Summary
Plant exosomes protect plants against infection; however, whether edible plant exosomes can protect mammalian hosts against infection is not known. In this study, we show that ginger exosome-like nanoparticles (GELNs) are selectively taken up by the periodontal pathogen Porphyromonas gingivalis in a GELN phosphatidic acid (PA) dependent manner via interactions with hemin-binding protein 35 (HBP35) on the surface of P. gingivalis. Compared with PA (34:2), PA (34:1) did not interact with HBP35, indicating that the degree of unsaturation of PA plays a critical role in GELN-mediated interaction with HBP35. On binding to HBP35, pathogenic mechanisms of P. gingivalis were significantly reduced following interaction with GELN cargo molecules, including PA and miRs. These cargo molecules interacted with multiple pathogenic factors in the recipient bacteria simultaneously. Using edible plant exosome-like nanoparticles as a potential therapeutic agent to prevent/treat chronic periodontitis was further demonstrated in a mouse model.
Subject Areas: Bacteriology, Biochemistry, Biological Sciences, Immunology, Microbiology, Molecular Biology, Oral Microbiology
Graphical Abstract
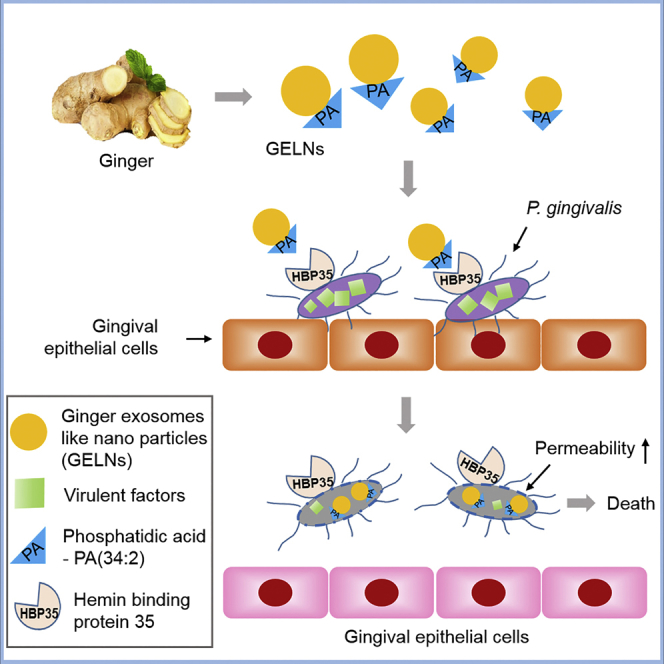
Highlights
-
•
Plant exosomes-like nanoparticles (ELNs) are selectively taken up by P. gingivalis
-
•
The degree of unsaturation of ELNs phosphatidic acid determines uptake specificity
-
•
ELN PA and miRs target multiple pathogenic factors of P. gingivalis
-
•
Orally taking ginger ELNs ameliorates bone loss induced by P. gingivalis
Bacteriology; Biochemistry; Biological Sciences; Immunology; Microbiology; Molecular Biology; Oral Microbiology
Introduction
Chronic infectious disease commonly involves large numbers of virulence factors that target several host factors in multiple pathways (Pan et al., 2014). Developing an effective therapeutic strategy that can inhibit a plurality of virulence factors without causing side effects requires a change from the current focus of delivering individual therapeutic agents to delivery of a package of therapeutic agents that can target multiple virulence factors simultaneously (Baron, 2013, Kauppi et al., 2003). No such ideal delivering vehicle that can selectively target pathogenic organisms and carry multiple therapeutic agents without causing toxicity is available. Recently, edible plant-derived exosome-like nanoparticles (ELNs) have been identified, and these consist of a large numbers of lipids, RNA including microRNAs (miRNAs), and proteins (Mu et al., 2014, Wang et al., 2013). It is well documented that edible plants are beneficial for human health and can prevent/treat chronic infectious diseases (Alvarez-Erviti et al., 2011, Zhuang et al., 2011). However, the cellular and molecular mechanism underlying the therapeutic effect on infectious disease by plants is not known. Since ELNs carry a large number and variety of molecules naturally (Mu et al., 2014, Xiao et al., 2018), we hypothesize that, upon ELNs being taken up by infectious agents, the ELN molecules can target multiple virulence factors simultaneously to prevent disease development.
Porphyromonas gingivalis, a Gram-negative anaerobe, is a major pathogen in chronic periodontitis, an inflammatory disease associated with dysbiotic host responses (Olsen et al., 2017). Periodontitis and periodontal pathogens have also been associated with serious systemic conditions including cardiovascular disease, type 2 diabetes mellitus, and adverse pregnancy outcomes (Liu et al., 2018, Reyes et al., 2018). P. gingivalis produces a number of virulence factors that enable colonization of oral surfaces, degradation of periodontal tissues, induction of destructive immune responses, and growth in the peptide- and hemin-rich inflammatory microenvironment. These include the FimA- and Mfa1-component fimbrial adhesins, arginine (Rgp)- and lysine (Kgp)-specific gingipain proteases, lipopolysaccharides, and hemin transport systems (Zenobia and Hajishengallis, 2015) (Lamont et al., 2018) (Bao et al., 2014).
Oral delivery of ginger exosome-like nanoparticles (GELNs) in mice leads to protection against alcohol-induced liver damage (Zhuang et al., 2015). Moreover, GELNs alter the gut microbiome composition and host physiology (Teng et al., 2018) leading us to test whether GELNs could be applied to treat/prevent oral infectious disease. In this study, ginger-derived ELNs (GELNs) were tested for antagonism of Porphyromonas gingivalis virulence factors and for inhibition of pathogenicity in a chronic periodontitis mouse model. Our data suggest that GELNs are selectively taken up by P. gingivalis whereupon pathogenicity of the organism is reduced. Pathogenic processes impacted by GELNs include growth, attachment, entry, and proliferation in host cells, with consequent reduced virulence in a mouse model of periodontal disease.
Results
Ginger Exosome-like Nanoparticles Are Selectively Taken up by P. gingivalis Leading to Inhibition of P. gingivalis Growth
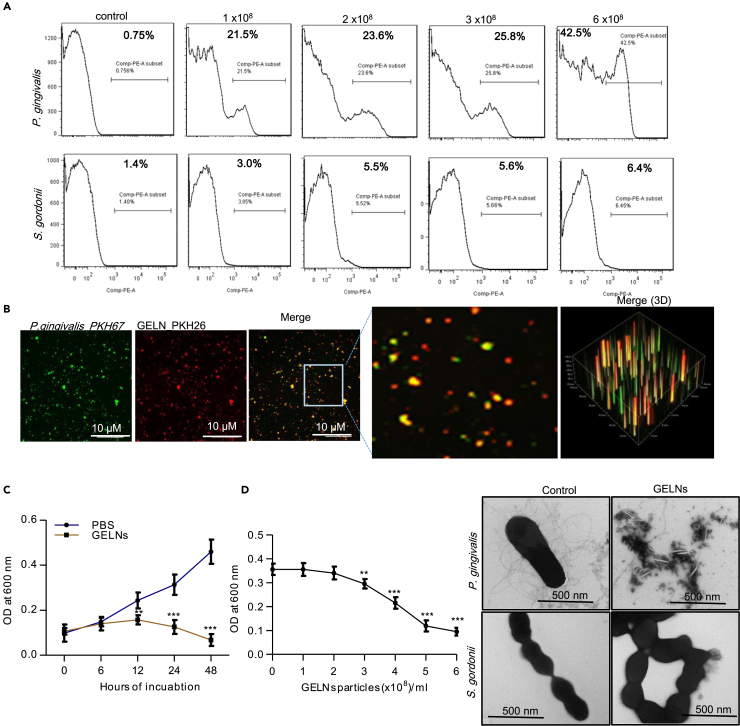
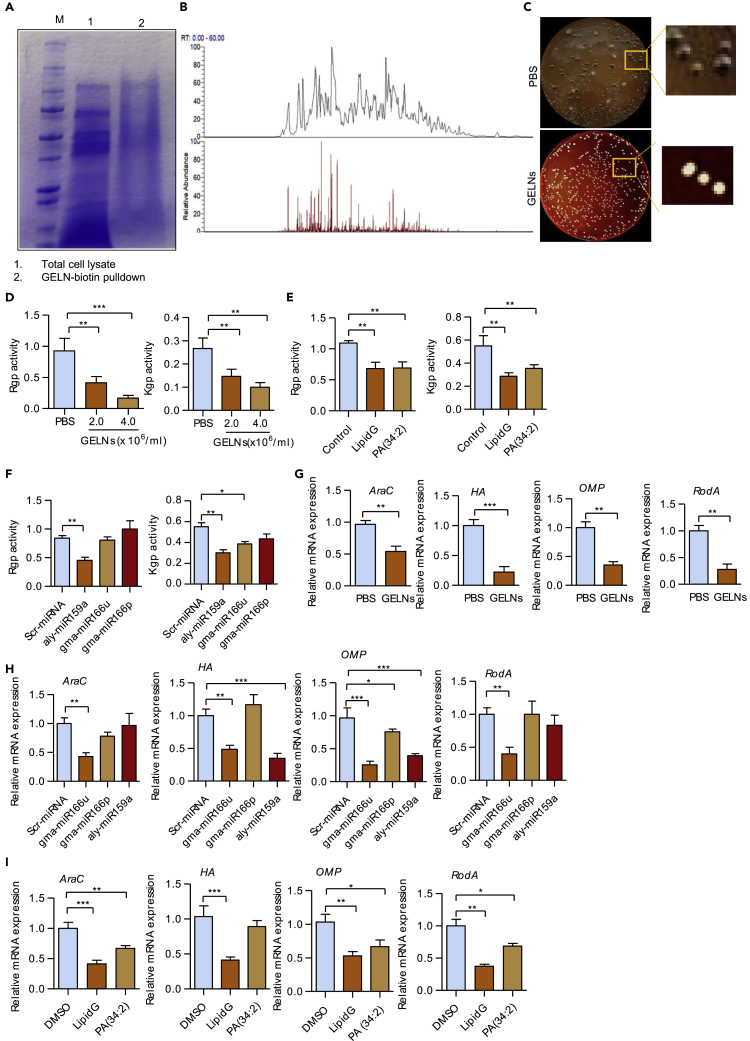
Our previous reports have shown that GELNs have anti-inflammatory effects (Mu et al., 2014) via interaction with host hepatocytes (Zhuang et al., 2015), and moreover GELN miRNAs selectively promote beneficial bacterial growth in the intestine (Teng et al., 2018). Whether GELNs have a direct effect on pathogenic oral bacteria such as P. gingivalis is not known. To test this, P. gingivalis, along with the oral commensal Streptococcus gordonii, was incubated with different concentrations (0–6.0 × 108 particles/mL) of PKH26-labeled GELNs for 1 h. fluorescence-activated cell sorting (FACS) analysis indicated that the GELNs were selectively taken up by P. gingivalis in a dose-dependent manner, whereas uptake of GELNs by S. gordonii was negligible (Figure 1A). P. gingivalis uptake of GELNs was further confirmed by confocal microscopy with fluorescently labeled P. gingivalis and GELNs (Figure 1B).
Figure 1.
Ginger Exosome-like Nanoparticles (GELNs) Selectively Inhibit Growth of the Pathogenic P. gingivalis but Not the Commensal S. gordonii
(A) P. gingivalis and S. gordonii were incubated with different concentrations (0–6.0 × 108 particles/mL) of PKH26-labeled GELNs for 1 h in an anaerobic chamber. GELN uptake by P. gingivalis and S. gordonii was quantified by flow cytometry.
(B) P. gingivalis and GELNs were labeled with fluorescent dyes PKH67 and PKH26, respectively. Then, P. gingivalis and GELNs were incubated at 37°C for 1 h in anaerobic chamber and fluorescence images were taken by confocal microscopy.
(C) P. gingivalis was incubated with GELNs (4.0 × 108/mL) for the indicated times. The growth of P. gingivalis was determined by measuring optical density at 600 nm.
(D) P. gingivalis was treated with different concentrations (0–6 × 108/mL) of GELNs and incubated at 37°C for 24 h. The growth of P. gingivalis was determined by measuring optical density at 600 nm. P. gingivalis and S. gordonii were treated with or without GELNs (6 × 108/mL) for 3 h and negatively stained with ammonium molybdate. The images were taken by transmission electron microscopy.
Results are expressed as mean ± standard deviation from three independent experiments. **p < 0.01, ***p < 0.001 compared with the untreated group using one-way ANOVA with Turkey’s Multiple comparison test.
Uptake of GELNs led to inhibition of the growth of P. gingivalis in a dose- and time-dependent manner (Figures 1C and 1D). At a higher dose (6×108 particles/mL), no growth of P. gingivalis was observed. Electron microscopy images further suggested that GELN treatment at the higher dose completely disrupted the morphology of P. gingivalis but not of S. gordonii (Figure 1D). Additionally, GELNs neither were taken up by S. gordonii nor inhibited the growth of this commensal. However, GELNs did inhibit the in vitro growth of other bacteria including F. nucleatum, P. intermedia, and A. actinomycetemcomitans (Figures S1C–S1E), which are associated with periodontitis.
Membrane depolarization has a profound impact on bacterial viability and signal transduction (Goldberg et al., 2013). Thus, we measured GELN effect on cytoplasmic membrane depolarization of P. gingivalis and S. gordonii using the membrane potential-sensitive dye diSC3-5 (Nusslein et al., 2006). The results showed that GELNs increased the depolarization of P. gingivalis. In contrast, GELNs did not affect the cytoplasmic membrane depolarization of S. gordonii (Figures S2A and S2B). In addition, we measured the P. gingivalis outer membrane barrier function by an ethidium bromide (EtBr) influx assay (Miki and Hardt, 2013). Our results showed that GELNs significantly increased fluorescence intensity in a dose-dependent manner (Figure S2C). Furthermore, we collected the supernatant from GELN-treated P. gingivalis and S. gordonii along with untreated control and analyzed these by SDS-PAGE electrophoresis. A large amount of proteins was released into the external milieu by GELN-treated P. gingivalis but not by GELN-treated S. gordonii (Figure S2D), indicating that interaction of GELNs with the P. gingivalis membrane leads to external release of cytoplasmic proteins. We also quantified metabolic products released from GELN-treated P. gingivalis (Figure S3), and the predicted role of these metabolic products is listed in Table S1. Collectively, the data indicate that GELNs are selectively taken up by pathogenic P. gingivalis, but not by commensal S. gordonii, leading to membrane perturbations and inhibition of P. gingivalis growth.
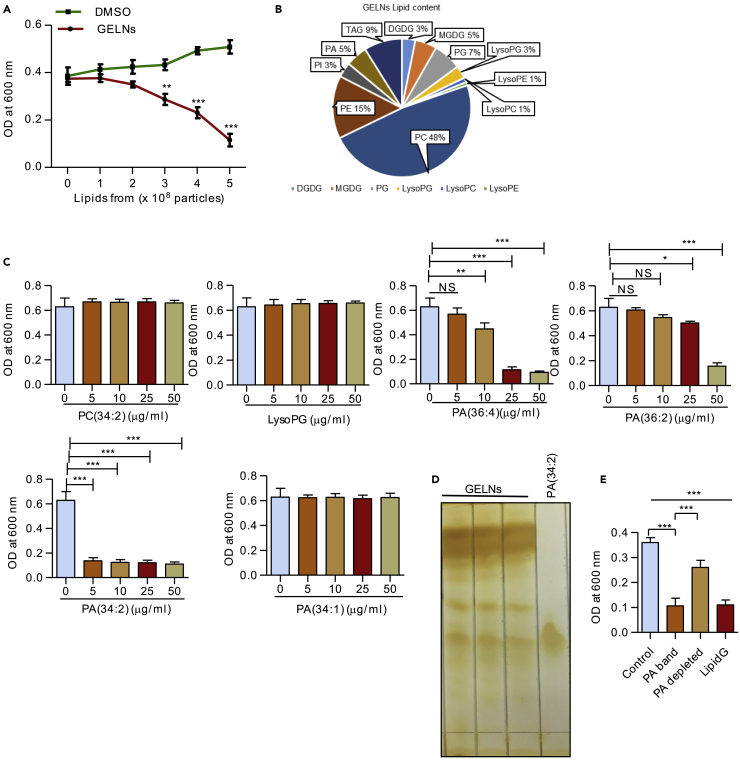
Edible plant exosomes, including GELNs, consist of a number of proteins, lipids, and RNAs including miRNA (Mu et al., 2014). Next, we determined which GELN-derived factor(s) specifically inhibits P. gingivalis growth. P. gingivalis was treated with different concentrations of total lipids derived from GELNs (lipidG) (0–5.0 × 108 particles/mL) for 24 h and growth measured. LipidG significantly decreased P. gingivalis growth in a dose-dependent manner (Figure 2A). RNA sequencing analysis of GELN RNAs showed that miRNAs are enriched (Figure S4). Target sequencing analysis showed that these GELN miRNAs have potential target sequences in a variety of genes in P. gingivalis (Table S2). Based on the target genes in P. gingivalis, we selected miRNAs aly-miR159a-3p, gma-miR166u, and gma-miR166p to further determine whether miRNAs play a role in inhibition of P. gingivalis growth. P. gingivalis was transduced with GELN-derived miRNAs, and P. gingivalis growth was measured. As shown in Figure S5, these miRNAs did not affect the growth of P. gingivalis, supporting the notion that GELN lipids play a critical role in inhibition of P. gingivalis growth. Thus, we next determined whether GELN-derived total lipids (lipidG) and miRNA have synergetic effects on the growth of P. gingivalis. However, as shown in Figure S6, lipidG and miRNA from GLENs do not have a synergetic effect. Next, we investigated which GELN lipid(s) inhibits P. gingivalis growth. Extracted total lipids from GELNs (5.0 ×108 particles) were subjected to mass spectrometry analysis. The lipid profile of LipidG is listed in Table S3, and the percentage of each lipid in is presented in Figure 2B. P. gingivalis was treated with different concentrations (0–50 μg/mL) of 1-palmitoyl-2-linoleoyl-sn-glycero-3-phosphocholine (PC 34:2), 1-oleoyl-2-hydroxy-sn-glycero-3-phospho-(1′-rac-glycerol) (LysoPG 18:1), 1,2-dilinoleoyl-sn-glycero-3-phosphate (PA 36:4), 1,2-dioleoyl-sn-glycero-3-phosphate (PA 36:2), 1-palmitoyl-2-linoleoyl-sn-glycero-3-phosphate (PA 34:2), and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphate (PA 34:1). Among these lipids tested, PA (34:2) inhibited P. gingivalis growth at a very low concentration (5 μg/mL, 2.5 × 109 particles have 1 μg of lipid that contains 325.5 nM of PA (34:2) compared with other lipid compounds) (Figure 2C). Next, we sought to determine the role of phosphatidic acid (PA) in the context of GELNs by depletion of PA. Total GELN lipids were extracted and separated by TLC that included a standard PA (34:2) (Figure 2D). The corresponding PA band from the TLC was excised, and the majority of excised lipids is PA (Lipid profile, Table S4); remaining lipids were pooled together as PA-depleted lipids. P. gingivalis was incubated with lipidG, the PA-containing band, and the PA-depleted lipids for 24 h. Interestingly, the PA-containing band alone inhibited P. gingivalis growth as potently as the total lipids, and in contrast to the PA-depleted lipids, which had minimal growth inhibition (Figure 2E). Since oxidation may alter the biological activity of the lipids, we further determined the lipid oxidation of GELN lipids before and after TLC extraction. No significant changes in lipid oxidation were observed after TLC extraction (Figure S7). Collectively, these results suggest that PA is the major active molecule in GELN lipids that inhibits P. gingivalis growth.
Figure 2.
PA-Containing Lipid of GELNs Inhibits P. gingivalis Growth
(A) GELN total lipids (LipidG) isolated from different concentrations of GELNs (0–5.0 × 108 particles) were reacted with P. gingivalis for 24 h in an anaerobic chamber. Bacterial growth was determined by measuring optical density at 600 nm.
(B) Total lipids extracted from GELNs was subjected to mass spectrophotometry for lipid identification. The concentrations of each lipid were calculated as nmol/mg of GELNs.
(C) P. gingivalis was treated with different concentrations (0–50 μg/mL) of PC (34:2), LysoPG (18:1), PA (36:4), PA (36:2), PA (34:2), and PA (34:1) for 24 h in an anaerobic chamber. Bacterial growth was determined by measuring optical density at 600 nm.
(D) Total lipids were separated on a silica gel-coated thin-layer chromatography plate.
(E) Depletion of PA lipids in the total GELN lipids (LipidG) by eluting the PA band on a TLC plate and mixing the remaining lipids. P. gingivalis was treated with lipids from the PA-containing band, PA depleted and LipidG incubated in anaerobic chamber for 24 h. Bacterial growth was measured at 600 nm.
Results are expressed as mean ± standard deviation from three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001 compared with an untreated group using a one-way ANOVA with the Turkey's Multiple Comparison Test.
GELN PA Directly Interacts with HBP35 Protein in P. gingivalis, Leading to Inhibition of P. gingivalis Growth
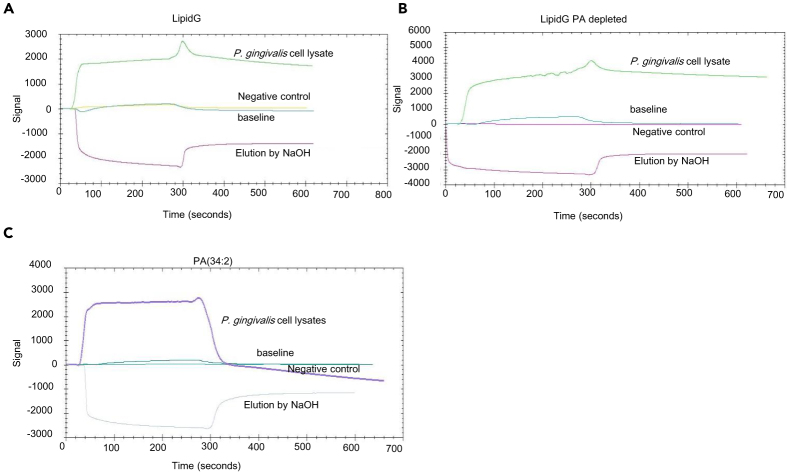
We hypothesized that GELN PA lipids interact with an outer membrane protein of P. gingivalis that modulates growth. To test this hypothesis, we used Surface Plasmon Resonance (SPR) to identify the P. gingivalis proteins that may interact with GELN lipids. We made lipid nanoparticles (Figure S8A) from GELNs PA (34:2) (Figure S8B). In terms of diameter, lipid nanoparticles cover a range between 100 and 500 nm and contain a negligible amount of TAG compared with total lipids derived from GELNs (Figures S8C and S8D). These lipid nanoparticles were immobilized on a LIP-1 sensor. P. gingivalis total cell lysates were run over the immobilized nanoparticles as analyte. As shown in Figures 3A–3C, the sensogram of SPR peaks revealed that P. gingivalis proteins can interact with lipid nanoparticles with/without depletion of PA lipid and PA (34:2). We eluted the lipid-binding protein from the immobilized nanoparticles by injection of NaOH (200 μM), which causes dissociation of protein from the nanoparticles, and eluted protein samples were subjected to tandem mass spectrometry (MS/MS) for protein identification. Interestingly, we identified several proteins (listed in Table 1) that bound to the GELN lipid nanoparticles and PA (34:2) but not with the PA-depleted lipid nanoparticles. From this result, we identified PA binding with P. gingivalis proteins/peptides, including the C-terminal domain of Arg and Lys-gingipain proteases, hemin-binding protein (35 kDa, HBP35), an electron transfer flavoprotein, an esterase, and an outer membrane lipoprotein. These proteins specifically bind with both GELN nanoparticles and PA.
Figure 3.
Identification of PA-Binding Proteins in P. gingivalis
Lipid nanoparticles were prepared from LipidG, PA-depleted GELN lipids, and PA (34:2), and these lipid nanoparticles were immobilized on a LIP-1 sensor. Total P. gingivalis cell lysates were used as analyte as described in the Transparent Methods. The lipid-protein interaction was determined by SPR sensograms.
(A) Sensogram of GELNs lipid nanoparticles.
(B and C) (B) Sensogram of PA-depleted GELN lipid nanoparticles and (C) Sensogram of PA (34:2) lipid nanoparticles. Protein bound with lipid nanoparticles was eluted by NaOH (200 μM), and the fraction was collected for MS/MS analysis for protein identification. The lipid-binding proteins in P. gingivalis are listed in Table 1.
Table 1.
PA-Binding Protein in P. gingivalis.
| Identified Proteins | Gene Symbol | Accession Number | Quantitative Value (iBAQ) |
||
|---|---|---|---|---|---|
| GELN Lipid Nanoparticles | PA Depleted GELN | PA (34:2) Lipid Nanoparticles |
|||
| CTD of Arg- and Lys-gingipain proteinase | rgpA | B2RHG9_PORG3 | 8,269,700 | 0 | 5.49E+07 |
| Uncharacterized protein | PGN_1182 | B2RK06_PORG3 | 1,974,600 | 0 | 4,816,600 |
| Exo-glucosaminidase LytG muramidase | lytG | B2RME7_PORG3 | 1,870,800 | 0 | 628,530 |
| 35 kDa hemin-binding protein | HBP35 | B2RII3_PORG3 | 540,420 | 0 | 1.11E+07 |
| Probable transcriptional regulator | asnC | B2RK48_PORG3 | 428,670 | 0 | 0 |
| Electron transfer flavoprotein beta subunit | carD | B2RJZ7_PORG3 | 280,750 | 0 | 3,750,900 |
| META domain lipoprotein implicated in motility | PGN_0740 | B2RIR4_PORG3 | 215,870 | 0 | 1,347,800 |
| Methylmalonyl-CoA decarboxylase-α subunit | pccB | B2RI24_PORG3 | 195,390 | 0 | 2,510,400 |
| 30S ribosomal protein S4 | rpsD | RS4_PORG3 | 190,850 | 0 | 2,002,700 |
| Oxygen-insensitive NADPH nitroreductase | rdxA | B2RIT9_PORG3 | 80,440 | 0 | 0 |
| Lys-gingipain protease | kgp | B2RMI8_PORG3 | 72,555 | 0 | 0 |
| Esterase | estD | B2RM02_PORG3 | 38,153 | 0 | 1,953,400 |
| Outer membrane lipoprotein 42 kDa antigen PG33 | ompA | B2RKC0_PORG3 | 36,170 | 0 | 0 |
| Uncharacterized protein | PGN_1697 | B2RLH1_PORG3 | 34,827 | 0 | 0 |
| Protein translocase SecA | secA | SECA_PORG3 | 30,589 | 0 | 0 |
| NAD-dependent 4-hydroxybutyrate dehydrogenase | adh | B2RIP8_PORG3 | 13,268 | 0 | 2,250,200 |
| L-erythro-35-diaminohexanoate dehydrogenase | kdd | B2RJY9_PORG3 | 10,490 | 0 | 1,263,500 |
| Chaperone protein DnaK | dnaK | DNAK_PORG3 | 6,316.60 | 0 | 23,177 |
| Long-chain-fatty-acid-CoA ligase | fadD | B2RK69_PORG3 | 4,928.30 | 0 | 0 |
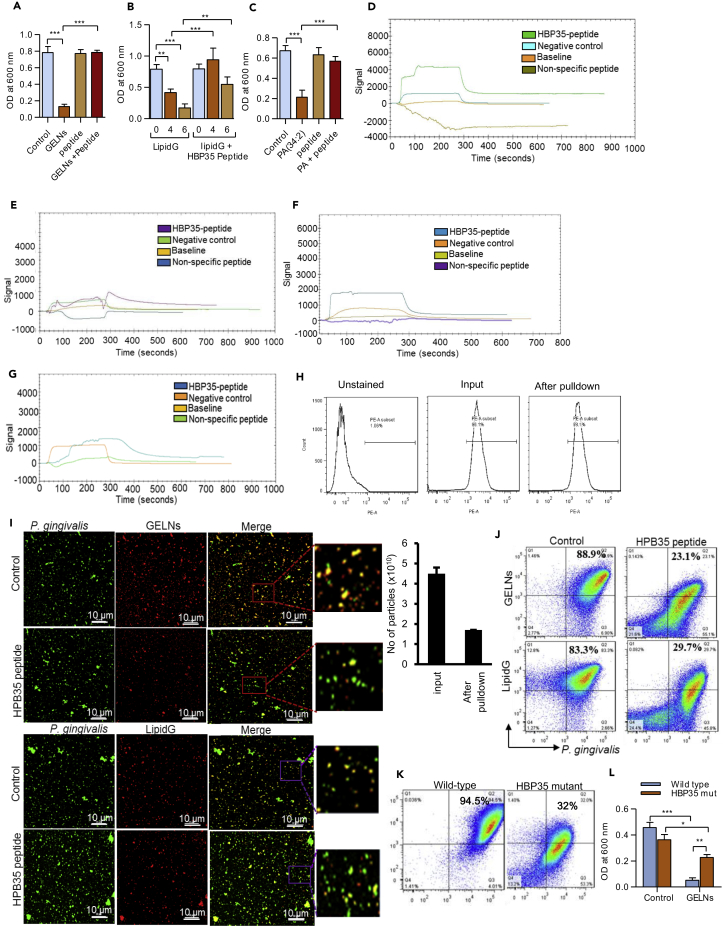
We next wanted to determine which proteins directly interacting with GELNs play an inhibitory role in P. gingivalis growth. HBP35 has been shown to be essential for growth of P. gingivalis and survival in hemin-depleted conditions (Shibata et al., 2011, Shoji et al., 2010) (Hiratsuka et al., 2010). Therefore, we next examined the interaction of GELNs and its lipids with HBP35. The functional domain of HBP35 is WPRVGQLFIALDQTLGIPGFPTFSVCRME, which plays a critical role in the hemolytic activity of P. gingivalis (Hiratsuka et al., 2010). To block the interaction of GELNs with HBP35, we utilized a synthetic peptide (29 amino acids) corresponding to the functional domain. GELNs (4.0 × 108 particles) were pre-incubated with the functional domain peptide (10 μM), then P. gingivalis was reacted with the GELNs and growth was measured. Interestingly, P. gingivalis growth was not affected by GELNs pre-incubated with HBP35 peptide (Figure 4A). Similarly, LipidG made from total lipid of 4.0 and 6.0 × 108 GELNs (Figure 4B) and PA (34:2) (Figure 4C) were pre-incubated with HBP35 synthetic peptide and treated with P. gingivalis. The synthetic peptide significantly blocked GELN lipids and PA-mediated inhibition of P. gingivalis growth (Figures 4B and 4C). To test whether the HBP35 synthetic peptide directly binds with PA (34:2) and PA (34:1) of GELN lipids, we again utilized SPR. Lipid nanoparticles from GELN total lipids, PA-depleted lipid, PA (34:2), and PA (34:1) were immobilized on the LIP-1 sensor. The HBP35 synthetic peptide and a non-specific peptide were used as analyte. The SPR sensogram peaks show that the HBP35 synthetic peptide directly interacts with GELNs and PA (34:2) nanoparticles but not with PA-depleted lipid nanoparticles and PA (34:1) nanoparticles (Figures 4D–4G). This result indicates that the degree of unsaturation of PA plays a critical role in GELN-mediated interaction with HBP35. In addition to using SPR, we further confirmed PA binding to HBP35 by a pull-down assay. PA (34:2) nanoparticles were fluorescently labeled with PKH26 (PE channel) and incubated with biotin-HBP35 peptide for 2 h. The lipid-peptide complex was pulled down by streptavidin beads, and the complex was washed thoroughly. The presence of PA (34:2) in the complex was confirmed by flow cytometry (Figure 4H, top panel). The number of lipid nanoparticles (Figure 4H, bottom, left panel) and fluorescent intensity (Figure 4H, bottom, right panel), including GELNs with/without depletion of PA, PA band from GELN, and PA (34:2) in the complex was determined. The results generated from all three independent assays demonstrated that PA lipid binds to HBP35. We further tested inhibition of GELN uptake by HBP35 peptide. P. gingivalis was fluorescently labeled with PKH67 (green), whereas GELNs and lipid nanoparticles were labeled with PKH26 (red). GELNs and GELN lipid nanoparticles were pre-incubated with HBP35 peptide (10 μM) and then reacted with P. gingivalis for 1 h at 37°C. P. gingivalis uptake of GELNs and lipid nanoparticles was visualized by confocal microscopy (Figure 4I) and quantified by flow cytometry (Figure 4J). Pre-incubation of synthetic peptide with GELNs and lipid nanoparticles significantly decreased P. gingivalis uptake of GELNs and lipid nanoparticles. HBP35-dependent GELN uptake by P. gingivalis was further demonstrated by deletion of the gene encoding HBP35 in P. gingivalis. The wild-type and HBP35 mutant of P. gingivalis were incubated with PKH26-labeled GELNs for 1 h at 37°C, and GELN uptake was quantified by flow cytometry. As shown in Figure 4K, the HBP35 mutant had a significantly decreased uptake of GLENs. Furthermore, the mutation of the gene HPB35 in P. gingivalis led to less inhibition of its growth by GELNs compared with wild-type P. gingivalis (Figure 4L). Taken together, these results suggest that the lipid moiety of GELNs, specifically PA (34:2), interacts with HBP35 on the outer membrane of P. gingivalis, and this interaction followed by lipid uptake leads to inhibition of bacterial growth.
Figure 4.
GELN Lipids Bind to Hemin-Binding Protein 35 (HBP35) on the Outer Membrane of P. gingivalis and Inhibit Its Growth
(A–E) (A–C) A synthetic peptide representing the functional domain of HBP35 WPRVGQLFIALDQTLGIPGFPTFSVCRME (10 μM) was pre-incubated with GELNs (4.0 × 108 particles), LipidG (4.0–6.0 × 108 particles), and 5 μg of PA (34:2) for 1 h at 37°C. The pre-treated GELNs and lipids were reacted with P. gingivalis for 24 h in anaerobic chamber, and growth of P. gingivalis was measured at 600 nm. Determination of direct binding of PA with HBP35 by surface plasmon resonance. Lipid nanoparticles were made from (D) LipidG (E) PA depleted GELN lipids.
(F and G) (F) PA (34:2) and (G) PA (34:1). These lipid nanoparticles were immobilized on a LIP-1 sensor, and HBP35 synthetic peptide was used as an analyte and non-specific peptide used as a negative control. The lipid-protein interaction was determined by SPR sensograms.
(H) Lipid nanoparticles were made from GELN lipids, PA-depleted GELNs, the PA-containing band, and PA (34:2). These nanoparticles were labeled with PKH26 red fluorescent dye and incubated with biotin-HBP35 peptide for 2 h at room temperature with rotation. The lipid nanoparticles and peptide complex were precipitated with streptavidin beads. After washing with PBS, the presence of lipid nanoparticles in the complex was confirmed by flow cytometry. The number of lipid nanoparticles in the complex was measured using a NanoSight 300, and the quantity of lipid nanoparticles was determined by fluorescence intensity as described in Transparent Methods.
(I) P. gingivalis was labeled with PKH67 (Green), and GELNs and GELN lipid nanoparticles were labeled with PKH26 (Red). The labeled particles were pre-incubated with synthetic peptide of HBP35 (10 μM) for 1 h at 37°C. Then, particles were treated with labeled P. gingivalis for 1 h at 37°C in an anaerobic chamber. Uptake of particles by P. gingivalis were visualized by confocal microscopy.
(J) P. gingivalis uptake of particles were quantified by flow cytometry.
(K) P. gingivalis wild-type and Δhbp35 mutant were labeled with PKH67, and GELNs were labeled with PKH26. GELNs were reacted with P. gingivalis for 1 h at 37°C in an anaerobic chamber. P. gingivalis and Δhbp35 mutant uptake of particles was quantified by flow cytometry.
(L) P. gingivalis wild-type and hbp35 mutant was reacted with GELNs for 24 h in an anaerobic chamber. Bacterial growth was determined by measuring optical density at 600 nm.
Results are expressed as mean ± standard deviation from three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001 compared with an untreated group using a one-way ANOVA with the Turkey's Multiple Comparison Test.
GELN miRNAs and Lipid PA (34:2) Inhibit the Virulence Activity of Gingipain and Type IX Secretion System of P. gingivalis
Besides growth, there are many specific virulence factors that which contribute to the pathogenicity of P. gingivalis (Lamont et al., 2018). GELNs are complex nanoparticles that could interact with a multiple of P. gingivalis virulence factors in addition to HBP35. To search for P. gingivalis factors that could interact with GELNs, biotin-labeled GELNs were incubated with P. gingivalis total cell lysates. After pull-down with streptavidin beads, the P. gingivalis factors that bind with GELNs were separated by SDS-PAGE, and analyzed by MS/MS (Figures 5A and 5B). The GELN-binding proteins which included HBP35, lysine and arginine gingipain, hemagglutinin (HagA), outer membrane protein A (OmpA), and type IX secretion system (T9SS) were identified (Table S5). Interestingly, we identified using two independent assays that GELN interacts with HBP35: the biotin pull-down and SPR assays.
Figure 5.
Identification of GELN-Binding Proteins in P. gingivalis
GELNs were labeled with biotin and incubated with total cell lysates of P. gingivalis for 1 h at room temperature. GELN-binding proteins were pulled down by streptavidin-conjugated beads.
(A) Protein was separated by SDS PAGE.
(B) Pull-down proteins were subjected to MS/MS analysis.
(C) P. gingivalis was incubated with GELNs (4.0 × 108 particles/mL) for 6 h in an anaerobic chamber; the bacteria was spread on blood agar plates and incubated anaerobically for 1 week.
(D) P. gingivalis was incubated with different concentrations (2.0 or 4.0 × 108 particles/mL) of GELNs for 6 h. P. gingivalis was lysed with Bugbuster lysis reagent, and gingipain activities were measured using specific substrates for Arg-specific protease (Rgp) and Lys-specific protease (Kgp).
(E) P. gingivalis was treated with total lipids derived from GELNs and PA (34:2) for 6 h. Gingipain activity was measured in P. gingivalis cell lysates.
(F) P. gingivalis was transduced with scrambled miRNA, aly-miR159a-3p, gma-miR166u, or gma-miR166p for 24 h and gingipain activity measured in total cell lysates.
(G) P. gingivalis was treated with GELNs (4 × 108 particles/mL) for 6 h.
(H) Transduced with scrambled miRNA, aly-miR159a, gma-miR166u, or gma-miR166p for 24 h. Expression of mRNA for araC, hagA, and rodA was determined by RT-qPCR.
(I) P. gingivalis was treated with total lipids derived from GELNs (4 × 108 particles) or 5 μg of PA (34:2) for 6 h. Expression of mRNA for araC, hagA, and rodA was determined by RT-qPCR.
Results are expressed as mean ± standard deviation from three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001 compared with untreated group using one-way ANOVA with Turkey's Multiple Comparison Test.
As an asaccharolytic organism P. gingivalis is highly proteolytic and the arginine- and lysine-specific gingipain are responsible for nearly 85% of the total proteolytic activity (Potempa et al., 2003). Gingipain are involved in a variety of pathogenic functions, including colonization, nutrition, and neutralization of host defenses, and contribute to extensive periodontal tissue destruction. Both the lysine-specific (Kgp) and arginine-specific (RgpA and RgpB) gingipain are also involved in the accumulation of hemin derivatives of the cell surface giving rise to black pigmentation of P. gingivalis colonies on blood agar (Lasica et al., 2016) (Smalley and Olczak, 2017). GELN treatment inhibited the formation of black-pigmented colonies of P. gingivalis (Figure 5C). This result was further supported by the finding that activity of both Rgp and Kgp was significantly decreased in P. gingivalis treated with GELNs (Figure 5D). GELN lipids, PA (34:2) (Figure 5E), and GELNs aly-miR159a (Figure 5F) all contributed to the inhibition of Rgp and Kgp activities.
We also examined the expression of genes encoding other P. gingivalis proteins that have the potential to contribute to the pathophysiology of the organism. GELNs strongly inhibited expression of mRNA encoding the AraC transcription factor; HagA, hemagglutinin; OmpA, outer membrane protein A; and the RodA, rod shape determining protein (Figure 5G). Furthermore, GELN aly-miR159a, gma-miR166u/p differentially regulated mRNA expression of AraC, HagA, OmpA, and RodA (Figure 5H). In addition, we found an aly-miR159a-3p-binding site in hagA (PGN_1733) and araC (PGN_0082), and gma-166p has a potential binding site in ompA (PGN_0299) (Figure S9). Indeed, miRNAs derived from GELNs have putative binding sites in several P. gingivalis genes, indicating the potential for broadly based inhibition of P. gingivalis function. Additionally, lipids derived from GELNs and PA (34:2) inhibited araC, hagA, ompA, and rodA mRNA expression (Figure 5I). Collectively, these results indicate that GELN-derived lipids and miRNAs target several virulence genes expressed in P. gingivalis.
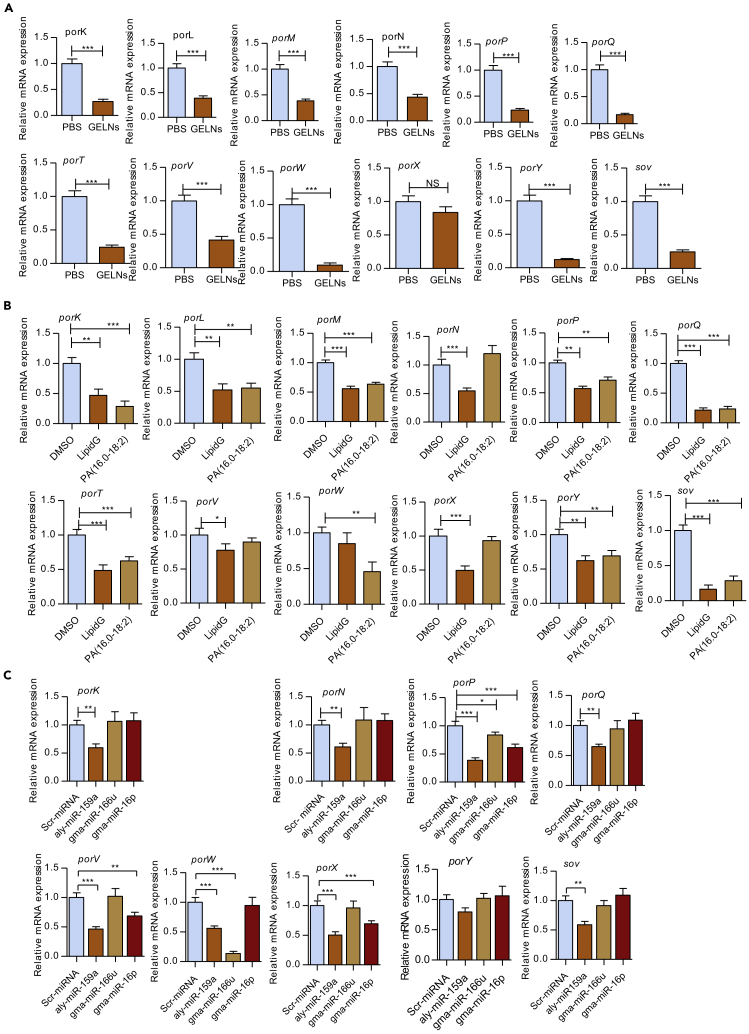
The translocation of gingipain to the bacterial surface requires the type IX secretion system (T9SS) (de Diego et al., 2016). T9SS also secrete a variety of other potential virulence factors that possess a conserved C-terminal domain (Lasica et al., 2017). Hence, we examined the role of GELNs and GELN lipids and miRNAs in modulating the expression of genes related to the T9SS. P. gingivalis was treated with GELNs (4.0 × 108 particles/mL) and GELN lipids derived from 4.0 × 108 particles, or 5 μg of PA (34:2) for 6 h miRNAs aly-miR159a, gma-miR166u, and gma-miR166p were transduced individually into P. gingivalis and incubated for 24 h. The expression of the T9SS family of genes (Vincent et al., 2017) including porK, porL, porN, porP, porQ, porT, porV, porW, porX, porY, and sov was quantified using RT-qPCR. GELNs and GELN total lipids significantly inhibited the expression of 11 of the 12 T9SS family of genes compared with the control (Figures 6A and 6B). In addition, PA (34:2) and miRNAs differentially modulated the expression of the T9SS family of genes (Figures 6B and 6C). Furthermore, we found aly-miR159a-3p has a binding site in the T9SS C-terminal target domain containing protein (PGN_0152) (Figure S9). Taken together, GELNs and total lipids derived from GELNs inhibit expression of the T9SS family of genes. GELN miRNAs and PA (34:2) preferentially inhibited expression of some of the T9SS family of genes, which can be predicted to impact the ability of the organism to secrete important virulence factors.
Figure 6.
GELNs Inhibit T9SS mRNA Expression in P. gingivalis
(A) P. gingivalis treated with GELNs (4 × 108 particles/mL).
(B) P. gingivalis treated with lipids extracted from GELNs (4 × 108 particles) or 5 μg of PA (34:2) for 6 h.
(C) P. gingivalis was transduced with scrambled miRNA, aly-miR159a, gma-miR166u, or gma-miR166p for 24 h T9SS family mRNA expression was measured by RT-qPCR.
Results are expressed as mean ± standard deviation from three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001 compared with untreated group using the one-way ANOVA with Turkey's Multiple Comparison Test and Student’s t test.
GELNs Inhibits P. gingivalis Attachment to and Invasion in Oral Epithelial Cells
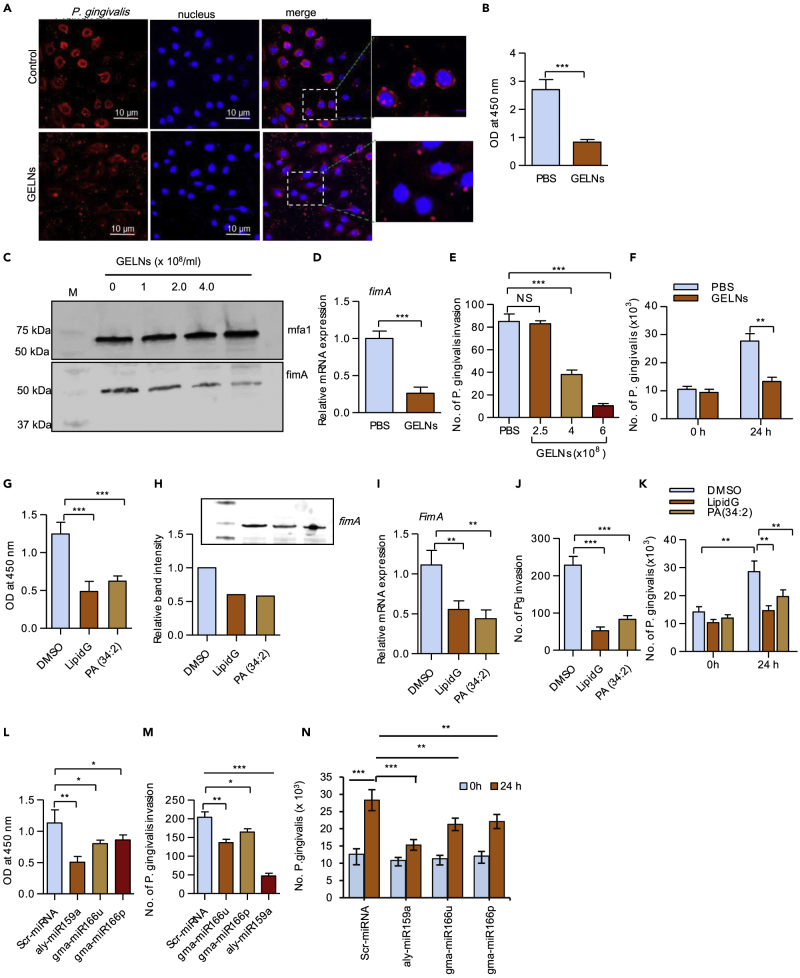
The early stages of P. gingivalis colonization involve attachment and invasion of gingival epithelial cells (Lamont et al., 1995); therefore, we determined whether GELNs have an effect on these processes. P. gingivalis was treated with GELNs (4 ×108 particles/mL) for 3 h, and then gingival epithelial cells (human telomerase immortalized gingival keratinocytes [TIGKs]) were infected with P. gingivalis (MOI 10) for 1 h. Visualization of P. gingivalis attachment to TIGKs by confocal microscopy showed that GELN treatment significantly decreased the level of P. gingivalis binding (Figure 7A). Consistent with this, when attachment of P. gingivalis to TIGK cells was determined by enzyme-linked immunosorbent assay (ELISA), GELN treatment significantly decreased the surface binding of P. gingivalis to TIGK cells (Figure 7B).
Figure 7.
GELNs Inhibit P. gingivalis Attachment, Invasion, and Proliferation in Oral Epithelial Cells
(A) P. gingivalis was treated with or without GELNs (4.0 × 108 particles/mL) for 3 h and reacted with TIGK cells at an MOI of 10 for 1 h. P. gingivalis was stained with whole-cell antibody, and the nucleus was stained with DAPI.
(B) P. gingivalis was treated with or without GELNs (4.0 × 108 particles/mL) for 3 h and reacted with TIGK cells at an MOI of 10 for 1 h. Binding was determined by ELISA.
(C) P. gingivalis was treated with or without GELNs (0–4.0 × 108 particles/mL) for 6 h, and total cell lysates were subjected to western blotting with the antibodies as indicated.
(D) P. gingivalis was treated with or without GELNs (4.0 × 108 particles/mL) for 24 h, and fimA mRNA expression was determined by RT-qPCR.
(E) P. gingivalis was treated with different concentrations of GELNs (0–4.0 × 108 particles/mL) for 3 h and invasion of TIGK cells determined by an antibiotic protection assay as described in the Transparent Methods section. The results are expressed as number of P. gingivalis invasion into TIGK cells.
(F) P. gingivalis was treated with or without GELNs (4.0 × 08 particles/mL) and infected into TIGK cells at an MOI of 10. After further culturing for 24 h, numbers of intracellular bacteria were determined by qPCR.
(G) P. gingivalis was treated with or without GELN (from 6 ×108 particles/mL) derived LipidG or PA (34:2) (5 μg/mL) and surface attachment to TIGK cells measured.
(H) P. gingivalis was treated with or without LipidG (from 6 ×108 particles/mL) or PA (34:2) (5 μg/mL) for 6 h, and cell lysates were examined by western blotting with FimA antibodies.
(I–K) (I) P. gingivalis was treated with or without LipidG (from 6 ×108 particles/mL) or PA (34:2) (5 μg/mL) and fimA mRNA determined by RT-qPCR (J) P. gingivalis invasion into oral epithelial cells and (K) P. gingivalis proliferation in oral epithelial cells as described in the Transparent Methods.
(L) P. gingivalis was transduced with miRNA from GELN aly-miR159a, gma-miR166u, gma-miR166p, or non-specific scrambled miRNA for 24 h and attachment to TIGK cells measured by ELISA.
(M and N) (M) P. gingivalis invasion into oral epithelial cells and (N) P. gingivalis proliferation in oral epithelial cells were determined as described in the Transparent Methods.
Results are expressed as mean ± standard deviation from three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001 compared with untreated group using one-way ANOVA with Turkey's Multiple Comparison Test.
The FimA component fimbriae are required for attachment of P. gingivalis to oral epithelial cells (Tribble and Lamont, 2010). Therefore, we next sought to determine the effect of GELNs on expression of FimA, as well as on expression of the Mfa1 component fimbriae. As presented in Figures 7C and 7D, expression FimA at both mRNA and protein levels was decreased by GELN treatment. However, GELNs did not change expression of Mfa1. Attachment of P. gingivalis to epithelial surfaces can also induce internalization of the organism, which can survive intracellularly. Pre-treatment of P. gingivalis with different concentrations of GELNs (0–6.0 ×108 particles/mL) for 3 h decreased intracellular invasion of TIGKs in an antibiotic protection assay (Figure 7E). After 24 h within the TIGKs, quantitation of intracellular P. gingivalis by qPCR analysis using 16S rRNA expression showed that GELNs significantly inhibit intracellular proliferation (Figure 7F). These results indicate that GELNs significantly inhibit attachment and invasion of P. gingivalis into oral epithelial cells.
Next, we determined which GELN factors contribute to the inhibition of the attachment and invasion of P. gingivalis in oral epithelial cells. We pretreated P. gingivalis with GELN-derived total lipids (from 4.0 × 108 GELNs) and 50 nM of PA (34:2), which is equivalent to lipids extracted from 4.0 × 108 GELNs for 3 h. Under these conditions P. gingivalis was not killed; however, there was a significant inhibition of attachment to TIGK cells (Figure 7G). Furthermore, P. gingivalis was treated with the same concentration of lipids for 6 h and FimA expression was determined by western blot and RT-qPCR analysis. FimA expression was decreased significantly (p = 0.002) in both LipidG- and PA (34:2)-treated P. gingivalis (Figures 7H and 7I). Furthermore, we next examined the effect of GELN total lipids and PA (34:2) on P. gingivalis invasion and proliferation in TIGK cells. A significant decrease in P. gingivalis invasion and proliferation was observed with both GELN total lipids and PA-treated P. gingivalis (Figures 7J and 7K). Collectively, these results suggest that GELN PA (34:2) inhibits the growth, attachment, and invasion of P. gingivalis.
To test whether GELN miRNAs play an inhibitory role in attachment and invasion of P. gingivalis, we transduced miRNAs gma-miR166u, gma-miR166p, or aly-miR159a into P. gingivalis and used scrambled miRNA as a control. Among these three miRNAs tested, aly-miR159a significantly decreased the attachment of P. gingivalis to TIGK cells and gma-miR166u and gma-166p moderately inhibited attachment (Figure 7L). Next, we determined the effect of these miRNAs on P. gingivalis invasion (Figure 7M) and proliferation (Figure 7N). As with attachment, inhibition of invasion by aly-miR159a was most pronounced (Figure 7M) and all of the miRNAs inhibited proliferation (Figure 7N). Collectively, these results suggest that GELNs and total lipids derived from GELNs, as well as constituent miRNAs, can impede colonization of P. gingivalis by antagonizing epithelial cell attachment and invasion.
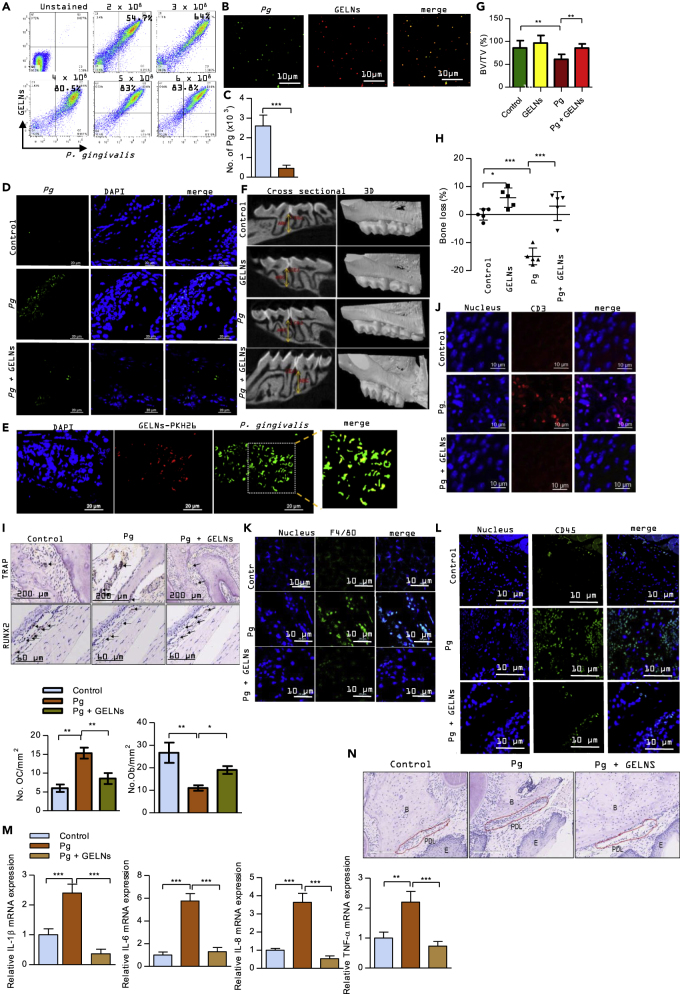
GELNs Inhibit P. gingivalis-Induced Bone Loss in an In Vivo Mouse Model
Consistent with its role as a periodontal pathogen, P. gingivalis can induce alveolar bone loss in rodent models (Baker et al., 2000). P. gingivalis, along with other pathogenic bacteria in the oral cavity, reside primarily in biofilm structures, which contribute to the alveolar bone loss. GELNs dose dependently decreased P. gingivalis biofilm formation in vitro (Figure S10). Therefore, we next wanted to test the effect of GELNs on P. gingivalis-induced alveolar bone loss. To confirm uptake of GELNs by P. gingivalis in the mouse oral cavity we inoculated fluorescent labeled (PKH67) P. gingivalis followed by fluorescent labeled (PKH26) GELNs. After 1 h incubation, the oral cavity was washed with PBS. As shown in Figures 8A and 8B, flow cytometry and confocal microscopy analyses of this PBS wash showed that P. gingivalis in the mouse oral cavity can take up GELNs. Next, the ability of GELNs to prevent/inhibit bone loss was examined. P. gingivalis was inoculated into the murine oral cavity every 2 days for a period of 10 days, and GELNs were given in drinking water ad libitum continuously until the end of the experiment. After 3 weeks, the colonization of P. gingivalis in the oral cavity was determined by qPCR. Oral samples were collected along the gingiva of the upper molars using a 15-cm sterile polyester-tipped applicator, and total genomic DNA was purified and amplified by qPCR with primers to 16 S rRNA. Numbers of P. gingivalis were calculated by comparison with a standard curve derived from known amounts of P. gingivalis. As shown in Figure 8C, the number of P. gingivalis in the oral cavity was significantly decreased with GELN treatment. This result indicates that GELN treatment given in drinking water had significant inhibition of colonization of P. gingivalis. In addition, the presence of P. gingivalis in the oral tissue was confirmed by confocal microscopy. It showed that P. gingivalis in the oral tissue was decreased in GELNs treated group compared with P. gingivalis alone infected tissue (Figure 8D). Furthermore, the in vivo uptake of GELNs by P. gingivalis was determined by confocal microscopy (Figure 8E). After 48 days, the mice were sacrificed and μCT analysis was used to determine alveolar bone loss by measuring the distance from the cementoenamel junction to the alveolar bone crest. In addition, bone volume was measured. A region of interest (ROI) was drawn manually on the axial planes, between the medial root surface of the first molar and distal root surface of the third molar. A three-dimensional image was generated from the ROI. All root volumes were excluded from the ROI to calculate the total volume (TV). The bone volume fraction (BV/TV) was calculated for each sample. GELNs significantly decreased P. gingivalis-induced bone loss, demonstrating that GELNs can inhibit P. gingivalis pathogenicity in vivo (Figures 8F–8H). Interestingly, GELNs alone significantly increased the bone density of alveolar bone compared with the sham condition (Figure 8H). Next, we sought to determine the effect of GELNs on osteoclast differentiation and osteoblast function in infected mice. GELNs treatment significantly decreased TRAP+ osteoclast number in P. gingivalis-infected mice compared with untreated mice (Figure 8I). Osteoblast cells were stained by RUNX2 expression and immunohistochemistry. As shown in Figure 8I, RUNX2 expression was significantly decreased in P. gingivalis-infected oral tissue sections, whereas RUNX2 expression was significantly increased by GELN treatment in P. gingivalis-infected mice. These results suggest that GELNs differentially regulate osteoclasts and osteoblasts in P. gingivalis-infected mice. Furthermore, we screened the cytokine profile in mouse plasma modulated by P. gingivalis and GELNs. Cytokine array analysis revealed that GELNs significantly decreased P. gingivalis induction of pro-inflammatory cytokines and documented bone resorptive cytokines, such as TNF-α, IL-1α, IL-1β, INF-γ, IL-6, IL-13, and IL-22 (Figure S11). These cytokines play an important role in recruiting inflammatory Th17 T cells and macrophages into periodontal tissues, leading to bone loss (Baker et al., 1999) (Lam et al., 2014, Tzach-Nahman et al., 2017, Zhuang et al., 2018). Next, we investigated the effect of GELNs on T cell, macrophage, and leukocyte recruitment by using immunofluorescent labeled CD3, F4/80, and CD45 antibodies to detect T cells, macrophages, and leukocytes, respectively. As shown in Figures 8J–8L, P. gingivalis infection enhanced infiltration of T cells, macrophages, and leukocytes in periodontal tissues and this infiltration was decreased significantly by GELNs. Furthermore, qPCR analysis showed that GELNs significantly decreased P. gingivalis-induced expression of mRNA for the inflammatory and bone resorptive cytokines IL-1β, IL-6, IL-8, and TNF-α in oral tissue (Figure 8M). Also, histology of oral sections showed increased cellular infiltration in the periodontal ligament region in P. gingivalis-infected mice compared with control, and this cellular infiltration was significantly decreased in GELN-treated mice (Figure 8N). Taken together, these results suggest that GELNs can target P. gingivalis and inhibit the expression of virulence factors and thereby decrease alveolar bone loss and inflammation induced by the organism in vivo.
Figure 8.
Effect of GELNs on P. gingivalis-Induced Bone Loss In Vivo
(A) P. gingivalis and GELNs were fluorescently labeled with PKH67 and PKH26, respectively. P. gingivalis (108) was inoculated into the oral cavity, and GELNs (6.0 × 108 particles) were applied to the oral cavity for 1 h. After washing, bacteria were collected with a sterile swab and uptake of GELNs was quantified by flow cytometry.
(B) P. gingivalis uptake of GELNs in the oral cavity was determined by confocal microscopy.
(C) P. gingivalis (108) was inoculated into the mouse oral cavity. The mice were given GELNs (4.0 × 108 particles/mL) ad libitum in drinking water. Three weeks after the final inoculation, the numbers of P. gingivalis were determined by qPCR.
(D) The presence of P. gingivalis in the oral tissue was determined by confocal microscopy. Oral tissue sections were stained with anti-P. gingivalis antibody followed by fluorescent Alexa Fluor 488-labeled secondary antibody. The presence of P. gingivalis was visualized by confocal microscopy. The nucleus was stained with DAPI.
(E) In vivo GELNs taken up by P. gingivalis were determined by confocal microscopy. P. gingivalis was inoculated in the mouse oral cavity three times on alternative days. Then, PKH26 labeled GELNs were added into the drinking water for 24 h. GELNs taken up by P. gingivalis in oral tissue were stained with anti-P. gingivalis followed by fluorescent Alexa Fluor 488-labeled secondary antibody. The GELN taken up by P. gingivalis was visualized by confocal microscopy.
(F) Linear measurements were taken from the cementoenamel junction (CEJ) to the alveolar bone crest (ABC) in the interdental space to measure alveolar bone loss. Images on the left are representative of 14 cross sections measured across the maxillary molars. On the right, three-dimensional reconstructed renderings are shown for representative specimens in each group, such as control (uninfected), GELNs (control mice drank GELNs), Pg (P. gingivalis infected), and Pg + GELNs (P. gingivalis infected mice continuously drank GELNs). The P. gingivalis-treated group shows clear exposure of the tooth root that is not observed when treated with GELNs.
(G) Quantification of alveolar bone fraction from the micro-CT three-dimensional image of alveolar bone crest of region of interest (ROI).
(H) Alveolar bone loss was measured by the distance between the ABC and CEJ at 14 predetermined points on the maxillary molars sites. Each symbol represents an individual mouse, and the short horizontal lines indicates the mean.
(I–L) (I) TRAP staining of an oral tissue section to determine osteoclast (OC) number, and RUNX2 expression for osteoblasts (OB) in an oral tissue section was determined by immunohistochemistry. The number of TRAP-positive multinucleated OCs and RUNX2-positive OBs was counted manually. (J–L) Oral tissue sections were stained with anti-CD3, anti-F4/80, and anti-CD45 antibodies followed by fluorescent labeled secondary antibodies. The expression of CD3, F4/80, and CD45 was visualized by confocal microscopy.
(M) Total RNA was isolated from the control and experimental mice oral tissue and analyzed by RT-qPCR for expression of mRNA for inflammatory and bone resorptive cytokines IL-1β, IL-6, IL-8, and TNF-α. Results are expressed as mean ± standard deviation from three independent experiments.
(N) Hematoxylin staining of oral section of control, P. gingivalis-infected mice, and GELN-treated mice. B, alveolar bone crest; PDL, periodontal ligament; E, epithelial cells.
Results are expressed as mean ± standard deviation from three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001 compared with an untreated group using a one-way ANOVA with the Turkey's Multiple Comparison Test.
Discussion
In this study we show that GELNs are selectively taken up by the oral pathogen P. gingivalis. This selectivity is determined by GELN-derived lipid PA, which interacts with hemin-binding protein 35 (HBP35) expressed on the surface of P. gingivalis. HBP35 is a functionally versatile protein involved in hemin uptake, colonization of epithelial surfaces, and biofilm development through binding to various oral Gram-positive and Gram-negative bacteria (Hiratsuka et al., 2010) (Hiratsuka et al., 2008). Our results showed that binding of HBP35 with GELN PA leads to inhibition of P. gingivalis growth. This conclusion is further supported by the finding that depletion of PA from GELNs or loss of HBP35 in P. gingivalis leads to no HBP35/PA interaction and consequently no disruption of P. gingivalis membrane. In addition, we found that pre-incubation of GELN with HBP35 peptide comprising the active binding domain prevents GELN-mediated inhibition of growth of P. gingivalis. At the molecular level, we further defined that the specificity of PA binding to HBP35 is dependent on the degree of unsaturation of PA. It has been shown that PA recruits and activates effector molecules that change the biophysical properties of the mammalian cell membrane and directly induce membrane destabilization (Athenstaedt and Daum, 1999, Kooijman et al., 2003). However, the effect of PA on the bacterial membrane has not been reported before. Collectively, these results suggest that GELN PA is required for selective uptake by P. gingivalis via interaction with the HBP35-binding domain. This finding is significant since the selectively of uptake could be exploited not only for targeting specific bacteria but also for delivery of therapeutic agents to specific pathogens for treatment.
Previous studies have shown that both secreted and surface-associated proteins contribute to the virulence of P. gingivalis. Arg-gingipain, Lys-gingipain, and hemagglutinins are among the major virulence factors of P. gingivalis (Savett and Progulske-Fox, 1995, Veith et al., 2002). In this study, we showed that GELN and its component lipids and miRNAs decreased significantly gingipain activities and hemagglutinin expression in P. gingivalis. Furthermore, we found that miR-159a-3p has several potential binding sites on the 3′-UTR of genes encoding gingipain and hemagglutinin. This finding provides a foundation for further study of the molecular mechanisms underlying plant exosome-like nanoparticle inhibition of oral bacterial pathogenicity via plant miRNA interaction with pathogenic factors such as gingipain and hemagglutinin. Furthermore, other GELN molecules can also participate in inhibition of bacterial pathogenicity. We identified GELN PA binding to the proteins containing the T9SS conserved C-terminal domain (CTD), including gingipain and hemagglutinin. The Type IX secretion system is composed of several outer membrane, periplasmic, and inner membrane proteins that play a role in gingipain secretion and transport other virulence factors to the host environment. Mutation of these proteins leads to accumulation of gingipain in the periplasm and non-pigmented colony morphology (de Diego et al., 2016, Lasica et al., 2016, Vincent et al., 2017). In the present study, we observed that GELN and its lipids and miRNAs decreased significantly the expression of T9SS in P. gingivalis. In addition to these virulence factors, GELNs, lipid, and miRNA also inhibited expression of other important P. gingivalis components such as OmpA, rod shape determining protein A (RodA), and the AraC transcriptional regulators. The AraC family of transcription regulators is one of the largest group of regulatory proteins in bacteria and can control the expression of several virulence factors in a variety of organisms (Yang et al., 2011). Collectively, our findings suggest that GELNs carry a broad spectrum of molecules, including lipids and miRNAs, that can inhibit multiple pathways in P. gingivalis.
Besides GELNs, the fact that all other types of edible plant exosome-like nanoparticles (ELNs) carry a broad spectrum of molecules supports further examination as to whether other ELNs have a role in inhibition of pathogenic factor(s) or/and promoting beneficial factor(s) from oral bacteria to regulating oral bacterial homeostasis. Over 700 bacterial species may be found in the oral cavity of humans (Paster et al., 2006). The diverse community that makes up the oral microbiome is of great importance in maintaining homeostasis. Oral dysbiosis contributes to the development of periodontitis. P. gingivalis is considered a keystone pathogen (Hajishengallis et al., 2012), and thus targeting of this organism may be sufficient to ameliorate or prevent disease. In this regard, in non-human primates where P. gingivalis is a natural inhabitant of the subgingival biofilm, immunization against P. gingivalis gingipain proteases causes a reduction in P. gingivalis numbers, total subgingival bacterial load, and bone loss (Page et al., 2007). Nonetheless, as periodontitis involves a community of organisms (Lamont et al., 2018), the ability of GELNs to modulate the pathogenic potential of other potential pathogens (e.g., Tannerella forsythia, Prevotella intermedia, Aggregatibacter actinomycetemcomitans, Treponema denticola, Filifactor alocis) alone and in the context of a heterotypic biofilm community also requires further study. The results presented in our previous report (Teng et al., 2018) imply that ginger edible exosome-like nanoparticles are taken up by a group of gut bacteria and have a role in maintaining gut microbiota homeostasis by inhibiting the growth of potential pathogens as well as increasing beneficial bacterial survival. The current study opens up a new avenue for studying whether a combination of other types of ELNs with GELNs can target more species than P. gingivalis to have greater potency in inhibiting the development of periodontitis. Collectively, this study further supports the hypothesis that edible plant exosome-like nanoparticles packaging various agents can target multiple virulence factors of infectious agents simultaneously for the prevention/treatment of infectious diseases. Since pathogenicity in general is multifactorial, the action of edible plant exosome-like nanoparticles (ELNs) is likely to be more efficient than that of any single molecule. ELNs have the potential to be further developed as a new source of prebiotics. In addition, currently, only a subset of gut and oral bacteria can be grown in the laboratory in pure culture (Stewart, 2012), and systematic approaches to identifying growth conditions for as yet uncultivable bacteria have been challenging (Vartoukian et al., 2010). Our findings also suggest the possibility of in vitro co-culturing of oral or gut bacteria with ELNs for enhancing growth.
The interaction of bacterial factors with host cells contributes to P. gingivalis colonization and pathogenicity. Several P. gingivalis proteins, including the FimA component fimbriae (fimA) and the Mfa1 component fimbriae, play a role in attachment to host surfaces (Lin et al., 2006) (Hamada et al., 1996) (Zheng et al., 2011). Our study showed that GELNs and their component lipids and miRNAs significantly reduced FimA expression and further inhibited attachment of P. gingivalis to oral epithelial cells. We also showed that treatment with GELNs significantly diminished P. gingivalis-induced alveolar bone loss in a mouse model. Moreover, GELNs impacted the immune response and we observed a decrease in the expression of bone resorptive cytokines, including IL-1β, IL-6, Il-8, and TNF-α, and decreased recruitment of macrophages, leukocytes, and CD3 cells into the oral tissue microenvironment. The data published also indicate that GELNs also have anti-inflammatory properties. Whether this GELN-mediated immune modulation is through direct interaction of GELNs with CD3 T cells and macrophages or through metabolites released by GELN-responsive oral bacteria cannot be distinguished in the standard in vivo periodontal bone loss model utilized in this study, and the relative importance of GELN effects on bacteria or host immunity requires further investigation. Indeed, GELNs may provide a two-hit effect, i.e., antibacterial and anti-inflammatory. Interestingly, we also found that naive mice treated with GELNs in the drinking water have higher bone mineral density than control naive mice. This finding will open up a new avenue to study how GELNs can improve the bone mineral density in general since disruption of bone metabolism is associated with many diseases.
Limitations of the Study
One limitation of this study is that we examined the effect of GELNs on in vitro monospecies biofilm formation, which is not representative of the influence of GELNs on established polymicrobial biofilms. Therefore, further studies are needed to determine if GELNs can penetrate an established microbial biofilm and have an influence of polymicrobial interactions and on the antibiotic susceptibility of biofilms. Additionally, the impact of GELNs on pre-existing multispecies biofilms, such as those that occur in human in vivo, requires further study.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We thank Dr. J. Ainsworth for editorial assistance. Funding: This work was supported by a grant from the NIH (UH3TR000875, R01AT008617), United States. H.-G.Z. is supported by a Research Career Scientist (RCS) Award, United States and P20GM125504. D.P.M is supported by K99DE028346, R.J.L is supported by DE011111 and DE012505, X.Z. is supported by 1S10OD020106, and M.L.M. is supported by P50 AA024337 and P20 GM113226.
Author Contributions
K.S., D.P.M., X.Z., Y.T., R.J.L., and H.-G.Z. designed the study, analyzed, interpreted data, and prepared the manuscript; K.S. and D.P.M. performed experiments and interpreted data; J.W.P. and M.S. provided bioinformatics analysis and quantitation; C.L., L.H., and Y.F. provided HPLC analysis; J.M. provided histological analysis; A.K., M.K.S., C.L., and K.S. prepared GELNs and bacteria; M.L.M. did protein analysis; L.Z. provided technical support; S.Q.Z, Y.J., and X.Z interpreted the findings.
Declaration of Interests
The subject matter of this work is covered by a United States provisional patent application, serial no. 62/812,644.
Published: November 22, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2019.10.032.
Supplemental Information
References
- Alvarez-Erviti L., Seow Y., Yin H., Betts C., Lakhal S., Wood M.J. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- Athenstaedt K., Daum G. Phosphatidic acid, a key intermediate in lipid metabolism. Eur. J. Biochem. 1999;266:1–16. doi: 10.1046/j.1432-1327.1999.00822.x. [DOI] [PubMed] [Google Scholar]
- Baker P.J., Dixon M., Evans R.T., Dufour L., Johnson E., Roopenian D.C. CD4(+) T cells and the proinflammatory cytokines gamma interferon and interleukin-6 contribute to alveolar bone loss in mice. Infect. Immun. 1999;67:2804–2809. doi: 10.1128/iai.67.6.2804-2809.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker P.J., Dixon M., Roopenian D.C. Genetic control of susceptibility to Porphyromonas gingivalis-induced alveolar bone loss in mice. Infect. Immun. 2000;68:5864–5868. doi: 10.1128/iai.68.10.5864-5868.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao K., Belibasakis G.N., Thurnheer T., Aduse-Opoku J., Curtis M.A., Bostanci N. Role of Porphyromonas gingivalis gingipains in multi-species biofilm formation. BMC Microbiol. 2014;14:258. doi: 10.1186/s12866-014-0258-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron C. A novel strategy to target bacterial virulence. Future Microbiol. 2013;8:1–3. doi: 10.2217/fmb.12.120. [DOI] [PubMed] [Google Scholar]
- de Diego I., Ksiazek M., Mizgalska D., Koneru L., Golik P., Szmigielski B., Nowak M., Nowakowska Z., Potempa B., Houston J.A. The outer-membrane export signal of Porphyromonas gingivalis type IX secretion system (T9SS) is a conserved C-terminal beta-sandwich domain. Sci. Rep. 2016;6:23123. doi: 10.1038/srep23123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg K., Sarig H., Zaknoon F., Epand R.F., Epand R.M., Mor A. Sensitization of gram-negative bacteria by targeting the membrane potential. FASEB J. 2013;27:3818–3826. doi: 10.1096/fj.13-227942. [DOI] [PubMed] [Google Scholar]
- Hajishengallis G., Darveau R.P., Curtis M.A. The keystone-pathogen hypothesis. Nat. Rev. Microbiol. 2012;10:717–725. doi: 10.1038/nrmicro2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada N., Sojar H.T., Cho M.I., Genco R.J. Isolation and characterization of a minor fimbria from Porphyromonas gingivalis. Infect. Immun. 1996;64:4788–4794. doi: 10.1128/iai.64.11.4788-4794.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsuka K., Hayakawa M., Kiyama-Kishikawa M., Sasaki Y., Hirai T., Abiko Y. Role of the hemin-binding protein 35 (HBP35) of Porphyromonas gingivalis in coaggregation. Microb. Pathog. 2008;44:320–328. doi: 10.1016/j.micpath.2007.10.006. [DOI] [PubMed] [Google Scholar]
- Hiratsuka K., Kiyama-Kishikawa M., Abiko Y. Hemin-binding protein 35 (HBP35) plays an important role in bacteria-mammalian cells interactions in Porphyromonas gingivalis. Microb. Pathog. 2010;48:116–123. doi: 10.1016/j.micpath.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Kauppi A.M., Nordfelth R., Uvell H., Wolf-Watz H., Elofsson M. Targeting bacterial virulence: inhibitors of type III secretion in Yersinia. Chem. Biol. 2003;10:241–249. doi: 10.1016/s1074-5521(03)00046-2. [DOI] [PubMed] [Google Scholar]
- Kooijman E.E., Chupin V., de Kruijff B., Burger K.N. Modulation of membrane curvature by phosphatidic acid and lysophosphatidic acid. Traffic. 2003;4:162–174. doi: 10.1034/j.1600-0854.2003.00086.x. [DOI] [PubMed] [Google Scholar]
- Lam R.S., O'Brien-Simpson N.M., Lenzo J.C., Holden J.A., Brammar G.C., Walsh K.A., McNaughtan J.E., Rowler D.K., Van Rooijen N., Reynolds E.C. Macrophage depletion abates Porphyromonas gingivalis-induced alveolar bone resorption in mice. J. Immunol. 2014;193:2349–2362. doi: 10.4049/jimmunol.1400853. [DOI] [PubMed] [Google Scholar]
- Lamont R.J., Chan A., Belton C.M., Izutsu K.T., Vasel D., Weinberg A. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect. Immun. 1995;63:3878–3885. doi: 10.1128/iai.63.10.3878-3885.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont R.J., Koo H., Hajishengallis G. The oral microbiota: dynamic communities and host interactions. Nat. Rev. Microbiol. 2018;16:745–759. doi: 10.1038/s41579-018-0089-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasica A.M., Goulas T., Mizgalska D., Zhou X., de Diego I., Ksiazek M., Madej M., Guo Y., Guevara T., Nowak M. Structural and functional probing of PorZ, an essential bacterial surface component of the type-IX secretion system of human oral-microbiomic Porphyromonas gingivalis. Sci. Rep. 2016;6:37708. doi: 10.1038/srep37708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasica A.M., Ksiazek M., Madej M., Potempa J. The type IX secretion system (T9SS): highlights and recent insights into its structure and function. Front. Cell. Infect. Microbiol. 2017;7:215. doi: 10.3389/fcimb.2017.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Wu J., Xie H. Porphyromonas gingivalis minor fimbriae are required for cell-cell interactions. Infect. Immun. 2006;74:6011–6015. doi: 10.1128/IAI.00797-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.S., Gkranias N., Farias B., Spratt D., Donos N. Differences in the subgingival microbial population of chronic periodontitis in subjects with and without type 2 diabetes mellitus-a systematic review. Clin. Oral Invest. 2018;22:2743–2762. doi: 10.1007/s00784-018-2660-2. [DOI] [PubMed] [Google Scholar]
- Miki T., Hardt W.D. Outer membrane permeabilization is an essential step in the killing of gram-negative bacteria by the lectin RegIIIbeta. PLoS One. 2013;8:e69901. doi: 10.1371/journal.pone.0069901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu J., Zhuang X., Wang Q., Jiang H., Deng Z.B., Wang B., Zhang L., Kakar S., Jun Y., Miller D. Interspecies communication between plant and mouse gut host cells through edible plant derived exosome-like nanoparticles. Mol. Nutr. Food Res. 2014;58:1561–1573. doi: 10.1002/mnfr.201300729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusslein K., Arnt L., Rennie J., Owens C., Tew G.N. Broad-spectrum antibacterial activity by a novel abiogenic peptide mimic. Microbiology. 2006;152:1913–1918. doi: 10.1099/mic.0.28812-0. [DOI] [PubMed] [Google Scholar]
- Olsen I., Lambris J.D., Hajishengallis G. Porphyromonas gingivalis disturbs host-commensal homeostasis by changing complement function. J. Oral Microbiol. 2017;9:1340085. doi: 10.1080/20002297.2017.1340085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page R.C., Lantz M.S., Darveau R., Jeffcoat M., Mancl L., Houston L., Braham P., Persson G.R. Immunization of Macaca fascicularis against experimental periodontitis using a vaccine containing cysteine proteases purified from Porphyromonas gingivalis. Oral Microbiol. Immunol. 2007;22:162–168. doi: 10.1111/j.1399-302X.2007.00337.x. [DOI] [PubMed] [Google Scholar]
- Pan X., Yang Y., Zhang J.R. Molecular basis of host specificity in human pathogenic bacteria. Emerg. Microbes Infect. 2014;3:e23. doi: 10.1038/emi.2014.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster B.J., Olsen I., Aas J.A., Dewhirst F.E. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol. 2000. 2006;42:80–87. doi: 10.1111/j.1600-0757.2006.00174.x. [DOI] [PubMed] [Google Scholar]
- Potempa J., Sroka A., Imamura T., Travis J. Gingipains, the major cysteine proteinases and virulence factors of Porphyromonas gingivalis: structure, function and assembly of multidomain protein complexes. Curr. Protein Pept. Sci. 2003;4:397–407. doi: 10.2174/1389203033487036. [DOI] [PubMed] [Google Scholar]
- Reyes L., Phillips P., Wolfe B., Golos T.G., Walkenhorst M., Progulske-Fox A., Brown M. Porphyromonas gingivalis and adverse pregnancy outcome. J. Oral Microbiol. 2018;10:1374153. doi: 10.1080/20002297.2017.1374153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savett D.A., Progulske-Fox A. Restriction fragment length polymorphism analysis of two hemagglutinin loci, serotyping and agglutinating activity of Porphyromonas gingivalis isolates. Oral Microbiol. Immunol. 1995;10:1–7. doi: 10.1111/j.1399-302x.1995.tb00110.x. [DOI] [PubMed] [Google Scholar]
- Shibata Y., Okano S., Shiroza T., Tahara T., Nakazawa K., Kataoka S., Ishida I., Kobayashi T., Yoshie H., Abiko Y. Characterization of human-type monoclonal antibodies against reduced form of hemin binding protein 35 from Porphyromonas gingivalis. J. Periodontal Res. 2011;46:673–681. doi: 10.1111/j.1600-0765.2011.01389.x. [DOI] [PubMed] [Google Scholar]
- Shoji M., Shibata Y., Shiroza T., Yukitake H., Peng B., Chen Y.Y., Sato K., Naito M., Abiko Y., Reynolds E.C. Characterization of hemin-binding protein 35 (HBP35) in Porphyromonas gingivalis: its cellular distribution, thioredoxin activity and role in heme utilization. BMC Microbiol. 2010;10:152. doi: 10.1186/1471-2180-10-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley J.W., Olczak T. Heme acquisition mechanisms of Porphyromonas gingivalis - strategies used in a polymicrobial community in a heme-limited host environment. Mol. Oral Microbiol. 2017;32:1–23. doi: 10.1111/omi.12149. [DOI] [PubMed] [Google Scholar]
- Stewart E.J. Growing unculturable bacteria. J. Bacteriol. 2012;194:4151–4160. doi: 10.1128/JB.00345-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng Y., Ren Y., Sayed M., Hu X., Lei C., Kumar A., Hutchins E., Mu J., Deng Z., Luo C. Plant-derived exosomal MicroRNAs shape the gut microbiota. Cell Host Microbe. 2018;24:637–652.e8. doi: 10.1016/j.chom.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribble G.D., Lamont R.J. Bacterial invasion of epithelial cells and spreading in periodontal tissue. Periodontol. 2000. 2010;52:68–83. doi: 10.1111/j.1600-0757.2009.00323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzach-Nahman R., Mizraji G., Shapira L., Nussbaum G., Wilensky A. Oral infection with Porphyromonas gingivalis induces peri-implantitis in a murine model: evaluation of bone loss and the local inflammatory response. J. Clin. Periodontol. 2017;44:739–748. doi: 10.1111/jcpe.12735. [DOI] [PubMed] [Google Scholar]
- Vartoukian S.R., Palmer R.M., Wade W.G. Strategies for culture of 'unculturable' bacteria. FEMS Microbiol. Lett. 2010;309:1–7. doi: 10.1111/j.1574-6968.2010.02000.x. [DOI] [PubMed] [Google Scholar]
- Veith P.D., Talbo G.H., Slakeski N., Dashper S.G., Moore C., Paolini R.A., Reynolds E.C. Major outer membrane proteins and proteolytic processing of RgpA and Kgp of Porphyromonas gingivalis W50. Biochem. J. 2002;363:105–115. doi: 10.1042/0264-6021:3630105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent M.S., Canestrari M.J., Leone P., Stathopulos J., Ize B., Zoued A., Cambillau C., Kellenberger C., Roussel A., Cascales E. Characterization of the Porphyromonas gingivalis Type IX secretion trans-envelope PorKLMNP core complex. J. Biol. Chem. 2017;292:3252–3261. doi: 10.1074/jbc.M116.765081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhuang X., Mu J., Deng Z.B., Jiang H., Zhang L., Xiang X., Wang B., Yan J., Miller D. Delivery of therapeutic agents by nanoparticles made of grapefruit-derived lipids. Nat. Commun. 2013;4:1867. doi: 10.1038/ncomms2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J., Feng S., Wang X., Long K., Luo Y., Wang Y., Ma J., Tang Q., Jin L., Li X. Identification of exosome-like nanoparticle-derived microRNAs from 11 edible fruits and vegetables. PeerJ. 2018;6:e5186. doi: 10.7717/peerj.5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Tauschek M., Robins-Browne R.M. Control of bacterial virulence by AraC-like regulators that respond to chemical signals. Trends Microbiol. 2011;19:128–135. doi: 10.1016/j.tim.2010.12.001. [DOI] [PubMed] [Google Scholar]
- Zenobia C., Hajishengallis G. Porphyromonas gingivalis virulence factors involved in subversion of leukocytes and microbial dysbiosis. Virulence. 2015;6:236–243. doi: 10.1080/21505594.2014.999567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C., Wu J., Xie H. Differential expression and adherence of Porphyromonas gingivalis FimA genotypes. Mol. Oral Microbiol. 2011;26:388–395. doi: 10.1111/j.2041-1014.2011.00626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X., Deng Z.B., Mu J., Zhang L., Yan J., Miller D., Feng W., McClain C.J., Zhang H.G. Ginger-derived nanoparticles protect against alcohol-induced liver damage. J. Extracell. Vesicles. 2015;4:28713. doi: 10.3402/jev.v4.28713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X., Xiang X., Grizzle W., Sun D., Zhang S., Axtell R.C., Ju S., Mu J., Zhang L., Steinman L. Treatment of brain inflammatory diseases by delivering exosome encapsulated anti-inflammatory drugs from the nasal region to the brain. Molecular therapy. J. Am. Soc. Gene Ther. 2011;19:1769–1779. doi: 10.1038/mt.2011.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Z., Yoshizawa-Smith S., Glowacki A., Maltos K., Pacheco C., Shehabeldin M., Mulkeen M., Myers N., Chong R., Verdelis K. Induction of M2 macrophages prevents bone loss in murine periodontitis models. J. Dental Res. 2018;98:200–208. doi: 10.1177/0022034518805984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.