Abstract
Background
Timely initiation of appropriate antimicrobial can improve the outcome in terms of reduced morbidity and mortality in addition to reduced health-care costs. Availability of early preliminary Antimicrobial Susceptibility Test (AST) report will be useful in directing antimicrobial therapy. The aim of the study was to correlate AST by disc diffusion method, directly from positively flagged blood culture bottles, with the AST by automated method.
Methods
A total of 144 aerobic blood culture bottles flagged positive by the automated blood culture system were processed. The bacteria were pelleted by two-step centrifugation of the broth from the bottle and used to make a smear for Gram stain as well as an inoculum for antimicrobial sensitivity testing by Kirby Bauer disc diffusion method. Automated identification and AST were also carried out.
Results
On direct staining, 94 samples showed gram-negative bacilli, 39 showed gram-positive cocci, and 11 showed yeasts or polymicrobial growth. In the case of gram-negative bacteria, there was 99% categorical agreement between direct sensitivity testing and automated sensitivity testing with 1% disagreement. Among the gram-positive cocci, there was 96% categorical agreement with 4% disagreement between the two methods.
Conclusion
High degree of agreement between the two methods is promising and applicable to situations where automated sensitivity testing is not available. Even if the systems are available, this method would prove useful as an adjunct to standard AST reporting. This sensitivity report can be generated earlier than the conventional AST, enabling choice of appropriate antimicrobial.
Keywords: Disk diffusion antimicrobial tests, Microbial sensitivity test, Blood culture, Bacteria
Introduction
Availability of culture and sensitivity results in patients with infections is of importance to the clinicians in guiding them to select the most appropriate antimicrobial for treatment, thereby increasing the chances of maximal therapeutic effect. To this end, it is incumbent on the microbiology laboratory to provide such information in a timely manner, especially with reference to cases of blood stream infections. Timely initiation of appropriate antimicrobial along with supportive management may improve the outcome in terms of reduced morbidity and mortality in addition to reduced health-care costs.1, 2 With the advent of automated blood culture methods, the time to detection of the organism has been reduced from 3–4 days to 2–3 days. However, even with the automation in place, a subculture is required to obtain a pure growth, so that Antimicrobial Susceptibility Testing (AST) can be carried out either by the Kirby Bauer method or an automated method. Owing to this inherent delay, the empirical therapy started initially with broad spectrum antimicrobials perforce continues till the sensitivity results are made available. However, it is to be emphasized that about 20–50% of all the prescribed antimicrobials are inappropriate.3 Patients getting these inappropriate antimicrobials get no extra clinical benefits while being at risk of suffering from adverse effects.4 The most serious and ever-increasing public health problem is emergence of antimicrobial resistance due to the misuse and abuse of antimicrobials.5 These drug-resistant pathogens pose a threat to health of patients in a health-care setup. Various reports from around the world indicate that there is an increase in the incidence of infections with multidrug-resistant organisms along with increased mortality being seen in both developing and developed countries.6, 7, 8
One of the useful inputs in implementation of antimicrobial stewardship is early availability of AST, which can help the clinician to de-escalate the antimicrobial, thereby reducing the chances of emergence of resistant organisms. The disc diffusion method for AST takes 48 h for the result to be generated. This includes the 24-h time taken for subculture from the positively flagged culture bottle onto solid culture media to obtain a pure growth, in addition to AST, which takes another day to complete. Even the automated methods for AST take another half to one day for the results to be available.
In this study, we have carried out AST by disc diffusion method, directly from the positively flagged blood culture bottles, and correlated it with the AST by automated method.
Material and methods
This study was carried out in a large tertiary care center between September 2016 and February 2017. Study population included patients admitted to acute care facility with suspected bacterial infection. A total of 144 nonrepeat BacT/ALERT® aerobic blood culture bottles flagged positive by the automated blood culture system (BacT/ALERT3D; bioMerieux, France) were processed. The blood culture bottle flagged positive by the system was taken out, and after gentle shaking, 1.5 mL of the broth was drawn using a sterile syringe. This was centrifuged in a 1.5-mL microcentrifuge tube at 600 ×g for 10 min to pellet the resin and the red blood cells (MiniSpin centrifuge; Eppendorf, Germany). The supernatant was taken into another 1.5-mL microcentrifuge tube and centrifuged at 3000 ×g for 10 min (MiniSpin centrifuge; Eppendorf, Germany) to pellet the bacteria.9 A smear for Gram stain was prepared from the deposit. The rest of the sediment was processed for preparation of inoculum for direct AST. Samples showing only single organism on Gram staining were further processed for direct AST.
AST by Kirby Bauer disc diffusion method
This pellet was resuspended in sterile saline to make the turbidity equivalent to 0.5 McFarland. This suspension was used for making the lawn culture for AST on Mueller-Hinton Agar (HiMedia, India) (Fig. 1). Antimicrobial panels for testing were chosen based on the Clinical & Laboratory Standards Institute (CLSI) guidelines 2016 depending on whether the organism was gram positive or gram negative on staining.10 In the case of gram-negative organisms, the panel chosen included antimicrobials covering both Enterobacteriaceae and nonfermenters, whereas the gram-positive panel included antimicrobials against both Staphylococcus and Enterococcus (Table 1). The antimicrobial discs were procured from HiMedia Labs, India. After overnight incubation at 37°C, the results were interpreted as per CLSI guidelines. Simultaneously, subcultures from the positive-flagged bottle broth were performed on blood and MacConkey agar (HiMedia, India).
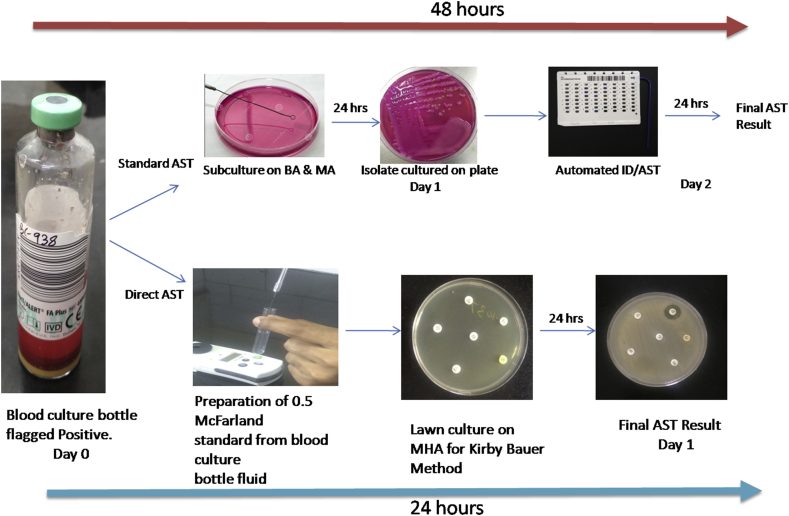
Fig. 1.
Flow chart for direct AST by disc diffusion and automated method. BA, blood agar; MA, MacConkey agar; MHA, Mueller-Hinton agar; ID/AST, identification/antimicrobial susceptibility test. Images are photographed by the authors.
Table 1.
List of antimicrobials tested against the isolates.
| Against GNB | Against GPC |
|---|---|
| Ampicillin 10 μg | Penicillin 10 units |
| Amoxicillin/clavulanic acid 20/10 μg | Cefoxitin 30 μg |
| Piperacillin/tazobactam 100/10 μg | Ciprofloxacin 5 μg |
| Cefuroxime 30 μg | Gentamicin 10 μg |
| Ceftazidime 30 μg | Erythromycin 15 μg |
| Ceftriaxone 30 μg | Clindamycin 2 μg |
| Cefepime 30 μg | Trimethoprim/sulfamethoxazole 1.25/23.75 μg |
| Ertapenem 10 μg | Linezolid 30 μg |
| Imipenem 10 μg | Teicoplanin 30 μg |
| Meropenem 10 μg | Vancomycin 30 μg |
| Amikacin 30 μg | |
| Gentamicin 10 μg | |
| Ciprofloxacin 5 μg | |
| Trimethoprim/sulfamethoxazole 1.25/23.75 μg |
GNB, gram-negative bacilli; GPC, gram-positive cocci.
For the interpretation of the direct AST results, the growth on the plates which had been subcultured from the positive blood culture bottle was classified based on the following biochemical reactions; catalase-positive and oxidase-negative gram-negative bacilli (GNB) along with colony morphology were presumptively identified as Enterobacteriaceae. Similarly, catalase-positive and oxidase-positive GNB were taken as nonfermenters. However, catalase-positive, oxidase-negative, gram-negative coccobacilli were presumed to be acinetobacter spp.
For the presumptive identification of gram-positive cocci (GPC), the following parameters were used. Typical colony morphology followed by catalase test and slide coagulase was used for classifying the organism as either Staphylococci, Enterococci, or Streptococci. The results of the direct AST were interpreted on the basis of oxidase test from the growth obtained on the subcultured plates.
Automated identification and AST
Simultaneously the positively flagged blood culture was subcultured on to blood agar and MacConkey agar. The growth on the solid media was further used for bacterial identification and AST using an automated identification (ID) and AST system (VITEK-2 Compact; bioMerieux, France) using appropriate Vitek ID cards and AST cards (N280/N281 for GNB and P628 for GPC). The N280 card was used for oxidase test–negative GNB and gram-negative Coccobacilli. The N281 card was used for oxidase test–positive GNB. The P628 card was used for Staphylococci spp. and Enterococci spp. No Streptococci were isolated during the period of study. VITEK-2 agreement of minimum 90% for the ID and green/yellow colors for confidence for advanced expert system (AES) were taken into consideration for the study.
After matching the results of the two methods, four interpretations were given11:
-
1.
Categorical agreement: when the results of AST by the two methods were in concordance.
-
2.
Very major errors (VMEs) (false susceptibility): when the isolate was sensitive to a drug by direct AST but turned out to be resistant by the standard automated AST method.
-
3.
Major errors (MEs) (false resistance): when the isolate was resistant to a drug by direct AST but turned out to be sensitive in the standard automated AST.
-
4.
Minor errors (mE): when the isolate was intermediate to a drug by direct AST but turned out to be either sensitive or resistant by the automated AST system.
Result
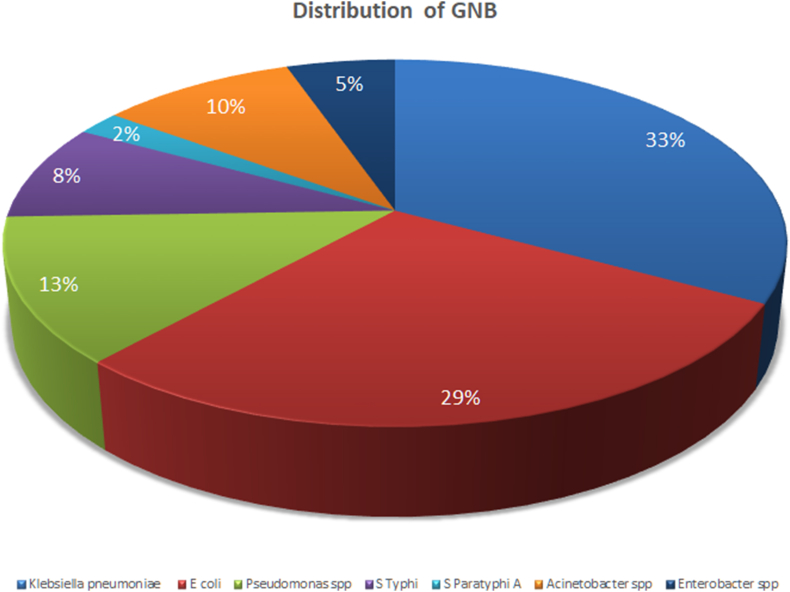
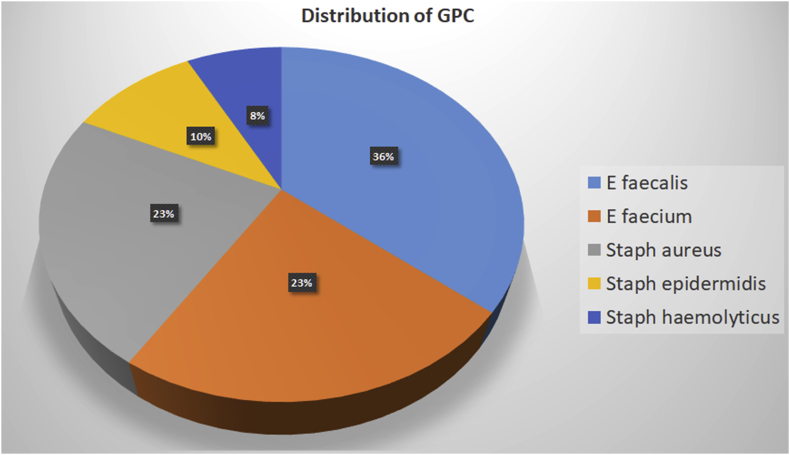
A total of 144 positive-flagged aerobic blood culture bottles were processed. On direct gram staining from the culture bottle fluid pellet, 94 samples showed gram-negative organisms, and 39 were GPC. Seven bottles showing yeasts and four bottles showing more than one organism were excluded from the study. There was a complete match between direct Gram stain result from the positive bottle and Gram stain from subcultures from the bottles obtained after overnight culture on solid media. The distribution of organisms identified by the automated system is given in Fig. 2, Fig. 3.
Fig. 2.
Distribution of gram-negative organisms isolated from positive blood cultures. GNB, gram-negative bacilli.
Fig. 3.
Distribution of gram-positive organisms isolated from positive blood cultures. GPC, gram-positive cocci.
A total of 949 isolate and antimicrobial agent combinations were generated from the sensitivity results in respect of Enterobacteriaceae group of organisms. Out of these, 939 (98.95%) combinations showed categorical agreement, whereas 10 combinations showed disagreement of which 2 (0.21%) were VMEs, 4 (0.42%) were MEs, and 4 (0.42%) were mEs (Table 2a). The categorical agreement for individual antimicrobials ranged from 98.63% for meropenem to 100% for ampicillin, piperacillin/tazobactam, ceftriaxone, ertapenem, amikacin, and ciprofloxacin.
Table 2a.
Agreement between direct AST and automated VITEK-2 AST among enterobacteriaceae (n = 73).
| Antimicrobial agent | Number of isolates tested |
|||
|---|---|---|---|---|
| Agreement | VME | ME | mE | |
| Ampicillin | 73 | |||
| Amoxicillin/clavulanic acid | 72 | 1 | ||
| Cefuroxime | 72 | 1 | ||
| Piperacillin/tazobactam | 73 | |||
| Ceftriaxone | 73 | |||
| Cefepime | 72 | 1 | ||
| Ertapenem | 73 | |||
| Imipenem | 72 | 1 | ||
| Meropenem | 72 | 1 | ||
| Amikacin | 73 | |||
| Gentamicin | 70 | 1 | 2 | |
| Ciprofloxacin | 73 | |||
| Trimethoprim/sulfamethoxazole | 71 | 1 | 1 | |
| Overall agreement for isolate antimicrobial combinations (%)a | 939 (98.95) | 2 (0.21) | 4 (0.42) | 4 (0.42) |
AST, antimicrobial susceptibility test; VME, very major error; ME, major error; mE, minor error.
Total number of isolate antimicrobial combinations = 949 (73 isolates × 13 antimicrobials).
In the case of nonfermenters that included the pseudomonas and the Acinetobacter group of organisms, 165 out of 168 (98.21%) combinations showed complete agreement. Only 3 out of these 168 combinations showed disagreement, of which 2 (1.19%) were MEs and 1 (0.60%) was mE. No VME was noted (Table 2b). The categorical agreement for individual antimicrobials ranged from 96.8% for meropenem to 100% for piperacillin/tazobactam, ceftazidime, ceftriaxone, cefepime, amikacin, trimethoprim, and ciprofloxacin.
Table 2b.
Agreement between direct AST and automated VITEK-2 Compact AST among gram-negative nonfermenter bacilli (n = 21, including 9 Acinetobacter spp. and 12 Pseudomonas spp.).
| Antimicrobial agent | Number of isolates tested |
|||
|---|---|---|---|---|
| Agreement | VME | ME | mE | |
| Piperacillin/tazobactam | 21 | |||
| Ceftazidimea | 12 | |||
| Ceftriaxonec | 09 | |||
| Cefepime | 21 | |||
| Imipenem | 20 | 1 | ||
| Meropenem | 19 | 1 | 1 | |
| Amikacina,b | 12 | |||
| Gentamicin | 21 | |||
| Ciprofloxacin | 21 | |||
| Trimethoprim/sulfamethoxazolec | 09 | |||
| Overall agreement for isolate antimicrobial combinations (%)d | 165 (98.21) | 2 (1.19) | 1 (0.60) | |
AST, antimicrobial susceptibility test; VME, very major error; ME, major error; mE, minor error.
Ceftazidime and amikacin tested only for Pseudomonas spp.
Sensitivity result for amikacin was not available from the automated system for comparison with disc diffusion for acinetobacter.
Ceftriaxone and trimethoprim/sulfamethoxazole tested only for Acinetobacter spp.
Total number of isolate antimicrobial combinations = 168 (21 isolates × 6 antimicrobials = 126; 9 isolates [acinetobacter] × 2 antimicrobials = 18; 12 isolates [pseudomonas] × 2 antimicrobials = 24).
Among the GPC, of the 144 isolate and antimicrobial agent combinations generated for Staphylococcus spp., 136 (94.44%) combinations showed complete agreement with a total of 5.56% errors, which included 2 (1.39%) combinations with VME, 2 (1.39%) combinations with ME, and 4 (2.78%) combinations with mE (Table 3a). Four isolates among these were methicillin-resistant Staphylococcus aureus (MRSA) by both the direct and the conventional AST methods. Of the 138 isolate and antimicrobial agent combinations tested for Enterococci spp., 135 (97.83%) combinations showed complete agreement with a total of 2.17% errors, which included 2 (1.45%) combinations with MEs and 1 (0.72%) combination with mEs (Table 3b). One isolate of Enterococci was vancomycin-resistant Enterococcus by both the direct and the conventional AST methods. Higher errors observed among Staphylococci spp. could be because of the lower number of isolates tested. This needs further study with more number of isolates. Complete agreement was seen for penicillin, cefoxitin, clindamycin, linezolid, and teicoplanin for Staphylococci spp. Similarly, complete agreement was seen for penicillin, ciprofloxacin, linezolid, and teicoplanin for Enterococci spp.
Table 3a.
Agreement between direct AST and automated VITEK-2 Compact AST among Staphylococcus spp (n = 16).
| Antimicrobial agent | Number of isolates tested |
|||
|---|---|---|---|---|
| Agreement | VME | ME | mE | |
| Penicillin | 16 | |||
| Cefoxitina | 16 | |||
| Ciprofloxacin | 15 | 1 | ||
| Gentamicin | 14 | 1 | 1 | |
| Erythromycin | 15 | 1 | ||
| Clindamycin | 16 | |||
| Trimethoprim/sulfamethoxazole | 12 | 1 | 1 | 2 |
| Linezolid | 16 | |||
| Teicoplanin | 16 | |||
| Overall agreement for isolate antimicrobial combinations (%)b | 136 (94.44) | 2 (1.39) | 2 (1.39) | 4 (2.78) |
AST, antimicrobial susceptibility test; VME, very major error; ME, major error; mE, minor error.
Four isolates were methicillin resistant.
Total number of isolate and antimicrobial combinations = 144 (16 isolates × 9 antimicrobials).
Table 3b.
Comparison of result between direct AST and automated VITEK-2 Compact AST among Enterococcus spp. (n = 23).
| Antimicrobial agent | Number of isolates tested |
|||
|---|---|---|---|---|
| Agreement | VME | ME | mE | |
| Penicillin | 23 | |||
| Ciprofloxacin | 23 | |||
| Erythromycin | 22 | 1 | ||
| Linezolid | 23 | |||
| Teicoplanin | 23 | |||
| Vancomycina | 21 | 1 | 1 | |
| Overall agreement for isolate antimicrobial combinations (%)b | 135 (97.83%) | 2 (1.45%) | 1 (0.72%) | |
AST, antimicrobial susceptibility test; VME, very major error; ME, major error; mE, minor error.
One was vancomycin resistant.
Total isolate and antimicrobial combinations = 138 (23 isolate × 6 antimicrobials).
Discussion
Our study showed a very good categorical agreement for the gram-negative organisms. In the case of gram-positive organisms, the percentage of errors for some of the antimicrobials such as gentamicin, cotrimoxazole, and vancomycin appears high possibly because of the low numbers of isolate-antimicrobial combinations tested in case of these antimicrobials. Testing with more number of such combinations is needed before arriving at a firm conclusion for these antimicrobials. The errors may be attributed to the limitations of the AES of VITEK-2 and will require further evaluation.12 However, limitations of the Vitek AES have not been considered for this study.
Various studies have compared the direct AST with the standard AST from blood culture bottles using different automated culture systems.13, 14 Most of these studies have found very good categorical agreement for the gram-negative organisms and not so good agreement for gram-positive organisms.14, 15 However, good categorical agreement for gram-positive organisms has been reported by Lupetti et al.13
Overall the high degree of agreement seen between the direct AST and the standard AST gives us an indication that these results can be useful to the clinician in deciding or modifying the specific antimicrobial therapy at the earliest saving as much as 24 critical hours. The present study was carried out targeting the acutely ill patients. However, it can be extrapolated to acute cases from the Out Patient Department (OPD) who are blood culture positive. Smaller hospitals lacking automated facilities will find this method highly useful for directing therapy in critically ill patients.
In routine practice, the subculture of the aspirate is made from the blood culture bottle once it is flagged positive by the automated culture system, and it takes 18–24 h for the growth to appear. Further processing for biochemical reactions and AST is carried out from the growth obtained on subculture. The method used in this study calls for direct inoculation onto appropriate media for AST which can result in saving of these 18–24 hours of delay and provide an idea of the sensitivity pattern in an earlier time frame especially with reference to acute care cases. Some workers have tried to inoculate the aspirate of the blood culture bottle directly onto Mueller-Hinton Agar (MHA) for performing the AST by disc diffusion method.16 However, this is a crude method as the final AST result can vary to a large extent depending on this single factor, i.e., the inoculum used for making the lawn culture. So, the method used in the present study, the differential centrifugation method, is the most accurate method that can be adapted from the available literature. This same inoculum prepared from the differential centrifugation method can also be used to perform the biochemical reactions for the identification of the organisms. Although we have not tried to perform the biochemical reactions from this inoculum in our study, if standardized, this can be a breakthrough achievement in early identification of the organisms that can prove very useful in centers where other options such as CHROMagar are not available. MHA with 5% sheep blood should be used instead of plain MHA for fastidious organisms such as Streptococcus spp. as per the CLSI guidelines. As the burden of these fastidious organisms in our center is very low, we have not used this media in our study. Some of the limitations of this study are the lack of definitive identification of the infecting bacteria and the exclusion of yeasts and polymicrobial organisms on Gram stain. Although the causative organism cannot be definitively identified by our method, it still enables preliminary AST testing, offering a chance for early institution of appropriate antimicrobial therapy. Interpretative criteria for some antimicrobials have not been defined for some of the organisms in the CLSI guidelines. In the case of S. aureus, no disc diffusion criteria have been described for vancomycin and teicoplanin; however, the same is available for Enterococcus spp. This necessitates testing of minimum inhibitory concentration to determine the susceptibility of all isolates of Staphylococci to vancomycin. The disk test neither does differentiate vancomycin-susceptible isolates of S. aureus from vancomycin-intermediate isolates, nor does it differentiate vancomycin-susceptible, intermediate, and vancomycin-resistant isolates of CoNS, all of which give similar size zones of inhibition. There were 4 strains of MRSA and 1 vancomycin-resistant Enterococcus isolated. However, vancomycin-intermediate or vancomycin-resistant S. aureus cannot be commented upon as there are no guidelines for disc diffusion testing of vancomycin against Staphylococcus spp. Inducible clindamycin resistance was not correlated in this study. Similarly, for colistin, no disc diffusion criteria are available for enterobacteriaceae group of organisms, Pseudomonas spp., and Acinetobacter spp. There is no disc diffusion criteria of cotrimoxazole available for Pseudomonas spp. Correlation of disc diameters with minimum inhibitory concentration values could have given a better idea; however, this was not attempted in the present study. We have also not compared the resistance phenotypes of GNB-like ESBL and Carbapenem Resistant Organisms (CRO) which are mentioned in the VITEK-2 results with that of the results of the disc diffusion. This is planned in further studies to be carried out presently.
Blondel-Hill et al have commented on the limitations of the VITEK-2 AES that at times incompatible results may be suggested, and thus, once the specific phenotypes are identified, there is no comparison of antibiotics within the same class; when biological corrections are more than one, the AES fails to identify the antibiotics with inconsistent results, which would be helpful to infer resistance mechanisms; and variability in susceptibility patterns of in different geographic regions and patient populations.17
Researchers have attempted to identify the microorganisms directly from the flagged blood culture bottles by detection methods such as matrix-assisted laser desorption ionization time of flight mass spectrometry, but the cost of the equipment is prohibitive especially in the peripheral health-care setup.18, 19 Chakravorty et al have tried using real-time polymerase chain reaction with molecular beacons for direct detection and speciation; however, this method is time-consuming, expensive, and labor-intensive.20 Others have attempted to perform AST directly from the positive-flagged culture bottle with the help of automated systems such as VITEK-2 system and Phoenix system (BD, USA) with good amount of correlation.11, 21 Chromogenic media (CHROMagar, France) have been used for direct identification of MRSA from blood cultures, whereas CHROMagar Mueller-Hinton Orientation medium has been developed subsequently and used for identification of organisms from urinary tract infection.22, 23 This medium can be used to presumptively carry out direct AST too.
In this present era with practically a limited number of antimicrobials in the development pipeline, optimum use of the existing antimicrobials is crucial. This misuse or abuse of antimicrobials has a direct relationship with the emergence and dissemination of resistant strains in health-care setups.24
All effort must be focused toward modifying the therapy to an appropriate antimicrobial at the earliest. The tagline for antimicrobial therapy is “Start Smart, and Then Focus”, which translates to starting empirically with a broad spectrum antimicrobial and then changing to a narrow spectrum when the culture AST results are available.25 Centre for Disease Control and Prevention, USA, Infectious Diseases Society of America, and European Centre for Disease Control have emphasized the judicious use of antimicrobials from time to time and have published guidelines for implementation of antimicrobial stewardship programs.25, 26, 27, 28 Taking cue, National Centre for Disease Control in India and the Indian Council of Medical Research have also published guidelines on the same subject.29, 30
Conclusion
This study has demonstrated good concordance between the direct AST and the automated VITEK-2 AST results. The method described here in this study will be useful in hospitals with laboratories lacking automated sensitivity testing systems. Even where automated AST systems are available, this method would prove useful as an adjunct to standard AST reporting and help contributing to the implementation of antimicrobial stewardship adhering to the principle of “Start Smart and Then Focus”.
Conflicts of interest
The authors have none to declare.
References
- 1.Gaieski D.F., Mikkelsen M.E., Band R.A. Impact of time to antibiotics on survival in patients with severe sepsis or septic shock in whom early goal-directed therapy was initiated in the emergency department. Crit Care Med. 2010;38(4):1045–1053. doi: 10.1097/CCM.0b013e3181cc4824. [DOI] [PubMed] [Google Scholar]
- 2.Sterling S.A., Miller W.R., Pryor J., Puskarich M.A., Jones A.E. The impact of timing of antibiotics on outcomes in severe sepsis and septic shock: a systematic review and meta-analysis. Crit Care Med. 2015;43(9):1907–1915. doi: 10.1097/CCM.0000000000001142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.US Department of Health and Human Services Core elements of hospital antibiotic stewardship programs. US Dep Health Hum Serv CDC. 2014:1–25. [Google Scholar]
- 4.Hensgens M.P.M., Goorhuis A., Dekkers O.M., Kuijper E.J. Time interval of increased risk for Clostridium difficile infection after exposure to antibiotics. J Antimicrob Chemother. 2011;67(3):742–748. doi: 10.1093/jac/dkr508. [DOI] [PubMed] [Google Scholar]
- 5.Huttner A., Harbarth S., Carlet J. Antimicrobial resistance: a global view from the 2013 world healthcare-associated infections forum. Antimicrob Resist Infect Control. 2013;2(1):31. doi: 10.1186/2047-2994-2-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paramythiotou E., Routsi C. Association between infections caused by multidrug-resistant gram-negative bacteria and mortality in critically ill patients. World J Crit Care Med. 2016;5(2):111. doi: 10.5492/wjccm.v5.i2.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colomb-Cotinat M., Lacoste J., Brun-Buisson C., Jarlier V., Coignard B., Vaux S. Estimating the morbidity and mortality associated with infections due to multidrug-resistant bacteria (MDRB), France, 2012. Antimicrob Resist Infect Control. 2016;5:56. doi: 10.1186/s13756-016-0154-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim C., Takahashi E., Hongsuwan M. Epidemiology and burden of multidrug-resistant bacterial infection in a developing country. Elife. 2016;5(September):1–18. doi: 10.7554/eLife.18082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gherardi G., Angeletti S., Panitti M. Comparative evaluation of the Vitek-2 compact and phoenix systems for rapid identification and antibiotic susceptibility testing directly from blood cultures of Gram-negative and Gram-positive isolates. Diagn Microbiol Infect Dis. 2012;72(1):20–31. doi: 10.1016/j.diagmicrobio.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute . 26th ed. 2016. Performance Standards for Antimicrobial Susceptibility Testing Supplement M100S. Wayne, Pennsylvania, USA. 252 p. [Google Scholar]
- 11.Machen A., Drake T., Wang Y.F.W. Same day identification and full panel antimicrobial susceptibility testing of bacteria from positive blood culture bottles made possible by a combined lysis-filtration method with MALDI-TOF VITEK mass spectrometry and the VITEK2 system. PLoS One. 2014;9(2) doi: 10.1371/journal.pone.0087870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bobenchik A.M., Hindler J.A., Giltner C.L., Saeki S., Humphries R.M. Performance of Vitek 2 for antimicrobial susceptibility testing of Staphylococcus spp. and Enterococcus spp. J Clin Microbiol. 2014;52(2):392–397. doi: 10.1128/JCM.02432-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lupetti A., Barnini S., Castagna B., Nibbering P.H., Campa M. Rapid identification and antimicrobial susceptibility testing of Gram-positive cocci in blood cultures by direct inoculation into the BD phoenix system. Clin Microbiol Infect. 2010;16(7):986–991. doi: 10.1111/j.1469-0691.2009.03006.x. [DOI] [PubMed] [Google Scholar]
- 14.Quesada M.D., Giménez M., Molinos S. Performance of VITEK-2 compact and overnight MicroScan panels for direct identification and susceptibility testing of Gram-negative bacilli from positive FAN BacT/ALERT blood culture bottles. Clin Microbiol Infect. 2010;16(2):137–140. doi: 10.1111/j.1469-0691.2009.02907.x. [DOI] [PubMed] [Google Scholar]
- 15.De Cueto M., Ceballos E., Martinez-Martinez L., Perea E.J., Pascual A. Use of positive blood cultures for direct identification and susceptibility testing with the Vitek 2 system. J Clin Microbiol. 2004;42(8):3734–3738. doi: 10.1128/JCM.42.8.3734-3738.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goel G., Das D., Mukherjee S. A method for early detection of antibiotic resistance in positive blood cultures: experience from an oncology centre in eastern India. Indian J Med Microbiol. 2015;33(5):53. doi: 10.4103/0255-0857.150883. [DOI] [PubMed] [Google Scholar]
- 17.Blondel-Hill E., Hetchler C., Andrews D., Lapointe L. Evaluation of VITEK 2 for analysis of enterobacteriaceae using the Advanced Expert System (AES) versus interpretive susceptibility guidelines used at Dynacare Kasper Medical Laboratories, Edmonton, Alberta. Clin Microbiol Infect. 2003;9(11):1091–1103. doi: 10.1046/j.1469-0691.2003.00697.x. [DOI] [PubMed] [Google Scholar]
- 18.Christner M., Rohde H., Wolters M., Sobottka I., Wegscheider K., Aepfelbacher M. Rapid identification of bacteria from positive blood culture bottles by use of matrix-assisted laser desorption-ionization time of flight mass spectrometry fingerprinting. J Clin Microbiol. 2010;48(5):1584–1591. doi: 10.1128/JCM.01831-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fothergill A., Kasinathan V., Hyman J., Walsh J., Drake T., Wang Y.F.W. Rapid identification of bacteria and yeasts from positive-blood-culture bottles by using a lysis-filtration method and matrix-assisted laser desorption ionization--time of flight mass spectrum analysis with the SARAMIS database. J Clin Microbiol. 2013;51(3):805–809. doi: 10.1128/JCM.02326-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chakravorty S., Aladegbami B., Burday M. Rapid universal identification of bacterial pathogens from clinical cultures by using a novel sloppy molecular beacon melting temperature signature technique. J Clin Microbiol. 2010;48(1):258–267. doi: 10.1128/JCM.01725-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hazelton B., Thomas L.C., Olma T. Rapid and accurate direct antibiotic susceptibility testing of blood culture broths using MALDI sepsityper combined with the BD phoenix automated system. J Med Microbiol. 2014;63:1590–1594. doi: 10.1099/jmm.0.075580-0. [DOI] [PubMed] [Google Scholar]
- 22.Chihara S., Hayden M.K., Minogue-Corbett E., Singh K. Shortened time to identify Staphylococcus species from blood cultures and methicillin resistance testing using CHROMAgar. Int J Microbiol. 2009;2009:5–8. doi: 10.1155/2009/636502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manickam K., Karlowsky J.A., Adam H. CHROMagar orientation medium reduces urine culture workload. J Clin Microbiol. 2013;51(4):1179–1183. doi: 10.1128/JCM.02877-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sievert D.M., Ricks P., Edwards J.R. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–2010. Infect Control Hosp Epidemiol. 2013;34(1):1–14. doi: 10.1086/668770. [DOI] [PubMed] [Google Scholar]
- 25.Public Health England Start Smart – then focus antimicrobial stewardship toolkit for English hospitals. Public Health Engl. 2015:1–26. https://www.gov.uk/government/publications/antimicrobial-stewardship-start-smart-then-focus Available from: [Google Scholar]
- 26.National Institute for Health and Care Excellence (NICE Guideline) 2015. Antimicrobial Stewardship: Systems and Processes for Effective Antimicrobial Medicine Use. [Internet] [cited 2017 Jul 26] [Google Scholar]
- 27.Public Health England . 2015. Start Smart Then Focus Appendix 1 Resource Materials: Examples of Audit Tools, Review Stickers and Drug Charts.www.nice.org.uk/guidance/ng15 [Internet] [cited 2017 Jul 26]. p. 1–28. Available from: [Google Scholar]
- 28.Infectious Diseases Society of America . 2016. New Antibiotic Stewardship Guidelines Focus on Practical Advice for Implementation; pp. 1–4.http://www.idsociety.org/New_Antimicrobial_Stewardship_Guidline_2016 [Internet] [cited 2017 Jul 30]. Available from: [Google Scholar]
- 29.Indian Council of Medical Research . 2017. Treatment Guidelines for Antimicrobial Use in Common Syndromes.http://www.icmr.nic.in/guidelines/treatment guidelines for antimicrobial.pdf New Delhi. Available from: [Google Scholar]
- 30.National Centre for Disease Control Ministry of Health and Family welfare Govt of India . 2016. National Treatment Guidelines for Antimicrobial Use in Infectious Diseases.http://www.ncdc.gov.in/writereaddata/linkimages/AMR_guideline7001495889.pdf Available from: [Google Scholar]