Abstract
Background
Resistance to oxaliplatin-based chemotherapy is a major cause of recurrence in colorectal cancer (CRC) patients. There is increasing evidence indicating that circHIPK3 is involved in the development and progression of tumours. However, little is known about the potential role of circHIPK3 in CRC chemotherapy and its molecular mechanisms in chemoresistance also remain unclear.
Methods
Quantitative real-time PCR was performed to detect circHIPK3 expression in tissues of 2 cohorts of CRC patients who received oxaliplatin-based chemotherapy. The chemoresistant effects of circHIPK3 were assessed by cell viability, apoptosis, and autophagy assays. The relationship between circHIPK3, miR-637, and STAT3 mRNA was confirmed by biotinylated RNA pull-down, luciferase reporter, and western blot assays.
Findings
In the pilot study, increased circHIPK3 expression was observed in chemoresistant CRC patients. Functional assays showed that circHIPK3 promoted oxaliplatin resistance, which was dependent on inhibition of autophagy. Mechanistically, circHIPK3 sponged miR-637 to promote STAT3 expression, thereby activating the downstream Bcl-2/beclin1 signalling pathway. A clinical cohort study showed that circHIPK3 was upregulated in tissues from recurrent CRC patients and correlated with tumour size, regional lymph node metastasis, distant metastasis, and survival.
Interpretation
circHIPK3 functions as a chemoresistant gene in CRC cells by targeting the miR-637/STAT3/Bcl-2/beclin1 axis and might be a prognostic predictor for CRC patients who receive oxaliplatin-based chemotherapy.
Funding
National Natural Science Foundation of China (81301506), Shandong Medical and Health Technology Development Project(2018WSB20002), Shandong Key Research and Development Program (2016GSF201122), Natural Science Foundation of Shandong Province (ZR2017MH044), and Jinan Science and Technology Development Plan(201805084, 201805003).
Keywords: Colorectal cancer, Chemoresistance, circHIPK3, Autophagy, miR-637, STAT3
Abbreviations: CRC, colorectal cancer; CircRNAs, circular rnas; ceRNA, competing endogenous RNA; RECIST, response evaluation criteria in solid tumours; DFS, disease free survival; OS, overall survival; 5FU, 5-fluorouracil; OXA, oxaliplatin; RT-qPCR, reverse transcription quantitative real-time PCR; ROC, Receiver operating characteristic; AUC, area under the ROC curve
Research in context.
Evidence before this study
Most colorectal cancer (CRC) patients respond poorly to oxaliplatin (OXA)-based chemotherapy regimen, but the molecular mechanisms underlying chemoresistance remain unclear. Recent studies have demonstrated that autophagy regulates the effects of anticancer drugs, indicating a new mechanism of chemoresistance. Recently, it has been reported that circular RNA, circHIPK3, plays a critical role in the development and progression of various cancers. This study aims to clarify the underlying mechanisms regarding the association of circHIPK3 and autophagy, and to assess the potential of circHIPK3 as a biomarker for the prediction of chemotherapeutic efficacy of CRC.
Added value of this study
In this study, we showed for the first time that increased circHIPK3 expression accompanied OXA chemoresistance in human CRC patients. The functional investigation demonstrated that circHIPK3 promoted OXA resistance, dependent on inhibition of autophagy-related cell death. Mechanistically, circHIPK3 functioned as a ceRNA to activate the STAT3/Bcl-2/beclin1 signalling pathway by sponging miR-637 during cellular autophagy regulation. A clinical cohort study showed that increased circHIPK3 expression could predict recurrence and poor survival in CRC patients who received OXA-based chemotherapy.
Implications of all the available evidence
This study provides evidence that circHIPK3 contributes to chemoresistance through inhibiting autophagy, and it can serve as a promising prognostic predictor and therapeutic target for CRC patients treated with OXA-based chemotherapy.
CRediT authorship contribution statement
Zhang Yanli: Conceptualization, Investigation, Project administration, Resources, Supervision, Writing - original draft, Writing - review & editing. Li Chen: Investigation, Writing - original draft, Writing - review & editing. Liu Xinfeng: Investigation, Resources, Writing - review & editing. Wang Yanlei: Investigation, Writing - review & editing. Zhao Rui: Investigation, Writing - review & editing. Yang Yongmei: Formal analysis, Writing - review & editing. Zheng Xin: Project administration, Supervision, Writing - original draft, Writing - review & editing. Zhang Yi: Conceptualization, Resources, Writing - review & editing. Zhang Xin: Conceptualization, Formal analysis, Project administration, Resources, Writing - review & editing.
Alt-text: Unlabelled Box
1. Introduction
Colorectal cancer (CRC) is the most common gastrointestinal malignant tumour, and is the second cause of cancer-related mortality worldwide [1]. Surgical resection combined with standard adjuvant chemotherapy strategies, such as FOLFOX (folinic acid, fluorouracil, and oxaliplatin), remain the most popular treatment of choice for CRC patients and have significantly improved disease-free survival (DFS) and overall survival (OS) [2]. However, only 50% of patients respond to chemotherapy regimen, and ultimately develop recurrent disease [2,3]. Moreover, a subgroup of patients might only experience drug toxicity without achieving chemotherapeutic benefit, and the prognosis is poor, with a median survival of 19•5 months [3,4]. Therefore, it is of utmost importance to further understand the molecular mechanisms underlying chemoresistance and identify molecules to predict chemotherapy efficacy, which might improve clinical outcomes in CRC patients.
Autophagy is an evolutionarily conserved intracellular process that can degrade dysfunctional cellular organelles and provide energy and biological components, thus maintaining cellular homeostasis [5]. Several autophagy-related genes (ATGs), which were first identified in yeast, are known to be involved in the progression of CRC. LC3 (microtubule-associated protein 1 light chain 3) is the mammalian homolog of ATG8, encoding 3 isoforms, LC3A, LC3B, and LC3C. During autophagy, LC3B is cleaved into the soluble protein LC3B-I, is conjugated to phosphatidylethanolamine to form LC3B-II, which accumulates on the surface of neonatal autophagosomes, and is one of the reliable markers of autophagy flux [6]. Wu et al. [7] showed that LC3B-II expression was significantly increased in CRC patients, especially in those with metastasis, suggesting that high levels of autophagy might improve the survival and invasiveness of tumour cells. p62 is an adaptor protein that recognises ubiquitinated proteins, interacts with LC3, and is subsequently degraded upon fusion with lysosomes [8]. Therefore, p62 can also be used as a marker for autophagy. Kim et al. [9] found that the levels of p62 decreased and the levels of LC3B-II increased in tumour cells along with the accumulation of a large number of isolated autophagosomes, in relapsed CRC patients. Cytoplasmic p62 expression predicted a favourable tumour-specific survival of CRC patients, particularly those with the KRAS-mutated subtype [10]. BECN1, the mammalian homologue of ATG6, encodes the beclin1 protein that regulates autophagosome formation [11]. Beclin1 is a component of the class III PI3K complex and is involved in autophagosome formation, thereby promoting autophagy, while it can also bind to Bcl-2 to inhibit autophagy [12]. In colorectal adenocarcinoma, the expression level of beclin1 is increased by more than 50%, while its decreased expression is associated with aggressive clinical behaviour [13].
Recently, mounting evidence demonstrates that autophagy can regulate the effects of anticancer drugs, which might be a new mechanism for chemoresistance [14]. O'Donovan et al. [15] showed that lithium upregulated autophagy and improved the efficacy of chemotherapeutic drugs in apoptosis deficient cancer cells. The autophagy inducer rapamycin, enhanced the tumour-suppressing ability of chemotherapeutic agents through the autophagy/p62/Nrf2 pathway [16]. In addition, miRNA-125b expression causes oxaliplatin (OXA) resistance by down-regulating EVA1A-mediated autophagy [17]. However, in some cellular conditions, autophagy can remove or mitigate harmful cytotoxic stimuli and delay CRC cell death, leading to therapeutic resistance [18]. Pan et al. [19] found that TRIM65 knockdown attenuated autophagy and improved cisplatin-induced apoptosis in non-small-cell lung cancer cell by regulating miR-138–5p. Till date, the mechanisms of autophagy involved in regulating CRC chemoresistance have not been well-documented.
Circular RNAs (circRNAs) are a novel class of non-coding regulatory RNAs, with covalently closed loop structures through the joining of 3′ and 5′ terminals. With the development of high-throughput sequencing technology and bioinformatics, several new circRNAs have been identified that participate in the pathology of disease, including cancer [20]. Among them, circHIPK3 is one of the most well-known circRNAs that functions as a cell growth modulator not only in endothelial cells but also in tumour cells [21,22]. Recently, Zeng et al. [23] found that circHIPK3 was upregulated in CRC tissues and correlated with metastasis and advanced clinical stage, while knockdown of its expression in vitro or in vivo markedly inhibited CRC growth and metastasis, indicating that it was involved in the development of CRC. Until now, little is known about the potential role of circHIPK3 in CRC chemotherapy, and its molecular mechanisms in chemoresistance also remain unclear.
In our pilot study, expression levels of circHIPK3 showed significant association with the efficacy of OXA-based chemotherapy. To verify this hypothesis, the biological role of circHIPK3-mediated autophagy in CRC chemoresistance was explored. In addition, the clinical role of circHIPK3 in CRC patients who received OXA-based chemotherapy was further investigated.
2. Materials and methods
2.1. Ethics statement
This study was approved by the Ethics Committee of Shandong Provincial Third Hospital and Qilu Hospital of Shandong University, and written informed consent was obtained from each patient.
2.2. Clinical samples
In the pilot study, 49 samples of primary tumour tissues were obtained from CRC patients who received 5-fluorouracil (5FU) and OXA-based first-line chemotherapy in Shandong Provincial Third Hospital between January 2017 and May 2018. These patients were classified according to Response Evaluation Criteria in Solid Tumours (RECIST) criteria, wherein 31 were responders with complete response (19 cases) and partial response (12 cases), and 18 were non-responders with stable disease (13 cases) and progressive disease (5 cases). In another independent cohort study of CRC patients, 179 of them, who received postoperative OXA-based adjuvant chemotherapy, were enrolled from Qilu Hospital of Shandong University between January 2010 and December 2012. All CRC patients underwent pathological diagnosis by two experienced pathologists and were followed up regularly for up to 5 years. The detailed clinical information is shown in Table S1. Relapsers were defined as patients who had either recurrence or metastasis during the follow-up, and patients with DFS more than 5 years were defined as non-relapsers. DFS or OS was defined as the interval between the date of radical surgery and the date of recurrence or death censoring at the time of last contact with the survivors. All tissues were stored at −150 °C until RNA extraction.
2.3. Cell lines and culture
The human CRC cell lines (HT29 and HCT116), and HEK293T were purchased from American Type Culture Collection (Manassas, VA, USA). The 5FU-resistant and OXA-resistant CRC cell lines were previously established in our lab by exposing cells sequentially to increasing drug concentrations [24,25]. The cells were maintained in RPMI 1640 or MEM (Thermo Fisher Scientific, Wilmington, DE, USA) supplemented with 10% foetal bovine serum (Sigma-Aldrich, St. Louis, MO, USA) and cultured in a humidified environment of 5% CO2 at 37 °C.
2.4. Vector construction and transfections
For circHIPK3 overexpression, the full-length sequence of circHIPK3 was synthesised and cloned into the pcDNA3.1-CMV-circRNA vector (Hanbio Biotechnology, Shanghai, China). siRNA specifically targeting the circHIPK3 junction site, siRNA control, miR-637 mimic, and mimic control were all synthesised by RiboBio (Guangzhou, China). Lipofectamine 2000 (Invitrogen, Eugene, OR, USA) was used for transfection according to the manufacturer's protocol.
2.5. Xenograft experiments
To evaluate the effects of circHIPK3 in vivo, 5-week-old male BALB/c athymic nude mice were randomly divided into four groups (n = 5 for each group), and subcutaneously injected with HCT116/HT29 (5 × 106/200 μl PBS) cells stably transfected with circHIPK3 overexpression/negative control vector. After 1 week, the mice were intraperitoneally injected with OXA (3 mg/kg) every week, and sacrificed on Day 28. Tumour volume was estimated every 3 days, and calculated by the formula: tumour volume = 0•5 × (length × width2). All animal experiments were performed in compliance with the guidelines of Animal Ethics Committee of Qilu Hospital of Shandong University.
2.6. RNA extraction and reverse transcription quantitative real-time PCR (RT-qPCR)
Total RNA was isolated from tissues and cells using the standard TRIzol method (Invitrogen, Carlsbad, CA, USA). For detection of circRNAs and mRNA, cDNA was synthesised using High Capacity cDNA Reverse Transcription Kit (Takara, Dalian, China), and qPCR was performed using SYBR Premix Ex Taq™ (Tli RNaseH Plus) (Takara) with GAPDH as an internal control. The primers were synthesised by BioSune Biotechnology (Shanghai, China) and are listed in Table S2. For miRNAs, SYBR PrimeScript miRNA RT-qPCR kit (Takara) was used as described previously, with U6 snRNA as an internal control. The miDETECT Track™ miRNA/U6 Forward Primers were provided by RiboBio Biotechnology (Guangzhou, China). Each experiment was performed in triplicates on CFX-96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA), and the relative expression levels were calculated using 2−ΔCT method.
2.7. Cell viability assay
Cell viability was analysed by Cell Counting Kit (CCK)−8 assay (Dojindo Laboratories, Kumamoto, Japan). After 24 h of transfection, cells (5000 cells per well) were seeded in 96-well plates in triplicates, and then treated under the indicated conditions. Next, 10 μl of CCK-8 solution was added at the end of the treatment and incubated for another 2 h at 37 °C. Finally, the absorbance was measured at 450 nm using Multiskan FC microplate reader (Thermo Fisher Scientific, Waltham, MA, USA).
2.8. Cell apoptosis assay
Cell apoptosis was assessed using Annexin V-FITC/PI staining kit (BD Bioscience, San Diego, CA, USA). After 24 h of transfection, 1 × 104 cells were incubated with 3 μM OXA for 48 h. Then, cells were collected and stained with Annexin V‑fluorescein isothiocyanate (FITC) for 15 min and propidium iodide (PI) for 5 min. The percentage of apoptotic cells was measured using FACSCanto II flow cytometer (BD, Bedford, MA, USA).
2.9. Cell autophagy assay
HCT116oxR cells, stably transfected with lentiviral vector mRFP-GFP-LC3B (Hanbio) were used to detect autophagic flux at 3 μM OXA. Cells were treated at the indicated conditions, and then fixed with 4% paraformaldehyde. The autophagosomes (yellow dots) and autolysosomes (red dots) were counted using Olympus FSX100 microscope (Olympus, Tokyo, Japan), and the images were captured using a Leica SP5 confocal microscope (Leica Micosystems, Mannheim, Germany).
2.10. Biotinylated RNA pull-down assay
The biotinylated RNA pull-down assay was performed as described previously [26]. To obtain probe-coated beads, circHIPK3 probe/oligo probe (RiboBio, Guangzhou, China) was incubated with C-1 magnetic beads (Life Technologies, Carlsbad, CA, USA) at 25 °C for 2 h. Then, the coated beads were incubated with sonicated HCT116 and HT29 cells at 4 °C overnight. For pull-down assay with biotinylated miR-637, 20 nM biotinylated miR-637 mimic or control RNA (RiboBio) was transfected into HCT116 and HT29 cells for 48 h, and then cells were lysed, sonicated, and incubated with streptavidin-coated magnetic beads (Life Technologies, Carlsbad, CA, USA). The bound RNA complexes were eluted from beads and purified using RNeasy Mini Kit (Qiagen, Valencia, CA, USA). The abundance of transcripts (circHIPK3 and miR-637) was evaluated by RT-qPCR analysis.
2.11. Luciferase reporter assay
The circHIPK3/STAT3 sequences with wild type (WT) or mutant (MUT) miR-637 binding sites were inserted between the hRluc and the hLuc gene of pmiR-REPORT™ vectors (RiboBio). HEK293T cells were seeded in 96-well plates at a density of 5000 cells/well, and then co-transfected with reporter vectors and miR-637 mimics / negative control using Lipofectamine 2000 (Invitrogen) for 48 h. Firefly and Renilla luciferase activities were detected using the Dual-Luciferase Assay System (Promega, Madison, WI, USA), and relative luciferase activities were calculated.
2.12. Western blot analysis
Western blotting was performed according to the standard protocols, using antibodies against human LC3B (#3868, Cell Signaling, Danvers, MA, USA, 1:1000), p62 (#16177, Cell Signaling, 1:1000), STAT3 (#ab68153, Abcam, Cambridge, MA, USA 1:1000), phospho-STAT3(#ab76315, Tyr705; Abcam, 1:1000), beclin1 (#ab207612, Abcam, 1:1000), Bcl-2 (#4223, Cell Signaling, 1:1000), and β-actin antibody (#4970, Cell Signaling, 1:5000). The bands were visualised using an enhanced chemiluminescence kit (Amersham Pharmacia Biotech, Piscataway, NJ, USA) on FluorChem E Chemiluminescent Western Blot Imaging System (Cell Biosciences, Santa Clara, CA, USA). Cell Signaling Technology, Inc. (Danvers, MA, USA).
3. Statistical analysis
The circHIPK3 expression in tissue samples was non-normal distribution and was compared using Mann-Whitney Utest or Kruskal-Wallis test. Student's t-test was performed to determine the significance of results in cell assays represented as mean ± standard deviation. Differences in group proportions were determined by Chi-square test. The correlation analysis was analysed by Spearman test. Survival curves were generated using Kaplan-Meier method and compared by log-rank test. Independent prognostic factors were identified by the Cox model. The above statistical analyses were performed using SPSS software, 22.0 for Windows (IBM Corporation, Armonk, NY, USA). The power of differential diagnosis was evaluated by Receiver operating characteristic (ROC) curves using MedCalc 9.3.9.0.
4. Results
4.1. circHIPK3 contributes to OXA chemoresistance in CRC
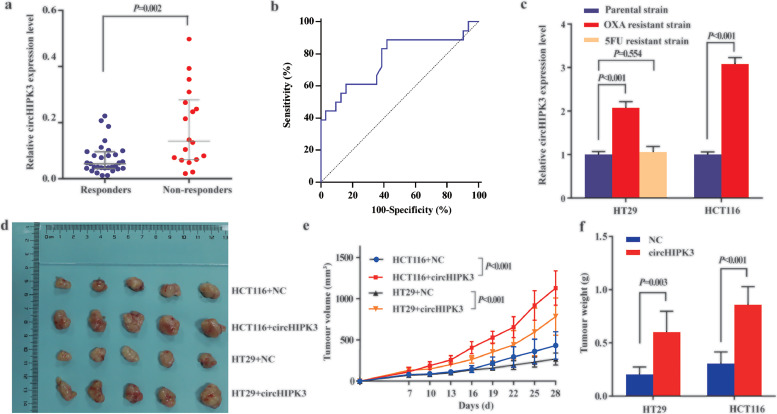
In the pilot study, we performed RT-qPCR to detect expression levels of circHIPK3 in CRC tissues. Sanger sequencing showed the PCR products contained the backsplice junction expecting in the circHIPK3 (Figure S1). As shown in Fig. 1a, expression levels of circHIPK3 in the non-responder group were significantly higher than in the responder group. When responders were included (n = 31) as the end point for detection compared to non-responders (n = 18), circHIPK3 yielded an area under the ROC curve (AUC) of 0•768 (95%CI = 0•625 to 0•876), which was more accurate than guessing for differential diagnosis (P = 0•0003, Fig. 1b). To determine the effect of circHIPK3 in chemoresistance, we employed the 5FU and OXA resistant CRC cell lines established in our laboratory. The concentration-effect curves were shown Figure S2. The IC50 values of OXA on HCT116oxR and HT29oxR cells was 13•3 μM and 6•0 μM, while 3•4 μM and 3•1 μM on their parental cells. The IC50 values of 5FU on HT29FuR cell and the parental cell was 68•2 μM and 15•1 μM, respectively. As shown in Fig. 1c, the expression of circHIPK3 was increased in OXA resistant HT29oxR and HCT116oxR, but not in 5FU resistant cell lines compared to their respective parental cell lines. To assess whether circHIPK3 exerts OXA resistance function in vivo, we established the xenograft mouse models, and treated with OXA once weekly. As shown in Fig. 1d–f, circHIPK3 overexpression group displayed larger tumour size and weight than those of NC group.
Fig. 1.
circHIPK3 is associated with CRC chemoresistance. (a) circHIPK3 expression was higher in tissues of non-responder group (n = 18) than in responder group (n = 31); Relative circHIPK3 expression levels were calculated using 2−ΔCT method and is represented as the median (interquartile range); [Mann–Whitney U test]. (b) ROC curve for discriminating responders from non-responders based on circHIPK3 expression; AUC = 0•768 (95%CI = 0•625 to 0•876). (c) Expression of circHIPK3 was increased in oxaliplatin-resistant HT29oxR and HCT116oxR (P < 0•001) cell lines while not in 5-FU resistant cell lines compared to their respective parental cell lines (P > 0•05) [student's t-test]. (d) Images of tumour mass of each group (n = 5) on the 28th day. (e and f) The tumour volumes and weights from circHIPK3 overexpression cells were significantly larger than those from negative control cells. Data are presented as mean ±standard deviation. [student's t-test].
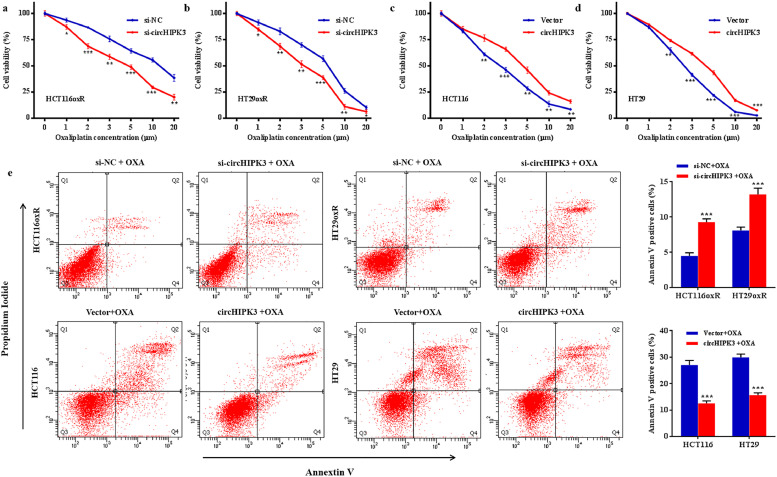
To further confirm the role of circHIPK3 in OXA chemoresistance of CRC cells, we transfected HT29oxR and HCT116oxR cells with siRNA specifically targeting the circHIPK3 junction site, and their respective parental cell lines with overexpression vector. The transfection effects were confirmed by RT-qPCR (Figure S3). CCK8 assay demonstrated that knockdown of circHIPK3 sensitised HT29oxR and HCT116oxR cells to OXA (Fig. 2a and b), while ectopic expression of circHIPK3 reduced the effect of OXA cytotoxicity in HT29 and HCT116 cells in a dose-dependent manner (Fig. 2c and d). Flow cytometry analysis showed that OXA-induced apoptotic cells were significantly increased after treatment with si-circHIPK3, whereas overexpression of circHIPK3 greatly inhibited the apoptosis of HT29 and HCT116 cells treated with 3 μM OXA (Fig. 2e).
Fig. 2.
circHIPK3 leads to oxaliplatin resistance in CRC cell lines. (a and b) Cell proliferative ability was assessed by CCK8 assay after circHIPK3 knockdown in HT29oxR (a) and HCT116oxR (b) cells at the indicated oxaliplatin concentration. (c and d) Cell proliferative ability was assessed by CCK8 assay after circHIPK3 overexpression, in HT29 (c) and HCT116 (d) cells at the indicated oxaliplatin concentration. (e) Oxaliplatin-induced apoptotic cell death was analysed by flow cytometry after treatment with si-circHIPK3 or overexpression vector in CRC cells. Data are presented as mean ±standard deviation from at least 3 independent experiments. *P < 0•05, ⁎⁎P < 0•01, ⁎⁎⁎P < 0•001 [student's t-test].
4.2. Silencing of circHIPK3 sensitises CRC to OXA via autophagy induction
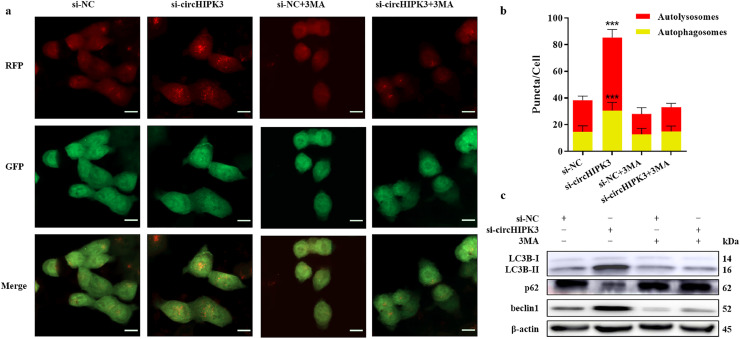
Autophagy flux was monitored in OXA resistant CRC cells transfected with lentiviral vector mRFP-GFP-LC3B. Since GFP signal was quenched in the acidic environment of lysosomes, only mRFP puncta(red) without GFP fluorescence indicates autolysosomes, and yellow puncta with both GFP and mRFP fluorescence corresponds to autophagosomes. As shown in Fig. 3a and b, silencing of circHIPK3 significantly enhanced the formation of autophagosomes and autolysosomes compared to cells transfected with control oligonucleotides. Western blotting analysis showed that circHIPK3 knockdown increased the ratio of LC3B-II to LC3B-I, and beclin1 expression, and decreased p62 expression (Fig. 3c), confirming that downregulation of circHIPK3 induced autophagy.
Fig. 3.
Silencing of circHIPK3 induces autophagy and sensitises CRC cells to oxaliplatin. (a) Transfection of mRFP-GFP-LC3B lentiviral vector into HCT116oxR cell line. Red puncta represent autolysosomes, and yellow puncta represent autophagosomes as visualised by confocal microscopy. Scare bar = 20 μm (b) Quantification of autophagic flux. Puncta were counted in 100 cells; ⁎⁎⁎P < 0•001 [student's t-test]. (c) Western blot analysis to determine LC3B- II/I ratio, p62 and, beclin1 expression in HCT116oxR cells.
To confirm whether autophagy was responsible for OXA sensitisation, we treated cells with 5 mM 3-MA (an autophagy inhibitor), and found that autophagy induction due to circHIPK3 silencing (as observed by the formation of autophagosomes and autolysosomes), was inhibited (Fig. 3a and b), and the ratio of LC3B-II to LC3B-I, and expressions of p62 and beclin1 were also restored to basal levels (Fig. 3c). We further showed that the proliferative and anti-apoptotic effects of circHIPK3 were counteracted by inhibition of autophagy (Figure S4). Taken together, silencing of circHIPK3 might sensitise the resistant CRC cells to OXA through induction of autophagy.
4.3. circHIPK3 sponges miR-637, and reverses the miR-637-induced promotion of OXA chemosensitivity
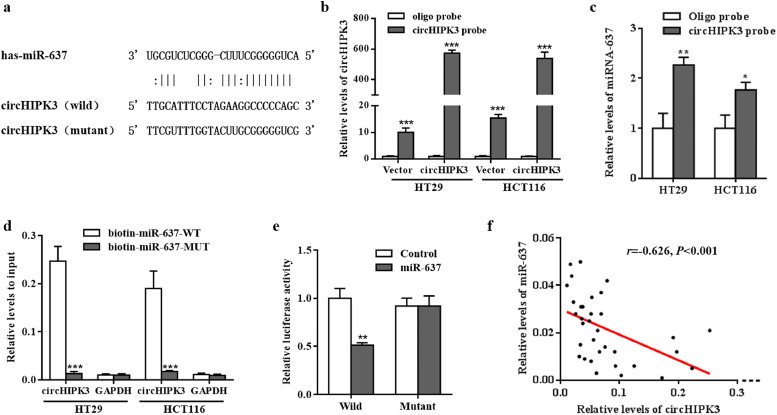
It has been reported that circRNAs act as miRNA sponges [27]. As shown in Fig. 4a, miR-637 was found to specifically target circHIPK3 as indicated by overlapping prediction results from RNAhybrid-2.1.2 (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid/) and microRNA.org 2010 Release (http://www.microrna.org/microrna/getDownloads.do) tools. To verify our prediction, we designed a biotinylated-circHIPK3 probe and the pull-down efficiency was confirmed in CRC cells over-expressing circHIPK3 (Fig. 4b). We found that miR-637 was abundantly pulled down by circHIPK3 probe in both HT29 and HCT116 cells (Fig. 4c). Moreover, biotinylated miR-637 mimics could capture additional circHIPK3 compared to control RNA (Fig. 4d). Next, we used the dual-reporter luciferase assay to test the binding site and found that the luciferase activity was significantly decreased in circHIPK3-wt plus (+) miR-637 group while not in circHIPK3-mut plus (+) miR-637 group (Fig. 4e). Consistent with the above results, there was also a significant inverse association between circHIPK3 and miR-637 expression in CRC tissues (r = −0•626, P<0•001; Fig. 4f).
Fig. 4.
circHIPK3 functions as an efficient miR-637 sponge. (a) The putative miR-637 binding site in circHIPK3 and the corresponding mutant motif. (b) The pull-down efficiency of biotinylated-circHIPK3 probe tested by RT-qPCR in HT29 and HCT116 cells; ⁎⁎⁎P < 0•001 [student's t-test]. (c) miR-637 pull-down by biotinylated-circHIPK3 probe was tested by RT-qPCR in HT29 and HCT116 cells transfected with circHIPK3 overexpression vector; Oligo probe was used as a control; *P<0•05, ⁎⁎P < 0•01 [student's t-test]. (d) circHIPK3 pull -down by biotinylated wild-type/mutant miR-637 was tested by RT-qPCR in CRC cells with circHIPK3 overexpression. Relative levels of circHIPK3 were normalised to input; GAPDH was used as an internal control; ⁎⁎⁎P < 0•001 [student's t-test]. (e) Relative luciferase activity of wild type or mutant circHIPK3 in miR-637 mimics or controls; ⁎⁎P < 0•01 [student's t-test]. (f) Correlation between circHIPK3 and miR-637 expression in CRC tissues; r = −0•626, P < 0•001[Spearman test].
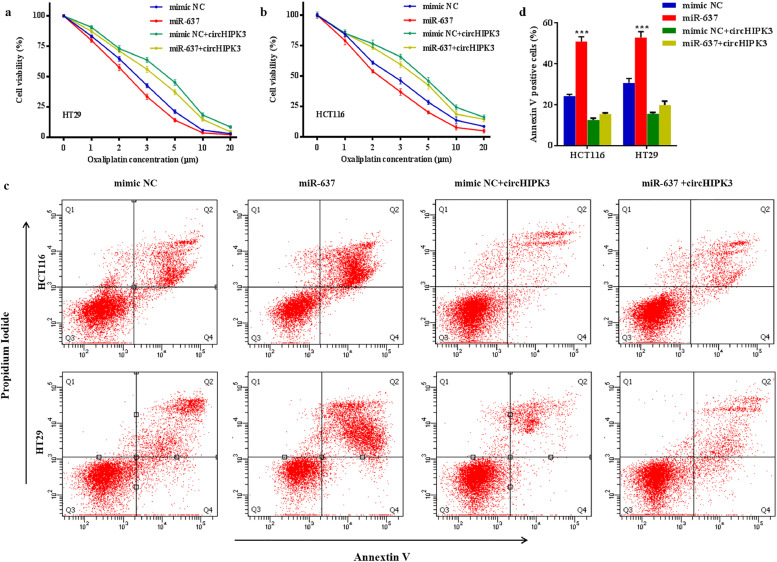
We then determined the role of miR-637 in OXA chemosensitivity in CRC cells. Transfection of miR-637 mimics significantly increased the chemosensitivity of OXA (Fig. 5a and b) and increased the number of apoptotic cells (Fig. 5c and d). When HCT116 and HT29 cells were co-transfected with miR-637 mimics and circHIPK3 vector, OXA chemosensitivity promoted by miR-637 was reversed (Fig. 5a and b). Moreover, miR-637 plus (+) circHIPK3 group showed less cell apoptotic rate compared to miR-637 group (Fig. 5c and d). These results indicated that circHIPK3 served as a sponge for miR-637, which promoted OXA chemosensitivity in CRC cells.
Fig. 5.
circHIPK3 reverses miR-637-induced elevation of oxaliplatin chemosensitivity. (a and b) Cell proliferative ability was assessed by CCK8 assay in HT29 (a) and HCT116 (b) cells after transfection with mimic NC, miR-637 mimics, mimic NC plus (+) circHIPK3 overexpression vector, and miR-637 mimics+ circHIPK3 overexpression vector at the indicated OXA concentration. (c and d) Oxaliplatin-induced apoptotic cell death was analysed by flow cytometry in the above mentioned four groups. ⁎⁎⁎P < 0•001 [student's t-test].
4.4. circHIPK3-miR-637-STAT3 is involved in the regulation of autophagy
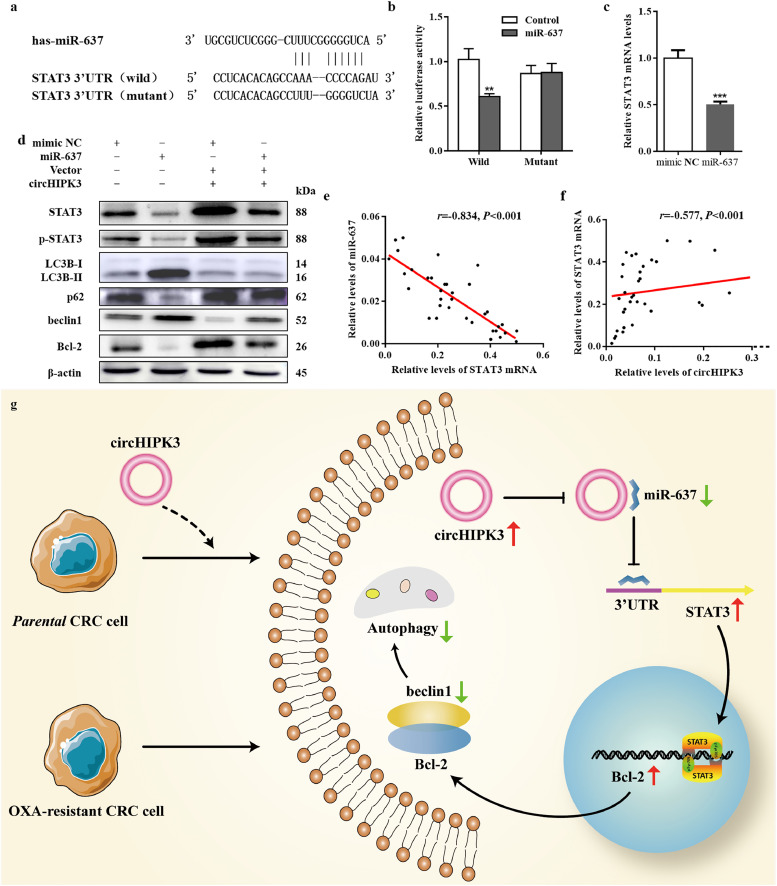
We examined the Targetscan and miRanda databases to predict the probable target gene of miR-637. One of the downstream genes, Stat3(Fig. 6a), was potentially interesting due to its role in the regulation of cellular autophagy [28]. To test whether miR-637 directly targeted STAT3, we performed dual-reporter luciferase assay and found that miR-637 decreased the luciferase activity of STAT3 3′UTR constructs while not affecting mutant 3′UTR constructs (Fig. 6b). Moreover, miR-637 could inhibit the expression of STAT3 at both RNA and protein levels (Fig. 6c and d). The relative levels of STAT3 mRNA were negatively related to miR-637 expression (Fig. 6e), while positively associated with circHIPK3 levels (Fig. 6f). Therefore, we speculated that circHIPK3 might function as a ceRNA to regulate STAT3 by sponging miR-637.
Fig. 6.
Identification and verification of circHIPK3/miR-637/STAT3/Bcl-2/beclin1 axis. (a) The putative miR-637 binding site in Stat3, and the corresponding mutant motif. (b) Relative luciferase activity of wild type or mutant Stat3 in miR-637 mimics or controls; ⁎⁎P < 0•01 [student's t-test]. (c) Levels of STAT3 mRNA were significantly down-regulated in HCT116 cells transfected with miR-637 mimics, compared to cells transfected with mimic NC; ⁎⁎⁎P < 0•001 [student's t-test]. (d) Western blot analysis of STAT3, p-STAT3, LC3BII/I, p62, beclin1, Bcl-2 protein expression in HCT116 cells transfected with mimic NC, miR-637 mimics, mimic NC+ circHIPK3 overexpression vector, and miR-637 mimics plus (+) circHIPK3 overexpression vector. (e) Correlation between STAT3 mRNA and miR-637 expression in CRC tissues; n = 49, r = −0•834, P < 0•001 [Spearman test]. (f) Correlation between circHIPK3 and STAT3 mRNA expression in CRC tissues; n = 49, r = 0•577, P < 0•001 [Spearman test]. (g) The proposed working model.
Immunoblotting analysis also showed that transfection of cells with miR-637 mimics could inhibit the expression of p-STAT3 and enhance autophagy by increasing LC3B II / I, beclin1 levels and decreasing p62 levels (Fig. 6d). In order to better understand the molecular mechanism of miR-637 in autophagy, STAT3-Bcl-2-beclin1 signalling pathway was analysed. As shown in Fig. 6d, Bcl-2 expression was decreased and beclin1 expression was increased after transfection with miR-637 mimics. Next, we co-transfected with circHIPK3 and miR-637 mimics and found that circHIPK3 could counteract the effect of miR-637 in CRC cells (Fig. 6d). The summarised diagram illustrating these results has been shown in Fig. 6g.
4.5. Increased circHIPK3 expression predicts recurrence and poor survival in CRC patients
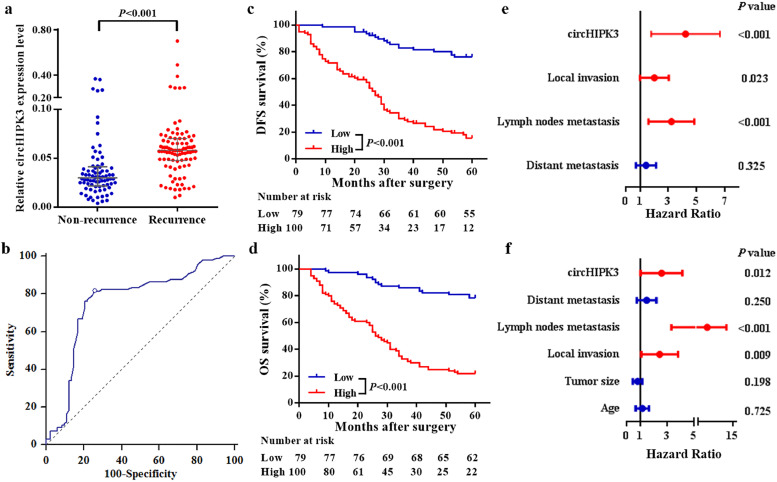
A clinical study of 179 patients who received postoperative OXA-based adjuvant chemotherapy showed that levels of circHIPK3 were closely related to tumour size, regional lymph node metastasis and distant metastasis (all values of P<0•05, Figure S5). More importantly, CRC patients with recurrence had higher levels of circHIPK3 than patients with non-recurrence (Fig. 7a). ROC curve analysis illustrated that it could distinguish patients with recurrence from those without recurrence, with AUC of 0•758 (95%CI = 0•689 to 0•819) (Fig. 7b), and the optimal cut-off value was 0•040, providing a sensitivity of 81•4% and a specificity of 74•4%.
Fig. 7.
Expression of circHIPK3 in CRC patients who received oxaliplatin-based chemotherapy after surgery. (a) circHIPK3 expression was higher in tissues of CRC recurrence group than in non-recurrence group; Data represents the median (interquartile range); P < 0•001 [Mann–Whitney U test]. (b) ROC curve for discriminating CRC patients with recurrence from those without recurrence based on circHIPK3 expression; AUC = 0•758 (95%CI = 0•689–0•819). (c and d) Kaplan-Meier curves for DFS (c) and OS (d) based on circHIPK3 expression; CRC patients were classified as high and low circHIPK3 expression according to the optimal cut off value (0•040); P < 0•001 [log-rank test]. (e and f), Multivariate Cox analysis for DFS (e) and OS (f) of CRC patients.
Then, CRC patients were divided into high and low circHIPK3 expression groups based on the optimal cut-off value (0•040). Similarly, the expression of circHIPK3 was significantly correlated with tumour size, regional lymph node metastasis, distant metastasis, and recurrence (all values of P < 0•05, Table 1). Kaplan-Meier curves showed that patients in the high circHIPK3 expression group had significantly worse 5-year DFS and OS rates compared to patients in the low expression group (Fig. 7c and d). Kaplan-Meier curves for other clinicopathologic characteristics are shown in figure S6. Cox model showed that circHIPK3 expression was an independent factor for both DFS and OS (Fig. 7e and f, Table S3).
Table 1.
Associations between circHIPK3 levels and clinicopathological characteristics.
| Parameters | Expression of circHIPK3a |
||
|---|---|---|---|
| high | low | P-valueb | |
| Age | 0•324 | ||
| <61 | 47 | 43 | |
| ≥61 (median) | 53 | 36 | |
| Gender | 0•373 | ||
| Male | 49 | 44 | |
| Female | 51 | 35 | |
| Tumor location | 0•091 | ||
| Colon | 52 | 51 | |
| Rectum | 48 | 28 | |
| Tumor size | 0•033 | ||
| <4cm | 54 | 55 | |
| ≥4cm | 46 | 24 | |
| Differentiation | 0•795 | ||
| Well | 29 | 20 | |
| Moderate | 52 | 45 | |
| Poor | 19 | 14 | |
| Local invasion | 0•072 | ||
| T1-T2 | 21 | 26 | |
| T3-T4 | 79 | 53 | |
| Regional lymph nodes metastasis | <0•001 | ||
| No | 31 | 69 | |
| Yes | 69 | 10 | |
| Distant metastasis | <0•001 | ||
| No | 75 | 75 | |
| Yes | 25 | 4 | |
| Recurrence | <0•001 | ||
| No | 21 | 61 | |
| Yes | 79 | 18 | |
CRC patients were classified as high or low circHIPK3 based on the optimal cut off value (0•040).
P-value was estimated by Chi-square test.
5. Discussion
Emerging studies have shown that circRNAs are involved in cancer development and progression and serve as tumour biomarkers, such as circHIPK3 [21]. However, the role of circHIPK3 in tumours is still controversial. For instance, Li et al. [26] found that circHIPK3 expression was decreased in bladder cancer tissues, and functioned as a tumour suppressor gene to inhibit invasion, migration, and angiogenesis of bladder cancer cells. Down-regulated expression of circHIPK3 was also observed in tissues, plasma and cell lines of osteosarcoma, which correlated with poor prognosis of patients [29]. Conversely, circHIPK3 was highly expressed in patients with nasopharyngeal carcinoma, predicting poor prognosis, while silencing its expression inhibited cell proliferation and migration [30]. Similar phenomena were also found in hepatocellular carcinoma [31] and colorectal cancer [23]. Moreover, the transcription factor c-myb, also known as a protooncogene is closely associated with resistance to chemotherapeutic drugs [32,33], and could promote the transcription of circHIPK3 [23]. However, the function of circHIPK3 in CRC chemotherapy was still unknown. In this study, we enrolled 49 CRC patients who received 5FU and OXA-based first-line chemotherapy and found that circHIPK3 expression was increased in patients with stable or progressive disease compared to those with complete or partial response. Further, we observed that circHIPK3 might contribute to the resistance of OXA, but not 5FU. Data from in vivo and vitro experiments showed overexpression of circHIPK3 promoted resistance to OXA. circHIPK3 was derived from exon2 of the HIPK3 gene, and is formed by direct back-splicing with the help of complementary ALU repeats [34]. Thus, it is interesting to study its relationship with its host gene. Zheng et al. [21] showed that the circular transcript of HIPK3 gene, and not the linear transcript, promoted human cell proliferation. In lung cancer, circHIPK3 and HIPK3 mRNA have opposite effects on autophagy [35]. In this study, we analysed the expression of HIPK3 mRNA in The Cancer Genome Atlas (TCGA) dataset, and found it was significantly decreased in 471 colon adenocarcinoma samples compared with its expression in 41 normal samples (Figure S7a), suggesting HIPK3 might function as a tumour suppressor gene. This is different from the cancer-promoting effect of the circHIPK3 in CRC [23]. Moreover, HIPK3 mRNA expression has no significant relationship with OS of colon adenocarcinoma patients (Figure S7b). Yao et al. [36] reported overexpression of HIPK3 reduced OXA induced CRC cell death, consistent with the phenomenon caused by circHIPK3 in this study. Taken together, although circular and linear transcripts of HIPK3 come from the same host gene, they have their respective roles in cancer.
Oncogenic mutations of KRAS, BRAF, and PI3KCA frequently occur in CRC tissues [37]. In this study, we employed the HCT116 cell line with mutant KRAS and wild type BRAF, and the HT29 cell line with mutant BRAF and wild type KRAS to evaluate circHIPK3-mediated autophagy. Several studies have found that mutant KRAS tumour cells have high basal levels of autophagy and respond poorly to chemotherapy [38,39]. Suppression of KRAS alone (RAF → MEK → ERK inhibition) further increased autophagic flux, and showed no clinical benefit, while concurrent treatment with an autophagy inhibitor displayed synergistic anti-proliferative effects [40,41]. Similar observations were also reported in BRAF-mutant tumours [41], [42], [43], [44]. Of note, both cell lines have mutated PIK3CA that constantly activates that AKT/mTOR signalling pathway, which is a well-known autophagy inhibiting pathway [45]. We found that knockdown of circHIPK3 resulted in enhanced autophagy and reversed the resistance of HT29oxR and HCT116oxR cells. 3-MA is a classic autophagy inhibitor that inhibits the formation of autophagosomes through suppression of class III PI3K activity [46]. Since 3-MA might promote autophagic flux under nutrient-rich conditions for extended periods of time [46], we treated HCT116 and HT29 cells with 5 mM 3-MA for 1 h in serum-free media. Upon inhibition of autophagy by 3-MA, apoptosis of both CRC cells lines was also reduced. These results were in agreement with a previous study [47], which demonstrated that anticancer agents induced autophagy-related cell death (type II programmed cell death), independent of or in parallel with apoptosis, thereby enhancing their therapeutic effects. circRNAs have been proven to function as ceRNAs to protect oncogenes or tumour suppressors through binding to functional miRNAs [27]. In this study, we also demonstrated that circHIPK3 could bind to miR-637, and a single miR-637 binding element present in circHIPK3 was found to be essential for their interaction by both miRanda and RNAhybrid tools. Moreover, RNA pull down and luciferase assays confirmed that circHIPK3 could be directly bound by miR-637 in CRC cells. miR-637 has been reported to inhibit cell proliferation and induce apoptosis in multiple cancers [48], [49], [50]. Wang et al. [51] found that blockage of miR-637 promoted viability, proliferation, migration and invasion capacity of CRC cells, suggesting miR-637 played a tumour suppressor role in CRC. Consistently, we found that overexpression of miR-637 mimicked the effect of circHIPK3 knockdown in CRC cell viability and apoptosis. Moreover, the effects caused by miR-637 also could be counteracted by circHIPK3 overexpression. Interestingly, miR-7 that found to be sponged by circHIPK3 in CRC has been reported down-regulated in OXA resistant tumour cells [23,52]. Since circHIPK3 serves as a sponge for multiple miRNAs, we speculated it might not cause chemotherapy resistance only through one miRNA.
It has been well established that miRNAs regulate gene expression by binding to the complementary sequences in 3′-UTRs of target genes. Our data revealed that miR-637 was able to directly target the 3′-UTR of STAT3, resulting in down-regulated expression of STAT3 at the post-transcriptional level. Moreover, there was a negative correlation between miR-637 and STAT3 mRNA in CRC tissues. Thus, we hypothesised that STAT3 might be one of the important targets of miR-637, in agreement with a previous study [53], which reported that miR-637 disrupted the activation of STAT3 and exhibited growth suppressive and apoptotic effects. Recently, STAT3 expression has been frequently correlated with autophagy and chemoresistance in CRC [54,55]. In addition to its direct effects, we found that miR-637 also decreased phosphorylation levels of STAT3, and subsequently repressed the transcriptional expression of Bcl-2, which could physically bind to the BH3-only domain within beclin1, that played a central role in autophagy [56]. In this study, activation of STAT3 was inhibited by miR-637 down-regulated Bcl-2 expression, thereby releasing beclin1 from the Bcl-2-beclin-1 complex, to induce autophagy, similar to the effects caused by circHIPK3 silencing. Conversely, these effects induced by miR-637 could also be rescued by circHIPK3 overexpression. Furthermore, a positive relationship was observed between circHIPK3 expression and STAT3 mRNA in CRC tissues. Taken together, circHIPK3 could function as a ceRNA via sponging miR-637 to activate the STAT3 signalling pathway, thus enhancing Bcl-2 expression and blocking beclin1 dissociation. These events finally resulted in reduced autophagic cell death, which contributes to the development of OXA resistance.
To test whether circHIPK3 could be used as a chemotherapy predictor, a large cohort of CRC patients who received OXA-based treatment were enrolled. Our results showed that circHIPK3 was significantly increased in patients with recurrent disease, and its high expression correlated with poor survival, which was an independent prognostic factor for these patients. In previous studies, circHIPK3 was also found to be an independent predictor of prognosis in glioma [57] and epithelial ovarian cancer [58]. Because there was no suitable method to accurately evaluate the autophagy levels on human samples [59], the relationship between circHIPK3 expression and autophagy levels in CRC tissues was unclear.
In summary, our findings suggest that circHIPK3 promotes OXA resistance in CRC by sponging miR-637, which is dependent on the inhibition of autophagy-related cell death via STAT3/Bcl-2/beclin1 signalling pathway. This study also provides evidence that circHIPK3 functions as a chemoresistance gene, and could be a promising prognostic predictor in CRC patients treated with OXA-based chemotherapy.
Funding sources
This study was supported by National Natural Science Foundation of China (81301506), Shandong Medical and Health Technology Development Project(2018WSB20002), Shandong Key Research and Development Program (2016GSF201122), Natural Science Foundation of Shandong Province (ZR2017MH044), Jinan Science and Technology Development Plan(201805084, 201805003). The funders had no role in study design, data collection, data analysis, interpretation, and writing of the manuscript.
Declaration of Competing Interest
There are no relevant conflicts of interest.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2019.09.051.
Appendix. Supplementary materials
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Andre T., Boni C., Navarro M., Tabernero J., Hickish T., Topham C. Improved overall survival with oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment in stage II or III colon cancer in the MOSAIC trial. J Clin Oncol. 2009;27(19):3109–3116. doi: 10.1200/JCO.2008.20.6771. [DOI] [PubMed] [Google Scholar]
- 3.Goldberg R.M., Sargent D.J., Morton R.F., Fuchs C.S., Ramanathan R.K., Williamson S.K. A randomized controlled trial of fluorouracil plus leucovorin, irinotecan, and oxaliplatin combinations in patients with previously untreated metastatic colorectal cancer. J Clin Oncol. 2004;22(1):23–30. doi: 10.1200/JCO.2004.09.046. [DOI] [PubMed] [Google Scholar]
- 4.O'Connell J.B., Maggard M.A., Ko C.Y. Colon cancer survival rates with the new American Joint Committee on Cancer sixth edition staging. J Natl Cancer Inst. 2004;96(19):1420–1425. doi: 10.1093/jnci/djh275. [DOI] [PubMed] [Google Scholar]
- 5.Mehrpour M., Esclatine A., Beau I., Codogno P. Overview of macroautophagy regulation in mammalian cells. Cell Res. 2010;20(7):748–762. doi: 10.1038/cr.2010.82. [DOI] [PubMed] [Google Scholar]
- 6.Schaaf M.B., Keulers T.G., Vooijs M.A., Rouschop K.M. LC3/GABARAP family proteins: autophagy-(un)related functions. FASEB J. 2016;30(12):3961–3978. doi: 10.1096/fj.201600698R. [DOI] [PubMed] [Google Scholar]
- 7.Wu S., Sun C., Tian D., Li Y., Gao X., He S. Expression and clinical significances of Beclin1, LC3 and mTOR in colorectal cancer. Int J Clin Exp Pathol. 2015;8(4):3882–3891. [PMC free article] [PubMed] [Google Scholar]
- 8.Liu X., Li Y., Wang X., Xing R., Liu K., Gan Q. The BEACH-containing protein WDR81 coordinates p62 and LC3C to promote aggrephagy. J Cell Biol. 2017;216(5):1301–1320. doi: 10.1083/jcb.201608039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim Y.S., Shin J.H., Bae M.K., Lee C.Y., Kim D.J., Chung K.Y. Autophagy activity in pulmonary metastatic tumor tissues from colorectal cancer: a pilot study. Yonsei Med J. 2014;55(6):1484–1488. doi: 10.3349/ymj.2014.55.6.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmitz K.J., Ademi C., Bertram S., Schmid K.W., Baba H.A. Prognostic relevance of autophagy-related markers LC3, p62/sequestosome 1, beclin-1 and ULK1 in colorectal cancer patients with respect to KRAS mutational status. World J Surg Oncol. 2016;14(1):189. doi: 10.1186/s12957-016-0946-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pattingre S., Espert L., Biard-Piechaczyk M., Codogno P. Regulation of macroautophagy by mTOR and Beclin 1 complexes. Biochimie. 2008;90(2):313–323. doi: 10.1016/j.biochi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 12.Sahni S., Merlot A.M., Krishan S., Jansson P.J., Richardson D.R. Gene of the month: BECN1. J Clin Pathol. 2014;67(8):656–660. doi: 10.1136/jclinpath-2014-202356. [DOI] [PubMed] [Google Scholar]
- 13.Choi J.H., Cho Y.S., Ko Y.H., Hong S.U., Park J.H., Lee M.A. Absence of autophagy-related proteins expression is associated with poor prognosis in patients with colorectal adenocarcinoma. Gastroenterol Res Pract. 2014;2014 doi: 10.1155/2014/179586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galluzzi L., Bravo-San Pedro J.M., Demaria S., Formenti S.C., Kroemer G. Activating autophagy to potentiate immunogenic chemotherapy and radiation therapy. Nat Rev Clin Oncol. 2017;14(4):247–258. doi: 10.1038/nrclinonc.2016.183. [DOI] [PubMed] [Google Scholar]
- 15.O'Donovan T.R., Rajendran S., O'Reilly S., O'Sullivan G.C., McKenna S.L. Lithium modulates autophagy in esophageal and colorectal cancer cells and enhances the efficacy of therapeutic agents in vitro and in vivo. PLoS ONE. 2015;10(8) doi: 10.1371/journal.pone.0134676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li S., Yang G., Zhu X., Cheng L., Sun Y., Zhao Z. Combination of rapamycin and garlic-derived S-allylmercaptocysteine induces colon cancer cell apoptosis and suppresses tumor growth in xenograft nude mice through autophagy/p62/Nrf2 pathway. Oncol Rep. 2017;38(3):1637–1644. doi: 10.3892/or.2017.5849. [DOI] [PubMed] [Google Scholar]
- 17.Ren W.W., Li D.D., Chen X., Li X.L., He Y.P., Guo L.H. MicroRNA-125b reverses oxaliplatin resistance in hepatocellular carcinoma by negatively regulating EVA1A mediated autophagy. Cell Death Dis. 2018;9(5):547. doi: 10.1038/s41419-018-0592-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang S., Wang K., Zhang C., Zhang W., Xu Q., Wang Y. Overaccumulation of p53-mediated autophagy protects against betulinic acid-induced apoptotic cell death in colorectal cancer cells. Cell Death Dis. 2017;8(10):e3087. doi: 10.1038/cddis.2017.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan X., Chen Y., Shen Y., Tantai J. Knockdown of TRIM65 inhibits autophagy and cisplatin resistance in A549/DDP cells by regulating miR-138-5p/ATG7. Cell Death Dis. 2019;10(6):429. doi: 10.1038/s41419-019-1660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y., Li C., Tan C., Liu X. Circular RNAs: a new frontier in the study of human diseases. J Med Genet. 2016;53(6):359–365. doi: 10.1136/jmedgenet-2016-103758. [DOI] [PubMed] [Google Scholar]
- 21.Zheng Q., Bao C., Guo W., Li S., Chen J., Chen B. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shan K., Liu C., Liu B.H., Chen X., Dong R., Liu X. Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation. 2017;136(17):1629–1642. doi: 10.1161/CIRCULATIONAHA.117.029004. [DOI] [PubMed] [Google Scholar]
- 23.Zeng K., Chen X., Xu M., Liu X., Hu X., Xu T. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9(4):417. doi: 10.1038/s41419-018-0454-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Li P., Zhang X., Wang L., Du L., Yang Y., Liu T. lncRNA HOTAIR contributes to 5FU resistance through suppressing miR-218 and activating NF-kappaB/TS signaling in colorectal cancer. Mol Ther Nucleic Acids. 2017;8:356–369. doi: 10.1016/j.omtn.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu T., Zhang X., Du L., Wang Y., Liu X., Tian H. Exosome-transmitted miR-128-3p increase chemosensitivity of oxaliplatin-resistant colorectal cancer. Mol Cancer. 2019;18(1):43. doi: 10.1186/s12943-019-0981-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Li Y., Zheng F., Xiao X., Xie F., Tao D., Huang C. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18(9):1646–1659. doi: 10.15252/embr.201643581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Z.B., Huang G.X., Fu Q., Han B., Lu J.J., Chen A.M. circRNA.33186 contributes to the pathogenesis of osteoarthritis by sponging miR-127-5p. Mol Ther. 2019;27(3):531–541. doi: 10.1016/j.ymthe.2019.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.You L., Wang Z., Li H., Shou J., Jing Z., Xie J. The role of STAT3 in autophagy. Autophagy. 2015;11(5):729–739. doi: 10.1080/15548627.2015.1017192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao-Long M., Kun-Peng Z., Chun-Lin Z. Circular RNA circ_HIPK3 is down-regulated and suppresses cell proliferation, migration and invasion in osteosarcoma. J Cancer. 2018;9(10):1856–1862. doi: 10.7150/jca.24619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ke Z., Xie F., Zheng C., Chen D. CircHIPK3 promotes proliferation and invasion in nasopharyngeal carcinoma by abrogating miR-4288-induced ELF3 inhibition. J Cell Physiol. 2019;234(2):1699–1706. doi: 10.1002/jcp.27041. [DOI] [PubMed] [Google Scholar]
- 31.Chen G., Shi Y., Liu M., Sun J. circHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis. 2018;9(2):175. doi: 10.1038/s41419-017-0204-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pekarcikova L., Knopfova L., Benes P., Smarda J. c-Myb regulates NOX1/p38 to control survival of colorectal carcinoma cells. Cell Signal. 2016;28(8):924–936. doi: 10.1016/j.cellsig.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 33.Del Bufalo D., Cucco C., Leonetti C., Citro G., D'Agnano I., Benassi M. Effect of cisplatin and c-myb antisense phosphorothioate oligodeoxynucleotides combination on a human colon carcinoma cell line in vitro and in vivo. Br J Cancer. 1996;74(3):387–393. doi: 10.1038/bjc.1996.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J. Circular RNAs are abundant, conserved, and associated with alu repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen X., Mao R., Su W., Yang X., Geng Q., Guo C. Circular RNA circHIPK3 modulates autophagy via MIR124-3p-STAT3-PRKAA/AMPKalpha signaling in STK11 mutant lung cancer. Autophagy. 2019:1–13. doi: 10.1080/15548627.2019.1634945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yao H., Xia D., Li Z.L., Ren L., Wang M.M., Chen W.S. MiR-382 functions as tumor suppressor and chemosensitizer in colorectal cancer. Biosci Rep. 2019;39(8) doi: 10.1042/BSR20180441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shiovitz S., Grady W.M. Molecular markers predictive of chemotherapy response in colorectal cancer. Curr Gastroenterol Rep. 2015;17(2):431. doi: 10.1007/s11894-015-0431-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alves S., Castro L., Fernandes M.S., Francisco R., Castro P., Priault M. Colorectal cancer-related mutant KRAS alleles function as positive regulators of autophagy. Oncotarget. 2015;6(31):30787–30802. doi: 10.18632/oncotarget.5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo J.Y., Chen H.Y., Mathew R., Fan J., Strohecker A.M., Karsli-Uzunbas G. Activated RAS requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25(5):460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bryant K.L., Stalnecker C.A., Zeitouni D., Klomp J.E., Peng S., Tikunov A.P. Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat Med. 2019;25(4):628–640. doi: 10.1038/s41591-019-0368-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kinsey C.G., Camolotto S.A., Boespflug A.M., Guillen K.P., Foth M., Truong A. Protective autophagy elicited by RAF –> MEK –> ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nat Med. 2019;25(4):620–627. doi: 10.1038/s41591-019-0367-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma X.H., Piao S.F., Dey S., McAfee Q., Karakousis G., Villanueva J. Targeting er stress-induced autophagy overcomes BRAF inhibitor resistance in melanoma. J Clin Invest. 2014;124(3):1406–1417. doi: 10.1172/JCI70454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goulielmaki M., Koustas E., Moysidou E., Vlassi M., Sasazuki T., Shirasawa S. BRAF associated autophagy exploitation: BRAF and autophagy inhibitors synergise to efficiently overcome resistance of BRAF mutant colorectal cancer cells. Oncotarget. 2016;7(8):9188–9221. doi: 10.18632/oncotarget.6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strohecker A.M., White E. Autophagy promotes BrafV600E-driven lung tumorigenesis by preserving mitochondrial metabolism. Autophagy. 2014;10(2):384–385. doi: 10.4161/auto.27320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aoki M., Fujishita T. Oncogenic roles of the PI3K/AKT/mTOR axis. Curr Top Microbiol Immunol. 2017;407:153–189. doi: 10.1007/82_2017_6. [DOI] [PubMed] [Google Scholar]
- 46.Wu Y.T., Tan H.L., Shui G., Bauvy C., Huang Q., Wenk M.R. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J Biol Chem. 2010;285(14):10850–10861. doi: 10.1074/jbc.M109.080796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fulda S., Kogel D. Cell death by autophagy: emerging molecular mechanisms and implications for cancer therapy. Oncogene. 2015;34(40):5105–5113. doi: 10.1038/onc.2014.458. [DOI] [PubMed] [Google Scholar]
- 48.Li J.X., Ding X.M., Han S., Wang K., Jiao C.Y., Li X.C. mir-637 inhibits the proliferation of cholangiocarcinoma cell QBC939 through interfering CTSB expression. Eur Rev Med Pharmacol Sci. 2018;22(5):1265–1276. doi: 10.26355/eurrev_201803_14467. [DOI] [PubMed] [Google Scholar]
- 49.Xu R.L., He W., Tang J., Guo W., Zhuang P., Wang C.Q. Primate-specific miRNA-637 inhibited tumorigenesis in human pancreatic ductal adenocarcinoma cells by suppressing Akt1 expression. Exp Cell Res. 2018;363(2):310–314. doi: 10.1016/j.yexcr.2018.01.026. [DOI] [PubMed] [Google Scholar]
- 50.Que T., Song Y., Liu Z., Zheng S., Long H., Li Z. Decreased miRNA-637 is an unfavorable prognosis marker and promotes glioma cell growth, migration and invasion via direct targeting Akt1. Oncogene. 2015;34(38):4952–4963. doi: 10.1038/onc.2014.419. [DOI] [PubMed] [Google Scholar]
- 51.Wang L., Jiang F., Xia X., Zhang B. LncRNA FAL1 promotes carcinogenesis by regulation of miR-637/NUPR1 pathway in colorectal cancer. Int J Biochem Cell Biol. 2019;106:46–56. doi: 10.1016/j.biocel.2018.09.015. [DOI] [PubMed] [Google Scholar]
- 52.Hu H., Yang L., Li L., Zeng C. Long non-coding RNA KCNQ1OT1 modulates oxaliplatin resistance in hepatocellular carcinoma through miR-7-5p/ ABCC1 axis. Biochem Biophys Res Commun. 2018;503(4):2400–2406. doi: 10.1016/j.bbrc.2018.06.168. [DOI] [PubMed] [Google Scholar]
- 53.Zhang J.F., He M.L., Fu W.M., Wang H., Chen L.Z., Zhu X. Primate-specific microRNA-637 inhibits tumorigenesis in hepatocellular carcinoma by disrupting signal transducer and activator of transcription 3 signaling. Hepatology. 2011;54(6):2137–2148. doi: 10.1002/hep.24595. [DOI] [PubMed] [Google Scholar]
- 54.Yu H., Yin S., Zhou S., Shao Y., Sun J., Pang X. Magnolin promotes autophagy and cell cycle arrest via blocking LIF/Stat3/Mcl-1 axis in human colorectal cancers. Cell Death Dis. 2018;9(6):702. doi: 10.1038/s41419-018-0660-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spitzner M., Roesler B., Bielfeld C., Emons G., Gaedcke J., Wolff H.A. STAT3 inhibition sensitizes colorectal cancer to chemoradiotherapy in vitro and in vivo. Int J Cancer. 2014;134(4):997–1007. doi: 10.1002/ijc.28429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sinha S., Levine B. The autophagy effector Beclin 1: a novel BH3-only protein. Oncogene. 2008;27(Suppl 1):S137–S148. doi: 10.1038/onc.2009.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jin P., Huang Y., Zhu P., Zou Y., Shao T., Wang O. CircRNA circHIPK3 serves as a prognostic marker to promote glioma progression by regulating miR-654/IGF2BP3 signaling. Biochem Biophys Res Commun. 2018;503(3):1570–1574. doi: 10.1016/j.bbrc.2018.07.081. [DOI] [PubMed] [Google Scholar]
- 58.Liu N., Zhang J., Zhang L.Y., Wang L. CircHIPK3 is upregulated and predicts a poor prognosis in epithelial ovarian cancer. Eur Rev Med Pharmacol Sci. 2018;22(12):3713–3718. doi: 10.26355/eurrev_201806_15250. [DOI] [PubMed] [Google Scholar]
- 59.Shibata M., Yoshimura K., Furuya N., Koike M., Ueno T., Komatsu M. The MAP1-LC3 conjugation system is involved in lipid droplet formation. Biochem Biophys Res Commun. 2009;382(2):419–423. doi: 10.1016/j.bbrc.2009.03.039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.