Abstract
Antisense oligomers and their analogs have been successfully utilized to silence gene expression for the treatment of many human diseases; however, the control of yeast’s virulence determinants has never been exploited before. In this sense, this work is based on the key hypothesis that if a pathogen’s genetic sequence is a determinant of virulence, it will be possible to synthesize a nucleic acid mimic based on antisense therapy (AST) that will bind to the mRNA produced, blocking its translation into protein and, consequently, reducing the pathogen virulence phenotype. EFG1 is an important determinant of virulence that is involved in the regulation of the Candida albicans switch from yeast to filamentous form. Thus, our main goal was to design and synthesize an antisense oligonucleotide (ASO) targeting the EFG1 mRNA and to validate its in vitro applicability. The results show that the anti-EFG1 2′-OMethylRNA (2′OMe) oligomer was able to significantly reduce the levels of EFG1 gene expression and of Efg1p protein translation (both approximately 60%), as well as effectively prevent filamentation of C. albicans cells (by 80%). Moreover, it was verified that anti-EFG1 2′OMe keeps the efficacy in different simulated human body fluids. Undeniably, this work provides potentially valuable information for future research into the management of Candida infections, regarding the development of a credible and alternative method to control C. albicans infections, based on AST methodology.
Keywords: Candidiasis, filamentation, nucleic acid mimics, 2′-OMethylRNA modification
Introduction
Candidiasis is the primary fungal disease, with a mortality rate of about 30%–50% and with costs associated with hospitalized patients that range from €5,700 to €85,000 (in U.S. dollars, approximately $6,286 to $93,752) per episode.1, 2 This important clinical, social, and economic problem is due to the recognized phenomenon of Candida species antifungal resistance, associated with the indiscriminate use of traditional antifungal agents.1, 2, 3 Candida albicans remains the most prevalent of all Candida species in Europe, with a range of incidence of around 40%.2, 4, 5 The pathogenicity of C. albicans is supported by a series of virulence factors, one of the most alarming being its ability to switch from yeast to filament forms, a tightly regulated process by a network of genes known as dimorphic switching.6 This virulence factor requires C. albicans to sense and respond to the host environment and is essential for its pathogenicity.7, 8, 9 EFG1 is one of the most important and well-studied regulator genes involved in C. albicans filamentation.10, 11, 12, 13, 14, 15
As a consequence of the rising levels of C. albicans multi-resistance to the traditional antifungal treatments, new alternative therapies, with novel mechanisms of action, enhanced therapeutic potential, improved pharmacokinetics, and less toxicity, are urgently needed.16, 17 Antisense therapy (AST) holds great promise for the treatment of many human chronic non-infectious diseases;18, 19, 20, 21, 22, 23, 24 however, for controlling Candida species growth, the knowledge is scarce.23, 25 Moreover, the control of yeast virulence determinants has never been exploited before with AST. The concept underlying AST is relatively straightforward: the use of a complementary sequence to a specific mRNA that can inhibit gene expression, inducing a blockage in the transfer of genetic information from DNA to protein.26
Antisense oligomers (ASOs) are simply short strands of nucleic acids that have a sequence that is complementary to the target mRNA and that bind to this target by means of standard Watson-Crick base pairing.26 Up to now, there have been three generations of ASOs24, 25, 26 with several chemical modifications in order to increase its nuclease resistance, reduce its toxicity, and enhance its affinity and half-life.22 The 2′-OMethylRNA (2′OMe) sugar modification belongs to the second generation of acid mimics; however, these do not support RNase H activity (a specific degradation mechanism cleaving the target mRNA).27, 28 An insertion of a longer central unmodified region, known as gapmers, has been used as a popular strategy to allow that RNase to join and activate the degradation of the mRNA target.29, 30
Thus, this work is based on the key hypothesis that, if a pathogen’s genetic sequence of a specific gene is a determinant of virulence, as is the case with the EFG1 gene, it will be possible to synthesize a nucleic acid mimic that will bind to the mRNA produced and degrade it, blocking its translation into protein and, consequently, reducing its virulence phenotype (which, in this case, would be the filament development). Our data confirm that ASOs, including 2′OMe, can control a virulence determinant of C. albicans. The results show that an anti-EFG1 2′OMe significantly reduced EFG1 gene expression and effectively prevented C. albicans cell filamentation in simulated human body fluids.
Results and Discussion
Despite an increasing number of successful applications of AST for the treatment of human chronic non-infectious diseases,20, 21, 22, 24, 31, 32, 33, 34, 35, 36, 37 and, more recently, to manage infectious bacteria,37, 38, 39 this methodology was never exploited to control Candida species virulence factors. Moreover, EFG1 has been reported as one of the most relevant virulence determinants involved in C. albicans filamentation and, consequently, in its pathogenicity.6, 10, 11, 12, 13, 14, 15 This makes EFG1 an ideal target for validating an AST approach against Candida, not only because its role has been repeatedly proved but also because morphological changes can be easily examined.
Thus, based on our key hypothesis, we intend, with this work, to validate in vitro the application of a 2′OMe oligomer to control C. albicans switching from yeast to filamentous form, reducing the mRNA produced by the EFG1 gene, and inactivate its translation into Efg1p.
Anti-EFG1 2′-OMe Characterization
Nucleic acid mimics in particular, the 2′OMe were the base for the design of the anti-EFG1 oligomer.26 It has been described that the nucleic acid mimics must be designed with melting temperatures (Tms) around 39°C–42°C and a guanine-cytosine (GC) content of approximately 50% to 60% in order to increase the binding affinity for target mRNA and stability in the human body.26, 30 Furthermore, several studies have shown that ASOs with sizes between 12 and 20 nt (nucleotides) usually present a good hybridization performance.40 Taking into account these features, the anti-EFG1 2′OMe sequence was the 5′-mG mG mC mA TACCGTTA mU mU mG mU-3′ (m-2′OMe), with a theoretical Tm of 41.1°C, 43.8% of GC, and a total of 16 nt (Table 1). Four 2′OMe chemical modifications were added to each end of the sequence to increase the stability of the ASO while maintaining the ability to recruit RNase H to degrade the mRNA.34 Being a synthetic molecule, 2′OMe is not recognized by the RNase H, but a small DNA gap in the middle of the ASO ensures the enzyme binding. This way, the ASO will act not only by directly blocking the protein synthesis but also by promoting the degradation of the target mRNA.
Table 1.
Sequence of Anti-EFG1 2′OMe (m) and Scramble ASO, with the Respective Size, Theoretical Tm, and GC Content
| ASO | Sequence (5′–3′) | Size | Tm (°C) | % of GC |
|---|---|---|---|---|
| Anti-EFG1 2′OMe | 5′-mG mG mC mA TACCGTTA mU mU mG mU-3′ | 16 nt | 41.1 | 43.8 |
| Scramble ASO | 5′-mG mG mC mA TTCCAGTA mU mU mG mU-3′ | 41 |
A scramble ASO with the same number of nucleotides and chemical modifications (5′-mG mG mC mA TTCCAGTA mU mU mG mU-3′) was also synthetized with three mismatches, resulting in a theoretical Tm of 41°C and 43.8% of GC (Table 1). Furthermore, the anti-EFG1 2′OMe was labeled with TYE563 at the 5′ end to investigate its cellular uptake, sensitivity, and specificity.
Anti-EFG1 2′OMe Cellular Uptake, Sensitivity, and Specificity
Sensitivity and specificity of the nucleic acid mimics are two important factors in the success of the ASO applicability.41, 42, 43 In this study, we used fluorescence in situ hybridization (FISH), a standard methodology used to identify microorganisms that makes use of nucleic acid coupled with fluorochromes,44, 45, 46, 47 and epifluorescence analysis to evaluate the anti-EFG1 2′OMe cellular uptake, sensitivity, and specificity against C. albicans cells.
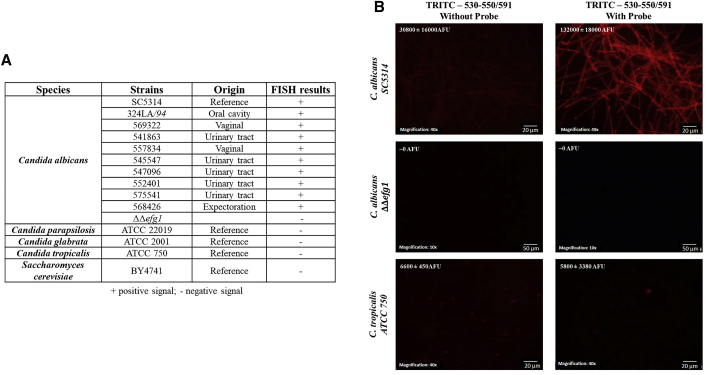
The anti-EFG1 2′OMe specificity was tested against 10 strains of C. albicans and 4 strains of other fungi (Figure 1). Anti-EFG1 2′OMe binding in C. albicans was confirmed by the positive signal (presence of fluorescence) observed for all C. albicans strains tested (n = 10) (Figures 1A and 1B; Figure S1). The negative signal (absence of fluorescence) obtained for the other fungi tested and for C. albicans ΔΔefg1 reinforces ASO specificity for C. albicans cells (Figure 1).
Figure 1.
Anti-EFG1 2′OMe Sensitivity and Specificity Obtained by FISH
(A) List of strains and species used and their origin, as well as the respective results obtained by FISH at 37°C, during 3 h. (B) Illustrative images obtained by epifluorescence microscopy. The exposure time was the same for each strain: Candida albicans SC5314 was obtained with 218.7 ms; Candida albicans HLC52 (ΔΔefg1 mutant strain) was obtained with 713.2 ms, and Candida tropicalis ATCC750 was obtained with 293.9 ms of exposure. Negative controls were prepared only with 20 μL hybridization solution without probe.
These studies demonstrate the anti-EFG1 2′OMe Candida cellular uptake without carriers or transfection agents for instance, by adsorptive endocytosis as in other microorganisms,48, 49, 50 and its ability to hybridize with the respective target with high specificity for C. albicans cells.
Anti-EFG1 2′OMe Oligomer Behavior
In order to determine the concentration of anti-EFG1 2′OMe to be used in vitro validation studies, C. albicans SC5314 was incubated with different concentrations of ASO (10–60 nM). Additionally, the same was applied to investigate the cytotoxic effect of the ASO on the 3T3 cell line.
Cytotoxicity Evaluation
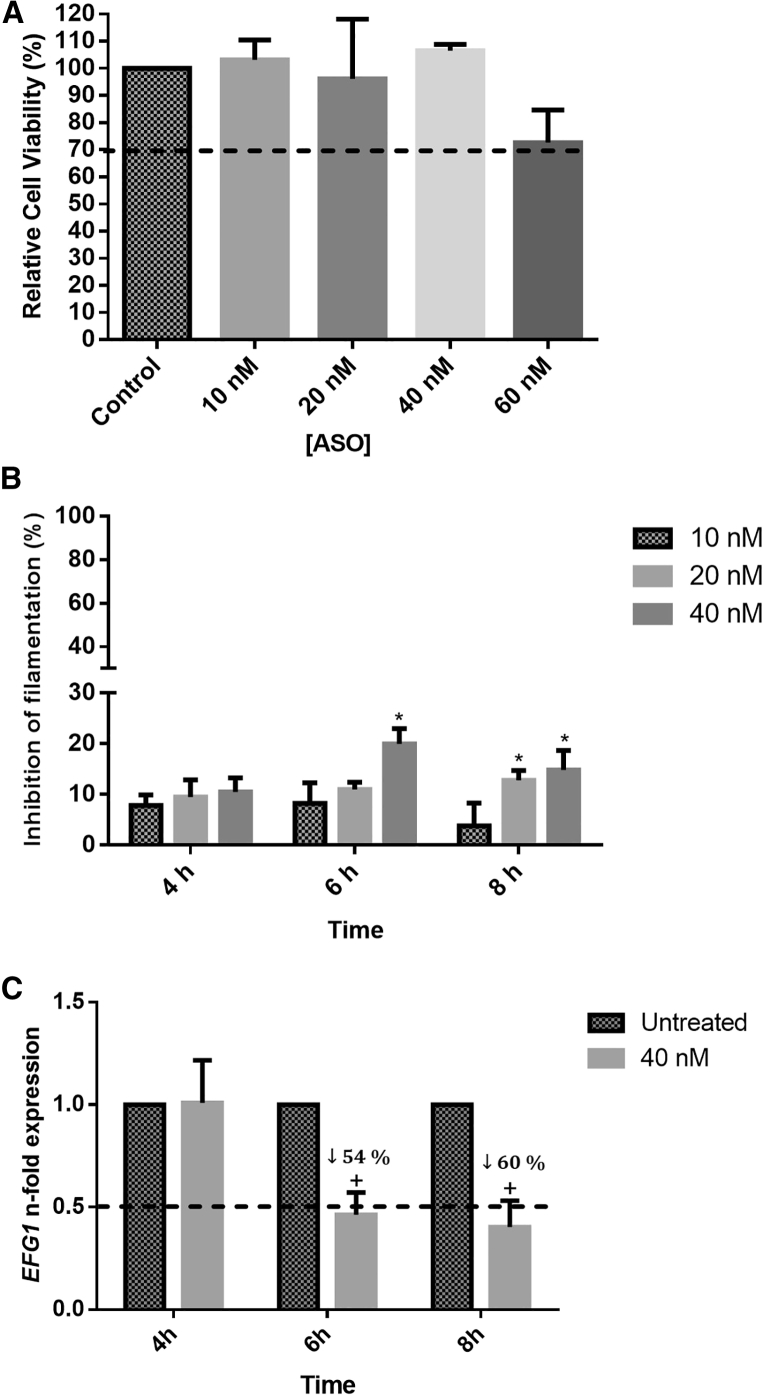
Figure 2 presents the results of ASO cytotoxicity on the 3T3 cell line for the determination of the minimal ASO concentration capable to inhibit C. albicans filamentation and EFG1 gene expression.
Figure 2.
Anti-EFG1 2′OMe Effect on Candida albicans Filamentation
(A) Relative cell viability (%) determined by the absorbance values (Abs; 490 nm cm−2) of formazan product obtained from 3T3 cells treated with different concentrations of ASO (10, 20, 40, and 60 nM). The control is related to the cells without ASO treatment. (B) Percentage of inhibition (%) of filamentous forms, after treatment with different concentrations of ASO (10, 20, and 40 nM). (C) Levels of EFG1 gene expression obtained by the Pfaffl method, after application of 40 nM ASO, at different time points (4, 6, and 8 h) in RPMI. Error bars represent SD. *Significant differences among 10 nM and the other concentrations of ASO tested (p < 0.05). +Significant difference between untreated and treated cells (p < 0.05).
MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium solution) assays were performed to infer about the anti-EFG1 2′OMe cytotoxicity against 3T3 cells. The results demonstrated that the ASO concentrations of 10, 20, and 40 nM tested were not cytotoxic, since the relative cell viability is higher than 70% of the control (absence of ASO) (Figure 2A).51 However, the relative cell viability for 60 nM is approximately 70%, so it could be considered a cytotoxic concentration. Therefore, it was decided to use 40 nM ASO for the next experimental assays.
Effect on Filamentation and Gene Expression
Concerning the anti-EFG1 2′OMe effect on C. albicans filamentation, it was possible to verify a reduction for all the concentrations tested (Figure 2B). As expected, the percentage of filamentation of C. albicans without ASO increased from 4 h to 8 h, reaching 80% filamentation (Figure S2A). In the presence of ASO, after 4 h of incubation, approximately 10% reduction was observed (Figure 2B) without statistical differences among the ASO concentrations tested (p > 0.05). Additionally, the results revealed a more pronounced effect after 6 h, specifically with 40 nM ASO, with approximately 20% reduction (p < 0.05). After 8 h of incubation, a similar performance was observed with 15% reduction, even for the lower concentration (20 nM) of ASO. Additionally, the ASO scramble was unable to reduce C. albicans filamentation (Figure S2C).
The EFG1 expression levels were determined for C. albicans SC5314 cells growing in the presence and the absence of 40 nM ASO in order to evaluate the effect of anti-EFG1 2′OMe in the blockage of the expression of the respective gene. As expected, this strain expresses the EFG1 gene, and a 3-fold increase on its expression levels was noticed from 4 h to 8 h (Figure S2B). Regarding ASO treatment, qRT-PCR studies revealed a decrease on the levels of EFG1 expression after 6 h and 8 h (p < 0.05) (Figure 2C). Indeed, a reduction of 54% at 6 h and 60% at 8 h on the EFG1 levels of expression was demonstrated (p < 0.05).
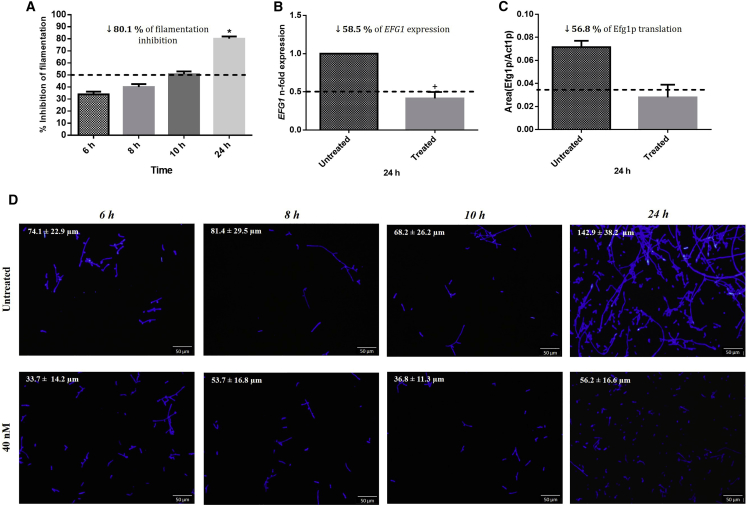
After defining the most appropriate concentration of anti-EFG1 2′OMe to be used (40 nM), we evaluated the performance of the ASO on longer periods (Figure 3). In terms of C. albicans filamentation reduction (Figure 3A), the results showed an increase of inhibition over time, reaching 80% after 24 h of treatment (p < 0.05) compared to the absence of ASO. It is important to address that the dimorphic switching in C. albicans is dependent on a network of genes.12, 14, 52, 53, 54, 55 Thus, it was not expected a total reduction on C. albicans filamentation. Subsequent examination of epifluorescence microscopy images confirms these results and also revealed a significant and relevant decrease in terms of the filaments’ lengths (74 μm to 34 μm at 6 h, 81 μm to 54 μm at 8 h, 68 μm to 37 μm at 10 h, and 143 μm to 56 μm at 24 h of treatment) (Figure 3D). This is an important result once C. albicans filamentation is considered one of the most problematic virulence factors, increasing its capability to invade human cells and causing tissue damage.56, 57
Figure 3.
Anti-EFG1 2′OMe Effect on Efg1p Translation
(A) Percent inhibition (%) of filamentous forms at different time points (6, 8, 10, and 24 h). (B) Levels of EFG1 gene expression obtained by the Pfaffl method at 24 h. (C) Levels of Efg1p translation normalized with the translation of Act1p at 24 h. (D) Epifluorescence microscopy images of Candida cells stained with Calcofluor after treatment with 40 nM ASO (control was prepared only with cells in RPMI; without ASO). The assays were performed for C. albicans SC5314. Error bars represent SD. *Significant difference between 6 h and the other times tested (p < 0.05). +Significant difference between untreated and treated cells (p < 0.05).
As mentioned earlier, ASOs affect cellular functions through transcription attenuation and protein translation inhibition.58, 59, 60, 61 The effects of anti-EFG1 2′OMe on EFG1 gene expression and Efg1 protein translation were determined at 24 h of treatment (Figures 3 B and 3C), as data from filamentation indicated this treatment time as quite effective. The results obtained showed a significant reduction in the levels of EFG1 expression (around 59%) (Figure 3B) and in Efg1 protein translation (around 57%) (Figure 3C), corroborating the morphological data (Figures 3A and 3D).
Performance on Simulated Human Body Fluids
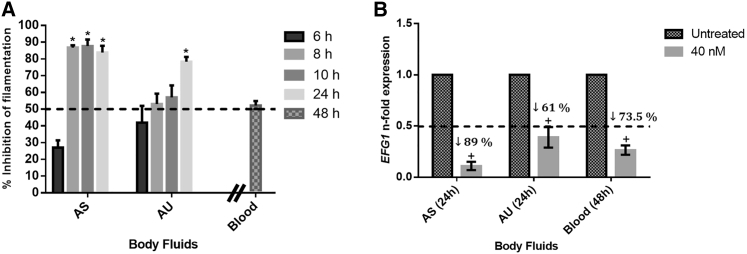
To mimic human body environments, the performance of anti-EFG1 2′OMe was also evaluated on different simulated human body fluids (artificial saliva [AS] and artificial urine [AU]) and horse blood (Figure 4). It is important to highlight that C. albicans was able to grow and filament in all simulated human body fluids tested, but in a time- and fluid-dependent manner (Figures S3A and S3B).
Figure 4.
Anti-EFG1 2′OMe Effect on Simulated Human Body Fluids (AS, AU, and Horse Blood)
(A) Percent inhibition (%) of filamentous forms at different time points (6, 8, 10, and 24 h for AS and AU; 48 h for horse blood). (B) Levels of EFG1 gene expression for C. albicans SC5314 obtained by the Pfaffl method, after treatment with 40 nM ASO in the presence of different simulated human body fluids (AS and AU at 24 h and horse blood at 48 h). Error bars represent SD. *Significant difference between 6 h and the other times tested (p < 0.05); +Significant difference between untreated and treated cells (p < 0.05).
Interestingly, it can be noticed that anti-EFG1 2′OMe maintains its performance in simulated human body fluids, reducing C. albicans filamentation and EFG1 gene expression. In fact, it was verified that the ASO was able to reduce 90% and 80% of C. albicans filamentation after 24 h of incubation in AS and AU (p < 0.05), respectively, and 50% after 48 h of incubation in horse blood (Figure 4A).
Figure 4B shows the levels of EFG1 gene expression and demonstrates decreases in the levels of expression of 89% in AS, 61% in AU, and 74% in horse blood (p < 0.05). It is important to highlight that the levels of EFG1 expression in the absence of ASO were different in all simulated human body fluids tested (Figure S2C), which justifies the different levels of reduction observed.
Considering any possible future clinical applications of the anti-EFG1 2′OMe in the control of local candidiasis (oral and urinary), as well as of systemic infections (blood), these are important results once the ASO maintains its performance in human fluids, inhibiting C. albicans filamentation and the EFG1 gene expression.
Materials and Methods
Microorganisms
A total of 11 clinical strains (Figure 1A), including Candida albicans (n = 10) and Saccharomyces cerevisiae (n = 1), recovered from different body sites, were used during this study. All isolates were recovered from vaginal, urinary, and oral tracts and were obtained from the Candida collection of the Biofilm group of the Centre of Biological Engineering, University of Minho, Braga, Portugal. Four reference strains—Candida albicans (SC5314), Candida parapsilosis (ATCC 22019), Candida tropicalis (ATCC 750), and Candida glabrata (ATCC 2001)—were included in this study. The mutant strain C. albicans ΔΔefg1 (HLC52) was also tested.62
Growth Conditions
For all experiments, yeast strains were subcultured on sabouraud dextrose agar (SDA; Merck, Darmstadt, Germany) and incubated for 24 h at 37°C. Cells were then inoculated in sabouraud dextrose broth (SDB; Merck, Darmstadt, Germany) and incubated overnight at 37°C, at 120 rpm. After incubation, the cells’ suspensions were centrifuged for 10 min at 3,000 g at 4°C and washed twice with PBS (pH 7, 0.1 M). Pellets were suspended in 5 mL RPMI medium (Sigma-Aldrich, St. Louis, MO, USA), and the cellular density was adjusted for each experiment using a Neubauer chamber (Paul Marienfeld, Lauda-Königshofen, Germany) to 1 × 105 or 1 × 106 cells per milliliter (mL−1). All experiments of this work were performed in triplicate and in a minimum of three independent assays.
Design and Synthesis
To design a specific ASO for C. albicans EFG1, the target region of the gene was selected based on a search conducted at the Candida Genome Database (http://www.candidagenome.org/cgi-bin/compute/blast_clade.pl). Several EFG1 gene sequences were aligned to make sure that conserved regions were used for the design. Also, a BLAST search was performed to ensure that the sequences were not targeting any sequence of the human genome or a similar region in another C. albicans gene. The EFG1 sequence 5′-ACAATAACGGTATGCC-3′ was selected as the target, taking into account its high specificity to the C. albicans genome, its non-binding against the Homo sapiens genome, and the number of nucleotides.26 Specific ASOs were then designed for the use of 2′ ribose modification. 2′OMe was selected, since it is one of the most used for antisense applications.28, 29, 30, 63 A gapmer was introduced to increase the odds of activating RNase H activity.64 The calculator from Integrated DNA Technologies (IDT; https://eu.idtdna.com/calc/analyzer) was used to determine the theoretical Tm and the GC content of the possible ASO for that target region. The selected ASO was produced according to the user’s own specifications at EXIQON and purified by high-pressure liquid chromatography (HPLC). The same ASO was synthetized with an orange-fluorescent fluorophore (TYE563). A scrambled ASO, similar to the EFG1 ASO, was also synthesized to be used as negative control.
Sensitivity and Specificity Tests
The sensitivity and specificity of anti-EFG1 2′OMe was determined against different yeast strains (Figure 1A) by FISH.65 For that, 20 μL of an inoculum of Candida cells adjusted to 1 × 106 cells mL−1 was transferred to a slide and fixated with 30 μL 4% (v/v) paraformaldehyde (Sigma-Aldrich) for 10 min, and the excess was removed. After that, cells were permeabilized with 30 μL 50% (v/v) ethanol for an additional 10 min and allowed to air dry. The hybridization step was performed with 20 μL ASO (200 nM) coupled with orange-fluorescent fluorophore diluted in hybridization solution (900 nM NaCl [PanReac Applichem, Barcelona, Spain], 30% formamide [Sigma-Aldrich, Sintra, Portugal], 20 mM Tris-HCl [Sigma-Aldrich, Sintra, Portugal], and 0.01% SDS [Sigma-Aldrich, Sintra, Portugal]). Negative controls were prepared only with 20 μL hybridization solution without probe. Samples were then covered with coverslips and incubated at 37°C for 3 h in dark conditions. After hybridization, slides were submerged in wash solution (20 mM Tris-HCl, 0.01% SDS, and 900 mM NaCl) and incubated for 30 min at the same temperature.
The images from cells were acquired with an epifluorescence microscope (Olympus Portugal, Porto, Portugal). Cells were observed using a 40× objective. The exposure time, gain, and saturation values were fixed for each sample. The TRITC filter (530-550/591) was used for image acquisition.
Cytotoxicity
In order to select the concentration of anti-EFG1 2′OMe without cytotoxicity to be used during this study, the ASO cytotoxicity was determined against 3T3 cell line (fibroblast cells, embryonic tissue, mice from the CCL 163 line, American Type Culture Collection). For that, 3T3 cells were grown in DMEM (Biochrom, Berlin, Germany) supplied by 10% fetal bovine serum (FBS; Sigma-Aldrich) and 1% antibiotic containing P/S (penicillin and streptomycin; Biochrom, Berlin, Germany). After detachment, a suspension with 1 × 105 cells mL−1 was added to a 96-well plate, and cells grew until attaining 80% confluence. Prior to the cytotoxicity assay, the wells were washed twice with PBS. Different concentrations of ASO (10, 20, 40, and 60 nM) were prepared in DMEM, and 50 μL of each concentration was added to each well. Negative control was performed by adding 50 μL DMSO to the cells, and positive control was performed by adding 50 μL DMEM. The plates were incubated for 24 h at 37°C and 5% CO2.
After incubation, 10 μL MTS (CellTiter 96 Aqueous One Solution Cell Proliferation Assay, Promega) and 1% DMEM without phenol were added to each well and incubated for 1 h. Lastly, the absorbance was measured at 490 nm in a microplate reader (Biochrom EZ Read 800 Plus, Biochrom, Cambridge, England). The cytotoxicity results were expressed as the percentage of viable cells corresponding the optical density 490 (OD490) of cells grown without ASO as 100% cell viability.
Effect on C. albicans Filamentation
In parallel, to determine the effect on C. albicans filamentation, the similar concentrations of ASOs (10, 20, and 40 nM) were incubated with C. albicans SC5314, and the effects were evaluated in terms of filament number. For this, 100 μL ASO at the different concentrations prepared in RPMI medium were added to each well of a 96-well plate (Orange Scientific, Braine-I’Alleud, Belgium) together with 100 μL of 1 × 106 cells mL−1 of Candida cell suspensions. The positive controls were prepared with 200 μL of cells in RPMI medium without the addition of ASO, and the negative controls were prepared only with RPMI medium. In addition, the scrambled ASO was used as control. The ASO effects were evaluated at 4, 6, and 8 h of incubation.
To determine the percentage of filamentation, Candida cells were scraped from each well, and the filaments were enumerated using a Neubauer chamber by optic microscopy. The results were presented as percentage of filamentation reduction through the following formula: percentage of filamentation inhibition = [(percentage of cells in filament on control) − (percentage of cells in filament in the presence of ASO)]/(percentage of cells in filament on control).
Effect on EFG1 Gene Expression
qRT-PCR studies were performed to determine the effect of 40 nM ASO on EFG1 gene expression. For that, in 24-well plates (Orange Scientific, Braine-I’Alleud, Belgium), 500 μL C. albicans cells at 1 × 106 cells mL−1 of were incubated with 500 μL ASO for the same periods of time. After each time point, the cells were collected from each well, recovered by centrifugation for 5 min at 7,000 × g and 4°C, and washed once with sterile water. RNA extraction was performed using the PureLink RNA Mini Kit (Invitrogen, Carlsbad, CA, USA).66, 67 Then, to avoid potential DNA contamination, samples were treated with DNase I (DNase I, Amplification Grade, Invitrogen), and RNA concentration was determined by optical density measurement with the NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific). The cDNA was synthetized using the iScript Reverse Transcriptase (Bio-Rad) in accordance with the manufacturer’s instructions. qRT-PCR (CFX96, Bio-Rad) was performed on a 96-well microtiter plate using EvaGreen Supermix (Bio-Rad, Berkeley, CA, USA). The expression of the EFG1 gene was normalized with the ACT1 Candida reference gene.68 No-reverse transcriptase controls (NRTs) and no-template controls (NTCs) were included in each run. Each reaction was performed in triplicate, and mean values of relative expression were determined for each gene. The primers were designed using the Primer 3 web-based software (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) and are described in Table 2.
Table 2.
Primers Used for qRT-PCR, with the Respective Theoretical Tm Obtained from the Calculator from IDT and Amplification Product
| Candida albicans Gene Name | Systematic Name | Sequence (5′–3′) | Primer | Tm (°C) | AP (BP) |
|---|---|---|---|---|---|
| EFG1 | CR_07890W_A/Orf19.610 | 5′-TTCTGGTGCAGGTTCCAC-3′ | forward | 57 | 168 |
| 5′-CCTGGTTGTGATGCAGGT-3′ | reverse | ||||
| ACT1 | C1_13700W_A/Orf19.5007 | 5′-AATGGGTAGGGTGGGAAAAC-3′ | forward | 57 | 150 |
| 5′-AGCCATTTCCATTGATCGTC-3′ | reverse |
AP, amplification product; BP, base pairs.
Performance
In order to evaluate the performance of the anti-EFG1 2′OMe throughout the study, C. albicans SC5314 was incubated with 40 nM ASO for 24 h. For that, 5 mL anti-EFG1 2′OMe at 40 nM prepared in RPMI medium was added to 5 mL of a C. albicans SC5314 suspension at 1 × 105 cells mL−1 and incubated at 37°C under gentle agitation (120 rpm). The positive control was prepared only with 10 mL of the same concentration of cells. At pre-determined time points (6, 8, 10, and 24 h) aliquots were recovered, and three complementary criteria were evaluated: percentage of filamentation reduction at 6, 8, 10, and 24 h; levels of EFG1 expression; and Efg1 protein translation at 24 h of incubation.
The ASO effect on C. albicans filamentation and on levels of EFG1 gene expression was evaluated as described previously. For that, the number of filaments of cells grown in the presence and absence of ASO was enumerated, and RNA was extracted from those cells to quantify the levels of EFG1 gene expression. Epifluorescence microscopy images were used to confirm the levels of filamentation and to determine the length of the filaments. C. albicans cells grown in the presence and absence of ASOs were stained with 20 μL calcofluor (2 g/L; Sigma-Aldrich, St. Louis, MO, USA) for 15 min in dark conditions. Consequently, the cells were centrifuged for 5 min and washed twice with ultra-pure water. All samples were observed with an Olympus BX51 microscope (Olympus Portugal, Porto, Portugal) coupled with a DP71 digital camera. The laser DM 405/488/559/635 and the emission filters BA 430-470 (blue channel) were used, and images were acquired with the program FluoView FV100 (Olympus). The length of the filaments was determined using ImageJ plug-in software.
Effect on Efg1p Translation
Liquid chromatography (LC)-MALDI-TOF-mass spectrometry (MS) (Q-Exactive Orbitrap, Thermo Fisher Scientific) was used to infer about the effect of ASO in the translation of respective genes into a protein (Efg1p).69 For that, the proteome of C. albicans cells grown in the presence and absence of the ASO was obtained as described previously,70 with some modifications. Cells were obtained by centrifugation for 5 min, at 8,000 × g and 4°C, and washed twice with sterile ice-cold ultrapure water. Then, cells were washed with lysis buffer (10 mM Tris-HCl [pH 7.4], 1 mM phenylmethylsulfonyl fluoride [PMSF]) and resuspended in ice-cold lysis buffer in order to lyse mechanically, with an equal volume of glass beads in a cell homogenizer (FastPrep, MP Biomedicals), four times. Lysed cells were separated by centrifugation at 1,000 × g for 10 min at 4°C, and the pellet was washed twice with each of the following ice-cold solutions: 1 mM PMSF and 5% NaCl (Thermo Fisher Scientific, Waltham, MA, USA) and 1 mM PMSF in ultrapure water. Then, it was resuspended in washing buffer (50 mM Tris-HCl [pH 8], 1 mM PMSF) and extracted by boiling with SDS extraction buffer (50 mM Tris-HCl [pH 8.0], 0.1 M EDTA, 2% SDS, 10 mM DTT) for 10 min. Finally, the supernatant was transferred to fresh tubes, and the protein concentration was determined using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific).
Protein samples were analyzed by nano-LC-MS/MS (tandem MS) in order to identify and quantify Efg1p and Act1p. First, protein samples were digested based on the filter-aided sample preparation (FASP) procedure described by Wiśniewski and colleagues,71 with some modifications. Protein digestion with a trypsin/Lys-C mix was performed overnight at 37°C (Promega, Madison, WI, USA), and each reaction was stopped with 1% (w/v) trifluoroacetic acid (TFA). Peptides were then recovered by centrifugation, followed by an additional centrifugation step with 0.1% TFA. Next, peptide samples were cleaned up and concentrated by SPE-C18 chromatography. Nano-LC-MS/MS equipment, composed of an Ultimate 3000 LC system coupled to a Q-Exactive Hybrid Quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific, Bremen, Germany), was used to identify and quantify the proteins.72 Samples were loaded onto a trapping cartridge (Acclaim PepMap 100 C18 [pore size, 100 Å, 5 mm × 300 μm i.d.], catalog no. 160454, Thermo Fisher Scientific) in a mobile phase of 2% acetonitrile (ACN), 0.1% formic acid (FA) at 10 μL/min. Data acquisition was controlled by Xcalibur 4.0 and Tune 2.8 software (Thermo Fisher Scientific, Bremen, Germany).
The raw data were processed using Proteome Discoverer v2.2.0.388 software (Thermo Fisher Scientific) and searched using the UniProt database for the taxonomic selection Candida albicans (November 2017 release). The Sequest HT search engine was used to identify tryptic peptides. The percentage of Efg1p translation was determined by the ratio of media of peptides area corresponding to Efg1p and media of peptides area corresponding to Act1p (used as reference protein).
Performance on Simulated Human Body Fluids
To mimic the human body fluids, AU, AS, and horse blood were used during this work. AU (pH 5.8) and AS (pH 6.8) were prepared with slight modifications to that previously described by Silva et al. in their 2010 and 2013 studies, respectively.73, 74 The composition of AU was CaCl2 (0.65 g/L), MgCl2 (0.65 g/L), NaCl (4.6 g/L), Na2SO4 (2.3 g/L), Na3C3H5O (CO2)3 (0.65 g/L), Na2C2O4 (0.02 g/L), KH2PO4 (2.8 g/L), KCl (1.6 g/L), NH4Cl (1.0 g/L), urea (25 g/L), creatinine (1.1 g/L), and glucose (3 g/L); and the composition of AS was peptone (5 g/L), glucose (2 g/L), mucin (1 g/L), NaCl (0.35 g/L), CaCl2 (0.2 g/L), and KCl (0.2 g/L). The blood used was defibrinated horse blood (Probiológica-Empresa de Produtos Biológicos, Belas, Portugal) supplemented with 50% of 0.9% NaCl.
Pellets obtained as described earlier were resuspended in 10 mL of each body fluid after adjusting the cellular density to 1 × 106 cells mL−1, using a Neubauer hemocytometer (Paul Marienfeld, Lauda-Königshofen, Germany) and incubated for 24 h (AS and AU) and 48 h (horse blood) at 37°C in Erlenmeyer flasks under gentle agitation (120 rpm). RPMI medium was used as positive control. All experiments were performed in triplicate and in a minimum of three independent assays.
To determine whether anti-EFG1 2′OMe maintains its performance on different simulated human body fluids, its ability to inhibit C. albicans filamentation and reduce EFG1 expression was determined. For that, at pre-determined time points (6, 8, 10, and 24 h for AS and AU and 48 h for horse blood), aliquots of each suspension were recovered, and the cells were harvested by centrifugation at 3000 × g for 10 min at 4°C and washed twice with PBS. To determine the percentage of filamentation, Candida cells that formed filaments were enumerated using a Neubauer chamber, as described earlier.
To determine the effect on C. albicans EFG1 gene expression, qRT-PCR studies were evaluated at 24 h (for AS, AU, and RPMI medium) and 48 h (for horse blood) of incubation. The extraction of RNA and qRT-PCR were performed as described earlier. Simultaneously, we evaluated C. albicans capability for growing on the different simulated human body fluids by colony-forming unit (CFU) determination methodology. For that, 1 mL of each suspension was recovered, and cells were harvested by centrifugation at 3,000 × g for 10 min at 4°C and washed twice with PBS. Serial dilutions were performed on PBS, inoculated onto SDA, and incubated for an additional 24 h at 37°C. The results are presented as the log of CFUs cm−2 (Figure S3A).
Statistical Analysis
Data are expressed as the mean ± SD of at least three independent experiments. Results were compared using two-way ANOVA using GraphPad Prism (GraphPad Software, San Diego, CA, USA). All tests were performed with a confidence level of 95%.
Conclusions
Our data demonstrate, for the first time, that it is possible to use antisense oligomers with 2′OMe chemical modifications to control virulence determinants of C. albicans. The anti-EFG1 2′OMe that we have projected has significantly reduced EFG1 gene expression and effectively prevented C. albicans cell filamentation in different simulated human body fluids. Undeniably, this work provides potentially valuable information for future research into the management of Candida infections. Thus, in the future, it will be possible to develop a credible and alternative method to control oral and urinary candidiasis, as well as systemic infections, based on AST methodology.
Author Contributions
D.A. and S.S. conceived and designed the study. D.A. the conducted the experiments and wrote the paper. N.A. conducted the RT-PCR experiments. A.B. conducted the body fluids experiments. C.A. helped to design the ASO. M.E.R. conducted the cytotoxicity assays. M.H. and S.S. performed the analysis and read the paper. All authors read and approved the manuscript.
Acknowledgments
This study was supported by the Portuguese Foundation for Science and Technology (FCT), under the scope of the strategic funding of the UID/BIO/04469 unit and COMPETE 2020 (POCI-01-0145-FEDER-006684) and the BioTecNorte operation (NORTE-01-0145-FEDER-000004), funded by the European Regional Development Fund under the scope of a Norte 2020 – Programa Operacional Regional do Norte and Daniela Eira Araújo (SFRH/BD/121417/2016) PhD grant. The authors also acknowledge the project funding by the “02/SAICT/2017 – Projetos de Investigação Científica e Desenvolvimento Tecnológico (IC&DT; POCI-01-0145-FEDER-028893).”
The mass spectrometry technique was performed at the Proteomics i3S Scientific Platform with the assistance of Hugo Osório. This work had support from the Portuguese Mass Spectrometry Network, integrated in the National Roadmap of Research Infrastructures of Strategic Relevance (ROTEIRO/0028/2013 and LISBOA-01-0145-FEDER-022125).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.09.016.
Supplemental Information
References
- 1.Quindós G. Epidemiology of invasive mycoses: a landscape in continuous change. Rev. Iberoam. Micol. 2018;35:171–178. doi: 10.1016/j.riam.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 2.Koehler P., Stecher M., Cornely O.A., Koehler D., Vehreschild M.J.G.T., Bohlius J., Wisplinfhoff H., Vehreschild J.J. Morbidity and mortality of candidaemia in Europe: an epidemiologic meta-analysis. Clin. Microbiol. Infect. 2019;25:1200–1212. doi: 10.1016/j.cmi.2019.04.024. [DOI] [PubMed] [Google Scholar]
- 3.Negri M., Henriques M., Svidzinski T.I., Paula C.R., Oliveira R. Correlation between Etest, disk diffusion, and microdilution methods for antifungal susceptibility testing of Candida species from infection and colonization. J. Clin. Lab. Anal. 2009;23:324–330. doi: 10.1002/jcla.20337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yapar N. Epidemiology and risk factors for invasive candidiasis. Ther. Clin. Risk Manag. 2014;10:95–105. doi: 10.2147/TCRM.S40160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonçalves B., Ferreira C., Alves C.T., Henriques M., Azeredo J., Silva S. Vulvovaginal candidiasis: epidemiology, microbiology and risk factors. Crit. Rev. Microbiol. 2016;42:905–927. doi: 10.3109/1040841X.2015.1091805. [DOI] [PubMed] [Google Scholar]
- 6.Araújo D., Henriques M., Silva S. Portrait of Candida species biofilm regulatory network genes. Trends Microbiol. 2017;25:62–75. doi: 10.1016/j.tim.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 7.Mayer F.L., Wilson D., Hube B. Candida albicans pathogenicity mechanisms. Virulence. 2013;4:119–128. doi: 10.4161/viru.22913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casadevall A., Pirofski L. Host-pathogen interactions: the attributes of virulence. J. Infect. Dis. 2001;184:337–344. doi: 10.1086/322044. [DOI] [PubMed] [Google Scholar]
- 9.Saville S.P., Lazzell A.L., Monteagudo C., Lopez-Ribot J.L. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot. Cell. 2003;2:1053–1060. doi: 10.1128/EC.2.5.1053-1060.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nobile C.J., Fox E.P., Nett J.E., Sorrells T.R., Mitrovich Q.M., Hernday A.D., Tuch B.B., Andes D.R., Johnson A.D. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell. 2012;148:126–138. doi: 10.1016/j.cell.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nobile C.J., Mitchell A.P. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr. Biol. 2005;15:1150–1155. doi: 10.1016/j.cub.2005.05.047. [DOI] [PubMed] [Google Scholar]
- 12.Connolly L.A., Riccombeni A., Grózer Z., Holland L.M., Lynch D.B., Andes D.R., Gácser A., Butler G. The APSES transcription factor Efg1 is a global regulator that controls morphogenesis and biofilm formation in Candida parapsilosis. Mol. Microbiol. 2013;90:36–53. doi: 10.1111/mmi.12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stoldt V.R., Sonneborn A., Leuker C.E., Ernst J.F. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 1997;16:1982–1991. doi: 10.1093/emboj/16.8.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ernst J.F. Transcription factors in Candida albicans – environmental control of morphogenesis. Microbiology. 2000;146:1763–1774. doi: 10.1099/00221287-146-8-1763. [DOI] [PubMed] [Google Scholar]
- 15.Ramage G., VandeWalle K., López-Ribot J.L., Wickes B.L. The filamentation pathway controlled by the Efg1 regulator protein is required for normal biofilm formation and development in Candida albicans. FEMS Microbiol. Lett. 2002;214:95–100. doi: 10.1111/j.1574-6968.2002.tb11330.x. [DOI] [PubMed] [Google Scholar]
- 16.Eggimann P., Garbino J., Pittet D. Epidemiology of Candida species infections in critically ill non-immunosuppressed patients. Lancet Infect. Dis. 2003;3:685–702. doi: 10.1016/s1473-3099(03)00801-6. [DOI] [PubMed] [Google Scholar]
- 17.Negri M., Silva S., Henriques M., Oliveira R. Insights into Candida tropicalis nosocomial infections and virulence factors. Eur. J. Clin. Microbiol. Infect. Dis. 2012;31:1399–1412. doi: 10.1007/s10096-011-1455-z. [DOI] [PubMed] [Google Scholar]
- 18.Van Hauwermeiren F., Vandenbroucke R.E., Grine L., Puimège L., Van Wonterghem E., Zhang H., Libert C. Antisense oligonucleotides against TNFR1 prevent toxicity of TNF/IFNγ treatment in mouse tumor models. Int. J. Cancer. 2014;135:742–750. doi: 10.1002/ijc.28704. [DOI] [PubMed] [Google Scholar]
- 19.Costerton W.J., Montanaro L., Balaban N., Arciola C.R. Prospecting gene therapy of implant infections. Int. J. Artif. Organs. 2009;32:689–695. doi: 10.1177/039139880903200919. [DOI] [PubMed] [Google Scholar]
- 20.Kenney S.P., Meng X.-J. Therapeutic targets for the treatment of hepatitis E virus infection. Expert Opin. Ther. Targets. 2015;19:1245–1260. doi: 10.1517/14728222.2015.1056155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warren T.K., Whitehouse C.A., Wells J., Welch L., Charleston J.S., Heald A., Nichols D.K., Mattix M.E., Palacios G., Kugleman J.R. Delayed time-to-treatment of an antisense morpholino oligomer is effective against lethal marburg virus infection in cynomolgus macaques. PLoS Negl. Trop. Dis. 2016;10:e0004456. doi: 10.1371/journal.pntd.0004456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng Y.B., Nong L.G., Huang W., Pang G.G., Wang Y.F. Inhibition of hepatitis B virus (HBV) replication using antisense LNA targeting to both S and C genes in HBV. Zhonghua Gan Zang Bing Za Zhi. 2009;17:900–904. [PubMed] [Google Scholar]
- 23.Testa S., Disney M., Gryaznov S., Turner D. 2000. Methods and compositions for inhibition of RNA splicing. U.S. pub. no. WO/2000/055374, filled March 15, 2005, and granted April 19, 2017. [Google Scholar]
- 24.Sully E.K., Geller B.L. Antisense antimicrobial therapeutics. Curr. Opin. Microbiol. 2016;33:47–55. doi: 10.1016/j.mib.2016.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Disney M.D., Haidaris C.G., Turner D.H. Uptake and antifungal activity of oligonucleotides in Candida albicans. Proc. Natl. Acad. Sci. USA. 2003;100:1530–1534. doi: 10.1073/pnas.0337462100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan J.H., Lim S., Wong W.S. Antisense oligonucleotides: from design to therapeutic application. Clin. Exp. Pharmacol. Physiol. 2006;33:533–540. doi: 10.1111/j.1440-1681.2006.04403.x. [DOI] [PubMed] [Google Scholar]
- 27.Kurreck J. Antisense technologies. Improvement through novel chemical modifications. Eur. J. Biochem. 2003;270:1628–1644. doi: 10.1046/j.1432-1033.2003.03555.x. [DOI] [PubMed] [Google Scholar]
- 28.Khvorova A., Watts J.K. The chemical evolution of oligonucleotide therapies of clinical utility. Nat. Biotechnol. 2017;35:238–248. doi: 10.1038/nbt.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dias N., Stein C.A. Antisense oligonucleotides: basic concepts and mechanisms. Mol. Cancer Ther. 2002;1:347–355. [PubMed] [Google Scholar]
- 30.DeVos S.L., Miller T.M. Antisense oligonucleotides: treating neurodegeneration at the level of RNA. Neurotherapeutics. 2013;10:486–497. doi: 10.1007/s13311-013-0194-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stein C.A., Castanotto D. FDA-approved oligonucleotide therapies in 2017. Mol. Ther. 2017;25:1069–1075. doi: 10.1016/j.ymthe.2017.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zamecnik P.C., Stephenson M.L. Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proc. Natl. Acad. Sci. USA. 1978;75:280–284. doi: 10.1073/pnas.75.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizuta T., Fujiwara M., Hatta T., Abe T., Miyano-Kurosaki N., Shigeta S., Yokota T., Takaku H. Antisense oligonucleotides directed against the viral RNA polymerase gene enhance survival of mice infected with influenza A. Nat. Biotechnol. 1999;17:583–587. doi: 10.1038/9893. [DOI] [PubMed] [Google Scholar]
- 34.Hou J., Liu Z., Gu F. Epidemiology and prevention of hepatitis B virus infection. Int. J. Med. Sci. 2005;2:50–57. doi: 10.7150/ijms.2.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holden K.L., Stein D.A., Pierson T.C., Ahmed A.A., Clyde K., Iversen P.L., Harris E. Inhibition of dengue virus translation and RNA synthesis by a morpholino oligomer targeted to the top of the terminal 3′ stem-loop structure. Virology. 2006;344:439–452. doi: 10.1016/j.virol.2005.08.034. [DOI] [PubMed] [Google Scholar]
- 36.Warren T.K., Warfield K.L., Wells J., Swenson D.L., Donner K.S., Van Tongeren S.A., Garza N.L., Dong L., Mourich D.V., Crumley S. Advanced antisense therapies for postexposure protection against lethal filovirus infections. Nat. Med. 2010;16:991–994. doi: 10.1038/nm.2202. [DOI] [PubMed] [Google Scholar]
- 37.Deng Y.B., Qin H.J., Luo Y.H., Liang Z.R., Zou J.J. Antiviral effect of hepatitis B virus S/C gene loci antisense locked nucleic acid on transgenic mice in vivo. Genet. Mol. Res. 2015;14:10087–10095. doi: 10.4238/2015.August.21.16. [DOI] [PubMed] [Google Scholar]
- 38.Singh P., Panda D. FtsZ inhibition: a promising approach for antistaphylococcal therapy. Drug News Perspect. 2010;23:295–304. doi: 10.1358/dnp.2010.23.5.1429489. [DOI] [PubMed] [Google Scholar]
- 39.Sawyer A.J., Wesolowski D., Gandotra N., Stojadinovic A., Izadjoo M., Altman S., Kyriakides T.R. A peptide-morpholino oligomer conjugate targeting Staphylococcus aureus gyrA mRNA improves healing in an infected mouse cutaneous wound model. Int. J. Pharm. 2013;453:651–655. doi: 10.1016/j.ijpharm.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stein C.A. The experimental use of antisense oligonucleotides: a guide for the perplexed. J. Clin. Invest. 2001;108:641–644. doi: 10.1172/JCI13885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawasaki A.M., Casper M.D., Freier S.M., Lesnik E.A., Zounes M.C., Cummins L.L., Gonzalez C., Cook P.D. Uniformly modified 2′-deoxy-2′-fluoro phosphorothioate oligonucleotides as nuclease-resistant antisense compounds with high affinity and specificity for RNA targets. J. Med. Chem. 1993;36:831–841. doi: 10.1021/jm00059a007. [DOI] [PubMed] [Google Scholar]
- 42.Partridge M., Vincent A., Matthews P., Puma J., Stein D., Summerton J. A simple method for delivering morpholino antisense oligos into the cytoplasm of cells. Antisense Nucleic Acid Drug Dev. 1996;6:169–175. doi: 10.1089/oli.1.1996.6.169. [DOI] [PubMed] [Google Scholar]
- 43.Cohen J.S., Devor E., Goodchild J. Designing antisense oligonucleotides as pharmaceutical agents. Trends Pharmacol. Sci. 1989;10:435–437. doi: 10.1016/S0165-6147(89)80004-5. [DOI] [PubMed] [Google Scholar]
- 44.Almeida C., Azevedo N.F., Santos S., Keevil C.W., Vieira M.J. Discriminating multi-species populations in biofilms with peptide nucleic acid fluorescence in situ hybridization (PNA FISH) PLoS ONE. 2011;6:e14786. doi: 10.1371/journal.pone.0014786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rocha R., Santos R.S., Madureira P., Almeida C., Azevedo N.F. Optimization of peptide nucleic acid fluorescence in situ hybridization (PNA-FISH) for the detection of bacteria: The effect of pH, dextran sulfate and probe concentration. J. Biotechnol. 2016;226:1–7. doi: 10.1016/j.jbiotec.2016.03.047. [DOI] [PubMed] [Google Scholar]
- 46.Vilas Boas D., Almeida C., Sillankorva S., Nicolau A., Azeredo J., Azevedo N.F. Discrimination of bacteriophage infected cells using locked nucleic acid fluorescent in situ hybridization (LNA-FISH) Biofouling. 2016;32:179–190. doi: 10.1080/08927014.2015.1131821. [DOI] [PubMed] [Google Scholar]
- 47.Ferreira A.M., Cruz-Moreira D., Cerqueira L., Miranda J.M., Azevedo N.F. Yeasts identification in microfluidic devices using peptide nucleic acid fluorescence in situ hybridization (PNA-FISH) Biomed. Microdevices. 2017;19:11. doi: 10.1007/s10544-017-0150-y. [DOI] [PubMed] [Google Scholar]
- 48.Asami Y., Yoshioka K., Nishina K., Nagata T., Yokota T. Drug delivery system of therapeutic oligonucleotides. Drug Discov. Ther. 2016;10:256–262. doi: 10.5582/ddt.2016.01065. [DOI] [PubMed] [Google Scholar]
- 49.Juliano R.L. Intracellular trafficking and endosomal release of oligonucleotides: what we know and what we don’t. Nucleic Acid Ther. 2018;28:166–177. doi: 10.1089/nat.2018.0727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen C., Yang Z., Tang X. Chemical modifications of nucleic acid drugs and their delivery systems for gene-based therapy. Med. Res. Rev. 2018;38:829–869. doi: 10.1002/med.21479. [DOI] [PubMed] [Google Scholar]
- 51.International Organization for Standardization . third edition. 2009. ISO 10993-5:2009. Biological evaluation of medical devices. Part 5: tests for in vitro cytotoxicity.https://www.iso.org/standard/68936.html (2009-06-01) [Google Scholar]
- 52.Liu H., Köhler J., Fink G.R. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- 53.Nobile C.J., Andes D.R., Nett J.E., Smith F.J., Yue F., Phan Q.T., Edwards J.E., Filler S.G., Mitchell A.P. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2006;2:0636–0649. doi: 10.1371/journal.ppat.0020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wan Harun W.H., Jamil N.A., Jamaludin N.H., Nordin M.A. Effect of Piper betle and Brucea javanica on the differential expression of hyphal wall protein (HWP1) in non-Candida albicans Candida (NCAC) species. Evid. Based Complement. Alternat. Med. 2013;2013:397268. doi: 10.1155/2013/397268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Orsi C.F., Borghi E., Colombari B., Neglia R.G., Quaglino D., Ardizzoni A., Morace G., Blasi E. Impact of Candida albicans hyphal wall protein 1 (HWP1) genotype on biofilm production and fungal susceptibility to microglial cells. Microb. Pathog. 2014;69-70:20–27. doi: 10.1016/j.micpath.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 56.Biswas S., Van Dijck P., Datta A. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol. Mol. Biol. Rev. 2007;71:348–376. doi: 10.1128/MMBR.00009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sudbery P.E. Growth of Candida albicans hyphae. Nat. Rev. Microbiol. 2011;9:737–748. doi: 10.1038/nrmicro2636. [DOI] [PubMed] [Google Scholar]
- 58.Smith R.A., Miller T.M., Yamanaka K., Monia B.P., Condon T.P., Hung G., Lobsiger C.S., Ward C.M., McAlonis-Downes M., Wei H. Antisense oligonucleotide therapy for neurodegenerative disease. J. Clin. Invest. 2006;116:2290–2296. doi: 10.1172/JCI25424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Keiser M.S., Kordasiewicz H.B., McBride J.L. Gene suppression strategies for dominantly inherited neurodegenerative diseases: lessons from Huntington’s disease and spinocerebellar ataxia. Hum. Mol. Genet. 2016;25(R1):R53–R64. doi: 10.1093/hmg/ddv442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harada T., Matsumoto S., Hirota S., Kimura H., Fujii S., Kasahara Y., Gon H., Yoshida T., Itoh T., Haraguchi N. Chemically modified antisense oligonucleotide against ARL4C inhibits primary and metastatic liver tumor growth. Mol. Cancer Ther. 2019;18:602–612. doi: 10.1158/1535-7163.MCT-18-0824. [DOI] [PubMed] [Google Scholar]
- 61.Luna Velez M.V., Verhaegh G.W., Smit F., Sedelaar J.P.M., Schalken J.A. Suppression of prostate tumor cell survival by antisense oligonucleotide-mediated inhibition of AR-V7 mRNA synthesis. Oncogene. 2019;38:3696–3709. doi: 10.1038/s41388-019-0696-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lo H.J., Köhler J.R., DiDomenico B., Loebenberg D., Cacciapuoti A., Fink G.R. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- 63.Altmann K.H., Fabbro D., Dean N.M., Geiger T., Monia B.P., Müller M., Nicklin P. Second-generation antisense oligonucleotides: structure-activity relationships and the design of improved signal-transduction inhibitors. Biochem. Soc. Trans. 1996;24:630–637. doi: 10.1042/bst0240630. [DOI] [PubMed] [Google Scholar]
- 64.Vickers T.A., Crooke S.T. The rates of the major steps in the molecular mechanism of RNase H1-dependent antisense oligonucleotide induced degradation of RNA. Nucleic Acids Res. 2015;43:8955–8963. doi: 10.1093/nar/gkv920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Almeida C., Azevedo N.F., Iversen C., Fanning S., Keevil C.W., Vieira M.J. Development and application of a novel peptide nucleic acid probe for the specific detection of Cronobacter genomospecies (Enterobacter sakazakii) in powdered infant formula. Appl. Environ. Microbiol. 2009;75:2925–2930. doi: 10.1128/AEM.02470-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fonseca E., Silva S., Rodrigues C.F., Alves C.T., Azeredo J., Henriques M. Effects of fluconazole on Candida glabrata biofilms and its relationship with ABC transporter gene expression. Biofouling. 2014;30:447–457. doi: 10.1080/08927014.2014.886108. [DOI] [PubMed] [Google Scholar]
- 67.Rodrigues C.F., Henriques M. Portrait of matrix gene expression in Candida glabrata biofilms with stress induced by different drugs. Genes (Basel) 2018;9:205. doi: 10.3390/genes9040205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nailis H., Coenye T., Van Nieuwerburgh F., Deforce D., Nelis H.J. Development and evaluation of different normalization strategies for gene expression studies in Candida albicans biofilms by real-time PCR. BMC Mol. Biol. 2006;7:25. doi: 10.1186/1471-2199-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Almeida A., Ferreira J.A., Teixeira F., Gomes C., Cordeiro M.N., Osório H., Santos L.L., Reis C.A., Vitorino R., Amado F. Challenging the limits of detection of sialylated Thomsen-Friedenreich antigens by in-gel deglycosylation and nano-LC-MALDI-TOF-MS. Electrophoresis. 2013;34:2337–2341. doi: 10.1002/elps.201300148. [DOI] [PubMed] [Google Scholar]
- 70.Pitarch A., Sánchez M., Nombela C., Gil C. Sequential fractionation and two-dimensional gel analysis unravels the complexity of the dimorphic fungus Candida albicans cell wall proteome. Mol. Cell. Proteomics. 2002;1:967–982. doi: 10.1074/mcp.m200062-mcp200. [DOI] [PubMed] [Google Scholar]
- 71.Wiśniewski J.R., Zougman A., Nagaraj N., Mann M. Universal sample preparation method for proteome analysis. Nat. Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 72.Michalski A., Damoc E., Hauschild J.P., Lange O., Wieghaus A., Makarov A., Nagaraj N., Cox J., Mann M., Horning S. Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole orbitrap mass spectrometer. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M111.011015. M111.011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Silva S., Negri M., Henriques M., Oliveira R., Williams D., Azeredo J. Silicone colonization by non-Candida albicans Candida species in the presence of urine. J. Med. Microbiol. 2010;59:747–754. doi: 10.1099/jmm.0.017517-0. [DOI] [PubMed] [Google Scholar]
- 74.Silva S., Pires P., Monteiro D.R., Negri M., Gorup L.F., Camargo E.R., Barbosa D.B., Oliveira R., Williams D.W., Henriques M., Azeredo J. The effect of silver nanoparticles and nystatin on mixed biofilms of Candida glabrata and Candida albicans on acrylic. Med. Mycol. 2013;51:178–184. doi: 10.3109/13693786.2012.700492. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.