Abstract
Background
Glioma is the most common primary malignant tumor in the central nervous system with frequent hypoxia and angiogenesis. Limb-Bud and Heart (LBH) is a highly conserved transcription cofactor that participates in embryonic development and tumorigenesis.
Methods
The conditioned media from LBH regulated human glioma cell lines and patient-derived glioma stem cells (GSCs) were used to treat the human brain microvessel endothelial cells (hBMECs). The function of LBH on angiogenesis were examined through methods of MTS assay, Edu assay, TUNEL assay, western blotting analysis, qPCR analysis, luciferase reporter assay and xenograft experiment.
Findings
Our study found for the first time that LBH was overexpressed in gliomas and was associated with a poor prognosis. LBH overexpression participated in the angiogenesis of gliomas via the vascular endothelial growth factor A (VEGFA)-mediated extracellular signal-regulated kinase (ERK) signalling pathway in human brain microvessel endothelial cells (hBMECs). Rapid proliferation of gliomas can lead to tissue hypoxia and hypoxia inducible factor-1 (HIF-1) activation, while HIF-1 can directly transcriptionally regulate the expression of LBH and result in a self-reinforcing cycle.
Interpretation
LBH may be a possible treatment target to break the vicious cycle in glioma treatment.
Keywords: Glioma, Glioma stem cells, LLimb-Bud and Heart, Angiogenesis, ERK signalling pathway
Research in context
Evidence before this study
Glioma is the major primary tumor in the central nervous system, which constitutes nearly 80% of all malignant brain tumors, with a median survival of no more than 15 months. Furthermore, the hypoxic tumor microenvironment can promote angiogenesis, proliferation and invasion in gliomas via recruiting and activating glioma stem cells (GSCs). Therefore, intervention in the processes of angiogenesis and GSC activation is of great therapeutic significance in gliomas. As a transcription cofactor, LBH is expressed in the central nervous system and brain, and is associated with ischemic stroke, Parkinson's disease, and other neurological diseases. LBH also participates in tumorigenesis and the development of several cancers, such as hepatocellular, gastric and breast cancer. However, whether LBH can participate in the functional regulation of glioma and GSCs is still not completely clear.
Added value of this study
Our study found for the first time that LBH was overexpressed in gliomas and was associated with a poor prognosis. GSEA analysis revealed that higher LBH expression participates in the angiogenesis of glioma under hypoxia. Our results found LBH-mediated glioma conditioned medium (GCM) can promote the proliferation, invasion and angiogenesis of hBMECs, and HIF-1 can directly upregulate the expression of LBH under hypoxia. Our research demonstrated that the expression and secretion of VEGFA was regulated by LBH in glioma cell lines and GSCs, and VEGFA-mediated ERK signaling was activated. In summary, LBH is a novel oncogene in glioma and mainly participates in the angiogenesis of glioma via the VEGFA-mediated ERK signaling pathway.
Implications of all the available evidence
We confirmed that LBH is overexpressed in glioma, and can activate the VEGFA-mediated ERK signaling pathway under transcriptionally regulated by HIF-1 under hypoxia, which provided a novel mechanism by which the hypoxic microenvironment promotes the tumorigenesis and angiogenesis of glioma via LBH upregulation. LBH is a possible treatment target to improve the prognosis of glioma patients.
1. Introduction
Glioma is the major primary tumor in the central nervous system and is characterized by rapid proliferation and invasion [1,2]. Despite an abundant blood supply in the tumor tissues [3], frequent occurrences of ischaemia, hypoxia and necrosis are reported [4,5]. The hypoxic tumor microenvironment can promote proliferation, invasion, angiogenesis and therapeutic resistance in gliomas via recruiting glioma stem cells (GSCs) and activating various cellular pathways [6,7]. Hypoxia-inducible factors (HIFs) are the major mediators of the transcriptional hypoxic response, and the accumulation of HIF-1α can lead to the transcription of hypoxic-related genes, which participate in tumorigenesis and angiogenesis [5,8]. GSCs have been identified as a subpopulation of tumor cells that play a key role in tumorigenesis, angiogenesis, therapy resistance and recurrence [9,10]. These cells thereby lead to a vicious cycle and aggravate the malignancy of the glioma. Therefore, intervention in the processes of angiogenesis and GSC activation is of great therapeutic significance in gliomas.
Limb-Bud and Heart (LBH) is a highly conserved transcription cofactor in vertebrates and is mainly expressed in embryonic tissues and several adult tissues, including the gut, spleen, kidney, brain and peripheral nervous system [11]. LBH has multiple functions in embryogenesis, especially in the development of limbs and the heart [12]. LBH can also regulate angiogenesis during foetal bone development through VEGF [13]. Moreover, LBH is reported to be involved in the tumorigenesis of several cancers. For example, LBH plays a carcinogenic role in hepatocellular, gastric and breast cancers [11,14,15]. However, LBH was also reported to have anticancer effects in human lung adenocarcinomas, nasopharyngeal carcinomas and prostate carcinomas [16], [17], [18].
Although LBH is expressed in the normal brain and associated with ischaemic stroke, Parkinson's disease, and other neurological diseases [19], there is no research about the possible roles of LBH in gliomas. It is unknown whether LBH participates in the tumorigenesis of gliomas. Moreover, LBH can maintain the stemness of multipotent retinal progenitor cells and mammary stem cells and inhibit the differentiation of these cells [20,21]. This suggests that LBH can participate in the functional regulation of GSCs.
Through bioinformatic analyses, our study first detected abnormally high LBH expression levels in gliomas, which were associated with poor prognosis. We then confirmed these findings in clinical specimens. We further demonstrated that the hypoxic microenvironment can promote the transcription and expression of LBH and lead to angiogenesis in gliomas via the vascular endothelial growth factor A (VEGFA)-mediated extracellular signal-regulated kinase (ERK) signalling pathway. Therefore, LBH is a promising target for glioma therapy.
2. Materials and methods
2.1. Cell culture and cell treatment
The human glioma cell lines, U87, U118 and U251 were purchased from the Chinese Academy of Sciences Cell Bank (Shanghai, China). T98G was purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). LN229, H4 and U178 cells were purchased from iCell Bioscience Inc. (Shanghai, China). All of the glioma cell lines were maintained in Dulbecco's Modified Eagle's Medium (DMEM, HyClone, Logan, UT, USA), supplemented with 10% fetal bovine serum (FBS; Gibco, Carlsbad, CA, USA) and 1% penicillin/streptomycin (Gibco) at 37 °C with 5% CO2. Normal human astrocytes (HA) and human brain microvessel endothelial cells (hBMECs) were purchased from ScienCell Research Laboratories (San Diego, CA, USA). The HA were maintained in astrocyte medium (ScienCell Research Laboratories) and hBMECs were maintained in endothelial cell medium (ScienCell Research Laboratories). Six patient-derived primary glioma cells from grade II to IV tumors, according to the World Health Organization (WHO) classification guidelines (grade II: GSC2A, GSC2B; III: GSC3A, GSC3B; IV: GSC4A, GSC4B), were also used, as previously described [22]. The detailed clinicopathological information is presented in supplementary table 1. The stemness of GSCs was detected by immunofluorescence using anti-CD133 (1: 100, #ab16518 Abcam, Cambridge, UK) or nestin antibodies (1: 100, #ab6320), and the multi-lineage differentiation capacity of GSCs was demonstrated via immunofluorescence staining of GFAP (1: 100, #ab16977) and β III tubulin (1: 100, #ab18207).
To induce hypoxia, the glioma cell lines or GSCs were cultured in a hypoxia chamber with 94% N2, 5% CO2, and 1% O2 at 37 °C. VEGFA-neutralizing antibody (#ab36424) was used at a concentration of 0.1 μg/mL. All of the above cells were cultured in vitro for no more than 10 passages.
2.2. Preparation of the glioma conditioned medium (GCM)
The glioma cell lines or GSCs were cultured to 80% confluence under different conditions. Then serum-free DMEM was used to wash these cells three times and then cells were cultured in serum-free DMEM for 24 h. The supernatants were collected and centrifuged at 3000 × g for 15 min at 4 °C to remove cell debris. The GCM was prepared and immediately used to treat hBMECs before performing the subsequent experiments, or was stored at ˗80 °C for no more than one week [23].
2.3. Patients and samples
Clinical samples from glioma patients and the normal control group were the same as previously described [24]. Briefly, 70 clinical samples from glioma patients were collected from January 2007 to December 2011 in the First Affiliated Hospital of China Medical University [24]. Among these, there were 20 cases of grade II, 25 cases of grade III and 25 cases of grade IV tumors, according to the WHO classification guidelines. The detailed clinicopathological information was presented in supplementary table 2. Samples from another 10 patients without any previous neurological diseases, suffered serve brain trauma and received the operation immediately were collected as the control group during the same period. This study was approved by the ethics committee of the First Affiliated Hospital of China Medical University and written informed consent was obtained from all patients.
2.4. Lentiviral vector construction and transfection
The lentivirus-based vectors for LBH overexpression and RNAi-mediated knockdown of LBH were obtained from Gene-Chem (Shanghai, China). Two siRNA sequences were designed for LBH silencing as follows: LBH-KD1: forward 5′-GGAUCGAGUUUGAGACUAAAG-3′, reverse 5′-UUAGUCUCAAACUCGAUCCCA-3′; LBH-KD2: forward 5′-CUGUGACAGUUGUAAAUAAAG-3′, reverse 5′-UUAUUUACAACUGUCACAGUG-3′. Lentivirus transfection was performed as previously described [22].
2.5. Real-time PCR
Real-time PCR was performed as previously described [22]. The Mini-BEST Universal RNA Extraction kit (#9767, TaKaRa, Kyoto, Japan) was used to isolate the total RNA of glioma cells, following first-strand cDNA synthesis by Prime-Script RT Master Mix (#6110A, TaKaRa) and qPCR detection by SYBR Green Master Mix (#RR420Q, TaKaRa) on a PCR LightCycler 480 (Roche Diagnostics Ltd., Basel, Switzerland). The sequences of PCR primer pairs were as follows: LBH, forward 5ʹ-GCCCCGACTATCTGAGATCG-3ʹ and reverse 5ʹ-GCGGTCAAAATCTGACGGGT-3ʹ; VEGFA, forward 5ʹ-AGGGCAGAATCATCACGAAGT-3ʹ and reverse 5ʹ-AGGGTCTCGATTGGATGGCA-3ʹ; β-actin, forward 5ʹ-CATGTACGTTGCTATCCAGGC-3ʹ and reverse 5ʹ-CTCCTTAATGTCACGCACGAT-3ʹ.
2.6. Western blot analysis
Western blotting was performed as previously described [22]. Briefly, the total protein of cells was isolated using a total cell protein extraction kit (KeyGen Biotechnology, Nanjing, China), followed by electrophoresis of equivalent amounts of protein. Proteins were then transferred to a nitrocellulose membrane, and the membrane was blocked with 2% bovine serum albumin (KeyGen Biotechnology). Relevant primary antibodies against LBH (1:500, #ab122223), HIF-1 (1:500, #ab1), VEGFA (1:1000, #ab52917), VEGFR2 (1:1000, #ab11939), p-VEGFR2 (1:1000, #ab194806), MEK1/2 (1:1000, #ab178876), p-MEK1/2 (1:1000, #ab96379), ERK1/2 (1:500, #ab17942), p-ERK1/2 (1:500, #ab223500) and β-actin (1:2000, #66009–1-Ig, ProteinTech, Chicago, IL, USA) were incubated with the membrane at 4 °C overnight. Following this, incubation with appropriate secondary antibodies (ProteinTech) was also performed. The chemiluminescence ECL kit (Beyotime Biotechnology, Beijing, China) was used to detect the protein bands of interest, and band density was quantified by Image J software (National Institutes of Health, Bethesda, MD, USA).
2.7. Immunohistochemistry
Immunohistochemistry was performed as previously described [22]. The tumor tissues were embedded in paraffin, sliced into 4-μm sections and labelled with primary antibody against LBH (1:100, #ab122223), VEGFA (1:100, #ab52917) and CD31 (1:100, #ab28364). Then the slices were stained with an immunohistochemical labeling kit (MaxVision Biotechnology, Fuzhou, Fujian, China) and imaged under a light microscope (Olympus). The staining intensity and expression levels were evaluated according to the German immunohistochemical score [25].
2.8. Immunofluorescence
Immunofluorescence staining was performed as previously described [22]. Briefly, the glioma cells were fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100, blocked with 5% BSA, and probed with primary antibodies against CD133, nestin, GFAP and βIII-tubulin at 4 °C overnight, followed with FITC- or rhodamine- conjugated secondary antibodies. The cells were then counterstained with DAPI (Sigma, Santa Clara, CA, USA). Staining was visualized via a laser scanning confocal microscope (Olympus).
2.9. Cell viability assay
The hBMECs treated under different conditions were plated in 96-well plates at 1 × 103 cells / well and cultured for 5 days. The CellTiter 96Ⓡ AQueous Non-Radioactive cell proliferation assay kit (MTS, # G3582, Promega, Madison, WI, USA) was used to detect the cell viability, according to the manufacturer's instructions, as previously reported [26].
2.10. EDU assay
The hBMECs treated under different conditions were seeded into 24-well plates at 1 × 105 cells / well and incubated for 24 h. The 5-ethynyl-20-deoxyuridine (EDU) incorporation assay was performed with an EDU assay kit (#C0075S, Beyotime Biotechnology), according to the manufacturer's instructions, as previously reported [22]. Images were taken under a laser scanning confocal microscope (Olympus) and the percentage of EDU-positive cells was calculated.
2.11. Transwell invasion assay
The transwell invasion assay was performed as previously described [27]. The HBMECs treated under different conditions were applied to Matrigel-coated filters at 1 × 105 cells / well for 20 h. The cells that invaded the lower compartments were stained with crystal violet (Beyotime Biotechnology), photographed, and counted under a microscope (Olympus).
2.12. Tube formation assay
First, precooled 96-well plates were coated with 70 μl of Matrigel (#354248, Corning Technology, Corning City, NY, USA) at 37 °C for 30 min. The HBMECs treated under different conditions were seeded at 2 × 104 cells / well for 3 h. Tube formation was photographed under a microscope (Olympus) and the number of branches and tube lengths were calculated using Image J software (National Institutes of Health).
2.13. Luciferase reporter assays
Luciferase reporter assays were performed as previously described [24,26]. Briefly, LBH reporter plasmids were constructed by Gene-Chem (Shanghai, China). The different types of glioma cells were seeded into 96-well plates at a density of 5 × 103 cells / well and were transfected with LBH wild-type or mutant reporter plasmids for 48 h. The Dual-Luciferase Reporter Assay System (#E1910, Promega) was then used to detect the luciferase activities.
2.14. Chromatin immunoprecipitation (ChIP) assays
ChIP assays were performed as previously described [22]. Briefly, the glioma cells treated under different conditions were plated in a 100-mm dish and immunoprecipitated with anti-HIF-1 antibodies (#ab1). The ChIP Assay Kit (#P2078, Beyotime Biotechnology) was used to purify DNA samples and the purified DNA was analyzed by qPCR. The primer pairs used to amplify the HIF-1 binding site in the LBH promoter was: f 5′-ACCCAGGACGACACCACA-3′ and r 5′-AATCCTCGCCCGCAGCCTTG-3′.
2.15. Enzyme-linked immunosorbent assay (ELISA)
ELISA was performed as previously described [22]. The Human VEGF Quantikine ELISA kit (#DVE00, R&D Systems, Minneapolis, MN, USA) was used to assess the concentration of VEGFA in the supernatant of the media of the glioma cells treated under different conditions. All readings were normalized to the protein concentration in the control groups.
2.16. Xenograft experiments
Xenograft experiments were performed as previously described [22]. Six-week-old female BALB/c nude mice (Beijing Vital River Laboratory Animal Technology Co., Ltd., Beijing, China) were bred in the Laboratory Animal Center of China Medical University under specific-pathogen-free conditions. Briefly, the glioma cells treated under different conditions were injected (5 × 104 cells per mouse) orthotopically into the mouse brain 2 mm lateral and 2 mm anterior to the bregma using stereotaxic apparatus. Each group with five mice was observed daily for signs of distress or death, and tumor volumes were calculated according to the formula: V = (D × d × d)/2, where D represents the longest diameter and d represents the shortest diameter. All animal experiments were performed in accordance with the Animal Care Committee of China Medical University.
2.17. Bioinformatics analysis
The data on LBH mRNA expression, WHO grades, isocitrate dehydrogenase (IDH 1/2) status and survival times of glioma patients were obtained from the Chinese Glioma Genome Atlas (CGGA, http://www.cgga.org.cn) including the mRNAseq_325 dataset and mRNAseq_693 dataset, and the Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov) in the HG-U133A platform. Gene set enrichment analysis (GSEA, http://www.broadinstitute.org/gsea/ index.jsp) was used to identify any enrichments in signalling pathways between the higher and lower LBH expression groups.
2.18. Statistical analysis
All experiments were repeated at least three times and the results were presented as the mean ± sd. The chi-square test was used to evaluate the relationship between LBH expression and clinicopathological characteristics. The two-tailed Student's t-test was used for comparisons of two independent groups. One-Way ANOVA was used to evaluate the statistical significance among three or more groups. The log-rank test and Kaplan–Meier analysis were used to analyze the survival rates of each group. Statistical analysis was performed using SPSS 23.0 software (IBM, Armonk, NY, USA). Two-tailed P values < 0.05 were considered statistically significant.
3. Results
-
1
LBH is overexpressed in gliomas and correlates with poor patient survival
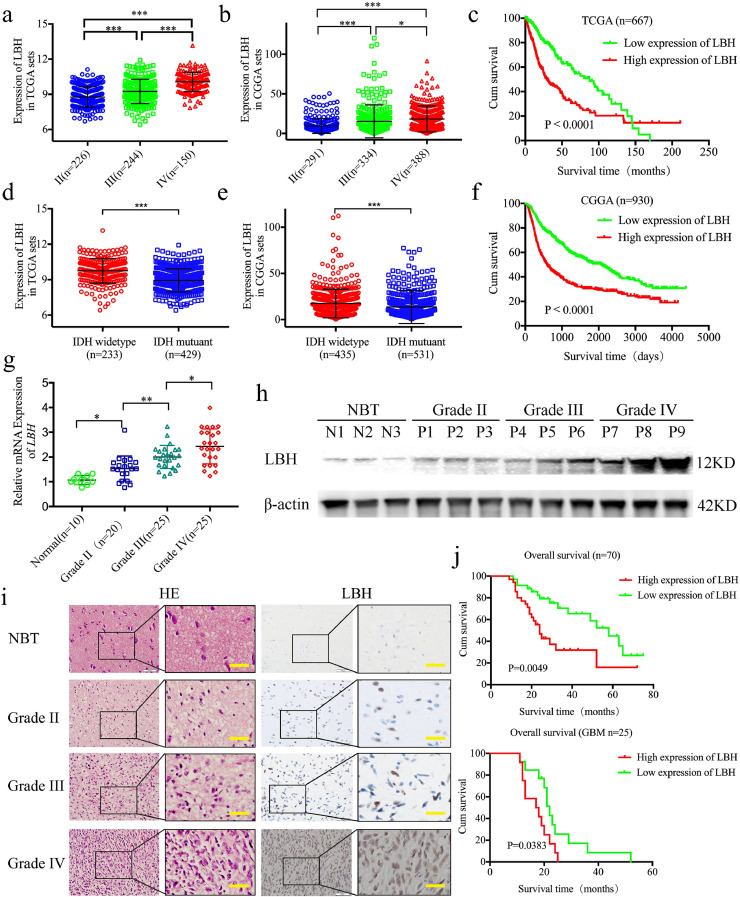
To investigate the possible functions of LBH in gliomas, we first searched the expression of LBH in the TCGA and CGGA datasets. Both results showed that expression was significantly increased with higher glioma WHO grades (TCGA: Fig. 1a, p <0.0001, CGGA: Fig. 1b, p <0.0001, One-Way ANOVA). Furthermore, higher expressions of LBH were observed in IDH-wild-type gliomas than in IDH-mutants in both the TCGA and CGGA datasets (TCGA: Fig. 1d, p <0.0001, CGGA: Fig. 1e, p < 0.0001, Student's t-test). LBH overexpression was associated with decreased survival rates in both TCGA and CGGA gliomas (TCGA: Fig. 1c, p <0.001, CGGA: Fig. 1f, log-rank test). Considering these bioinformatic results, we further detected the expression of LBH in 70 glioma patients and 10 normal brain tissues. All of the qPCR, western blot, and immunohistochemistry results showed higher expressions of LBH in glioma tissues than in normal brain tissues (Fig. 1g–i). Furthermore, the expression of LBH was significantly increased in higher glioma WHO grades (Fig. 1g–i). Kaplan–Meier survival analyses showed that the median survival time of total glioma patients and GBM patients with higher LBH expressions were all shorter than in patients with lower LBH expression levels (Fig. 1j).
Fig. 1.
LBH is expressed at higher levels in gliomas and is correlated with poor patient survival
a, b: The mRNA expression of LBH is shown according to WHO grades in the TCGA (a) and CGGA (b) databases.
d, e: The mRNA expression of LBH is shown according to IDH status in the TCGA (d) and CGGA (e) databases.
c, f: The prognostic significance of LBH was confirmed in the TCGA (c) and CGGA (f) databases.
g: LBH is expressed at higher levels in glioma patients with different grades of disease, compared with normal brain tissue (NBT) as measured by qPCR. (II vs. NBT: p = 0.0104, III vs. NBT: p = 0.0001, IV vs. NBT: p<0.0001, III vs. II p = 0.0023, VI vs. III: p = 0.0156, One-Way ANOVA).
h: Representative western blots showing higher LBH expression in glioma patients with different tumor grades, compared with NBT.
i: Representative immunohistochemical staining for LBH in NBT and glioma patients with different tumor grades (grade II, n = 20; grade III, n = 25; grade IV, n = 25; NBT n = 10). Scale bar = 50 μm. (II vs. NBT, p = 0.0026; III vs. NBT, p<0.0001; IV vs. NBT, p< 0.0001; One-Way ANOVA).
j: Kaplan–Meier analysis of the total 70 cases of glioma patients and 25 cases of GBM patients with high LBH expression versus low LBH expression by immunohistochemistry. (median survival times: 70 cases: 29 and 58 months, respectively, p = 0.0049; GBM: 17.5 and 22 months, respectively, p = 0.0383)
All data are shown as the mean ± SD (three independent experiments). *P < 0.05; **P < 0.01; ***P < 0.001.
We further isolated GSCs from six glioma patients with different WHO pathological diagnoses (e.g., WHO grade II, III or IV). The six GSCs were designated GSC-2A and GSC-2B (WHO grade II), GSC-3A and GSC-3B (WHO grade III), and GSC-4A and GSC-4B (WHO grade IV). All GSCs were confirmed via immunofluorescence staining of the stem cell markers, CD133 and nestin, and the differentiation markers, GFAP and β-III tubulin (Fig. S1a, b). qPCR and western blotting showed higher expressions of LBH in WHO grade IV GSCs (GSC-4A and GSC-4B) than in WHO grade II GSCs (GSC-2A and GSC- 2B) (Fig. S2a, d, p < 0.0001, One-Way ANOVA). We also analyzed seven glioma cell lines and one normal human astrocyte (HA) cell line. qPCR and western blot analyses revealed a higher expression of LBH in the glioma cell lines than in HAs (Fig. S2b, c, p < 0.0001, One-Way ANOVA). Therefore, we conclude that the higher expression of LBH in gliomas is correlated with poor patient survival.
-
1
LBH-mediated GCM regulates the proliferation, invasion and angiogenesis of hBMECs
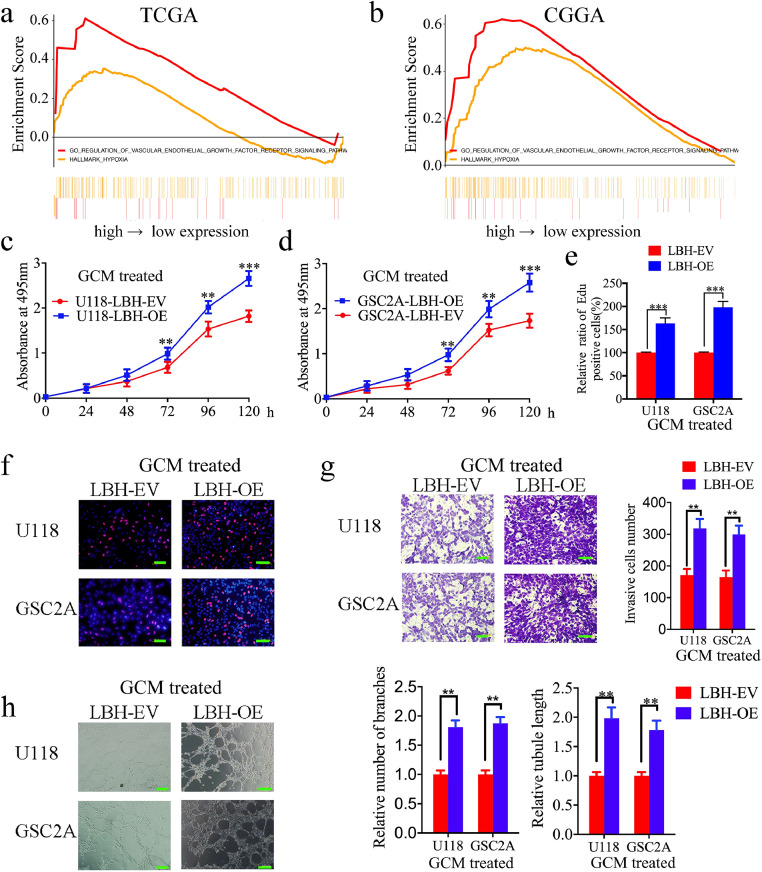
To investigate the function of LBH in gliomas, we performed gene set enrichment analysis (GSEA) based on LBH expression. The results showed that a higher LBH expression was associated with enrichment of the regulation of the VEGF receptor signalling pathway and hypoxia in both the TCGA and CGGA datasets (Fig. 2a, b). This suggested that higher LBH expression may promote glioma angiogenesis, especially under hypoxia. According to the expression of LBH in patient-derived GSCs and glioma cell lines in Fig. S2a–d, GSC4B and T98G expressed the highest levels of LBH, while GSC2A and U118 expressed the lowest levels of LBH. Therefore, GSC4B and T98G were chosen for LBH silencing and GSC2A and U118 were chosen for LBH overexpression. Lentiviral-based transfection and the effects on LBH overexpression or silencing were validated by both qPCR and western blotting (Fig. S2e–h). The supernatants of these GSCs and glioma cell lines were then collected and used to treat hBMECs.
Fig. 2.
LBH overexpression in glioma conditioned media (GCM) can promote the proliferation, invasion and angiogenesis of hBMECs
a, b: Gene set enrichment analysis (GSEA) indicates that high expression of LBH is associated with the VEGF signalling pathway and hypoxia in both the TCGA (a) and CGGA (b) databases.
c, d: Vascular endothelial cell viability increased after treatment with LBH-overexpressed U118 (c) and GSC2A (d) conditioned media, as measured by the MTS assay. (U118: p = 0.0019, GSC2A: p = 0.0053, One-Way ANOVA).
e, f: The proliferation of vascular endothelial cells increased following treatment with LBH-overexpressed U118 and GSC2A conditioned media, as measured by the EDU incorporation assay, scale bar = 100 μm. (U118: p <0.0001, GSC2A: p <0.0001, Student's t-test)
g: Representative transwell assay showing the increase in invasion of vascular endothelial cells after treatment with LBH-overexpressed U118 and GSC2A conditioned media. Scale bar = 100 μm. (U118: p = 0.0087, GSC2A: p = 0.0093, Student's t-test)
h: Representative tube formation assay showing the increase in tubulogenesis of vascular endothelial cells after treatment with LBH-overexpressed U118 and GSC2A conditioned media. Scale bar = 100 μm. (number of branches: U118: p = 0.0071, GSC2A: KD1, p = 0.0089; tubule length: U118, p = 0.0025, GSC2A: p = 0.0039, Student's t-test)
All data are shown as the mean ± SD (three independent experiments). *P < 0.05; **P < 0.01; ***P < 0.001.
We first evaluated the positive effects on the proliferation of LBH via MTS and EDU assays. The results showed that treatment with the conditioned media from LBH-overexpressed GSC2A and U118 cells increased cell viability and the rates of EDU-positive hBMECs (Fig. 2c–f). However, the opposite results were obtained after treatment with LBH-silenced GSC4B and T98G conditioned media (Fig. S3a–c). Transwell assays then showed that treatment with the LBH-overexpressed conditioned media increased the number of invading cell numbers of hBMECs (Fig. 2g), whereas treatment with LBH-silenced conditioned media decreased the invasive cell numbers (Fig. S3d). Finally, tube formation assays were performed to detect the effect of LBH on angiogenesis regulation. Treatment with LBH-overexpressed conditioned media increased the number of branches and tubule length of hBMECs, compared with GCM treatment of the empty vector groups (Fig. 2h). However, the opposite results were obtained after treatment with LBH-silenced conditioned media (Fig. S3e). Taken together, these results clearly demonstrated that LBH overexpression in glioma cells and GSCs can promote the proliferation, invasion and angiogenesis of hBMECs.
-
1
HIF-1 can directly induce the expression of LBH under hypoxia
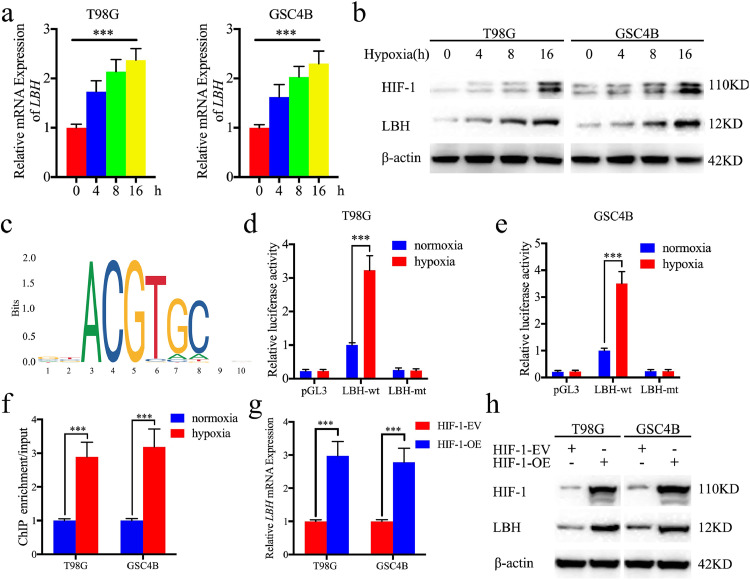
Next, we investigated whether there is a functional relationship between LBH and hypoxia. The T98G and GSC4B cells were treated under hypoxia and the expression of LBH was detected by both qPCR and western blotting. Extension of the hypoxia treatment time increased the expression of LBH, accompanied by higher HIF-1 expression (Fig. 3a, b). Since HIF-1 is one of the most important transcription factors under hypoxia, we investigated whether HIF-1 can bind to and induce the expression of LBH using JASPAR (Fig. 3c). Using luciferase reporter assays and ChIP-qPCR assays, hypoxia treatment was shown to increase the relative luciferase activity and LBH was enriched in both T98G and GSC4B cells (Fig. 3d–f). qPCR and western blot also showed the expression of LBH was obviously upregulated after HIF-1 overexpression (Fig. 3g, h). We therefore concluded that HIF-1 can directly induce the expression of LBH under hypoxia.
-
1
LBH knockdown abolished hypoxia-induced hBMEC proliferation, invasion and angiogenesis
Fig. 3.
HIF1 can directly induce the expression of LBH under hypoxia
a: LBH mRNA expression of T98G (left) and GSC4B (right) gradually increased during prolonged treatment under hypoxia as measured by qPCR. (T98G: p <0.0001, GSC4B: p <0.0001, One-Way ANOVA)
b: LBH protein expression of T98G and GSC4B was gradually increased during prolonged treatment under hypoxia as measured by western blotting.
c: Sequence motif representing the consensus HIF-1 binding motif (JASPAR database).
d, e: Luciferase reporter assays showed hypoxia can upregulate the luciferase promoter activities of LBH in T98G (left) and GSC4B (right) cells. (T98G: p <0.0001, GSC4B: p <0.0001, Student's t-test)
e: ChIP qPCR showed HIF-1 binding to the promoter of LBH under hypoxia. (T98G: p <0.0001, GSC4B: p <0.0001, Student's t-test)
g, h: qPCR (g) and western blot (h) showed the expression of HIF-1 overexpression can upregulate the expression of LBH. (T98G: p <0.0001, GSC4B: p <0.0001, Student's t-test)
All data are shown as the mean ± SD (three independent experiments). *P < 0.05; **P < 0.01; ***P < 0.001.
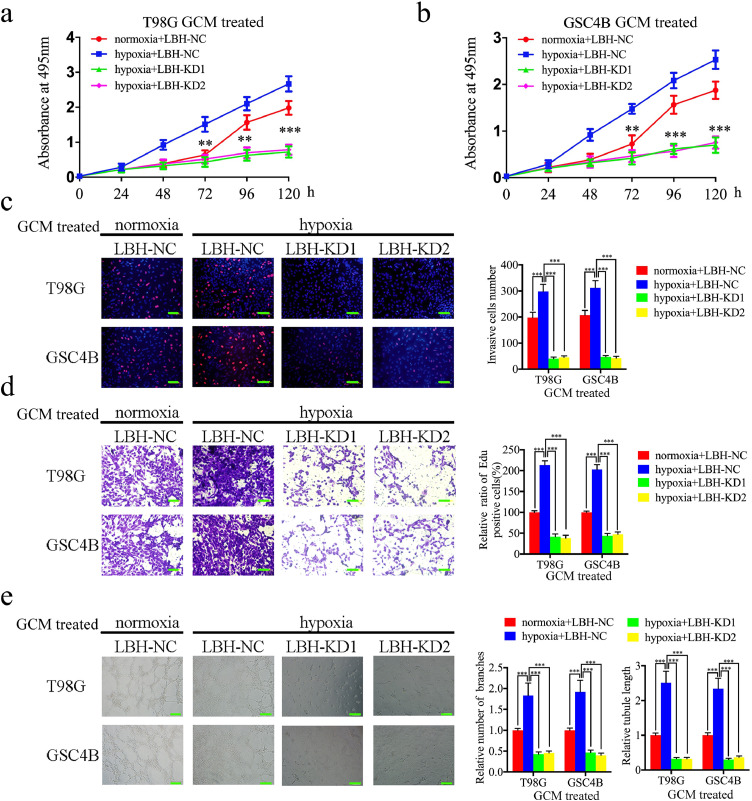
We next detected the function of LBH under hypoxia. Conditioned media collected from T98G and GSC4B cells under hypoxia or normoxia were used to treat hBMECs. MTS and EDU assays showed that hypoxic GCM treatment increased cell viability and the rates of EDU-positive hBMECs, compared with normoxic GCM treatment (Fig. 4a–c). However, after LBH silencing, the increase in proliferation of hBMECs induced by hypoxic GCM treatment was abolished (Fig. 4a–c). Furthermore, the transwell assay showed that hypoxic GCM treatment promoted the invasion of hBMECs, while LBH silencing abolished these effects (Fig. 4d). Similar results were obtained in the tube formation assays with an increased number of branches and greater tubule lengths observed for hBMECs under hypoxic GCM treatment than under normoxic GCM treatment. Such findings were reversed after LBH silencing (Fig. 4e). These findings suggested that gliomas under hypoxic conditions can promote hBMEC proliferation, invasion and angiogenesis, possibly via a mechanism involving LBH expression, since LBH silencing abolished the hypoxia-induced functions of hBMECs.
-
1
LBH activates VEGFA-mediated ERK signalling in hBMECs under hypoxia
Fig. 4.
LBH knockdown can abolish the hypoxia-induced hBMECs proliferation, invasion and angiogenesis
a, b: Hypoxia-induced vascular endothelial cell viability was reversed after treatment with LBH-silenced T98G (a) and GSC4B (b) conditioned media, as measured by an MTS assay. (T98G: p <0.0001, GSC4B: p <0.0001, One-Way ANOVA)
c: Hypoxia-induced proliferation of vascular endothelial cells was reversed after treatment with LBH-silenced T98G and GSC4B conditioned media, as measured by an EDU incorporation assay, scale bar = 100 μm. (T98G: p <0.0001, GSC4B: p <0.0001, One-Way ANOVA)
d: Representative transwell assay showing that hypoxia-induced invasion of vascular endothelial cells was reversed after treatment with LBH-silenced T98G and GSC4B conditioned media. Scale bar = 100 μm. (T98G: p <0.0001, GSC4B: p <0.0001, One-Way ANOVA)
e: Representative tube formation assay showing that hypoxia-induced tubulogenesis of vascular endothelial cells was reversed after treatment with LBH-silenced T98G and GSC4B conditioned media. Scale bar = 100 μm. (number of branches: T98G: p <0.0001, GSC4B: p <0.0001; tubule length: T98G: p <0.0001, GSC4B: p <0.0001, One-Way ANOVA)
All data are shown as the mean ± SD (three independent experiments). *P < 0.05; **P < 0.01; ***P < 0.001.
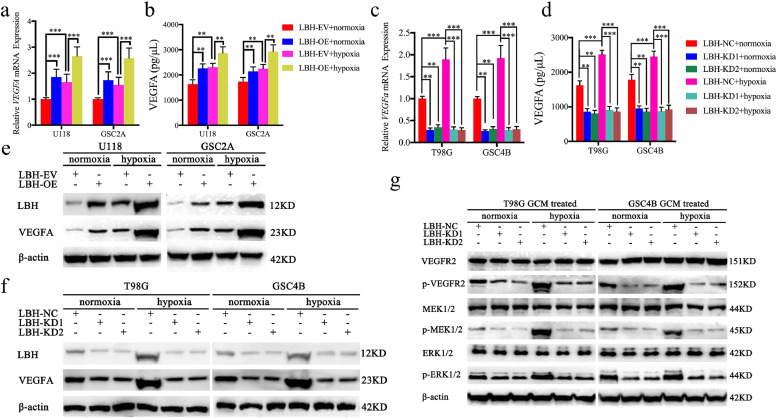
Since higher LBH expression showed VEGF signalling pathway enrichment according to GSEA (Fig. 2a, b), we detected both the expression and secretion of VEGFA after LBH regulation. The qPCR, western blot and ELISA assays all confirmed that LBH overexpression increased the mRNA levels, protein levels, and the secretion of VEGFA under both normoxia and hypoxia (Fig. 5a, b, e). However, the opposite results were obtained following LBH knockdown (Fig. 5c, d, f). Furthermore, compared with normoxia, hypoxia treatment led to higher expression and secretion of VEGFA in all normal control glioma cells (Fig. 5a–f), while LBH silencing reversed this phenomenon (Fig. 5c, d, f). LBH overexpression further promoted the expression and secretion of VEGFA (Fig. 5a, b, e). Finally, GCMs collected under different conditions were used to treat hBMECs and the downstream signalling pathways were analyzed by western blotting. The results showed that LBH-silenced GCM treatment can downregulate the expression of p-VEGFR2, p-MEK1/2 and p-ERK1/2 under both hypoxia and normoxia (Fig. 5g). Similarly, compared with normoxic GCMs, hypoxic GCM treatment can also upregulate the expression of p-VEGFR2, p-MEK1/2 and p-ERK1/2 (Fig. 5g). This phenomenon was also abolished after LBH silencing (Fig. 5g). These findings suggested that gliomas can regulate LBH expression under hypoxia and activate VEGFA-mediated ERK signalling in hBMECs.
-
1
Anti-VEGFA treatment can abolish LBH-induced hBMEC proliferation, invasion and angiogenesis under hypoxia
Fig. 5.
LBH can activate VEGFA-mediated ERK signalling in hBMECs under hypoxia
a, b: The VEGFA mRNA expression and secretion level of U118 and GSC2A cells was upregulated under hypoxia and further upregulated after LBH overexpression, as measured by qPCR (a) and ELISA (b). (qPCR: U118: p <0.0001, GSC2A: p <0.0001; ELISA: U118: p = 0.0019, GSC2A: p = 0.0011, One-Way ANOVA)
c, d: The VEGFA mRNA expression and secretion level of T98G and GSC4B cells was upregulated under hypoxia and decreased after LBH knockdown, as measured by qPCR (c) and ELISA (d). (qPCR: T98G: p <0.0001, GSC4B: p = 0.0012; ELISA: T98G: p = 0.0023, GSC4B: p = 0.0029, One-Way ANOVA)
e: The VEGFA protein expression of U118 and GSC2A cells was upregulated under hypoxia and further upregulated after LBH overexpression, as shown by western blotting.
f: The VEGFA protein expression of T98G and GSC4B cells was upregulated under hypoxia and decreased after LBH knockdown, as shown by western blotting.
g: The VEGFR-ERK signalling pathway in vascular endothelial cells following treatment with LBH-silenced T98G and GSC4B conditioned media under normoxia or hypoxia was measured by western blotting.
All data are shown as the mean ± SD (three independent experiments). *P < 0.05; **P < 0.01; ***P < 0.001.
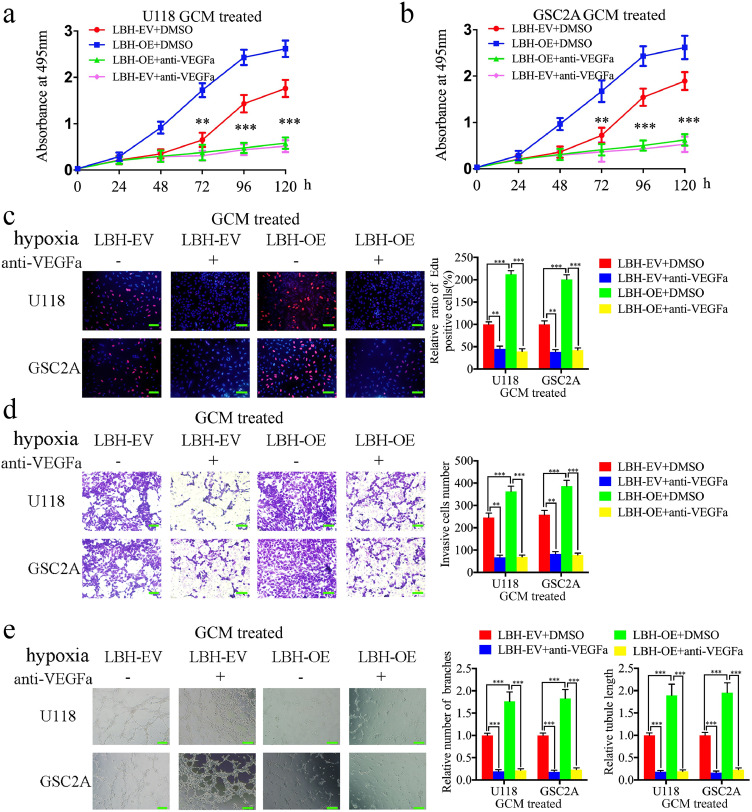
Since LBH may promote the secretion of VEGFA in glioma cells, a VEGFA-neutralizing antibody was used to block the function of VEGFA. MTS and EDU assays showed that treatment with VEGFA-neutralizing antibodies decreased cell viability and the rates of EDU-positive hBMECs in all hypoxic GCM treatment groups (Fig. 6a–c). hBMEC proliferation induced by LBH overexpression under hypoxia was also abolished by treatment with VEGFA-neutralizing antibodies (Fig. 6a–c). A transwell assay showed that treatment with VEGFA-neutralizing antibodies decreased the number of invading hBMECs in all hypoxic GCM treatment groups (Fig. 6d). Similarly, hBMEC invasion induced by LBH overexpression under hypoxia was also abolished after treatment with VEGFA-neutralizing antibodies (Fig. 6d). Tube formation assays revealed that VEGFA-neutralizing antibody treatment decreased the number of branches and the tubule length of hBMECs in all hypoxic GCMs (Fig. 6e), and angiogenesis induced by LBH overexpression under hypoxia was abolished by VEGFA-neutralizing antibody treatment (Fig. 6e). Taken together, these data provide further evidence that LBH induces hBMEC proliferation, invasion and angiogenesis under hypoxia via VEGFA upregulation, and these functions are abolished by anti-VEGFA treatment.
-
1
LBH regulates glioma tumorigenesis and angiogenesis in vivo
Fig. 6.
Anti-VEGFA treatment can abolish LBH-induced hBMECs proliferation, invasion and angiogenesis under hypoxia
a, b: The induction of vascular endothelial cell viability following treatment with LBH-overexpressed U118 (a) and GSC2A (b) conditioned media was reversed following anti-VEGFA treatment, as measured by an MTS assay. (U118: p = 0.0016, GSC2A: p = 0.0011, One-Way ANOVA)
c: The proliferation of vascular endothelial cells following treatment with LBH-overexpressed U118 and GSC2A conditioned media was reversed following anti-VEGFA treatment, as measured by an EDU incorporation assay. Scale bar = 100 μm. (U118: p = 0.0008, GSC2A: p < 0.0001, One-Way ANOVA)
d: Representative transwell assay showing that treatment with LBH-overexpressed U118 and GSC2A conditioned media induced invasion of vascular endothelial cells that was reversed after anti-VEGFA treatment. Scale bar = 100 μm.
(U118: p <0.0001, GSC2A: p = 0.0018, One-Way ANOVA)
e: Representative tube formation assay showing that treatment with LBH-overexpressed U118 and GSC2A conditioned media induced tubulogenesis of vascular endothelial cells that was reversed after anti-VEGFA treatment. Scale bar = 100 μm. (number of branches: U118: p <0.0001, GSC2A: p < 0.0001, tubule length: U118: p = 0.0012, GSC2A: p = 0.0019, One-Way ANOVA)
All data are shown as the mean ± SD (three independent experiments). *P < 0.05; **P < 0.01; ***P < 0.001.
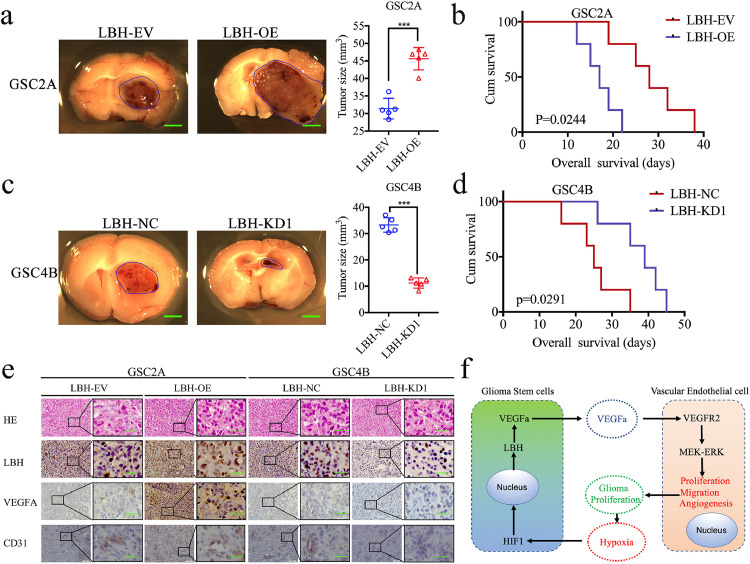
The effects of LBH on gliomas were further evaluated using orthotopic xenograft models. The tumor volumes were obviously enlarged after LBH overexpression (Fig. 7a, p < 0.0001, Student's t-test), and were accompanied by shorter survival times and a decrease in the median survival time (MST) from 28 to 17 days (Fig. 7b, p = 0.0244, log-rank test). However, the tumor volumes were obviously decreased after LBH silencing (Fig. 7c, p < 0.0001, Student's t-test), and were accompanied by longer survival times and an increase in the MST from 25 to 39 days (Fig. 7d, p = 0.0291, log-rank test). Immunohistochemistry was performed to detect the effects of LBH overexpression or silencing on tumor tissues. In the LBH overexpression group, the staining intensity and expression levels of LBH, VEGFA and the endothelial marker CD31 were all increased, while the opposite results were obtained in the LBH silenced group (Fig. 7e). Taken together, these results suggested that LBH can promote tumorigenesis and angiogenesis in nude mice.
Fig. 7.
LBH regulates glioma tumorigenesis and angiogenesis in vivo
a, c: Representative photographs showing the size of intracranial tumors in the coronal position. LBH overexpression in GSC2A cells increased the intracranial tumor size (a), whereas LBH knockdown in GSC4B cells decreased the intracranial tumor size (c). Scale bar = 10 mm.
b, d: LBH-overexpressed GSC2A cells implanted into tumor-bearing mice showed shorter survival times as measured by Kaplan–Meier survival curves (b), compared with longer survival times when LBH-silenced GSC4B cells were implanted into tumor bearing mice (d). For each group, n = 5.
e: Representative immunohistochemical staining showing the changes in LBH, VEGFA and CD31 in LBH overexpression and knockdown orthotopic xenograft models. Scale bar = 50 μm.
f: Schematic diagram to illustrate that overexpression of LBH promotes angiogenesis in human glioma via VEGFA-mediated ERK signalling under hypoxia.
4. Discussion
Since the clinical therapeutic effects of surgery, chemotherapy and radiotherapy are all limited for gliomas, molecular targeted therapy is regarded as a new promising method for glioma treatment [2]. Studies on the mechanisms and modes of action of glioma-related genes may provide important information for future molecular targeted therapies [28,29]. In this study, we found LBH to be a novel oncogene in the development of gliomas. The bioinformatics analyses based on the TCGA and CGGA datasets and experiments on clinical specimens demonstrated that LBH expression in higher grade gliomas was higher than that in lower grade gliomas. Patients with higher LBH expression also showed shorter survival times than those with lower LBH expression. Moreover, LBH expression was higher in IDH-wild-type gliomas than in IDH-mutants, indicating that higher LBH-expressing patients may suffer poor temozolomide treatment effects [30].
The published literature indicates that LBH plays contradictory roles in several cancers. LBH is overexpressed in human hepatocellular carcinomas and is associated with poor prognosis [31]. Elevated LBH levels promote the proliferation and invasion of gastric cancer via the integrin/FAK/Akt pathway and indicate a poor prognosis [14]. Loss of LBH attenuates mammary hyperplasia and tumor development in MMTV-Wnt1 transgenic mice [15]. However, LBH overexpression can also inhibit the growth and invasion of human lung adenocarcinoma cells and predict better survival outcomes [16]. LBH can function as a tumor suppressor in nasopharyngeal carcinomas by inducing G1/S cell cycle arrest [17]. LBH can also inhibit prostate cancer cell proliferation and tumor growth by inducing cell cycle arrest through downregulating cyclin D1 and cyclin E2 expression [18]. No previous studies reported the function of LBH in gliomas; however, our results demonstrated that LBH is an oncogene in gliomas.
The published studies suggest that LBH is involved in neurodevelopment and neurological diseases [32]. LBH can regulate neural crest cell development and migration [33], maintain the stemness of multipotent mammary stem cells and inhibit their differentiation [20,21]. LBH was also found to be downregulated in neurodegenerative diseases, such as stroke, Parkinson's disease and Alzheimer's disease [11,19,34,35]. These studies all indicated that LBH may participate in the survival and function of neurons and neural stem cells. We may further infer that LBH overexpression regulates the stemness, survival and invasion of GSCs and participates in gliomagenesis. These findings all require further study in the future.
According to GSEA analysis, higher LBH expression participates in the angiogenesis of gliomas under hypoxia. As a transcription factor, we found that HIF-1 can directly upregulate the expression of LBH under hypoxia. Hypoxia and active capillary endothelial proliferation are typical features of high-grade gliomas [36]. The frequent hypoxia caused by the rapid growth of tumors can activate HIF-1 [36,37], which can promote the proliferation, migration and angiogenesis of gliomas and the self-renewal of GSCs [36,38,39], and even possibly lead to the transcription and expression of LBH. Our study provides a possible explanation for LBH overexpression in gliomas.
A study reported that LBH can inhibit angiogenesis during foetal bone development by downregulating the expression of VEGFA [13]. However, our experiments performed on glioma cell lines and patient-derived GSCs showed that LBH-mediated GCM can promote the proliferation, invasion and angiogenesis of hBMECs via upregulating the expression of VEGFA. This finding indicates that LBH may play a contradictory role in the angiogenesis of tumors and foetal bone development. VEGFA is the most important growth factor in angiogenesis and is synthesized and secreted by most cancer cells [40,41]. Secreted VEGFA can bind to and activate the receptors on endothelial cells, especially VEGF receptor 2 (VEGFR2), leading to the proliferation, invasion and tube formation of endothelial cells [42], [43], [44].
VEGFA-neutralizing antibody was used to block the function of VEGFA, and the LBH-mediated GCM treatment effects on hBMECs were consequently abolished. As a conserved transcription cofactor, LBH can mediate the transcription and activation of molecules involved in downstream pathways, such as the MAPK signalling pathway, and the development of limbs and the heart [31,34,45]. Although our research demonstrated that the expression and secretion of VEGFA was regulated by LBH in glioma cell lines and GSCs, the exact transcriptional mechanism of LBH on VEGFA requires further study. Moreover, VEGF is a vital treatment target for anti-tumor therapy, and there are several VEGF-targeting drugs, such as bevacizumab [46,47]. It is possible that patients with increased LBH expression may be sensitive to anti-VEGF therapy.
We further detected the downstream signalling pathways induced by LBH-mediated GCM treatment in hBMECs, and the results showed that VEGFA-mediated ERK signalling was activated. It has been reported that VEGFA can lead to the phosphorylation of VEGFR2 and further activate the ERK signalling pathway in endothelial cells [48], [49], [50]. The ERK signalling pathway plays a key role in the regulation of cell proliferation, differentiation, migration and anti-apoptosis functions in various tumor and normal cells [51,52].
Orthotopic xenograft studies confirmed that LBH can promote the tumorigenesis of patient-derived GSCs and decrease the mean survival time of nude mice. Immunohistochemical staining also showed that the expression of VEGFA and CD31 was upregulated after LBH overexpression. These results further demonstrate the possible mechanism of LBH-mediated tumorigenesis and angiogenesis in gliomas under hypoxia.
5. Conclusion
In summary, LBH is a novel oncogene in glioma and mainly participates in the angiogenesis of glioma via the VEGFA-mediated ERK signalling pathway. LBH can also be transcriptionally regulated by HIF-1 under hypoxia. Therefore, our findings provided a novel mechanism by which the hypoxic microenvironment promotes the tumorigenesis and angiogenesis of glioma via LBH upregulation. The overgrowth of gliomas can further lead to a hypoxic microenvironment, resulting in a self-reinforcing cycle and a poor prognosis (Fig. 7f). LBH is a possible treatment target to break this vicious cycle and improve the prognosis of glioma patients.
Funding
This work was supported by National Natural Science Foundation of China (No. 81101917, 81270036, 30901736), the Plan to Focus on Research and Development from Science and Technologyproject of Liaoning Province (No. 2017225029), Natural Science Foundation of Liaoning Province (No. 20170541022), Science and Technology Plan Project of Shenyang City(No. 18–014–4–11), and Fund for Scientific Research of The First Hospital of China Medical University(No. FHCMU- FSR), and Shanghai Sailing Program(No. 19YF1439000). The funders did not participate in study design, data collection, data analysis, interpretation, and writing of the report.
Authors' contributions
YZ and ZTJ conceived and designed the study; YJ and JPZ performed the experiments and collected the data; DQH, HYZ, JSZ and LL performed Bioinformatics analysis and analyzed the data. YJ, JPZ, DZ and JFH interpreted results and wrote the manuscript. YJ and JPZ contributed equally to this work. All authors read and approved the final version of the manuscript.
Declaration of Competing Interest
The authors have declared that no conflict of interest exists.
Acknowledgements
We would like to acknowledge our lab colleagues for their support in the development of this article.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2019.09.037.
Contributor Information
Ye Zhang, Email: yzhang21@cmu.edu.cn.
Zhitao Jing, Email: jingzhitao@hotmail.com.
Appendix. Supplementary materials
Reference
- 1.Behnan J., Finocchiaro G., Hanna G. The landscape of the mesenchymal signature in brain tumours. Brain. 2019;142(4):847–866. doi: 10.1093/brain/awz044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behnan J., Stangeland B., Hosainey S.A., Joel M., Olsen T.K., Micci F. Differential propagation of stroma and cancer stem cells dictates tumorigenesis and multipotency. Oncogene. 2017;36(4):570–584. doi: 10.1038/onc.2016.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu H., Zheng J., Liu X., Xue Y., Shen S., Zhao L. Transcription factor NFAT5 promotes glioblastoma cell-driven angiogenesis via SBF2-AS1/miR-338-3p-mediated EGFL7 expression change. Front Mol Neurosci. 2017;10:301. doi: 10.3389/fnmol.2017.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo X., Qiu W., Liu Q., Qian M., Wang S., Zhang Z. Immunosuppressive effects of hypoxia-induced glioma exosomes through myeloid-derived suppressor cells via the miR-10a/Rora and miR-21/Pten pathways. Oncogene. 2018;37(31):4239–4259. doi: 10.1038/s41388-018-0261-9. [DOI] [PubMed] [Google Scholar]
- 5.Joseph J.V., Conroy S., Pavlov K., Sontakke P., Tomar T., Eggens-Meijer E. Hypoxia enhances migration and invasion in glioblastoma by promoting a mesenchymal shift mediated by the HIF1alpha-ZEB1 axis. Cancer Lett. 2015;359(1):107–116. doi: 10.1016/j.canlet.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 6.Yue X., Lan F., Xia T. Hypoxic glioma cell-secreted exosomal miR-301a activates Wnt/beta-catenin signaling and promotes radiation resistance by targeting TCEAL7. Mol Therapy. 2019 doi: 10.1016/j.ymthe.2019.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bar E.E., Lin A., Mahairaki V., Matsui W., Eberhart C.G. Hypoxia increases the expression of stem-cell markers and promotes clonogenicity in glioblastoma neurospheres. Am J Pathol. 2010;177(3):1491–1502. doi: 10.2353/ajpath.2010.091021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chouaib S., Noman M.Z., Kosmatopoulos K., Curran M.A. Hypoxic stress: obstacles and opportunities for innovative immunotherapy of cancer. Oncogene. 2017;36(4):439–445. doi: 10.1038/onc.2016.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schonberg D.L., Lubelski D., Miller T.E., Rich J.N. Brain tumor stem cells: molecular characteristics and their impact on therapy. Mol Aspects Med. 2014;39:82–101. doi: 10.1016/j.mam.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheray M., Begaud G., Deluche E., Nivet A., Battu S., Lalloue F. Cancer stem-like cells in glioblastoma. In: De Vleeschouwer S, editor. Glioblastoma. Codon Publications; Brisbane (AU): 2017. editor. Chapter 4. [PubMed] [Google Scholar]
- 11.Rieger M.E., Sims A.H., Coats E.R., Clarke R.B., Briegel K.J. The embryonic transcription cofactor LBH is a direct target of the WNT signaling pathway in epithelial development and in aggressive basal subtype breast cancers. Mol Cell Biol. 2010;30(17):4267–4279. doi: 10.1128/MCB.01418-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Briegel K.J., Baldwin H.S., Epstein J.A., Joyner A.L. Congenital heart disease reminiscent of partial trisomy 2p syndrome in mice transgenic for the transcription factor LBH. Development. 2005;132(14):3305–3316. doi: 10.1242/dev.01887. [DOI] [PubMed] [Google Scholar]
- 13.Conen K.L., Nishimori S., Provot S., Kronenberg H.M. The transcriptional cofactor LBH regulates angiogenesis and endochondral bone formation during fetal bone development. Dev Biol. 2009;333(2):348–358. doi: 10.1016/j.ydbio.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu R., Li Z., Zhang C., Song H., Deng M., Sun L. Elevated Limb-Bud and Heart development (LBH) expression indicates poor prognosis and promotes gastric cancer cell proliferation and invasion via upregulating Integrin/FAK/Akt pathway. PeerJ. 2019;7:e6885. doi: 10.7717/peerj.6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ashad-Bishop K., Garikapati K., Lindley L.E., Jorda M., Briegel K.J. Loss of Limb-Bud-and-Heart (LBH) attenuates mammary hyperplasia and tumor development in MMTV-Wnt1 transgenic mice. Biochem Biophys Res Commun. 2019;508(2):536–542. doi: 10.1016/j.bbrc.2018.11.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng M., Yu R., Wang S., Zhang Y., Li Z., Song H. Limb-Bud and Heart attenuates growth and invasion of human lung adenocarcinoma cells and predicts survival outcome. Cell Physiol Biochem. 2018;47(1):223–234. doi: 10.1159/000489801. [DOI] [PubMed] [Google Scholar]
- 17.Liu Q., Guan X., Lv J., Li X., Wang Y., Li L. Limb-Bud and Heart (LBH) functions as a tumor suppressor of nasopharyngeal carcinoma by inducing G1/S cell cycle arrest. Sci Rep. 2015;5:7626. doi: 10.1038/srep07626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Q., Li E., Huang L., Cheng M., Li L. Limb-Bud and Heart overexpression inhibits the proliferation and migration of PC3M cells. J Cancer. 2018;9(2):424–432. doi: 10.7150/jca.21375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lang W., Wang J., Ma X., Zhang N., Li H., Cui P. Identification of shared genes between ischemic stroke and Parkinson's disease using genome-wide association studies. Front Neurol. 2019;10:297. doi: 10.3389/fneur.2019.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li W.H., Zhou L., Li Z., Wang Y., Shi J.T., Yang Y.J. Zebrafish LBH-like is required for Otx2-mediated photoreceptor differentiation. Int J Biol Sci. 2015;11(6):688–700. doi: 10.7150/ijbs.11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindley L.E., Curtis K.M., Sanchez-Mejias A., Rieger M.E., Robbins D.J., Briegel K.J. The WNT-controlled transcriptional regulator LBH is required for mammary stem cell expansion and maintenance of the basal lineage. Development. 2015;142(5):893–904. doi: 10.1242/dev.110403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang Y., Han S., Cheng W., Wang Z., Wu A. NFAT1-regulated IL6 signalling contributes to aggressive phenotypes of glioma. Cell Commun Signal. 2017;15(1):54. doi: 10.1186/s12964-017-0210-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jia P., Cai H., Liu X., Chen J., Ma J., Wang P. Long non-coding RNA H19 regulates glioma angiogenesis and the biological behavior of glioma-associated endothelial cells by inhibiting microRNA-29a. Cancer Lett. 2016;381(2):359–369. doi: 10.1016/j.canlet.2016.08.009. [DOI] [PubMed] [Google Scholar]
- 24.Jiang Y., Zhou J., Luo P., Gao H., Ma Y., Chen Y.S. Prosaposin promotes the proliferation and tumorigenesis of glioma through toll-like receptor 4 (TLR4)-mediated NF-kappaB signaling pathway. EBioMedicine. 2018;37(1):78–90. doi: 10.1016/j.ebiom.2018.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han S., Zhang C., Li Q., Dong J., Liu Y., Huang Y. Tumour-infiltrating CD4(+) and CD8(+) lymphocytes as predictors of clinical outcome in glioma. Br J Cancer. 2014;110(10):2560–2568. doi: 10.1038/bjc.2014.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang Y., Song Y., Wang R., Hu T., Zhang D., Wang Z. NFAT1-mediated regulation of NDEL1 promotes growth and invasion of glioma stem-like cells. Cancer Res. 2019;79(10):2593–2603. doi: 10.1158/0008-5472.CAN-18-3297. [DOI] [PubMed] [Google Scholar]
- 27.Jiang Y., Zhou J., Hou D., Luo P., Gao H., Ma Y. Prosaposin is a biomarker of mesenchymal glioblastoma and regulates mesenchymal transition through the TGF-beta1/Smad signaling pathway. J Pathol. 2019;249(1):26–38. doi: 10.1002/path.5278. [DOI] [PubMed] [Google Scholar]
- 28.Chai Z., Ran D., Lu L., Zhan C., Ruan H., Hu X. Ligand-modified cell membrane enables the targeted delivery of drug nanocrystals to glioma. ACS Nano. 2019;13(5):5591–5601. doi: 10.1021/acsnano.9b00661. [DOI] [PubMed] [Google Scholar]
- 29.Tomiyama A., Ichimura K. Signal transduction pathways and resistance to targeted therapies in glioma. Semin Cancer Biol. 2019;58:118–129. doi: 10.1016/j.semcancer.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 30.Louis D.N., Perry A., Reifenberger G., von Deimling A., Figarella-Branger D., Cavenee W.K. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 31.Chen J., Huang C., Chen K., Li S., Zhang X., Cheng J. Overexpression of LBH is associated with poor prognosis in human hepatocellular carcinoma. Onco Targets Ther. 2018;11:441–448. doi: 10.2147/OTT.S152953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamazaki T., Muramoto M., Okitsu O., Morikawa N., Kita Y. Discovery of a novel neuroprotective compound, AS1219164, by high-throughput chemical screening of a newly identified apoptotic gene marker. Eur J Pharmacol. 2011;669(1–3):7–14. doi: 10.1016/j.ejphar.2011.07.027. [DOI] [PubMed] [Google Scholar]
- 33.Powder K.E., Cousin H., McLinden G.P., Craig Albertson R. A nonsynonymous mutation in the transcriptional regulator lbh is associated with cichlid craniofacial adaptation and neural crest cell development. Mol Biol Evol. 2014;31(12):3113–3124. doi: 10.1093/molbev/msu267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Briegel K.J., Joyner A.L. Identification and characterization of LBH, a novel conserved nuclear protein expressed during early limb and heart development. Dev Biol. 2001;233(2):291–304. doi: 10.1006/dbio.2001.0225. [DOI] [PubMed] [Google Scholar]
- 35.Yamaguchi-Kabata Y., Morihara T., Ohara T., Ninomiya T., Takahashi A., Akatsu H. Integrated analysis of human genetic association study and mouse transcriptome suggests LBH and SHF genes as novel susceptible genes for amyloid-beta accumulation in Alzheimer's disease. Hum Genet. 2018;137(6–7):521–533. doi: 10.1007/s00439-018-1906-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y., Jin G., Zhang J., Mi R., Zhou Y., Fan W. Overexpression of STAT1 suppresses angiogenesis under hypoxia by regulating VEGFA in human glioma cells. Biomed Pharmacother. 2018;104:566–575. doi: 10.1016/j.biopha.2018.05.079. [DOI] [PubMed] [Google Scholar]
- 37.Xue H., Yuan G., Guo X., Liu Q., Zhang J., Gao X. A novel tumor-promoting mechanism of IL6 and the therapeutic efficacy of tocilizumab: hypoxia-induced IL6 is a potent autophagy initiator in glioblastoma via the p-STAT3-MIR155-3p-CREBRF pathway. Autophagy. 2016;12(7):1129–1152. doi: 10.1080/15548627.2016.1178446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Almiron Bonnin D.A., Havrda M.C., Lee M.C., Liu H., Zhang Z., Nguyen L.N. Secretion-mediated STAT3 activation promotes self-renewal of glioma stem-like cells during hypoxia. Oncogene. 2018;37(8):1107–1118. doi: 10.1038/onc.2017.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ji X., Wang H., Zhu J., Zhu L., Pan H., Li W. Knockdown of Nrf2 suppresses glioblastoma angiogenesis by inhibiting hypoxia-induced activation of HIF-1alpha. Int J Cancer. 2014;135(3):574–584. doi: 10.1002/ijc.28699. [DOI] [PubMed] [Google Scholar]
- 40.Wang X., Fu X., Zhao S., Fu X., Zhang H., Shao L. Antiangiogenic properties of caudatin in vitro and in vivo by suppression of VEGF-VEGFR2-AKT/FAK signal axis. Mol Med Rep. 2017;16(6):8937–8943. doi: 10.3892/mmr.2017.7730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cai H., Liu X., Zheng J., Xue Y., Ma J., Li Z. Long non-coding RNA taurine upregulated 1 enhances tumor-induced angiogenesis through inhibiting microRNA-299 in human glioblastoma. Oncogene. 2017;36(3):318–331. doi: 10.1038/onc.2016.212. [DOI] [PubMed] [Google Scholar]
- 42.Ferrara N. Vascular endothelial growth factor: basic science and clinical progress. Endocr Rev. 2004;25(4):581–611. doi: 10.1210/er.2003-0027. [DOI] [PubMed] [Google Scholar]
- 43.Ye J., Zhu J., Chen H., Qian J., Zhang L., Wan Z. A novel lncRNA-LINC01116 regulates tumorigenesis of glioma by targeting VEGFA. Int J Cancer. 2019 doi: 10.1002/ijc.32483. [DOI] [PubMed] [Google Scholar]
- 44.Tan Z., Chen K., Wu W., Zhou Y., Zhu J., Wu G. Overexpression of HOXC10 promotes angiogenesis in human glioma via interaction with PRMT5 and upregulation of VEGFA expression. Theranostics. 2018;8(18):5143–5158. doi: 10.7150/thno.27310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ai J., Wang Y., Tan K., Deng Y., Luo N., Yuan W. A human homolog of mouse LBH gene, hLBH, expresses in heart and activates SRE and AP-1 mediated MAPK signaling pathway. Mol Biol Rep. 2008;35(2):179–187. doi: 10.1007/s11033-007-9068-4. [DOI] [PubMed] [Google Scholar]
- 46.Diaz R.J., Ali S., Qadir M.G., De La Fuente M.I., Ivan M.E., Komotar R.J. The role of bevacizumab in the treatment of glioblastoma. J Neurooncol. 2017;133(3):455–467. doi: 10.1007/s11060-017-2477-x. [DOI] [PubMed] [Google Scholar]
- 47.Rahat M.A. Targeting angiogenesis with peptide vaccines. Front Immunol. 2019;10:1924. doi: 10.3389/fimmu.2019.01924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhong Z., Huang M., Lv M., He Y., Duan C., Zhang L. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–317. doi: 10.1016/j.canlet.2017.06.027. [DOI] [PubMed] [Google Scholar]
- 49.Lin J., Cao S., Wang Y., Hu Y., Liu H., Li J. Long non-coding RNA UBE2CP3 enhances HCC cell secretion of VEGFA and promotes angiogenesis by activating ERK1/2/HIF-1alpha/VEGFA signalling in hepatocellular carcinoma. J Exp Clin Cancer Res. 2018;37(1):113. doi: 10.1186/s13046-018-0727-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Z., Jin C., Li X., Ding K. Sulfated polysaccharide JCS1S2 inhibits angiogenesis via targeting VEGFR2/VEGF and blocking VEGFR2/Erk/VEGF signaling. Carbohydr Polym. 2019;207:502–509. doi: 10.1016/j.carbpol.2018.11.091. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Q., Lu S., Li T., Yu L., Zhang Y., Zeng H. ACE2 inhibits breast cancer angiogenesis via suppressing the VEGFA/VEGFR2/ERK pathway. J Exp Clin Cancer Res. 2019;38(1):173. doi: 10.1186/s13046-019-1156-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mu N., Gu J., Liu N., Xue X., Shu Z., Zhang K. PRL-3 is a potential glioblastoma prognostic marker and promotes glioblastoma progression by enhancing MMP7 through the ERK and JNK pathways. Theranostics. 2018;8(6):1527–1539. doi: 10.7150/thno.22699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.