Abstract
Cercospora kikuchii (Tak. Matsumoto & Tomoy.) M.W. Gardner 1927 is an ascomycete fungal pathogen that causes Cercospora leaf blight and purple seed stain on soybean. Here, we report the first draft genome sequence and assembly of this pathogen. The C. kikuchii strain ARG_18_001 was isolated from soybean purple seed collected from San Pedro, Buenos Aires, Argentina, during the 2018 harvest. The genome was sequenced using a 2 × 150 bp paired-end method by Illumina NovaSeq 6000. The C. kikuchii protein-coding genes were predicted using FunGAP (Fungal Genome Annotation Pipeline). The draft genome assembly was 33.1 Mb in size with a GC-content of 53%. The gene prediction resulted in 14,856 gene models/14,721 protein coding genes. Genomic data of C. kikuchii presented here will be a useful resource for future studies of this pathosystem. The data can be accessed at GenBank under the accession number VTAY00000000 https://www.ncbi.nlm.nih.gov/nuccore/VTAY00000000.
Keywords: Cercospora kikuchii, Draft genome, Next generation sequencing (NGS), Cercospora leaf blight (CLB), Purple seed stain (PSS), Agriculture, Bioinformatics, Fungal pathogens
Specifications table
| Subject | Biology |
| Specific subject area | Bioinformatics (Genomics) |
| Type of data | Raw sequencing reads, draft genome assembly, gene prediction and phylogenetic position of C. kikuchii strain ARG_18_001 |
| How data were acquired | Whole genome sequencing was performed using an Illumina NovaSeq 6000 sequencing system |
| Data format | Raw sequencing reads, draft genome assembly and gene prediction |
| Parameters for data collection | Reads were filtered and merged with Trimmomatic (v 0.39) and FLASH (v 1.2.11). The genome was assembled with Celera Assembler (v 8.3) and Spades (v 3.11.1). Gene prediction was performed with FunGAP (v 1.0.1), tRNAscan-SE (v 2.0.3), rnammer (v 1.2) and mfannot (v 1.35). Protein-coding gene annotation was performed with hmmsearch (v 3.1b2), ncbi-blast (v 2.2.25+) and Blast2GO (v 2.5) using the ragp R package (v 0.3.0.0001). RepeatMasker (v 4.0.9) was used to identify and filter repetitive regions. |
| Description of data collection | Strain ARG_18_001 was isolated from soybean seeds of variety DM62R63 sampled during the 2018 harvest that exhibited symptoms of purple seed stain. |
| Data source location | Samples were originally collected from Gobernador Castro, San Pedro, Buenos Aires, Argentina (33°39′26.37″S, 59°49′36.00″O) |
| Data accessibility | This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession VTAY00000000 https://www.ncbi.nlm.nih.gov/nuccore/VTAY00000000. The version described in this paper is version VTAY00000000.1 https://www.ncbi.nlm.nih.gov/nuccore/VTAY00000000. |
Value of the Data
|
1. Data
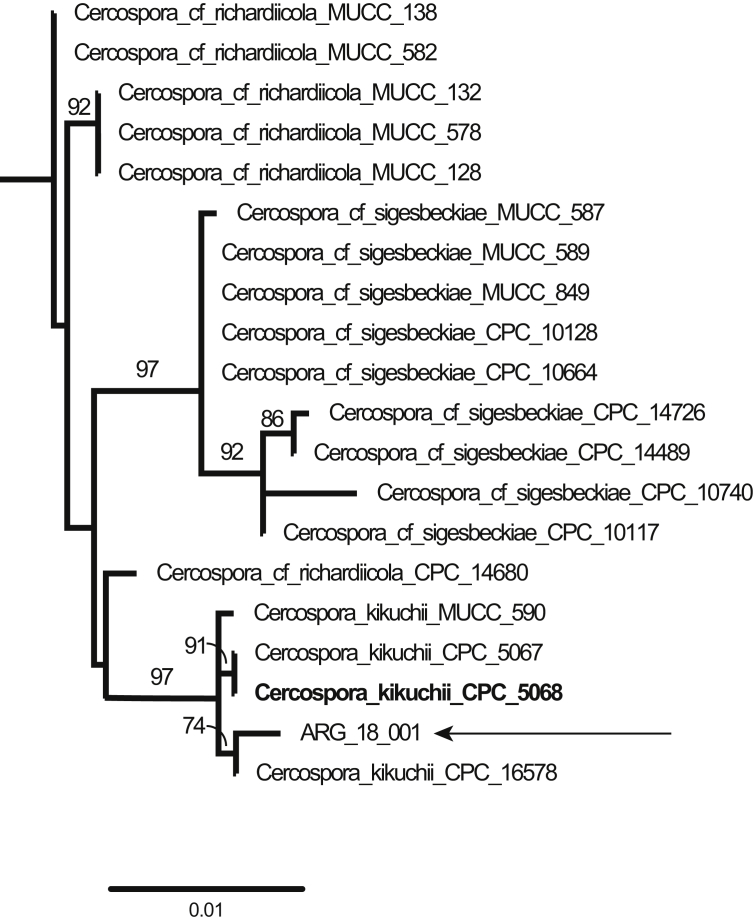
We present the draft genome assembly and gene prediction of the fungus C. kikuchii, causal agent of Cercospora leaf blight (CLB) and purple seed stain (PSS) of soybean. Recently, multi-locus phylogenetic studies confirmed that CLB and PSS is a disease complex caused by several Cercospora species. Phylogenetic analyses of cercosporoid fungi isolated from infected soybean in Argentina, Brazil and the USA determined that the species C. kikuchii, C. cf. flagellaris and C. cf. sigesbeckiae are causal agents of these diseases [1,2]. More recently, C. cf. nicotianae isolated from soybean leaves in Bolivia has been identified as a species in association with CLB [3]. A maximum-likelihood phylogenetic tree of Cercospora species was inferred in RAxML using seven nuclear loci, with data from isolate ARG_18_001 sliced from the genome assembly. The strain ARG_18_001 nested within the clade that includes other isolates of C. kikuchii, including the ex-type, with 97% bootstrap support (Fig. 1).
Fig. 1.
Subtree from a maximum-likelihood phylogenetic analysis of Cercospora species. The complete phylogeny was inferred in RAxML assuming the GTRGAMMA model by integrating data sliced from the genome of ARG_18_001 with the following seven loci from 379 other isolates in Groenewald et al. (2013) and Bakhshi et al. (2018): actA, cmdA, nrITS, gapdh, histone 3, tef1-alpha, and tub2. The subtree that includes ARG_18_001 was pruned from the rest of the tree for ease of reference. Branches are labeled with bootstrap support values ≥ 70%. Bold font indicates the placement of the ex-type of C. kikuchii. Arrow indicates the placement of the isolate for which the genome was sequenced. The scale bar indicates the estimated number of substitutions per site.
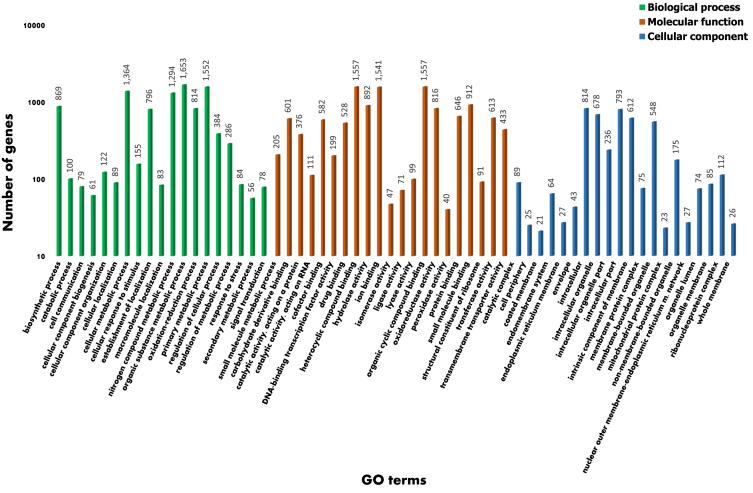
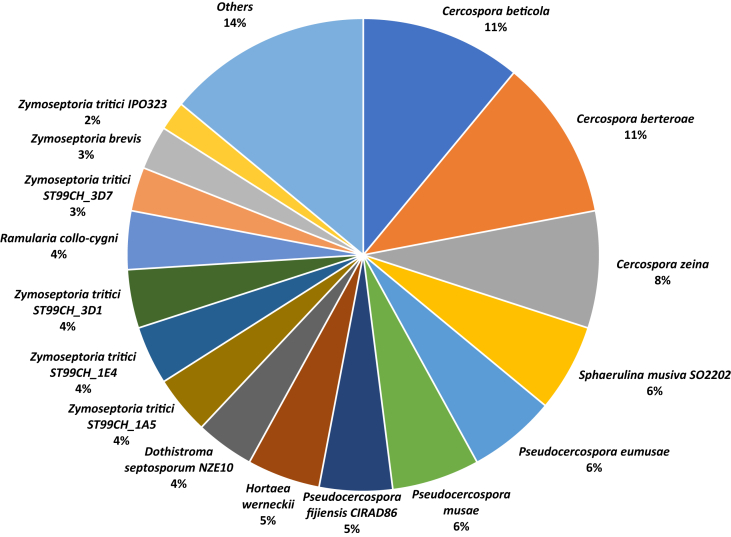
A total of 33,107,531 reads were assembled de novo, resulting in 136 scaffolds of at least 500 bp with the largest scaffold 3,211,885 bp and an N50 value of 898,622 bp. The mean coverage of the total assembly was 196.72-fold. The G + C content was 53.04%. The gene prediction resulted in 14,856 gene models with 14,721 protein coding genes and 135 non coding RNAs, including the mitochondrial genome (Table 1). The distribution of protein annotations are summarized in Table 2, and Table 3 provides the summary statistics of the identified repetitive elements. The distribution of functional gene ontology (GO) terms from the annotated C. kikuchii ARG_18_001 genes are illustrated in Fig. 2. The distribution of species from the top BLAST hit of the predicted protein coding genes is shown in Fig. 3.
Table 1.
Genome features of C. kikuchii strain ARG_18_001.
| Features | C. kikuchii ARG_18_001 |
|---|---|
| Assembled length | 33,197,932 |
| Scaffold length (≥ 50,000 bp) | 32,541,287 |
| Number of scaffolds (>500 bp) | 136 |
| Number of scaffolds (>1 kb) | 107 |
| Number of scaffolds (>50 kb) | 71 |
| Sequencing read coverage depth (fold) | 196.72 |
| GC-Content | 53.04 |
| No. of predicted protein-coding genes | 14,721 |
| Gene density (genes/Mb) | 447.5 |
| Average length of transcripts | 1468.7 |
| Average CDS length | 1354.2 |
| Average protein length | 451.4 |
| Average exon length | 568.6 |
| Average intron length | 82.9 |
| Spliced genes | 9702 (66.0%) |
| Number of total introns | 20,309 |
| Median number of introns per gene | 2.0 |
| Number of total exons | 35,010 |
| Median number of exons per gene | 2.0 |
Table 2.
Genome annotation summary of C. kikuchii strain ARG_18_001.
| Summary | Number |
|---|---|
| Number of protein-coding gene models | 14,721 |
| Number of models with BLAST hit | 13,015 (88.4%) |
| Blast2GO annotation | 6296 (42.8%) |
| PFAM annotation | 5684 (38.6%) |
Table 3.
Summary of repetitive elements in the assembled genome of C. kikuchii strain ARG_18_001.
| Summary | Number |
|---|---|
| Total of bases masked | 178,815 (0.54%) |
| Number of simple repeats | 3131 |
| Number of low complexity repeats | 358 |
| Number of DNA transposons | 68 |
| Number of LTRs | 2 |
| Number of LINEs | 254 |
| Number of SINEs | 21 |
Fig. 2.
Histogram representing the gene ontology distribution of the annotated Cercospora kikuchii ARG_18_001 genes. The functionally annotated genes were assigned to three main GO categories: Biological Process (BP), Molecular Function (MF) and Cellular Component (CC).
Fig. 3.
Pie chart denoting the species distribution based on the top BLAST hit of the Cercospora kikuchii ARG_18_001 genes queried against the nr database with an E-value cut-off of 1E-10. The category “Others” includes species with less than 1% representation.
2. Experimental design, materials, and methods
2.1. Genomic DNA extraction and sequencing
Cercospora kikuchii strain ARG_18_001 was isolated from a single conidium from soybean seeds of variety DM62R63 sampled that exhibited symptoms of purple seed stain during the 2018 harvest in San Pedro, Buenos Aires, Argentina. The isolation technique is described in [4]. This strain was deposited in the fungal culture collection of the Department of Plant Pathology, School of Agriculture, University of Buenos Aires (FAUBA, Argentina). Genomic DNA was isolated from hyphal tissue grown in potato dextrose broth for four days in darkness and constant agitation. The DNA extraction was carried out at the Institute of Microbiology and Agricultural Zoology (IMYZA -INTA) using a modified cetyltrimethylammonium bromide (CTAB) extraction protocol developed by [5]. Total DNA was quantified by fluorometry using a Picogreen dsDNA dye kit (Quant-iT, Invitrogen, by Life Technologies, CA, USA) with a Victor 3 plate reader.
Paired-end whole-genome shotgun libraries were constructed using the TruSeq Nano DNA (insert size 350 bp) library preparation kit following Illumina (San Diego, CA) protocols. Sequencing was performed using a NovaSeq 6000 sequencing system (Illumina) and yielded 65,202,278 reads.
2.2. Phylogenetic species identification
The isolate ARG_18_001 was identified by aligning seven nuclear loci (actin (actA), calmodulin (cmdA), nuclear ribosomal internal transcribed spacer region (nrITS), glyceraldehyde-3-phosphate dehydrogenase (gapdh), histone H3 (his 3), translation elongation factor 1-a (tef1-alpha) and beta tubulin (tub2)) with data from [6,7]. A maximum-likelihood phylogeny was then inferred in RAxML (Randomized Axelerated Maximum Likelihood) [8] assuming a GTRGAMMA model with Septoria provencialis CPC_12226 as an outgroup.
2.3. Genome assembly and annotation
Read trimming and filtering was performed using Trimmomatic [9] and merging of paired-end reads from shorter fragments was made using FLASH [10]. De novo assembly was carried out using the Celera Assembler [11] and then completed with Spades [12] using a wide range of k-mer values from 21 to 111 with a step of 2. The genome was annotated using FunGAP [13], tRNAscan-SE [14], rnammer [15] and MFannot (http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl) [16]. For predicting genes with FunGAP, the C. kikuchii ARG_18_001 genome assembly and the C. beticola 10.73.4 (Bioproject PRJNA294383) RNA-seq reads were used as inputs. To perform the functional annotation, we used hmmsearch [17] against PFAM database (v32.0) (e-value cut off ≤ 10e-5) and BLASTP [18] (e-value cut off ≤ 10e-10) against the NCBI nr database. To assign Gene Ontology [19] terms we used Blast2GO [20] and pfam2go table (http://www.geneontology.org/external2go/pfam2go) with the ragp R package (https://rdrr.io/github/missuse/ragp/). The repetitive regions, including tandem repeats and transposable elements, were detected using the repeat identification tool RepeatMasker [21].
Acknowledgments
This work was financially supported by the University of Buenos Aires, Project UBACyT 20020170100147BA and partially by BASF Argentina S.A.
We specially thank Dr. Norma Paniego and the Bioinformatics Unit at IB/IABIMO INTA for technical assistance and support.
Conflict of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Soares A.P.G., Guillin E.A., Borges L.L., Silva A.C.Td., Almeida Á.M.Rd., Grijalba P.E., Gottlie A.M., Bluhm B.H., Oliveira L.O. More Cercospora species infect soybeans across the Americas than meets the eye. PLoS One. 2015;10 doi: 10.1371/journal.pone.0133495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albu S., Schneider R.W., Price P.P., Doyle V.P. Cercospora cf. flagellaris and Cercospora cf. sigesbeckiae are associated with Cercospora leaf blight and purple seed stain on soybean in North America. Phytopathology. 2016;106:1376–1385. doi: 10.1094/PHYTO-12-15-0332-R. [DOI] [PubMed] [Google Scholar]
- 3.Sautua F.J., Searight J., Doyle V.P., III Price P.P., Scandiani M.M., Carmona M.A. The G143A mutation confers azoxystrobin resistance to soybean Cercospora leaf blight in Bolivia. Plant Health Prog. 2019;20:2–3. [Google Scholar]
- 4.Price P., Purvis M.A., Cai G., Padgett G.B., Robertson C.L., Schneider R.W., Albu S. Fungicide resistance in Cercospora kikuchii, a soybean pathogen. Plant Dis. 2015;99:1596–1603. doi: 10.1094/PDIS-07-14-0782-RE. [DOI] [PubMed] [Google Scholar]
- 5.Berretta M.F., Lecuona R.E., Zandomeni R.O., Grau O. Genotyping isolates of the entomopathogenic fungus Beauveria bassiana by RAPD with fluorescent labels. J. Invertebr. Pathol. 1998;71:145–150. doi: 10.1006/jipa.1997.4727. [DOI] [PubMed] [Google Scholar]
- 6.Groenewald J.Z., Nakashima C., Nishikawa J., Shin H.D., Park J.H., Jama A.N., Groenewald M., Braun U., Crous P.W. Species concepts in Cercospora: spotting the weeds among the roses. Stud. Mycol. 2013;75:115–170. doi: 10.3114/sim0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bakhshi M., Arzanlou M., Babai-Ahari A., Groenewald J.Z., Crous P.W. Novel primers improve species delimitation in Cercospora. IMA Fungus. 2018;9:299–332. doi: 10.5598/imafungus.2018.09.02.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magoc T., Salzberg S. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Myers E.W., Sutton G.G., Delcher A.L., Dew I.M., Fasulo D.P., Flanigan M.J., Kravitz S.A., Mobarry C.M., Reinert K.H., Remington K.A., Anson E.L., Bolanos R.A., Chou H.H., Jordan C.M., Halpern A.L., Lonardi S., Beasley E.M., Brandon R.C., Chen L., Dunn P.J., Lai Z., Liang Y., Nusskern D.R., Zhan M., Zhang Q., Zheng X., Rubin G.M., Adams M.D., Venter J.C. A whole-genome assembly of Drosophila. Science. 2000;287:2196–2204. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- 12.Bankevich A., Nurk S., Antipov D., Gurevich A., Dvorkin M., Kulikov A.S., Lesin V., Nikolenko S., Pham S., Prjibelski A., Pyshkin A., Sirotkin A., Vyahhi N., Tesler G., Alekseyev M.A., Pevzner P.A. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Min B., Grigoriev I.V., Choi I.-G. FunGAP: fungal Genome Annotation Pipeline using evidence-based gene model evaluation. Bioinformatics. 2017;33(18):2936–2937. doi: 10.1093/bioinformatics/btx353. [DOI] [PubMed] [Google Scholar]
- 14.Chan P.P., Lowe T.M. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol. Biol. 2019;1962:1–14. doi: 10.1007/978-1-4939-9173-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lagesen K., Hallin P.F., Rødland E., Stærfeldt H.H., Rognes T., Ussery D.W. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beck N., Lang B. MFannot, Organelle Genome Annotation Websever. 2010. http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl
- 17.Eddy S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011;7(10) doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Harris M.A., Hill D.P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J.C., Richardson J.E., Ringwald M., Rubin G.M., Sherlock G. Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conesa A., Götz S., García-Gómez J.M., Terol J., Talón M., Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(Suppl. 18):3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 21.Tarailo-Graovac M., Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinform. 2004;5:4–10. doi: 10.1002/0471250953.bi0410s25. [DOI] [PubMed] [Google Scholar]