Abstract
Objective
To develop a deep convolutional neural network (DCNN) that can automatically detect laryngeal cancer (LCA) in laryngoscopic images.
Methods
A DCNN-based diagnostic system was constructed and trained using 13,721 laryngoscopic images of LCA, precancerous laryngeal lesions (PRELCA), benign laryngeal tumors (BLT) and normal tissues (NORM) from 2 tertiary hospitals in China, including 2293 from 206 LCA subjects, 1807 from 203 PRELCA subjects, 6448 from 774 BLT subjects and 3191 from 633 NORM subjects. An independent test set of 1176 laryngoscopic images from other 3 tertiary hospitals in China, including 132 from 44 LCA subjects, 129 from 43 PRELCA subjects, 504 from 168 BLT subjects and 411 from 137 NORM subjects, was applied to the constructed DCNN to evaluate its performance against experienced endoscopists.
Results
The DCCN achieved a sensitivity of 0.731, a specificity of 0.922, an AUC of 0.922, and the overall accuracy of 0.867 for detecting LCA and PRELCA among all lesions and normal tissues. When compared to human experts in an independent test set, the DCCN’ s performance on detection of LCA and PRELCA achieved a sensitivity of 0.720, a specificity of 0.948, an AUC of 0.953, and the overall accuracy of 0.897, which was comparable to that of an experienced human expert with 10–20 years of work experience. Moreover, the overall accuracy of DCNN for detection of LCA was 0.773, which was also comparable to that of an experienced human expert with 10–20 years of work experience and exceeded the experts with less than 10 years of work experience.
Conclusions
The DCNN has high sensitivity and specificity for automated detection of LCA and PRELCA from BLT and NORM in laryngoscopic images. This novel and effective approach facilitates earlier diagnosis of early LCA, resulting in improved clinical outcomes and reducing the burden of endoscopists.
Research in context
Evidence before this study
Laryngeal cancer (LCA) is the most common form of malignant tumor of the head and neck region. The prognosis of patients with LCA depends on the cancer stage at diagnosis. Although endoscopy is the standard procedure for diagnosis of LCA, detection of LCA at an early stage depends on the experience of the endoscopists with routine white-light endoscopy. Recently, endoscopic systems with narrow band imaging (NBI), which enhances the visualization of epithelial and subepithelial microvascular patterns, achieves both a high sensitivity and specificity in early detection of LCA. However, advanced magnifying endoscopes, specific training time and well-trained endoscopists limits the clinical application of NBI endoscopy, especially in low-income areas and countries.
Added value of this study
In the present study, we developed and validated a deep convolutional neural network (DCNN) that can automatically detect LCA in laryngoscopic images. To the best of our knowledge, this is the first study that uses a deep learning algorithm to detect LCA with laryngoscopic images. The well-trained DCNN's performance in diagnosing LCA was comparable to that of a human expert with significant clinical experience (20 years of experience) in laryngeal diseases.
Implications of the available evidence
This novel and effective approach has great potential for raising the diagnosis rates of early LCA, reducing the burden of endoscopists, and telemedicine in regions and countries where there are shortage of advanced endoscopes and experienced endoscopists.
1. Introduction
Laryngeal cancer (LCA) is the most frequent and predominant malignant tumor of the head and neck region. Treatment outcomes of LCA at an early stage are favorable, by which 5-year-survival rate of patients with Tis, T1, T2 LCA ranges from 80 to 90% [1]. Although endoscopy is the primary tool for detection of LCA in clinical practice, endoscopy with conventional white light is limited in both resolution and contrast, which may results in overlook or misdiagnosis of superficial mucosal cancer and its precursor lesions, even by experienced endoscopists [2]. On the contrary, false cancer detection and unnecessary biopsy are other major problems in clinical practice, which is due to a natural anxiety of endoscopists to avoid overlooking early cancer [3]. Therefore, most patients receive their diagnoses at an advanced stage and often suffer loss of vocal function causing deterioration of the quality of life.
Recently, endoscopic systems with narrow band imaging (NBI), which enhances the visualization of epithelial and subepithelial microvascular patterns, play a critical role in early detection of LCA with a high sensitivity of 88.9–97.0% at a high specificity of 84.6–96.0% [[4], [5], [6], [7], [8]]. Nevertheless, the use of NBI for diagnosis requires advanced magnifying endoscopes, specific training time and experienced endoscopists, which limits the clinical application of NBI endoscopy in many developing countries, including China. Therefore, the use of traditional white light and non-magnifying images for LCA diagnosis is practically meaningful and even crucial for less developed regions or countries which are faced with a shortage of advanced imaging endoscopes and experienced endoscopists.
Due to the particular physiological structures and characteristics, it is usually difficult for human eyes to capture insignificant laryngeal cancer lesions from non-magnified endoscopy. Fortunately, deep convolutional neural network (DCNN) based machine learning (or called deep learning) techniques have recently demonstrated remarkable abilities in diagnosing a variety of diseases, such as skin cancers and diabetic retinopathy [9,10]. Fed with a large number of manually-labeled images of the target diseases for training, a DCNN model is learned via certain optimization algorithms, which, at the testing stage, can automatically predict the category label of a given test image [11,12]. Benefitted from the strong feature representation capability of DCNN as well as the use of large datasets for training, the learned DCNN model can be generalized well to unseen testing images, achieving comparable or even higher classification accuracy than experienced human experts.
Inspired by the success of previous works that detection and classification of skin and retinal diseases was performed by image recognition through DCNN [9,13,14], in the present study, we assumed the clinical diagnosis of LCA could also benefit from deep learning techniques. Towards this end, we acquired a large collection of laryngoscopic images to build a DCNN model and assessed its performance.
2. Materials and methods
2.1. Data preparation
Our raw laryngoscopic images were collected from the clinical cases at five tertiary hospitals in China, including Sun Yat-sen Memorial Hospital of Sun Yat-sen University, the Third Affiliated Hospital of Sun Yat-sen University, Peking University Shenzhen Hospital, Puning People's Hospital and Taizhou First People‘s Hospital. These images were captured using standard endoscopes (ENF-V2, Olympus Medical Systems Corp., Tokyo, Japan) and endoscopic video systems (OTV-S7Pro; Olympus Medical Systems Corp., Tokyo, Japan) with white light source (CLV-S40Pro; Olympus Medical Systems Corp., Tokyo, Japan). We considered the diagnosis of four classes of human subjects in this study, including LCA, precancerous laryngeal lesions (PRELCA), benign laryngeal tumors (BLT) and normal tissues (NORM). PRELCA were defined as keratosis with various degrees of dysplasia based on histological diagnoses. An experienced endoscopist from Sun Yat-sen Memorial Hospital manually scanned the raw images to exclude the ones out of focus or of low quality, and selected 5 to 10 images captured from different perspectives for each human subject. After the manual scan, we finally retained three independent sets of images, referred to as DS1, DS2 and DS3 for the clarity of presentation. DS1 had 10,892 images from 1451 subjects at Sun Yat-sen Memorial Hospital, including 1776 from 164 LCA subjects, 1476 from 162 PRELCA subjects, 5127 from 619 BLT subjects and 2513 from 506 NORM subjects. DS2 had 2829 images from 365 subjects at the Third Affiliated Hospital of Sun Yat-sen University, including 517 from 42 LCA subjects, 331 from 41 PRELCA subjects, 1321 from 155 BLT subjects and 660 from 127 NORM subjects. DS3 was another smaller set of 1200 laryngoscopic images from 407 subjects (132 from 51 LCA subjects, 153 from 51 PRELCA subjects, 504 from 168 BLT subjects and 411 from 137 NORM subjects) at other 3 hospitals (Peking University Shenzhen Hospital, Puning People's Hospital and Taizhou First People‘s Hospital). A summary of the image sets was provided in Table 1. We used these three image sets to construct and evaluate our DCNN, and compare the performance against experienced endoscopitsts with various work years (see Section 2.2.4 for details of evaluation protocols). It was noted that all the images in our dataset were biopsy-proven and thereby had highly reliable class labels, which was crucial to our study.
Table 1.
Details of the image sets used for experiments.
| Image sets | LCA | PRELCA | BLT | NORM | Total |
|---|---|---|---|---|---|
| DS1 | 1776 (164) | 1476 (162) | 5127 (619) | 2513 (506) | 10,892 (1451) |
| DS2 | 517 (42) | 331 (41) | 1321 (155) | 660 (127) | 2829 (365) |
| DS1 + DS2 | 2293 (206) | 1807 (203) | 6448 (774) | 3191 (633) | 13,721 (1816) |
| DS3 | 132 (44) | 129 (43) | 504 (168) | 411 (137) | 1176 (392) |
| Total | 2425 (250) | 1936 (246) | 6952 (942) | 3602 (770) | 14,897 (2208) |
2.2. Methods
2.2.1. Deep learning architecture
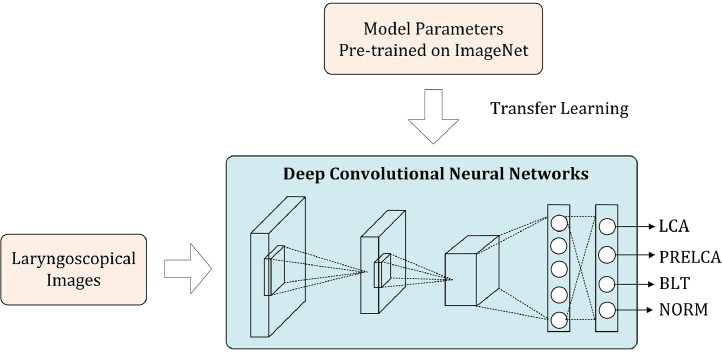
Despite the availability of a large number of laryngoscopic images, these images were still insufficient to train a high-quality DCNN model from scratch. In such a case, an effective scheme is the so-called transfer learning, which resorts to external data for boosting the model training over the target data. More specifically, as illustrated in Fig. 1, we adopt a DCNN model pre-trained on the ImageNet dataset, which consists of over one million natural images belonging to 1000 visual object categories, to initialize the DCNN model to be learned. Since we were dealing with a four-class problem (instead of 1000 classes), we needed firstly to adapt the pre-trained network by modifying the last softmax layer to be one with four output nodes. Then, during the training, we used our own laryngoscopic image dataset to fine-tune the pre-trained DCNN model.
Fig. 1.
Overview of the deep learning architecture. Parameters pre-trained on the external ImageNet dataset are used to initialize the deep convolutional neural network, which is then fine-tuned on the target dataset.
2.2.2. Training details
We adopt the GoogLeNet Inception v3 network as the backbone network. Data augmentation was performed at the factor of 720, and the cross-entropy loss was used as the loss function. We used the SGD optimizer with the following parameter settings: batch_size = 64, learning_rate = 0.001, decay = 0.9, momentum = 0, epillon = 1e-10. During the fine-tuning, we progressively unfroze the parameters from back to front layers. All the experiments throughout this study were carried out on a deep learning workstation with 4 Titan XP 12 GB GPU.
2.2.3. Attention map generation
To interpret the learned model intuitively, we calculated saliency maps for the test images using the method. The pixel-wise values in a saliency map reflected the contributions of the corresponding image pixels to the classification for a specific class, which was quantified by the gradients of the class-specific loss with respect to the input pixels. We visualized a saliency map by using a heat map overlaid upon the input image to observe whether the salient regions in the saliency maps indeed correspond to the ROI for decision making.
2.2.4. Evaluation protocols
We carried out two experiments to evaluate our DCNN model on the three image sets DS1, DS2 and DS3. First, we took DS1 as the training set to construct the DCNN model and generally evaluated its performance on DS2. Second, we took DS1 and DS2 together as the training set to train the DCNN model, and compare its performance against human endoscopitsts as DS3. Note that such settings can guarantee the independency between the training set and the testing set. For each setting, we were concerned with the ability of the model in making both 4-class and binary predictions. Here by binary prediction we meant the judgment between Urgent versus Non-urgent cases, which the former included LCA and PRELCA, and the latter included BLT and NORM. Binary classification scores were generated by combining the scores of LCA and PRELCA into Urgent cases and those of BLT and NORM into Non-urgent cases.
2.2.5. Performance metrics
We evaluated the effectiveness of our model by using several different metrics.
2.2.5.1. Sensitivity-specificity curve
For a particular category, when a testing image is fed into a learned DCNN network, it can output the probability of the image belonging to this categoryp, and one can make a hard binary classification by thresholding this probabilityp ≥ twheretis a threshold value. Over the whole testing set, one can calculate a population-level sensitivity and specificity defined by
where TP,TN, FP and FN are the numbers of true positives, true negatives, false positives and false negatives. By varying the threshold valuet, a sensitivity-specificity curve can be generated.
2.2.6. Confusion matrix
A confusion matrix can be simply obtained by comparing the predicted results with the ground truth labels, and counting the correct and incorrect predictions in each class.
2.2.7. Overall accuracy
The overall accuracy is the ratio between the number of correctly categorized images and the total number of testing images, that is,
2.3. Ethics
The study was approved by the ethical review board of Sun Yat-sen Memorial Hospital, Sun Yat-sen University.
3. Results
3.1. Performance of model
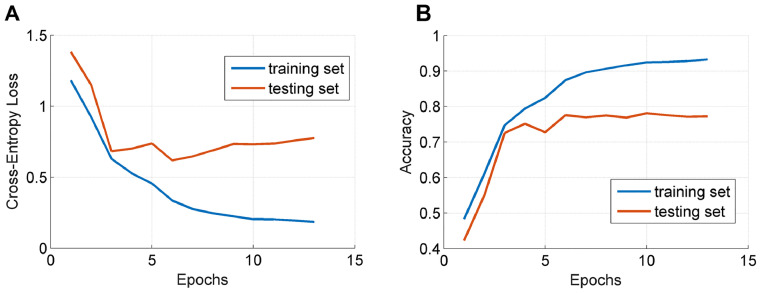
We first took DS1 as the training set and DS2 as the testing set to evaluate the DCNN model. As previously stated, our model was trained on the training set following a transfer learning scheme. An example demonstrating the changes of the loss function value and the classification accuracy over the training and testing sets respectively were depicted in Fig. 2. As can be observed, the model can converge after running the optimization for a number of epochs.
Fig. 2.
Illustration of the changes of the loss function value (A) and the classification accuracy (B) over the training and testing sets.
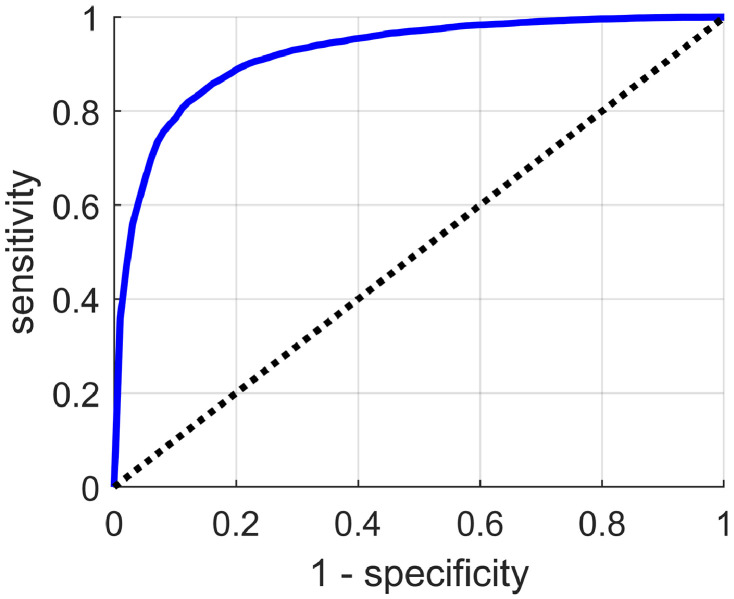
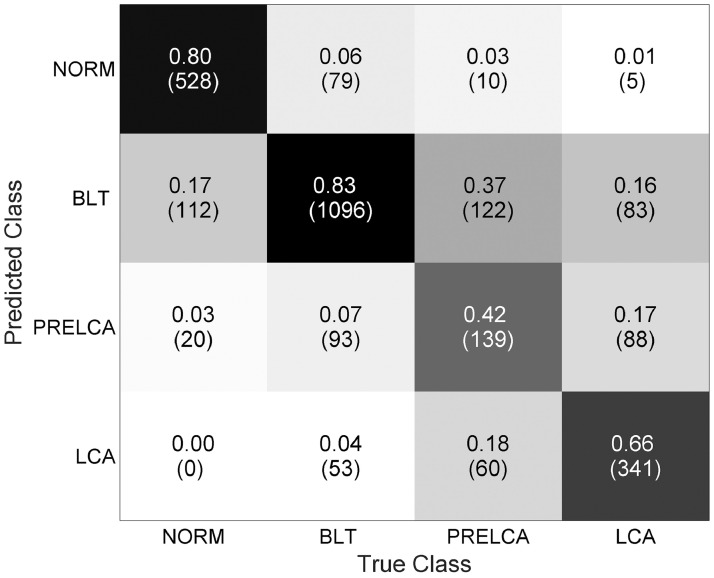
We assessed the model in terms of its ability in making binary predictions on Urgent versus Non-urgent subjects. Clinically, such a binary classification task makes great sense because urgent cases demand immediate treatment, and any delay, caused by misclassification for instance, will increase the risk of death. The Urgent versus Non-urgent performance in terms of the sensitivity-specificity curve was plotted in Fig. 3. Our model overall can achieve a sensitivity of 0.731, a specificity of 0.922, an AUC of 0.922, and the overall accuracy of 0.867. We also reported the confusion matrix of the original four-class classifiers in Fig. 4, where the overall accuracy was 0.745.
Fig. 3.
The sensitivity-specificity curve for Urgent versus Non-urgent binary classification.
Fig. 4.
Confusion matrix for 4-class categorization.
3.2. Comparison with human experts
To further validate the effectiveness of our algorithm, we compared its performance against human experts. Towards this end, we took DS1 and DS2 together as the training set to train the DCNN model and DS3 as the testing set. We chose three endoscopists with different work experience levels (~3, 3–10 and 10–20 years of experience, respectively) in laryngeal diseases diagnosis from Sun Yat-sen Memorial Hospital, who were given the laryngoscopic images in the testing set and instructed to independently classify these images into the four classes aforementioned, without using any other additional information.
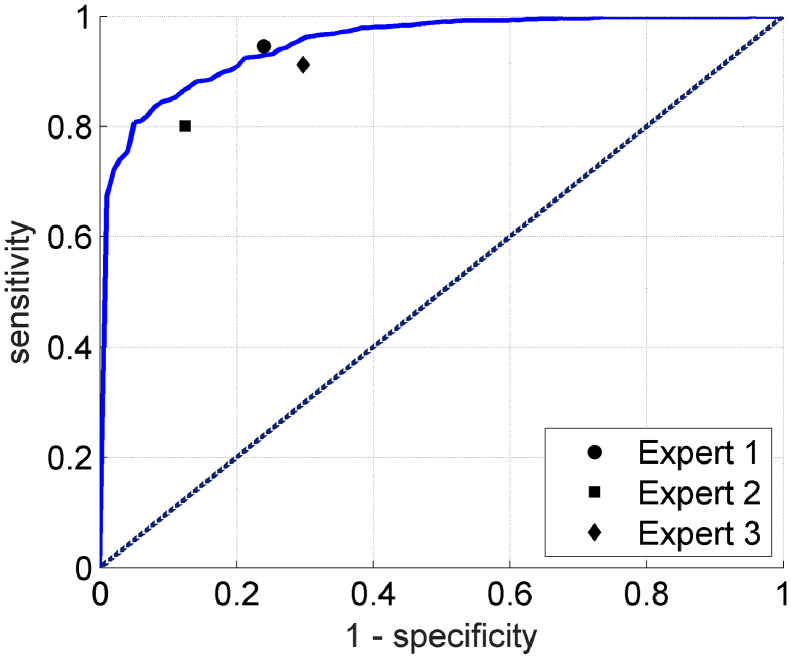
We first compared the performance of our algorithm and human experts in making Urgent versus Non-urgent binary predictions. Similar to the evaluation protocol in Section 2.2.4, for human experts we also combined the 4-class decisions into two classes by counting Urgent cases for LCA or PRELCA, and Non-urgent cases for BTL or NORM. The results were shown in Fig. 5, which demonstrated that our algorithm can perform significantly better than two (~3 and 3–10 years of experience) out of the three experts and comparably to the other expert with 10–20 years of experience. More precisely, the DCNN model can achieve a sensitivity of 0.720, a specificity of 0.948, an AUC of 0.953, and the overall accuracy of 0.897.
Fig. 5.
Comparison between the deep learning algorithm and three human experts in Urgent versus Non-urgent binary classification. Expert 1: expert with 10–20 years of experience. Expert 2: expert with ~3 years of experience. Expert 3: expert with 3–10 years of experience.
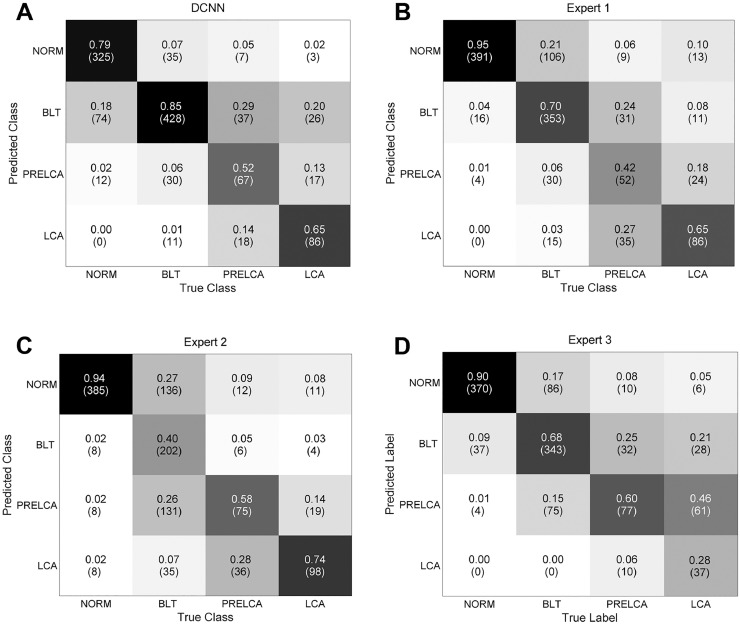
We also compared our algorithm against the human experts in terms of the confusion matrices of four-class classification as shown in Fig. 6, where the overall accuracy of the model was 0.773 while that of experts was 0.750 (10–20 years of experience), 0.647 (~3 years of experience) and 0.704 (3–10 years of experience) respectively.
Fig. 6.
Four-class confusion matrices obtained by our DCNN model (A) and the three human experts (B-D). Expert 1: expert with 10–20 years of experience. Expert 2: expert with ~3 years of experience. Expert 3: expert with 3–10 years of experience.
3.3. Comparison with existing works
To our knowledge, this is the first study which deploys deep learning methods to the task of laryngoscopic image classification. Therefore, we chose the representative traditional method presented by Verikas and his colleagues [15] for comparison. This approach adopts multiple hand-crafted features, including color, texture, frequency-domain characteristics, et al., and exploits the SVM classifier for classification. Identical to the setting above, i.e., taking DS1 and DS2 together as the training set and DS3 as the testing set, the results were reported in Table 2, which demonstrated DCNN overwhelmingly outperformed the method that Verikas and his colleagues utilized.
Table 2.
Summary of the quantitative experimental results.
| Evaluation settings/Methods | 2-Class |
4-Class |
|||
|---|---|---|---|---|---|
| Sensitivity | Specificity | Accuracy | Accuracy | ||
| DS1 training, DS2 testing | DCNN | 0.731 | 0.922 | 0.867 | 0.745 |
| DS1 + DS2 training, DS3 testing | DCNN | 0.720 | 0.948 | 0.897 | 0.773 |
| Verikas et al. [20] | 0.603 | 0.820 | 0.651 | 0.460 | |
| Expert 1 | 0.761 | 0.946 | 0.906 | 0.750 | |
| Expert 2 | 0.875 | 0.801 | 0.817 | 0.647 | |
| Expert 3 | 0.702 | 0.902 | 0.858 | 0.704 | |
3.4. Attention maps
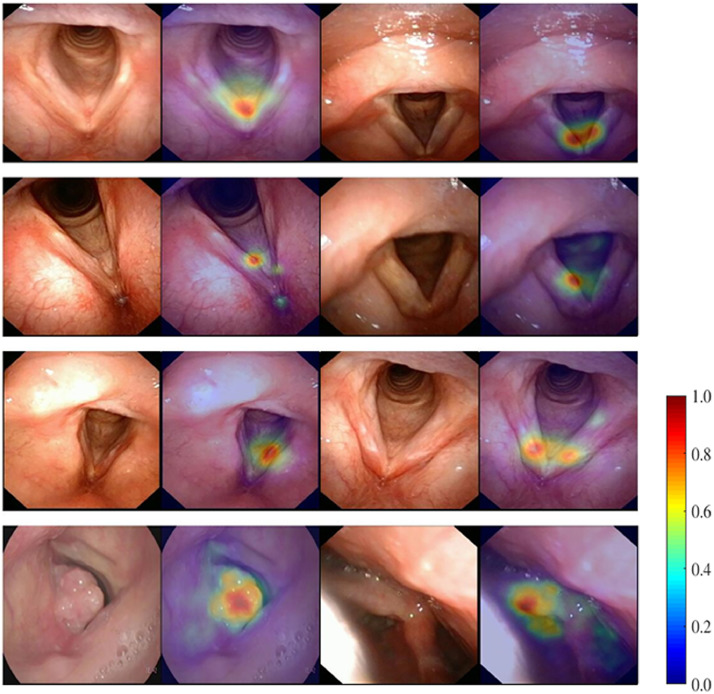
Attention map is a common way to visualize the contribution of each pixel in the image to the classification. Some typical attention maps of the four classes were visualized in Fig. 7. Ideally, if a DCNN model is reasonably trained, the attention map should be localized to lesions or other key structures which contribute to decision-making, which was the case for our model as can be observed in Fig. 7.
Fig. 7.
Representative attention maps obtained by the DCNN model on the classes of NORM, BLT, PRELCA and LCA from top to bottom respectively. Attention maps are displayed as heat maps overlay upon the original images, where warmer colors indicate higher saliency, i.e., higher contribution to the classification decision.
4. Discussion
In this study, we developed and validated a deep learning algorithm through DCNN for the diagnosis of LCA. To the best of our knowledge, this is the first study that uses a deep learning algorithm to detect laryngeal cancer with laryngoscopic images. Our results indicate that a deep learning algorithm through DCNN can be trained to differentiate LCA from multiple benign or nonneoplastic lesions in laryngoscopic images with high sensitivity and high specificity. Strikingly, our deep learning algorithm's performance in diagnosing LCA was comparable to that of a human expert with significant clinical experience (20 years of experience) with laryngeal diseases. Therefore, this deep-learning algorithm for the detection of LCA offers many advantages, such as consistency of interpretation, high sensitivity and specificity, and high rate of speed.
Deep learning is a novel and promising avenue of machine learning, which allows machines to analyze multiple images for training and extract specific features via a back-propagation algorithm [16]. After training, machines could analyze and recognize newly acquired images prospectively, especially through DCNN. DCNN has been developed as an effective class of models for understanding image content, giving advanced results on image recognition, segmentation, detection and retrieval [17,18]. The key points behind these results are techniques for scaling up the networks to millions of parameters and big labeled datasets which can support the learning process. Under such conditions, DCNN learn powerful and interpretable image features and have been widely employed as an excellent technology for image recognition and classification [19,20]. Recently, image recognition by deep learning through DCNN has been applied to detection and classification of multiple diseases increasingly. Gulshan and his colleagues reported a sensitivity of 97.5% and a specificity of 93.4% for detecting referable diabetic retinopathy [13]. A study by Hirasawa and his colleagues reported an overall sensitivity of 92.2% for detecting gastric cancer lesions [16]. In a recent deep learning competition for classification of skin cancer, deep learning through DCNN achieved performance on par with 21 board-certified dermatologists across tasks that differentiate keratinocyte carcinomas, seborrheic keratoses, malignant melanomas and benign nevi [9]. The present study extends such work by using deep learning through DCNN to generate an algorithm with a sensitivity of 0.731 and a specificity of 0.922 for LCA and PRELCA detection. Even for four-class classifiers, DCNN also achieved an overall accuracy of 0.745. Then, we compared the performance of this well-trained DCNN with experienced endoscopists on detection of different laryngeal lesions in laryngoscopic images. Strikingly, the DCNN demonstrated an artificial intelligence capable of detecting laryngeal cancer with a level of competence comparable to an experienced endoscopist with work experience of 10–20 years. Therefore, we believe that such a well-trained DCNN has a great potential to assist those less-experienced endoscopists from rural areas or developing countries in the earlier diagnosis of LCA.
There are limitations to this DCNN system. First, although the DCNN achieved excellent overall performance comparable to the experienced human expert with work experience of 10–20 years, the prediction accuracy of the DCNN for LCA and PRELCA was relatively low. In addition, LCA and PRELCA were also difficult to be differentiated from each other by the DCNN according to the relatively low sensitivity. Similarly, even experienced endoscopists were bothered by the low accuracy of early detection of LCA and PRELCA in clinic practice as well through routine white-light endoscopy, especially for those intraepithelial and submucosal changes of the lesions [2,6]. Therefore, performance of current trained DCNN is inferior to that of NBI in detection of LCA and PRELCA, because the latter has a relatively high sensitivity and specificity in early detection of LCA by highlighting microsurface patterns and microvascular morphologies on the mucosal surface [2,5,8]. However, NBI is not always available in most rural areas of developing countries due to high costs of the equipments. Moerover, LCA usually grade-increasing progresses gradually from a low-grade dysplasia to a high-grade intraepithelial neoplasia and finally develops carcinoma in situ. The slow progression is considered to last several years and those small LCA cases that were missed at initial screening would be detected when performing annual endoscopy. Therefore the clinical applicability of the DCNN in detection of LCA and PRELCA might not be considerably compromised. Furthermore, we believe the performance of DCNN in detection of LCA or PRELCA can be further improved by simply increasing training images numbers or using better algorithm design. For instance, one can introduce an alignment module at the front of the pipeline, or an attention mechanism into the deep network for improvement. Second, the DCNN in the present study was only trained to differentiate 4 classes of laryngeal lesions and normal tissues. In the future study, it is essential to evaluate the performance in a real-world, clinical setting, for validating this technique across the full distribution and spectrum of laryngeal lesions encountered in typical practice. Third, all tested laryngoscopic images were obtained by the same type of standard endoscopes and endoscopic video systems. Further study should test the performance of DCNN in detection of lesions in laryngoscopic images of different quality from various endoscopic devices.
5. Conclusion
In summary, we developed an endoscopic image based-deep learning algorithm through DCNN which had high sensitivity and specificity for automated detection of LCA. This novel and effective approach holds the potential for substantial clinical impact, including improving the diagnosis rates of early LCA, reducing the burden of endoscopists, and telemedicine in regions and countries where there are shortage of advanced endoscopes and experienced endoscopists.
Author contributions
Hao Xiong, Peiliang Lin, Jin-Gang Yu, Yuanqing Li, Yiqing Zheng and Haidi Yang: designed the study, collected the data, analyzed the data and wrote the manuscript. Jin Ye, Lichao Xiao, Yuan Tao, Zebin Jiang, Wei Lin, Mingyue Liu, Jingjing Xu, Wenjie Hu, Yuewen Lu, and Huaifeng Liu collected the data. All authors read and approved the final version of the manuscript.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Acknowledgments
This work was supported by National Key R&D Program of China (2017YFB1002505), Guangzhou Science and Technology Program (201904010299 and 201903010088) and National Natural Science Foundation of China (81570916, 81771018, 61703166 and 81873699, 81970887).
Contributor Information
Yuanqing Li, Email: auyqli@scut.edu.cn.
Yiqing Zheng, Email: zhengyiq@mail.sysu.edu.cn.
Haidi Yang, Email: yanghd@mail.sysu.edu.cn.
References
- 1.Marioni G., Marchese-Ragona R., Cartei G., Marchese F., Staffieri A. Current opinion in diagnosis and treatment of laryngeal carcinoma. Cancer Treat Rev. 2006;32:504–515. doi: 10.1016/j.ctrv.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 2.Ni X.G., Zhang Q.Q., Wang G.Q. Narrow band imaging versus autofluorescence imaging for head and neck squamous cell carcinoma detection: a prospective study. J Laryngol Otol. 2016;130:1001–1006. doi: 10.1017/S0022215116009002. [DOI] [PubMed] [Google Scholar]
- 3.Barbalata C., Mattos L.S. Laryngeal tumor detection and classification in endoscopic video. IEEE J Biomed Health Inform. 2016;20:322–332. doi: 10.1109/JBHI.2014.2374975. [DOI] [PubMed] [Google Scholar]
- 4.Dai Z.H., Chen L.B. Yi chuan = Hereditas. vol. 32. 2010. The impact of microRNAs on the evolution of metazoan complexity; pp. 105–114. [DOI] [PubMed] [Google Scholar]
- 5.Kraft M., Fostiropoulos K., Gurtler N., Arnoux A., Davaris N., Arens C. Value of narrow band imaging in the early diagnosis of laryngeal cancer. Head Neck. 2016;38:15–20. doi: 10.1002/hed.23838. [DOI] [PubMed] [Google Scholar]
- 6.De Vito A., Meccariello G., Vicini C. Narrow band imaging as screening test for early detection of laryngeal cancer: a prospective study. Clin Otolaryngol. 2017;42:347–353. doi: 10.1111/coa.12728. [DOI] [PubMed] [Google Scholar]
- 7.Sun C., Han X., Li X., Zhang Y., Du X. Diagnostic performance of narrow band imaging for laryngeal Cancer: a systematic review and meta-analysis. Otolaryngol Head Neck Surg. 2017;156:589–597. doi: 10.1177/0194599816685701. [DOI] [PubMed] [Google Scholar]
- 8.Yang Y., Liu J., Song F., Zhang S. The clinical diagnostic value of target biopsy using narrow-band imaging endoscopy and accurate laryngeal carcinoma pathologic specimen acquisition. Clin Otolaryngol. 2017;42:38–45. doi: 10.1111/coa.12654. [DOI] [PubMed] [Google Scholar]
- 9.Esteva A., Kuprel B., Novoa R.A., Ko J., Swetter S.M., Blau H.M. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sempere L.F., Cole C.N., McPeek M.A., Peterson K.J. The phylogenetic distribution of metazoan microRNAs: insights into evolutionary complexity and constraint. J Exp Zool B Mol Dev Evol. 2006;306:575–588. doi: 10.1002/jez.b.21118. [DOI] [PubMed] [Google Scholar]
- 11.Rose J.M., Novoselov S.S., Robinson P.A., Cheetham M.E. Molecular chaperone-mediated rescue of mitophagy by a parkin RING1 domain mutant. Hum Mol Genet. 2011;20:16–27. doi: 10.1093/hmg/ddq428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khosravi P., Kazemi E., Imielinski M., Elemento O., Hajirasouliha I. Deep convolutional neural networks enable discrimination of heterogeneous digital pathology images. EBioMedicine. 2018;27:317–328. doi: 10.1016/j.ebiom.2017.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gulshan V., Peng L., Coram M., Stumpe M.C., Wu D., Narayanaswamy A. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316:2402–2410. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 14.Kermany D.S., Goldbaum M., Cai W., Valentim C.C.S., Liang H., Baxter S.L. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172:1122–1131. doi: 10.1016/j.cell.2018.02.010. [e1129] [DOI] [PubMed] [Google Scholar]
- 15.Verikas A., Gelzinis A., Valincius D., Bacauskiene M., Uloza V. Multiple feature sets based categorization of laryngeal images. Comput Methods Prog Biomed. 2007;85:257–266. doi: 10.1016/j.cmpb.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 16.Hirasawa T., Aoyama K., Tanimoto T., Ishihara S., Shichijo S., Ozawa T. Application of artificial intelligence using a convolutional neural network for detecting gastric cancer in endoscopic images. Gastric Cancer. 2018;21:653–660. doi: 10.1007/s10120-018-0793-2. [DOI] [PubMed] [Google Scholar]
- 17.Liu Z., Gao J., Yang G., Zhang H., He Y. Localization and classification of paddy field pests using a saliency map and deep convolutional neural network. Sci Rep. 2016;6 doi: 10.1038/srep20410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chmelik J., Jakubicek R., Walek P., Jan J., Ourednicek P., Lambert L. Deep convolutional neural network-based segmentation and classification of difficult to define metastatic spinal lesions in 3D CT data. Med Image Anal. 2018;49:76–88. doi: 10.1016/j.media.2018.07.008. [DOI] [PubMed] [Google Scholar]
- 19.Hay E.A., Parthasarathy R. Performance of convolutional neural networks for identification of bacteria in 3D microscopy datasets. PLoS Comput Biol. 2018;14 doi: 10.1371/journal.pcbi.1006628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strodthoff N., Strodthoff C. Detecting and interpreting myocardial infarction using fully convolutional neural networks. Physiol Meas. 2018;40 doi: 10.1088/1361-6579/aaf34d. [DOI] [PubMed] [Google Scholar]