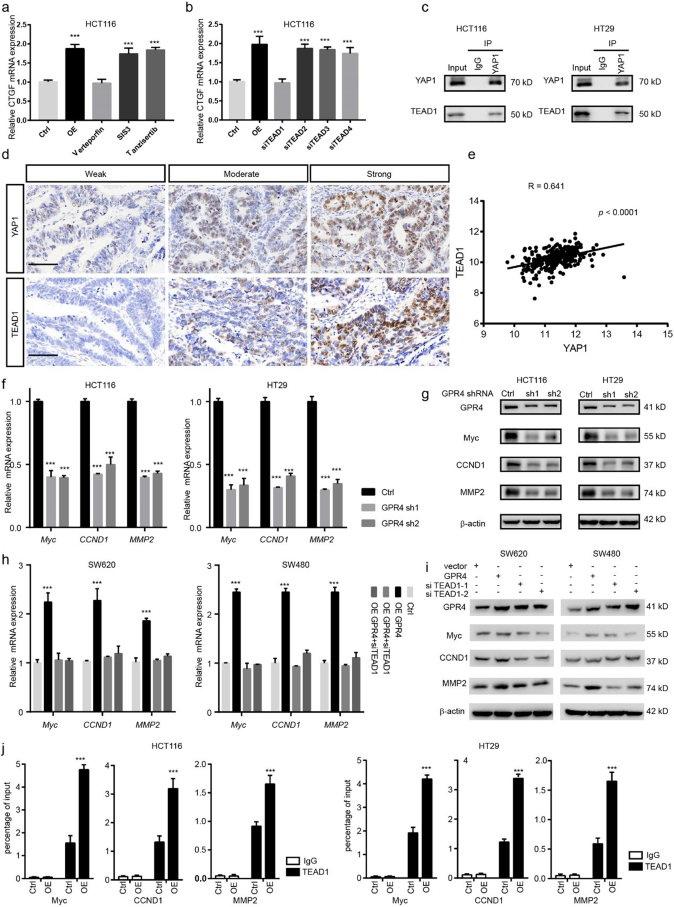
Given the evidence that YAP1 works as a transcriptional coactivator, it cannot bind to DNA independently. Next, we aimed to determine which transcription factors may cooperate with YAP1 to mediate the tumour-promoting effect of GPR4. SMAD, C-jun and TEAD transcription factors have been widely reported to interact with YAP1 and activate downstream gene expression under different conditions. Notably, only the YAP-TEAD inhibitor verteporfin could greatly inhibit the upregulation of CTGF induced by forced GPR4 expression; the SMAD inhibitor SIS3 and the C-jun inhibitor tanzisertib did not have this effect (Fig. 4a). The TEAD family includes four members, TEAD1, TEAD2, TEAD3 and TEAD4. To further determine which one mediates this effect, these molecules were knocked down by siRNA in HCT116 cells. The results showed that TEAD1 might be the dominant effector (Fig. 4b). In line with this finding, co-immunoprecipitation results showed that TEAD1 could physically interact with endogenous YAP1 in both HCT116 and HT29 CRC cancer cell lines (Fig. 4c). In addition, the results of IHC performed on CRC primary tissue serial sections indicated that YAP1 and TEAD1 expression are positively associated (Fig. 4d and Supplementary Fig. 5a), which was further confirmed by a CRC TCGA data analysis showing that the Pearson correlation coefficient for TEAD1 and YAP1 is the highest compared with that for YAP1 and other TEAD family members (Fig. 4e and Supplementary Fig. 5b–d). Furthermore, siRNA-mediated TEAD1 depletion could reduce the growth-promoting effect of GPR4. Next, we examined the target genes of the YAP1-TEAD1 complex under the regulation of GPR4. Analyses were performed using CHIP-seq data and the Cistrome Data Browser. We focused on genes related to invasion and growth. Matrix metallopeptidase family member, pro-oncogenes and cyclins (MYC, CCND1, and MMP2) were the potential candidates. We validated their expression changes by Q-PCR and WB in GPR4 knockdown HCT116 and HT29 cells (Fig. 4f–g). Indeed, MYC, CCND1, MMP2 were shown to be downregulated in GPR4-depleted cancer cells. Additionally, siRNA-mediated TEAD1 silencing blocked the increase in MYC, CCND1, and MMP2 triggered by forced GPR4 expression in SW620 and SW480 cells (Fig. 4h–i). For further verification, a ChIP-PCR assay was conducted in both the corresponding control and GPR4-overexpressing CRC cancer cells. Consistently, the data indicated that TEAD1 could bind to the promoters of MYC, CCND1 and MMP2 (Fig. 4j). Together, these analyses support the idea that YAP1 cooperates with TEAD1 to activate oncogene expression under the regulation of GPR4.
Fig. 4.
YAP1 cooperates with TEAD1 to activate oncogene expression under the regulation of GPR4.
(a) Q-PCR analysis of the relative mRNA levels of CTGF under conditions of GPR4 overexpression, GPR4 overexpression plus verteporfin, GPR4 overexpression plus SIS3, or GPR4 overexpression plus tanzisertib in HCT116 cells. (b) Q-PCR analysis of the relative mRNA levels of CTGF in GPR4-overexpressing HCT116 cells treated with siTEAD1, siTEAD2, siTEAD3 or siTEAD4, respectively. (c) Co-immunoprecipitation (coIP) was performed in HCT116 and HT29 cells with anti-YAP1, anti-TEAD1, or immunoglobulin G (IgG) antibodies, following immunoblotting. (d) IHC analysis of YAP1 and TEAD1 in human colorectal cancer tissue. Scale bars, 50 μm (e) Pearson correlation coefficient of YAP1 and TEAD1 in the COAD-TCGA dataset. (f) Q-PCR analysis of the relative mRNA levels of Myc, CCND1, and MMP2 upon GPR4 silencing by shRNA in HCT116 and HT29 cells. (g) Immunoblot analysis of the protein levels of Myc, CCND1, and MMP2 upon GPR4 silencing by shRNA. (h) Q-PCR analysis of the relative mRNA levels of Myc, CCND1, and MMP2 upon GPR4 overexpression and TEAD1 depletion in SW620 and SW480 cells. (i) Immunoblot analysis of the protein levels of Myc, CCND1, and MMP2 upon GPR4 overexpression and TEAD1 depletion. (j) ChIP-PCR assay was used to measure the recruitment of TEAD1 to the Myc, CCND1 and MMP9 promoters in HCT116 and HT29 cells. Differences between the groups in (a, b, f, h and j) were analysed by unpaired Student's t-test (* P < .05; ** P < .01; *** P < .001).
3.5. The RhoA-F-actin axis mediated the signalling between GPR4 and the hippo pathway
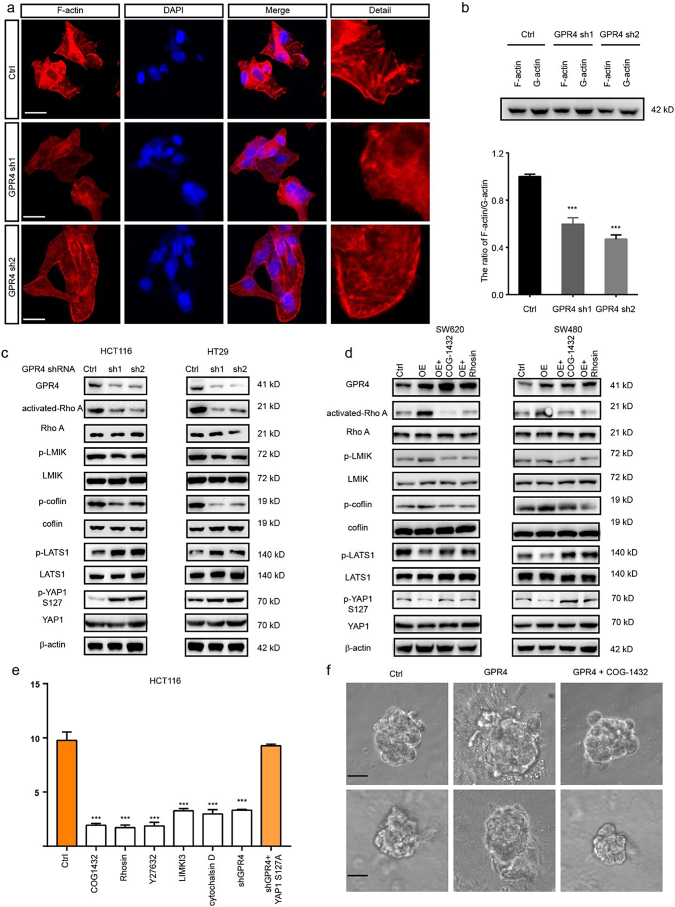
Next, we sought to determine the mechanism by which GPR4 activated the hippo pathway. Given the evidence that p-MST1 could not be affected by GPR4 and that F-actin is reported to be a regulator of p-LATS1/2, we measured f-actin levels in GPR4 WT and knockdown HCT116 cells (Supplementary Fig. 6a) by phalloidin staining. The immunofluorescence results showed that GPR4-depleted cells presented filamentous actin (hereafter, F-actin) rearrangement of the cytoskeleton and formed fewer stress fibres (Fig. 5a). Thus, the levels of F-actin and G-actin were detected by immunoblotting. Similarly, the ratio of F-actin/G-actin was greatly decreased in cells lacking GPR4 (Fig. 5b). Considering that RhoA acts as one of the main regulators of actin polymerization, the levels of GTP bound-on RhoA and its downstream effectors LIMK and cofilin were detected in GPR4-deficient HCT116 and HT29 cells. Activated-RhoA, p-LIMK and p-cofilin levels were significantly reduced, and p-LATS1, p-YAP1 levels were greatly increased (Fig. 5c). On the other hand, RhoA inhibitors COG-1432 and Rhosin were applied to CRC cells with ectopic GPR4 expression to validate the regulation of LAST1 by RhoA signalling. The results showed that both COG-1432 and Rhosin could recover the p-LATS1, p-YAP1 levels suppressed by GPR4 expression, indicating that RhoA was necessary for GPR4-related LAST regulation (Fig. 5d). Next, we aimed to determine whether Rho and its regulation of F-actin formation were necessary for GPR4 regulation of the YAP1-TEAD1 complex. The ROCK inhibitor Y27362, the LIMK inhibitor LIMKi3 and the actin polymerization inhibitor cytochalasin D were applied to HCT116 cells. These treatments caused a sharp decrease in luciferase activity. However, ectopically expressed constitutively active YAP1-S127A was resistant to the above inhibitor interference (Fig. 5e). In addition, the involvement of RhoA in GPR4 tumour promotive roles also confirmed by 3D on-top growth under the administration with COG1432 (Fig. 5f and Supplementary Fig. 6b). These data clarified that the RhoA-F-actin axis was necessary downstream of GPR4 to govern the hippo pathway.
Fig. 5.
The RhoA-F-actin axis mediated the signalling between GPR4 and the hippo pathway.
(a) Immunofluorescence (IF) staining analysis of F-actin labelled with TRITC phalloidin in GPR4-depleted HCT116 cells or control cells. Scale bars, 20 μm. (b) Representative western blot quantification of the G-actin/F-actin ratio in GPR4-depleted HCT116 cells or control cells. (c) Western blot quantification the level of GPR4, activated RhoA, p-LMIK, p-cofilin, p-LATS1 and p-YAP1 in GPR4-depleted HCT116 and HT29 cells. (d) Western blot quantification of the level of GPR4, activated RhoA, p-LMIK, p-cofilin, p-LATS1 and p-YAP1 in SW620 and SW480 cells with GPR4 overexpression and RhoA inhibition by COG1432 and Rhosin or the corresponding control cells. (e) Luciferase report assay of the CTGF promotor in HCT116 cells with the indicated treatment. (f) 3D on-top growth assay of SW480 and SW620 cells with GPR4 overexpression or GPR4 overexpression plus COG-1432. Scale bars, 100 μm. Differences between the groups in (b, e) were analysed by unpaired Student's t-test (* P < .05; ** P < .01; *** P < .001).
3.6. Genetic inhibition of GPR4 suppressed CRC progression in vivo
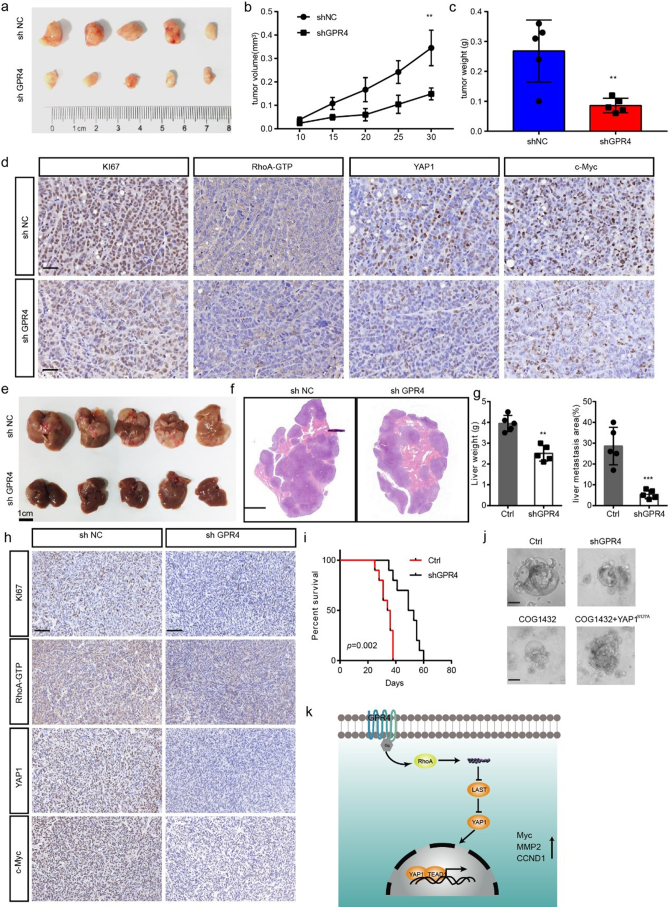
To determine the roles of GPR4 in tumour progression in vivo, a subcutaneous xenograft model and CRC liver metastasis model were created. First of all, we detected the pH value in the tumour microenvironment by measuring the tumour interstitial fluid. Consistent with previous reports [4], the results showed that the pH value in the tumour microenvironment is much lower than physiological pH. Knockdown of GPR4 in CRC cancer cells faintly increased pH value but the difference was not statistically significant (Supplementary Fig. 7a). In line with our in vitro findings, suppression of GPR4(Supplementary Fig. 7b) attenuated tumour growth, as evidenced by reduced tumour weight, tumour volume and tumour growth ability (Fig. 6a–c). IHC results revealed that staining for Ki-67, Active RhoA -GTP, nuclear YAP1 and Myc was significantly reduced in GPR4-deficient CRC cells (Fig. 6d), but not angiogenesis (Supplementary Fig. 7c). Considering to the metastasis promotive effect of GPR4 and liver metastasis is the most common metastasis destination organ, we knocked down GPR4 in MC38 cells (Supplementary Fig. 7d) to provide insight into whether GPR4 is involved in CRC liver metastasis. The results showed that shRNA-mediated knockdown of GPR4 did not reduce tumour angiogenesis (Supplementary Fig. 7e), but strongly suppressed the colonization of these cells (Fig. 6e–g), greatly reduced the downstream signalling (Fig. 6h) and significantly prolonged the survival of tumour-bearing mice (Fig. 6i). In addition, GPR4 over-expression significantly facilitated the proliferation and metastasis in the in vivo (Supplementary Fig. 7f–g), implying that GPR4 is indeed involved in CRC tumour growth progression. To further validated the mechanism and potentiality of targeting GPR4 in CRC, primary CRC cells derived organoids [21] that were established. The results showed that GPR4 depletion or RhoA inhibition mediated by COG-1432 administration greatly suppressed the development of organoids. In addition, the introduction of YAP1S127 could restored the growth ability suppressed by GPR4 and RhoA inhibition (Fig. 6j). Together, our data indicate that GPR4 promotes tumour progression by activating the hippo pathway and downstream oncogenes.
Fig. 6.
Genetic GPR4 inhibition suppressed CRC progression in vivo.
(a) Subcutaneous xenografts derived from shNC or shGPR4 HCT116 cells. (b) Tumour volume growth curves of the subcutaneous xenografts (n = 5). (c) Tumour weights of the subcutaneous xenografts (n = 5). (d) IHC staining for Ki67, Active RhoA -GTP, YAP1 and Myc in subcutaneous tumour sections. Scale bar, 100 μm. (e) Liver metastasis burden of mice injected with shNC or shGPR4 MC38 cells. (f) H&E staining of livers from shNC and shGPR4 MC38 mice. Scale bar, 500 μm. (g) Statistical analysis of liver weights and percentage of tumor invasive area (n=5). (h) IHC staining for Ki67, Active RhoA -GTP, YAP1 and Myc in liver metastasis tumour sections. (i) Survival of liver metastasis mice with shNC MC38 and shGPR4 MC38 tumours. (j) Organoid culture of colorectal cancer with the indicated treatment. (k) Model proposing that upregulated GPR4 promotes colorectal cancer progression in an acid tumour microenvironment. Differences between the groups in (b) were analysed by one-way ANOVA test (* P < .05; ** P < .01; *** P < .001). Differences between the groups in (c, g) were analysed by unpaired Student's t-test (* P < .05; ** P < .01; *** P < .001). Differences between two groups in (i) were analysed by log-rank test. (* P < .05; ** P < .01; *** P < .001).
4. Discussion
The tumour microenvironment has been widely reported to support cancer initiation and progression [24]. Extensive research has focused on the components of those microenvironments, including infiltrated immune cells, tumour-associated fibroblasts and extracellular cytokines/chemokines. However, the effects of the physical characteristics of the tumour microenvironment, for example, its high acidity, on tumour progression are not well known, especially in CRC. In this study, we found that the proton sensor receptor GPR4 is overexpressed in colorectal cancer, which promotes the proliferation and migration of CRC cancer cells upon activation in an acidic context. Mechanistic studies reveal that upregulated GPR4 in CRC cancer cells is activated by extracellular protons (H+) and leads to RhoA activation and F-actin rearrangement, which further trigger the hippo pathway and promote YAP1 nuclear localization, which then interacts with TEAD1 and enhances Myc, CCND1 and MMP2 expression, finally facilitating the proliferation and metastasis of colorectal cancer (Fig. 6k).
GPR4 is a member of the GPCR superfamily. In line with our observations, proceeding researches also reported that GPR4 expression upregulated in breast tumours, ovarian tumours, colon tumours, liver tumours, kidney tumours and head and neck cancer [14,25,26]. However, the role of GPR4 in cancer progression is conversive. GPR4 overexpression could significantly inhibit the tumour cell metastasis ability in B16F10 cell and TRAMP-C1 cell [27,28]. Nonetheless, ectopically expressed GPR4 can greatly enhance NIH3T3 fibroblast cell growth and facilitate angiogenesis in squamous cells and head and neck cancer cells. Additionally, proceeding research reported that GPR4 depletion impaired angiogenesis in host mice, leading to a reduction in the growth of tumour xenografts [28]. In line with these findings, our results showed that upregulated GPR4 could significantly promote tumour cell proliferation and metastasis. However, GPR4 depletion seems not impair the angiogenesis of CRC according our in vivo results. Taken together, the function and roles of GPR4 may differ depending on the cell type and specific tumour context.
In early research, GPR4 was reported to elicit cyclic AMP formation and trigger downstream PKA signalling [29]. Recently, research in endothelial cells indicated that notch signalling could also be activated by the GPR4 receptor [30]. In our study, suppressed LAST1/2 phosphorylation and enhanced YAP1 nuclear localization were observed upon GPR4 activation, indicating that GPR4 could affect the hippo pathway. Consistently, YAP1 was shown to be a downstream effector of GPR4 signalling in bone marrow-derived mesenchymal stem cells [31]. Although multiple signalling pathways could be activated by GPR4, our study showed that disruption of the interaction between YAP1 and TEAD1 or TEAD1 depletion could block the GPR4 overexpression-induced growth-promoting effect in colorectal cancer cells, suggesting that the tumorigenesis role of GPR4 is dependent on hippo pathway activation and downstream oncogene expression. According to the dataset analysis and CHIP-PCR results, we revealed that Myc, CCND1 and MMP2 are target genes of YAP1/TEAD1. In line with this finding, Myc, CCND1 and MMP2 were also reported as YAP1 target genes in previous studies in other cancers [[32], [33], [34]]. However, we cannot rule out other potential target genes involved in the tumour-promoting role of GPR4. More studies are needed to reveal all of the target genes of the YAP1/TEAD1 complex in colorectal cancer.
Acidosis has been reported as one of the early events in solid tumours and is defined as a characteristic of the tumour microenvironment [35]. Given that acidosis is a stressor and select cancer cells are suited for an acidic microenvironment, targeting tumour acid homeostasis would be potential strategies. We found that GPR4 are the dominative expressed proton sensor receptor. We explored the potential of targeting GPR4 in colorectal cancer. Genetic-mediated GPR4 inhibition greatly relieved the tumour burden in xenograft mice, as well as reduced liver metastasis and prolonged the survival of liver metastasis mice. And, the preliminary data from tumour organoid that can recapitulate the characteristic parental tumours indicated GPR4 might be a perspective target in CRC. In addition, pharmacological inhibition of GPR4 was reported to remediates intestinal inflammation in a mouse colitis model [36]. Unfortunately, the selective GPR4 antagonist GPR4 antagonist 13 (NE-52-QQ57) is commercially unavailable, and we could not evaluate its effect on CRC progression [37]. Our study just laid an early foundation for targeting GPR4 in CRC. More efforts are needed for GPR4 transform medical research.
In summary, our studies revealed that GPR4, whose expression is positively associated with poor prognosis in CRC, responded to extracellular H+ and promoted the growth and migration of CRC cancer cells. Mechanistic studies indicated that activated GPR4 triggers YAP1 nuclear localization and downstream target gene expression via the RhoA-F-actin axis. Furthermore, mouse model experiments showed that GPR4 deletion suppressed CRC tumour progression, indicating that it may be a promising therapeutic target. In fact, we established a primary foundation for GPR4 translational research, and future efforts will attempt to elucidate the effect of GPR4 in non-tumour cells and evaluate its potential for translational applications.
The following are the supplementary data related to this article.
Q-PCR primers used in this study.
CHIP-PCR primers used in this study.
Supplementary figures.
Author contributions
M.Z. and M.H·Y. designed the research; M.H·Y, R.C., Y.L., S.L.Q., performed the research; M.H·Y, performed main experiments on the two models. R.C. contributed to the in vivo homing study and helped with animal experiments. M.H.Y, R.C., Y.L., S.L.Q. analysed and interpreted the data; M.Z. and M.H.Y. wrote the paper.
Acknowledgment and funding sources
This work was supported by the grant from the National Natural Science Foundation of China No.81702300 (to M.H·Y) & No.81873555 to (M.Z.).
Declaration of Competing Interest
The authors declare no potential conflicts of interest.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Connell L.C., Mota J.M., Braghiroli M.I., Hoff P.M. The rising incidence of younger patients with colorectal cancer: questions about screening, biology, and treatment. Curr Treat Options Oncol. 2017;18(4):23. doi: 10.1007/s11864-017-0463-3. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R.L., Miller K.D., Fedewa S.A., Ahnen D.J., Meester R.G.S., Barzi A. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67(3):177–193. doi: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 4.Hashim A.I., Zhang X.M., Wojtkowiak J.W., Martinez G.V., Gillies R.J. Imaging pH and metastasis. NMR Biomed. 2011;24(6):582–591. doi: 10.1002/nbm.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barathova M., Takacova M., Holotnakova T., Gibadulinova A., Ohradanova A., Zatovicova M. Alternative splicing variant of the hypoxia marker carbonic anhydrase IX expressed independently of hypoxia and tumour phenotype. Brit J Cancer. 2008;98(1):129–136. doi: 10.1038/sj.bjc.6604111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gatenby R.A., Gillies R.J. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4(11):891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 7.Ludwig M.-G., Vanek M., Guerini D., Gasser J.A., Jones C.E., Junker U. Proton-sensing G-protein-coupled receptors. Nature. 2003;425(6953):93–98. doi: 10.1038/nature01905. [DOI] [PubMed] [Google Scholar]
- 8.Radu C.G., Nijagal A., McLaughlin J., Wang L., Witte O.N. Differential proton sensitivity of related G protein-coupled receptors T cell death-associated gene 8 and G2A expressed in immune cells. Proc Natl Acad Sci U S A. 2005;102(5):1632–1637. doi: 10.1073/pnas.0409415102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar N.N., Velic A., Soliz J., Shi Y., Li K., Wang S. PHYSIOLOGY. Regulation of breathing by CO₂ requires the proton-activated receptor GPR4 in retrotrapezoid nucleus neurons. Science. 2015;348(6240):1255–1260. doi: 10.1126/science.aaa0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guyenet P.G., Bayliss D.A. Neural control of breathing and CO2 homeostasis. Neuron. 2015;87(5):946–961. doi: 10.1016/j.neuron.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun X., Yang L.V., Tiegs B.C., Arend L.J., McGraw D.W., Penn R.B. Deletion of the pH sensor GPR4 decreases renal acid excretion. J Am Soc Nephrol: JASN. 2010;21(10):1745–1755. doi: 10.1681/ASN.2009050477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang F., Mehta D., Predescu S., Kim K.S., Lum H. A novel lysophospholipid- and pH-sensitive receptor, GPR4, in brain endothelial cells regulates monocyte transmigration. Endothel: J Endothel Cell Res. 2007;14(1):25–34. doi: 10.1080/10623320601177288. [DOI] [PubMed] [Google Scholar]
- 13.Okajima F. Regulation of inflammation by extracellular acidification and proton-sensing GPCRs. Cell Signal. 2013;25(11):2263–2271. doi: 10.1016/j.cellsig.2013.07.022. [DOI] [PubMed] [Google Scholar]
- 14.Sin W.C., Zhang Y., Zhong W., Adhikarakunnathu S., Powers S., Hoey T. G protein-coupled receptors GPR4 and TDAG8 are oncogenic and overexpressed in human cancers. Oncogene. 2004;23(37):6299–6303. doi: 10.1038/sj.onc.1207838. [DOI] [PubMed] [Google Scholar]
- 15.Wyder L., Suply T., Ricoux B., Billy E., Schnell C., Baumgarten B.U. Reduced pathological angiogenesis and tumor growth in mice lacking GPR4, a proton sensing receptor. Angiogenesis. 2011;14(4):533–544. doi: 10.1007/s10456-011-9238-9. [DOI] [PubMed] [Google Scholar]
- 16.Hosford P.S., Mosienko V., Kishi K., Jurisic G., Seuwen K., Kinzel B. CNS distribution, signalling properties and central effects of G-protein coupled receptor 4. Neuropharmacology. 2018;138:381–392. doi: 10.1016/j.neuropharm.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tobo M., Tomura H., Mogi C., Wang J.Q., Liu J.P., Komachi M. Previously postulated "ligand-independent" signaling of GPR4 is mediated through proton-sensing mechanisms. Cell Signal. 2007;19(8):1745–1753. doi: 10.1016/j.cellsig.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 18.Liu J.P., Nakakura T., Tomura H., Tobo M., Mogi C., Wang J.Q. Each one of certain histidine residues in G-protein-coupled receptor GPR4 is critical for extracellular proton-induced stimulation of multiple G-protein-signaling pathways. Pharmacol Res. 2010;61(6):499–505. doi: 10.1016/j.phrs.2010.02.013. [DOI] [PubMed] [Google Scholar]
- 19.Zhang T., Li J., Yin F., Lin B., Wang Z., Xu J. Toosendanin demonstrates promising antitumor efficacy in osteosarcoma by targeting STAT3. Oncogene. 2017;36(47):6627–6639. doi: 10.1038/onc.2017.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van de Wetering M., Francies H.E., Francis J.M., Bounova G., Iorio F., Pronk A. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161(4):933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujii M., Shimokawa M., Date S., Takano A., Matano M., Nanki K. A colorectal tumor organoid library demonstrates progressive loss of niche factor requirements during tumorigenesis. Cell Stem Cell. 2016;18(6):827–838. doi: 10.1016/j.stem.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 22.Lee J.V., Berry C.T., Kim K., Sen P., Kim T., Carrer A. Acetyl-CoA promotes glioblastoma cell adhesion and migration through Ca(2+)-NFAT signaling. Genes Dev. 2018;32(7–8):497–511. doi: 10.1101/gad.311027.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Justus C.R., Dong L., Yang L.V. Acidic tumor microenvironment and pH-sensing G protein-coupled receptors. Front Physiol. 2013;4:354. doi: 10.3389/fphys.2013.00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jing Z., Xu H., Chen X., Zhong Q., Huang J., Zhang Y. The proton-sensing G-protein coupled receptor GPR4 promotes angiogenesis in head and neck cancer. PLoS One. 2016;11(4) doi: 10.1371/journal.pone.0152789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ren J., Jin W., Gao Y.E., Zhang Y., Zhang X., Zhao D. Relations between GPR4 expression, microvascular density (MVD) and clinical pathological characteristics of patients with epithelial ovarian carcinoma (EOC) Curr Pharm Des. 2014;20(11):1904–1916. doi: 10.2174/13816128113199990530. [DOI] [PubMed] [Google Scholar]
- 27.Justus C.R., Yang L.V. GPR4 decreases B16F10 melanoma cell spreading and regulates focal adhesion dynamics through the G13/Rho signaling pathway. Exp Cell Res. 2015;334(1):100–113. doi: 10.1016/j.yexcr.2015.03.022. [DOI] [PubMed] [Google Scholar]
- 28.Castellone R.D., Leffler N.R., Dong L., Yang L.V. Inhibition of tumor cell migration and metastasis by the proton-sensing GPR4 receptor. Cancer Lett. 2011;312(2):197–208. doi: 10.1016/j.canlet.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 29.Xue C., Bahn Y.S., Cox G.M., Heitman J. G protein-coupled receptor Gpr4 senses amino acids and activates the cAMP-PKA pathway in cryptococcus neoformans. Mol Biol Cell. 2006;17(2):667–679. doi: 10.1091/mbc.E05-07-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ren J., Zhang Y., Cai H., Ma H., Zhao D., Zhang X. Human GPR4 and the Notch signaling pathway in endothelial cell tube formation. Mol Med Rep. 2016;14(2):1235–1240. doi: 10.3892/mmr.2016.5380. [DOI] [PubMed] [Google Scholar]
- 31.Tao S.C., Gao Y.S., Zhu H.Y., Yin J.H., Chen Y.X., Zhang Y.L. Decreased extracellular pH inhibits osteogenesis through proton-sensing GPR4-mediated suppression of yes-associated protein. Sci Rep. 2016;6 doi: 10.1038/srep26835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mizuno T., Murakami H., Fujii M., Ishiguro F., Tanaka I., Kondo Y. YAP induces malignant mesothelioma cell proliferation by upregulating transcription of cell cycle-promoting genes. Oncogene. 2012;31(49):5117–5122. doi: 10.1038/onc.2012.5. [DOI] [PubMed] [Google Scholar]
- 33.Wei H., Wang F., Wang Y., Li T., Xiu P., Zhong J. Verteporfin suppresses cell survival, angiogenesis and vasculogenic mimicry of pancreatic ductal adenocarcinoma via disrupting the YAP-TEAD complex. Cancer Sci. 2017;108(3):478–487. doi: 10.1111/cas.13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cai J., Song X., Wang W., Watnick T., Pei Y., Qian F. A RhoA-YAP-c-Myc signaling axis promotes the development of polycystic kidney disease. Genes Dev. 2018;32(11−12):781–793. doi: 10.1101/gad.315127.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ji K., Mayernik L., Moin K., Sloane B.F. Acidosis and proteolysis in the tumor microenvironment. Cancer Metastasis Rev. 2019;38(1-2):103–112. doi: 10.1007/s10555-019-09796-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanderlin E.J., Marie M., Velcicky J., Loetscher P., Yang L.V. Pharmacological inhibition of GPR4 remediates intestinal inflammation in a mouse colitis model. Eur J Pharmacol. 2019;852:218–230. doi: 10.1016/j.ejphar.2019.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Velcicky J., Miltz W., Oberhauser B., Orain D., Vaupel A., Weigand K. Development of selective, orally active GPR4 antagonists with modulatory effects on nociception, inflammation, and angiogenesis. J Med Chem. 2017;60(9):3672–3683. doi: 10.1021/acs.jmedchem.6b01703. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Q-PCR primers used in this study.
CHIP-PCR primers used in this study.
Supplementary figures.