Abstract
Fragile X-associated tremor ataxia syndrome (FXTAS) is a rare disorder associated to the presence of the fragile X premutation, a 55–200 CGG repeat expansion in the 5′ UTR of the FMR1 gene. Two main neurological phenotypes have been described in carriers of the CGG premutation: (1) neurodevelopmental disorders characterized by anxiety, attention deficit hyperactivity disorder (ADHD), social deficits, or autism spectrum disorder (ASD); and (2) after 50 years old, the FXTAS phenotype. This neurodegenerative disorder is characterized by ataxia and a form of parkinsonism. The molecular pathology of this disorder is characterized by the presence of elevated levels of Fragile X Mental Retardation 1 (FMR1) mRNA, presence of a repeat-associated non-AUG (RAN) translated peptide, and FMR1 mRNA-containing nuclear inclusions. Whereas in the past FXTAS was mainly considered as a late-onset disorder, some phenotypes of patients and altered learning and memory behavior of a mouse model of FXTAS suggested that this disorder involves neurodevelopment. To better understand the physiopathological role of the increased levels of Fmr1 mRNA during neuronal differentiation, we used a small interfering RNA (siRNA) approach to reduce the abundance of this mRNA in cultured cortical neurons from the FXTAS mouse model. Morphological alterations of neurons were rescued by this approach. This cellular phenotype is associated to differentially expressed proteins that we identified by mass spectrometry analysis. Interestingly, phenotype rescue is also associated to the rescue of the abundance of 29 proteins that are involved in various pathways, which represent putative targets for early therapeutic approaches.
Keywords: FXTAS, premutation, Fmr1 mRNA, dendritic arborization, Tia1, ROAA, Hnrnpll, Aldh4a1/P5CDH, dendrtic spine
Introduction
The Fragile X Mental Retardation 1 (FMR1) gene encodes the fragile X mental retardation protein (FMRP), an RNA binding protein whose functional absence causes fragile X syndrome (FXS), the most common form of intellectual disability (ID) and autism spectrum disorder (ASD). The mutation in the FMR1 gene causing FXS is the presence of a repeated sequence encompassing >200 CGG repeats in its 5′ UTR. Hypermethylation of this sequence determines gene promoter inactivation, causing the silencing of the FMR1 gene.1 Although 6–54 CGG repeats in the 5′ UTR of FMR1 is a polymorphism in normal individuals, a repeat sequence of variable length (55–200 CGG repeats) represents the premutation1 that can cause fragile X tremor ataxia syndrome (FXTAS) in patients over 50 years of age.2, 3 This is an adult-onset progressive neurodegenerative disorder leading to a variable combination of ataxia, essential tremor, gait imbalance, parkinsonism, peripheral neuropathy, anxiety, and cognitive decline, occurring predominantly in older men carrying the premutation. It is known that people carrying the premutation have a reduced hippocampal volume that correlates with impaired performance in standardized tests of memory.3 At the cellular level, this disorder is characterized by the presence of eosinophilic, ubiquitin-positive nuclear inclusions, which have been observed throughout the brain, with a high percentage being located in the hippocampus of patients, as well as in the animal model of the disease.3, 4 Inclusions are negative for tau isoforms, alpha-synuclein, or polyglutamine peptides, reflecting a new class of inclusion disorder.3 At the molecular level, FXTAS is characterized by an elevated (2- to 8-fold) level of FMR1 mRNA, whereas the level of FMRP is normal or slightly reduced in patients, as well in the CGG-KI mouse model.2, 3, 4 The FMR1 mRNA is a component of nuclear inclusions3. The product of repeat-associated non-ATG (RAN) translation of the FMR1 mRNA was also reported to be involved in the generation of inclusions when overexpressed.5, 6 Although FXTAS is a late-onset disorder, it is also characterized by a set of developmental hallmarks, such as self-reported memory problems, autism-related traits, attention deficit hyperactivity disorder (ADHD), executive functioning, and psychopathology.7 Knockin (KI) mouse models have been generated displaying both neurodegenerative and neurodevelopmental phenotypes. It was reported that neuronal abnormalities and behavioral alterations in the animal model are present before the appearance of neuronal inclusions. Indeed, cultured hippocampal neurons obtained from a CGG-KI display shorter dendritic length and reduced dendritic complexity.4, 8, 9, 10 In CGG-KI mice, cortical migration is also affected. Indeed, at embryonic day (E) 17, a 2-fold higher percentage of migrating neurons was detected to be oriented toward the ventricle in wild-type (WT) compared with CGG-KI mice.11 All of these abnormalities were observed in inclusion-free cells, because CGG-KI mice display ubiquitin-positive intranuclear inclusions in neurons and astrocytes of the hippocampus and cerebellar internal cell layer starting at 12 weeks of age.4, 8, 9, 11 Here we investigated the impact of normalization of Fmr1 mRNA levels on the morphology and proteomics of cortical cultured neurons obtained from a FXTAS mouse model (CGG-KI)8, 9 before the generation of nuclear inclusions. By this analysis we have obtained a deeper insight into the physiopathological role of Fmr1 mRNA levels in FXTAS and identified putative targetable pathways for early treatments.
Results
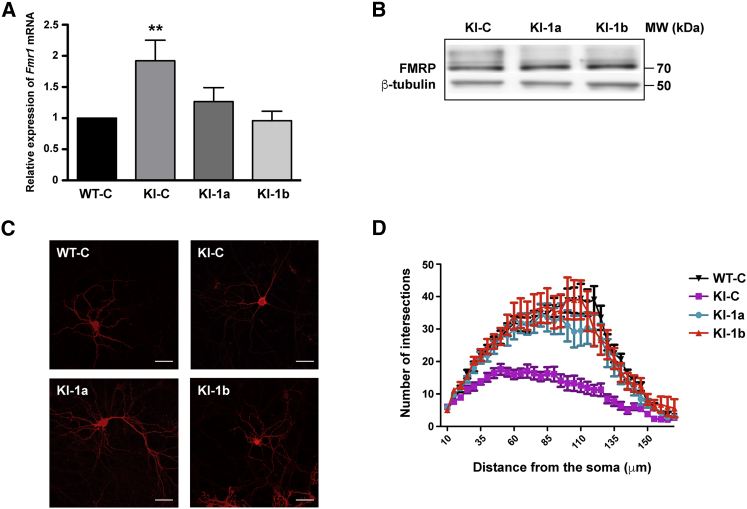
Hippocampal neurons were isolated from knockin-CGG (KI-CGG) E15.5 mice harboring the CGG premutated allele. It has been shown that these mice display abnormal cortical neuron migration patterns in utero.10, 11 Furthermore, abnormal dendritic architecture and reduced cellular viability have been observed in hippocampal primary neurons of KI-CGG mice, where increased expression of Fmr1 is already present even if nuclear inclusions are not detected.10 Indeed, nuclear inclusions appear first in the hippocampus of these mice at the age of 3 months, whereas the same hallmark appears later in the parietal neocortex.8, 9 We decided to explore whether cultured cortical neurons obtained from these mice have an abnormal morphology of dendrites and axons during development before the appearance of nuclear inclusions. We studied the dendritic arborization of wild-type (WT) and KI-CGG cortical neurons by Sholl analysis, as previously described,12 and observed that KI-CGG neurons have a reduced arborization compared with controls (Figures S1A and S1B). To assess whether Fmr1 mRNA levels impact the morphology of these cultured neurons, we produced lentiviruses expressing two different shRNAs selectively reducing the expression of Fmr1 mRNA (sh-1a and sh-1b) and one shRNA control (sh-C),13 and we used them to transduce cortical cultured neurons obtained from WT and from KI-CGG mice. The infection was performed at 5 DIV (days in vitro), and RNA and proteins were prepared from these cultures at 20 DIV. As expected,4, 13 Fmr1 mRNA levels were elevated 2-fold in cultured KI cortical neurons compared with WT. sh-1a reduced Fmr1 transcript levels by 30%, whereas sh-1b reduced them by 50%, (Figure 1A). FMRP levels are not changed in KI cultured neurons compared with WT,10, 13 and as previously shown in patients.2, 3 Similarly, the expression level of FMRP was not affected by Fmr1 knockdown in KI neurons (Figure 1B), as we already observed by transfecting fibroblasts obtained from FXTAS patients with the same shRNAs.13 We then analyzed the arborization of WT and KI cortical neurons transduced with Fmr1 shRNAs. We confirmed that KI neurons transduced with the lentivirus expressing the control shRNA (KI-C) are less arborized than WT neurons transduced with the same lentivirus (Figures 1C and 1D). However, KI neurons transduced with lentivirus expressing either sh-1a or sh-1b displayed a normal dendritic arborization (Figures 1C and 1D).
Figure 1.
Role of Fmr1 mRNA Levels in Dendritic Arborization
(A) RNA was prepared from cultured WT and knockin-CGG (KI-CGG) neurons transduced with the control (C) shRNA or two different shRNAs directed against Fmr1 mRNA (1a and 1b). The level of Fmr1 mRNA was measured by qRT-PCR using specific primers. Ten different experiments have been carried out for each transduced lentivirus, and a reduction of Fmr1 levels was observed by using both shRNAs. Results are presented as mean ± SEM; one-way ANOVA with Tukey’s multiple comparisons test, **p < 0.01. (B) Representative western blot analysis of cell cultures of cortical neurons transduced with C, 1a, or1b shRNAs. FMRP (70 kDa) and β-tubulin (50 kDa) were revealed with specific antibodies. (C) Image of WT and KI-CGG cortical neurons transduced with lentiviruses expressing C, 1a, or 1b shRNAs. Scale bars: 20 μm. (D) Sholl analysis of WT and KI cultured mouse cortical neurons transduced with C or 1a or 1b shRNAs. Reduced arborization of KI-CGG neurons is rescued by Fmr1 knockdown. Two-way ANOVA was used to compare KI-C and 1a treatments: genotype F(2; 1,527) = 227.9, p < 0.0001; treatment (34; 1,527) = 34.12, p < 0.0001; interaction F(68; 1,527) = 3.067, p < 0.0001; two-way ANOVA was used to compare KI-C and 1b treatments: genotype F(2; 1,318) = 293.6, ****p < 0.0001; treatment F(34; 1,318) = 36.21, ****p < 0.0001; interaction F(68; 1,318) = 3,803, p < 0.0001.
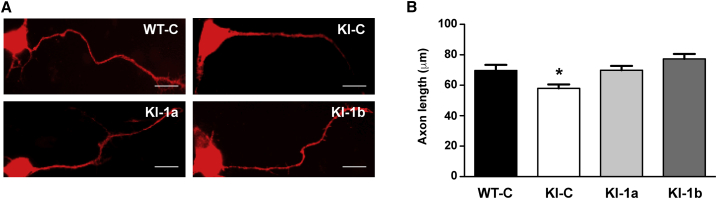
In 2 DIV neurons we measured the axon length, and we found that they are significantly shorter in Fmr1-KI neurons compared with WT (Figure S2). We confirmed that KI-C neurons have shorter axons than WT-C, but the length of these axons was normalized in KI-1a and KI-1b neurons (Figures 2A and 2B).
Figure 2.
Fmr1 mRNA Levels Are Associated to Axon Growth
(A) Representative pictures of 2 days in vitro cultured WT-C, KI-C, KI-1a, and KI-1b primary cortical neurons. Scale bars: 10 μm. (B) Histogram of axon length of WT and KI cultured neurons transfected with C, 1a, or 1b shRNAs. Results show mean axon length ± SEM of 150 randomly selected cells for each condition from three independent cultures. One-way ANOVA with Tukey’s multiple comparisons test, *p < 0.05.
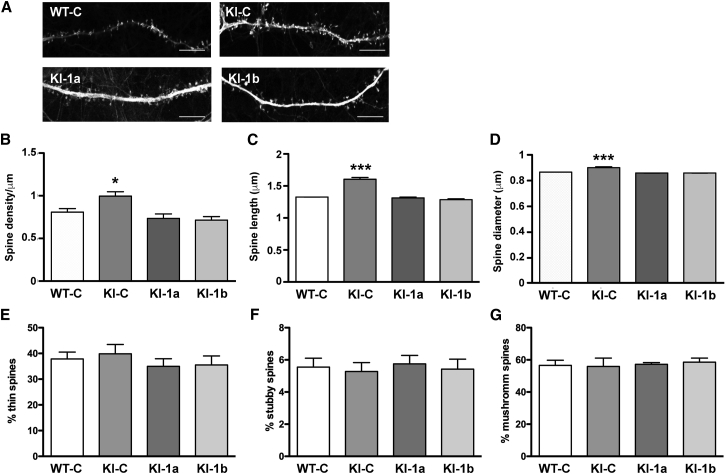
We then focused on dendritic spine abnormalities. The absence of FMRP causes a peculiar morphology of dendritic spines that has been described in FXS patients and in the mouse model, where it is particularly evident in young animals.14, 15 Conversely, the study of dendritic spines in the brain of FXTAS patients was never reported, and in Fmr1 KI mice it was studied only in hippocampi and in the visual cortex from adult animals, displaying an increased length of spines.4, 16 Here we studied the morphology of dendritic spines of 20 DIV cultured cortical neurons. KI-C spines are longer, denser, and larger than controls. All of these hallmarks are rescued in KI-1a and KI-1b neurons (Figures 3A–3D), indicating that the elevated level of Fmr1 mRNA causes a set of morphological alterations in KI-CGG neurons. However, the percentage of the different types of spines (thin, stubby, and mushroom) is not significantly different in the two genotypes (Figures 3E–3G).
Figure 3.
Reduction of Fmr1 mRNA Levels Normalizes Dendritic Spine Morphology in KI Cortical Neurons
(A) Representative high-resolution confocal images showing dendritic spine morphology were assessed using NeuronStudio software and compared with the measurement obtained from control transduced neurons. Scale bars: 2 μm. All histograms present mean ± SEM values. Statistical significance was assessed by one-way ANOVA with Tukey’s multiple comparisons test. (B–G) Histograms showing the mean ± SEM values of (B) protrusion frequency in the various cultures, *p < 0.05; (C) spine length, ***p < 0.001; (D) spine head size, ***p < 0.001; (E) percentage of thin spines; (F) percentage of stubby spines; and (G) percentage of mushroom spines.
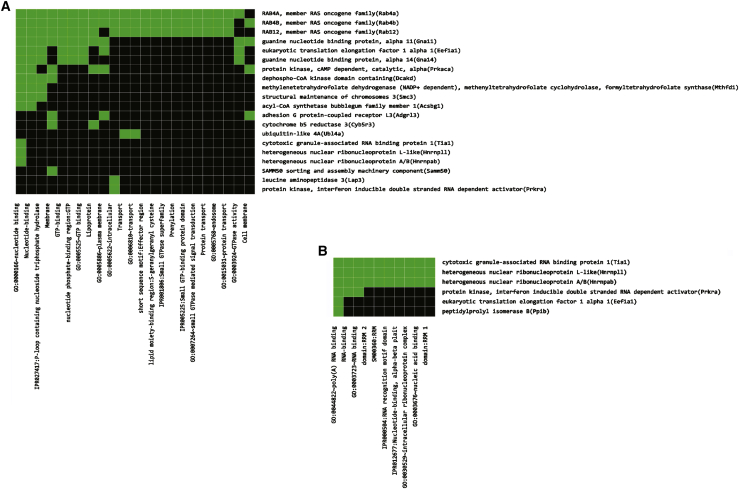
To get deeper insight into the molecular pathology of FXTAS cultured neurons, we performed a proteomic analysis of neurons that have been transduced with control shRNA or with the shRNAs targeting the Fmr1 mRNA. Protein extracts of three replicates for each condition were analyzed by nano-liquid chromatography-tandem mass spectrometry (nano-LC-MS/MS) after tryptic digestion.17 A total of 487 proteins were identified (Table S1), among which 251 have a ratio >2 (upregulation) or <0.5 (downregulation) for at least one of the three comparisons (WT-C versus KI-C, WT-C versus KI-1a, and WT-C versus KI-1b) with a p value <0.10. Interestingly, the abundance of 29 differentially expressed proteins is rescued after reduction of Fmr1 mRNA by using both specific shRNAs (Table 1). Gene Ontology analysis of these proteins reveals a significant enrichment of proteins involved in different biochemical pathways, among which are GTP binding and RNA binding proteins (Figure 4; Table S2).
Table 1.
Proteins Differentially Expressed in the Different Samples Studied
| Name | UniProt Accession Number | WT-C | KI-C | WT-C/Ki-C | Log (Fold Change) | p Value | KI-1a | WT-C/Ki-1a | Log (Fold Change) | p Value | KI-1b | WT-C/KI-1b | Log (Fold Change) | p Value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sp|O89023|TPP1_MOUSE | O89023 | 0.00 | 0.93 | 0.00 | NA | 0.00001 | 0.00 | NA | NA | 1.00000 | 0.00 | NA | NA | 1.00000 |
| sp|P05132|KAPCA_MOUSE | P05132 | 5.16 | 10.17 | 0.51 | −0.29 | 0.00132 | 5.83 | 0.89 | −0.05 | 0.81905 | 4.79 | 1.08 | 0.03 | 0.81905 |
| sp|P21126|UBL4A_MOUSE | P21126 | 0.33 | 1.53 | 0.21 | −0.67 | 0.04762 | 0.66 | 0.49 | −0.31 | 0.51266 | 0.38 | 0.85 | −0.07 | 0.51266 |
| sp|P21278|GNA11_MOUSE | P21278 | 7.43 | 4.06 | 1.83 | 0.26 | 0.03470 | 6.49 | 1.14 | 0.06 | 0.69780 | 7.98 | 0.93 | −0.03 | 0.69780 |
| sp|P24369|PPIB_MOUSE | P24369 | 8.43 | 12.31 | 0.68 | −0.16 | 0.03540 | 6.73 | 1.25 | 0.10 | 0.68137 | 9.91 | 0.85 | −0.07 | 0.68137 |
| sp|P30677|GNA14_MOUSE | P30677 | 2.57 | 0.93 | 2.77 | 0.44 | 0.04928 | 2.40 | 1.07 | 0.03 | 0.88974 | 3.24 | 0.79 | −0.10 | 0.88974 |
| sp|P35283|RAB12_MOUSE | P35283 | 5.82 | 3.08 | 1.89 | 0.28 | 0.00979 | 5.87 | 0.99 | 0.00 | 0.95886 | 5.51 | 1.06 | 0.02 | 0.95886 |
| sp|P52912|TIA1_MOUSE | P52912 | 0.00 | 0.93 | 0.00 | NA | 0.00001 | 0.00 | NA | NA | 1.00000 | 0.32 | 0.00 | NA | 1.00000 |
| sp|P56371|RAB4A_MOUSE | P56371 | 5.81 | 3.08 | 1.89 | 0.28 | 0.00560 | 5.87 | 0.99 | 0.00 | 0.94159 | 5.19 | 1.12 | 0.05 | 0.94159 |
| sp|Q80TS3|AGRL3_MOUSE | Q80TS3 | 0.97 | 2.16 | 0.45 | −0.35 | 0.01678 | 0.37 | 2.64 | 0.42 | 0.17665 | 0.38 | 2.53 | 0.40 | 0.17665 |
| sp|Q8BGH2|SAM50_MOUSE | Q8BGH2 | 2.28 | 0.29 | 7.74 | 0.89 | 0.01329 | 2.07 | 1.10 | 0.04 | 0.79778 | 2.13 | 1.07 | 0.03 | 0.79778 |
| sp|Q8BHC4|DCAKD_MOUSE | Q8BHC4 | 0.00 | 0.93 | 0.00 | NA | 0.00001 | 0.31 | 0.00 | NA | 0.37390 | 0.00 | NA | NA | 0.37390 |
| sp|Q8CHT0|AL4A1_MOUSE | Q8CHT0 | 2.94 | 0.63 | 4.64 | 0.67 | 0.02926 | 2.43 | 1.21 | 0.08 | 0.54007 | 1.95 | 1.51 | 0.18 | 0.54007 |
| sp|Q91ZR1|RAB4B_MOUSE | Q91ZR1 | 5.82 | 3.68 | 1.58 | 0.20 | 0.02923 | 5.51 | 1.05 | 0.02 | 0.71442 | 5.19 | 1.12 | 0.05 | 0.71442 |
| sp|Q921F4|HNRLL_MOUSE | Q921F4 | 0.97 | 0.00 | ∞ | NA | 0.00000 | 0.98 | 0.99 | 0.00 | 0.98875 | 0.78 | 1.24 | 0.09 | 0.98875 |
| sp|Q922D8|C1TC_MOUSE | Q922D8 | 1.28 | 2.78 | 0.46 | −0.34 | 0.00809 | 0.92 | 1.40 | 0.15 | 0.72353 | 1.29 | 1.00 | 0.00 | 0.72353 |
| sp|Q99020|ROAA_MOUSE | Q99020 | 1.63 | 0.31 | 5.34 | 0.73 | 0.04589 | 1.69 | 0.97 | −0.02 | 0.90543 | 2.19 | 0.74 | −0.13 | 0.90543 |
| sp|Q99PU5|ACBG1_MOUSE | Q99PU5 | 2.92 | 1.55 | 1.88 | 0.27 | 0.01399 | 2.73 | 1.07 | 0.03 | 0.62466 | 2.61 | 1.12 | 0.05 | 0.62466 |
| sp|Q9CPY7|AMPL_MOUSE | Q9CPY7 | 1.61 | 3.38 | 0.48 | −0.32 | 0.00921 | 2.00 | 0.81 | −0.09 | 0.53465 | 1.43 | 1.13 | 0.05 | 0.53465 |
| sp|Q9CW03|SMC3_MOUSE | Q9CW03 | 0.33 | 1.53 | 0.21 | −0.67 | 0.04762 | 0.71 | 0.46 | −0.34 | 0.64890 | 0.38 | 0.85 | −0.07 | 0.64890 |
| sp|Q9CYR6|AGM1_MOUSE | Q9CYR6 | 0.33 | 1.86 | 0.18 | −0.75 | 0.01091 | 0.00 | ∞ | ∞ | 0.37390 | 0.32 | 1.04 | 0.02 | 0.37390 |
| sp|Q9DCN2|NB5R3_MOUSE | Q9DCN2 | 4.53 | 6.79 | 0.67 | −0.18 | 0.00385 | 4.79 | 0.95 | −0.02 | 0.62926 | 4.48 | 1.01 | 0.00 | 0.62926 |
| sp|Q9WTX2|PRKRA_MOUSE | Q9WTX2 | 0.33 | 1.53 | 0.21 | −0.67 | 0.04762 | 0.00 | ∞ | ∞ | 0.37390 | 0.00 | ∞ | ∞ | 0.37390 |
| sp|Q9Z0H8|CLIP2_MOUSE | Q9Z0H8 | 4.51 | 8.09 | 0.56 | −0.25 | 0.04003 | 4.68 | 0.96 | −0.02 | 0.88427 | 5.62 | 0.80 | −0.10 | 0.88427 |
| tr|A0A087WPM2|A0A087WPM2_MOUSE | A0A087WPM2 | 1.94 | 2.78 | 0.70 | −0.16 | 0.00099 | 1.64 | 1.18 | 0.07 | 0.61624 | 1.12 | 1.73 | 0.24 | 0.61624 |
| tr|A0AUM9|A0AUM9_MOUSE | A0AUM9 | 0.00 | 0.93 | 0.00 | NA | 0.00001 | 0.00 | NA | NA | 1.00000 | 0.00 | NA | NA | 1.00000 |
| tr|D3YZ68|D3YZ68_MOUSE | D3YZ68 | 39.80 | 47.10 | 0.84 | −0.07 | 0.03447 | 38.80 | 1.03 | 0.01 | 0.89853 | 36.50 | 1.09 | 0.04 | 0.89853 |
| tr|E9Q7C9|E9Q7C9_MOUSE | E9Q7C9 | 7.46 | 9.91 | 0.75 | −0.12 | 0.03009 | 5.75 | 1.30 | 0.11 | 0.10120 | 7.97 | 0.94 | −0.03 | 0.10120 |
| tr|S4R232|S4R232_MOUSE | S4R232 | 5.50 | 3.04 | 1.81 | 0.26 | 0.03279 | 5.50 | 1.00 | 0.00 | 0.99389 | 5.17 | 1.06 | 0.03 | 0.99389 |
NA, not applicable.
Figure 4.
Gene Ontology Classification of Rescued Proteins in KI-CGG Neurons
(A and B) The DAVID Gene Ontology Functional Annotation Clustering tool40 was used to show functional classification of rescued proteins in Ki-CGG neurons involved in (A) nucleotide-GTP binding and (B) RNA binding.
Discussion
Two main neurological phenotypes have been described in carriers of the CGG premutation: those exhibiting neurodevelopmental disorders characterized by anxiety, ADHD, social deficits, or ASD, and after 50 years old, FXTAS, a neurodegenerative disorder.3 For some time, nuclear inclusions have been considered as the cause of neurodegeneration, whereas more recent studies suggest that nuclear inclusions may represent a mechanism used by neurons to protect themselves from toxic events.4, 18, 19, 20 So far, two other main physiopathological elements are known to underpin the FXTAS phenotype: the elevated abundance of FMR1 mRNA2 and the presence of a RAN polypeptide.5, 6 We considered that it is crucial to unravel the role of each element in the molecular pathology of FXTAS to understand the progression of the disorder and its role in pathophysiology. Indeed, it is interesting to remind that neuronal abnormal dendritic morphology (e.g., reduced length and number of dendrites that display longer spines) has been observed in FXTAS neurons at a time of development when nuclear inclusions are not detectable.3, 4, 8, 9, 10, 11 These findings suggest that some developmental abnormalities may contribute to the late manifestation of neurodegenerative process and/or that the disease appears, with subtle phenotypes, earlier than predicted up to date. So far, no specific treatment is available for FXTAS patients. A therapy based on allopregnanolone was shown to improve cognitive functioning in patients with FXTAS and to partially alleviate some aspects of neurodegeneration, but no data are known for neurodevelopmental hallmarks,21 opening the possibility to search for new targetable pathways in young patients. In this study, we used a murine model of FXTAS to investigate the impact that the reduced level of Fmr1 mRNA, but not of its encoded protein, has on the morphological and molecular phenotypes of FXTAS cultured neurons. Certainly, by reducing Fmr1 mRNA levels, we are also supposed to reduce RAN levels, which is possibly involved in the pathophysiology of FXTAS.6 However, that peptide was never detected at endogenous levels in neurons so far.3, 22
First, we defined phenotypes that were never considered before in cultured cortical neurons obtained from the FXTAS mouse model: axon length and dendrite morphology. All of these new phenotypes are detected in cells not yet displaying nuclear inclusions, which are known to first appear in hippocampal neurons of 12-week-old KI-CGG mice.9 Interestingly, these animals have mild learning and memory deficits,4, 9 and this behavioral phenotype is consistent with the phenotype that we observed in cultured cortical neurons. Importantly, we confirm that these phenotypes are associated with the levels of Fmr1 mRNA because they are rescued after the expression of shRNAs specifically targeting this mRNA. We can deduce that an elevated abundance of Fmr1 mRNA may interfere with normal RNA metabolism. We can predict that the overexpression of Fmr1 mRNA may also likely interfere with the normal activity of RNA binding proteins or microRNAs (miRNAs) in neuronal soma, and at the synapse by competing for their binding at specific sites or at miRNA response elements (MREs).13 All of these considerations suggest that Fmr1 mRNA metabolism can be regulated by a large number of RNA binding proteins and can be co-regulated with a plethora of other RNAs. For this reason, as the second step of our study, we performed a differential proteomic analysis of neurons expressing control or Fmr1-specific shRNAs, and we observed that a set of proteins, whose expression is deregulated in KI-CGG neurons, is normalized after treatment. Consistently, several proteins rescued by Fmr1-specific shRNAs are RNA binding proteins (e.g., Tia1, Hnrnpll, and ROAA). In addition, some proteins whose levels were rescued by the reduction of Fmr1 mRNA abundance are involved in the regulation of actin cytoskeleton dynamics, which is known to modulate the morphology of neurons and, in particular, of dendritic spines. Defective synaptic actin regulation seems to be involved in different neurodevelopmental and psychiatric disorders.23 Interestingly, we have shown here that in KI-CGG neurons, dendritic spines are more numerous, and they appear overall longer and with a larger head, but the percentage of the various types of spines is unchanged. These results are consistent with a previous study displaying longer, but not immature, spines in the CGG-KI visual cortex.16 In addition, our findings suggest that, at least at 20 DIV, the elevated levels of Fmr1 mRNA do not interfere with the spine maturation process but cause subtle abnormalities of their features, which may have an impact on synaptic transmission. Other rescued proteins belong to the Rab-GTPase family, a sub-class of the RAS superfamily, which spatially and temporally orchestrates specific vesicular trafficking that is critical for synaptic function in neurons in brain developmental disorders,24 as well as in Parkinson’s disease.25 Along the same direction, another interesting protein is CLIP2, a cytoplasmic linker factor that is considered as a mediator between organelles and the cytoskeleton.26 Consistent with their role in neurons, mutants of this class of proteins are associated with impaired cognitive functions.21, 24, 27 Importantly, we also found mitochondrial proteins (e.g., pyrroline-5-carboxylate dehydrogenase [Aldh4a1/P5CDH] and Samm50) that are deregulated in CGG-KI neurons, confirming the importance of mitochondria in the pathophysiology of FXTAS, as previously reported by independent studies.28, 29 This result suggests that an altered mitochondrial function is probably involved in early phenotypes of this disorder. Remarkably, the levels of those proteins were normalized by the reduction of Fmr1 mRNA abundance. Thus, our data show molecular alterations that may contribute to explain the neurodevelopmental phenotypes of the mouse model and patients carrying the CGG premutation, thus providing an indication for future early therapies to treat CGG-premutation carriers. For instance, Aldh4a1/P5CDH is a mitochondrial matrix NAD(+)-dependent dehydrogenase that catalyzes the second step of the proline degradation pathway, converting pyrroline-5-carboxylate to glutamate. Altered function of this enzyme can result in a deregulation of the glutamate signaling, which is at the basis of various forms of brain disorders, as well as of Parkinson’s disease.30, 31 Thus, Aldh4a1/P5CDH could be a therapeutic target for FXTAS patients during different periods of life and phases of the disorder associated with the CGG premutation. Other targets may be small GTPases by using, in this case, a strategy based on the modulation of their regulators, as recently proposed.32, 33 In conclusion, by our approach, we defined altered pathways in FXTAS neurons that are due to the increased levels of Fmr1 mRNA and impact on neuronal morphology. They represent the molecular pathology underpinning the late FXTAS phenotype. In particular, differentially expressed proteins between WT and CGG-KI neurons are promising pharmacological targetable molecules for early therapeutic intervention for FXTAS. On the other side, future gene therapies could also target the CGG-repeat by excising it, as already shown in an induced pluripotent stem cell (iPSC) line.34
Materials and Methods
Animals
The experiments were performed following the Animals in Research: Reporting In Vivo Experiments (ARRIVE) guidelines.35 Fmr1 knockin (KI) and wild-type (WT) mice on a C57BL/6J congenic background were produced as described previously.36 All animals were generated and housed in groups of four in standard laboratory conditions (22°C, 55% ± 10% humidity, and 12-h light/12-h dark diurnal cycles) with food and water provided ad libitum. Animal care was conducted in accordance with the European Community Directive 2010/63/EU. All experiments were approved by the local ethics committee (Comité d’Ethique en Expérimentation Animale CIEPAL-AZUR N. 00788.01).
Lentivirus Generation
Lentivirus particles were produced as previously described.37
Axon Length Measurements
Dissociated neurons were transfected with lentivirus plasmids by using Nucleofector as previously described.13 Neurons were cultured for 48 h and then fixed. Axons were labeled with anti-Tuj1 antibody, and the size of individual growing axons (distance from the soma to the tip of the axon) was manually measured using the ImageJ software.15
Neuronal Culture and Dendritic Spine Morphology Analysis
Primary neurons were prepared from E15.5 pregnant C57BL/6 Fmr1CGG/y and WT mice as previously described.12, 15 Neurons (5 days in vitro) were transduced with lentivirus as previously described.12 Transduced neurons (20 days in vitro) were rinsed twice in PBS at room temperature (RT) after 19 h of transduction and then fixed with 4% paraformaldehyde and Triton-permeabilized. Sequential confocal images (512 × 220 pixels; zoom 3.0; average 4; speed 7) of GFP-expressing neurons were acquired with a 63× oil-immersion lens (NA 1.4) on an inverted Zeiss LSM780 confocal microscope. z series of seven to eight images of randomly selected secondary dendrites were analyzed using the NeuronStudio software, which allows for the automated detection and quantification of dendrite parameters and morphological classification, as previously performed.15
Protein Extraction and Western Blot Analysis
Immunoblotting was performed as follows: cells or grinded tissues were homogenized in lysis buffer, and debris was removed by centrifugation (20,000 × g, 10 min, 4°C).38 Protein content in the supernatant was measured using the Bradford assay (Bio-Rad), and samples were separated on NuPAGE Bis-Tris 4%–12% gels in MOPS buffer. Separated proteins were transferred to nitrocellulose membranes (Bio-Rad). Membranes were blocked with PBS-Tween (0.05%) and milk (5%), and incubated with primary antibodies overnight.
Antibodies
The monoclonal 1C3 anti-FMRP antibody39 was used at a 1:1,000 dilution, the anti-Tuj1 (BioLegend; TUJ1 1-15-56) antibody was used following the manufacturer’s instructions, and the monoclonal anti-β-tubulin antibody (Sigma) was used at a 1:10,000 dilution.
qRT-PCR
qPCR was performed on a LightCycler 480 (Roche) with MasterMix SYBRGreen (Roche) as previously described.13 Primers used to amplify Fmr1 mRNA were previously reported.37
Protein Identification and Analysis
Proteomic analysis was performed as previously described17 and resulted in the identification of around 2,000 proteins for each condition. These proteins were then compared to highlight the proteins identified under only one condition or under several conditions (“on/off” effect) and to highlight proteins predominantly identified in one condition relative to another (up if ratio >2, down if ratio <0.5). In order to compile a short list of differentially regulated proteins, a Student’s t test was performed after normalization of the spectral count quantification data. This allowed us to obtain a p value used to make this short list: from the global alignment table of the 2,888 different proteins, we focused on proteins with a p value threshold less than 0.10.
Statistical Analysis
Results are expressed as mean ± SEM. All statistical analyses were based on biological replicates. Appropriate statistical tests used for each experiment are described in the corresponding figure legends. All statistical analyses were carried out using GraphPad Prism version 6.0e.
Author Contributions
M.D., S.D., T.M., S.M., M.G., F.B., M.J., and M.C. performed the experiments; R.W. and R.K.H. provided material; T.M., E.L., and B.B. designed the experiments; M.C., E.L., R.K.H., and B.B. wrote the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
The authors are grateful to S. Zongaro, P. Hammann, L. Khunn, and S. Abekhoukh for help. This study was supported by CNRS, INSERM, Association Française contre les Myopathies (AFM), Fondation Recherche Médicale (FRM) grant DEQ20140329490, Fondation Recherche sur le Cerveau (FRC), and Agence Nationale de la Recherche grants ANR-15-CE16-0015 and ANR-11-LABX-0028-01. M.D. and S.C. were recipients of a fellowship from the international PhD LabEx “Signalife” Program. S.D. was a recipient of a MRES fellowship.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.09.018.
Supplemental Information
References
- 1.Bardoni B., Mandel J.L., Fisch G.S. FMR1 gene and fragile X syndrome. Am. J. Med. Genet. 2000;97:153–163. doi: 10.1002/1096-8628(200022)97:2<153::aid-ajmg7>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 2.Tassone F., Hagerman R.J., Taylor A.K., Gane L.W., Godfrey T.E., Hagerman P.J. Elevated levels of FMR1 mRNA in carrier males: a new mechanism of involvement in the fragile-X syndrome. Am. J. Hum. Genet. 2000;66:6–15. doi: 10.1086/302720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hagerman P. Fragile X-associated tremor/ataxia syndrome (FXTAS): pathology and mechanisms. Acta Neuropathol. 2013;126:1–19. doi: 10.1007/s00401-013-1138-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berman R.F., Buijsen R.A., Usdin K., Pintado E., Kooy F., Pretto D., Pessah I.N., Nelson D.L., Zalewski Z., Charlet-Bergeurand N. Mouse models of the fragile X premutation and fragile X-associated tremor/ataxia syndrome. J. Neurodev. Disord. 2014;6:25. doi: 10.1186/1866-1955-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kearse M.G., Green K.M., Krans A., Rodriguez C.M., Linsalata A.E., Goldstrohm A.C., Todd P.K. CGG Repeat-Associated Non-AUG Translation utilizes a cap-dependent scanning mechanism of initiation to produce toxic proteins. Mol. Cell. 2016;62:314–322. doi: 10.1016/j.molcel.2016.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sellier C., Buijsen R.A.M., He F., Natla S., Jung L., Tropel P., Gaucherot A., Jacobs H., Meziane H., Vincent A. Translation of Expanded CGG Repeats into FMRpolyG Is Pathogenic and May Contribute to Fragile X Tremor Ataxia Syndrome. Neuron. 2017;93:331–347. doi: 10.1016/j.neuron.2016.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hessl D., Grigsby J. Fragile X-associated tremor/ataxia syndrome: another phenotype of the fragile X gene. Clin. Neuropsychol. 2016;30:810–814. doi: 10.1080/13854046.2016.1186661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hunsaker M.R., Arque G., Berman R.F., Willemsen R., Hukema R.K. Mouse models of the fragile x premutation and the fragile X associated tremor/ataxia syndrome. Results Probl. Cell Differ. 2012;54:255–269. doi: 10.1007/978-3-642-21649-7_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hunsaker M.R., Wenzel H.J., Willemsen R., Berman R.F. Progressive spatial processing deficits in a mouse model of the fragile X premutation. Behav. Neurosci. 2009;123:1315–1324. doi: 10.1037/a0017616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y., Tassone F., Berman R.F., Hagerman P.J., Hagerman R.J., Willemsen R., Pessah I.N. Murine hippocampal neurons expressing Fmr1 gene premutations show early developmental deficits and late degeneration. Hum. Mol. Genet. 2010;19:196–208. doi: 10.1093/hmg/ddp479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cunningham C.L., Martínez Cerdeño V., Navarro Porras E., Prakash A.N., Angelastro J.M., Willemsen R., Hagerman P.J., Pessah I.N., Berman R.F., Noctor S.C. Premutation CGG-repeat expansion of the Fmr1 gene impairs mouse neocortical development. Hum. Mol. Genet. 2011;20:64–79. doi: 10.1093/hmg/ddq432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abekhoukh S., Sahin H.B., Grossi M., Zongaro S., Maurin T., Madrigal I., Kazue-Sugioka D., Raas-Rothschild A., Doulazmi M., Carrera P. New insights into the regulatory function of CYFIP1 in the context of WAVE- and FMRP-containing complexes. Dis. Model. Mech. 2017;10:463–474. doi: 10.1242/dmm.025809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zongaro S., Hukema R., D’Antoni S., Davidovic L., Barbry P., Catania M.V., Willemsen R., Mari B., Bardoni B. The 3′ UTR of FMR1 mRNA is a target of miR-101, miR-129-5p and miR-221: implications for the molecular pathology of FXTAS at the synapse. Hum. Mol. Genet. 2013;22:1971–1982. doi: 10.1093/hmg/ddt044. [DOI] [PubMed] [Google Scholar]
- 14.Nimchinsky E.A., Oberlander A.M., Svoboda K. Abnormal development of dendritic spines in FMR1 knock-out mice. J. Neurosci. 2001;21:5139–5146. doi: 10.1523/JNEUROSCI.21-14-05139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maurin T., Melancia F., Jarjat M., Castro L., Costa L., Delhaye S., Khayachi A., Castagnola S., Mota E., Di Giorgio A. Involvement of Phosphodiesterase 2A Activity in the Pathophysiology of Fragile X Syndrome. Cereb. Cortex. 2018;29:3241–3252. doi: 10.1093/cercor/bhy192. [DOI] [PubMed] [Google Scholar]
- 16.Berman R.F., Murray K.D., Arque G., Hunsaker M.R., Wenzel H.J. Abnormal dendrite and spine morphology in primary visual cortex in the CGG knock-in mouse model of the fragile X premutation. Epilepsia. 2012;53(Suppl 1):150–160. doi: 10.1111/j.1528-1167.2012.03486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rottloff S., Miguel S., Biteau F., Nisse E., Hammann P., Kuhn L., Chicher J., Bazile V., Gaume L., Mignard B. Proteome analysis of digestive fluids in Nepenthes pitchers. Ann. Bot. 2016;117:479–495. doi: 10.1093/aob/mcw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chonchaiya W., Schneider A., Hagerman R.J. Fragile X: a family of disorders. Adv. Pediatr. 2009;56:165–186. doi: 10.1016/j.yapd.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schluter E.W., Hunsaker M.R., Greco C.M., Willemsen R., Berman R.F. Distribution and frequency of intranuclear inclusions in female CGG KI mice modeling the fragile X premutation. Brain Res. 2012;1472:124–137. doi: 10.1016/j.brainres.2012.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Entezam A., Biacsi R., Orrison B., Saha T., Hoffman G.E., Grabczyk E., Nussbaum R.L., Usdin K. Regional FMRP deficits and large repeat expansions into the full mutation range in a new Fragile X premutation mouse model. Gene. 2007;395:125–134. doi: 10.1016/j.gene.2007.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J.Y., Trivedi A.M., Carrillo N.R., Yang J., Schneider A., Giulivi C., Adams P., Tassone F., Kim K., Rivera S.M. Open-Label Allopregnanolone Treatment of Men with Fragile X-Associated Tremor/Ataxia Syndrome. Neurotherapeutics. 2017;14:1073–1083. doi: 10.1007/s13311-017-0555-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma L., Herren A.W., Espinal G., Randol J., McLaughlin B., Martinez-Cerdeño V., Pessah I.N., Hagerman R.J., Hagerman P.J. Composition of the Intranuclear Inclusions of Fragile X-associated Tremor/Ataxia Syndrome. Acta Neuropathol Commun. 2019;7:143. doi: 10.1186/s40478-019-0796-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan Z., Kim E., Datta D., Lewis D.A., Soderling S.H. Synaptic Actin Dysregulation, a Convergent Mechanism of Mental Disorders? J. Neurosci. 2016;36:11411–11417. doi: 10.1523/JNEUROSCI.2360-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mignogna M.L., D’Adamo P. Critical importance of RAB proteins for synaptic function. Small GTPases. 2018;9:145–157. doi: 10.1080/21541248.2016.1277001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao Y., Wilson G.R., Stephenson S.E.M., Bozaoglu K., Farrer M.J., Lockhart P.J. The emerging role of Rab GTPases in the pathogenesis of Parkinson’s disease. Mov. Disord. 2018;33:196–207. doi: 10.1002/mds.27270. [DOI] [PubMed] [Google Scholar]
- 26.Vandeweyer G., Van der Aa N., Reyniers E., Kooy R.F. The contribution of CLIP2 haploinsufficiency to the clinical manifestations of the Williams-Beuren syndrome. Am. J. Hum. Genet. 2012;90:1071–1078. doi: 10.1016/j.ajhg.2012.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramakers G.J. Rho proteins, mental retardation and the cellular basis of cognition. Trends Neurosci. 2002;25:191–199. doi: 10.1016/s0166-2236(00)02118-4. [DOI] [PubMed] [Google Scholar]
- 28.Hukema R.K., Buijsen R.A., Raske C., Severijnen L.A., Nieuwenhuizen-Bakker I., Minneboo M., Maas A., de Crom R., Kros J.M., Hagerman P.J. Induced expression of expanded CGG RNA causes mitochondrial dysfunction in vivo. Cell Cycle. 2014;13:2600–2608. doi: 10.4161/15384101.2014.943112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alvarez-Mora M.I., Rodriguez-Revenga L., Madrigal I., Guitart-Mampel M., Garrabou G., Milà M. Impaired Mitochondrial Function and Dynamics in the Pathogenesis of FXTAS. Mol. Neurobiol. 2017;54:6896–6902. doi: 10.1007/s12035-016-0194-7. [DOI] [PubMed] [Google Scholar]
- 30.Naaijen J., Lythgoe D.J., Zwiers M.P., Hartman C.A., Hoekstra P.J., Buitelaar J.K., Aarts E. Anterior cingulate cortex glutamate and its association with striatal functioning during cognitive control. Eur. Neuropsychopharmacol. 2018;28:381–391. doi: 10.1016/j.euroneuro.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 31.Carrillo-Mora P., Sila-Adaya D., Villasenor-Aguayo K. Glutamate in Parkinson’s disease: Role of antiglutamatergic drugs. Basal Ganglia. 2013;3:147–157. [Google Scholar]
- 32.O’Gorman Tuura R.L., Baumann C.R., Baumann-Vogel H. Beyond Dopamine: GABA, Glutamate, and the Axial Symptoms of Parkinson Disease. Front. Neurol. 2018;9:806. doi: 10.3389/fneur.2018.00806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gray J.L., von Delft F., Brennan P. Targeting the Small GTPase Superfamily through their Regulatory Proteins. Angew. Chem. Int. Ed. Engl. 2019 doi: 10.1002/anie.201900585. Published online March 14, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie N., Gong H., Suhl J.A., Chopra P., Wang T., Warren S.T. Reactivation of FMR1 by CRISPR/Cas9-Mediated Deletion of the Expanded CGG-Repeat of the Fragile X Chromosome. PLoS ONE. 2016;11:e0165499. doi: 10.1371/journal.pone.0165499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kilkenny C., Browne W.J., Cuthill I.C., Emerson M., Altman D.G. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol. 2010;8:e1000412. doi: 10.1371/journal.pbio.1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bontekoe C.J., Bakker C.E., Nieuwenhuizen I.M., van der Linde H., Lans H., de Lange D., Hirst M.C., Oostra B.A. Instability of a (CGG)98 repeat in the Fmr1 promoter. Hum. Mol. Genet. 2001;10:1693–1699. doi: 10.1093/hmg/10.16.1693. [DOI] [PubMed] [Google Scholar]
- 37.Khalfallah O., Jarjat M., Davidovic L., Nottet N., Cestèle S., Mantegazza M., Bardoni B. Depletion of the Fragile X mental retardation protein in embryonic stem cells alters the kinetics of neurogenesis. Stem Cells. 2017;35:374–385. doi: 10.1002/stem.2505. [DOI] [PubMed] [Google Scholar]
- 38.Maurin T., Lebrigand K., Castagnola S., Paquet A., Jarjat M., Popa A., Grossi M., Rage F., Bardoni B. HITS-CLIP in various brain areas reveals new targets and new modalities of RNA binding by fragile X mental retardation protein. Nucleic Acids Res. 2018;46:6344–6355. doi: 10.1093/nar/gky267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bardoni B., Castets M., Huot M.E., Schenck A., Adinolfi S., Corbin F., Pastore A., Khandjian E.W., Mandel J.-L. 82-FIP, a novel FMRP (fragile X mental retardation protein) interacting protein, shows a cell cycle-dependent intracellular localization. Hum. Mol. Genet. 2003;12:1689–1698. doi: 10.1093/hmg/ddg181. [DOI] [PubMed] [Google Scholar]
- 40.Huang D.W., Sherman B.T., Tan Q., Collins J.R., Alvord W.G., Roayaei J., Stephens R., Baseler M.W., Lane H.C., Lempicki R.A. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.