Abstract
Long noncoding RNAs (lncRNAs) have emerged as key regulators of cell differentiation and development. However, potential roles for lncRNAs in chondrogenic differentiation have remained poorly understood. Here we identify lncRNA ADAMTS9 antisense RNA 2, ADAMTS9-AS2, which controls the chondrogenic differentiation by acting as a competing endogenous RNA (ceRNA) in human mesenchymal stem cells (hMSCs). We screen out ADAMTS9-AS2 of undifferentiated and differentiated cells during chondrogenic differentiation by microarrays. Suppression or overexpression of lncRNA ADAMTS9-AS2 correlates with inhibition and promotion of hMSC chondrogenic differentiation, respectively. We find that ADAMTS9-AS2 can sponge miR-942-5p to regulate the expression of Scrg1, a transcription factor promoting chondrogenic gene expression. Finally, we confirm the function of ADAMTS9-AS2 to cartilage repair in the absence of transforming growth factor β (TGF-β) in vivo. In conclusion, ADAMTS9-AS2 plays an important role in chondrogenic differentiation as a ceRNA, so that it can be regarded as a therapy target for cartilage repair.
Keywords: LncRNA ADAMTS9-AS2, MiR-942-5p, chondrogenesis, hMSCs, cartilage regeneration, chondrogenic differentiation, cartilage defect
Introduction
Cartilage plays an important role in human growth and development, and the formation and growth of limb bones are derived from the endochondral ossification.1 Chondrogenesis involves condensation and arrangement of mesenchymal cells in the embryo.2 This process depends on precise control of key signal pathways and core transcription factors, such as Indian hedgehog signal pathway3 and Sox9.4 Osteoarthritis and other degenerative diseases can cause cartilage tear and progressive loss of chondrocytes. Articular cartilage has weak regeneration potential due to its avascularity.5 Chondrogenic differentiation from mesenchymal stem cells (MSCs) in vitro is a promising approach for cartilage repair, but the fibrosis and hypertrophy of chondrocytes has affected this process.6 Thus, knowledge of the molecular switch that controls chondrogenic differentiation is critical to acquiring a better understanding of cartilage development and to designing new strategies for cartilage degenerative disease.
MSCs show excellent tissue regeneration ability by their intrinsic capacity for self-renewal and multipotent differentiation.7 MSCs also display immunomodulatory capacities for T cell, B cell, and natural killer cell proliferation and function.8, 9 MSCs have been identified as attractive cell sources for cartilage repair for their chondrogenesis ability. So far, several transcription factors and growth factors are reported to promote MSC chondrogenesis, such as TGF-β10 and the insulin growth factor (IGF)11 superfamily. In addition, a variety of scaffolds combined with MSCs is used to boost cartilage regeneration.12 Thus, identifying additional factors that promote chondrogenic differentiation may provide new insights into cartilage repair.
LncRNAs are broadly classified as transcripts longer than 200 nt, and they have limited protein-coding potential.13, 14 lncRNAs are emerging as important players in cell differentiation and determination, such as controlling muscle differentiation15 and cardiovascular lineage commitment16 and driving thermogenic adipocyte differentiation,17 indicating that they have the potential ability to determine cell destiny. A new regulatory circuitry has been recently focused on that lncRNA can crosstalk with mRNA by competing for shared microRNAs (miRNAs).15 Such competing endogenous RNAs (ceRNAs) regulate the distribution of miRNA molecules on their targets and, thereby, impose an additional level of post-transcriptional regulation. This finding has prompted the in-depth studies of the circuitries that are regulated by this molecular mechanism.
Recent studies have shown that cartilage development and homeostasis are not only controlled by protein-coding genes but also regulated by specific miRNAs. For example, miR-140 shows dual effect in cartilage development and homeostasis,18 miR-146a facilitates osteoarthritis progression,19 and miR-221 promotes cartilage repair.20 Meanwhile, it has been reported that lncRNAs exert their roles as ceRNAs by repressing the functions of miRNAs in various research fields, such as lncARSR in renal cancer,21 lncRNA ODRUL that contributes to osteosarcoma progression,22 and lncRNA MD1 that controls muscle differentiation.15 Despite these inspiring findings, our knowledge of lncRNAs that function in chondrogenic differentiation is limited, and a detailed understanding of circuitries they regulate is lacking.
In this study, we report the identification of ADAMTS9 antisense RNA 2, ADAMTS9-AS2, an lncRNA in humans that is necessary for chondrogenic differentiation. By inducing human MSCs (hMSCs) for chondrogenic differentiation, we found that the expression of ADAMTS9-AS2 increased during chondrogenesis by microarrays. Then we explored the function of ADAMTS9-AS2 for hMSC chondrogenic differentiation in vitro. Further experiments showed that ADAMTS9-AS2 acted as a ceRNA for miR-942-5p. Moreover, miR-942-5p also played a critical role to regulate chondrogenic differentiation, and it exerted its function by regulating its target gene Scrg1. At last we confirmed that ADAMTS9-AS2 could facilitate cartilage repair in an in vivo microenvironment by seeding hMSCs into cartilage defects implanted subcutaneously into nude mice. Together, these results indicate that ADAMTS9-AS2 functions as a ceRNA that promotes hMSC differentiation toward chondrocytes. More broadly, our work identifies the capacity of ADAMTS9-AS2 for cartilage regeneration, and it suggests that ADAMTS9-AS2 may present a promising therapy target for cartilage degeneration diseases.
Results
ADAMTS9-AS2 Is Upregulated during Chondrogenic Differentiation
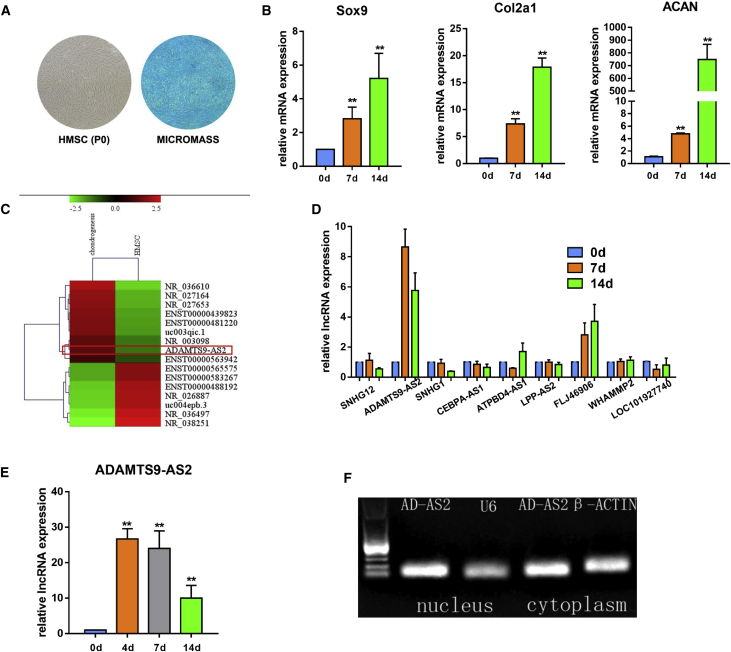
To test the chondrogenic differentiation ability of our hMSCs, we cultured hMSCs in chondrogenic induced medium as a micromass model.23 After standard chondrogenic induction, we tested the glycosaminoglycan expression in the extracellular matrix by Alcian blue staining, and we detected chondrocyte marker gene expression by qPCR. The content of glycosaminoglycan in the extracellular matrix increased after differentiation, and chondrocyte marker genes were continuously upregulated during chondrogenic differentiation (Figures 1A and 1B).
Figure 1.
lncRNA ADAMTS9-AS2 Is Upregulated during hMSC Chondrogenic Differentiation
(A) Isolated hMSCs from bone marrow and Alcian blue staining for hMSC micromass chondrogenesis on 14 days. (B) mRNA level of chondrogenic genes (Sox9, Col2α1, and ACAN) during chondrogenic differentiation on 14 days. (C) Heatmaps of lncRNA differentially expressed during hMSC chondrogenesis. (D) Expression of the upregulated and downregulated lncRNAs by qPCR. (E) qPCR for the expression of ADAMTS9-AS2 during chondrogenic differentiation at the indicated time points. (F) RT-PCR analysis indicating ADAMTS9-AS2 localization in the nucleus and the cytoplasm. Experiments were performed in triplicate and error bars represent SD of a triplicate set of experiments. Data are shown as mean ± SD; *p, 0.05; **p, 0.01; ns, non-significant.
To determine lncRNAs that affect differentiation, we used microarray analysis to compare undifferentiated and differentiated cells during chondrogenesis (Figure 1C). Next, we confirmed that the expression of ADAMTS9-AS2 was increased during differentiation by qPCR, which was consistent with the microarray result (Figure 1D). In addition, ADAMTS9-AS2 exhibited a gradual increase until 7 days and then it reduced expression in 14 days (Figure 1E). To determine the subcellular localization of ADAMTS9-AS2, we separated nuclear and cytoplasmic RNA, respectively, and ADAMTS9-AS2 was specifically amplified by reverse transcription PCR. Through standard denaturing agarose gel electrophoresis, it revealed that ADAMTS9-AS2 localized both in the nucleus and cytoplasm (Figure 1F).
ADAMTS9-AS2 Regulates hMSC Chondrogenic Differentiation In Vitro
To determine the biological effects of ADAMTS9-AS2 on the chondrogenic differentiation of hMSCs, we constructed lentiviruses for lncRNA knockdown and overexpression. To avoid potential off-target effects of small hairpin RNA (shRNA) plasmids, we constructed five shRNA plasmids and only one could inhibit the expression of ADAMTS9-AS2 (data not shown). Meanwhile, because of lncRNA ADAMTS9-AS2 existing both in the nucleus and cytoplasm, we also tested the antisense oligonucleotide (ASO) efficiency to knock down this lncRNA (Figure S1). We further tested the efficiency of the two lentiviruses to change lncRNA expression in hMSCs (Figures 2A and 3A).
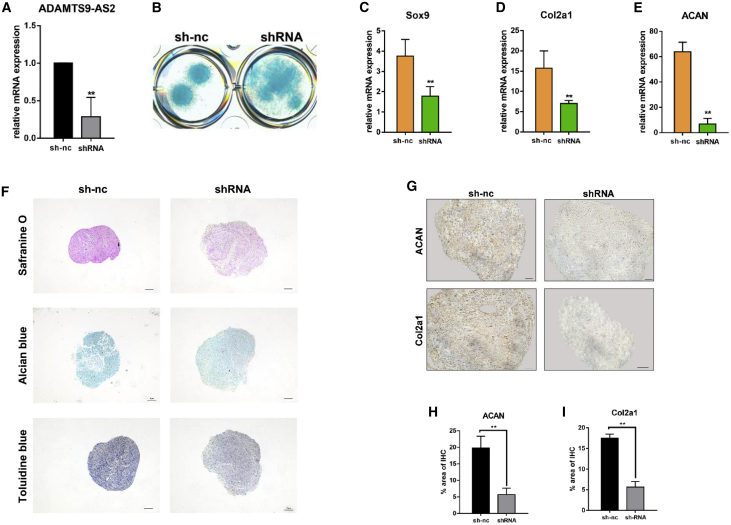
Figure 2.
ADAMTS9-AS2 Is Required for Proper Chondrogenic Differentiation
(A) Targeting of ADAMTS9-AS2 in hMSCs by small hairpin RNA-constructed lentivirus led to significant depletion of the transcript, as measured by qPCR. (B) ADAMTS9-AS2 depletion affects cell condensation morphology and Alcian blue staining on cells on day 7. (C–E) Expression of chondrogenic genes measured by qPCR in ADAMTS9-AS2-depleted hMSCs after 14 days of chondrogenic differentiation in negative control and shRNA treatment group. (F) Synthetic glycosaminoglycan in ADAMTS9-AS2-knockdown hMSCs compared to negative controls on day 14, assessed by Safranine O staining, Alcian blue staining, and toluidine blue staining. Scale bar, 100 μm. (G) Antibody staining of paraffin-embedded sections of day 14 micromass showed Col2α1 and ACAN abundance between negative control and shRNA treatment group. Scale bar, 50 μm. (H and I) Quantitative statistics for dyeing results of (G) between negative control and shRNA treatment group. Experiments were performed in triplicate and error bars represent SD of a triplicate set of experiments. Data are shown as mean ± SD; *p, 0.05; **p, 0.01; ns, non-significant.
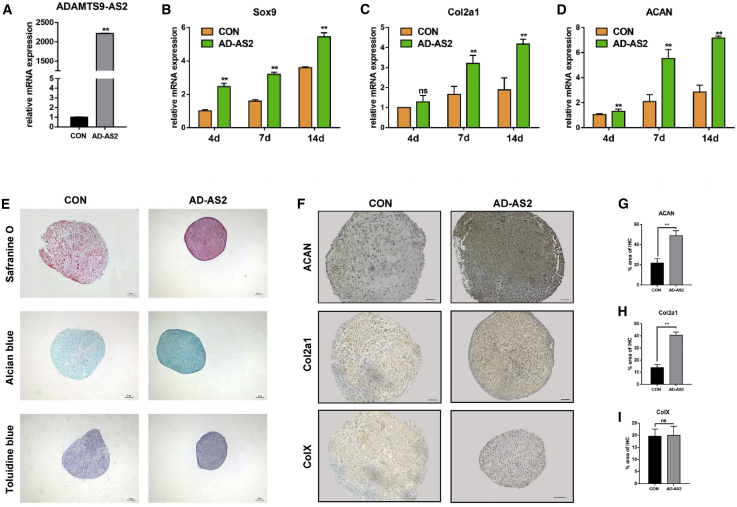
Figure 3.
ADAMTS9-AS2 Promotes hMSC Chondrogenic Differentiation
(A) Forced expression of ADAMTS9-AS2 in hMSCs led to a significant increase of the transcript, as measured by qPCR. (B–D) Expression of chondrogenic genes measured by qPCR at representative time points in hMSCs during chondrogenic differentiation between control and ADAMTS9-AS2 overexpression group. (E) Synthetic glycosaminoglycan in ADAMTS9-AS2-overexpressed hMSCs compared to controls on day 14, assessed by Safranine O staining, Alcian blue staining, and toluidine blue staining. Scale bar, 100 μm. (F) Antibody staining of paraffin-embedded sections of day 14 micromass showed Col2α1, ACAN, and ColX abundance between control and ADAMTS9-AS2 overexpression group. Scale bar, 50 μm. (G–I) Quantitative statistics for dyeing results of (F) between control and ADAMTS9-AS2 overexpression group. Experiments were performed in triplicate and error bars represent SD of a triplicate set of experiments. Data are shown as mean ± SD; *p, 0.05; **p, 0.01; ns, non-significant.
Chondrogenesis of MSCs requires the ordered arrangement and condensation of the cells for subsequent differentiation,2 so that the condensation process is a critical step during stem cell chondrogenesis. Therefore, we used low-density micromass culture to detect the influence of ADAMTS9-AS2 on hMSC condensation. It revealed that knockdown of ADAMTS9-AS2 interfered with hMSC condensation ability during chondrogenic differentiation (Figure 2B). We found that ADAMTS9-AS2 inhibition significantly decreased the mRNA levels of chondrogenic genes, including Sox9, Col2α1, and ACAN, in hMSCs (Figures 2C–2E). Conversely, ADAMTS9-AS2 overexpression increased the mRNA levels of these genes in hMSCs (Figures 3B–3D). Safranine O staining, Alcian blue staining, and toluidine blue staining showed that ADAMTS9-AS2 overexpression increased the synthesis of glycosaminoglycan and formed a more aggregated shape, whereas chondrocyte extracellular matrix decreased after shRNA lentivirus treatment (Figures 2F and 3E). Then we performed immunohistochemistry staining to detect the protein levels of chondrogenic genes in micromass. It also revealed that ADAMTS9-AS2 overexpression increased Col2α1 and ACAN protein levels (Figure 2G) and suppression of ADAMTS9-AS2 limited their expression (Figure 3F).
It is well known that the expression of collagen type X (ColX), a marker gene for chondrocyte hypertrophy and apoptotic death, is an undesired outcome of a cell-based approach to cartilage regeneration.24 We tested the protein level of ColX by immunohistochemistry staining. It showed that, although forced expression of ADAMTS9-AS2 promoted chondrogenic differentiation, it did not increase ColX expression (Figure 3F). Furthermore, the dye depth of the staining may cause false-positive results depending on the cell quantity, so we examined the effect of ADAMTS9-AS2 on cell proliferation. We used the PrestoBlue Cell Viability Reagent to test cell proliferation for 4 consecutive days, and we found no significant difference after the hMSCs were treated with ADAMTS9-AS2-overexpressing lentivirus (data not shown).
ADAMTS9-AS2 Acts as a Sponge of miR-942-5p, and miR-942-5p Can Negatively Regulate Chondrogenic Differentiation
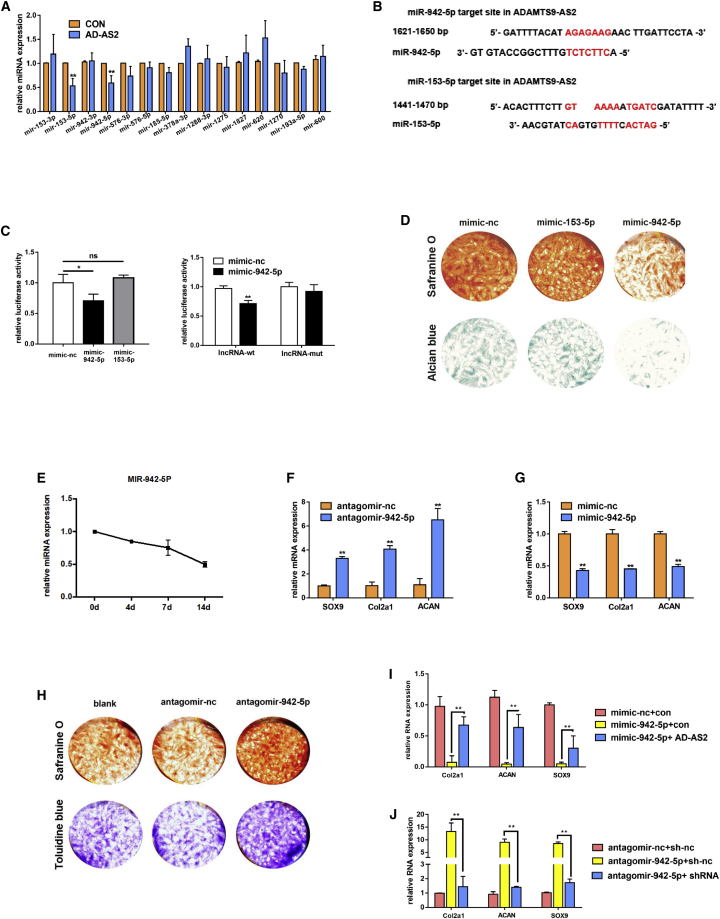
Recent studies have focused on lncRNA function as a ceRNA by sponging miRNA and regulating its target transcription factor.15 According to the target prediction websites (miRDB, http://mirdb.org/custom.html; TargetScan, www.targetscan.org; and Computational Medicine Center, https://cm.jefferson.edu/), we collected 15 potential miRNAs that exist in the above three databases simultaneously. Referring to some research articles, we found that lncRNAs could regulate the sponging miRNAs.15, 25, 26 We detected these 15 miRNA levels after ADAMTS9-AS2 overexpression, and only two miRNAs (miR-153-5p and miR-942-5p) changed (Figure 4A). Furthermore, we found that ADAMTS9-AS2 contained complementary sequences with miR-153-5p and miR-942-5p (Figure 4B).
Figure 4.
ADAMTS9-AS2 Acts as a Sponge of miR-942-5p, and miR-942-5p Can Negatively Regulate Chondrogenic Differentiation
(A) Expression of miRNAs measured by qPCR after overexpressing ADAMTS9-AS2 in hMSCs. (B) Schematic of the miR-153-5p and the miR-942-5p putative target site on ADAMTS9-AS2. (C) The luciferase reporter assay for the wide-type ADAMTS9-AS2 in the presence of miR-153-5p and miR-942-5p and the mutant ADAMTS9-AS2 in the presence of miR-942-5p. The values are showed as Renilla luciferase activity to firefly luciferase activity. (D) Representative images of hMSCs transfected with mimic-nc, mimic-153-5p, and mimic-942-5p after 14 days of chondrogenic differentiation. (E) qPCR for the expression of miR-942-5p during chondrogenic differentiation at the indicated time points. (F) Expression of chondrogenic genes measured by qPCR in transfected hMSCs with mimic-942-5p after 14 days of chondrogenic differentiation. (G) Expression of chondrogenic genes measured by qPCR in transfected hMSCs with antagomir-942-5p after 14 days of chondrogenic differentiation. (H) Representative images of hMSCs transfected with blank, antagomir-nc, and antagomir-942-5p after 14 days of chondrogenic differentiation. (I and J) qPCR for mRNA levels of chondrogenic genes in different groups as shown in the figure. Experiments were performed in triplicate and error bars represent SD of a triplicate set of experiments. Data are shown as mean ± SD; *p, 0.05, **p, 0.01; ns, non-significant.
To determine whether ADAMTS9-AS2 directly regulates the two miRNAs, we generated a luciferase reporter construct. It showed that the lncRNA reporter was suppressed by miR-942-5p rather than miR-153-5p. Meanwhile, the mutant lncRNA reporter was not affected by miR-942-5p (Figure 4C). In addition, to prove the accuracy of miR-942-5p, we detected the two miRNAs’ function in chondrogenic differentiation. It showed that only miR-942-5p could regulate hMSC chondrogenic differentiation (Figure 4D). Thus, we believed that miR-942-5p could interact with ADAMTS9-AS2 and it also played a critical role during chondrogenic differentiation.
We examined the change of miR-942-5p during chondrogenic differentiation to further explore its specific function on chondrogenesis. miR-942-5p exhibited a gradual decrease after chondrogenic induction (Figure 4E). We further detected the mRNA levels of chondrogenic genes under miR-942-5p knockdown and overexpression after chondrogenic induction. It revealed that suppression of miR-942-5p increased the chondrogenic genes’ expression (Figure 4F). Additionally, overexpression of miR-942-5p inhibited their expression (Figure 4G). The staining for the extracellular matrix by Safranine O and toluidine blue also showed the same trend results (Figure 4H).
Now that ADAMTS9-AS2 could sponge miR-942-5p, we predicted that ADAMTS9-AS2 could reverse the miR-942-5p effect on hMSC chondrogenic differentiation. We found that inhibition of chondrogenic differentiation causing by miR-942-5p could be rescued though overexpression of ADAMTS9-AS2 (Figure 4I). Meanwhile, after suppression of ADAMTS9-AS2, the promotion effect of hMSCs chondrogenic differentiation by downregulating miR-942-5p was reversed (Figure 4J).
ADAMTS9-AS2 Modulates Transcription Factor SCRG1 through Regulating miR-942-5p
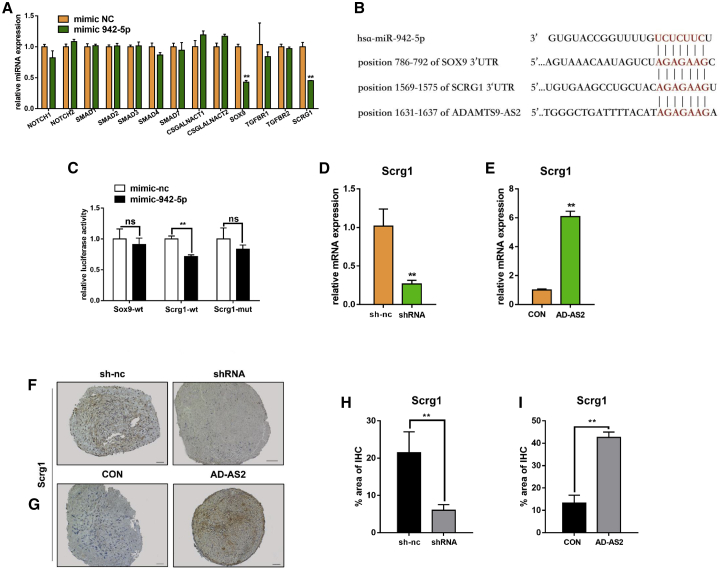
MiRNAs can direct the RNA-induced silencing complex (RISC) to downregulate gene expression by two posttranscriptional mechanisms: mRNA cleavage or translational repression.27 We sought the target genes of miR-942-5p from miRNA prediction website miRBase (www.mirbase.org), and we obtained large amounts of predicted target genes for miR-942-5p. Chondrogenesis was controlled by several key signaling pathways along with some transcriptional factors, such as Notch, Indian hedgehog, TGF-β signaling pathway, Sox9, and a series of genes. Among various putative targets for the miR-942-5p, we screened out 13 mRNAs encoding for proteins with a relevant function in chondrogenesis. Numerous miRNAs possess the ability to regulate mRNA decay.28 Hence, we tested the mRNA levels of these genes after miR-942-5p overexpression by qPCR. It revealed that only two genes (Sox9 and Scrg1) were inhibited after overexpressing miR-942-5p (Figure 5A). Then we found that ADAMTS9-AS2 contains the same complementary sequence with miR-942-5p as Scrg1 and Sox9 (Figure 5B), which further proved that ADAMTS9-AS2 functioned as a ceRNA to miR-942-5p target genes.
Figure 5.
ADAMTS9-AS2 Modulates Transcription Factor SCRG1 through Regulating miR-942-5p
(A) Expression of mRNAs measured by qPCR after hMSCs were treated with mimic-942-5p. (B) Schematic of the miR-942-5p putative target site on ADAMTS9-AS2, Sox9, and Scrg1. (C) The luciferase reporter assay for the wide-type Sox9 and Scrg1 in the presence of miR-942-5p and the mutant Scrg1 in the presence of miR-942-5p. The values are shown as Renilla luciferase activity to firefly luciferase activity. (D and E) qPCR for mRNA levels of Scrg1 in hMSCs after downregulating and overexpressing ADAMTS9-AS2. (F and G) Immunohistochemistry assay for sections of day 14 micromass for Scrg1 protein abundance in hMSCs after downregulating and overexpressing ADAMTS9-AS2. Scale bar, 50 μm. (H and I) Quantitative statistics for dyeing results of (F) and (G) after downregulating and overexpressing ADAMTS9-AS2. Experiments were performed in triplicate and error bars represent SD of a triplicate set of experiments. Data are shown as mean ± SD; *p, 0.05, **p, 0.01; ns, non-significant.
Furthermore, to determine whether miR-942-5p directly regulated the two genes, we constructed the two luciferase reporter plasmids. The result revealed that only the Scrg1 reporter was inhibited by miR-942-5p and the Sox9 reporter was not influenced by miR-942-5p. Additionally, the mutant Scrg1 reporter was not affected by miR-942-5p (Figure 5C). The miR-942-5p had a direct effect on Scrg1, and it could influence Sox9 expression without interacting with it directly. Consistent with ADAMTS9-AS2 being a decoy for miR-942-5p, we proved that ADAMTS9-AS2 depletion reduced the levels of Scrg1 (Figure 5D) while its overexpression produced an increase in mRNA level (Figure 5E). The same trend results were demonstrated on protein levels in micromass by immunohistochemistry staining (Figures 5F and 5G).
ADAMTS9-AS2 Also Can Promote hMSC Chondrogenic Differentiation In Vivo
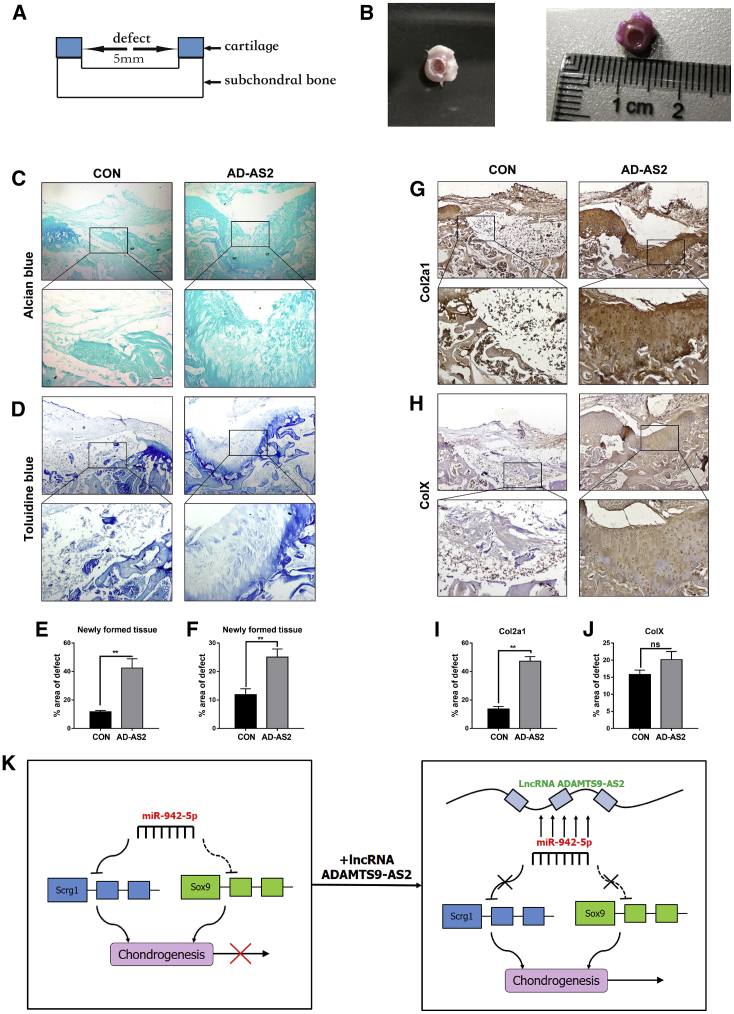
To validate the promoting effect on chondrogenic differentiation of ADAMTS9-AS2 in vivo, hMSCs were cultured in an osteochondral microenvironment presented by a cartilage defect model (Figures 6A and 6B). For in vivo experiments, hMSCs treated with ADAMTS9-AS2-overexpressing lentivirus or control lentivirus compounding with alginate were seeded in the osteochondral defects, and then we implanted the defects subcutaneously into nude mice. After 16 weeks, ADAMTS9-AS2-overexpressing hMSCs generated a cartilage-characterized region by extensive production of glycosaminoglycans, as evidenced by the presence of a large Alcian blue- and toluidine blue-positive area throughout the entire region of the defect. This phenomenon was different from the newly formed tissue on the cartilage defect when control hMSCs were used (Figures 6C and 6D). Consistent with these results, immunohistochemistry staining analysis revealed a significantly stronger staining for the chondrogenic marker gene, Col2α1, in the osteochondral defects filled with ADAMTS9-AS2-overexpressing hMSCs compared with osteochondral defects filled with control hMSCs (Figure 6G).
Figure 6.
Evaluation of the Ability of ADAMTS9-AS2-Overexpressing hMSCs to Stimulate Cartilage Repair In Vivo
(A) Scheme graph for osteochondral defects with a diameter of 5 mm. (B) Photograph of the osteochondral defects from rats (8–10 weeks old) femur heads. (C and D) Cartilage formation was assessed by Alcian blue staining and toluidine blue staining in control and ADAMTS9-AS2 overexpression group. (G) Immunohistochemistry assay for sections of Col2α1 abundance after 3 months in control and ADAMTS9-AS2 overexpression group. (H) Antibody staining of paraffin-embedded sections of ColX expression after 3 months in control and ADAMTS9-AS2 overexpression group. (E, F, I, and J) Quantitative statistics for dyeing results of (C), (D), (G), and (H) between different groups. (K) Schematic image of the circuitry linking lncRNA-ADAMTS9-AS2, miR-942-5p, and Scrg1 and chondrogenic differentiation. Data are shown as mean ± SD; *p, 0.05, **p, 0.01; ns, non-significant. Experiments were performed in triplicate and error bars represent SD of a triplicate set of experiments. NT, newly formed tissue. Scale bars, 100 and 50 μm for the lower and higher magnification photomicrographs, respectively.
The expression of ColX was slightly increased in the regeneration group but showed a non-significant difference (Figure 6H). This phenomenon was consistent with the results in vitro. It presented once again that upregulation of collagen type II is not correlated with an increased deposition of ColX after ADAMTS9-AS2 overexpression in hMSCs.
Discussion
It is now widely accepted that the noncoding portion of the genome accounts for significant roles in biology development, cell fate determination, and disease therapy. Various new noncoding RNAs with different functions have been identified and studied in stem cell differentiation. In contrast to the extensive study of well-known small noncoding RNAs, LncRNAs are now attracting much interest. Recent studies have showed that lncRNAs can regulate the coding mRNA by competing to bind miRNAs; this class of lncRNAs is termed as ceRNA.29 ceRNAs can sponge miRNAs, so that they can protect the target genes from miRNA repression.
In this study, we identified a novel lncRNA ADAMTS9-AS2 that promoted chondrogenic differentiation in vitro and in vivo. (The detailed information about lncRNA ADAMTS9-AS2 can be viewed in Figure S1 and the Supplemental Material.) It regulated the key transcription factor Scrg1 by displaying its decoy activity for miR-942-5p to accomplish its function.
There is a growing demand for advanced experimental strategies and technology to accomplish intact and effective repair of damaged cartilage. hMSCs are broadly used in the regeneration of cartilage defects.6, 30 Different hMSC application methods have been exploited, such as the cells combining with Bio-printed smart scaffold material31 or growth factors like TGF-β32 and IGF.33 However, the most optimized conditions for cartilage repair are still not being identified. The appropriate growth conditions and growth density as well as the key transcription factors for hMSC chondrogenic differentiation should be further considered and explored. We demonstrated that lncRNA ADAMTS9-AS2 can promote hMSC chondrogenic differentiation in micromass in vitro and form cartilage without TGF-β induction in vivo.
Because lncRNA ADAMTS9-AS2 was limited to the human species, genetically modified mice could not be applied in this study. To prove the function of this lncRNA ADAMTS9-AS2 in an in vivo microenvironment for cartilage repair and to avoid immune rejections between the cells from different species, we used the nude mice as our experimental mouse model. According to previous studies, direct subcutaneous implantation of MSCs can cause cell fibrosis and ossification instead of differentiation to cartilage. The formation of ectopic cartilage requires the cooperation of chondrocytes or the microenvironment of the chondrocyte extracellular matrix.34 The differentiation of MSCs requires the secreted cytokines for chondrogenesis from surrounding chondrocytes and the interaction of MSCs and chondrocyte extracellular matrix.35
So, we used an in vivo method with subcutaneous implantation in mice derived from a recently reported osteochondral defect model.36 This model can supply soluble factors other than TGF-β and the IGF superfamily, which contribute to the microenvironment in vivo for chondrogenic differentiation. Indeed, implanting in the osteochondral defect with ADAMTS9-AS2-overexpressed hMSCs significantly improved cartilage repair compared with control hMSCs. Additionally, ColX was not discovered upregulated after implantation of modified cells. This is an exciting result because ColX is a marker gene for hypertrophic chondrocyte, which represents the beginning of cartilage aging and ossification. Therefore, lncRNA ADAMTS9-AS2 has the ability to be a candidate for applying to hMSCs for articular cartilage defect repair, and it might have a broad prospect in future application.
ADAMTS9-AS2 belongs to antisense lncRNA, which plays crucial roles in the regulation of gene expression and genomic integrity. Like BACE1-AS, it originates from the BACE1 gene in the antisense orientation, and it can enhance the expression of BACE1 via the formation of the RNA duplex.37 Also, CDKN2B-AS1 has the ability to repair DNA damage.38 So, when we started to explore the mechanism for ADAMTS9-AS2, we first detected the gene ADAMTS9 expression after lncRNA change. We found the expression of gene ADAMTS9 remaining unchanged during lncRNA fluctuation. Then we focused on the ceRNA function of ADAMTS9-AS2, and we discovered that antisense lncRNA also had the ability to sponge miRNA and repressed its target gene. Like HOTAIR, it is a kind of antisense lncRNA to the HOXC11 and HOXC12 genes, and it also can control the Rab22a gene expression by sponging miR-373.39 In this study, we confirmed that ADAMTS9-AS2 can sponge miR-942-5p and reverse its repression on chondrogenesis-promoting transcriptional factor stimulator of chondrogenesis 1 (Scrg1).
Scrg1 is a transcript originally discovered through identification from the genes associated with neurodegenerative changes observed in transmissible spongiform encephalopathies from mice.40 Then Scrg1 of the human species was found related to self-renewal, migration, osteogenic differentiation, and chondrogenic differentiation of MSCs.41 Previous study has reported that Scrg1 was induced by dexamethasone through the activation of glucocorticoid receptor and forced expression of Scrg1 stimulates in vitro chondrogenesis.42 In our study, we detected that miR-942-5p overexpression in hMSCs inhibited the expression of Sox9 and Scrg1. Meanwhile, we found miR-942-5p repressing the luciferase activity from Scrg1 reporter rather than Sox9 plasmid. So we believed there was a tight interaction between the miR-942-5p and Scrg1. Furthermore, forced expression of miR-942-5p could inhibit the expression of Sox9. It indicated that Sox9 might not be the direct target for miR-942-5p, but Sox9 was still regulated on the downstream of the regulatory circuitry. However, the specific molecular mechanism still needs to be explored and studied.
Stem cells have great potential for cell differentiation and function. Therefore, it’s a great challenge for us to regulate the stem cells precisely to maximize their capability. For example, subsets of stem cells expressing different surface markers own diverse multipotent differentiation ability.43 Meanwhile, metabolic ability also affects the function of stem cells.44, 45 Furthermore, differences between constant and intermittent hormonal stimulation affect the differentiation ability of stem cells.46 Hence, it requires us to analyze multiple complex factors before utilizing stem cells for chondrogenic differentiation. In the future, stem cells need to be classified according to the chondrogenic differentiation ability, so that they can be better used in clinical trials. Also, we should explore the methods from different ways of applying cytokines in cartilage repair.
Many studies have paid much attention to growth factors that are involved in promoting cartilage repair from hMSCs. These growth factors, particularly the members of the transforming growth factor-beta family, have the ability to induce hMSC chondrogenesis and synthesize specific chondrogenic extracellular matrix like collagen type II and aggrecan. However, the use of TGF-β produced contradictory results due to its side effect to drive stem cells toward hypertrophy or fibrosis.47 Additionally, several studies have reported that TGF-β plays a critical role in the progression of osteoarthritis, a kind of cartilage degenerative disease.48 For this purpose, we focused on the function of lncRNA ADAMTS9-AS2. Our study revealed that ADAMTS9-AS2 had the ability to promote hMSC chondrogenic differentiation in osteochondral defect in vivo, without the addition of TGF-β. In this case, ADAMTS9-AS2 can positively regulate the chondrogenic differentiation of hMSCs both in vitro and in vivo. Our findings provide a novel perspective for using lncRNA to induce hMSC chondrogenesis for cartilage defect therapy.
Materials and Methods
hMSC Cultures
Primary hMSCs were collected from the bone marrow of 6 individuals, aged 35–55 years, who were undergoing total hip replacement. None of these selected patients had any clinical evidence of recent infection or systemic disease, and none had a history of smoking or radiotherapy or chemotherapy. All samples were collected from the Shanghai Ninth People’s Hospital. Written informed consent was provided by all participants.
hMSCs from bone marrow aspirates were cultured in expansion medium (10% fetal calf serum, a-MEM) supplemented with 1% penicillin-streptomycin and 1 ng/mL basic fibroblast growth factor. Non-adherent cells were washed off after 2 days, and adherent cells were further expanded. At subconfluence, hMSCs were trypsinized for cell passages. The expanded cells at passage 3–6 were used for experiments.
Microarray and Data Analysis
Two groups of cells (hMSCs from the same patient undergoing expansion and chondrogenic induction to chondrogenesis), which each contained the mixture of cell sources from three parallel experiment, were prepared, and total RNA was isolated using TRIzol reagent. RNA quality and quantity were measured by Nanodrop 2000 (Thermo Fisher Scientific). RNA integrity was assessed by standard denaturing agarose gel electrophoresis. For microarray analysis, Agilent Array platform was employed. The sample preparation and microarray hybridization were performed based on the manufacturer’s standard protocols with minor modifications. All labeled lncRNAs were hybridized onto an Arraystar Human LncRNA Microarray version -3.0 (Arraystar, Kangcheng, Shanghai, China). The microarray screening data have been submitted to the GEO database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE128533). A heatmap was exported using Agilent Feature Extraction software (version 11.0.11, Agilent Technologies). Differentially expressed lncRNAs with statistical significance between the two samples were identified through fold change filtering.
Oligonucleotide and Cell Transfection
Both the miRNA mimics (mimic-nc) and miRNA antagomir (antagomir-nc) were synthesized by RiboBio Biotech (Guangzhou, China). Oligonucleotide transfection was conducted using Lipofectamine 2000 transfection reagent (Invitrogen), in accordance with the manufacturer’s recommendations. For induction of hMSC chondrogenic differentiation, the cells were transfected every 3 days to maintain the effectiveness of genetic changes during chondrogenic induction.
Lentiviruses and Plasmids
The shRNA oligonucleotides were designed from the website RNAiDesigner (http://rnaidesigner.thermofisher.com/rnaiexpress/). The shRNA and sh-nc plasmid were constructed by using the plasmid pLVX-shRNA, and the overexpressing plasmid was constructed by using the plasmid pLVX-puro. For lentivirus packaging, 293T cells were transfected with the above plasmids and the packaging plasmids pMD2.G and psPVX2. The harvested lentiviruses were concentrated, purified, and preserved at −80°C. The lentivirus titer was 1*108TU/mL. hMSCs were cultured at a concentration of 2 × 105 cells per well in six-well plates. After the cells had grown to 30%–40% confluence, they were infected with 20 μL lentiviruses in the presence of polybrene. When hMSCs were transfected with lentiviruses for 12 h, we replaced with fresh medium to grow the cells. Because the plasmids we used contained GFP fragments, so the lentivirus infection efficiency could be tested by GFP expression.
In Vitro Chondrogenic Differentiation
Micromass culture was performed with hMSCs at passage 3–6. hMSCs were infected with lentiviruses to inhibit and overexpress ADAMTS9-AS2, respectively. Then hMSCs were collected and plated at a density of 2 × 107 cells/mL as 10-μL drops on a dish. After cell adhesion of 3 h, we supplemented with chondrogenic induced medium to the plate. Then we replaced the chondrogenic induced medium every 3 days. The chondrogenic induced medium contained insulin-transferrin-selenium (100*), Thermo Fisher Scientific; L-proline (4 μg/mL), Sigma; dexamethasone (100 nM), Sigma; sodium L-ascorbate (10 μg/mL), Sigma; linoleic acid (470 ng/mL), Sigma; BSA (50 μg/mL, BSA), Sigma; and TGF-β1 (10 ng/mL), PeproTech.
To explore the condensation ability of hMSCs after inhibiting the expression of ADAMTS9-AS2, we performed semi-micromass culture. hMSCs at the density of 5 × 106 cells/mL as 10-μL drops were plated on a dish. Further induction methods were the same as those mentioned above.
Luciferase Report Assay
The putative miR-942-5p and miR-153-5p target-binding sequence in wild-type lncRNA ADAMTS9-AS2 and the mutant of binding sequence for miR-942-5p were synthesized and cloned downstream of luciferase gene in psi-check2 luciferase vectors. To determine whether miR-942-5p directly targets Scrg1 and Sox9, we constructed wild-type Scrg1 and Sox9 reporter plasmids and mutant plasmid by changing binding sequences of Scrg1 with psi-check2 plasmids.
These plasmids and miRNA mimics (mimic-nc and mimic-miRNA) were co-transfected into 293T cells. Firefly and Renilla luciferase activities were measured by a Dual Luciferase Assay (Promega) at 48 h after transfection. All experiments were repeated three times.
RNA Isolation and qPCR
Total RNA was extracted with TRIzol reagent (Invitrogen). cDNA was synthesized with a reverse transcript kit (Takara) according to the manufacturer’s instructions. qPCR was performed in a ViiA 7 real-time PCR system (Applied Biosystems) using the SYBR premix EX TAG (Takara). miRNAs were detected by using Bulge-loop primers designed from RiboBio Biotech (Guangzhou, China). mRNA primers were designed and synthesized from Sangon Biotech. β-actin and small nuclear RNA U6 were used as internal controls for mRNAs and miRNAs.
Histology and Immunohistochemistry
The presence of sulfated proteoglycans and glycosaminoglycan from the extracellular matrix indicative of cartilage formation was detected by Safranine O staining, Alcian blue staining, and toluidine blue staining. Micromass from hMSC differentiation was washed in PBS, fixed in 4% paraformaldehyde for 1 h, and rehydrated with graded ethanol. Safranine O was stained with 0.2% Safranine O solution (Sigma). Alcian blue was stained with 1% Alcian blue (Sigma) dissolved in acetic acid. Toluidine blue was stained with 0.5% toluidine blue solution (Sigma). Each experiment was repeated at least three times using triplicate samples.
Sections of hMSC micromass (5 mm) or osteochondral constructs (6 mm) were subjected to immunohistochemistry. With this aim, immunohistochemistry antibodies were used as follow: Col2α1 (1:200, Abcam), ACAN (1:100, Abcam), Sox9 (1:200, Abcam), and Scrg1 (1:200, Novus Biologicals). Pepsin (Sigma) was used to retrieve the antigen and 5% goat serum was used to block the unspecific background antigen. Sections were incubated with the primary antibodies overnight at 4°C. A streptavidin-horseradish peroxidase (HRP) detection system (MX Biotech) was used according to the manufacturer’s instructions. Sections were then counterstained with hematoxylin and sealed with neutral balsam. For the statistical analysis on immunohistochemistry of the sections, the positive immunostaining was expressed as percentage of positive area of the samples.
Osteochondral Defect Model
Osteochondral defects were created from rat osteochondral femur head, this method slightly modified from a previous study. The femur heads were taken from 8- to 10-week-old male rats. All of the rats were maintained in a specific pathogen-free animal facility of the Institute of Health Sciences (the Chinese Academy of Sciences). All of the animal experiments were approved by the Ethics Committee of the Shanghai Jiao Tong University School of Medicine. The tissues were then incubated overnight in 10% fetal calf serum DMEM high-glucose supplemented with 1.5 mg/mL fungizone and 50 mg/mL gentamicin to verify sterility. Osteochondral defects were produced using a 5-mm diameter electric drill, and the osteochondral defects were created according to the follow aspects: the cartilage sections were removed completely and the upper parts of the subchondral bone were damaged by scraping the surface. To prevent outgrowth of cells from the subchondral bone, the defect tissues were placed in 2% low-gelling agarose (Sigma), in such a way that the subchondral bone was coated by the agarose and the cartilage and supplemented hMSCs were above the agarose surface.
In Vivo Implantation of Osteochondral Defects with hMSCs
hMSCs were infected with overexpression ADAMTS9-AS2 lentivirus and control lentivirus as described above, and then the cells were collected at day 7 and resuspended in 1.2% low-viscosity alginate (Sigma), at a density of 2.5 × 107 cells/mL. Meanwhile, 40 mL alginate cell suspension and 60 mL 102 mM CaCI2 were added to the engineered osteochondral defects. The defect tissues were cultured overnight in a 37°C incubator to stabilize this system. The osteochondral defects were implanted subcutaneously on the backs of 10- to 12-week-old female nude mice under 4% chloral hydrate anesthesia. Each mouse was implanted with two osteochondral defect tissues, in such way that the two different condition hMSCs (treated with control lentivirus and ADAMTS9-AS2 overexpression lentivirus) were present in the same animal, which guaranteed the similar microenvironment. Before implantation the surface of the osteochondral defects was covered with a permeable membrane, which allowed the surrounded cytokines to pass through it. This membrane originated from Milli Hang Culture transwell (Millipore), the pore size 8 μm, was cut with scissors to cover the defect. In such a way, the membrane prevented the seeded cells from escaping from the defects and stopped cells from the outside of the defects entering the injury position for repair. After 12 weeks, mice were euthanized by cervical dislocation, and the osteochondral defects were taken out and fixed in 4% paraformaldehyde for 24 h, decalcified in 12.5% EDTA (pH 7.0) for 3 weeks, and embedded in paraffin. The samples were then sectioned and subjected to histological analysis. All of the animals were maintained in a specific pathogen-free animal facility of the Institute of Health Sciences (the Chinese Academy of Sciences). All of the animal experiments were approved by the Ethics Committee of the Shanghai Jiao Tong University School of Medicine.
Statistical Analysis
Statistical analysis was performed with SPSS 19.0 (IBM, Armonk, NY, USA). Data were presented as mean ± SD. A statistical difference between two groups was determined by a two-tailed Student’s t test. p < 0.05 was considered statistically significant.
Author Contributions
M.-J.H. and J.-Y. Z. conceived and designed the study; M.-J.H., J.-Y.Z., and J.-J.X. carried out experiments; J.L. and Y.-F.Z. analyzed the data; M.-J.H. drafted the paper; X.-L.Z. revised the paper. All authors have read and approved the paper.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81830078, 81772347, and 81572123), the Science and Technology Commission of Shanghai Municipality (19XD1434100 and 16430723500), Shanghai Municipal Education Commission-Gao feng Clinical Medicine Grant Support (20161314), the Shanghai Jiao Tong University International Collaboration Fund, and the Shanghai Jiao Tong University-The Chinese University of Hong Kong Joint Research Collaboration Fund.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.08.027.
Supplemental Information
References
- 1.Erlebacher A., Filvaroff E.H., Gitelman S.E., Derynck R. Toward a molecular understanding of skeletal development. Cell. 1995;80:371–378. doi: 10.1016/0092-8674(95)90487-5. [DOI] [PubMed] [Google Scholar]
- 2.Kronenberg H.M. Developmental regulation of the growth plate. Nature. 2003;423:332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 3.Vortkamp A., Lee K., Lanske B., Segre G.V., Kronenberg H.M., Tabin C.J. Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science. 1996;273:613–622. doi: 10.1126/science.273.5275.613. [DOI] [PubMed] [Google Scholar]
- 4.Dy P., Wang W., Bhattaram P., Wang Q., Wang L., Ballock R.T., Lefebvre V. Sox9 directs hypertrophic maturation and blocks osteoblast differentiation of growth plate chondrocytes. Dev. Cell. 2012;22:597–609. doi: 10.1016/j.devcel.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evans C.H. Catering to chondrocytes. Sci. Transl. Med. 2018;10:eaav7043. doi: 10.1126/scitranslmed.aav7043. [DOI] [PubMed] [Google Scholar]
- 6.Somoza R.A., Welter J.F., Correa D., Caplan A.I. Chondrogenic differentiation of mesenchymal stem cells: challenges and unfulfilled expectations. Tissue Eng. Part B Rev. 2014;20:596–608. doi: 10.1089/ten.teb.2013.0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson K., Zhu S., Tremblay M.S., Payette J.N., Wang J., Bouchez L.C., Meeusen S., Althage A., Cho C.Y., Wu X., Schultz P.G. A stem cell-based approach to cartilage repair. Science. 2012;336:717–721. doi: 10.1126/science.1215157. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y., Wang L., Kikuiri T., Akiyama K., Chen C., Xu X., Yang R., Chen W., Wang S., Shi S. Mesenchymal stem cell-based tissue regeneration is governed by recipient T lymphocytes via IFN-γ and TNF-α. Nat. Med. 2011;17:1594–1601. doi: 10.1038/nm.2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nauta A.J., Fibbe W.E. Immunomodulatory properties of mesenchymal stromal cells. Blood. 2007;110:3499–3506. doi: 10.1182/blood-2007-02-069716. [DOI] [PubMed] [Google Scholar]
- 10.Massagué J., Blain S.W., Lo R.S. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 11.Geiger B.C., Wang S., Padera R.F., Jr., Grodzinsky A.J., Hammond P.T. Cartilage-penetrating nanocarriers improve delivery and efficacy of growth factor treatment of osteoarthritis. Sci. Transl. Med. 2018;10:eaat8800. doi: 10.1126/scitranslmed.aat8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma B., Fermanian S., Gibson M., Unterman S., Herzka D.A., Cascio B., Coburn J., Hui A.Y., Marcus N., Gold G.E., Elisseeff J.H. Human cartilage repair with a photoreactive adhesive-hydrogel composite. Sci. Transl. Med. 2013;5:167ra6. doi: 10.1126/scitranslmed.3004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Batista P.J., Chang H.Y. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kung J.T., Colognori D., Lee J.T. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O., Chinappi M., Tramontano A., Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klattenhoff C.A., Scheuermann J.C., Surface L.E., Bradley R.K., Fields P.A., Steinhauser M.L., Ding H., Butty V.L., Torrey L., Haas S. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–583. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao X.Y., Li S., Wang G.X., Yu Q., Lin J.D. A long noncoding RNA transcriptional regulatory circuit drives thermogenic adipocyte differentiation. Mol. Cell. 2014;55:372–382. doi: 10.1016/j.molcel.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyaki S., Sato T., Inoue A., Otsuki S., Ito Y., Yokoyama S., Kato Y., Takemoto F., Nakasa T., Yamashita S. MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes Dev. 2010;24:1173–1185. doi: 10.1101/gad.1915510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X., Wang C., Zhao J., Xu J., Geng Y., Dai L., Huang Y., Fu S.C., Dai K., Zhang X. miR-146a facilitates osteoarthritis by regulating cartilage homeostasis via targeting Camk2d and Ppp3r2. Cell Death Dis. 2017;8:e2734. doi: 10.1038/cddis.2017.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lolli A., Narcisi R., Lambertini E., Penolazzi L., Angelozzi M., Kops N., Gasparini S., van Osch G.J., Piva R. Silencing of Antichondrogenic MicroRNA-221 in Human Mesenchymal Stem Cells Promotes Cartilage Repair In Vivo. Stem Cells. 2016;34:1801–1811. doi: 10.1002/stem.2350. [DOI] [PubMed] [Google Scholar]
- 21.Qu L., Ding J., Chen C., Wu Z.J., Liu B., Gao Y., Chen W., Liu F., Sun W., Li X.F. Exosome-Transmitted lncARSR Promotes Sunitinib Resistance in Renal Cancer by Acting as a Competing Endogenous RNA. Cancer Cell. 2016;29:653–668. doi: 10.1016/j.ccell.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 22.Zhu K.P., Ma X.L., Zhang C.L. LncRNA ODRUL Contributes to Osteosarcoma Progression through the miR-3182/MMP2 Axis. Mol. Ther. 2017;25:2383–2393. doi: 10.1016/j.ymthe.2017.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.DeLise A.M., Stringa E., Woodward W.A., Mello M.A., Tuan R.S. Embryonic limb mesenchyme micromass culture as an in vitro model for chondrogenesis and cartilage maturation. Methods Mol. Biol. 2000;137:359–375. doi: 10.1385/1-59259-066-7:359. [DOI] [PubMed] [Google Scholar]
- 24.Steinert A.F., Ghivizzani S.C., Rethwilm A., Tuan R.S., Evans C.H., Nöth U. Major biological obstacles for persistent cell-based regeneration of articular cartilage. Arthritis Res. Ther. 2007;9:213. doi: 10.1186/ar2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cazalla D., Yario T., Steitz J.A. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science. 2010;328:1563–1566. doi: 10.1126/science.1187197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li R., Fang L., Pu Q., Bu H., Zhu P., Chen Z., Yu M., Li X., Weiland T., Bansal A. MEG3-4 is a miRNA decoy that regulates IL-1β abundance to initiate and then limit inflammation to prevent sepsis during lung infection. Sci. Signal. 2018;11:eaao2387. doi: 10.1126/scisignal.aao2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 28.Fabian M.R., Sonenberg N., Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 29.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xian C.J., Foster B.K. Repair of injured articular and growth plate cartilage using mesenchymal stem cells and chondrogenic gene therapy. Curr. Stem Cell Res. Ther. 2006;1:213–229. doi: 10.2174/157488806776956904. [DOI] [PubMed] [Google Scholar]
- 31.Liao J., Shi K., Ding Q., Qu Y., Luo F., Qian Z. Recent developments in scaffold-guided cartilage tissue regeneration. J. Biomed. Nanotechnol. 2014;10:3085–3104. doi: 10.1166/jbn.2014.1934. [DOI] [PubMed] [Google Scholar]
- 32.Tang Q.O., Shakib K., Heliotis M., Tsiridis E., Mantalaris A., Ripamonti U., Tsiridis E. TGF-beta3: A potential biological therapy for enhancing chondrogenesis. Expert Opin. Biol. Ther. 2009;9:689–701. doi: 10.1517/14712590902936823. [DOI] [PubMed] [Google Scholar]
- 33.Patil A.S., Sable R.B., Kothari R.M. Role of insulin-like growth factors (IGFs), their receptors and genetic regulation in the chondrogenesis and growth of the mandibular condylar cartilage. J. Cell. Physiol. 2012;227:1796–1804. doi: 10.1002/jcp.22905. [DOI] [PubMed] [Google Scholar]
- 34.Xue J.X., Gong Y.Y., Zhou G.D., Liu W., Cao Y., Zhang W.J. Chondrogenic differentiation of bone marrow-derived mesenchymal stem cells induced by acellular cartilage sheets. Biomaterials. 2012;33:5832–5840. doi: 10.1016/j.biomaterials.2012.04.054. [DOI] [PubMed] [Google Scholar]
- 35.Gong Y.Y., Xue J.X., Zhang W.J., Zhou G.D., Liu W., Cao Y. A sandwich model for engineering cartilage with acellular cartilage sheets and chondrocytes. Biomaterials. 2011;32:2265–2273. doi: 10.1016/j.biomaterials.2010.11.078. [DOI] [PubMed] [Google Scholar]
- 36.de Vries-van Melle M.L., Mandl E.W., Kops N., Koevoet W.J., Verhaar J.A., van Osch G.J. An osteochondral culture model to study mechanisms involved in articular cartilage repair. Tissue Eng. Part C Methods. 2012;18:45–53. doi: 10.1089/ten.tec.2011.0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Faghihi M.A., Zhang M., Huang J., Modarresi F., Van der Brug M.P., Nalls M.A., Cookson M.R., St-Laurent G., 3rd, Wahlestedt C. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 2010;11:R56. doi: 10.1186/gb-2010-11-5-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Congrains A., Kamide K., Ohishi M., Rakugi H. ANRIL: molecular mechanisms and implications in human health. Int. J. Mol. Sci. 2013;14:1278–1292. doi: 10.3390/ijms14011278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Z., Cheng J., Wu Y., Qiu J., Sun Y., Tong X. LncRNA HOTAIR controls the expression of Rab22a by sponging miR-373 in ovarian cancer. Mol. Med. Rep. 2016;14:2465–2472. doi: 10.3892/mmr.2016.5572. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Dandoy-Dron F., Guillo F., Benboudjema L., Deslys J.P., Lasmézas C., Dormont D., Tovey M.G., Dron M. Gene expression in scrapie. Cloning of a new scrapie-responsive gene and the identification of increased levels of seven other mRNA transcripts. J. Biol. Chem. 1998;273:7691–7697. doi: 10.1074/jbc.273.13.7691. [DOI] [PubMed] [Google Scholar]
- 41.Aomatsu E., Takahashi N., Sawada S., Okubo N., Hasegawa T., Taira M., Miura H., Ishisaki A., Chosa N. Novel SCRG1/BST1 axis regulates self-renewal, migration, and osteogenic differentiation potential in mesenchymal stem cells. Sci. Rep. 2014;4:3652. doi: 10.1038/srep03652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ochi K., Derfoul A., Tuan R.S. A predominantly articular cartilage-associated gene, SCRG1, is induced by glucocorticoid and stimulates chondrogenesis in vitro. Osteoarthritis Cartilage. 2006;14:30–38. doi: 10.1016/j.joca.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 43.Pinho S., Lacombe J., Hanoun M., Mizoguchi T., Bruns I., Kunisaki Y., Frenette P.S. PDGFRα and CD51 mark human nestin+ sphere-forming mesenchymal stem cells capable of hematopoietic progenitor cell expansion. J. Exp. Med. 2013;210:1351–1367. doi: 10.1084/jem.20122252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ren R., Ocampo A., Liu G.H., Izpisua Belmonte J.C. Regulation of Stem Cell Aging by Metabolism and Epigenetics. Cell Metab. 2017;26:460–474. doi: 10.1016/j.cmet.2017.07.019. [DOI] [PubMed] [Google Scholar]
- 45.Yu Y., Newman H., Shen L., Sharma D., Hu G., Mirando A.J., Zhang H., Knudsen E., Zhang G.F., Hilton M.J., Karner C.M. Glutamine Metabolism Regulates Proliferation and Lineage Allocation in Skeletal Stem Cells. Cell Metab. 2019;29:966–978.e4. doi: 10.1016/j.cmet.2019.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fan Y., Hanai J.I., Le P.T., Bi R., Maridas D., DeMambro V., Figueroa C.A., Kir S., Zhou X., Mannstadt M. Parathyroid Hormone Directs Bone Marrow Mesenchymal Cell Fate. Cell Metab. 2017;25:661–672. doi: 10.1016/j.cmet.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Beuningen H.M., Glansbeek H.L., van der Kraan P.M., van den Berg W.B. Osteoarthritis-like changes in the murine knee joint resulting from intra-articular transforming growth factor-beta injections. Osteoarthritis Cartilage. 2000;8:25–33. doi: 10.1053/joca.1999.0267. [DOI] [PubMed] [Google Scholar]
- 48.van der Kraan P.M. The changing role of TGFβ in healthy, ageing and osteoarthritic joints. Nat. Rev. Rheumatol. 2017;13:155–163. doi: 10.1038/nrrheum.2016.219. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.