Abstract
Background
Metastasis is a major obstacle in the treatment of bladder cancer (BC). Circular RNAs exert various functions in the aggressive biological behaviour of cancers. In this study, we aimed to elucidate how circPICALM influences BC metastasis.
Methods
The expression of circPICALM was analysed by real-time PCR. The tumourigenic properties of BC cells were evaluated using in vitro migration, invasion, and wound healing assays and an in vivo footpad model. The interaction between circPICALM and miR-1265 was confirmed by pull-down and dual-luciferase reporter assays and biotin-labelled miRNA capture. The interaction of STEAP4 and focal adhesion kinase (FAK) was confirmed by co-immunoprecipitation.
Findings
CircPICALM was downregulated in BC tissues, and low circPICALM expression was related to advanced T stage, high grade, lymph node positivity and poor overall survival. Overexpression of circPICALM inhibited the metastasis of BC cells, and DHX9 negatively regulated circPICALM levels. CircPICALM colocalized with miR-1265 and acted as a sponge for this miRNA, and the pro-invasion effect of miR-1265 was abolished by circPICALM overexpression. STEAP4, a target of miR-1265, suppressed metastasis; it bound to FAK to prevent autophosphorylation at Y397 and influenced EMT in BC cells.
Interpretation
CircPICALM can inhibit BC metastasis and bind to miR-1265 to block its pro-invasion activity. STEAP4 is a target of miR-1265 and is related to FAK phosphorylation and EMT.
Fund
This research was supported by National Natural Science Foundation of China, No.81772728, National Natural Science Foundation of China, No.81772719, National Natural Science Foundation of China No.81572514.
Keywords: circPICALM, miR-1265, STEAP4, EMT, Metastasis, Bladder cancer
Research in context.
Evidence before this study
Circular RNAs (circRNAs) are newly discovered noncoding RNAs that play an emerging role in regulating gene expression and various biological processes. Metastasis is a pivotal aspect of BC malignancy that negatively influences clinical outcome. DHX9 regulates a subclass of circRNAs. There is accumulating evidence that EMT profoundly influences the mobility and dissemination potential of cancer cells, and FAK is closely related to EMT in BC. RNA expression data were obtained from dataset GSE97239 and TCGA.
Added value of this study
We first evaluated circPICALM expression in BC and found that low circPICALM expression predicted a poor outcome, evidence that circPICALM could be a promising predictive and therapeutic target. We clarified the molecular mechanism of circPICALM, which provided insight into the mechanisms by which FAK is regulated in BC. We described the pro-metastatic roles of miR-1265 and its direct target STEAP4, which influenced EMT, in BC.
Implications of all the available evidence
Our research identified a new circRNA contributing to the regulation of BC metastasis. Further investigations inspired by our study could focus on the value of circPICALM as a preoperative predictive biomarker of BC metastasis and the applicability of the identified molecular mechanisms to other cancers.
Alt-text: Unlabelled Box
1. Introduction
Bladder cancer (BC) is the ninth most prevalent cancer worldwide, causing an estimated 165,000 deaths each year [1]. It is predicted that in 2040, BC will cause 323,450 deaths annually [2]. Approximately three-fourths of BCs are classified as non–muscle-invasive BC (NMIBC), which has a high tendency to relapse [3]. Up to 50% of patients with muscle-invasive BC (MIBC) develop distant recurrence, at least half of whom will die from metastasis within two years [4,5]. Overall, visceral metastasis is an independent risk factor of BC [6], and lymphatic metastasis is associated with poor survival. Therefore, it is of great significance to thoroughly understand the mechanism of tumour invasion and metastasis.
Tumour metastasis is a complicated multi-step process whose molecular mechanism remains barely understood [7,8]. Current studies have found that epithelial–mesenchymal transition (EMT) exerts a profound influence on tumour invasion and metastasis [9]. BC patients with high EMT-related gene expression levels respond poorly to PD-1 blockage treatment and have shorter progression-free and overall survival [10]. Focal adhesion kinase (FAK) is a protein tyrosine kinase that plays a critical role in cellular morphology and motility [11]. Several studies have suggested that FAK activation promotes BC cell invasion through AKT or Src to influence EMT [[12], [13], [14]]. However, the functional role and interaction network of FAK have not been fully elucidated.
Circular RNAs (circRNAs) are a subclass of transcripts with covalently closed loop structures that lack classic 5′ caps or 3′ polyadenylated tails [15,16]; these RNAs used to be regarded as by-products of erroneous splicing [17,18]. However, with advanced computational analysis of sequencing data, thousands of circRNAs have been discovered in humans and have been shown to serve various functions in carcinogenesis, cardiac regeneration, immunomodulation, and myogenesis [[19], [20], [21], [22]]. Some circRNAs can act as microRNA (miRNA) sponges to regulate target genes in trans, while some circRNAs interfere with their parental genes in cis by competing with linear splicing [14,23]. CircRNAs can also interact directly with proteins or be translated into proteins [22,24].
In this study, we queried a published dataset and identified circPICALM as a differentially expressed circRNA. Overexpression of circPICALM inhibited BC cell invasion in vitro and in vivo through sponging miR-1265, a miRNA that promoted invasion and bound to the 3′ untranslated region (UTR) of STEAP4. Importantly, STEAP4 inhibited BC metastasis by modulating FAK activation and EMT.
2. Materials and methods
2.1. Patients and samples
A total of 168 BC samples were obtained at surgery and immediately stored in liquid nitrogen. And 40 corresponding adjacent normal tissue samples from the macroscopic tumour margin in the cohort were isolated and processed at the same time, which were obtained at a distance of over 3 cm from the edge of cancer tissues. The histological and pathological diagnoses were confirmed and the specimens were classified according to the 2004 World Health Organization Consensus Classification and Staging [25,26]. Patients underwent surgery from 2010 to 2016 at Sun Yat-sen Memorial Hospital, Sun Yat-sen University. All procedures were in accordance with the Declaration of Helsinki and approved by the Ethics Committee of Sun Yat-sen Memorial Hospital, Sun Yat-sen University. Written informed consent was obtained from each patient before the study. Clinical information of the patients was summarized in Table 1.
Table 1.
Correlation of circPICALM levels and clinical parameters.
| Characteristics | circPICALM expression |
|||
|---|---|---|---|---|
| No. (%) | Low (%) | High (%) | P-value | |
| Gender | ||||
| Male | 139 (82.7) | 39 (28.1) | 100 (71.9) | 0.290 |
| Female | 29 (17.3) | 11 (37.9) | 18 (62.1) | |
| Age | ||||
| <65 | 86 (51.2) | 21 (24.4) | 65 (75.6) | 0.121 |
| ≥65 | 82 (48.8) | 29 (35.4) | 53 (64.6) | |
| Tumour size | ||||
| <3 cm | 140 (83.3) | 44 (31.4) | 96 (68.6) | 0.291 |
| ≥3 cm | 28 (16.7) | 6 (21.4) | 22 (78.6) | |
| Pathology stage | ||||
| pTa-pT1 | 79 (47.0) | 12 (15.2) | 67 (84.8) | 0.000⁎⁎ |
| pT2-T4 | 89 (53.0) | 38 (42.7) | 51 (57.3) | |
| Grade | ||||
| Low | 35 (20.8) | 4 (11.4) | 31 (88.6) | 0.008⁎⁎ |
| High | 133 (79.2) | 46 (34.6) | 87 (65.4) | |
| Lymphatic metastasis | ||||
| Yes | 27 (16.1) | 20 (74.1) | 7 (25.9) | 0.000⁎⁎ |
| No | 141 (83.9) | 30 (21.3) | 111 (78.7) | |
| Total | 168 | 50 | 118 | |
Chi-square test.
P < .01.
2.2. Cell culture
Human BC cell lines (T24, RRID:CVCL_0554; UM-UC-3, RRID:CVCL_1783; J82, RRID:CVCL_0359; RT-4, RRID:CVCL_0036), a human immortalized uroepithelium cell line (SV-HUC-1, RRID:CVCL_3798), and human embryonic kidney cells (HEK-293 T, RRID:CVCL_0063) were obtained from American Type Culture Collection (Manassas, VA, USA). Human BC EJ cells were obtained from the Institute of Biochemistry and Cell Biology of Chinese Academy of Sciences (Shanghai, China). T24 and EJ cells were cultured with RPMI1640 (Gibco, USA). UM-UC-3, J82 and HEK-293 T cells were cultured with DMEM (Gibco, USA). RT-4 cells were cultured with McCoy's 5A (Gibco, USA) and SV-HUC-1 cells were cultured with F-12 K medium (Gibco, USA). All media were supplemented with 10% foetal bovine serum (BI, Israel) and 1% penicillin/streptomycin (Gibco, USA), and all cells were cultured in an incubator at 37 °C with 5% CO2.
2.3. RNA preparation, reverse transcription, real-time PCR (RT-PCR), and nuclear mass separation assay
RNA isolation, reverse transcription, RT-PCR, and nuclear mass separation assays were conducted according to the protocols reported before [27]. The cycle number for RT-PCR was 40. The primers used in this study are listed in Supplementary Table 1.
2.4. Nucleic acid electrophoresis
The PCR products were mixed with 6× loading buffer (TaKaRa, Japan) and GelRed (Biotium, US). Electrophoresis was performed in 1% agarose with TAE buffer. A 100-bp ladder was used as a molecular weight marker (CWBIO, China). Bands were visualized by UV irradiation.
2.5. Actinomycin D assay and RNase R treatment
For the actinomycin D assay, T24 and UM-UC-3 cells were cultured with 5 μg/ml actinomycin D (APE×BIO, USA) in RPMI1640 medium and DMEM medium, respectively. Cells were harvested at 0, 4, 8, 12, and 24 h after the addition of actinomycin D. The relative abundance was normalized to the expression of 0 h. For RNase R treatment, RNA was extracted and incubated with RNase R (Epicenter Technologies, USA) at 37 °C for 30 min. Then, the stability of circRNA and its linear transcript was measured by RT-PCR.
2.6. Florescence in situ hybridization (FISH)
Cells were seeded into confocal dishes and fixed at 50% confluence. Cy3-labelled circPICALM and FAM-labelled miR-1265 probes were designed and synthesized by GenePharma (Shanghai, China). Nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI). All processes were conducted using the Fluorescent In Situ Hybridization Kit (GenePharma, Shanghai, China) following the manufacturer's instructions. Images were obtained by a TCS SP9 STED 3× confocal microscope (Leica, USA). The probes used in this study are listed in Supplementary Table 2.
2.7. Oligonucleotide transfection, vector construction and stable transfection
Cells were transfected with small interfering RNAs (siRNAs), miRNA mimics, or miRNA inhibitor (GenePharma, China) using Lipofectamine RNAiMAX (Invitrogen, USA). The circPICALM sequence was cloned into the pLenti-ciR-GFP-T2A-puro vector (IGEbio, China), and the STEAP4 sequence was cloned into the pCDH-GFP + Puro-3xFLAG vector (IGEbio, China). To package lentivirus, HEK-293 T cells were transfected with overexpression vectors, psPAX2 (Addgene, USA, RRID:Addgene_12260) and PMD2.G (Addgene, USA, RRID:Addgene_12259) using X-tremeGENE (Sigma, USA). Lentiviruses were harvested, filtered and concentrated. Cells were infected with virus and selected with puromycin. The oligonucleotides transfected in this study are listed in Supplementary Table 3.
2.8. Wound healing, cell migration and cell invasion assays
Wound healing, cell migration, and cell invasion assays were performed as previously described [28].
2.9. Animal experiments and HE staining
Animal experiments were performed according to the guidelines of the National Institutes of Health and were approved by the Animal Ethics Committee of Sun Yat-sen University. A total of 5 × 106 circPICALM-overexpressing or control UM-UC-3 cells were injected into the footpads of 4-week-old male BALB/c nude mice. Four weeks later, the popliteal lymph nodes (LNs) were harvested, and their volume was calculated as volume = (length× width2)/2. For HE staining, tissues were paraffin-embedded, dewaxed, rehydrated, and stained with HE.
2.10. Western blot analysis
Cells were treated with radioimmunoprecipitation assay buffer (RIPA, Beyotime, China) supplemented with 1% protease and phosphatase inhibitors (CWBIO, China). Equal amounts of protein were electrophoresed and transferred onto polyvinylidene fluoride (PVDF) membranes, which were blocked by 5% nonfat milk in TBST buffer for one hour and then incubated with primary antibodies against the following proteins: DHX9 (ab26271, Abcam, UK, RRID:AB_777725), GAPDH (AC002, ABclonal, China, RRID:AB_2736879), STEAP4 (ab63967, Abcam, UK, RRID:AB_1143135), FAK (ab40794, Abcam, UK, RRID:AB_ 732,300), pFAK (phospho Y397) (ab81298, Abcam, UK, RRID:AB_1640500), ZO-1 (#8193, CST, US, RRID:AB_10898025), E-Cadherin (#3195, CST, US, RRID:AB_2291471), Vimentin (#5741, CST, US, RRID:AB_10695459), N-Cadherin (#13116, CST, US, RRID:AB_2687616), ZEB1 (#3396, CST, US, RRID:AB_1904164), Slug (#9585, CST, US, RRID:AB_2239535), and β-Catenin (#9562, CST, US, RRID:AB_331149). The next day, the membranes were washed with TBST buffer and incubated with HRP-conjugated secondary antibodies. Signal intensities were measured by Immobilon ECL substrate (Millipore, Germany), and images were acquired using an OPTIMAX X-ray Film Processor (Protec, Germany).
2.11. RNA immunoprecipitation (RIP)
RIP was conducted with a Magna RIP™ RNA-binding protein immunoprecipitation kit (Millipore, USA) according to the manufacturer's guidelines. Briefly, anti-Ago2 antibody (Millipore, USA) or rabbit IgG was incubated with magnetic beads at room temperature for 30 min to generate antibody-coated beads. Approximately 1 × 107 cells were lysed and mixed with beads on a rotator at 4 °C overnight. Then, the beads were washed, and co-immunoprecipitated RNA was extracted by RNAiso Plus (TaKaRa, Japan) and detected by RT-PCR.
2.12. Pull-down assay
Biotin-labelled circPICALM or oligo probes (GenePharma, China) were pre-incubated with Streptavidin-Dyna beads M-280 (Invitrogen, USA). CircPICALM-overexpressing and control cells were lysed and incubated with the beads at 4 °C overnight. Then, the beads were washed, and the crosslinking was reversed. RNA was extracted using RNAiso Plus (TaKaRa, Japan) and measured by RT-PCR.
2.13. Dual-luciferase reporter assay
Target sequences were synthesized and inserted into the psiCHECK-2 plasmid. Then, Renilla luciferase (Rluc) and firefly luciferase activities were detected using the Dual-Glo Luciferase Assay System (Promega, USA) following the method described before [28].
2.14. Biotin-labelled miRNA capture
Stable circPICALM-overexpressing cells were transfected with biotinylated miR-1265 mimics or control (GenePharma, China). After 48 h, the lysates were incubated with pre-blocked streptavidin magnetic beads. Then, the beads were washed, and circPICALM and the 3’UTR of STEAP4 abundance were determined by RT-PCR and electrophoresis.
2.15. Co-immunoprecipitation (Co-IP)
T24 and UM-UC-3 cells stably expressing STEAP4–3 × FLAG were established. Co-IP was performed using a Pierce™ Crosslink Magnetic IP/Co-IP Kit (Thermo Fisher Scientific, USA) with an anti-FLAG antibody (#14793, CST, US, RRID:AB_2572291) according to the manufacturer's instructions. Immunoprecipitated proteins were analysed by western blotting with the anti-FAK and anti-Flag antibodies described above.
2.16. Statistical analysis
Data were analysed by SPSS 22.0 (IBM, USA, RRID:SCR_002865), and a P-value<.05 was considered statistically significant. Student's t-test, the Wilcoxon rank-sum test, or the Mann-Whitney U test were applied to compare the means between groups. Spearman's rank correlation coefficient assays were used to analyse the expression correlation. A chi-square test and univariate and multivariate Cox proportional hazards regression model were used to analyse correlations between circPICALM levels and clinical parameters. The log-rank test and Kaplan-Meier survival curve was used to evaluate overall survival. Data are presented as the mean ± standard deviation (SD).
3. Results
3.1. Identifying circPICALM in BC
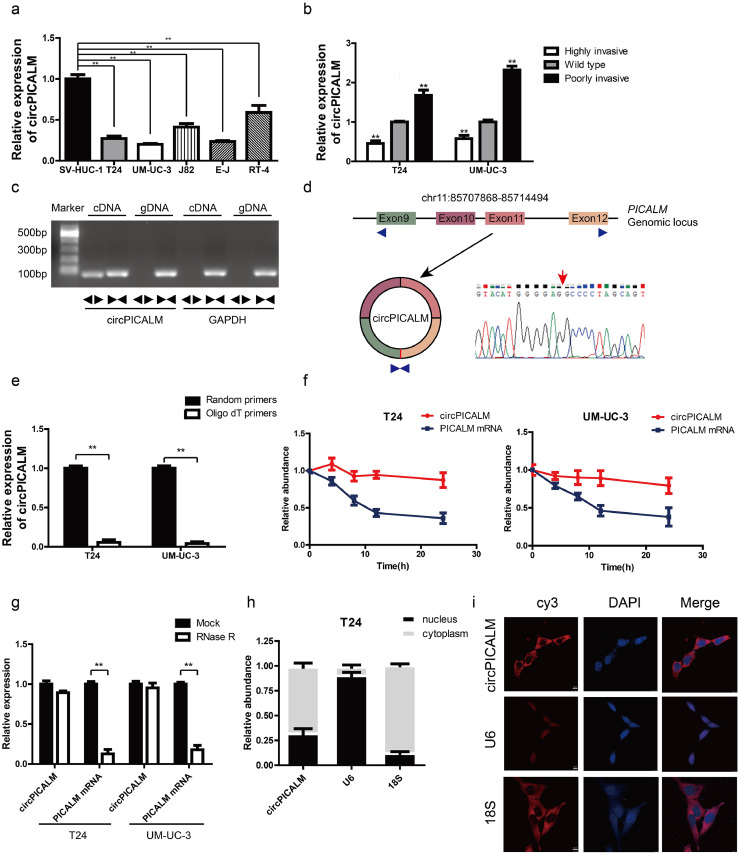
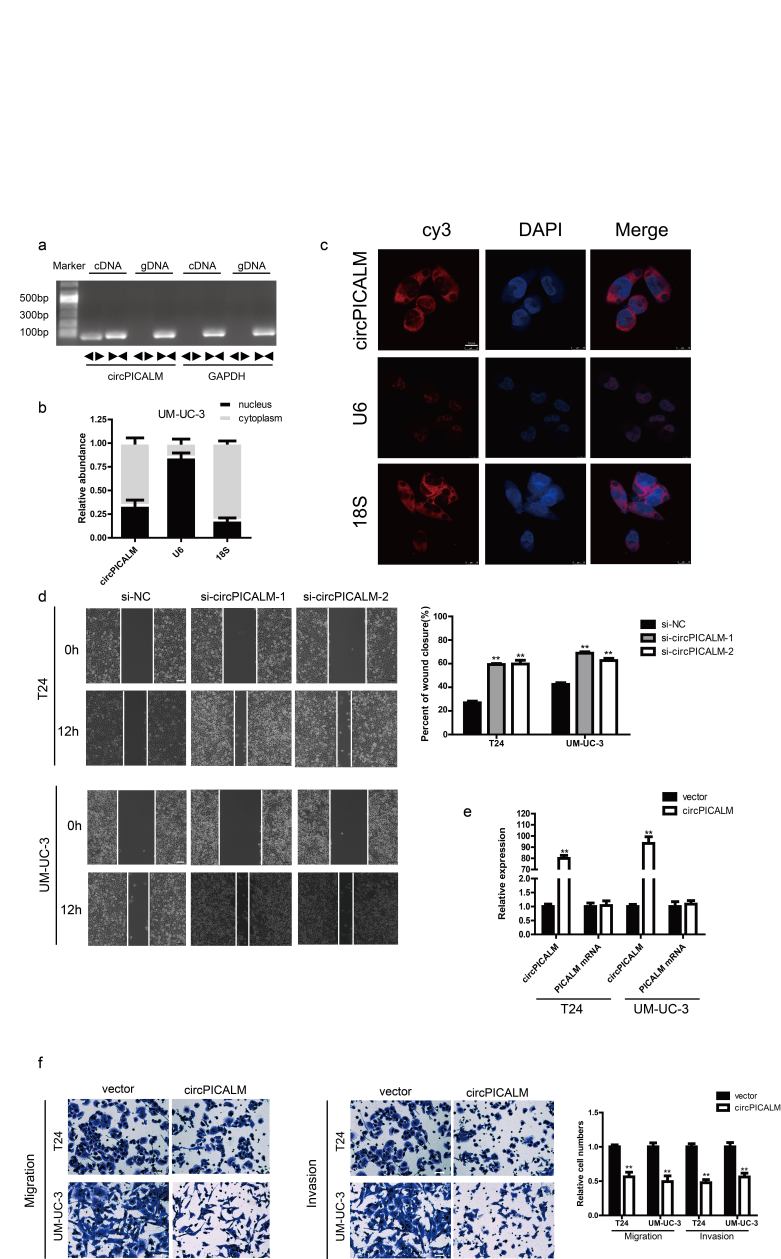
We used the published GSE97239 dataset to identify circRNAs differentially expressed between BC and adjacent normal tissues [29]. We chose to investigate downregulated hsa_circ_0023919 (circPICALM) for the following reasons: 1, circPICALM had relatively low expression levels in commonly used BC cell lines compared with the human immortalized uroepithelium cell line SV-HUC-1 (Fig. 1a); 2, circPICALM levels were downregulated in the highly invasive T24 and UM-UC-3 cells, models we established and described previously [14], and circPICALM levels were relatively higher in poorly invasive BC cells (Fig. 1b); and 3, functional studies of circPICALM are rare. CircRNAs and their linear counterparts have identical sequences, except at the junction of the transcript [15]. Therefore, we designed primers targeting the back-splice junction (divergent primers) and primers targeting the linear section (convergent primers). We performed RT-PCR using these two primer sets with cDNA and genomic DNA (gDNA) as templates. Not surprisingly, unique products of the expected length were amplified (Fig. 1c; Supplementary Fig. 1a). CircPICALM is derived from exon 9 to exon 12 of the PICALM gene, and the junction site was further verified by Sanger sequencing (Fig. 1d). CircPICALM levels were significantly lower when oligo-dT primers were used than when random primers were used in the reverse transcription system (Fig. 1e). We also found that circPICALM was more stable than its linear form after treatment with actinomycin D (Fig. 1f) and RNase R (Fig. 1g), as evidenced by RT-PCR detection. We examined the relative abundance of circPICALM in the nucleus and cytoplasm via nuclear mass separation assays (Fig. 1h; Supplementary Fig. 1b) and FISH (Fig. 1i; Supplementary Fig. 1c) and concluded that circPICALM was mainly located in the cytoplasm. These results indicate that circPICALM is a rather stable, mainly cytoplasmic transcript derived from exons 9 to 12 of the PICALM gene.
Fig. 1.
Characteristic of circPICALM.
a. Relative expression of circPICALM in SV-HUC-1, T24, UM-UC-3, J82, E-J, and RT-4 cells measured by RT-PCR. b. Relative abundance of circPICALM in our previously established invasion cell models. c. PCR products of divergent or convergent primers and T24 cDNA or gDNA were validated by gel electrophoresis. d. Schematic illustration of formation of circPICALM. The junction site was proved by Sanger sequence and indicated by red arrow. e. RT-PCR analysis of circPICALM expression in random primers or oligo dT primers reverse transcription systems. f. Relative abundance of circPICALM and linear PICALM in T24 and UM-UC-3 cells treated with 5μg/ml actinomycin D. RNAs were measured via RT-PCR at the indicated time. g. RT-PCR results of circPICALM and linear PICALM in BC cells treated with or without RNase R. h. Subcellular distribution of circPICALM was detected by nuclear mass separation assay in T24 cells. U6 and 18S acted as nuclear and cytoplasm controls, respectively. i. Subcellular location of circPICALM was illustrated by FISH in UM-UC-3 cells. U6 and 18S acted as nuclear and cytoplasm controls, respectively. Scale bar, 10 μm. (Data are presented as the mean ± SD, n = 3. Unpaired, two-tailed student's t-test, **P<0.01).
3.2. CircPICALM has significantly lower expression in BC and is related to survival
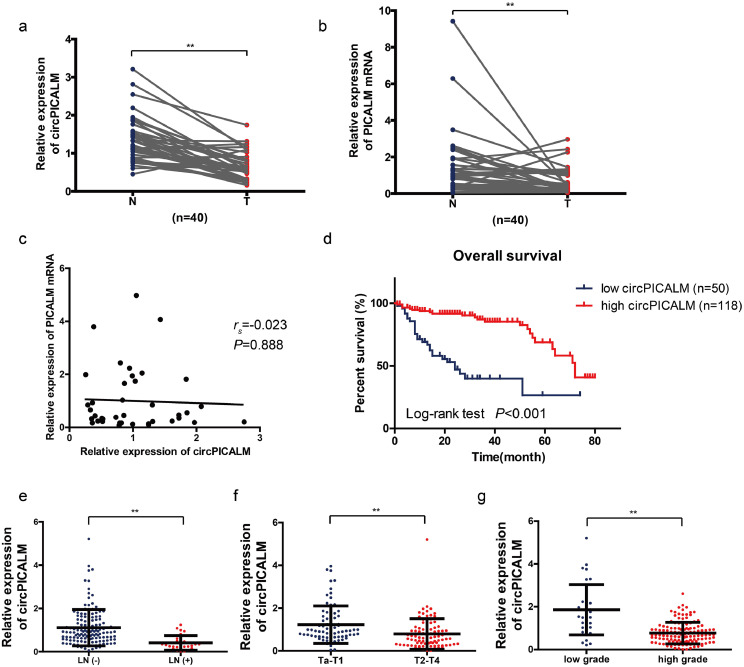
Next, we investigated whether circPICALM is differentially expressed in BC patients. We performed RT-PCR to ascertain circPICALM and PICALM mRNA expression in 40 paired cancer and adjacent normal tissue samples. In accordance with the GSE97239 dataset, circPICALM had markedly lower expression in BC than in adjacent noncancerous tissues (Fig. 2a). PICALM mRNA also had lower expression (Fig. 2b), but there was no correlation between circPICALM and PICALM mRNA expression in BC samples (Fig. 2c). Subsequently, we used RT-PCR to measure circPICALM expression in 168 BC cases with follow-up and clinical parameter data. Kaplan-Meier survival analysis indicated that patients with a relatively lower abundance of circPICALM had poorer outcomes (Fig. 2d). Additionally, patients with LN metastasis, advanced T stage and high grade had lower circPICALM expression (Fig. 2e-g). A chi-square test was conducted to elucidate the correlation between circPICALM status and clinical parameters. Pathological stage, grade, and LN metastasis, but not other features, showed a strong correlation with circPICALM levels (Table 1). We performed an univariate and multivariate Cox regression analysis and found that lymphatic metastasis and circPICALM expression were independent prognostic factors for BC patients (Table 2). Taken together, the data indicate that circPICALM overexpression suggests favourable survival of BC patients, suggesting a regulatory role for circPICALM.
Fig. 2.
Clinical values of circPICALM in BC.
a. Relative expression of circPICALM in 40 paired primary BC (T) and adjacent normal (N) tissues, measured by RT-PCR. b. Relative expression of PICALM mRNA in 40 paired primary BC (T) and adjacent normal (N) tissues, measured by RT-PCR. c. The correlation between circPICALM and PICALM expression in 40 BC samples. d. The Kaplan-Meier survival curve comparing patients' survival percentages with relatively high or low expression of circPICALM. e-g. Relative abundance of circPICALM in BC patients compared by LN status, T stage, and grade. (a, b, Paired, two-tailed, Wilcoxon rank-sum test; c. Spearman's rank correlation; d, Log-rank test; e-g, Mann-Whitney U test. **P<0.01).
Table 2.
Univariate and multivariate Cox regression analysis of circPICALM expression and survival of BC patients.
| Characteristics | Univariate analysis |
P-value |
Multivariate analysis |
P-value | ||
|---|---|---|---|---|---|---|
| HR | 95%CI | HR | 95%CI | |||
| Gender | 0.734 | 0.382–1.410 | 0.353 | |||
| Age | 1.024 | 0.999–1.049 | 0.061 | |||
| Tumour size | 0.858 | 0.403–1.825 | 0.691 | |||
| Pathology stage | 2.264 | 1.260–4.068 | 0.006⁎⁎ | 1.435 | 0.761–2.704 | 0.264 |
| Grade | 3.783 | 1.483–9.649 | 0.005⁎⁎ | 1.517 | 0.547–4.208 | 0.423 |
| Lymphatic metastasis | 4.026 | 2.253–7.194 | 0.000⁎⁎ | 2.034 | 1.043–3.963 | 0.042⁎ |
| circPICALM expression | 0.274 | 0.150–0.501 | 0.000⁎⁎ | 0.443 | 0.234–0.841 | 0.013⁎ |
Abbreviations: HR hazard ratio, CI confidence interval. Cox regression analysis.
P < .05.
P < .01.
3.3. CircPICALM inhibits BC cell invasion and can be regulated by DHX9
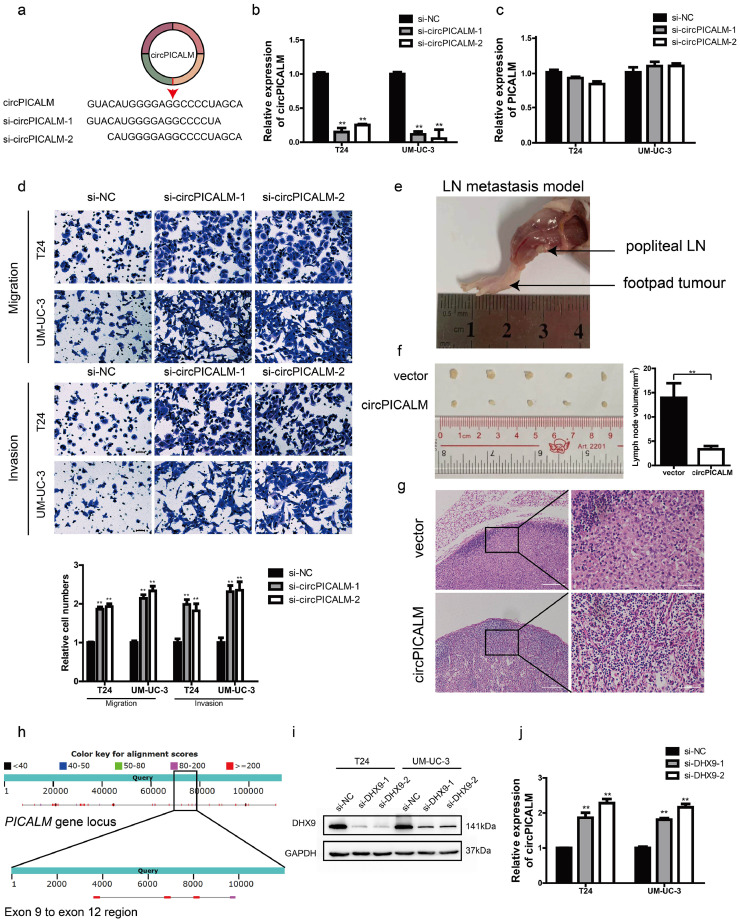
We investigated the biological role of circPICALM in BC via loss- and gain-of-function assays. We used siRNAs designed to target the junction site of circPICALM (Fig. 3a). RT-PCR analysis showed that these siRNAs successfully silenced circPICALM expression (Fig. 3b) with minimal impact on the linear form (Fig. 3c). T24 and UM-UC-3 cells with circPICALM knockdown had a more invasive phenotype and more rapid wound healing (Fig. 3d; Supplementary Fig. 1d). We then constructed plasmids to stably overexpress circPICALM in BC cell lines (Supplementary Fig. 1e). Overexpression of circPICALM inhibited T24 and UM-UC-3 cell migration, invasion, and wound healing in vitro (Supplementary Fig. 1f; Supplementary Fig. 2a). We further investigated the effect of circPICALM in vivo. UM-UC-3 cells overexpressing circPICALM or negative control vector were injected into the footpads of BALB/c nude mice (4–6 weeks old, five mice per group) to assess lymphatic metastasis. Popliteal LNs were harvested four weeks later (Fig. 3e). Not surprisingly, circPICALM potently reduced the volume of popliteal LNs (Fig. 3f) and lymphatic invasion (Fig. 3g). DHX9 is a well-known RNA helicase that can interact with inverted Alu repeats (IARs) to reduce the expression of a subset of circRNAs [30]. Abundant IARs can be found within the PICALM gene locus, some of which are located around exons 9 to 12 (Fig. 3h). Data confirmed that circPICALM abundance increased after silencing DHX9 (Fig. 3i-j), suggesting that DHX9 could be a potential regulator.
Fig. 3.
Effects of circPICALM in BC in vitro and in vivo.
a. Schematic illustration of two siRNAs targeting the junction site of circPICALM. b. and c. Expression of circPICALM and PICALM mRNA in T24 and UM-UC-3 cells transfected with si-NC or si-circPICALM. d. Cell migratory and invasive abilities of BC cells treated with circPICALM siRNAs were measured by migration and invasion assays. Scale bar, 100 μm. e. A representative image of in vivo footpad model. f. BALB/c nude mice were injected with circPICALM overexpression or control UM-UC-3 cells into the footpads. Four weeks later, the popliteal LNs were harvested and measured. (n = 5) g. HE staining of popliteal LNs. Scale bar, 200 μm for the left, 50 μm for the right. h. Distribution of IARs within PICALM gene locus. Upper band showed the whole length of PICALM, while the lower band showed the ±500 bp of exon 9 to exon 12 region. Alignment scores were indicated with specific colours. i. Expression levels of DHX9 were evaluated by western blotting after transfected with DHX9 siRNAs in T24 and UM-UC-3 cells. i. Relative levels of circPICALM in T24 and UM-UC-3 cells were measured after DHX9 was silenced. (Data are presented as the mean ± SD, n = 3. Unpaired, two-tailed student's t-test, **P<0.01).
3.4. CircPICALM binds to and sponges miR-1265
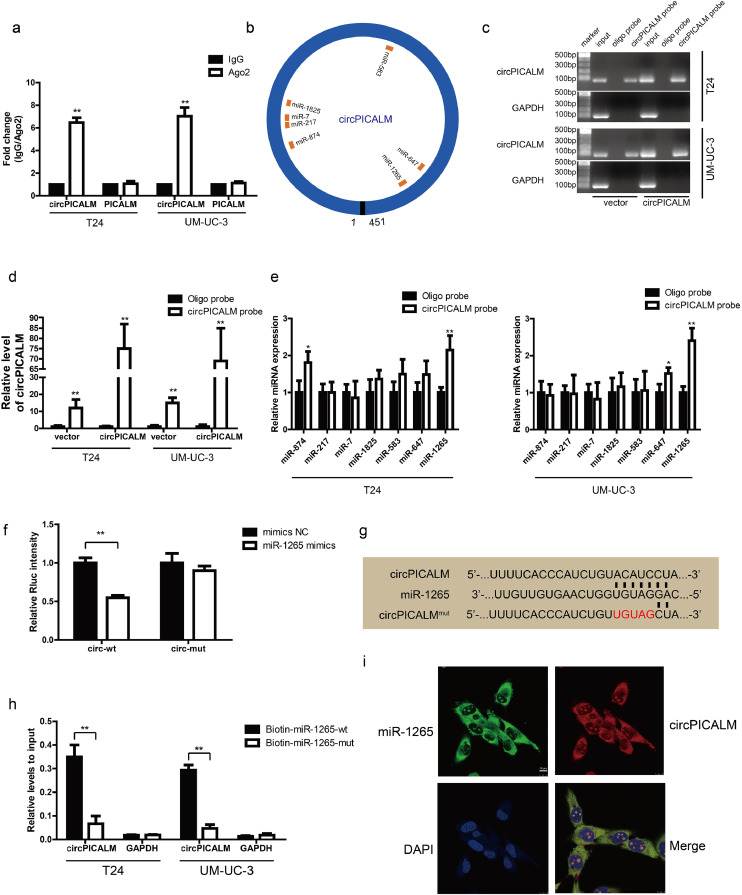
Considering the subcellular location of circPICALM in BC cells, we hypothesized that it may function by sponging miRNA. We measured the abundance of circPICALM and linear PICALM using anti-Ago2 and IgG RIP assays in T24 and UM-UC-3 cells and found that circPICALM was enriched by the anti-Ago2 antibody (Fig. 4a), which preliminarily supported our theory. Next, we identified potential target miRNAs by cross-referencing the predictions from CircInteractome, miRanda, and RNAhybrid. Seven candidate miRNAs (miR-874, miR-217, miR-7, miR-1825, miR-583, miR-647, and miR-1265) were chosen for the following study (Fig. 4b). The biotin-labelled circPICALM probe robustly enriched circPICALM compared with the oligo probe in BC cells, verifying the efficiency of the pull-down assay (Fig. 4c-d). RNA extracted from the pull-down assay was analysed by RT-PCR, and miR-1265 was the most promising miRNA due to consistent results in T24 and UM-UC-3 cells (Fig. 4e). We then constructed a psiCHECK-2 plasmid containing the wild-type circPICALM sequence. Compared with the NC group, the miR-1265 mimics group showed a significant reduction in relative Rluc activity in HEK-293 T cells (Fig. 4f); however, mutating the putative circPICALM binding site (Fig. 4 g) abolished this reduction (Fig. 4f). To validate the physical interaction, we performed RNA capture with biotin-labelled miR-1265 mimic probes in stable circPICALM-overexpressing T24 and UM-UC-3 cells; considerably more circPICALM was captured by the miR-1265 wild-type probe than the mutant probe (Fig. 4 h), confirming the role of circPICALM as a miR-1265 sponge. Finally, FISH assays revealed that circPICALM and miR-1265 highly colocalized in the cytoplasm of T24 and UM-UC-3 cells (Fig. 4i; Supplementary Fig. 2b). In conclusion, the results above prove our hypothesis that circPICALM interacts with and sponges miR-1265.
Fig. 4.
circPICALM binds to and sponges miR-1265.
a. RIP assay for circPICALM and PICALM mRNA fold changes in BC cells using anti-Ago2 or IgG antibodies. IgG was used as a negative control. b. Schematic illustration of the predicted binding sites between circPICALM and seven candidate miRNAs. c. and d. RNA pull-down assay with circPICALM or oligo probes, validated by gel electrophoresis and RT-PCR. e. Relative levels of seven candidate miRNAs in cell lysate pulled down by oligo or circPICALM probes were detected by RT-PCR. f. A dual-luciferase reporter assay in HEK-293 T cells to prove the interaction between circPICALM and miR-1265. Wild-type or mutant circPICALM sequences were cloned into psiCHECK-2 plasmids. Rluc intensity was normalized to firefly luciferase activity. g. Part of wild-type and mutant circPICALM sequences for the dual-luciferase reporter assay and the binding site for miR-1265. h. Biotin-labelled miR-1265 probe capture in circPICALM overexpression T24 and UM-UC-3 cells. The RNA abundance was measured by RT-PCR and normalized to input. i. Colocalization of miR-1265 and circPICALM in UM-UC-3 cells, demonstrated by FISH. Probe for miR-1265 was labelled by FAM and probe for circPICALM was labelled by cy3. Scale bar, 10 μm. (Data are presented as the mean ± SD, n = 3. Unpaired, two-tailed student's t-test, *P<0.05, **P<0.01).
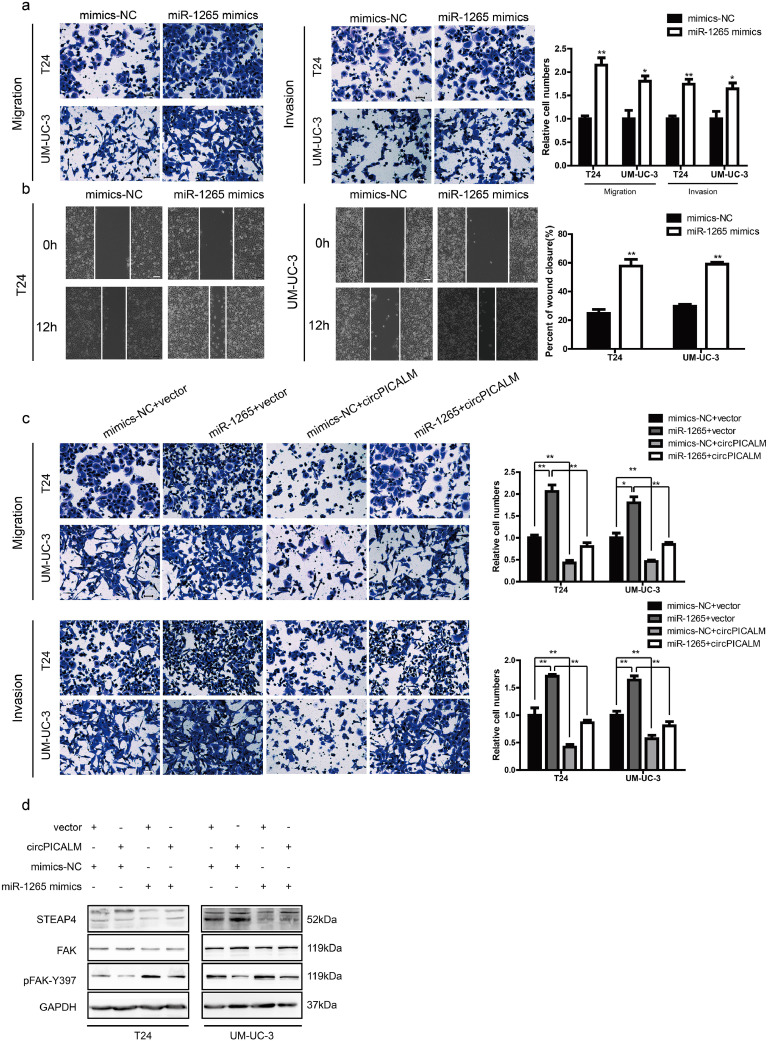
3.5. miR-1265 promotes invasion, which is inhibited by circPICALM overexpression
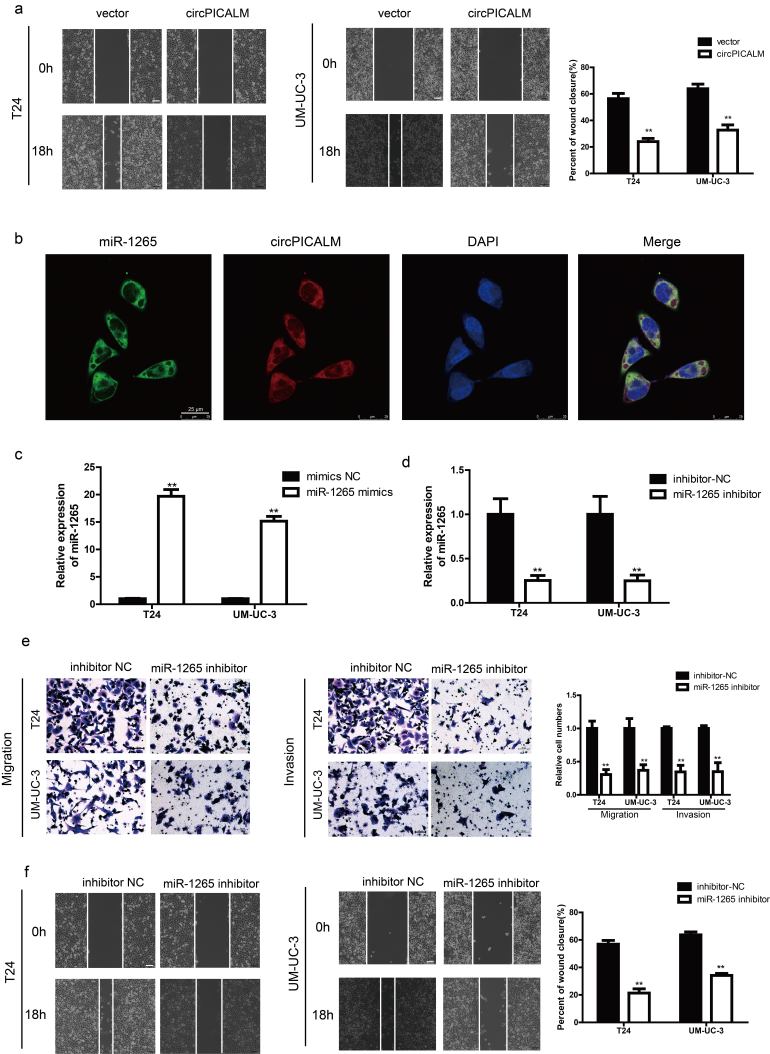
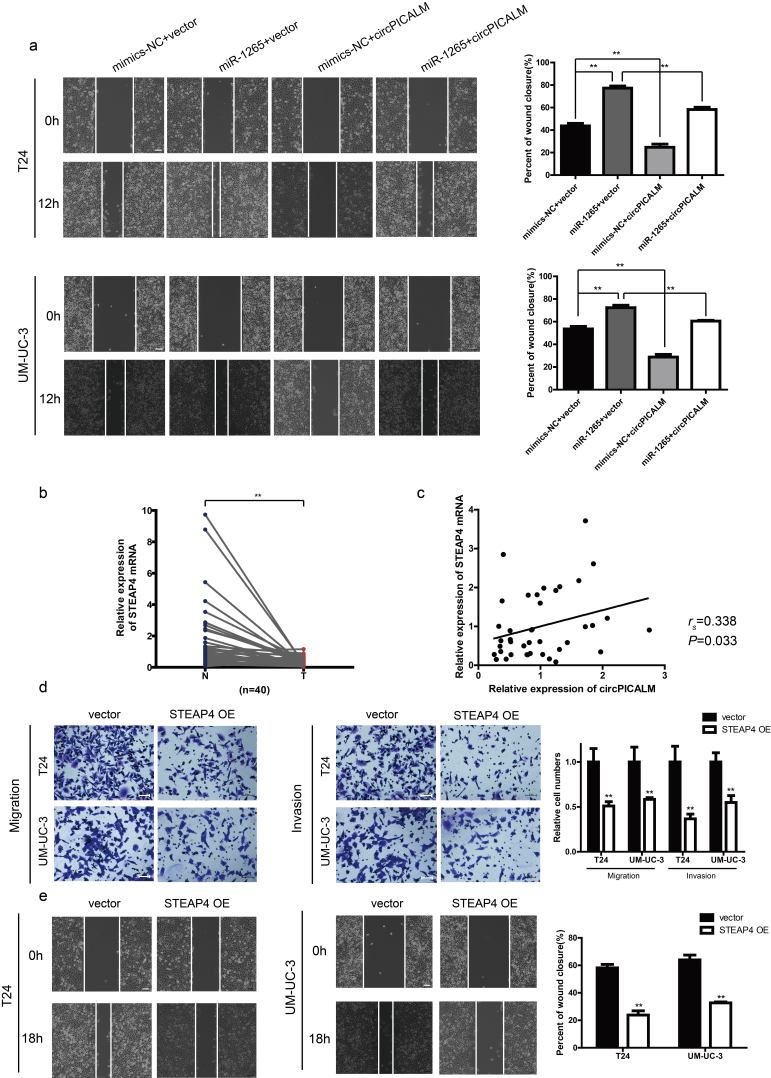
There are few studies on the functions of miR-1265 in cancer, especially in BC. We investigated T24 and UM-UC-3 cell invasion after transfection with NC, miR-1265 mimics, or miR-1265 inhibitor via migration, Matrigel invasion, and wound healing assays (Supplementary Fig. 2c-d). As expected, miR-1265 promoted BC cell migration, invasion and wound healing (Fig. 5a-b), while knockdown of miR-1265 had the opposite effect (Supplementary Fig. 2e-f). Based on the competing endogenous RNA (ceRNA) mechanism, circRNAs can act as sponges to influence miRNA function [14]. Overexpression of circPICALM abrogated, at least partly, the pro-metastasis effect of miR-1265 on BC cells (Fig. 5c; Supplementary Fig. 3a). Besides, overexpression of circPICALM could also impair the effect of miR-1265 on downstream molecules (Fig. 5d). Collectively, these data indicate that miR-1265 promotes BC metastasis and is potentially regulated by circPICALM in a classic ceRNA mechanism.
Fig. 5.
miR-1265 promotes invasion, which is inhibited by circPICALM overexpression.
a. and b. T24 and UM-UC-3 cells transfected with miR-1265 mimics had more invasive phonotype, detected by migration, invasion, and wound healing assays. c. and d. Migration and invasion assays showed that overexpression of circPICALM could eliminate the pro-metastasis function of miR-1265 in BC cells, and the expression of STEAP4, FAK, and pFAK-Y397. Scale bar, 100 μm. (Data are presented as the mean ± SD, n = 3. Unpaired, two-tailed student's t-test, *P<0.05, **P<0.01).
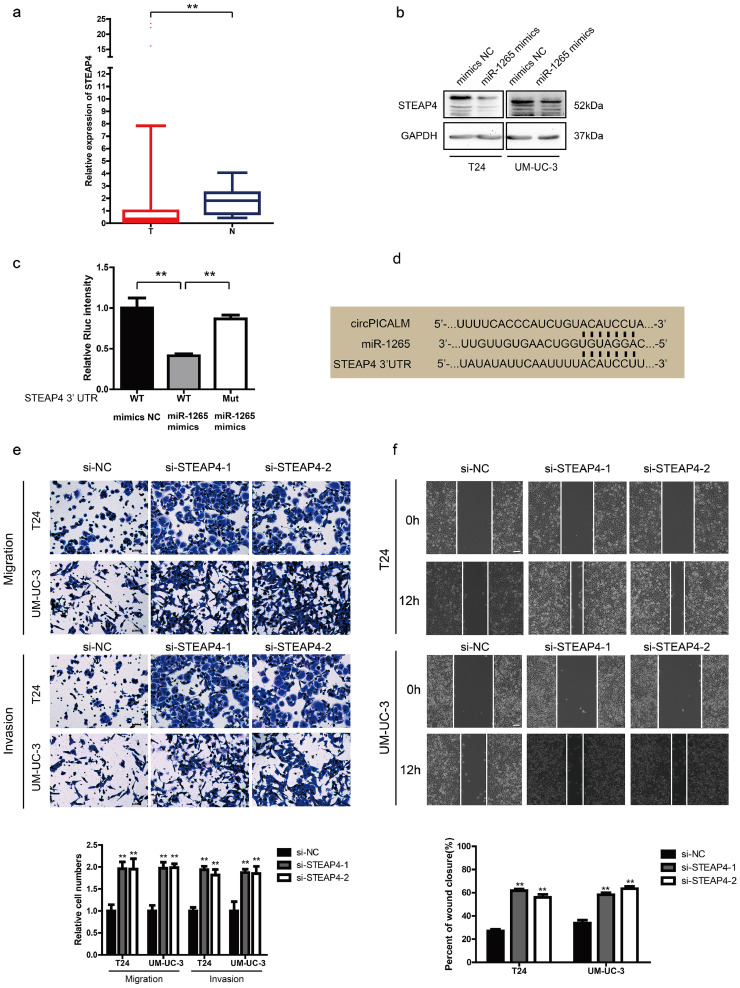
3.6. STEAP4 is a direct target of miR-1265 and dysregulates metastasis
It is acknowledged that miRNAs can function by degrading mRNA or obstructing transcription [31]. Therefore, we investigated how miR-1265 affects downstream factors. Upon synthesis of the predictions in miRWalk, TargetScan and miRDB, we found a series of candidates, among which STEAP4 drew our attention. We checked STEAP4 expression levels in TCGA data and found that STEAP4 mRNA expression was markedly lower in BC tissues than in normal uroepithelium (Fig. 6a). The trend in our samples was consistent with that in TCGA data (Supplementary Fig. 3b), and the expression of STEAP4 mRNA had a positive correlation with circPICALM expression (Supplementary Fig. 3c). When T24 and UM-UC-3 cells were transfected with miR-1265 mimics, STEAP4 levels decreased (Fig. 6b), and the function of miR-1265 mimics could be abolished by circPICALM overexpression (Fig. 5d). A dual-luciferase reporter assay was conducted; relative Rluc intensity decreased notably when HEK-293 T cells were co-infected with the psiCHECK-2 plasmid containing the STEAP4 3’ UTR and miR-1265 mimics (Fig. 6c). Meanwhile, mutation of the miR-1265 binding site abolished the repression of Rluc activity (Fig. 6c-d). STEAP4 is a member of the human 6-transmembrane epithelial antigen of prostate family; it is considered a diagnostic marker in prostate and hepatic cancer, and it has been validated to play an important role in androgen-responsive/androgen receptor-positive prostate cancer, whereas little is known about its function in BC cells or the invasive phenotype of cancer [[32], [33], [34], [35]]. T24 and UM-UC-3 cells were exposed to STEAP4 siRNAs, which increased migration, invasion, and wound healing capacities (Fig. 6e-f). And overexpression of STEAP4 exerted an opposite function (Supplementary Fig. 4d-e). These findings provide evidence that STEAP4 is a target of miR-1265.
Fig. 6.
STEAP4 is a direct target of miR-1265 and dysregulates metastasis.
a. STEAP4 mRNA expression of 408 BC tissues (T) and 19 normal tissues (N) in TCGA. b. STEAP4 abundance in T24 and UM-UC-3 cells transfected with miR-1265 mimics, detected by western blotting. GAPDH was used as an internal control. c. The dual-luciferase reporter assay to prove interaction of miR-1265 and the STEAP4 3’ UTR. d. The predicted binding sites of miR-1265 with circPICALM and the 3’ UTR of STEAP4. e. and f. Effects of knocking down STEAP4 on migration, invasion, and wound healing capacities of BC cells. Scale bar, 100 μm. (a. Error bars indicate 1–99 percentile. c. and e-g. Data are presented as the mean ± SD, n = 3. a. Unpaired two-tailed, Wilcoxon rank-sum test. c. and e-g. Unpaired, two-tailed student's t-test, **P<0.01).
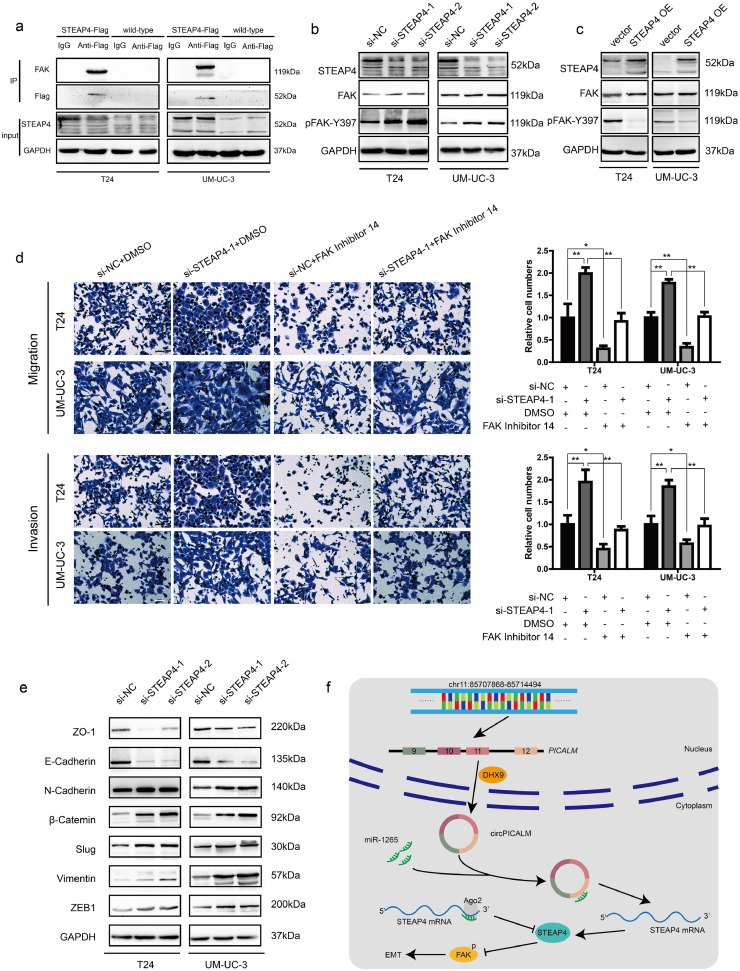
3.7. STEAP4 decreases the pFAK/FAK ratio and influences EMT
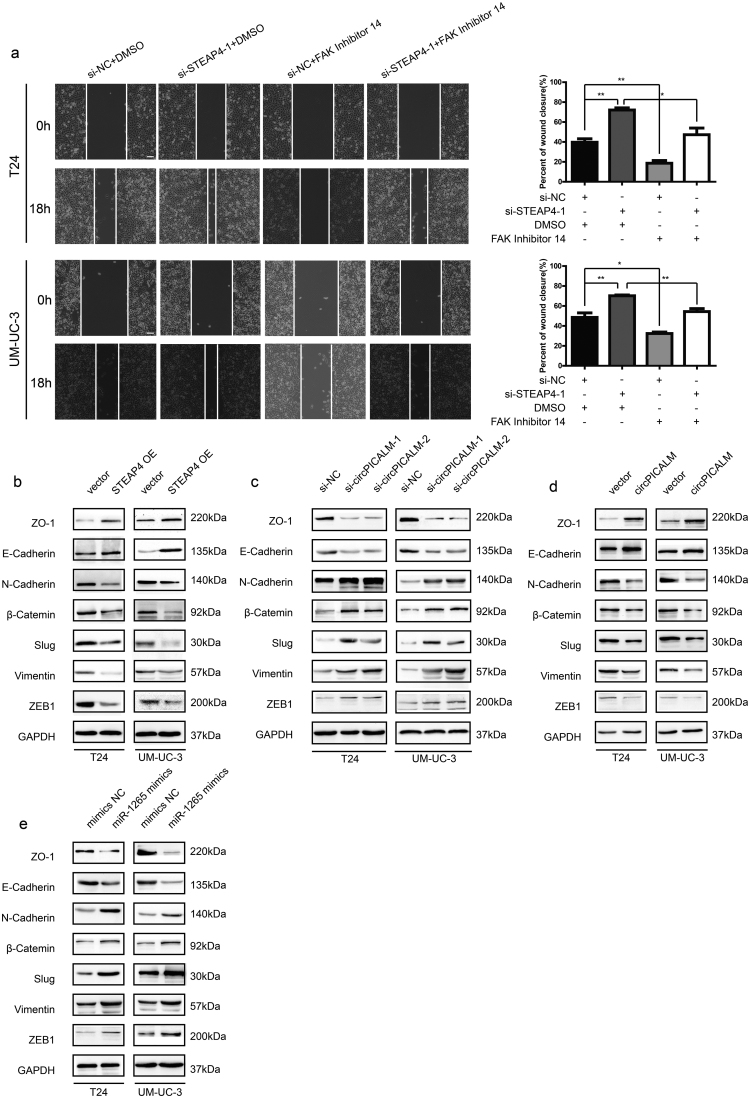
It has been reported that STEAP4 can interact with and negatively regulate the activation of FAK, a key regulator of cell migration, through effects on Y397 phosphorylation and, therefore, inhibit anchorage-independent cell growth [35]. The physical interaction between STEAP4 and FAK was verified by co-IP in T24 and UM-UC-3 cells (Fig. 7a). Moreover, we performed western blotting to determine the expression levels of total FAK and phosphorylated FAK (pFAK-Y397) in BC cells. In T24 and UM-UC-3 cells, silencing STEAP4 led to increased pFAK-Y397 levels but no change in total FAK levels (Fig. 7b), whereas ectopic expression of STEAP4 decreased the pFAK/FAK ratio (Fig. 7c). Interestingly, miR-1265 mimics could increase the pFAK/FAK ratio, and overexpression of circPICALM abrogated that effect (Fig. 5d). FAK Inhibitor 14 is a selective FAK inhibitor that can block FAK phosphorylation at Y397 [36]. Migration, invasion, and wound healing assays revealed that FAK Inhibitor 14 robustly counteracted the pro-metastasis effects of STEAP4 knockdown (Fig. 7d; Supplementary Fig. 4a). EMT is a crucial step in cancer development and relapse, as it helps cells acquire the different characteristics of stemness, drug resistance, invasion, and metastasis [9]. After STEAP4 knockdown, the levels of β-Catenin, Vimentin, ZEB1, Slug, and N-Cadherin were upregulated, while E-Cadherin and ZO-1 were downregulated, evidence of an increasing mesenchymal phenotype (Fig. 7e). Moreover, western blotting showed that epithelial characteristics were more evident upon STEAP4 overexpression (Supplementary Fig. 4b). When circPICALM was silenced, mesenchymal markers were increased, and overexpression of circPICALM reversed the BC cells towards epithelial phenotype (Supplementary Fig. 4c-d). Elevation of miR-1265 also drove the EMT process (Supplementary Fig. 4e). Collectively, we clarified that circPICALM influences BC metastasis via the miR-1265/STEAP4/FAK/EMT axis (Fig. 7f).
Fig. 7.
STEAP4 decreases the pFAK/FAK ratio and influences EMT.
a. Co-IP using anti-Flag antibody or IgG in wild-type BC cells or cells stably expressing STEAP4–3 × Flag. Proteins immunoprecipitated were detected by western blotting with anti-FAK and anti-Flag antibodies. b. and c. Western blotting showed FAK and its phosphorylation form in T24 and UM-UC-3 cells when silencing or overexpressing STEAP4. d. FAK Inhibitor 14 could eliminate the pro-metastasis effect of STEAP4 silencing, indicated by migration and Matrigel invasion assays. Scale bar, 100 μm. e. Western blotting illustrated EMT markers expression in BC cells transfected with STEAP4 siRNAs. f. Model we propose that circPICALM sponges miR-1265 to influence the metastasis of BC. STEAP4 is a direct miR-1265 target and exerts its function on EMT through binding and inhibiting the phosphorylation of FAK. (Data are presented as the mean ± SD, n = 3. Unpaired, two-tailed student's t-test, *P<0.05, **P<0.01).
4. Discussion
CircRNAs compose a large population of noncoding RNAs derived from various genomic locations and are expressed in tissue-, cell type- and developmental stage-specific manners [37]. Due to their covalent loop structure without 5′ caps or 3′ polyadenylated tails, circRNAs are difficult to be reverse transcribed with oligo-dT primers and more stable than linear mRNAs, which gives them specific advantages as biomarkers of various cancers [38,39]. Abundant circRNAs have been reported to exert diverse functions in many cancers, including BC [14,23]. In the present study, we identified circPICALM as a differentially expressed circRNA between cancer and normal uroepithelium. We further verified this finding in BC cell lines and highly or poorly invasive cell subpopulations. Low expression of circPICALM predicted advanced T stage, high grade, LN positivity, and poor outcome. Moreover, circPICALM expression and LN metastasis were independent prognostic factors in our cohort.
Alu elements are well-known retrotransposons, members of the short interspersed nuclear element (SINE) family of repetitive elements, and IARs around exons promote circRNA formation [16,40]. DHX9, an abundant nuclear RNA helicase, binds specifically to IARs, and loss of DHX9 can increase the formation of a subclass of circRNAs [30]. We found that multiply IARs were located within PICALM gene, some of which were around exons 9 to12, where circPICALM was formed. CircPICALM expression was further elevated by silencing DHX9, suggesting that DHX9 can affect the circularization of the PICALM gene in an Alu element-dependent manner.
Metastasis and invasion indicate BC malignancy and restrict treatment options [41]. There is an urgent need to identify invasion-related molecules and elucidate the associated mechanisms. We found that silencing circPICALM robustly promoted the migration, invasion, and wound healing capacities of BC cells, whereas circPICALM overexpression had the opposite effects in vivo and in vitro.
The majority of circRNAs derived from back-splicing exons are located in the cytoplasm [37,41]. However, only a few cytoplasmic circRNAs contain binding sites to trap particular miRNAs, known as the ceRNA mechanism [15,42]. Previous studies discovered that miRNAs are complementary to the 3’ UTR of protein-coding mRNAs and mediate negative post-transcriptional regulation through RNA duplex formation [31]. Ago2, a protein in the RNA-induced silencing complex (RISC), can facilitate the repression and degradation of target mRNAs by miRNAs [43]. Recent studies suggest that circRNAs trap miRNAs to prevent them from downregulating target genes [14]. CircPICALM is generated from exons 9–12 of PICALM, and we performed FISH and cytoplasm and nuclear mass separation assays to ascertain the predominantly cytoplasmic localization of circPICALM, which led us to test its capacity for sponging miRNAs. A pull-down assay indicated that circPICALM was substantially enriched by an anti-Ago2 antibody, suggesting its potential function in the AGO-RISC complex in BC cells. We cross-referenced the predicted candidates from CircInteractome, miRanda, and RNAhybrid, and performed a series of pull-down, miRNA capture, and dual-luciferase reporter assays to confirm that circPICALM directly interacts with miR-1265. Furthermore, circPICALM could, at least partially, reverse the pro-metastatic role of miR-1265, providing another line of evidence that circPICALM acts as a miR-1265 trap.
STEAP4 is a six transmembrane metalloreductase and predicted to be a target gene of miR-1265 by miRWalk, TargetScan and miRDB [44]. STEAP4 levels decreased when BC cells were treated with miR-1265 mimics, suggesting its potential as a target of miR-1265. More interestingly, STEAP4 expression is positively related to circPICALM expression. We validated the interaction between miR-1265 and STEAP4 using a dual-luciferase reporter assay. Besides, consistent with ceRNA hypothesis, the binding site between miR-1265 and circPICALM is identical to the one between miR-1265 and the STEAP4 3’ UTR. STEAP4 is hypermethylated and downregulated in hepatocellular carcinoma and is elevated in breast cancer cells that obtain oncogenic features after long-term exposure to ethanol [33,45]; however, little is known about its function in BC. TCGA and our data revealed relatively low expression of STEAP4 in BC. We found that knocking down STEAP4 led to a more invasive phenotype of BC cells, while overexpression STEAP4 exerted an opposite function.
FAK is a well-studied tyrosine kinase that is abnormally elevated in many cancers, such as lung, ovarian, breast, cervical, and bladder cancer [[46], [47], [48], [49], [50]]. FAK can influence fundamental oncogenic cellular behaviours, including adhesion, migration, proliferation and angiogenesis, and it is closely related to metastasis and poor survival [51]. Autophosphorylation of Y397 is vital for FAK activation, as it forms the binding site for Src homology 2 (SH2) domain-containing molecules, and several reports have proposed pFAK-Y397 as a potential therapeutic target for pancreatic cancer, malignant pleural mesothelioma, lung adenocarcinoma and metastatic osteosarcoma [[51], [52], [53], [54]]. We discovered that STEAP4 interacted with FAK and that pFAK-Y397 levels increased in response to STEAP4 silencing in BC cells. Overexpression of miR-1265 could both increase the pFAK/FAK ratio and decrease STEAP4 expression; however, this effect could be eliminate by upregulating circPICALM. Moreover, the selective FAK Inhibitor 14, which prevents FAK autophosphorylation at Y397, abolished the pro-invasive effect of STEAP4 knockdown. These results indicate that pFAK-Y397 is a pivotal regulator of STEAP4 that influences BC cell invasion.
Numerous studies have reported that FAK promotes EMT through FAK/PI3K/AKT and FAK/Src pathways, which profoundly influences the metastatic potential of BC [[12], [13], [14],55]. Consequently, we detected EMT markers by western blotting and found that inhibition of STEAP4 increased epithelial markers and decreased mesenchymal markers, while the trend was reversed when STEAP4 or circPICALM was overexpressed. Overexpression of miR-1265 promoted EMT. Our results show that circPICALM influences EMT to inhibit BC metastasis via the miR-1265/STEAP4/FAK axis.
In summary, we identified circPICALM as an invasion-related and differentially expressed circRNA in BC. Low circPICALM levels were observed in BC tissues and predicted to be related to poor survival and LN positivity in BC patients. Mechanistically, circPICALM acted as a miR-1265 sponge to regulate STEAP4 and further influenced pFAK-Y397 status and EMT. Our research provides insights into the prognostic and therapeutic potential of circPICALM.
Funding
This work was supported by National Natural Science Foundation of China, No. 81772728, National Natural Science Foundation of China, No.81772719, National Natural Science Foundation of China No.81572514.
Author contributions
Tianxin Lin and Jian Huang conceived and designed the study. Dong Yan, Wei Dong and Qingqing He performed the in vitro experiments. Meihua Yang conducted in vivo experiments. Lifang Huang provided bioinformatics and statistical analysis. Jianqiu Kong collected and organized specimens with clinical parameters.
Declaration of Competing Interest
Authors declare to conflicts of interest.
Acknowledgments
The authors thanked the support of National Natural Science Foundation of China, No. 81772728, National Natural Science Foundation of China, No.81772719, National Natural Science Foundation of China No.81572514. The funders in this study didn't have any role in study design, data collection, data analysis, interpretation or writing of the report.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.08.074.
Contributor Information
Tianxin Lin, Email: lintx@mail.sysu.edu.cn.
Jian Huang, Email: huangj8@mail.sysu.edu.cn.
Appendix A. Supplementary data
Primers used in this study.
Probes used in this study.
The oligonucleotides transfected in this study are listed as follows.
Supplementary Fig. 1.
a. PCR products of divergent or convergent primers and UM-UC-3 cDNA or gDNA were validated by gel electrophoresis. b. Subcellular distribution of circPICALM was detected by nuclear mass separation assay in UM-UC-3 cells. U6 and 18S acted as nuclear and cytoplasm controls, respectively. c. Subcellular location of circPICALM was illustrated by FISH in T24 cells. U6 and 18S acted as nuclear and cytoplasm controls, respectively. Scale bar, 10 μm. d. BC cells treated with circPICALM siRNAs were measured by the wound healing assay. Scale bar, 100 μm. e. Abundance of circPICALM and PICALM mRNA in BC cells infected with NC or circPICALM overexpression lentivirus. f. Invasive abilities of BC cells overexpressing circPICALM, measured by migration and invasion assays. Scale bar, 100 μm. (Data are presented as the mean ± SD, n = 3. Unpaired, two-tailed student's t-test, **P<0.01).
Supplementary Fig. 2.
a. Invasive abilities of BC cells overexpressing circPICALM, measured by wound healing assays. Scale bar, 100 μm. b. Colocalization of miR-1265 and circPICALM in T24 cells, demonstrated by FISH. Probe for miR-1265 was labelled by FAM and probe for circPICALM was labelled by cy3. Scale bar, 25 μm. c. and d. Relative expression of miR-1265 in BC cells transfected with mimics or inhibitor. e. and f. Migratory and Invasive abilities of BC cells treated with miR-1265 inhibitor, measured by migration, invasion, and wound healing assays. Scale bar, 100 μm. (Data are presented as the mean ± SD, n = 3. Unpaired, two-tailed student's t-test, **P<0.01).
Supplementary Fig. 3.
a. The wound healing assay showed that overexpression of circPICALM could eliminate the pro-metastasis function of miR-1265 in BC cells. Scale bar, 100 μm. b. Relative expression of STEAP4 mRNA in 40 paired primary BC (T) and adjacent normal (N) tissues, measured by RT-PCR. c. The correlation between circPICALM and STEAP4 expression in 40 BCE samples. d. and e. STEAP4 overexpression (OE) inhibited the metastasis of BC, detected by migration, invasion, and wound healing assays. Scale bar, 100 μm. (Data are presented as the mean ± SD, n = 3. a, d, and e. unpaired, two-tailed student's t-test; b, paired, two-tailed, Wilcoxon rank-sum test; c. Spearman's rank correlation. **P<0.01).
Supplementary Fig. 4.
a. FAK Inhibitor 14 could eliminate the pro-metastasis effect of STEAP4 silencing, indicated by the wound healing assay. Scale bar, 100 μm. b. Effect of STEAP4 overexpression on EMT in BC cells. c. and d. Effect of circPICALM silencing or overexpressing on EMT in BC cells. e. Effect of miR-1265 mimics on EMT in BC cells. (Data are presented as the mean ± SD, n = 3. Unpaired, two-tailed student's t-test, *P<0.05, **P<0.01).
References
- 1.Antoni S., Ferlay J., Soerjomataram I., Znaor A., Jemal A., Bray F. Bladder cancer incidence and mortality: a global overview and recent trends. Eur Urol. 2017;71(1):96–108. doi: 10.1016/j.eururo.2016.06.010. [DOI] [PubMed] [Google Scholar]
- 2.Foreman K.J., Marquez N., Dolgert A. Forecasting life expectancy, years of life lost, and all-cause and cause-specific mortality for 250 causes of death: reference and alternative scenarios for 2016–40 for 195 countries and territories. The Lancet. 2018;392(10159):2052–2090. doi: 10.1016/S0140-6736(18)31694-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abufaraj M., Dalbagni G., Daneshmand S. The role of surgery in metastatic bladder cancer: a systematic review. Eur Urol. 2018;73(4):543–557. doi: 10.1016/j.eururo.2017.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alfred Witjes J., Lebret T., Comperat E.M. Updated 2016 EAU guidelines on muscle-invasive and metastatic bladder cancer. Eur Urol. 2017;71(3):462–475. doi: 10.1016/j.eururo.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 5.Wu X.R. Urothelial tumorigenesis: a tale of divergent pathways. Nat Rev. Cancer. 2005;5(9):713–725. doi: 10.1038/nrc1697. [DOI] [PubMed] [Google Scholar]
- 6.Bajorin D.F., Dodd P.M., Mazumdar M. Long-term survival in metastatic transitional-cell carcinoma and prognostic factors predicting outcome of therapy. J Clin Oncol. 1999;17:3173–3181. doi: 10.1200/JCO.1999.17.10.3173. [DOI] [PubMed] [Google Scholar]
- 7.Xie R., Chen X., Chen Z. Polypyrimidine tract binding protein 1 promotes lymphatic metastasis and proliferation of bladder cancer via alternative splicing of MEIS2 and PKM. Cancer Lett. 2019;449:31–44. doi: 10.1016/j.canlet.2019.01.041. [DOI] [PubMed] [Google Scholar]
- 8.Lambert A.W., Pattabiraman D.R., Weinberg R.A. Emerging biological principles of metastasis. Cell. 2017;168(4):670–691. doi: 10.1016/j.cell.2016.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pastushenko I., Brisebarre A., Sifrim A. Identification of the tumour transition states occurring during EMT. Nature. 2018;556(7702):463–468. doi: 10.1038/s41586-018-0040-3. [DOI] [PubMed] [Google Scholar]
- 10.Wang L., Saci A., Szabo P.M. EMT- and stroma-related gene expression and resistance to PD-1 blockade in urothelial cancer. Nat Commun. 2018;9(1):3503. doi: 10.1038/s41467-018-05992-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsia D.A., Mitra S.K., Hauck C.R. Differential regulation of cell motility and invasion by FAK. J Cell Biol. 2003;160(5):753–767. doi: 10.1083/jcb.200212114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li C., Yang Z., Du Y. BCMab1, a monoclonal antibody against aberrantly glycosylated integrin alpha3beta1, has potent antitumor activity of bladder cancer in vivo. Clin Cancer Res. 2014;20(15):4001–4013. doi: 10.1158/1078-0432.CCR-13-3397. [DOI] [PubMed] [Google Scholar]
- 13.Egawa H., Jingushi K., Hirono T. The miR-130 family promotes cell migration and invasion in bladder cancer through FAK and Akt phosphorylation by regulating PTEN. Sci Rep. 2016;6 doi: 10.1038/srep20574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu H., Bi J., Dong W. Invasion-related circular RNA circFNDC3B inhibits bladder cancer progression through the miR-1178-3p/G3BP2/SRC/FAK axis. Mol Cancer. 2018;17(1):161. doi: 10.1186/s12943-018-0908-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeck W.R., Sharpless N.E. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X.O., Wang H.B., Zhang Y., Lu X., Chen L.L., Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159(1):134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Cocquerelle C., Mascrez B., Hétuin D., Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7(1):155–160. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- 18.Pasman Z., Been M.D., Garcia-Blanco M.A. Exon circularization in mammalian nuclear extracts. RNA. 1996;2(6):603–610. [PMC free article] [PubMed] [Google Scholar]
- 19.Glazar P., Papavasileiou P., Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20(11):1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang S., Li X., Zheng H. Loss of super-enhancer-regulated CircRNA Nfix induces cardiac regeneration after myocardial infarction in adult mice. Circulation. 2019 doi: 10.1161/CIRCULATIONAHA.118.038361. (undefined(undefined): undefined) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wesselhoeft R.A., Kowalski P.S., Parker-Hale F.C., Huang Y., Bisaria N., Anderson D.G. RNA circularization diminishes immunogenicity and can extend translation duration in vivo. Mol Cell. 2019;74(3):508–520. doi: 10.1016/j.molcel.2019.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Legnini I., Di Timoteo G., Rossi F. Circ-ZNF609 Is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66(1):22–37.e9. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang F., Fang E., Mei H. Cis-Acting circ-CTNNB1 promotes beta-catenin signaling and cancer progression via DDX3-mediated transactivation of YY1. Cancer Res. 2019;79(3):557–571. doi: 10.1158/0008-5472.CAN-18-1559. [DOI] [PubMed] [Google Scholar]
- 24.Du W.W., Yang W., Chen Y. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J. 2017;38(18):1402–1412. doi: 10.1093/eurheartj/ehw001. [DOI] [PubMed] [Google Scholar]
- 25.Palmieri F., Brunocilla E., Bertaccini A. Prognostic value of lymphovascular invasion in bladder cancer in patients treated with radical cystectomy. Anticancer Res. 2010;30(7):2973–2976. [PubMed] [Google Scholar]
- 26.Miyamoto H., Miller J.S., Fajardo D.A. Non-invasive papillary urothelial neoplasms: the 2004 WHO/ISUP classification system. Pathol Int. 2010;60(1):1–8. doi: 10.1111/j.1440-1827.2009.02477.x. [DOI] [PubMed] [Google Scholar]
- 27.Dong W., Bi J., Liu H. Circular RNA ACVR2A suppresses bladder cancer cells proliferation and metastasis through miR-626/EYA4 axis. Mol Cancer. 2019;18(1):95. doi: 10.1186/s12943-019-1025-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Z., Chen X., Xie R. DANCR promotes metastasis and proliferation in bladder cancer cells by enhancing IL-11-STAT3 signaling and CCND1 expression. Mol Ther. 2019;27(2):326–341. doi: 10.1016/j.ymthe.2018.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y., Zheng F., Xiao X. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18(9):1646–1659. doi: 10.15252/embr.201643581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aktaş T., Avşar Ilık İ., Maticzka D. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature. 2017;544(7648):115–119. doi: 10.1038/nature21715. [DOI] [PubMed] [Google Scholar]
- 31.Lai E.C. Micro RNAs are complementary to 3’ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30(4):363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 32.Wu H.C., Yang H.I., Wang Q., Chen C.J., Santella R.M. Plasma DNA methylation marker and hepatocellular carcinoma risk prediction model for the general population. Carcinogenesis. 2017;38(10):1021–1028. doi: 10.1093/carcin/bgx078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamada N., Yasui K., Dohi O. Genome-wide DNA methylation analysis in hepatocellular carcinoma. Oncol Rep. 2016;35(4):2228–2236. doi: 10.3892/or.2016.4619. [DOI] [PubMed] [Google Scholar]
- 34.Korkmaz C.G., Korkmaz K.S., Kurys P. Molecular cloning and characterization of STAMP2, an androgen-regulated six transmembrane protein that is overexpressed in prostate cancer. Oncogene. 2005;24(31):4934–4945. doi: 10.1038/sj.onc.1208677. [DOI] [PubMed] [Google Scholar]
- 35.Tamura T., Chiba J. STEAP4 regulates focal adhesion kinase activation and CpG motifs within STEAP4 promoter region are frequently methylated in DU145, human androgen-independent prostate cancer cells. Int J Mol Med. 2009;24(05) doi: 10.3892/ijmm_00000270. [DOI] [PubMed] [Google Scholar]
- 36.Huang W.K., Akçakaya P., Gangaev A. miR-125a-5p regulation increases phosphorylation of FAK that contributes to imatinib resistance in gastrointestinal stromal tumors. Exp Cell Res. 2018;371(1):287–296. doi: 10.1016/j.yexcr.2018.08.028. [DOI] [PubMed] [Google Scholar]
- 37.Memczak S., Jens M., Elefsinioti A. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 38.Li Y., Zhao J., Yu S. Extracellular vesicles long RNA sequencing reveals abundant mRNA, circRNA, and lncRNA in human blood as potential biomarkers for cancer diagnosis. Clin Chem. 2019 doi: 10.1373/clinchem.2018.301291. [DOI] [PubMed] [Google Scholar]
- 39.Sole C., Arnaiz E., Manterola L., Otaegui D., Lawrie C.H. The circulating transcriptome as a source of cancer liquid biopsy biomarkers. Semin Cancer Biol. 2019 doi: 10.1016/j.semcancer.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 40.Jeck W.R., Sorrentino J.A., Wang K. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bella A.J., Stitt L.W., Chin J.L., Izawa J.I. The prognostic significance of metastatic perivesical lymph nodes identified in radical cystectomy specimens for transitional cell carcinoma of the bladder. J Urol. 2003;170(6):2253–2257. doi: 10.1097/01.ju.0000095804.33714.ea. Pt 1. [DOI] [PubMed] [Google Scholar]
- 42.Guo J.U., Agarwal V., Guo H., Bartel D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15(7):409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sheu-Gruttadauria J., Xiao Y., Gebert L.F., MacRae I.J. Beyond the seed: structural basis for supplementary microRNA targeting by human Argonaute2. EMBO J. 2019 doi: 10.15252/embj.2018101153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oosterheert W., van Bezouwen L.S., Rodenburg R.N.P. Cryo-EM structures of human STEAP4 reveal mechanism of iron(III) reduction. Nat Commun. 2018;9(1):4337. doi: 10.1038/s41467-018-06817-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gelfand R., Vernet D., Bruhn K.W. Long-term exposure of MCF-7 breast cancer cells to ethanol stimulates oncogenic features. Int J Oncol. 2017;50(1):49–65. doi: 10.3892/ijo.2016.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu H., Wang L., Gao W. IGFBP2/FAK pathway is causally associated with dasatinib resistance in non-small cell lung cancer cells. Mol Cancer Ther. 2013;12(12):2864–2873. doi: 10.1158/1535-7163.MCT-13-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ciriello G., Gatza M.L., Beck A.H. Comprehensive molecular portraits of invasive lobular breast cancer. Cell. 2015;163(2):506–519. doi: 10.1016/j.cell.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oktay M.H., Oktay K., Hamele-Bena D., Buyuk A., Koss L.G. Focal adhesion kinase as a marker of malignant phenotype in breast and cervical carcinomas. Hum Pathol. 2003;34(3):240–245. doi: 10.1053/hupa.2003.40. [DOI] [PubMed] [Google Scholar]
- 50.Sulzmaier F.J., Jean C., Schlaepfer D.D. FAK in cancer: mechanistic findings and clinical applications. Nat Rev. Cancer. 2014;14(9):598–610. doi: 10.1038/nrc3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luo M., Guan J.L. Focal adhesion kinase: a prominent determinant in breast cancer initiation, progression and metastasis. Cancer Lett. 2010;289(2):127–139. doi: 10.1016/j.canlet.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kanteti R., Mirzapoiazova T., Riehm J.J. Focal adhesion kinase a potential therapeutic target for pancreatic cancer and malignant pleural mesothelioma. Cancer Biol Ther. 2018;19(4):316–327. doi: 10.1080/15384047.2017.1416937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou Q., Guo X., Choksi R. Activation of focal adhesion kinase and src mediates acquired sorafenib resistance in A549 human lung adenocarcinoma xenografts. J Pharmacol Exp Ther. 2017;363(3):428–443. doi: 10.1124/jpet.117.240507. [DOI] [PubMed] [Google Scholar]
- 54.Thanapprapasr K., Nartthanarung A., Thanapprapasr D., Jinawath A. pFAK-Y397 overexpression as both a prognostic and a predictive biomarker for patients with metastatic osteosarcoma. PLoS One. 2017;12(8) doi: 10.1371/journal.pone.0182989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tao Y.M., Huang J.L., Zeng S. BTB/POZ domain-containing protein 7: epithelial-mesenchymal transition promoter and prognostic biomarker of hepatocellular carcinoma. Hepatology. 2013;57(6):2326–2337. doi: 10.1002/hep.26268. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used in this study.
Probes used in this study.
The oligonucleotides transfected in this study are listed as follows.