Abstract
Background
USP11 is an ubiquitin-specific protease that plays an important role in tumor progression via different mechanisms. However, the expression and prognostic significance of USP11 in colorectal cancer (CRC) remain unknown.
Methods
Bioinformatics analyses, qRT-PCR, western blotting, and immunohistochemistry were applied for investigating USP11 expression in CRC tissues. Kaplan–Meier analysis with log-rank test was used for survival analyses. LC–MS/MS was performed for identifying potential protein interactions with USP11. In vitro and in vivo assays were used for exploring the function of USP11 during the progression of CRC.
Findings
USP11 was overexpressed in CRC tissues and functioned as an oncogene. Overexpression or knockdown of USP11 promoted or inhibited, respectively, the growth and metastasis of CRC cells in vitro and in vivo. Mechanically, USP11 stabilized PPP1CA by deubiquitinating and protecting it from proteasome-mediated degradation. Moreover, the USP11/PPP1CA complex promoted CRC progression by activating the ERK/MAPK signaling pathway.
Interpretation
USP11 promoted tumor growth and metastasis in CRC via the ERK/MAPK pathway by stabilizing PPP1CA, suggesting USP11 is a potential prognostic marker.
Fund
This work was supported by National Natural Science Foundation of China (NSFC81530044, NSFC81220108021, NSFC81802343), Technology Major Project of China Grants 2017ZX10203206, Shanghai Sailing Program (19YF1409600) and The project of Shanghai Jiaotong University (YG2017QN30).
Keywords: USP11, PPP1CA, Colorectal cancer, Deubiquitination, MAPK pathway
Research in context.
Evidence before this study
Tumor metastasis is one of the primary causes of colorectal cancer (CRC)-related deaths. Invasion and metastasis of CRC cells involves multiple genes and processes. The discovery of novel metastasis-associated biomarkers for predicting the efficiency of treatment and prognosis of CRC is essential. Although USP11 has been reported to be involved in several kinds of tumors via different mechanisms, its role in CRC progression remains unclear.
Added values of this study
Our in vitro and in vivo studies demonstrated that USP11 promotes CRC, and the USP11/PPP1CA/MAPK axis plays an important role in CRC progression.
Implications of all the available evidence
The USP11/PPP1CA complex promotes CRC growth and metastasis, and may represent a promising potential prognostic marker.
Alt-text: Unlabelled Box
1. Introduction
Colorectal cancer (CRC) is the third-most common cancer and third-leading cause of cancer-related death worldwide [1]. Advances in diagnostic detection and therapeutic strategies have led to improvements in the prognosis of CRC during the past decades [2]. However, the five-year relative survival rate of patients with distant tumor metastasis still remains at only 11.7% [3]. A lack of effective biomarkers to predict prognosis and guide therapy is one of the primary causes of poor prognosis. Accordingly, the discovery of novel metastasis-associated biomarkers and clarification of the mechanisms involved is necessary to improve the prognosis of CRC patients.
Recently, the integration of multiple cytotoxic agents and targeted therapies has improved the clinical outcomes of metastatic CRC (mCRC) [4]. Several angiogenesis-targeting agents and epidermal growth factor receptor (EGFR)–targeted antibodies have proved to be applicable in mCRC. Previous studies suggested that anti-EGFR therapy should be excluded from mCRC patients with RAS and RAF mutations, particularly in the K-RAS and B-RAF genes [4,5]. Reportedly, RAS gene mutations are frequently associated with CRC tumorigenesis and progression [6], and RAS proteins may activate the mitogen-activated protein kinase (MAPK) cascade in concert with RAF kinases [7]. The abnormal activation of the RAS/RAF/MAPK signal pathway may not only induce chemoresistance to anti-EGFR therapy in CRC patients, but also affect immune-targeted therapy in other cancers [8,9]. However, the RAS gene encodes a large number of proteins with various functions; therefore, directly targeting RAS remains a challenge [7]. Accordingly, discovering oncogenes or suppressors involved in the RAS/RAF/MAPK pathways, and developing novel drugs may be the best strategy for treating mCRC and improving the prognosis of patients with advanced stage CRC.
Ubiquitination and deubiquitination are the two main types of post-translational modification that can maintain protein homeostasis during biological process [10]. Abnormal ubiquitination or deubiquitination often results in disease. Increasing evidence indicates that deregulation of deubiquitinating enzymes (DUBs) plays significant roles in the progression of cancers [11]. Importantly, some drugs that target specific DUBs have shown good anti-tumor efficacy [12]. DUBs can be classified into different groups: ubiquitin C-terminal hydrolases, ubiquitin-specific proteases (USPs), Machado–Joseph disease protein domain proteases, ovarian tumor proteases, JAB1/MPN/MOV34 family, and the newly identified motif interacting with Ub-containing novel DUB family [13,14]. Among these, USPs are reported to be the most common DUBs, and are involved in protein deubiquitination and regulation of RAS activity [[15], [16], [17]]. Ubiquitin-specific protease 11 (USP11) locates at Xp11.23 and plays an important role in biological functions through stabilizing other proteins via deubiquitination [18,19]. USP11 has been shown to be relevant in tumorigenesis and prognosis in many solid tumors [[20], [21], [22], [23], [24], [25]]. However, the expression and prognostic significance of USP11 in CRC is not well understood, and it remains unclear whether it is involved in CRC progression.
In the present study, we demonstrated that USP11 was overexpressed at both protein and mRNA levels in CRC tissues. Increased expression of USP11 promoted proliferation and metastasis of CRC cells in vitro and in vivo via PPP1CA-mediated activation of the ERK/MAPK pathway. These findings contribute to improving our understanding of the molecular classification of mCRC, and may contribute to the development of a novel therapeutic strategy for mCRC.
2. Materials and methods
2.1. Tissue specimens
CRC tissues and matched normal colon tissues were collected at the Shanghai General Hospital from 2013 to 2015 and used in this study. Prior to surgery, no patients received radiotherapy, chemotherapy, or other related neoadjuvant therapies. This research was approved by the Ethics Committee of Shanghai General Hospital and informed consent was obtained from all patients enrolled in the study.
2.2. Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was isolated from tissues and CRC cell lines using TRIzol (Invitrogen, CA, USA) according to the manufacturer's instructions. cDNA was synthesized using a reverse transcription kit (Takara, Dalian, China). GAPDH was used as an internal control to calculate relative mRNA expression levels using the comparative Ct method. All experiments were performed in triplicate.
2.3. Cell culture and stable cell line construction
CRC cell lines (SW480, SW620, CaCO2, HCT116, RKO, LoVo, HT29, and HCT8) were maintained at 37 °C in a humidified incubator with 5% CO2.
Lentiviral preparations were generated by transiently transfecting HEK-293 T cells with psPAX2 and pMD2.G using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Negative control and empty vector (EV) were used as controls.
2.4. Western blotting
Total protein from tissue samples or cells was extracted and protein concentrations were quantified using a BCA assay kit (Beyotime Biotechnology, China). Equal amounts of protein were separated using SDS-PAGE and electrophoretically transferred to PVDF membranes (Millipore). Antibodies against GAPDH (AF0006, Beyotime Biotechnology), β-actin (AF0003, Beyotime Biotechnology), and β-tubulin (10094-1-AP, Proteintech) were used as loading controls. Horseradish peroxidase (HRP)-conjugated secondary antibodies were used and the protein signals were visualized using ECL detection reagents (Millipore). Antibodies used were: USP11 (ab109232, Abcam), ERK1/2 (#4695, CST), phospho-ERK1/2 (#4370, CST), p38 (#8690, CST), p-p38 (#4511, CST), JNK (#9252, CST), p-JNK (#4668, CST), HA-tag (#3724, CST), Myc-tag (05-724,Sigma-Aldrich), Flag-tag (F3165, Millipore), PPP1CA (sc-271762, Santa Cruz Biotechnology), MEK1/2 (sc-81504, Santa Cruz Biotechnology) and p-MEK1/2 (sc-81,503, Santa Cruz Biotechnology).
2.5. Hematoxylin and eosin (H&E) staining and immunohistochemistry (IHC)
For H&E staining, the slides were dewaxed, rehydrated, stained with H&E, and dehydrated.
For IHC staining, the slides were incubated at 65 °C for 45 min and then dewaxed. Next, we continued to retrieve the antigen using citrate buffer and blocking endogenous peroxidase with 3% H2O2. After non-specific antigen blocking, the slides were incubated with antibodies overnight at 4 °C. The antibody against USP11 for IHC was purchased from Abnova (PAB4191). The next day, slides were incubated with secondary antibody labeled with HRP at room temperature for 1 h. Finally, the reaction was visualized using DAB, and the slides were counterstained with hematoxylin. The IHC score was calculated according to the intensity and extent of the staining (staining intensity: negative = 0, weak = 1, moderate = 2, strong = 3, and staining extent: 0 = no staining, 1 = 0%–10%, 2 = 10%–50%, and 3 = 50%–100%). The total score was calculated as intensity score × extent score. Scores of 0–3 were considered negative expression; 4–6, weak expression; and 8–12, strong expression. IHC score was independently determined by two pathologists who were blinded to the patient characteristics.
2.6. In vitro assays
Cell proliferation was assessed using a CCK-8 kit (Cell Counting Kit-8, Dojindo) to measure cell number and viability. A total of 2000 cells per well were seeded in 96-well plates. The absorbance was determined using a spectrophotometer at 450 nm at 24, 48, 72, and 96 h. For colony formation, 1000 cells per well were seeded into six-well plates. After 2 weeks, cell clones were fixed and stained using crystal violet.
For the wound healing assay, cells were plated into six-well plates and cultured to nearly 90% confluence. A scratch was made in the cell layer using a sterile pipette tip, and the wounded cell layer was washed with phosphate-buffered saline. Cells were incubated with DMEM (Gibco, USA) and images of the wound closure were captured at 0, 24, and 48 h.
Transwell cell migration plates were used for migration and invasion assays. Cells were seeded in serum-free medium in the upper chamber, and 600 μL of DMEM supplemented with 10% FBS (Gibco, USA) was added to the lower chamber. For the invasion assay, the matrigel (Corning Cat. No. 356234) was overlaid in the upper chamber in advance. The cells were incubated for 24 h and fixed. Cells that adhered to the underside of the membrane were stained using 0.1% crystal violet and counted under microscope.
Flow cytometry was performed using an Annexin V PE apoptosis detection kit (BD Biosciences) according to the manufacturer's protocol.
2.7. In vivo assays
Animal experiments were approved by the Institutional Animal Care and Use Committee of the Shanghai Jiao Tong University School of Medicine. Procedures involving animal experiments complied with the guidelines of the Institutional Animal Care and Use Committee of Shanghai General Hospital. A total of 106 cells were injected subcutaneously into the right flanks of 6-week-old male nude mice (Shanghai SLAC Laboratory Animal Co., Ltd., Shanghai, China) to establish a tumor-bearing model (five mice per group). Tumor size was measured every 3 or 4 days using Vernier calipers. Tumor volume was calculated using the formula: volume = length × width2 × 0.5. All experimental mice were euthanized after 4 weeks, and tumors were dissected and fixed in formalin.
To explore the metastatic ability, cells (105/mL, 200 μL) were injected into the spleens of nude mice. All mice were sacrificed after 6 weeks. The tumor colonies in the livers were observed under a microscope using H&E staining.
2.8. Co-immunoprecipitation (IP) assays, mass spectrometry, and ubiquitination assays
Co-IP and ubiquitination assays were performed as previously described [26]. Candidate bands were subjected to mass spectrometry analysis for protein identification. Cells were treated with cycloheximide (CHX, 100 μg/ML, MedChemExpress, Cat. No. HY-12320) and/or MG-132 (15 μM, Selleck, Cat. No. S2619) for the indicated times, followed by Western blotting.
2.9. Statistics
Quantitative variables were analyzed by Student's t-test between groups. One-way ANOVA was used for multiple group comparisons. The association between USP11 expression and clinicopathological features of CRC patients was assessed by chi-square test. Log-rank test using the Kaplan–Meier method was used to assess patients' survival outcome. P < 0·05 was considered statistically significant.
3. Results
3.1. USP11 is overexpressed in CRC and is associated with poor prognosis
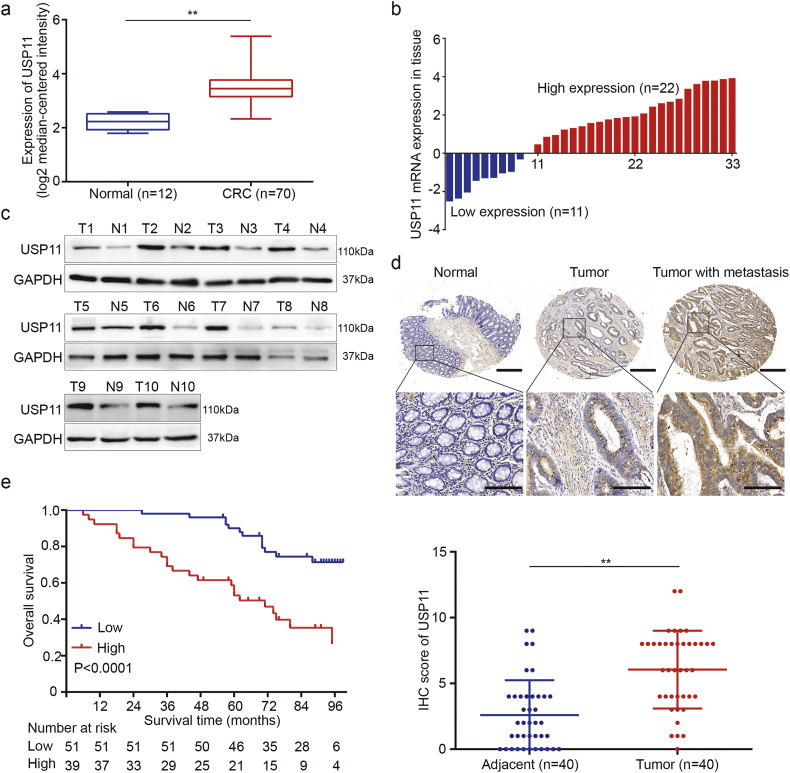
To assess expression levels of USP11 in CRC and normal tissues, we first analyzed the public mRNA data on the Oncomine platform (GSE9348). This platform adopts log2 median-centered intensity to quantify the data for subsequent analysis. We found that mRNA levels of USP11 were higher in CRC tissues (3·487 ± 0·070) compared with normal tissues (P < 0·01, Student's t-test) (Fig. 1a). The public data available in TCGA database also showed similar results (Fig. S1a). We also analyzed mRNA levels of USP11 in CRC and paired non-tumor tissue obtained at our center using qPCR. We found that 66·7% (22/33) of CRC tissues showed higher USP11 levels compared with the paired adjacent tissues (Fig. 1b). Western blot analyses showed that USP11 protein levels were also significantly overexpressed in CRC tissues compared with adjacent normal colon mucosa (Fig. 1c). Moreover, IHC staining analysis was conducted to determine USP11 protein expression in tissue microarray (TMA) containing 40 cases of CRC and paired adjacent normal mucosa. The protein expression levels of USP11 were higher in CRC compared with adjacent colon tissues (Fig. 1d). These results indicated increased expression of USP11 in CRC tissues at both mRNA and protein levels.
Fig. 1.
Expression of USP11 in CRC and its clinical significance. (a) USP11 expression levels of 70 CRC tissues and 12 normal tissues from the Oncomine platform (GSE9348). (b) qRT-PCR analysis of USP11 expression in 33 paired CRC tissues and adjacent normal tissues. (c) Western blot analysis of USP11 expression in 10 randomly selected paired specimens. (d) IHC staining scores of USP11 expression in 40 paired CRC samples. Representative images of different USP11 expression levels are shown, up panel, scale bar 500um; down panel, scale bar 200um. (e) Kaplan–Meier analysis with a log-rank test was performed in CRC patients with different USP11 expression levels. In remaining cases, **P < 0·01, Student's t-test.
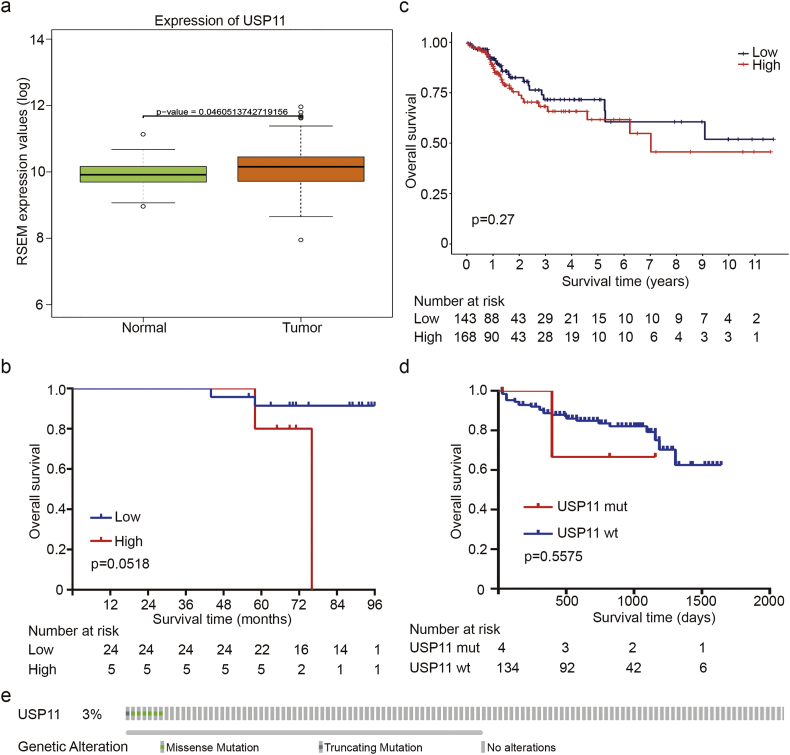
We further analyzed correlations between USP11 protein expression and CRC clinicopathologic features. We divided 90 CRC patients into USP11 high-expression (43.3%, 39/90) and low-expression (56.7%, 51/90) groups according to IHC staining. We discovered that high levels of USP11 expression were positively correlated with tumor stage, tumor T stage and M stage (P < 0·05, Pearson's chi-square test) (Table 1), while there was no correlation between USP11 expression and gender, age, tumor location, tumor N stage, or tumor differentiation (P > 0·05, Pearson's chi-square test). In addition, Kaplan–Meier survival analysis with log-rank testing was applied to evaluate the prognostic value of USP11 to predict survival in CRC patients, and showed that patients with high levels of USP11 protein expression had lower overall survival (OS) rates compared with those with low levels (P < 0·0001, log-rank test) (Fig. 1e). However, USP11 overexpression could not predict poor prognosis in early stage CRC patients (Fig. S1b). Furthermore, there was no significant relationship between OS rates and USP11 mRNA levels (Fig. S1c). We also investigated the genetic mutations of USP11 in CRC by using public data from TCGA. There were only few data regarding the mutations of USP11, what's more, USP11 mutations couldn't contribute to CRC progression (Fig. S1d and e). Taken together, these data indicate that USP11 overexpression may be relevant with the tumorigenesis and progression of CRC.
Table 1.
USP11 expression in CRC and clinicopathological characteristics (n = 90).
| USP11 expression |
|||
|---|---|---|---|
| Low(n = 51) | High(n = 39) | P value | |
| Gender | |||
| Male | 22 | 24 | 0·084 |
| Female | 29 | 15 | |
| Age, years | |||
| <65 | 21 | 15 | 0·794 |
| >65 | 30 | 24 | |
| Tumor location | |||
| Colon | 30 | 25 | 0·611 |
| Rectum | 21 | 14 | |
| Tumor T stage | |||
| 1 | 4 | 0 | 0·001 |
| 2 | 20 | 5 | |
| 3 | 18 | 13 | |
| 4 | 9 | 21 | |
| Tumor N stage | |||
| 0 | 44 | 29 | 0·164 |
| 1 | 7 | 8 | |
| 2 | 0 | 2 | |
| Tumor M stage | |||
| 0 | 50 | 32 | 0·008 |
| 1 | 1 | 7 | |
| Tumor stage | |||
| 1 | 22 | 5 | 0·003 |
| 2 | 21 | 21 | |
| 3 | 7 | 6 | |
| 4 | 1 | 7 | |
| Tumor differentiation | |||
| Well | 13 | 6 | 0·427 |
| Moderate | 33 | 27 | |
| Poor | 5 | 6 | |
Pearson's chi-square test was used and P < 0·05 was considered statistically significant.
3.2. USP11 promotes CRC cell proliferation, migration, and invasion in vitro
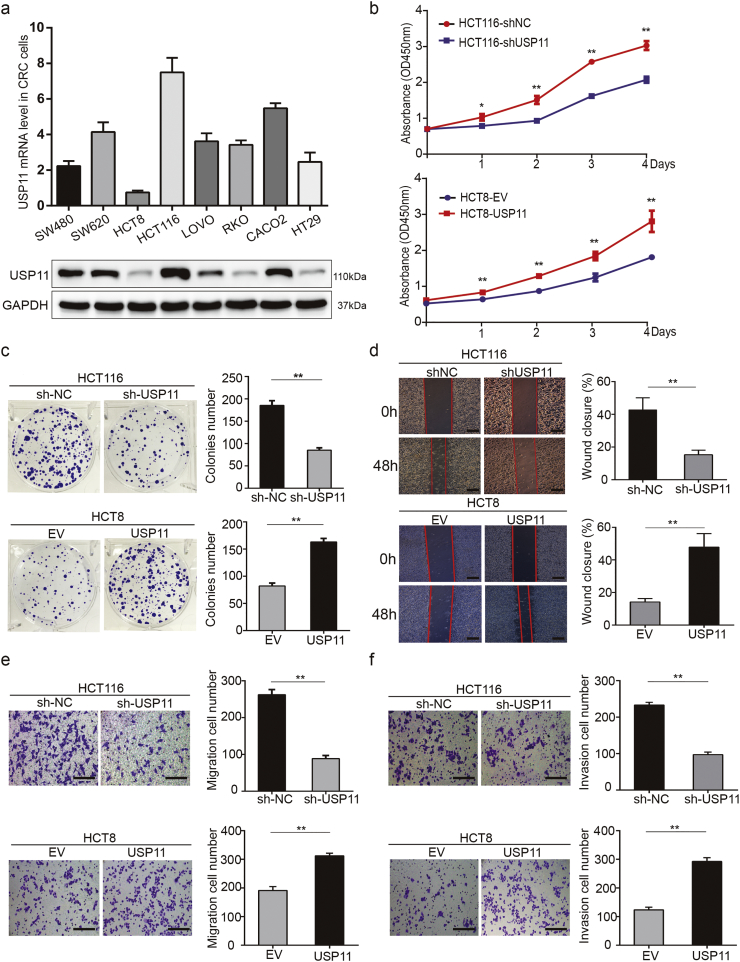
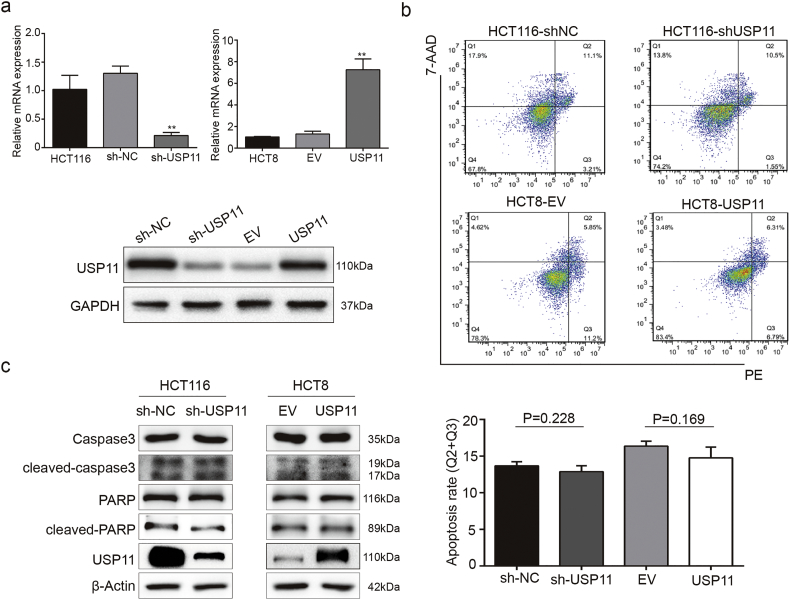
To explore the underlying roles of the USP11 gene on promoting the development of CRC, we first measured USP11 expression levels in eight different CRC cell lines. Among the eight CRC cell lines, HCT116 and HCT8 cells showed the highest and lowest USP11 expression levels, respectively, at both mRNA and protein levels (Fig. 2a). We selected the HCT116 and HCT8 cell lines for further experiments and constructed USP11 knockdown and overexpressing cells with appropriate controls (Fig. S2a). Interestingly, we found that the knockdown and overexpression experiments generated larger differences than those in cell lines. This may be partly due to the complicate biological functions of cells and novel experimental methods are urgent in the future. Cell Counting Kit 8 (CCK8) assay showed that USP11 knockdown significantly inhibited cell viability and proliferation in HCT116 cells compared with the control group, whereas the opposite effect was observed in USP11-overexpressing HCT8 cells (P < 0·01, Student's t-test) (Fig. 2b). These findings were further confirmed using colony formation assays (Fig. 2c). Moreover, wound healing assays showed that USP11 knockdown prohibited wound closure compared with the controls. Conversely, USP11 overexpression promoted wound closure in HCT8 cells (Fig. 2d). Transwell migration and invasion assays also showed that USP11 knockdown or overexpression resulted in lower or higher cell migration and invasion rates, respectively, compared with the controls (Fig. 2e and f).
Fig. 2.
USP11 promotes CRC cell growth, migration, and invasion in vitro. (a) Levels of USP11 mRNA and protein in eight CRC cell lines. (b) Cell proliferation in CRC cells with different USP11 expression levels was assessed using CCK-8 assays. (c) Colony formation assays were performed in CRC cells with different levels of USP11 expression. (d) Wound healing assays were performed to evaluate the migration of CRC cells and the percentage wound closure was calculated; scale bar 200um. (e, f) Transwell assays showed the effect of USP11 expression levels on migration (e) and invasion (f) abilities of CRC cells. Error bars indicate the standard deviation of triplicates; scale bar 200um. *P < 0·05, **P < 0·01, Student's t-test.
However, flow cytometry apoptosis assays showed that neither USP11 overexpression nor knockdown affected the apoptosis rate of CRC cell lines (Fig. S2b). Similarly, the protein expression levels of cleaved-caspase3 and cleaved-PARP showed no obvious changes compared with the controls (Fig. S2c). In summary, in vitro assays indicated that USP11 enhances CRC cell proliferation, migration, and invasion, but showed no effect on apoptosis.
3.3. USP11 enhances tumorigenesis and liver metastasis of CRC in vivo
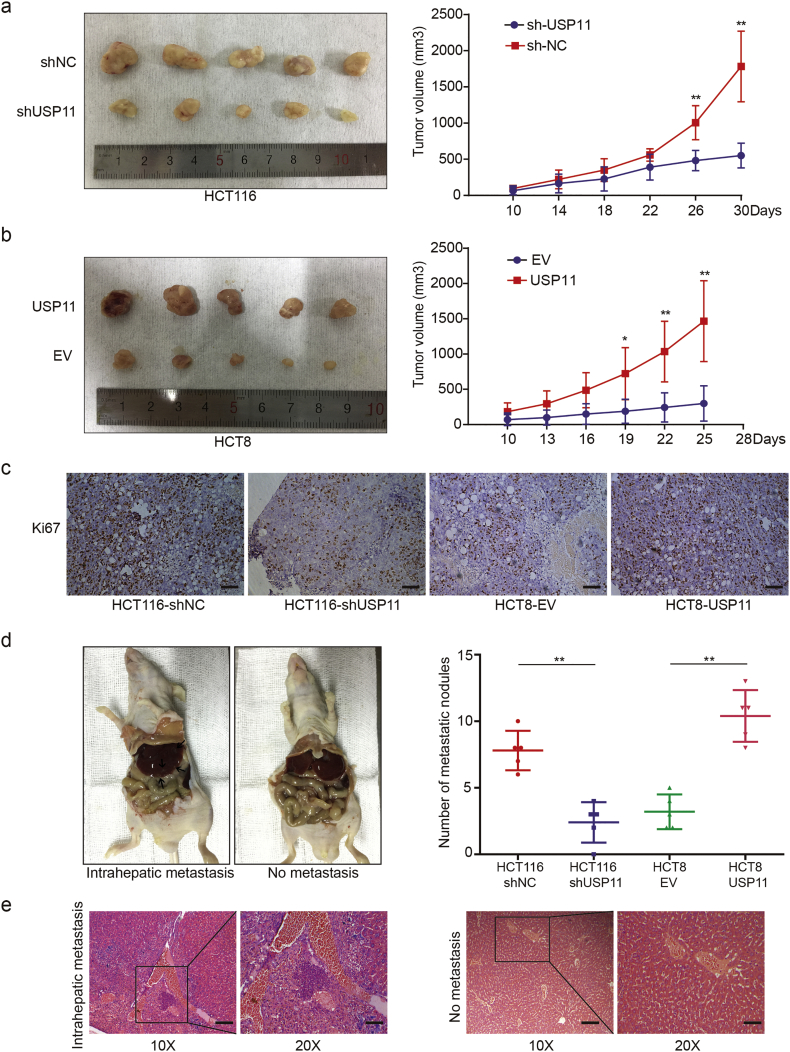
We developed a subcutaneous xenograft model to assess the tumorigenesis ability of cancer cells in vivo as well as the effects of USP11 on proliferation and tumorigenesis. As shown in Fig. 3a, the tumor volume generated by HCT116 cells transfected with shUSP11 plasmids was smaller than that of the controls. The average tumor volume derived from USP11-overexpressing HCT8 cells grew more rapidly than that of the controls (Fig. 3b). IHC staining showed that Ki-67 protein expression was weakened or enhanced in xenograft tumors with USP11 knockdown or overexpression, respectively, compared with the controls (Fig. 3c). As liver is the most common distant metastatic organ in CRC patients, we injected CRC cells into the spleens of nude mice and found that USP11 knockdown or overexpression induced fewer or more metastatic nodules in the liver, respectively (Fig. 3d and e). These results indicate that USP11 promotes the progression of CRC by increasing CRC cell proliferation and liver metastasis in vivo.
Fig. 3.
USP11 promotes CRC growth and metastasis in vivo. Effect of USP11 knockdown (a) or overexpression (b) on CRC tumorigenesis in vivo. The volume of subcutaneous tumors was measured (n = 5). (c) IHC staining of Ki67 in nude mouse xenograft tumors derived from HCT116–shUSP11 cells, HCT8–USP11 cells, and controls; scale bar 100um. (d) Effects of USP11 knockdown or overexpression on intrahepatic metastasis (black arrows) (n = 5). (e) Representative H&E images of intrahepatic metastasis, original magnification 10×, scale bar 200um; original magnification 20×, scale bar 100um. *P < 0·05, **P < 0·01, Student's t-test.
3.4. USP11 may interact with PPP1CA
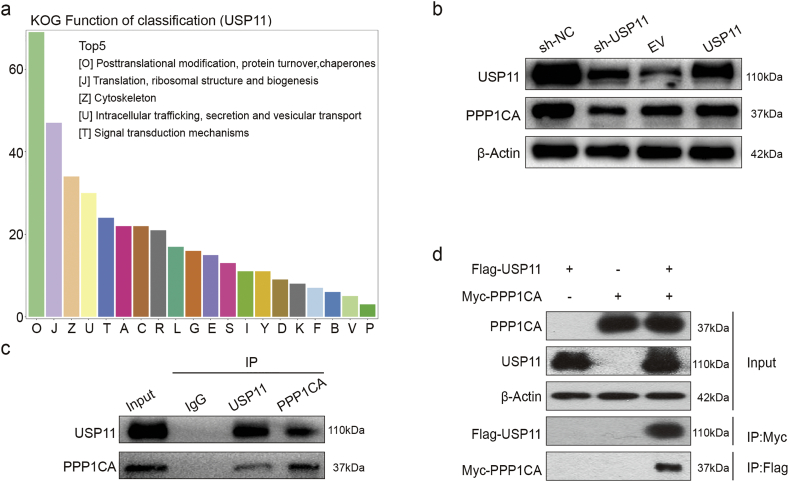
To explore how USP11 promotes the progression of CRC, we first transfected HEK293T cells with either Flag–USP11 plasmid or EV plasmid. We then used two-dimensional PAGE electrophoresis and liquid chromatography-tandem mass spectrometry (LC–MS/MS) to identify altered protein expression induced by USP11 overexpression. A total of 151 proteins were identified from 469 differential protein dots and classified according to biological function. KOG analysis showed a large proportion of post-translational modification of proteins (Fig. 4a). Enrichment analysis of differentially expressed proteins revealed alterations in proteasome, protein phosphatase, and MAPK pathway. These results suggested that USP11 may play a role in regulating protein phosphatase stabilization. During the altered expression of protein phosphatases, PPP1CA was shown to be involved in tumorigenesis via the MAPK pathway [27]. Therefore, we hypothesized that USP11 may promote the progression of CRC by stabilizing PPP1CA.
Fig. 4.
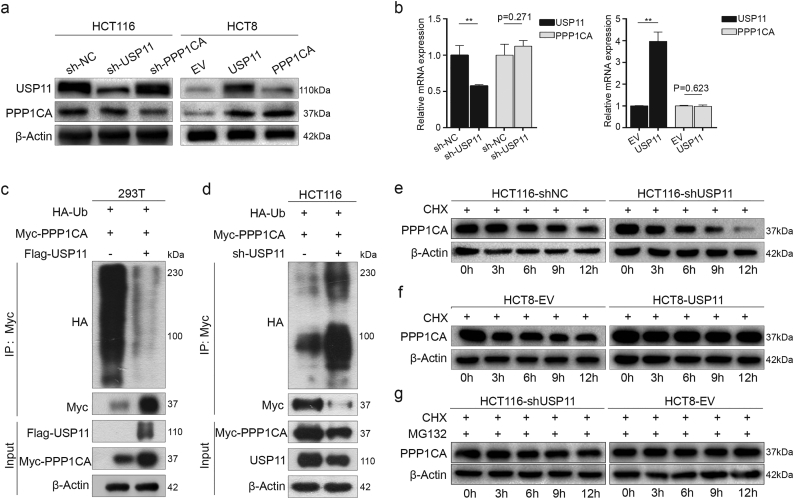
USP11 interacts with PPP1CA endogenously and exogenously. (a) KOG analysis using Co-IP and high-resolution LC–MS/MS assay data. (b) Western blotting was used to detect USP11 and PPP1CA expression in stably transfected cell lines. (c) Endogenous formation of the USP11/PPP1CA complex in HCT116 cells was validated by Co-IP assay. (d) Exogenous USP11/PPP1CA complex formation in 293 T cells transfected with Flag–USP11 and Myc–PPP1CA plasmids was confirmed by Co-IP assays.
To test our hypothesis, we investigated the potential biochemical interrelationship between USP11 and PPP1CA. We found that USP11 knockdown reduced PPP1CA expression, while USP11 overexpression slightly increased PPP1CA expression (Fig. 4b). We then performed an endogenous Co-IP assay in HCT116 cells, and found high USP11 protein expression levels. The results revealed that USP11 can form a complex with PPP1CA in HCT116 cells (Fig. 4c). Exogenous reciprocal Co-IP assays were further performed by co-transfecting Flag–USP11 and Myc–PPP1CA into 293T cells. The results showed Co-IP of Myc–PPP1CA with Flag–USP11 using an anti-Flag antibody. Similarly, Flag–USP11 was also precipitated using an anti-Myc antibody (Fig. 4d). These findings indicated that USP11 and PPP1CA could interact both endogenously and exogenously.
3.5. USP11 stabilizes PPP1CA and protects it from degradation by proteasomes
As a member of the DUB family, USP11 is predominantly involved in the post-translational modification of proteins, and stabilizes downstream targets via deubiquitination. Therefore, we investigated whether USP11 could stabilize PPP1CA expression via deubiquitination.
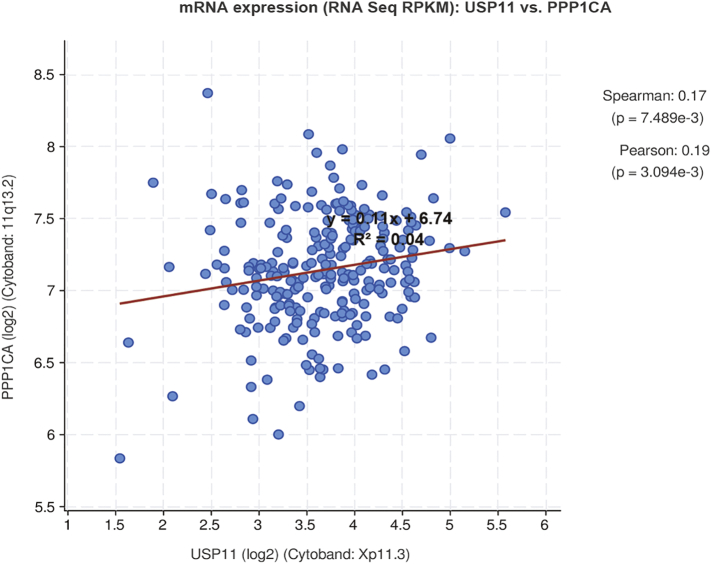
We found that transfection of overexpression or knockdown of USP11 in HCT8 or HCT116 cells increased or decreased PPP1CA expression, respectively. However, knockdown or overexpression of PPP1CA did not influence USP11 protein levels (Fig. 5a), although mRNA levels of USP11 and PPP1CA were positively correlated in CRC tissues (Fig. S3). Neither knockdown nor overexpression of USP11 caused significant changes in PPP1CA mRNA levels by qRT-PCR (Fig. 5b). These findings demonstrated that USP11 regulates PPP1CA post-translationally.
Fig. 5.
USP11 protects PPP1CA from proteasome-mediated degradation via deubiquitination. (a) Effect of USP11 and PPP1CA expression on each other's protein levels. (b) Effects of USP11 on PPP1CA mRNA levels. (c) Functions of USP11 on PPP1CA ubiquitination in 293 T cells. Western blotting analysis was used to detect poly-ubiquitination of PPP1CA. (d) Roles of USP11 on PPP1CA ubiquitination in HCT116 cells. Western blotting analysis showed poly-ubiquitination of PPP1CA. (e, f) Effects of USP11 on degradation of PPP1CA. HCT116–shNC/shUSP11 cells and HCT8–EV/USP11 cells were exposed to CHX (100 μg/mL) and harvested at the indicated times. PPP1CA was measured using Western blotting. (g) PPP1CA was stabilized when cells were co-treated with CHX and MG-132 (15 μM). **P < 0·01, Student's t-test.
Fig. S3.
Levels of USP11 and PPP1CA mRNA were positively correlated in CRC tissues. Correlation analysis (Pearson's correlation) of USP11 and PPP1CA mRNA expression was performed using cbioportal public data.
Ubiquitinaton and deubiquitination maintain the homeostasis of protein modification, and abnormal ubiquitination or deubiquitination frequently result in changes to cell biofunctions. However, it remains unclear whether USP11 could stabilize PPP1CA expression by deubiquitination. We co-transfected Myc–PPP1CA and HA–ubiquitin plasmids into 293T cells transfected with either Flag–USP11 plasmid or EV. Ubiquitination assay further revealed that USP11 overexpression significantly inhibited poly-ubiquitination of PPP1CA protein (Fig. 5c). Conversely, knockdown of USP11 by shRNA in HCT116 cells increased the poly-ubiquitination levels of PPP1CA protein (Fig. 5d).
To demonstrate that USP11 affected PPP1CA expression at the post-translational level but not protein synthesis level, cycloheximide (CHX, 100 μg/mL), a protein synthesis inhibitor, was used to study the degradation rate of PPP1CA. Interestingly, knockdown or overexpression of USP11 resulted in lower or higher protein stability of PPP1CA upon adding CHX, respectively, compared with the controls (Fig. 5e and f). These results suggested that the degradation rate of PPP1CA protein was increased or decreased due to USP11 knockdown or overexpression, respectively. As USP11 predominantly modifies protein expression at the ubiquitination level, we simultaneously treated CRC cells with CHX and the proteasome inhibitor, MG-132 (15 μM). We found that the PPP1CA degradation caused by low USP11 expression was rescued (Fig. 5g). Collectively, these findings suggest that USP11 could stabilize PPP1CA via deubiquitination and prevent ubiquitin-mediated proteasome degradation.
3.6. USP11 promotes MAPK signaling pathway in a PPP1CA-dependent manner
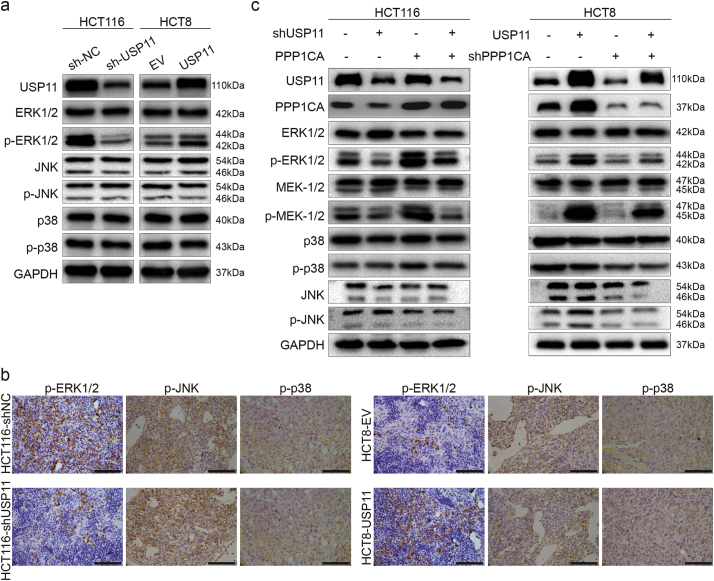
Our previous results revealed that the MAPK pathway was enriched by USP11 overexpression, and it is well known that abnormal activation of MAPK pathway plays significant roles in tumor development. To investigate whether USP11 regulates MAPK pathway activation during the progression of CRC, we further examined levels of key proteins involved in the MAPK pathway. Western blot analysis showed that knockdown of USP11 in HCT116 cells decreased protein levels of phosphorylated ERK1/2 (p-ERK1/2), but did not affect ERK1/2 levels. Conversely, overexpression of USP11 in HCT8 cells increased p-ERK1/2 protein levels. However, there were no significant changes in p38, phosphorylated p38 (p-p38), JNK, and phosphorylated JNK (p-JNK) protein levels (Fig. 6a). Furthermore, in vivo IHC staining showed that overexpression or knockdown USP11 could also enhance or decrease p-ERK1/2 staining in xenograft tumors generated by injecting USP11 overexpression or knockdown cells. (Fig. 6b). These results indicated that USP11 could specifically activate the ERK-dependent MAKP signaling pathway.
Fig. 6.
USP11 promotes the MAPK signaling pathway dependent on PPP1CA. (a) Effect of USP11 on the activation of the MAPK signaling pathway. (b) IHC staining showed p-ERK1/2 staining in xenograft tumors with different USP11 levels; scale bar 100um. (c) Involvement of PPP1CA on USP11-mediated activation of the ERK/MAPK pathway.
Next, we further explored the roles of PPP1CA in USP11-mediated ERK/MAPK pathway activation in vitro. We found that PPP1CA overexpression could partially rescue the decreased phosphorylation of ERK1/2 caused by transfection with the shUSP11 plasmid. Interestingly, transfection with shPPP1CA plasmids in USP11-overexpressing HCT8 cells could also partially alleviate phosphorylation of ERK1/2. Moreover, these similar results were also observed for MEK1/2 protein, which is upstream of ERK1/2. However, there were still no significant changes in p38, p-p38, JNK, and p-JNK protein levels (Fig. 6c). Taken together, our data indicate that USP11 positively regulates the ERK/MAPK signaling pathways in a PPP1CA-dependent manner.
3.7. PPP1CA is indispensable for USP11-mediated promotion of CRC
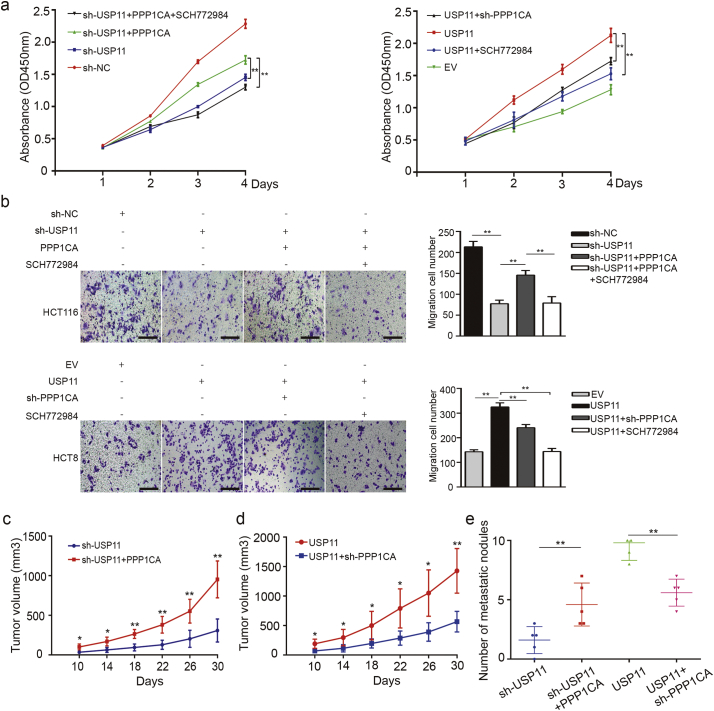
PPP1CA is reported to be one of three catalytic subunits of protein phosphatase 1 (PP1), and has been shown to be closely related to the development of malignant tumors [27]. To investigate whether PPP1CA affects the oncogenic roles of USP11 in CRC, we used CCK-8 and transwell assays to investigate the effects of PPP1CA on USP11-promoted proliferation and migration of CRC cell lines. We found that overexpression of PPP1CA could partially rescue the inhibiting effects on cell proliferation and migration via knockdown of USP11 in HCT116 cells. Conversely, knockdown of PPP1CA could also partially decrease the effects of overexpression of USP11-promoted cell proliferation and migration in HCT8 cells. Furthermore, these promoting effects on cell proliferation and migration caused by increased expression of USP11 or PPP1CA could be weakened by the ERK1/2 inhibitor, SCH772984 (Selleck, Cat. No.S7101, Fig. 7a and b).
Fig. 7.
PPP1CA is essential for USP11-mediated promotion of CRC. (a) The effects of PPP1CA on USP11-induced cell proliferation were investigated using CCK-8 assays. SCH772984 was added as an ERK1/2 inhibitor. (b) Transwell assays were used to evaluate the migration ability in response to changes in USP11 and PPP1CA. SCH772984 was also used as an ERK1/2 inhibitor; scale bar 200um. (c, d) Effect of PPP1CA overexpression or knockdown on the growth rate of subcutaneous tumor caused by USP11 knockdown or overexpression in vivo (n = 5). (e) An in vivo liver metastatic assay was used to clarify the function of PPP1CA during the USP11-mediated promotion of CRC metastasis. *P < 0·05, **P < 0·01. P values were calculated with one-way ANOVA in Fig. a-b and Student's t-test in other figures.
To further validate the effects of PPP1CA on the oncogenic role of USP11 in vivo, we transfected PPP1CA overexpressing plasmids into USP11 knockdown HCT116 cells and injected these cell subcutaneously into nude mice. Subcutaneous xenograft assays showed that overexpression of PPP1CA in HCT116 cells transfected with shUSP11 plasmids generated larger average volumes of xenograft tumors compared with the control group (Fig. 7c). Conversely, sh-PPP1CA decreased the xenograft tumor growth caused by USP11 overexpression (Fig. 7d). In addition, the in vivo metastatic assay also showed that PPP1CA was indispensable for the USP11-mediated promotion of CRC metastasis (Fig. 7e). Thus, these findings confirm that PPP1CA is essential for USP11-mediated CRC progression.
4. Discussion
The development of CRC is a complex process involving multiple molecular interactions that are regulated by key genes. Studies into the regulatory mechanisms involved in these key genes may provide promising strategies for precise treatment of CRC patients. USP11, a member of ubiquitin-specific proteases, has been shown to be involved in the tumorigenesis and prognosis of many solid tumors [[20], [21], [22], [23], [24], [25]]. It has been reported that USP4, USP11 and USP15 are highly paralogous and have partially redundant roles in human malignancies [28]. Another study also showed that there were unique peptide ligands for USP11 which were present in neither USP4 nor USP15 [29]. Based on these findings, we hypothesized that USP11 might have some unique uncovered functions. In the present study, we found that USP11 was upregulated in CRC tissues, and high expression of USP11 predicted poor prognosis in CRC patients. We also demonstrated that overexpression of USP11 promoted proliferation and metastasis of CRC cells in vitro and in vivo, indicating that USP11 plays an important role in the development of CRC. Mechanically, we revealed that USP11 could activate the ERK/MAPK signaling pathway by deubiquitinating and stabilizing PPP1CA, resulting in the malignant development of CRC. While our study highlights the significant roles of USP11 in the progression of CRC, further research and prospective clinical studies are required to identify novel biomarkers for the diagnosis and treatment of CRC patients.
The hallmarks of cancer include sustained proliferative signaling, resistance to cell death, induced angiogenesis, replicative immortality, evasion of growth suppressors, and activation of invasion and metastasis [30]. Among these features, increased proliferation ability or apoptosis inhibition of tumor cells plays significant roles in the development of cancer. In the present study, we demonstrated that USP11 could promote the proliferation of CRC cells via CCK8 and colony formation assays in vitro and subcutaneous xenograft assay in vivo. IHC staining also suggested that Ki67, a proliferation marker, showed strong staining in response to USP11 overexpression. These findings supported our clinical discovery that USP11 expression levels were positively correlated with tumor T stage. Similarly, USP11 was reported to control the proliferative capacity of progenitor cells in human breast cancer cells [31]. However, there was no effect on apoptosis, regardless of USP11 expression levels in CRC cells. This revealed that USP11 did not facilitate the malignant progression of CRC by inhibiting apoptosis, which was different from a previous study that reported that USP11 knockdown induced apoptosis [22]. This indicates that USP11 could play different roles in different cancers, and further highlights the specificity of USP11 in tumor malignant progression and warrants further investigation.
It is widely known that the limitless multiplication of cancer cells caused by promotion of proliferation and inhibition of apoptosis leads to direct invasion, lymph node metastasis, or blood metastasis, driving the progression of cancer cells and resulting in poor prognosis. In the present study, analysis of USP11 expression and clinical features also showed that USP11 expression levels were positively correlated with tumor M stage. This suggested that USP11 promotes CRC invasion and metastasis. In vitro assays also verified that USP11 overexpression could enhance the invasive and metastasis capabilities of CRC cells, especially to the liver in vivo. Liver metastasis is the leading cause of cancer-related death among CRC patients. Approximately 25% of all CRC patients present with liver metastases at first diagnosis, and a further 15%–25% of patients who undergo radical tumor resection also experience liver metastases, resulting in poor prognosis [32]. Therefore, our findings may provide a novel biomarker for predicting of liver metastasis as well as a new therapy target.
Finding novel biomarkers is the crux of precise diagnosis and treatment of CRC, and clarifying the mechanisms involved could promote translational application in clinic. Therefore, it is necessary to understand how USP11 is involved in the malignant progression of CRC as a DUB, based on analysis of clinical specimens and cell function. Maintaining a dynamic equilibrium of ubiquitination and deubiquitination is a key regulatory event during the post-translational modification of proteins. For protein ubiquitination, Ub-activating enzyme (E1), Ub-conjugating enzyme (E2), and Ub ligase (E3) are involved in this sequential process, and mediate the degradation of the targeted proteins. DUBs are able to competitively bind to ubiquitin binding sites and stabilize their targeted proteins, and play significant regulatory roles in a multitude of processes from cancer to neurodegeneration [14,17,33]. USPs are major members of the DUB family of protein, and are involved in many types of cancer and various signaling pathways [34]. USP11, a USP family member, has been proved to stabilize p53, p21, VGLL4 and PTEN as a tumor suppressor [18,19,35,36]. However, USP11 could also promote tumorgenesis by stabilizing Snail, eIF4B and cIAP2 [22,37,38]. The present study revealed that USP11 could promote CRC through stabilizing PPP1CA expression via deubiquitination. These findings suggested that USP11 might act as a double-edged sword in tumors. Furthermore, USP11 has never been reported to activate ERK/MAPK signaling pathway by stabilizing PPP1CA in CRC.
The PPP1CA gene encodes PP1α, an isoform of PP1 protein phosphatases specific for serine/threonine [39]. Protein phosphorylation at serine and threonine residues is ubiquitous in many cellular functions [40,41]. The level of protein phosphorylation is tightly controlled by the interaction between protein kinases and protein phosphatases. Other previous studies have reported that PPP1CA overexpression activates the downstream MAPK pathway. Moreover, our LC–MS/MS results also indicated enrichment of the MAPK pathway via USP11 overexpression. We hypothesize that deubiquitination modification mediated by USP11 could result in PPP1CA accumulation, and further induce the activation of the downstream kinase signaling pathway.
The MAPK signaling pathway plays a key role in the regulation of cell proliferation, differentiation, apoptosis, and innate immunity in numerous biological systems [42,43]. The MAPK signaling pathway comprises at least three sequential kinase components: MAP3K, MAP2K, and MAPK. MAP3Ks phosphorylate and activate MAP2Ks, which, in turn, phosphorylate and activate MAPKs which act to phosphorylate a diverse range of target proteins. There are several signaling markers such as JNK, p38, and ERK that play significant roles in the progression of cancers and drug resistance [43]. Interestingly, only the ERK/MAPK pathway was activated by high USP11 expression in the present study. Mechanically, the oncogene function of USP11 can be weakened by PPP1CA knockdown or ERK1/2 inhibitors. As reported, ERK1/2 is phosphorylated by MEK1/2, which is activated by RAF. The three isoforms of RAF (ARAF, BRAF, and CRAF) are activated by RAS [43]. The RAS/RAF/MEK/ERK signaling pathway can stimulate cancer cell proliferation and metastasis in cancer development [44]. Genetic deregulation can activate the MAPK signaling pathway during the induction and progression of the tumor. RAS and RAF are frequently mutated in mCRC, resulting in the activation of MAPK pathway, which can be treated as a therapeutic target using specific inhibitors [45]. The findings of the present study contribute to the search for novel biomarkers related to the RAS/RAF/MAPK pathway, and may provide useful information regarding the best strategy for mCRC treatment and may improve the prognosis of patients with advanced stage CRC.
In summary, the present study demonstrated that USP11 activated the ERK/MAPK signaling pathway by stabilizing PPP1CA during the development of CRC. USP11 is a promising biomarker for predicting the prognosis of CRC patients, and our findings may provide a new focus for CRC therapeutic strategies. However, CRC is a malignant tumor, and its initiation and development involve complex processes that include multiple genetic changes and epigenetic modifications. Therefore, the underlying molecular mechanisms of USP11 in CRC still need to be further explored.
The following are the supplementary data related to this article.
Fig. S1.
USP11 expression levels in CRC and its prognostic roles. (a) USP11 mRNA expression levels of CRC tissues and normal tissues from TCGA database, Student's t-test. (b) Kaplan–Meier analysis with a log-rank test was performed in stage I CRC patients with different USP11 expression levels. (c) Kaplan–Meier analysis with a log-rank test for OS according to USP11 mRNA expression levels. (d) Effects of genetic mutations in USP11 on overall survival of CRC patients from the TCGA dataset. (e) Genetic alteration analyses of USP11 in CRC by using public data.
Fig. S2.
USP11 has no effect on CRC cell apoptosis. (a) Western blotting and qRT-PCR were used to validate the efficacy of USP11 knockdown and overexpression. (b) Quantification of the apoptotic CRC cell population by flowcytometry. USP11 knockdown and overexpression showed no statistically significant changes compared with the control groups. (c) Western blotting assay was used to investigate the effect of USP11 expression on apoptosis-associated proteins. Error bars indicate the standard deviation of triplicates. **P < 0·01. P values were calculated with Student's t-test.
The primer sequences used for qRT-PCR in the study.
Sequences of shRNAs for different genes.
Sources of funding
This work was supported by National Natural Science Foundation of China (NSFC81530044, NSFC81220108021, and NSFC81802343), Technology Major Project of China Grants 2017ZX10203206, Shanghai Sailing Program (19YF1409600) and the project of Shanghai Jiaotong University (YG2017QN30). The funders had no role in study design and implementation.
Author contributions
Hongze Sun, Baochi Ou, Senlin Zhao, Zhihai Peng conceived and designed the research study; Hongze Sun, Xueni Liu, Liwei Song, Xisheng Liu, Rangrang Wang processed and analyzed the data. Hongze Sun, Baochi Ou, Senlin Zhao, Zhihai Peng wrote and revised the manuscript. All authors read and approved the final manuscript.
Declaration of Competing Interest
The authors have nothing to disclose.
Acknowledgements
We want to thank Shuang Xu (Shanghai General Hospital, School of medicine, Shanghai Jiao Tong University) for providing guidance and technical assistance in the study.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Fedewa S.A. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67(3):177–193. doi: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 3.Brenner H., Kloor M., Pox C.P. Colorectal cancer. Lancet. 2014;383(9927):1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 4.Fakih M.G. Metastatic colorectal cancer: current state and future directions. J Clin Oncol. 2015;33(16):1809–1824. doi: 10.1200/JCO.2014.59.7633. [DOI] [PubMed] [Google Scholar]
- 5.Amado R.G., Wolf M., Peeters M. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26(10):1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 6.Vogelstein B., Fearon E.R., Hamilton S.R. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319(9):525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 7.Simanshu D.K., Nissley D.V., McCormick F. RAS proteins and their regulators in human disease. Cell. 2017;170(1):17–33. doi: 10.1016/j.cell.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lois S., Dushyanthen S., Beavis P.A. RAS/MAPK activation is associated with reduced tumor-infiltrating lymphocytes in triple-negative breast Cancer: therapeutic cooperation between MEK and PD-1/PD-L1 immune checkpoint inhibitors. Clin Cancer Res. 2016;22(6):1499–1509. doi: 10.1158/1078-0432.CCR-15-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benvenuti S., Sartore-Bianchi A., Di Nicolantonio F. Oncogenic activation of the RAS/RAF signaling pathway impairs the response of metastatic colorectal cancers to anti-epidermal growth factor receptor antibody therapies. Cancer Res. 2007;67(6):2643–2648. doi: 10.1158/0008-5472.CAN-06-4158. [DOI] [PubMed] [Google Scholar]
- 10.Gu H., Shi X., Liu C. USP8 maintains embryonic stem cell stemness via deubiquitination of EPG5. Nat Commun. 2019;10(1):1465. doi: 10.1038/s41467-019-09430-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim S.O., Li C.W., Xia W. Deubiquitination and stabilization of PD-L1 by CSN5. Cancer Cell. 2016;30(6):925–939. doi: 10.1016/j.ccell.2016.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D'Arcy P., Linder S. Proteasome deubiquitinases as novel targets for cancer therapy. Int J Biochem Cell Biol. 2012;44(11):1729–1738. doi: 10.1016/j.biocel.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 13.Clague M.J., Heride C., Urbe S. The demographics of the ubiquitin system. Trends Cell Biol. 2015;25(7):417–426. doi: 10.1016/j.tcb.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 14.He M., Zhou Z., Wu G., Chen Q., Wan Y. Emerging role of DUBs in tumor metastasis and apoptosis: therapeutic implication. Pharmacol Ther. 2017;177:96–107. doi: 10.1016/j.pharmthera.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhattacharya S., Ghosh M.K. Cell death and deubiquitinases: perspectives in cancer. Biomed Res Int. 2014;2014 doi: 10.1155/2014/435197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim K.H., Ramakrishna S., Baek K.H. Molecular mechanisms and functions of cytokine-inducible deubiquitinating enzymes. Cytokine Growth Factor Rev. 2013;24(5):427–431. doi: 10.1016/j.cytogfr.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 17.D'Arcy P., Wang X., Linder S. Deubiquitinase inhibition as a cancer therapeutic strategy. Pharmacol Ther. 2015;147:32–54. doi: 10.1016/j.pharmthera.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 18.Deng T., Yan G., Song X. Deubiquitylation and stabilization of p21 by USP11 is critical for cell-cycle progression and DNA damage responses. Proc Natl Acad Sci U S A. 2018;115(18):4678–4683. doi: 10.1073/pnas.1714938115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park M.K., Yao Y., Xia W. PTEN self-regulates through USP11 via the PI3K-FOXO pathway to stabilize tumor suppression. Nat Commun. 2019;10(1):636. doi: 10.1038/s41467-019-08481-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burkhart R.A., Peng Y., Norris Z.A. Mitoxantrone targets human ubiquitin-specific peptidase 11 (USP11) and is a potent inhibitor of pancreatic cancer cell survival. Mol Cancer Res. 2013;11(8):901–911. doi: 10.1158/1541-7786.MCR-12-0699. [DOI] [PubMed] [Google Scholar]
- 21.Bayraktar S., Gutierrez Barrera A.M., Liu D. USP-11 as a predictive and prognostic factor following neoadjuvant therapy in women with breast cancer. Cancer J (Sudbury, Mass) 2013;19(1):10–17. doi: 10.1097/PPO.0b013e3182801b3a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee E.W., Seong D., Seo J., Jeong M., Lee H.K., Song J. USP11-dependent selective cIAP2 deubiquitylation and stabilization determine sensitivity to Smac mimetics. Cell Death Differ. 2015;22(9):1463–1476. doi: 10.1038/cdd.2014.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marquez J., Kohli M., Arteta B. Identification of hepatic microvascular adhesion-related genes of human colon cancer cells using random homozygous gene perturbation. Int J Cancer. 2013;133(9):2113–2122. doi: 10.1002/ijc.28232. [DOI] [PubMed] [Google Scholar]
- 24.Zhou Z., Luo A., Shrivastava I. Regulation of XIAP turnover reveals a role for USP11 in promotion of tumorigenesis. EBioMedicine. 2017;15:48–61. doi: 10.1016/j.ebiom.2016.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Al-Salihi M.A., Herhaus L., Macartney T., Sapkota G.P. USP11 augments TGFbeta signalling by deubiquitylating ALK5. Open Biol. 2012;2(6) doi: 10.1098/rsob.120063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma H., Chen H., Guo X. M phase phosphorylation of the epigenetic regulator UHRF1 regulates its physical association with the deubiquitylase USP7 and stability. Proc Natl Acad Sci U S A. 2012;109(13):4828–4833. doi: 10.1073/pnas.1116349109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen M., Wan L., Zhang J. Deregulated PP1alpha phosphatase activity towards MAPK activation is antagonized by a tumor suppressive failsafe mechanism. Nat Commun. 2018;9(1):159. doi: 10.1038/s41467-017-02272-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vlasschaert C., Xia X., Coulombe J., Gray D.A. Evolution of the highly networked deubiquitinating enzymes USP4, USP15, and USP11. BMC Evol Biol. 2015;15:230. doi: 10.1186/s12862-015-0511-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spiliotopoulos A., Blokpoel Ferreras L., Densham R.M. Discovery of peptide ligands targeting a specific ubiquitin-like domain-binding site in the deubiquitinase USP11. J Biol Chem. 2019;294(2):424–436. doi: 10.1074/jbc.RA118.004469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 31.Garcia D.A., Baek C., Estrada M.V. USP11 enhances TGFβ-induced epithelial-mesenchymal plasticity and human breast cancer metastasis. Mol Cancer Res. 2018;16(7):1172–1184. doi: 10.1158/1541-7786.MCR-17-0723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen D.L., Liu Y.X., Zhang J.X. Long non-coding RNA UICLM promotes colorectal cancer liver metastasis by acting as a ceRNA for microRNA-215 to regulate ZEB2 expression. Theranostics. 2017;7(19):4836–4849. doi: 10.7150/thno.20942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nijman S.M., Luna-Vargas M.P., Velds A. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123(5):773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 34.Singh N., Singh A.B. Deubiquitinases and cancer: a snapshot. Crit Rev Oncol Hematol. 2016;103:22–26. doi: 10.1016/j.critrevonc.2016.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ke J.Y., Dai C.J., Wu W.L. USP11 regulates p53 stability by deubiquitinating p53. J Zhejiang Univ Sci B. 2014;15(12):1032–1038. doi: 10.1631/jzus.B1400180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang E., Shen B., Mu X. Ubiquitin-specific protease 11 (USP11) functions as a tumor suppressor through deubiquitinating and stabilizing VGLL4 protein. Am J Cancer Res. 2016;6(12):2901–2909. [PMC free article] [PubMed] [Google Scholar]
- 37.Kapadia B., Nanaji N.M., Bhalla K. Fatty acid synthase induced S6Kinase facilitates USP11-eIF4B complex formation for sustained oncogenic translation in DLBCL. Nat Commun. 2018;9(1):829. doi: 10.1038/s41467-018-03028-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang W., Wang J., Yan H., Zhang K., Liu Y. Upregulation of USP11 promotes epithelial-to-mesenchymal transition by deubiquitinating snail in ovarian cancer. Oncol Rep. 2019;41(3):1739–1748. doi: 10.3892/or.2018.6924. [DOI] [PubMed] [Google Scholar]
- 39.Castro M.E., Ferrer I., Cascon A. PPP1CA contributes to the senescence program induced by oncogenic Ras. Carcinogenesis. 2008;29(3):491–499. doi: 10.1093/carcin/bgm246. [DOI] [PubMed] [Google Scholar]
- 40.Cohen P.T. Protein phosphatase 1--targeted in many directions. J Cell Sci. 2002;115:241–256. doi: 10.1242/jcs.115.2.241. Pt 2. [DOI] [PubMed] [Google Scholar]
- 41.Wies E., Wang M.K., Maharaj N.P. Dephosphorylation of the RNA sensors RIG-I and MDA5 by the phosphatase PP1 is essential for innate immune signaling. Immunity. 2013;38(3):437–449. doi: 10.1016/j.immuni.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arthur J.S., Ley S.C. Mitogen-activated protein kinases in innate immunity. Nat Rev Immunol. 2013;13(9):679–692. doi: 10.1038/nri3495. [DOI] [PubMed] [Google Scholar]
- 43.Kim E.K., Choi E.J. Compromised MAPK signaling in human diseases: an update. Arch Toxicol. 2015;89(6):867–882. doi: 10.1007/s00204-015-1472-2. [DOI] [PubMed] [Google Scholar]
- 44.Das Thakur M., Stuart D.D. Molecular pathways: response and resistance to BRAF and MEK inhibitors in BRAF(V600E) tumors. Clin Cancer Res. 2014;20(5):1074–1080. doi: 10.1158/1078-0432.CCR-13-0103. [DOI] [PubMed] [Google Scholar]
- 45.Burotto M., Chiou V.L., Lee J.M., Kohn E.C. The MAPK pathway across different malignancies: a new perspective. Cancer. 2014;120(22):3446–3456. doi: 10.1002/cncr.28864. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The primer sequences used for qRT-PCR in the study.
Sequences of shRNAs for different genes.