Abstract
Biopharmaceutical industrial processes are based on high yielding stable recombinant Chinese Hamster Ovary (CHO) cells that express monoclonal antibodies. However, the process and feeding regimes need to be adapted for each new cell line, as they all have a slightly different metabolism and product performance. A main limitation for accelerating process development is that the metabolic pathways underlying this physiological variability are not yet fully understood. This study describes the evolution of intracellular fluxes during the process for 4 industrial cell lines, 2 high producers and 2 low producers (n = 3), all of them producing a different antibody. In order to understand from a metabolic point of view the phenotypic differences observed, and to find potential targets for improving specific productivity of low producers, the analysis was supported by a tailored genome-scale model and was validated with enzymatic assays performed at different days of the process. A total of 59 reactions were examined from different key pathways, namely glycolysis, pentose phosphate pathway, TCA cycle, lipid metabolism, and oxidative phosphorylation. The intracellular fluxes did not show a metabolic correlation between high producers, but the degree of similitude observed between cell lines could be confirmed with additional experimental observations. The whole analysis led to a better understanding of the metabolic requirements for all the cell lines, allowed to the identification of metabolic bottlenecks and suggested targets for further cell line engineering. This study is a successful application of a curated genome-scale model to multiple industrial cell lines, which makes the metabolic model suitable for process platform.
Keywords: Genome-scale metabolic model, Chinese hamster ovary, Flux distribution, Mathematical modeling, Metabolic engineering
Abbreviations: CHO, Chinese Hamster Ovary; GSM, genome-scale metabolic model; pFBA, parsimonious flux balance analysis; FBA, flux balance analysis; PPP, pentose phosphate pathway; TCA, tricarboxylic acid cycle
Highlights
-
•
Successful application of a tailored genome-scale model to four different industrial cell lines.
-
•
Validation of intracellular predicted fluxes with enzymatic assays.
-
•
Suggestions of metabolic markers for industrial clone selection.
1. Introduction
The global market for biological products is driven by increased demand in research activity to develop biologics against high incidence rate diseases such as cancers, immunological or neurological disorders. Most of the treatments rely on protein-based therapies, which are produced essentially in Chinese Hamster Ovary (CHO) cells, due to their suitability for large and complex recombinant protein synthesis, and their high productivity. Today, CHO cells are used to produce more than half of all therapeutic proteins on the market. However, one of the challenges for CHO cells is the poor understanding of the sources of phenotypic variations during cell culture processes, despite a long and laborious cell line development step dedicated to the selection of the best candidate. Efforts in cell line engineering have been relatively few compared to the importance of the cell factory, and focused on energy metabolism (Irani et al., 1999; Jeong et al., 2006; Wlaschin and Hu, 2007), cell cycle mechanisms and especially apoptosis and cell death (Mastrangelo et al., 2000; Meents et al., 2002). To increase cell productivity, any of the steps involved in the production of complex proteins such as antibodies needs to be studied as any of them could be a bottleneck. When the mechanisms are properly understood, the ultimate goal is to identify new markers that would help cell line development teams to decide whether a clone is a high-producer or not. The efficiency in recombinant protein production indeed relies on different steps, such as efficient gene transcription, transport of messenger RNA to the membrane of the endoplasmic reticulum, protein translation and translocation, protein maturation by posttranslational modification, and secretion of the correctly assembled molecule (Reinhart et al., 2014). Also, as a direct observation in industrial CHO cell processes, the metabolism of the CHO cell is characterized by high consumption rates of substrates added in medium and feed, as well as accumulation of by-products and metabolic intermediates during the run (Pereira et al., 2018). This observation underlines that CHO cell metabolism could be further optimized, and that cells have the potential to increase their performances if correct metabolic targets can be identified. Bearing in mind the expectations for a more efficient metabolism, metabolic flux analysis has emerged as a powerful technique to provide quantitative information on the cellular machinery at the level of flux distribution. As such, it can be used to gain fundamental understanding in the metabolic pathways for product formation (Ahn and Antoniewicz, 2012).
Here we present an application of a curated genome-scale model to 4 different industrial cell lines, that have been characterized as high and low producers, in order to identify bottlenecks in a number of metabolic pathways. We will describe the mathematical approach, discuss the predictions and compare them to experimental observations in order to infer information on the metabolic traits of a high-producing cell line and identify specific metabolic targets.
2. Material and methods
2.1. Cell culture and extracellular metabolite analysis
2.1.1. Cell culture conditions
Twelve fed-batch cell culture experiments were run independently, for which cell phenotype, variation of extracellular metabolite concentrations, and process parameters were monitored. 4 CHO DG44 cell lines producing four different monoclonal antibodies (mAb1, mAb2, mAb3 and mAb4) were cultivated in 2L stirred tank glass bioreactors in triplicates, controlled by supply towers (C-DCUII, Sartorius Stedim Biotech) and monitored by a multi-fermenter control system (MFCS, Sartorius). The cells were grown under serum free conditions in a proprietary and chemically defined media. Precultures were cultivated in increased volume capacity of Erlenmeyer flasks (Corning Inc, Germany) on a shaking device at 37 °C and 5% CO2 in a humid atmosphere. The starting culture volume was identical for the different production runs and the bioreactors were inoculated at similar target seeding density. The cultivation temperature was kept constant at 36.8 °C and the impeller used was a 3-segment blade impeller. During the cultivation, the pH was fixed at 7.0, with an allowable variation of 0.2, and controlled by gassing CO2 and a sodium carbonate solution. Dissolved oxygen was maintained at 40% of the saturation concentration. Continuous nutrient feeding was started 72 hours after inoculation, with predetermined rate using a proprietary, chemically defined concentrated feed. The feed rate was adapted every day, following a predefined feeding profile. In addition to this continuous feed, a bolus feed addition also started 72 hours after inoculation. Samples were taken once a day, before feeding. When the glucose concentration was below 5.6 g/L, a glucose solution of 500 g/L was added as a bolus. Specific growth rate was calculated for each experimental condition as in equation (1):
| (1) |
The exponential growth equation is described by equation (2):
| ln(X) = ln(X0) + μt | (2) |
where μ is the specific growth rate, X is the cell density at time t (i.e. cell mass or number per unit volume of culture medium), and X0 is the initial cell density at the onset of exponential growth. This equation was used to determine the specific growth rate from linear regression of cell density measurements over time.
2.1.2. Analytical methods
Samples from the bioreactor were taken daily for cell density and viability analysis using the VI-CELL® XR (Beckman-Coulter, Inc., Brea, CA), based on the trypan blue exclusion method. Samples were centrifuged and supernatants were analyzed to quantify concentration of glucose, lactate, amino acid and monoclonal antibody. Glucose, lactate, glutamate, glutamine and ammonium concentrations were determined using a Cedex Bio HT Analyzer (Innovatis, Bielefeld, Germany) or a NOVA 400 BioProfile automated analyzer (Nova Biomedical, Waltham, MA). Cell culture supernatant samples were stored at −80 °C or directly analyzed for product titer with a ForteBio Octet model analyzer (ForteBio, Inc., Menlo Park, CA) or protein A high performance liquid chromatography (HPLC). Amino acids were analyzed by reversed-phase UPLC (Waters AccQ · Tagultra method) after ultra-filtration using Amicon Ultra-0.5 mL centrifugal filters (Merck Millipore, Billerica, MA). pH and DO were measured on-line and the measurement accuracy was verified through offline analysis of pH and partial pressure of CO2 (pCO2) using a BioProfile pHOx® blood gas analyzer (Nova Biomedical Corporation, Waltham, MA).
2.2. Modeling procedure
2.2.1. Genome-scale model
The genome-scale model used was tailored to high-yielding cell lines (Calmels et al., 2018; under review with Metabolic Engineering). For each cell line modeled, the reaction responsible for antibody production in the model was tailored to the antibody produced by the cell line chosen. The stoichiometric coefficients of the amino acids required to produce the IgG were changed to the one corresponding to the specific amino acid sequence of the antibody produced, thus generating 4 genome-scale models for the cell lines producing mAb1, mAb2, mAb3 and mAb4. For each cell line, three biological replicates were modeled independently. The model-predicted fluxes are shown with error bars which represent experimental uncertainty quantified from multiple replicate experiments.
As multiple predicted solutions are possible due to typical underdetermination of flux-balance-based models, the following considerations were addressed and are more detailed in the next sections: (1) additional constraints were introduced at each day of the culture, (2) a robust optimization method was employed, and (3) a set of predicted fluxes was compared to experimental values assessed with enzymatic assays.
2.2.2. Theoretical approach for modeling
The modeling framework can be represented by a stoichiometric matrix (S) and a vector of reaction fluxes (v) indicating the reaction rates. The basic steady-state mass balance constraint can be enforced by the linear equation (5). Additional constraints can be introduced by restricting fluxes with upper or lower bounds through inequality 6. To find the flux distribution, parsimonious enzyme usage Flux Balance Analysis (pFBA) (Lewis et al., 2010) was employed. This optimization method is based on the assumption that the cell is using a minimum amount of enzymes to reach a maximized objective, under the hypothesis of steady-state. The mathematical formulation of the objective function is given by equation (3), and the requirement for the minimum absolute values among all the alternatives optima is described by equation (4). To find the flux distributions, the network is constrained by imposing lower and upper bounds for each flux, and by assuming a steady-state condition, which leads to the following optimization problem:
| (3) |
| (4) |
| (5) |
and,
| (6) |
Where cv corresponds to the objective function and c is a vector of weights, indicating how much each reaction contributes to the objective function.
Linear programming was performed using Gurobi Optimizer (Gurobi Optimization Inc., Houston TX) in Python 2.7.12 (Python Software Foundation, Delaware, United States).
2.2.3. Data processing and transforming primary data into flux constraints
Substrate concentrations at each day of the cell culture were transformed into rates in mmol of product per gDW of cells per hour. The modeling time frame chosen was between day 2 and day 7 of cell culture, which corresponds to the exponential growth phase where the pseudo-steady-state is assumed. All the input flux rate values were calculated using an average dry cell weight of CHO cells as 330 pg/cell, as derived as an average of published values (Bonarius et al., 1996; Vriezen, 1998; Xie and Wang, 1994; Zupke and Stephanopoulos, 1995). Daily experimental uptake or production rates of 24 metabolites in medium were calculated from experimental measurements, as followed:
Where rA,i indicates the rate of metabolite A at day i, and i = [0, 14]; Q A,i+1,BF is the measured quantity of metabolite A at day i+1 before feeding; Q A,i,AF is the theoretical quantity of metabolite A at day i after feeding; and VCC is the cell number. Q A,i,AF is the sum of Q A,i+1,BF and the theoretical quantity of metabolite A added after feeding (Qth), calculated with the concentration of metabolite A in the feed and the volume of feed added:
The calculated rates were post-processed with a smoother function in order to reduce experimental noise and normalized with one value corresponding to a specific growth rate of the mAb 1 producing cell line. The experimental variation can be explained by several factors, including the sensibility and technical variability of the analytical quantification method, but also the degree of biological reproducibility inter-process.
Experimental production and consumption rates calculated were smoothed using a kernel smoother statistical function, in order to avoid irregular data points and noisy observations obtained from experimental results (Hastie et al., 2009). The Kernel smoother method was applied on calculated experimental flux rates with the support of SAS software JMP 11 ©.
The calculated and smoothed experimental values were then used to set the upper or lower limit of the consumption or production rate of the metabolites measured daily in the extracellular environment. Exceptions were made for ammonium, alanine, and lactate which were set to be equal to the experimental value in order to constrain the model with the metabolic switch between consumption and production.
The final set of constrained metabolite exchanges comprised the uptake and secretion of 24 metabolites. The limitations for uptake and production rates were set for each day of the time frame chosen for modeling the cell culture process. The objective function chosen was maximization of growth. Estimation of metabolic fluxes was performed using CobraPy (Ebrahim et al., 2013).
2.2.4. Pathway analysis
Metabolic fluxes predicted in selected pathways in the genome-scale model were analyzed. The pathways chosen were: glycolysis (glucose uptake rate, hexokinase, Glucose-6-phosphate isomerase, Phosphofructokinase, Triose phosphate isomerase, Glyceraldehyde-3-phosphate dehydrogenase, Enolase, Pyruvate kinase), pentose phosphate pathway (Glucose-6-phosphate dehydrogenase, Ribulose-5-phosphate-3-epimerase, Transketolase 1 and Transketolase 2), tricarboxylic acid cycle (Citrate synthase, Aconitate hydratase, Isocitrate dehydrogenase, 2-oxoglutarate dehydrogenase, Succinyl-CoA synthetase, Succinate-CoA ligase, Fumarase, Malate dehydrogenase, Malic enzyme, Pyruvate dehydrogenase, and Glutamate dehydrogenase), lipid (Acetyl-CoA carboxylase, Palmitoyl-CoA synthesis, Phosphatidylinositol synthase, Cardiolipin synthase, Diacylglycerol phosphate kinase, Glycogen synthase, Glycerol-3-phosphate acyltransferase, Lipase, Phosphatidylglycerol phosphate phosphatase, Phosphatidylserine decarboxylase, Sterol O-acyltransferase, and Glycerol-3-phosphate dehydrogenase). To complete the predictions, the analysis was read in conjunction with experimental metabolite exchanges (consumption rate of the 20 amino acids and secretion rate of lactate and ammonia), as well as specific growth rate and specific antibody productivity. A total of 59 reaction rates were collected for each individual replicate of the 4 cell lines at day 2–7.
2.3. Statistical analysis
In order to quantify comparisons between predicted and experimental enzymatic activities, a multiple linear regression (MLR) method was used.
As enzymatic activities and flux predictions were not expressed in the same units, each individual cell culture replicate (for both experimental and predicted) were normalized with the highest value inside the dataset. Thus, the statistic test was based on data ranging from 0 to 1.
The statistical analyses were performed using SAS software JMP 8.0 ©. The null hypothesis corresponded to the absence of significant differences between observations. This hypothesis was tested in the multiple regression model. The parameters tested were the time (days), the dataset type (experimental or predicted datasets) and cell clone (HP1 or HP2) with linear, quadratic and interactions effects (level 2) in order to assess statistical difference of datasets over cell culture time. If this test proved significant (p-value <0.05), the null hypothesis was rejected.
2.4. Enzymatic activities
2.4.1. Sample preparation
For each kit, the required amount of cells was collected depending on the test and according to the manufacturer protocol. Cells were immediately incubated in cold methanol (5 min, −20 °C), which decreases the temperature of the sample down to 4 °C in less than a minute, in order to stop enzymatic reactions while avoiding liquid freezing. Samples were then washed twice, by two centrifugation steps at 3 °C for 8 min, at 1400 rpm and resuspension in cold PBS. Cell pellets were put in dry ice for 5 min and stored at −80 °C until needed. Samples were thawed and pellets were resuspended with cold corresponding assay buffer. Cells were lysed by sonication with 3 cycles of 30s pulse, 5s interval, on ice (Branson Sonifier 250). Cells were centrifuged (10 min, 10 000 g, 3 °C) and supernatant were transferred to new tubes to be used immediately for enzyme quantification.
2.4.2. ELISA assay kits
2.4.2.1. Activities of mitochondrial ETC complexes I, II, and V
Respiratory complex activity was determined using the Complex I enzyme activity microplate assay kit (Abcam, ab109721), the Complex II enzyme activity microplate assay kit (Abcam, ab109908) and the Complex V enzyme activity microplate assay kit (Abcam, ab109714). All assays were performed following the manufacturer’s instructions.
For complex I, 60 μg proteins were added to the pre-coated wells. In this assay, complex I activity is measured by the oxidation of NADH to NAD+ and the simultaneous reduction of a provided dye, which leads to increased absorbance measured at 450 nm (Thermo Scientific Multiskan Go) every 30s for 30min.
60 μg proteins were added to the pre-coated wells with capture antibody for complex II. In this assay, the production of ubiquinol by complex II protein is related to the reduction of the 2,6-diclorophenolindophenol (blue) to DCPIPH2 (colorless), and the decreasing absorbance is measured at 600 nm every 20s for 60min.
Complex V or ATP synthase is immunocaptured within the wells of the microplate, which has a monoclonal antibody pre-bound to the wells. 50 μg proteins from the samples were used for this assay. In this assay, the conversion of ATP to ADP is coupled to the oxidation of NADH to NAD+, which is monitored as a decrease in absorbance at 340 nm every minute for 120min.
2.4.2.2. Activity of citrate synthase
Citrate synthase was quantified with enzymatic assay kit (Abcam, ab119692). Cell pellets were solubilized by adding extraction buffer to the samples, and 125 μg proteins were used for this assay. The activity of citrate synthase was determined in an immunocapture based manner by recording the color development of 5-thio-2-nitrobenzoic acid at 412 nm every 30 s for 30 min.
2.4.2.3. Activity of pyruvate dehydrogenase
PDH activity in samples was quantified using a microplate assay kit (Abcam, ab109902). Cell pellets were solubilized by addition of detergent, and 400 μg of total proteins were added to anti-PDH antibody–coated wells of a microplate. PDH activity was then determined spectrophotometrically by monitoring every 20 s for 15 min the reduction of NAD + to NADH, coupled to the reduction of a reporter dye at absorbance 450 nm.
2.4.2.4. Activity of malate dehydrogenase
Malate Dehydrogenase 2 activity assay (Abcam, ab119693) was used to determine mitochondrial malate dehydrogenase activity. Cells were lysed in extraction buffer, and 150 μg of proteins were added to the antibody capture plates. Buffer containing a reagent dye was added and the increased absorbance at 450 nm was recorded every 30 s for 30 min.
2.4.2.5. Colorimetric assay kits
Glyceraldehyde 3 Phosphate Dehydrogenase activity was assessed by assay kit (Abcam, ab204732), as well as glucose 6 phosphate dehydrogenase (Abcam, ab102529), isocitrate dehydrogenase (Abcam, ab102528), and glutamate dehydrogenase (Abcam, ab102527).
2.4.2.6. Total protein quantification
Total proteins were quantified with the RC DC™ protein assay manufactured by Bio-Rad. This colorimetric assay is based on a modification of the Lowry protocol (Lowry et al., 1951), and can determine protein concentration in the presence of both reducing agents and detergents. Fatty acid free bovine serum albumin was used as a standard.
3. Results
3.1. Unwanted phenotypic heterogeneity observed for industrial cell lines in upstream process development
Fig. 1 is showing the growth rate and Qp phenotypes of 4 different industrial production cell lines under pharmaceutical development, stably transfected with a vector-expressing recombinant protein, all derived from the DG44 lineage and cultivated under the same conditions.
Fig. 1.
Phenotype of 4 industrial cell lines cultivated in 2L stainless steel bioreactors in a fed-batch process. HP1 (n = 3), HP2 (n = 3), LP1 (n = 3), and LP2 (n = 3).
In the same growing conditions, cells exhibit different phenotypic features. All the cell lines, which produce a different therapeutic antibody, display a comparable growth rate but a significantly different antibody production rates (Qp). On day 7, the specific productivity of HPs is twice larger than LP1 and four times larger than LP2. We have categorized 2 cell lines as high producers (HP) and 2 as low producers (LP), given the evolution of the specific productivity over cell culture time. The first HP has a significantly higher Qp from day 2–7 which is at least twice as large as the other cell lines, and the second HP has a later increase of titer, starting from day 5 to exceed the average Qp of LPs. Despite a comparable cell growth, specific production rates of recombinant proteins for the 2 LPs remain on average around half of the HPs value.
This unwanted phenotypic heterogeneity is usually tackled by industrial CHO production platforms with media and process optimization. In this study, the metabolic status of these cells was evaluated at different days of the process, in order to identify specific metabolic traits for HP and potential bottlenecks that prevent a higher recombinant protein production.
3.2. Modeling performances of the industrial cell lines
A curated genome-scale model specific to CHO DG44 cell lines was used to predict intracellular metabolic flux distributions of the 4 cell lines. For each cell lines, all the biological replicates were modeled independently, for each day between 2 and 7. The model was semi-constrained with exometabolomics data, which means that the experimental consumption rates of amino acid and other metabolites were used to set the theoretical maximal bounds in the model. The IgG production rate was set strictly equal to the experimental observation. The predicted nutrient consumption rates and by-product production rates were then compared to the experimental setting, as a way to calibrate the model and to validate that the predictions correspond to the environmental conditions were the cells evolved.
As the objective function to maximize was the growth rate, in the defined modeling conditions the predicted growth rate was compared to the experimental specific cell growth. Suppl. Fig. 1 shows the correlation coefficient of all the independent bioreactor run, which is on average above 0.7 for all the cell lines. This apparent elevated correlation with in vivo observations promotes the use of the model to further analyze intracellular fluxes and to compare them for HP and LP cell lines.
We have decided to focus the next part of the study on comparing trends of intracellular predicted fluxes with experimental fluxes, for the two high producers, in order to validate the predictions of the model. To find the best straightforward approach for assessment of experimental fluxes, we have to consider flux definition. Flux rates predicted by the model are defined as the turnover enzymatic rate of molecules through a metabolic pathway, and the level of regulation is function of 3 factors, namely (i) the activity level of the enzyme catalyzing the reaction, (ii) the properties and affinities of the enzyme, and (iii) the concentrations of the reactants and products (Nielsen, 2003). Thus, we have decided to measure enzymatic activities of cell lines HP1 and HP2 in order to compare in silico flux rates to in vivo activities. In order to account for difference in cell growth, all the enzymatic activities measured were normalized according to the total amount of protein synthesized in cells.
3.3. Enzymatic activities comparable with predicted fluxes highlight specific metabolic traits for each HPs
The objectives of the following experiments were both to compare predicted fluxes to in vivo activities, and to compare the metabolism of cell lines HP1 and HP2 that have an elevated antibody productivity.
3.3.1. Experimental approach for model validation
Regarding the modeling approach, for the different cell lines intracellular metabolic fluxes were quantified based on the predictions from the tailored genome-scale model (details in section 2.2.4.). One genome-scale model was generated for each cell line with their specific antibody composition. They were individually constrained with experimental exometabolomics data sets, in triplicate for each cell line.
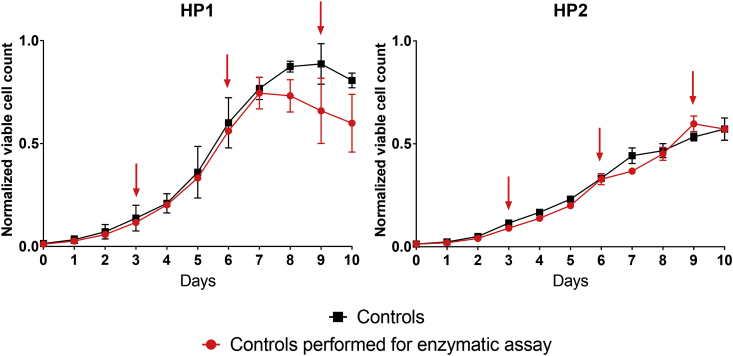
Regarding the experimental approach, the two HPs cell lines were cultivated under the same conditions as the one applied to generate the dataset included in the model, in order to be able to compare enzymatic activities to the predicted fluxes obtained from data collected from previous experiments. Sampling for enzyme quantification was performed on day 3, 6 and 9 for enzymatic activity measurements (Fig. 2).
Fig. 2.
Normalized viable cell count of the control 2L cell culture processes that were performed to collect samples for enzymatic activity measurements. Red dots: 2L control bioreactor runs (n = 2) repeated for collecting cell pellets; Black squares: 2L control bioreactor runs (n = 3) data that was used to model the cell lines; Red arrows: time points for performing enzymatic assays. Normalization according to maximum viable cell count reached. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
For cell line HP1 and HP2, cell growth obtained in the more recent experiment was comparable to historical data. Enzymatic results for HP1 on day 9 have to be analyzed with more caution as the average viable cell count is 35% lower than historical data. Except from this time point, observations made for all days for HP1 and HP2 regarding the performed enzymatic study can be extrapolated to the control conditions.
3.3.2. Enzymatic activity in electron transport chain confirms predictive trends and shows similar activities for both high producers
As electron transport chain catalyzes the phosphorylation of ADP to ATP by exploiting the transmembrane proton motive force, this is one of the most important cellular process that ensures cell survival and proliferation. The activity of protein complexes involved in oxidative phosphorylation was measured, for complexes I, II and V.
Our results show no visual difference in the activity of ATP synthase between the experimental and predicted values for HPs. The activity is on average higher for HP1 at the beginning of the process (Fig. 3), which is observed in the predicted flux rates at day 4.
Fig. 3.
Normalized experimental and predicted enzymatic activity. The detection was based on ELISA assay and the activity was measured in mOD/min/mg of proteins. The data are normalized to the highest activity measured for these tests. Error bars of predicted fluxes are standard deviation of predicted values for each replicate. Dashed lines highlight the potential higher error rate for HP1 at day 9, as average viable cell density was lower than expected. CS, Citrate synthase; MDH, Malate dehydrogenase; PDH, Pyruvate dehydrogenase.
However, the activities of complex I and complex II are very comparable and relatively constant during the process. According to the enzymatic activities, HP1 seems to have on average a more efficient metabolism at the beginning of the process, when HP2 seems to be at the highest energetical level at day 6. For complex II the trends are comparable between experimental and predicted values.
We can also note that experimental activity of ATP synthase is on average slightly higher than complex I and complex II, which is the case as well for predicted fluxes. However, predictions describe a lower activity in complex II compared to complex I, which is not confirmed experimentally.
3.3.3. Examination of pyruvate fluxes indicate a more active glycolysis for HP2 in the early days when HP1 seems to be transitioning earlier to efficient energy metabolism
Pyruvate obtained from glycolysis can be metabolized in several key metabolic pathways. Pyruvate can be reduced by lactate dehydrogenase leading to production of extracellular lactate, it can be converted into fatty acids, or to energy by pyruvate dehydrogenase (PDH). Indeed, when processed towards the TCA cycle, pyruvate decarboxylation produces Acetyl-CoA which is converted to Citrate, a tricarboxylic acid cycle intermediate (Fig. 4). Additionally, pyruvate can be converted to alanine and produced from cysteine.
Fig. 4.
Normalized experimental enzymatic activities and net influx and efflux measured during the process.
PDH activity is very comparable with in silico predictions. A higher activity is observed on day 3 for HP1 (Fig. 3), which confirms the trend predicted by the model. The activity decreases from day 3–6 for HP1, and increases on the day 6 for HP2 followed by a significant decrease on day 9.
Extracellular pyruvate available in the medium and feed starts to be consumed for HP1 by day 3 (Suppl. Fig. 2), which can explain the higher PDH activity for this cell line, whereas for HP2 pyruvate is secreted in the medium. On day 3, both cell lines have a similar glucose consumption rate and lactate is efflux is negligible. As a general observation, from day 0–3 HP1 has a lower glucose consumption, a lower lactate production rate and consumes pyruvate at a higher rate than HP2. Coupled with PDH activity on day 3, for HP1 these results highlight an earlier transition of fluxes from glycolysis to tricarboxylic acid cycle for ATP production.
As HP2 has a lower specific productivity than HP1 until day 5 of the process (Fig. 1), we could hypothesize that cells that are able to funnel pyruvate fluxes early in the process towards TCA cycle will obtain a higher specific productivity. PDH activity in the beginning of the process could be a marker to isolate high producers.
3.3.4. Bottlenecks identified in TCA cycle for both HPs
Malate dehydrogenase (MDH) catalyzes the oxidation of malate to oxaloacetate using the reduction of NAD + to NADH. The product of this reaction is then used as a substrate by citrate synthase (CS) to form citrate, which constitutes the first step of the TCA cycle (Fig. 4).
Quantifications of MDH and CS show a constant elevated activity during the run for both cell lines (Fig. 3). The trends are comparable with predictions, characterized by a slightly higher activity for HP1 in the first days. However, what was not predicted is that among all the enzymatic activities measured in the same units, these two enzymes show the highest activity. Moreover, constant and elevated experimental CS activity during the run is observed for the two cell lines. As in TCA cycle MDH reaction is preceding CS, these results give a new hint that malate dehydrogenase can be a bottleneck. Indeed, if MDH is a bottleneck, a constant amount of oxaloacetate is produced in the mitochondria, which limits CS activity. In another study (Chong et al., 2010), conducted exometabolomics analysis during a CHO cell fed-batch run, and revealed that extracellular malate accumulation was the most significant among all the metabolites they identified. MDH was identified as a bottleneck of TCA cycle, and its overexpression led to an increased cell growth. This could be applied to the cell lines used in the industrial production process to further improve their performances.
3.3.5. Higher PPP activity potentially linked to a better Qp
Cancer cells perform aerobic glycolysis at high rates and display high levels of glucose uptake and lactate production, as it has been observed by Otto Warburg more than 80 years ago (Kim and Dang, 2006). One enzyme in the glycolytic pathway has been quantified as a marker of glycolysis rate, the glyceraldehyde 3 phosphate dehydrogenase (GAPDH). This enzyme is the most concentrated among all the enzymes tested, both for the experimental and predicted activities (Fig. 5). The predicted trend for HP1 is similar to the experimental one, with an activity that is divided by 2 between day 3 and 6. The activity is correlated to the glucose uptake rate, HP1 has a lower glycolytic rate on day 6 whereas glucose uptake rate and thus GAPDH activity is similar on day 3 and 6 for HP2.
Fig. 5.
Normalized experimental and predicted enzymatic activity. The detection was based on indirect detection of NADH and the activity was measured in nmol/min/mg of proteins. The data are normalized to the highest activity measured for these tests. Dashed lines highlight the potential higher error rate for HP1 at day 9, as average viable cell density was lower than expected.
Glucose-6-phosphate dehydrogenase (G6PD) is the first step of the pentose phosphate pathway (PPP), generating reducing energy from the first glycolytic intermediate D-glucose 6-phosphate. Activity assay shows a negligible G6PD activity for HPs at the beginning of the process on day 3 (Fig. 5). For HP1, on day 6 the activity is seven times higher than on the day 3, whereas HP2 shows an activity as low as on day 3.
This observation can also be linked with GAPDH activity, as on day 6 a lower ratio of glucose 6 phosphate is addressed in glycolysis for HP1. In HP1, glucose 6 phosphate is funneled to PPP on day 6, which makes it less available for glycolysis and leads to a decrease of GAPDH concentration.
Given that HP1 is transitioning faster than HP2 towards a high Qp, the ability to overexpress G6PD at the middle of the exponential phase might be a marker for identification of HPs. The results suggest that recombinant protein production could be the cause or the consequence of an increased activity in pentose phosphate pathway. Overexpression of G6PD has often been used to improve protein production in many hosts (Davy et al., 2017).
The primary results of PPP are the generation of NADPH, which is used to prevent oxidative stress and also in fatty acid synthesis, and the production of precursors used in the synthesis of nucleotides and amino acids. Based on the NADPH outcome, two hypotheses could explain the difference of level of G6PD expression on day 6. Either HP1 has to cope with more oxidative stress than HP2, and then needs a higher amount of NAPDH on day 6, either HP1 is more efficient to orientate glucose 6 phosphate to PPP when oxidative stress increases in cells.
3.3.6. Isocitrate dehydrogenase requires more experimental data to interpret difference in early days
Isocitrate dehydrogenase (IDH) is an enzyme involved in the TCA cycle (Fig. 4). Three isoforms are present in CHO cells and have been quantified (Fig. 5). The IDH3 isoform catalyzes the oxidative decarboxylation of isocitrate, producing oxoglutarate, carbon dioxide and NADH. The isoforms 1 and 2 are NADP-dependent; IDH1 is located in the cytosol and peroxisome, and IDH2 in the mitochondrion.
A fair prediction of IDH fluxes can be confirmed with the enzymatic assay, as the only difference is noticed for the activity of HP2 on day 3, which is more elevated than HP1. On days 6 and 9, experimental IDH activity is similar for both cell lines, whereas on day 3 HP2 shows a higher activity. As citrate synthase is at constant high rate for both cell lines, this difference could be explained by a higher activity of the reaction upwards or downwards IDH for HP2 on day 3. Either aconitase activity concentration is higher, then producing higher amounts of IDH substrate, or oxoglutarate dehydrogenase is more concentrated and thus IDH’s product -oxoglutarate- turnover is higher than in HP1.
3.3.7. Glutamate dehydrogenase highlights a significant difference between HP1 and HP2 in terms of glutamate processing
Glutamate dehydrogenase (GLUD) is an enzyme that reversibly converts glutamate to α-ketoglutarate (Fig. 4). Experimental results show that the enzyme is not initially detected in HP1, and its concentration increases until day 9, while GLUD shows a constant elevated activity for HP2 (Fig. 5). This result is surprising because glutamine consumption rate is similar for both cell lines, and according to the high release of ammonium from day 0 to day 3 glutamine seems indeed to be converted to glutamate (Suppl. Fig. 3). The data suggest that for HP2 all glutamate is converted by GLUD and injected into TCA cycle, while for HP1 glutamate is addressed into another pathway.
3.3.8. Statistical summary
As seen in Table 1, statistically significant differences between experimental and predicted enzymatic activities over time were found for the following reactions: Complex II, CS, MDH, GAPD, GLUDH and G6PD. When combining data from both cell lines, 40% of the enzymatic reactions are statistically similar between predicted and experimental values. This result can be explained by the poor match between predicted and experimental obtained for HP1 (20% similitude), whereas 60% of the data are statistically similar for HP2.
Table 1.
Results from MLR statistical test.
| Fluxes |
p-value |
||
|---|---|---|---|
| Cell line | HP1 and HP2 | HP1 | HP2 |
| ATP Synthase | 0.0502 | 0.0311 | 0.1534 |
| Complex I | 0.08 | 0.211 | 0.0411 |
| Complex II | 0.0032 | 0.008 | 0.1157 |
| PDH | 0.5713 | 0.261 | 0.0485 |
| CS | 0.0137 | 0.0005 | 0.6287 |
| MDH | 0.0136 | 0.0005 | 0.6311 |
| GAPD | 0.0242 | 0.0527 | 0.1094 |
| GLUDH | 0.0001 | 0.0004 | 0.0022 |
| IDC | 0.4679 | 0.0051 | 0.1081 |
| G6PD | 0.0001 | 0.0001 | 0.0001 |
| % of reactions statistically similar | 40 | 20 | 60 |
The poor comparability obtained for HP1 can be explained by the uncertainty of the experimental measurements performed on day 9, as cell growth was lower than historical data which were used to predict enzymatic fluxes. However, as the enzymatic rates were normalized by the total protein quantity, it was assumed that the experimental quantification on day 9 could be taken into consideration.
As 60% of the experimental and predicted activities are proven statistically comparable for HP2, model’s predictions were considered as reliable.
3.4. Assessment of cell metabolism during exponential phase based on modeling predictions
The previous section has shown that modeled pathways are verified with enzymatic activities for 2 HPs. The predictions were assessed as reliable and were analyzed to highlight metabolic differences.
3.4.1. Predicted intracellular fluxes show comparable metabolic activities
First, a sensitivity analysis was performed on the predicted fluxes. Within one cell line, at each day of the culture the level of dispersion of the three predicted metabolic rates – that were independently computed from three biological replicates - was calculated in the form of a coefficient of variation. The CVs calculated for each day were then average between day 2 and 7, in order to obtain one single CV per metabolic rate and cell line. Suppl. Fig. 4 displays the average CV from day 2–7 for each reaction. Around 90% of the CVs are below 30% for each cell lines (89, 87, 92 and 92% of the reactions respectively for HP1, HP2, LP1 and LP2), which indicates a very low level of dispersion around the mean and a reproducible prediction (Reed et al., 2002). The average values of each predicted flux, calculated for each cell line, were then analyzed with a hierarchical clustering. Fig. 6 shows the average predicted value over the six chosen days. According to the model, the central metabolism of HP1 and LP2 is the most comparable, particularly due to higher glycolysis activity which can be explained by a high experimental glucose uptake rate on the first days.
Fig. 6.
A. Visualization of the metabolic rates for each cell lines (n = 3), clustered according to the similarity between the average predicted value from day 2 to day 7 of the 35 selected reactions; B. Cell line clustering according to the average value from day 2 to day 7 of the predicted reactions rates (n = 3) in electron transport chain. All the predicted flux rates are normalized. Hierarchical clustering follows the agglomerative strategy (Murtagh, 1983), where each observation starts in its own cluster, and at each step the Euclidian distance between each cluster is calculated to only merge the two clusters that are the closest together.
As pyruvate is synthesized at a higher rate, pyruvate dehydrogenase is overall more active and the TCA cycle activity is also higher for those cell lines. LP2 shows a TCA cycle which seems to be more active than in the other cultures, predicted to be more intense on the upper part on average (malate dehydrogenase, citrate synthase, aconitate hydratase, isocitrate dehydrogenase). Predicted lipid metabolism is less intense for cell line HP2 than the other on average. The rate of synthesis of palmitoyl, which leads to sphingolipid synthesis, a major constituent of cell membranes, is the lowest for HP2 and LP1. Biosynthesis rate of some lipids is reduced for HP2, such as phosphatidic acid (Diacylglycerol kinase), triacylglycerol (Glycerol-3-phosphate acyltransferase) and glycerol 3-phosphate mainly synthesized through glycolysis.
The activity of the electron transport chain is displayed on Fig. 6. B. Apart from LP2, which displays a slightly higher uptake rate of oxygen and activity of complex I and II, the four cell lines are predicted to have a very comparable activity on average from day 2–7. These predictions do not allow to infer a significant difference between high and low producers at the level of energy metabolism. This could mean that the low producers are either wasting energy, or a consequent part of this energy is used in a process that is not considered in the model, or cells are not producing as much ATP as predicted because of a bottleneck that is not considered in the model. The bottlenecks that are not included in this mathematical way of modeling cell metabolism could be at the level of transcription and translation regulation (Davy et al., 2017), protein processing (Reinhart et al., 2014), feedback regulation of enzymatic reactions, or missing constraints for the availability of other nutrients.
3.4.2. Predicted activity in lipid metabolism confirmed with difficulties encountered during harvest
In order to explain the predicted results obtained for lipid metabolism, we compared the different approaches employed for the harvest step of the industrial processes. Harvest can differ significantly from a cell line to another, depending on the physical properties of the cell culture fluid at the end of the process, and the techniques applied for enhancing clarification performances are diverse. Some additional pre-treatments are often required prior to the traditional continuous centrifugation, depth filtration and sterile filtration steps that are performed in the end of the culture. These additional treatments can facilitate separation of cells and cell debris and are often the only strategy for industrial companies to avoid early filter fouling. Among the techniques used, we can cite the addition of flocculants (Han et al., 2003; Kang et al., 2013; Roush and Lu, 2008) such as polyamines (Peram et al., 2010), chitosan (Riske et al., 2007), and polydiallyldimethylammonium chloride (McNerney et al., 2015) that bind to negatively charged surfaces of cell debris. Acid precipitation is also another improved method for clarifying a cell culture (Lydersen et al., 1994), which involves addition of concentrated phosphoric acid in order to decrease pH and thus solubility, leading to precipitation of cell debris but not the antibody.
During the harvest step of the process, the experimental difficulties observed for these cell lines can be interpreted with the predicted fluxes. HP2 is the only cell line that does not need any pre-treatment before harvest step at the end of the cell culture process. The other cell lines have shown early filter fouling due to increased amount of solids content, that are highly suspected to be lipids. In-house experiments have shown that after each pre-treatment, a significant reduction of total lipid quantification is observed (data not shown). The fact that HP2 does not require any pre-treatment may indicate that the cells total lipid content is less important than the other cell lines which is in agreement with the model’s predictions in terms of activity in lipid pathway.
3.4.3. Predicted clustering of cell lines was confirmed with analysis of amino acid consumption rate and antibody protein sequence
In order to investigate why the cell lines are clustered together, an analysis was performed on the input data of the model, namely the experimental flux rates that were used to constrain the model. The experimental amino acid consumption rates (Suppl. Fig. 5) reflect clustering results based on predicted fluxes (Fig. 6). On day 2, 4 and 5, HP1 and LP2 are clustered together, which shows an overall similarity in amino acid requirements, and justify a high degree of comparability of predicted reaction rates in different metabolic pathways.
To fully understand the clustering of the cell lines based on the predicted rates, the antibody amino acid sequence was analyzed for each cell line (Suppl. Fig. 6). It becomes apparent that HP1 and LP2 have the most similar amino acid composition of their antibody, and that HP2 is most comparable to LP1. This result is consistent with the clustering obtained for the predicted fluxes and the amino acid consumption for each cell line (Fig. 6). This whole analysis reveals that the cell lines can be divided into two groups based on their metabolism driven by a similar amino acid requirement.
As the intracellular predicted fluxes could not differentiate between high and low producers, a more detailed examination of the experimental amino acid uptake rates was performed to reveal any general feature of high or low producers. We observe that HP1 has a different behavior for aspartate and glutamate (Suppl. Fig. 7), which are produced until day 2 and a switch occurs at day 3 where HP1 starts to consume these amino acids. We could hypothesize that HPs have an overproduction of oxaloacetate and α-ketoglutarate in the first days in the TCA cycle, resulting in a net aspartate and glutamate secretion. As day 3 corresponds to glutamine depletion, the switch between production to secretion can be the time point where glutamine can no longer provide its anaplerotic role. For the other cell lines, their metabolism does not seem as efficient as HP1 when considering secretion of TCA cycle intermediates. According to the predicted fluxes, HP1 and LP2 have the highest activity in TCA cycle, however for LP2 on average none of these intermediates are secreted, thus the feed regime and composition are probably not overestimated in terms of nutrient concentration and rate of addition.
4. Discussion
For predicted ATP synthase activity, we evaluated the proportion of ATP that could potentially be involved in amino acid transporters, as 537 amino acid transporters were deleted to simplify the model. We considered if this action could have a significant impact on predicted ATP turnover. Actually, although cell lines do not probably express the same set of amino acid transporters to support their growth (Hyde et al., 2003), human cancer cells have been shown to express 4 amino acid transporters in high levels (Bhutia et al., 2015). None of these transporters are active transporters, they are either symporters or exchangers coupled with amino acid or sodium substrate. Thus, the ATP consumption linked to amino acid influx could be negligible. Consequently, we can assume that the simplification in the model regarding the amino acid transportation system, with the removal of active ATP-dependent transporters, does not lead to a significative underestimation of the ATP consumption.
Regarding PDH activity, we found it interesting to note that it is higher for HP1 than for HP2 on day 3, whereas the same day the activity of citrate synthase is similar for both cell lines. As PDH produces acetyl-CoA that is the substrate of citrate synthase, we would have expected a higher citrate synthase activity in HP1. Some hypotheses could explain this result. One hypothesis is connected to the fate of amino acids linked to pyruvate and acetyl-CoA metabolism, and Suppl. Fig. 2 shows the experimental variation of their fluxes during the process. The amino acid precursors of pyruvate production in CHO cells are cysteine (Hecklau et al., 2016), alanine which can be converted in a reversible reaction to pyruvate by alanine transaminase (Duarte, 2014, Li et al., 2012), and serine (Vacanti et al., 2014). As such, glycine and threonine can be also used for pyruvate production as threonine can be catalyzed into glycine by threonine aldolase, and glycine to serine by glycine hydroxymethyltransferase. On day 3, HP1 consumes more cysteine and serine than HP2, but HP1 releases more glycine and does not consume alanine on the opposite of HP2. Amino acid consumption and production rates that could explain fluctuations in pyruvate availability are balanced, and thus are not sufficient to explain the difference observed with PDH activity on day 3.
Another hypothesis formulated here is that a significant part of acetyl-CoA produced by PDH for HP1 can be converted to Malonyl-CoA and then redirected to fatty acid and steroid synthesis. This assumption is enforced with the experimental isoleucine uptake rate, that can be degraded to acetyl-CoA, which is similar for both cell line on day 3 (Suppl. Fig. 2). Either HP1 increases an internal lipid pool by deriving fluxes towards malonyl CoA production, or HP2 catabolizes lipids from an intracellular pool to fuel its TCA cycle. Assuming that an increased intracellular lipid pool would lead to a higher cell volume and thus a higher cell diameter (Pan et al., 2017), HP2 accumulates more fatty acids than HP1 from day 6–9, but seems to consumes this lipid pool from day 10 until the end of the process as the final diameter is on average smaller than HP1 on day 14 (Suppl. Fig. 8). As HP2 activates glycolytic pathway at high rate around day 6 of the process, the more likely hypothesis would be that glucose is converted to build up a fatty acid storage, which is catabolized later on to fuel TCA cycle with pyruvate. Based on the fact that CS is a bottleneck, this cell line defines a metabolic strategy with accumulation of energy in the form of lipid droplets while the substrates are available in high quantities, because anyway pyruvate cannot be processed more efficiently in the TCA cycle.
Enzymatic measurements of G6PD and GLUD reactions showed different trends than predictions. In order to understand the discrepancy, we compared our results to 13C labeling experiments and metabolic flux analysis performed with CHO cells in fed-batch culture.
Our experimental measurements were confirmed for the reaction catalyzed by G6PD by (Ahn and Antoniewicz, 2012; Sengupta et al., 2011; Templeton et al., 2013). Indeed, the trend of G6PD flux is exactly the same as the experimental one in this study. Minimal activity is observed during early exponential phase, and increased flux is measured at the late exponential and stationary phase. This raises the question about the source of NADPH required for growth in vivo in early process stage, as shown by a high predicted G6PD flux in the beginning of the process. During cell growth, NADPH is not only required to maintain reduced glutathione pools, but also to support reductive biosynthesis such as lipid synthesis (Xie and Wang, 2000). Another pathway that could be mainly responsible for generating NADPH for growth is the conversion of serine to glycine, which has been shown by isotopic labeling to contribute to NADPH production through the reaction catalyzed by methylene tetrahydrofolate dehydrogenase (MTHFD2 and MTHFD2L) in the mitochondria (Lewis et al., 2014).
As far as GLUD reaction is concerned, the trends reported in the literature with isotope labeling experiments confirm the predictions, not the experimental measurements (Nolan and Lee, 2011; Templeton et al., 2013). Although the measurement was performed with biological duplicates, we assumed that there was a loss of sample for the first time point measurement and thus that the first time point was very likely to be inaccurate.
5. Conclusion
This study demonstrates a generally reliable prediction of the intracellular fluxes based on a genome-scale model, which was only constrained with amino acids and by-products rates. As at least 70% of the predicted fluxes are in correlation with the enzymatic activities measured, the enzyme assays support the model flux predictions and validate the analysis performed on the predicted intracellular fluxes.
A total of 59 intracellular reactions were examined from day 2–7 in different key pathways, namely glycolysis, the pentose phosphate pathway, TCA cycle, lipid metabolism, and oxidative phosphorylation. The predicted fluxes were relatively robust within the replicates, and indicated that the metabolism of HP1 and LP2 were the most comparable, essentially characterized by an intense TCA cycle activity. The clustering of the predicted fluxes was in correlation with the clustering performed on experimental nutrient consumption rates and with the clustering of the relative composition of amino acids in the antibody produced. The in silico analysis indicates that the cells clustered together have a similar amino acid requirement despite a different efficiency in antibody production. One assumption made from the analysis of experimental amino acid metabolism, is that HPs have a higher activity in TCA cycle in the first days resulting in a net production of aspartate and glutamate.
HP2 was clustered separately than HP1 and LP2 based on an overall lower lipid activity from day 2–7. To summarize the experimental insights collected with enzymatic activities, industrial process adaptations and evolution of cell diameter during the process, the overall assumption for HP2 is divided into three phases: (i) at the beginning of the process, HP2 accumulates less lipid than the other cell lines, (ii) HP2 starts intracellular lipid storage around day 6, and (iii) from day 10 the intracellular pool starts to be consumed, as its final cell diameter is lower than HP1, and also because HP2 is less difficult to harvest than HP1 at the end of the process.
Analysis of enzymatic activities could explain the difference of Qp observed in the beginning of the process, which is higher for HP1 than HP2. HP1 has a more efficient transition of fluxes from glycolysis to TCA cycle, and also has a more active PPP activity in this time frame. As a suggestion for industrial clone selection, we recommend to use PDH and G6P as markers for HPs. These enzymes could also be valuable metabolic targets, and overexpressed for improving cell’s performances. Additionally, experimental measurements of MDH activity suggest that this enzyme is a bottleneck, and Qp could be potentially improved when overexpressing this enzyme.
Acknowledgments
This work was supported by an H2020 Marie Sklodowska-Curie Actions ITN, grant number 642663.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mec.2019.e00097.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Ahn W.S., Antoniewicz M.R. Towards dynamic metabolic flux analysis in CHO cell cultures. Biotechnol. J. 2012;7:61–74. doi: 10.1002/biot.201100052. [DOI] [PubMed] [Google Scholar]
- Bhutia Y.D., Babu E., Ramachandran S., Ganapathy V. Amino Acid transporters in cancer and their relevance to "glutamine addiction": novel targets for the design of a new class of anticancer drugs. Cancer Res. 2015;75:1782–1788. doi: 10.1158/0008-5472.CAN-14-3745. [DOI] [PubMed] [Google Scholar]
- Bonarius H.P.J., Hatzimanikatis V., Meesters K.P.H., de Gooijer C.D., Schmid G., Tramper J. Metabolic flux analysis of hybridoma cells in different culture media using mass balances. Biotechnol. Bioeng. 1996;50:299–318. doi: 10.1002/(SICI)1097-0290(19960505)50:3<299::AID-BIT9>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Chong W.P., Reddy S.G., Yusufi F.N., Lee D.Y., Wong N.S., Heng C.K., Yap M.G., Ho Y.S. Metabolomics-driven approach for the improvement of Chinese hamster ovary cell growth: overexpression of malate dehydrogenase II. J. Biotechnol. 2010;147:116–121. doi: 10.1016/j.jbiotec.2010.03.018. [DOI] [PubMed] [Google Scholar]
- Davy A.M., Kildegaard H.F., Andersen M.R. Cell factory engineering. Cell Syst. 2017;4:262–275. doi: 10.1016/j.cels.2017.02.010. [DOI] [PubMed] [Google Scholar]
- Duarte Metabolic responses of CHO cells to limitation of key amino acids. Biotechnol Bioeng. 2014 doi: 10.1002/bit.25266. [DOI] [PubMed] [Google Scholar]
- Ebrahim A., Lerman J.A., Palsson B.O., Hyduke D.R. COBRApy: constraints-based reconstruction and analysis for Python. BMC Syst. Biol. 2013;7:74. doi: 10.1186/1752-0509-7-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B., Akeprathumchai S., Wickramasinghe S.R., Qian X. Flocculation of biological cells: experiment vs. theory. AIChE J. 2003;49:1687–1701. [Google Scholar]
- Hastie T., Tibshirani R., Friedman J. Springer-Verlag; New York: 2009. The Elements of Statistical Learning. [Google Scholar]
- Heclau S-Sulfocysteine simplifies fed-batch processes and increases the CHO specific productivity via anti-oxidant activity. J Biotechnol. 2016 doi: 10.1016/j.jbiotec.2015.11.022. [DOI] [PubMed] [Google Scholar]
- Hyde R., Taylor P.M., Hundal H.S. Amino acid transporters: roles in amino acid sensing and signalling in animal cells. Biochem. J. 2003;373:1–18. doi: 10.1042/bj20030405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irani N., Wirth M., van Den Heuvel J., Wagner R. Improvement of the primary metabolism of cell cultures by introducing a new cytoplasmic pyruvate carboxylase reaction. Biotechnol. Bioeng. 1999;66:238–246. doi: 10.1002/(sici)1097-0290(1999)66:4<238::aid-bit5>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Jeong D.W., Cho I.T., Kim T.S., Bae G.W., Kim I.H., Kim I.Y. Effects of lactate dehydrogenase suppression and glycerol-3-phosphate dehydrogenase overexpression on cellular metabolism. Mol. Cell. Biochem. 2006;284:1–8. doi: 10.1007/s11010-005-9004-7. [DOI] [PubMed] [Google Scholar]
- Kang Y.K., Hamzik J., Felo M., Qi B., Lee J., Ng S., Liebisch G., Shanehsaz B., Singh N., Persaud K., Ludwig D.L., Balderes P. Development of a novel and efficient cell culture flocculation process using a stimulus responsive polymer to streamline antibody purification processes. Biotechnol. Bioeng. 2013;110:2928–2937. doi: 10.1002/bit.24969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.W., Dang C.V. Cancer’s molecular sweet tooth and the Warburg effect. Cancer Res. 2006;66:8927–8930. doi: 10.1158/0008-5472.CAN-06-1501. [DOI] [PubMed] [Google Scholar]
- Lewis C.A., Parker S.J., Fiske B.P., McCloskey D., Gui D.Y., Green C.R., Vokes N.I., Feist A.M., Vander Heiden M.G., Metallo C.M. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol. Cell. 2014;55:253–263. doi: 10.1016/j.molcel.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis N.E., Hixson K.K., Conrad T.M., Lerman J.A., Charusanti P., Polpitiya A.D., Adkins J.N., Schramm G., Purvine S.O., Lopez-Ferrer D., Weitz K.K., Eils R., Konig R., Smith R.D., Palsson B.O. Omic data from evolved E. coli are consistent with computed optimal growth from genome-scale models. Mol. Syst. Biol. 2010;6:390. doi: 10.1038/msb.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wong C.L., Vijayasankaran N., Hudson T., Amanullah A. Feeding lactate for CHO cell culture processes: Impact on culture metabolism and performance. Biotechnology and Bioengineering. 2012 doi: 10.1002/bit.24389. [DOI] [PubMed] [Google Scholar]
- Lowry O.H., Rosebrough N.J., Farr A.L., Randall R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Lydersen B.K., Brehm-Gibson T., Murel A. Acid precipitation of mammalian cell fermentation broth. Ann. N. Y. Acad. Sci. 1994;745:222–231. doi: 10.1111/j.1749-6632.1994.tb44375.x. [DOI] [PubMed] [Google Scholar]
- Mastrangelo A.J., Hardwick J.M., Zou S., Betenbaugh M.J. Part II. Overexpression ofbcl-2 family members enhances survival of mammalian cells in response to various culture insults. Biotechnol. Bioeng. 2000;67:555–564. doi: 10.1002/(sici)1097-0290(20000305)67:5<555::aid-bit6>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- McNerney T., Thomas A., Senczuk A., Petty K., Zhao X., Piper R., Carvalho J., Hammond M., Sawant S., Bussiere J. PDADMAC flocculation of Chinese hamster ovary cells: enabling a centrifuge-less harvest process for monoclonal antibodies. mAbs. 2015;7:413–428. doi: 10.1080/19420862.2015.1007824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meents H., Enenkel B., Eppenberger H.M., Werner R.G., Fussenegger M. Impact of coexpression and coamplification of sICAM and antiapoptosis determinants bcl-2/bcl-x(L) on productivity, cell survival, and mitochondria number in CHO-DG44 grown in suspension and serum-free media. Biotechnol. Bioeng. 2002;80:706–716. doi: 10.1002/bit.10449. [DOI] [PubMed] [Google Scholar]
- Murtagh F. A survey of recent advances in hierarchical clustering algorithms. Comput. J. 1983;26:354–359. [Google Scholar]
- Nielsen J. It is all about metabolic fluxes. J. Bacteriol. 2003;185:7031–7035. doi: 10.1128/JB.185.24.7031-7035.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan R.P., Lee K. Dynamic model of CHO cell metabolism. Metab. Eng. 2011;13:108–124. doi: 10.1016/j.ymben.2010.09.003. [DOI] [PubMed] [Google Scholar]
- Pan X., Dalm C., Wijffels R.H., Martens D. Metabolic characterization of a CHO cell size increase phase in fed-batch cultures. Appl Microbiol Biotechnol. 2017 doi: 10.1007/s00253-017-8531-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peram T., McDonald P., Carter-Franklin J., Fahrner R. Monoclonal antibody purification using cationic polyelectrolytes: an alternative to column chromatography. Biotechnol. Prog. 2010;26:1322–1331. doi: 10.1002/btpr.437. [DOI] [PubMed] [Google Scholar]
- Pereira S., Kildegaard H.F., Andersen M.R. Impact of CHO metabolism on cell growth and protein production: an overview of toxic and inhibiting metabolites and nutrients. Biotechnol. J. 2018;13(3) doi: 10.1002/biot.201700499. [DOI] [PubMed] [Google Scholar]
- Reed G.F., Lynn F., Meade B.D. Use of coefficient of variation in assessing variability of quantitative assays. Clin. Vaccine Immunol. 2002;9:1235–1239. doi: 10.1128/CDLI.9.6.1235-1239.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart D., Sommeregger W., Debreczeny M., Gludovacz E., Kunert R. In search of expression bottlenecks in recombinant CHO cell lines--a case study. Appl. Microbiol. Biotechnol. 2014;98:5959–5965. doi: 10.1007/s00253-014-5584-z. [DOI] [PubMed] [Google Scholar]
- Riske F., Schroeder J., Belliveau J., Kang X., Kutzko J., Menon M.K. The use of chitosan as a flocculant in mammalian cell culture dramatically improves clarification throughput without adversely impacting monoclonal antibody recovery. J. Biotechnol. 2007;128:813–823. doi: 10.1016/j.jbiotec.2006.12.023. [DOI] [PubMed] [Google Scholar]
- Roush D.J., Lu Y. Advances in primary recovery: centrifugation and membrane technology. Biotechnol. Prog. 2008;24:488–495. doi: 10.1021/bp070414x. [DOI] [PubMed] [Google Scholar]
- Sengupta N., Rose S.T., Morgan J.A. Metabolic flux analysis of CHO cell metabolism in the late non-growth phase. Biotechnol. Bioeng. 2011;108:82–92. doi: 10.1002/bit.22890. [DOI] [PubMed] [Google Scholar]
- Templeton N., Dean J., Reddy P., Young J.D. Peak antibody production is associated with increased oxidative metabolism in an industrially relevant fed-batch CHO cell culture. Biotechnol. Bioeng. 2013;110:2013–2024. doi: 10.1002/bit.24858. [DOI] [PubMed] [Google Scholar]
- Vacanti Regulation of substrate utilization by the mitochondrial pyruvate carrier. Mol Cell. 2014 doi: 10.1016/j.molcel.2014.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriezen N. Delft University of Technology; 1998. Physiology of Mammalian Cells in Suspension Culture; pp. 121–144. Doctoral thesis. [Google Scholar]
- Wlaschin K.F., Hu W.S. Engineering cell metabolism for high-density cell culture via manipulation of sugar transport. J. Biotechnol. 2007;131:168–176. doi: 10.1016/j.jbiotec.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Xie L., Wang D.I. Applications of improved stoichiometric model in medium design and fed-batch cultivation of animal cells in bioreactor. Cytotechnology. 1994;15:17–29. doi: 10.1007/BF00762376. [DOI] [PubMed] [Google Scholar]
- Xie L., Wang D.I.C. Energy metabolism and ATP balance in animal cell cultivation using a stoichiometrically based reaction network. Biotechnol. Bioeng. 2000;52:591–601. doi: 10.1002/(SICI)1097-0290(19961205)52:5<591::AID-BIT6>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Zupke C., Stephanopoulos G. Intracellular flux analysis in hybridomas using mass balances and in vitro (13)C nmr. Biotechnol. Bioeng. 1995;45:292–303. doi: 10.1002/bit.260450403. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.