Abstract
Mosquitoes are important vectors of pathogens due to their blood feeding behavior. Aedes aegypti (Diptera: Culicidae) transmits arboviruses, such as dengue, Zika, and Chikungunya. This species carries several bacteria that may be beneficial for its biological and physiological development. Therefore, studying the response of its microbiota to chemical products could result in vector control. Recently, imidazolium salts (IS) were identified as effective Ae. aegypti larvicides. Considering the importance of the mosquito microbiota, this study addressed the influence of IS on the bacteria of Ae. aegypti larvae. After exposition of larvae to different IS concentrations, the cultured microbiota was identified through culturomics and mass spectrometry, and the non-cultivated microbiota was characterized by molecular markers. In addition, the influence of the IS on axenic larvae was studied for comparison. There was an alteration in both cultivable species and in their diversity, including modifications in bacterial communities. The axenic larvae were less susceptible to the IS, which was increased after exposing these larvae to bacteria of laboratory breeding water. This highlights the importance of understanding the role of the larval microbiota of Ae. aegypti in the development of imidazolium salt-based larvicides. Such effect of IS towards microbiota of Ae. aegypti larvae, through their antimicrobial action, increases their larvicidal potential.
Subject terms: Bacteria, Microbiology
Introduction
Aedes aegypti (Diptera: Culicidae) is an important mosquito species that is involved in the maintenance, replication and transmission of pathogenic agents, such as dengue, urban yellow fever, Zika and Chikungunya1. The incidence of these arboviruses has increased gradually through the decades, causing a major impact on health worldwide2,3. Currently, there is only an effective vaccine against the yellow fever virus. In the case of the registered dengue vaccine, it has an efficiency of only 60%4. Therefore, controlling and preventing dengue and other arboviruses still depends on vector handling. Within this perspective, the World Health Organization established normatives for controlling larvae and adult populations of Ae. aegypti, recommending the use of a combination of chemical and biological products5.
Current control methods have problems such as a high cost for the simultaneous biological and chemical control, and a low specificity for target organisms, being, therefore, toxic for non-target organisms, including the human being. Besides, several Ae. aegypti populations distributed worldwide showed resistance to the most used insecticides like the organophosphate Temephos, carbamates and pyrethroids6. For these reasons, the search for new products specific for target organisms with low impact in the environment is crucial for effective vector control.
Recently, Goellner et al. demonstrated the efficacy of two imidazolium salts (IS), 1-n octadecil-3-methylimidazolium chloride (C18MImCl) and 1-n-hexadecil-3-methylimidazolium methanesulfonate (C16MImMeS), in the control of Ae. aegypti larvae (Fig. 1)7. A broad variety of biological properties have been reported for these IS, including antibacterial activities8–10. This has been ascribed to Coulombic interactions between IS ions and phosphatidylethanolamine groups of the bacterial cell membrane, causing a disjunction of the membrane that leads to lysis and cell death11. As a consequence, the understanding of the role of Ae. aegypti microbiota is of importance when treating the larvae with IS. This mosquito species can develop ecological interactions with other species, mostly with microorganisms. The microbiota has performed a crucial role in the evolutionary process of organisms, influencing in several biological functions, such as nutrition, reproduction, development and immunity. In addition, some bacteria can mediate or stop the settlement of pathogens and, consequently, their transmission12,13. Herein, we describe for the first time the effect of the Ae. aegypti larvae microbiota on the larvicidal potential of the IS presented in Fig. 1.
Figure 1.
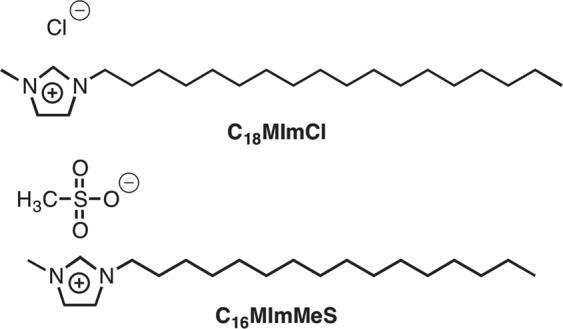
IS with previously identified larvicidal activity against Ae. aegypti.
Results
Identification of bacterial isolates in larvae treated with IS
Larvae were exposed to three previously defined concentrations (LC99, LC50 and LC20)7 of the two IS presented in Fig. 1. All IS treatments caused changes in the larvae microbiota. Besides, some species disappeared and others appeared, compared to the control group. The identified bacterial species are listed in Table 1.
Table 1.
Bacterial diversity identified by culturomics in Aedes aegypti larvae treated with LC99, LC50 and LC20 IS solutions. x Means occurrence and — absence of the species after treatment.
| Microbiota (Control) | C18MImCl | C16MImMeS | ||||
|---|---|---|---|---|---|---|
| LC99 | LC50 | LC20 | LC99 | LC50 | LC20 | |
| Serratia marcescens | — | — | x | x | — | x |
| Enterobacter asburiae | — | — | x | x | x | x |
| Stenotrophomonas altophilia | — | — | x | x | x | x |
| Klebsiella oxytoca | x | x | x | x | x | x |
| Delftia acidovorans | — | x | x | x | x | x |
| Enterobacter kobei | x | x | x | x | x | x |
| Providencia rettgeri | — | x | x | x | x | x |
| Aeromonas sp. | x | — | x | x | x | x |
| Pseudomonas sp. | x | x | x | — | x | x |
| Burkholderia sp. | — | — | — | — | — | x |
| Microbacterium sp. | x | x | — | — | — | — |
A total of eleven distinct bacterial morphotypes were isolated and identified by using MALDI-TOF mass spectrometry, being seven and four through species and genus level, respectively. C18MImCl decreased the number of bacterial species with an increasing IS concentration. When larvae were treated with C16MImMeS, the number of bacterial species was relatively concentration independent, showing only a slightly lower number of species than the control group.
Ribosomal Intergenic Space Analysis (RISA) enabled building phylogenetic trees based on the position and the number of amplified DNA fragments, and the Shannon’s (H′) diversity analysis through the intensity of DNA fragments. Larvae treated with LC99 and LC50 IS concentrations were evaluated and compared to the control group. To analyze the bacterial communities in larvae exposed to IS, the genetic similarity between samples was calculated through the Dice coefficient and the data grouped through the unweighted pair group method with arithmetic mean (UPGMA).
The Figs 2 and 3 show the phylogenetic trees after the exposition to C16MImMeS and C18MImCl, respectively. Larvae treated with the LC50 of C16MImMeS and the control group showed the same coefficient of dissimilarity. Its LC99 affected the bacterial community, observing a greater distance and the creation of a new branch in the phylogenetic tree (Fig. 2). Almost the same happened to larvae treated with C18MImCl. Now, although the coefficients of dissimilarity were different between the groups, the bacterial population did not distance itself from the main branch for the LC99 treatment, even though a new one was created (Fig. 3).
Figure 2.
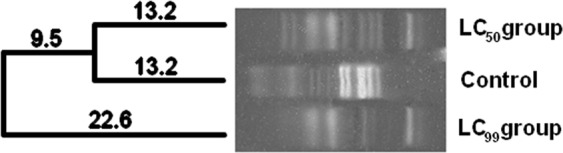
Phylogenetic tree generated through RISA analyses of the bacterial communities treated with C16MImMeS in different concentrations. Tree built using the Dice coefficient and grouping by the UPGMA algorithm. The distances represent dissimilarity. Samples derive from the same experiment. The full-length gel is presented in the Supplementary Information as Fig. S5.
Figure 3.
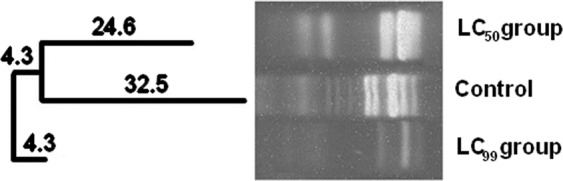
Phylogenetic tree generated through RISA analyses of bacterial communities treated with C18MImCl in different concentrations. Tree built using the Dice coefficient and grouping by the UPGMA algorithm. The distances represent dissimilarity. Samples derive from the same experiment. The full-length gel is presented in the Supplementary Information as Fig. S6.
In both IS treatments, the Shannon’s (H′) diversity decreased as its concentration was increased (Fig. 4). C16MImMeS presented a less pronounced difference in diversity when compared to C18MImCl.
Figure 4.
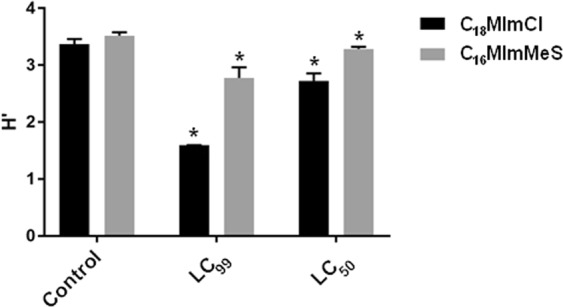
Shannon diversity index (H′) determined for the microbiota after the treatments of larvae with different IS concentrations. The bars represent standard deviations. *Represents significant difference compared to the control.
Effect of IS in axenic larvae
The effect of the IS and Temephos was verified in axenic larvae. This axenic condition was proven by the absence of any bacterial growth. Figure 5 shows the mortalities of the exposed non-axenic and axenic larvae. Although Temephos caused the same high mortality of ≥80% in both tested groups, C18MImCl and C16MImMeS showed a significant (p < 0.05) reduction in mortality in axenic larvae.
Figure 5.
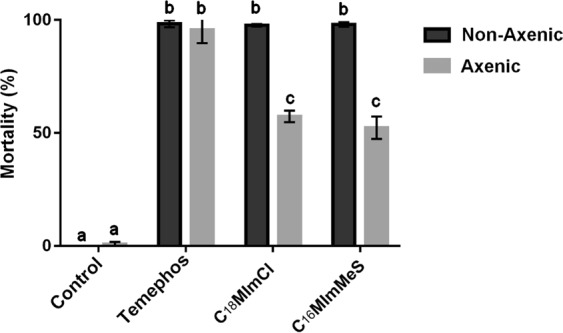
Mortality of larvae (axenic and non-axenic) after 48 h of exposure to IS (5 µg/mL) or Temephos (6 µg/L). The control is without IS and Temephos. Letters represent significant differences between treatments. The bars represent standard deviations.
Effect of IS over re-colonized axenic larvae
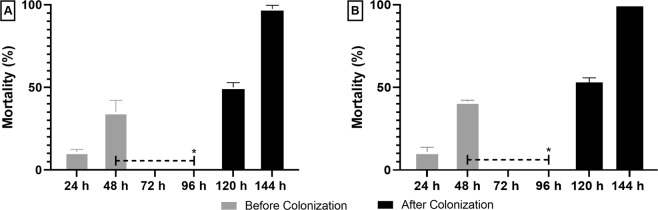
Exposure of axenic larvae to an IS LC99 resulted in the survival of half of the larvae after 48 h. Those axenic larvae, when exposed to the standard non-sterile laboratory breeding conditions, became colonized by bacteria. In addition, they became susceptible to the effect of IS. The axenia of the larvae was checked before and after the re-colonization. In the first case, no bacterial growth was observed and, after re-colonization, the turbidity in the medium after the maceration of larvae indicated the presence of bacteria. After 48 h of post-exposure and re-colonization, mortality rates higher than 95% were obtained for both treatments (Fig. 6).
Figure 6.
Mortality of Aedes aegypti larvae after being re-colonized and exposed to IS: (A) (C18MImCl); (B) (C16MImMeS). The dashed line (between 48 and 96 h) represents the time that axenic larvae that survived the first exposure to IS were re-colonized with microorganisms from standard laboratory breeding water. *(96 h) Represents the time that re-colonized larvae were re-exposed to IS. The bars represent standard deviations.
Discussion
The Ae. aegypti larvae treated with IS showed eleven species of bacteria, all of them belonging to the phylum Proteobacteria and being mostly gram-negative. Similar results have been described in the identification of cultivable normal microbiota of some culicide larvae14–17. A study conducted by Apte-Deshpande et al.18 demonstrated the presence of seven bacterial genera in the intestines of a laboratory reared Ae. aegypti larvae (Aeromonas, Burkholderia, Enterobacter, Microbacterium, Pantoea, Pseudomonas and Serratia). In our study, in addition to the mentioned genera, Stenotrophomonas, Delftia and Providencia were also identified. The identification of more bacterial genera could be attributed to the use of the culturomic approach, which used the entire larva instead of dissected intestines. This avoids the loss of bacterial species during the process and allows the evaluation of the complete larval microbiota.
The IS C18MImCl modified the cultivable microbiota of the exposed larvae in relation to the non-appearance of certain species, when evaluated at different concentrations. For example, regarding to Serratia marcescens, Enterobacter asburiae and Stenotrophomonas maltophila, these were only found when larvae were exposed to LC20. In the case of C16MImMeS, this was found only for Burkholdeira sp. Independent of the applied IS concentration, C16MImMeS did not exert any influence on the cultivable microbiota. Changes in the microbiota of culicids have been related to the food supply, habitat and genetic factor, among others19.
Nowadays, the functional and physiological role of bacterial species in larvae is still not fully understood, once the studies about microbiota were mainly focused on adult mosquitoes. In this way, some species of Serratia and Enterobacter produce hemolytic enzymes, being related to the digestion of blood in adult females and probably acquired during the larval stage20,21. Herein, Stenotrophomonas maltophila was found in larvae of a laboratory Ae. aegypti. This bacterial species was found in great abundance in the gut of adult Ae. aegypti caught in the field22, but its presence in larvae has not yet reported.
When verifying the action of IS C18MImCl through the cultivable microbiota, RISA demonstrated a Shannon’s diversity (H′) lower than that of the control group for both LC99 and LC50. For C16MImMeS, the LC99 resulted in a lower H′ and the LC50 presented a similar result as the control group.
The phylogenetic trees (Figs 2 and 3) suggested that there were changes in the bacterial community, since some species were eliminated and others emerged (Operational Taxonomic Unities does not always represent the same species)23. A total change in the microbiota might occur, which could promote larval death or increase the susceptibility towards IS which was confirmed by changes in the bacterial communities inside the larvae after exposure to IS.
As these alterations in the microbiota are not necessarily a determining factor in the larval death, the effect of IS on larvae without their microbiota was studied. After 48 h, significant differences were observed when axenic larvae were exposed to the IS, when compared with the group of non-axenic larvae. This suggested that bacterial communities interact with the IS, which potentializes their larvicidal action. The antibacterial IS could kill or reduce the proliferation of part of the microbiota and favor the multiplication of pathogenic bacteria. This hypothesis was further corroborated by the survival of axenic larvae after IS exposure, which became susceptible after being re-colonized by bacteria. In this way, one can speculate that the IS interact preferentially with bacteria acquired from the environment than with those that are transmitted vertically. Within this context, it is worth remembering that these results were obtained with larvae reared in laboratory. Therefore, this dynamic could change for populations of larvae from the field, once these mosquitoes have a more diversified microbiota than those from the laboratory24.
A benchmark larvicide was also tested for comparison, since it may be that all larvicides have some kind of interaction with the microbiota, thus triggering the death of the larvae. Temephos was chosen for this purpose, which showed high mortality in both non-axenic and axenic larvae. The mode of action of this molecule is well known, acting in the central nervous system of the larvae and inhibiting cholinesterase25. As far as we know, there is no study that relates chemical insecticides and mosquito microbiota. In the case of biological insecticides, larvae change their microbiota when intoxicated with the entomopathogenic bacterium Bacillus thuringiensis israelensis26. This study demonstrated that Bti-tolerant larvae (with death above 12 h of post-exposure) have low diversity and large inter-individual differences. It was also described that the gut bacteria of larvae aid in the degradation of toxins produced by Bti, therefore decreasing their effectiveness27. The fungus Beauveria bassiana also interacts with the Anopheles stephensi microbiota, accelerating mosquito death. In axenic mosquitoes, the death slowed down, as seen in our study28.
Thus, when larvae are treated with IS, the microbiota of mosquitoes can play an important role in their control. This report is the first study using MALDI-TOF mass spectrometry and culturomics for the identification of cultivable microbiota of larvae in Ae. aegypti. It is also the first report related to a potential chemical larvicide for the control of Ae. aegypti that can be potentialized by the microbiota, providing a first insight into the IS-microbiota interaction.
Material and Methods
Maintenance of mosquitoes
Larvae of the Ae. aegypti Rockefeller strain were used for all bioassays. They were reared at 28 °C, a 12-hour light/dark photoperiod, and an 80% relative humidity (RH). Cat cub food was offered twice a week for larvae development. After the development of pupae, they were daily removed and placed in cages for adult emergence. Adult males were fed on honey solution (10%), while females were fed on sheep’s blood (Newprov®) through a membrane for eggs development.
Imidazolium salts
Nuclear magnetic resonance (NMR) spectra were recorded on a Bruker (400 MHz) equipment at ambient temperature. The chemical shifts are given in parts per million (ppm) and referenced to the residual solvent signal (CDCl3 = 7.26 (1H), 77.16 (13C)). Attenuated total reflection Fourier transform infrared spectroscopy (ATR-FTIR), in the mid infrared range (4000–500 cm−1), was performed on an ALPHA-P compact Bruker FTIR spectrometer. High-resolution mass spectrometry spectra were recorded on an electrospray ionization (ESI) Q-Tof MicroTM equipment (Micromass, Manchester, UK) in the positive mode.
1-Methyl-3-n-octadecylimidazolium chloride (C18MImCl) was purchased at CJC CHINA JIE CHEMICAL. n-Hexadecyl methanesulfonate was synthesized according to previously reported procedures; and the spectral data were in agreement with those reported29. A modified literature procedure was used for the synthesis of 1-n-hexadecyl-3-methylimidazolium methanesulfonate (C16MImMeS)30. The mixture of 1-hexadecyl methanesulfonate (11.9 mmol, 1.00 equiv.) and 1-methylimidazole (11.9 mmol, 1.00 equiv.) was stirred and heated at 100 °C for 2 h. Next, the crude product was recrystallized from hot ethyl acetate.
C16MImMeS was obtained in 89% yield as a white solid. 1H NMR (400 MHz, CDCl3) δ 9.74 (s, 1H), 7.45 (t, J = 1.6 Hz, 1H), 7.30 (t, J = 1.7 Hz, 1H), 4.20 (t, J = 7.4 Hz, 2H), 4.00 (s, 3H), 2.72 (s, 3H), 1.83 (m, 2H), 1.26–1.20 (m, 26H), 0.83 (t, J = 6.8 Hz, 3H). 13C NMR (101 MHz, CDCl3) δ 137.98, 123.61, 121.76, 49.95, 39.65, 36.38, 31.88, 30.23, 29.89–29.22 (9C), 28.98, 26.23, 22.65, 14.09. ESI(+)-MS: C20H39N2+ - calculated: 307.3108, obtained: 307.4160 (See Supplementary Information).
Before use, the IS were dried under vacuum for 5 h at 60 °C to remove residual water.
Larvae exposition to IS
Based on the report of Goellner et al.7, the IS LC99 (5 µg/mL), LC50 (2.5 µg/mL) and LC20 (1.25 µg/mL) were tested to study the influence of IS on the larvae microbiota of Ae. aegypti. For the evaluation of cultivable microbiota; four groups of larvae were used for each IS: control (non-exposed), LC20, LC50 and LC99. For the diversity index experiments, the LC99 and LC50 were used. Eggs of Ae. aegypti were placed to hatch in 1 liter of distilled water. After hatching and visualizing the larvae, they were kept in an incubator (Tecnal TE-401) at 28 °C and 80% RH, and fed with previously sterilized cat puppy food. After reaching the earlier L2 and later L3 stages, the larvae were removed and used as control group (non-exposed). At the same stages, the larvae were subjected to IS treatments, using 25 larvae for each group (LC20, LC50 and LC99). Each group of 25 larvae was disposed in a sterile glass container with 100 mL of autoclaved distilled water. Only larvae that showed non-lethargy after IS treatment were used in further studies.
Isolating and identifying cultivable bacteria
This method comprehends a culturomic approach31, which was adapted for healthy adult mosquitos of Ae. aegypti (unpublished data) and then used in larvae. The experiments described below were carried out in triplicate, and each group consisted of five larvae (non-treated, LC99, LC50 and LC20). The larvae were washed in 70% ethanol (3 minutes), and rinsed three-times with ultra-pure water to assure a high level of disinfection. The larvae were macerated in 2 mL of sterile distilled water, submitted to a pre-incubation medium composed of brain-heart infusion broth (10 g/L), yeast extract (5 g/L), bacteriological peptone (5 g/L) and 5% of sheep’s blood (Newprov®) in 1000 mL of distilled water, and incubated at 28 °C during a 30-day period. Aliquots (1 mL) were removed from the pre-incubation at the beginning of the experiment and after every five days. All aliquots were serially diluted until 1 × 10−10, and each dilution was sown in Petri dishes containing trypticase soy agar. From the growth of bacterial colonies, each colony morphotype was transferred to a new Petri dish and subjected to its identification.
Each colony was submitted, in duplicate, to matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS). The samples were placed on the target plate (96 target MSP polished steel, Bruker Daltonics) and covered with the α-cyano-4-hydroxycinnamicacid matrix (Bruker Daltonics) diluted in 50% acetonitrile (v/v), 2.5% trifluoroacetic acid (v/v) and Milli-Q ultrapure water, followed by a drying period at room temperature. The calibration was performed with the bacterial test standard reagent (Bruker Daltonics). All spectra were recorded in the linear mode, within a range of 2,000 to 20,000 daltons. The peaks of each spectrum were compared to the database (containing 5,623 reference spectra) and the score considered ideal for this study was higher or equal to 2.0 (identification at the species level). When there was no identification of the species, the colonies were tested for at least three times. The spectra were generated using Biotyper Software (Bruker Daltonics, Germany).
Cultivation-independent analysis
For the microbial diversity assessment of larvae exposed to IS, the ribosomal intergenic space analysis (RISA) technique was applied, using the protocol of Solaiman & Marschner32. Three groups of five larvae were used: control (non-exposed), LC50 and LC99. All tests were made in triplicate. Finally, the larvae were washed subsequently in 70% ethanol for 3 min and three times in sterile ultra-pure water for a superficial disinfection. Each larva was macerated in 2 mL of sterile distilled water and subjected to the PowerSoil® kit for total DNA extraction, according to the manufacturer’s guidelines, and quantified through NanoDrop®. Extracted DNA samples were submitted to PCR, using the oligonucleotide primers 23S (5′ GGGTTBCCCATTCRG 3′) and 1406 (5′ TGYACACACCGCCCGT 3′). These primers amplify a gene rRNA operon region between small (16S) and large (23S) sub unities, termed intergenic spacer region. Each sample was prepared with 5 µL of the 5X tampon, 0.2 mM of dNTP, 10 µM of primers, Taq DNA polymerase 1U, 50 ng extracted DNA and Milli-Q sterile water to complete the volume of 50 µL. The process initiated with an initial denaturation at 94 °C for 2 min, followed by 35 denaturation cycles at 94 °C for 30 sec, followed by annealing at 51 °C for 30 sec, extension at 72 °C for 1 min, and at last finalextension at 72 °C for 5 min. The PCR products (30 µL of each PCR reaction) were placed on a 3% agarose gel in TBE 0.5X, and stained with GelRed® containing bromophenol. For the electrophoresis, 75 V and 120 mA were applied for 3 h.
Production of axenic larvae
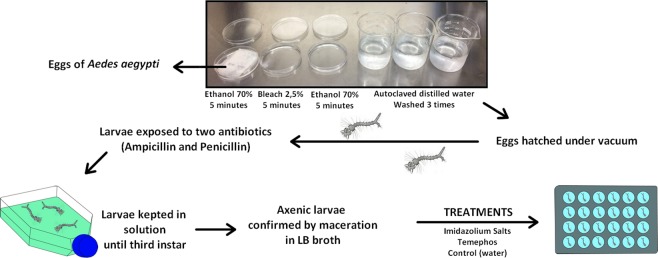
The protocol of Correa et al. was modified for the production of axenic larvae (Fig. 7)13. Thus, a filter paper strip with eggs of Ae. aegypti was disinfected by immersion for 5 min in 70% ethanol, followed by 5 min in aqueous 2.5% sodium hypochlorite and again in 70% ethanol for 5 min. Finally, the eggs were washed three times with autoclaved distilled water. For hatching, the eggs were immersed in 250 mL of phosphate-buffered saline solution (PBS) and exposed to vacuum at 25 Hz for 15 min at room temperature. After hatching, the larvae were exposed to two different antibiotics, supplementing the solution of hatching eggs with a final concentration of 75 µg/L of ampicillin and 125 µg/L of penicillin, following an incubation period of 4 h at 28 °C. Next, the larvae were transferred to a new medium containing 500 mL of distilled water and 4 g of E. coli agar, and then stored at a temperature of 28 °C, until reaching the third instar. For axenia control, 15 larvae were separated at the beginning of the treatment (first instar) and another 15 larvae at the end of the treatment (third instar), obtaining two control groups of triplicates of five larvae. They were then muddled, submitted to 14 mL of LB broth, and left for 48 h at 28 °C. For comparison purposes, the same was done with the laboratory larvae and the axenia was verified by bacterial growth in the broth by the turbidity of the medium.
Figure 7.
Experimental design for the production of axenic larvae of Aedes aegypti.
Axenic larvae susceptibility
Two groups of L3 axenic larvae were exposed to an IS LC99 concentration. Another group was exposed to Temephos to check if the absence of microorganisms in the larvae affected the efficacy of this larvicide. These bioassays were conducted in 24-well plates, in triplicate and repeated four times. For each well, 10 larvae were laid in 2 mL of the solutions (LC99 = 5 µg/mL for C18MImCl and C16MImMeS; and LC99 = 6 µg/L for Temephos), using also standard lab larvae as a control group. The larval mortality was determined after 24 and 48 h.
Susceptibility assays with re-colonized axenic larvae
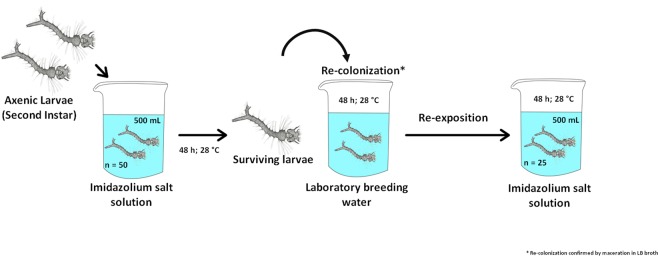
Axenic larvae that were exposed to the IS treatments and remained alive were exposed to usual larvae breeding water (Fig. 8). Subsequently, they were exposed again to the IS to check if the reintroduction of bacteria could influence in their mortality. The same procedures described above were followed for the production of axenic larvae. For both IS, fifty second-instar (to ensure a number of live larvae until the end of the experiment) larvae were exposed to 500 mL of an IS LC99 solution for 48 h at 28 °C (mortality was observed after 24 and 48 h). Afterwards, all the non-lethargic surviving larvae were laid into a breeding place with 500 mL of water, and incubated for 48 h at 28 °C (48 h–96 h). The number of re-exposed larvae was standardized to 25 before the second exposition to an IS solution, and the mortality was observed after 120 and 144 h. Axenia and re-colonization was confirmed by exposing 5 larvae to 14 mL of LB broth for 48 h at 28 °C. This test was performed when the larvae reached the second instar, before they were exposed for the first time to the IS, and after being fed for 48 h with the standard laboratory larvae breeding medium. All procedures were made in triplicate (50 axenic larvae; 25 recolonized per replicate).
Figure 8.
Experimental design for the treatment of axenic larvae of Aedes aegypti with IS, their re-colonization with bacteria, and subsequent second treatment with IS.
Statistical analysis
For the culture-independent assay, it was possible to verify the similarity between fingerprints generated by RISA. The bands were normalized creating a pattern of absence-presence of bands and generating a binary matrix, which was used to calculate the Dice coefficient. The results were grouped by the unweighted pair group method with arithmetic mean (UPGMA).
The diversity of bacteria communities was calculated through the diversity Shannon’s index (H′). For this, the intensity of the fingerprint bands was determined by densitometric analysis. The equation used was: H′ = −∑ (ni/N) log (ni/N), where ni is the amount related to the peak intensity of each band and N is the sum of all peak intensities, obtained through the densitometric curve33.
For the axenic larvae treatments and Shannon Index (H′), significant effects of the IS on larvae mortality were determined by two-way variance analysis (ANOVA). The differences between the groups were determined using Tukey’s test, considering the significance level P < 0.05.
Supplementary information
Acknowledgements
This study was financed in part by: the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) - Finance Code 001; the Conselho Nacional de Desenvolvimento Científico e Tecnológico - Brasil (CNPq); and the Fundação de Amparo à Pesquisa do Estado de Rio Grande do Sul - Brasil (FAPERGS); Institute of Petroleum and Natural Resources (PUCRS) for providing the primers used in this study.
Author contributions
C.E. Silva, G. Corção, H.S. Schrekker, and O.S. da Silva designed research; H.L. Pilz-Júnior, A.B. de Lemos, and K.N. de Almeida performed research; and H.L Pilz-Júnior, A.B. de Lemos, G. Corção, H.S. Schrekker, and O.S. da Silva wrote the paper.
Data availability
No datasets were generated or analysed during the current study.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Henri Stephan Schrekker, Email: henri.schrekker@ufrgs.br.
Onilda Santos da Silva, Email: onilda.silva@ufrgs.br.
Supplementary information
is available for this paper at 10.1038/s41598-019-52687-4.
References
- 1.Souza-Neto JA, Powell JR, Bonizzoni M. Aedes aegypti vector competence studies: A review. Infect. Genet. Evol. 2019;67:191–209. doi: 10.1016/j.meegid.2018.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ducheyne E, et al. Current and future distribution of Aedes aegypti and Aedes albopictus (Diptera: Culicidae) in WHO Eastern Mediterranean Region. Int. J. Health Geogr. 2018;17:1–13. doi: 10.1186/s12942-018-0125-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kraemer MUG, et al. Past and future spread of the arbovirus vectors Aedes aegypti and Aedes albopictus. Nat. Microbiol. 2019;4:854–863. doi: 10.1038/s41564-019-0376-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robinson M, Durbin A. Dengue vaccines: implications for dengue control. Curr. Opin. Infect. Dis. 2017;30:449–454. doi: 10.1097/QCO.0000000000000394. [DOI] [PubMed] [Google Scholar]
- 5.WHO. Global Alert and Response (GAR). Dengue/dengue haemorrhagic fever. Available at, http://www.who.int/csr/disease/dengue/en/ (2017).
- 6.Moyes CL, et al. Contemporary status of insecticide resistance in the major Aedes vectors of arboviruses infecting humans. PLoS Negl. Trop. Dis. 2017;11:e0005625. doi: 10.1371/journal.pntd.0005625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goellner E, et al. Larvicidal and residual activity of imidazolium salts against Aedes aegypti (Diptera: Culicidae) Pest Manag. Sci. 2018;74:1013–1019. doi: 10.1002/ps.4803. [DOI] [PubMed] [Google Scholar]
- 8.Furlani RE, Yeagley AA, Melander C. A flexible approach to 1,4-di-substituted 2-aminoimidazoles that inhibit and disperse biofilms and potentiate the effects of β-lactams against multi-drug resistant bacteria. Eur. J. Med. Chem. 2013;62:59–70. doi: 10.1016/j.ejmech.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 9.Raucci MG, et al. Antimicrobial imidazolium ionic liquids for the development of minimal invasive calcium phosphate-based bionanocomposites. ACS Appl. Mater. Interfaces. 2018;10:42766–42776. doi: 10.1021/acsami.8b12696. [DOI] [PubMed] [Google Scholar]
- 10.Schrekker CML, et al. Multitask imidazolium salt additives for innovative poly(l-lactide) biomaterials: morphology control, Candida spp. biofilm inhibition, human mesenchymal stem cell biocompatibility, and skin tolerance. ACS Appl. Mater. Interfaces. 2016;8:21163–21176. doi: 10.1021/acsami.6b06005. [DOI] [PubMed] [Google Scholar]
- 11.Egorova KS, Gordeev EG, Ananikov V. Biological activity of ionic liquids and their application in pharmaceutics and medicine. Chem. Rev. 2017;117(10):7132–7189. doi: 10.1021/acs.chemrev.6b00562. [DOI] [PubMed] [Google Scholar]
- 12.Rosso F, et al. Reduced diversity of gut microbiota in two Aedes mosquitoes species in areas of recent invasion. Sci. Rep. 2018;8:16091. doi: 10.1038/s41598-018-34640-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Correa MA, Matusovsky B, Brackney DE, Steven B. Generation of axenic Aedes aegypti demonstrate live bacteria are not required for mosquito development. Nat. Commun. 2018;9:4464. doi: 10.1038/s41467-018-07014-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong Y, Manfredini F, Dimopoulos G. Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog. 2009;5:e10000423. doi: 10.1371/journal.ppat.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez-Ceron L, Santillan F, Rodriguez MH, Mendez D, Hernandez-Avila JE. Bacteria in midguts of field-collected Anopheles albimanus block Plasmodium vivax sporogonic development. J. Med. Entomol. 2003;40:371–374. doi: 10.1603/0022-2585-40.3.371. [DOI] [PubMed] [Google Scholar]
- 16.Briones AM, Shililu J, Githure J, Novak R, Raskin L. Thorsellia anophelis is the dominant bacterium in a Kenyan population of adult Anopheles gambiae mosquitoes. ISME J. 2008;2:74–82. doi: 10.1038/ismej.2007.95. [DOI] [PubMed] [Google Scholar]
- 17.Joyce JD, Nogueira JR, Bales AA, Pittman KE, Anderson JR. Interactions between La Crosse virus and bacteria isolated from the digestive tract of Aedes albopictus (Diptera: Culicidae) J. Med. Entomol. 2011;48:389–394. doi: 10.1603/ME09268. [DOI] [PubMed] [Google Scholar]
- 18.Apte-Deshpande A, Paingankar M, Gokhale MD, Deobagkar DN. Serratia odorifera a midgut inhabitant of Aedes aegypti mosquito enhances its susceptibility to dengue-2 virus. PLoS One. 2012;7:e40401. doi: 10.1371/journal.pone.0040401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minard G, Mavingui P, Moro CV. Diversity and function of bacterial microbiota in the mosquito holobiont. Parasit Vectors. 2013;6:146. doi: 10.1186/1756-3305-6-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gusmão DS, et al. Culture-dependent and culture-independent characterization of microorganisms associated with Aedes aegypti (Diptera: Culicidae) (L.) and dynamics of bacterial colonization in the midgut. Acta Trop. 2010;115:275–281. doi: 10.1016/j.actatropica.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 21.Gaio ADO, et al. Contribution of midgut bacteria to blood digestion and egg production in Aedes aegypti (diptera: Culicidae) (L.) Parasit Vectors. 2011;4:105. doi: 10.1186/1756-3305-4-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yadav KK, et al. Molecular characterization of midgut microbiota of Aedes albopictus and Aedes aegypti from Arunachal Pradesh, India. Parasit Vectors. 2015;8:1–8. doi: 10.1186/s13071-014-0608-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dickson LB, et al. Carryover effects of larval exposure to different environmental bacteria drive adult trait variation in a mosquito vector. Sci. Adv. 2017;3:e1700585. doi: 10.1126/sciadv.1700585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Osei-Poku J, Mbogo CM, Palmer WJ, Jiggins FM. Deep sequencing reveals extensive variation in the gut microbiota of wild mosquitoes from Kenya. Mol. Ecol. 2012;21:5138–5150. doi: 10.1111/j.1365-294X.2012.05759.x. [DOI] [PubMed] [Google Scholar]
- 25.Pereira Lima JB, et al. Resistance of Aedes aegypti to organophosphates in several municipalities in the state of Rio de Janeiro and Espírito Santo, Brazil. Am. J. Trop. Med. Hyg. 2003;68:329–333. doi: 10.4269/ajtmh.2003.68.329. [DOI] [PubMed] [Google Scholar]
- 26.Tetreau G, et al. Bacterial microbiota of Aedes aegypti mosquito larvae is altered by intoxication with Bacillus thuringiensis israelensis. Parasit Vectors. 2018;11:121. doi: 10.1186/s13071-018-2741-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patil CD, Borase HP, Salunke BK, Patil SV. Alteration in Bacillus thuringiensis toxicity by curing gut flora: Novel approach for mosquito resistance management. Parasitol. Res. 2013;112:3283–3288. doi: 10.1007/s00436-013-3507-z. [DOI] [PubMed] [Google Scholar]
- 28.Wei G, et al. Insect pathogenic fungus interacts with the gut microbiota to accelerate mosquito mortality. Proc Natl Acad Sci USA. 2017;114:5994–5999. doi: 10.1073/pnas.1703546114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toppino A, et al. A carborane-derivative “Click” reaction under heterogeneous conditions for the synthesis of a promising lipophilic MRI/GdBNCT agent. Chem. Eur. J. 2013;19:721–728. doi: 10.1002/chem.201201634. [DOI] [PubMed] [Google Scholar]
- 30.Schrekker HS, et al. Preparation, cation-anion interactions and physicochemical properties of ether-functionalized imidazolium ionic liquids. J. Braz. Chem. Soc. 2008;19:426–433. doi: 10.1590/S0103-50532008000300009. [DOI] [Google Scholar]
- 31.Tandina F, et al. Use of MALDI-TOF MS and culturomics to identify mosquitoes and their midgut microbiota. Parasit Vectors. 2016;9:1–11. doi: 10.1186/s13071-016-1776-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Solaiman, Z. & Marschner, P. DGGE and RISA protocols for microbial community analysis in soil. In Advanced Techniques in Soil Microbiology (eds Varma, A. & Oelmüller, R.) 167–180 (Springer, Berlin, Heidelberg, 2007).
- 33.Ciesielski S, et al. Ribosomal intergenic spacer analysis as a tool for monitoring methanogenic archaea changes in an anaerobic digester. Curr. Microbiol. 2013;67:240–248. doi: 10.1007/s00284-013-0353-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.