Abstract
Rhodosporidium toruloides is a red, basidiomycetes yeast that can accumulate a large amount of lipids and produce carotenoids. To better assess this non-model yeast’s metabolic capabilities, we reconstructed a genome-scale model of R. toruloides IFO0880’s metabolic network (iRhto1108) accounting for 2204 reactions, 1985 metabolites and 1108 genes. In this work, we integrated and supplemented the current knowledge with in-house generated biomass composition and experimental measurements pertaining to the organism’s metabolic capabilities. Predictions of genotype-phenotype relations were improved through manual curation of gene-protein-reaction rules for 543 reactions leading to correct recapitulations of 84.5% of gene essentiality data (sensitivity of 94.3% and specificity of 53.8%). Organism-specific macromolecular composition and ATP maintenance requirements were experimentally measured for two separate growth conditions: (i) carbon and (ii) nitrogen limitations. Overall, iRhto1108 reproduced R. toruloides’s utilization capabilities for 18 alternate substrates, matched measured wild-type growth yield, and recapitulated the viability of 772 out of 819 deletion mutants. As a demonstration to the model’s fidelity in guiding engineering interventions, the OptForce procedure was applied on iRhto1108 for triacylglycerol overproduction. Suggested interventions recapitulated many of the previous successful implementations of genetic modifications and put forth a few new ones.
Highlights
-
•
A genome-scale metabolic model (2204 reactions and 1108 genes) was built.
-
•
The model accounts for organism-derived biomass composition and ATP maintenance.
-
•
Functional genomics data were used to improve gene-protein-reaction associations.
-
•
The model predicted increased lipid accumulation under nutrient limitation.
-
•
The model recapitulated successful interventions for triacylglycerol overproduction.
1. Introduction
Rhodotorula genus species are found in various habitats including soil, water, air, on animals and plants, and even in extreme environments such as arctic ice sheets (Starmer et al., 2005). Among them, Rhodosporidium toruloides (or Rhodotorula toruloides) is a basidiomycete yeast generally found in soil (Garay et al., 2016), containing carotenoid compounds giving the organism its characteristic red color (Buzzini et al., 2007; Zhu et al., 2012). R. toruloides is an attractive metabolic engineering host for producing lipid and fatty acid-derived products due to its ability to accumulate lipid (predominantly triacylglycerols (Beopoulos et al., 2011)) as high as 76% of the cell dry weight (Xue et al., 2018) and maintain lipid production in biomass hydrolysates containing growth inhibitory compounds (Hu et al., 2009). It can also grow in high density cell cultures (Li et al., 2007) and utilize a wide variety of substrates (Castañeda et al., 2018; Wiebe et al., 2012). It has a rather compact genome (i.e., haploid genome of 20 Mb with 20% being intergenic sequence) that is tractable for genetic interventions (Coradetti et al., 2018; Park et al., 2018; Zhang et al., 2016b). Extensive metabolic engineering efforts have been focused on lipid production in R. toruloides by exploiting the organism’s ability to accumulate lipid under NaCl-enriched glucose-based media (Tchakouteu et al., 2017), nitrogen-limitation (Shen et al., 2017), sulfur-limitation (Wu et al., 2011), and phosphate-limitation conditions (Wang et al., 2018). R. toruloides can accumulate lipids utilizing multiple substrates (Wiebe et al., 2012) and substrate mixtures (Bommareddy et al., 2015). Genetic interventions aimed at enhancing lipid accumulation have also been explored (Zhang et al., 2016b, 2016a) to overproduce fatty acid derived compounds (e.g., fatty alcohols and esters), used in surfactants, paints, and cosmetics (Adrio, 2017; Yu et al., 2014). In addition to lipids, R. toruloides has also been used as a host for carotenoid (Lee et al., 2016) and d-arabitol production (Jagtap and Rao, 2018).
R. toruloides has recently been the target of significant research efforts including genome (re)sequencing (Coradetti et al., 2018; Zhang et al., 2016b; Zhu et al., 2012), functional genomics analyses (Coradetti et al., 2018), differential ‘omics characterization (Zhu et al., 2012), determination of macromolecular composition (Shen et al., 2017), and growth kinetics in a continuous culture (Shen et al., 2013). Collectively, these experiments have ushered an improved understanding of R. toruloides metabolism and provided the basis for the reconstruction of a metabolic model with genome-wide coverage. A comprehensive genome-scale metabolic reconstruction for R. toruloides would facilitate the integration of various heterogeneous datasets (O’Brien et al., 2015; Thiele and Palsson, 2010) in making predictions of cellular phenotypes under various environmental and genetic perturbations and model-driven knowledge discovery (O’Brien et al., 2015), exploration of organism production potential (Feist et al., 2010; Lee and Kim, 2015; Maia et al., 2016), and extensions towards kinetic descriptions of metabolism (King et al., 2015b; Srinivasan et al., 2015). A successively improving sequence of metabolic models for S. cerevisiae have ushered significant insight into the organism’s physiology and offered many clues for re-engineering (Lopes and Rocha, 2017; Österlund et al., 2012). Metabolic reconstruction of non-model yeasts have recently received significant attention, starting with Pichia pastoris for use in the production of recombinant proteins (Lopes and Rocha, 2017) and the model oleaginous yeast Yarrowia lipolytica (Adrio, 2017; Shi and Zhao, 2017) for which five genome-scale models of iteratively higher level of detail have been reconstructed (Lopes and Rocha, 2017; Wei et al., 2017). They were used to suggest fed-batch strategies to improve lipid accumulation and elucidate the regulation mechanism of lipid accumulation (Kavšček et al., 2015; Kerkhoven et al., 2016). We anticipate that similar advances would be spearheaded for R. toruloides facilitated by the genome-scale model described herein. In addition, to the benefits for guiding re-engineering efforts, a genome-scale model for R. toruloides will fill in a significant knowledge gap as the Basidiomycota phylum is highly under-represented in terms of metabolic model reconstructions. As of today, only a small metabolic model containing 85 reactions (without gene associations) (Bommareddy et al., 2015; Castañeda et al., 2018), and a genome-scale model associating with 897 genes (Tiukova et al., 2019) exist. In contrast, there exist numerous genome-scale models (Lopes and Rocha, 2017; Monk et al., 2014) for eight organisms in the closely related Ascomycota phylum.
We hereby introduce the comprehensive genome-scale metabolic model of R. toruloides strain IFO0880, referred to hereafter as iRhto1108 spanning 1108 genes from the latest version of the genome (Coradetti et al., 2018). The strain IFO0880 has been shown to be a robust host for lipid overproduction (Zhang et al., 2016b). We used the model yeast 7.6 for S. cerevisiae (Aung et al., 2013) as the reconstruction process’ starting point and refined the draft reconstruction of the model by incorporating biochemical information from the latest genome annotation (Coradetti et al., 2018), biochemical (KEGG) information and the KBase database (Arkin et al., 2018; Kanehisa and Goto, 2000). iRhto1108 contains 326 unique metabolic reactions (out of a total of 1399 reactions) compared to both yeast 7.6 and the recent genome-scale model for R. toruloides strain NP11 version 1.1.1 (Tiukova et al., 2019). iRhto1108 was curated to achieve the recapitulation of 84.5% of the gene essentiality data (Coradetti et al., 2018) and growth on all thirteen carbon substrates and five amino acids (as nitrogen source). Quantitative model predictions are also enhanced by organism-derived biomass compositions determined in this study under both carbon and nitrogen limited conditions revealing a much higher proportion of lipids in R. toruloides biomass compared to S. cerevisiae under the same experimental condition. ATP maintenance requirements were derived from a study on R. toruloides’s growth kinetics (Shen et al., 2013). Curation of the biomass description results in an addition of three cell wall components, nine cofactor and prosthetic groups, and seven metal ions. Under nutrient starvation, iRhto1108 successfully captured R. toruloides’s lipid accumulation phenotype. As a demonstration of iRhto1108’s appropriateness to guide strain design, the OptForce procedure (Ranganathan et al., 2010) was used on the model to pinpoint genetic interventions that led to an triacylglycerol overproducing phenotypes. Strain design solutions were in line with in vivo implemented flux “push-pull” strategy (Tai and Stephanopoulos, 2013) that increased lipid production by approximately two-fold in R. toruloides (Zhang et al., 2016b). Overall, iRhto1108 has undergone a detailed range of testing and validation studies promising to aid in future investigations of R. toruloides.
2. Results and discussion
2.1. Model attributes and refinement of draft reconstruction
iRhto1108 is a comprehensive genome-scale model that integrates yeast biochemistry information from (i) previously built genome-scale models (S. cerevisiae yeast 7.6 (Aung et al., 2013), (ii) KBase fungal models (Arkin et al., 2018)), and (iii) R. toruloides specific information extracted from the primary literature (Coradetti et al., 2018; Jagtap and Rao, 2018; Kot et al., 2018) or generated herein. The model statistics are summarized in Table 1. Genes included in iRhto1108 cover 13% of the organism’s chromosomal (i.e., 51.5% of metabolic genes annotated by Eukaryotic orthologous group (KOG) and KEGG including poorly annotated genes) and 6% of mitochondrial genome. Throughout this article, genes named will be referred to by the corresponding S. cerevisiae’s homolog name (if available) (e.g., HOM6 for homoserine dehydrogenase) or otherwise using R. toruloides gene IDs (e.g., rt6880 for serine O-acetyltransferase). R. toruloides protein IDs are not used herein (e.g., RTO4_15248 for serine O-acetyltransferase) to retain consistency in gene identification. iRhto1108 shares 65% of genes, 70% of reactions, and 67% of metabolites with the S. cerevisiae yeast 7.6 model (Aung et al., 2013). KBase entries contributed 8% of genes, 5% of reactions, and 7% of metabolites of iRhto1108. KBase was used to identify additional homologous genes and extract reactions from metabolic reconstructions for non-model yeasts. The remainder of the model content (i.e., 27% of genes, 25% of reactions, and 26% of metabolites) was directly culled from the genome annotation and subsequently manually curated. Many of these model additions do not necessarily capture R. toruloides-specific biochemistry but instead unpack aggregated yeast 7.6’s reaction content or replace redundant yeast 7.6’s features. For example, using KBase, a lumped palmitoyl-CoA synthesis (fatty acid C16:0) reaction in yeast 7.6 is detailed into 28 steps catalyzed by fatty acid synthase in iRhto1108 (seven elongation cycles, each cycle contains four elementary steps). In addition, a set of aggregated reactions simplified lipid metabolism in yeast 7.6 (Aung et al., 2013; B. J.Sánchez et al., 2018b). For example, a single generalized reaction for diacylglycerol acyltransferase (DGAT_rm) replaced 32 copies of DGAT_rm in yeast 7.6 operating on 32 variants of triacylglycerol. Overall, gene-protein-reaction associations were assigned for 93% of metabolic reactions in iRhto1108. Not surprisingly, identification of genes coding for transporters remained a challenge as was the case for yeast 7.6. Missing GPR assignments in iRhto1108 are mostly in intracellular transport between compartments (i.e., 364 from a total of 456 GPR-lacking reactions). Throughout the reconstruction process, we manually curated the GPR of 543 reactions associated with 373 genes. Determination of metabolic role of a gene(s) in GPRs was assisted using NCBI’s Conserved Domain Database (NCBI’s CDD) (Marchler-Bauer et al., 2015). GPR rules that capture protein subunits and isozymes were inferred from yeast 7.6 model, R. toruloides’s genome annotation, and primary literature sources. Reactions ported from yeast 7.6 were assigned with yeast 7.6’s GPRs whereas new reactions were assigned GPRs based on the genome annotation. Literature information was used to reconstruct GPRs capturing multimeric proteins. Further manual curation of GPRs was performed to improve gene essentiality prediction (see section 2.3). NCBI’s CDD was used throughout the GPR curation process as a reliable source of protein’s functional assignment, as shown in the following case of functional elucidation of spermidine synthase. Initially no reaction enabling the synthesis of spermidine, a biomass constituent, was found as no spermidine synthase gene was identified via bidirectional BLAST. However, subsequent analysis by NCBI’s CDD revealed a catalytic domain on rt8465 (homolog of S. cerevisiae’s LYS9). Notably, this catalytic domain does not overlap with saccharopine dehydrogenase domain identified by bidirectional BLAST. Thus, the spermidine synthase reaction was subsequently determined to associate to rt8465 and thereby fill the gap in spermidine synthesis. Furthermore, using the more recent version of the consensus yeast model available at https://github.com/SysBioChalmers/yeast-GEM (yeast 8.3.3 (B. Sánchez et al., 2018a)), GPR assignments for 22 reactions in iRhto1108 were updated. For example, drawn from yeast 8.3.3’s GPR assignment, isozyme rt1542 (homologous to YOR283W) was added to the glycolysis reaction phosphoglycerate mutase’s GPR.
Table 1.
R. toruloides iRhto1108 genome-scale model statistics.
| Properties | Statistics |
|---|---|
| Genes | 1108 |
| Identified from yeast 7.6 | 717 |
| Identified from KBase | 86 |
| From chromosome (% of chromosomal ORFs) | 1087 (13%) |
| with S. cerevisiae homolog identified | 908 |
| From mitochondrial genome (% of mitochondrial ORFs) | 21 (6%) |
| with S. cerevisiae homolog identified | 7 |
| Reactions | 2204 |
| From yeast 7.6 | 1514 |
| From KBase | 117 |
| Metabolic reactions | 1399 |
| With GPR assigned | 1306 |
| Unique number of metabolic reactions | 1123 |
| Transport reactions | 619 |
| Extracellular transport | 163 |
| with GPR assigned | 75 |
| Intracellular transport | 456 |
| with GPR assigned | 92 |
| Exchange reactions | 186 |
| Metabolites | 1985 |
| From yeast 7.6 | 1328 |
| From KBase | 141 |
| Unique metabolites | 1044 |
| Formula and charge assignments from database | 1537 |
| Compartments | 14 |
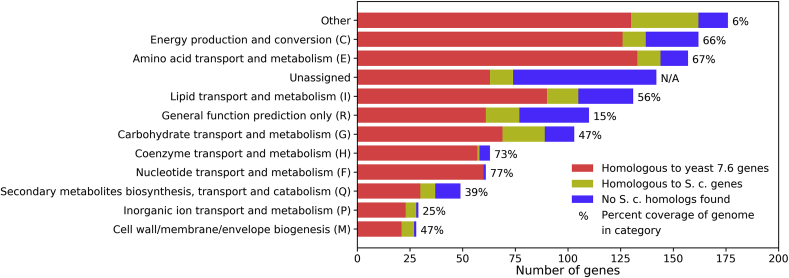
The classification of iRhto1108 genes shown in Table 1 quantifies the extent of contribution from previous yeast reconstructions and the significant expansion in iRhto1108 (Fig. 1). Eukaryotic orthologous group (KOG) assignments (Koonin et al., 2004), a Eukaryotic-specific Cluster of orthologous groups of protein (COG) provided by the updated genome annotations (Coradetti et al., 2018), were used in classifying genes and the associated reactions. Most of the genes in iRhto1108 are homologous to S. cerevisiae’s genes (Fig. 1), in agreement with the relative phylogenetic proximity between the two species of yeasts (i.e., their respective divisions, Ascomycota and Basidiomycota, are grouped to the sub-kingdom Dikarya). Highly conserved metabolic functions (i.e., 80–98% genes with homologs identified per KOG class) between the two are observed in all the core metabolic functions (listed in decreasing degree of conservation): nucleotide, inorganic ion, cell wall, coenzyme, amino acid, carbohydrate, energy production, and lipid metabolism. Nevertheless, R. toruloides has a number of unique metabolic capabilities compared to S. cerevisiae. These new functionalities, 244 metabolic reactions out of 1,399, are not predominantly localized in any specific pathway but rather span multiple KOG classifications. In terms of genome coverage, iRhto1108 is able to account for 66–77% of genes in KOG classes, namely energy nucleotide, coenzyme, amino acid, and energy production metabolism. The category with the lowest genome coverage (i.e., 25%) is inorganic ion transport and metabolism. A significant fraction of iRhto1108’s genes fell into the “Unassigned” category due to KOG’s inability to identify genes with non-homologous sequences that perform core metabolic functions. For example, R. toruloides fatty acid synthase subunit I and II were not classified by KOG annotation into the lipid metabolism group, possibly due to irregular arrangement of catalytic sequence motifs compared to S. cerevisiae and other types of yeast (Xue et al., 2018).
Fig. 1.
Classifications of genes in iRhto1108. Eukaryotic orthologous groups (KOG) annotations are provided in the genome annotation and used for classifying genes to the corresponding functions. Group abbreviations are in the parentheses. A gene with multiple KOG groups assignments were added to all the groups. A gene without KOG annotation was manually assigned to a KOG group. Other groups include A, B, D, J, K, L, N, O, S, T, U, V, W, Y, and Z (see https://genome.jgi.doe.gov/Tutorial/tutorial/kog.html).
The majority of the novel metabolic functions captured in iRhto1108 are extracted directly from the genome annotation or open literature. For example, included in the model are reactions and associated genes for both the carotenoid (Kot et al., 2018) and d-arabitol production pathways (Jagtap and Rao, 2018) which were absent in yeast 7.6. Most of the novel functions belong to lipid and carotenoid metabolism. Some examples in lipid metabolism are ATP citrate lyase in acetyl-CoA production (Evans and Ratledge, 1985), cytoplasmic malic enzyme (Zhang et al., 2016b), stearoyl-CoA desaturase in polyunsaturated acyl-CoA production (Zhang et al., 2016a), and mitochondrial beta-oxidation (Coradetti et al., 2018; Hiltunen et al., 2003). The carotenoid biosynthesis pathway whose products are responsible for the organism’s characteristic red color (Buzzini et al., 2007) is captured in iRhto1108. In addition, an NADH oxidoreductase reaction (complex I in electron transport chain) is included in iRhto1108 which is known to be absent in S. cerevisiae (Kerscher, 2000). Non-essential metabolic functions found in the genome annotation with an unclear physiological role such as a peroxisomal d-amino acid oxidase (Hsieh et al., 2009) are also recorded in the model.
Model annotation and network consistency are important properties for testing genome-scale model quality (Lieven et al., 2018). We evaluated iRhto1108 using standardized tests provided by the memote test suite and updated the model based on detected issues. Under the independent section (scored tests), iRhto1108 received a high score of 87% on biochemical annotation and network consistency tests. Since R. toruloides genome is recorded on JGI Mycocosm and not on memote-verified databases such as KEGG, we only provide the S. cerevisiae homologs as genes’ annotation. iRhto1108 also achieves a score of 99.7% on network consistency tests. Some lost points are from memote’s mistaken identification of reaction imbalances, namely generalized reactions that set the composition of generic acyl-CoA (using “Acyl” as a group in the formula) and biochemical reactions associated with that generic acyl-CoA group. Detected by memote, unbounded fluxes that can form a thermodynamically infeasible cycle (Schellenberger et al., 2011) were fixed by restricting the directionality of transporters and/or reactions. For example, the tyrosine importer (AVT1), exporter (AVT3), and efflux transporter (ATG22) from cytosol to vacuole can shuttle tyrosine in and out of the vacuole with no driving force leading to an unbounded flux. To remedy this, the efflux transporter was allowed to only import (not export) tyrosine to the vacuole. Export function can be re-activated if needed (e.g., under autophagy-induced protein degradation (Yang et al., 2006)). In addition, under network topology tests, memote reported a high number of blocked reactions (677 out of 2204) (see Supplementary Materials 1). These reactions and metabolites were retained in iRhto1108 as they are based on assignments from homologous genes and genome annotation. They cause no problems in flux balance analyses and may serve in the future as gap-filling targets. The gap-free network contains 1527 reactions (i.e., 69% of all reactions), 1221 metabolites (62%), and 806 genes (73%). In comparison, S. cerevisiae yeast 7.6 model includes 60% of reactions that are not blocked.
2.2. Update of biomass composition and revision of ATP maintenance requirements
In addition to differences in pathways, an important contribution in iRhto1108 is the expansion of the list of biomass constituents by 23 components from the original 45 taken from yeast 7.6 (Table 2). Four new fatty acid species, C18:2, C18:3, C20:0, and C24:0, are added to the biomass reaction based on their detection by mass spectrometry measurements (see section 4.7). Seven metal ions are added based on biomass measurements for S. cerevisiae (Lange and Heijnen, 2001). We identified three cell wall components and nine cofactors and prosthetic groups that must be added to match measured phenotypes (Coradetti et al., 2018) (see section 4.2). For instance, lethal knockouts of dephospho-CoA kinase (gene: CAB5) or GPI anchor biosynthesis (gene: GPI13) (KEGG reaction R05923) are unresolvable without the additions of coenzyme-A and GPI anchor to the biomass reaction, respectively. These validations are provided in Supplementary Materials 2. The revised list of biomass constituents and the experimentally determined macromolecular composition are provided in Table 2. The full description of the biomass reaction is detailed in the Supplementary Materials 1. Moreover, we experimentally determined R. toruloides macromolecular composition separately under both carbon and nitrogen limitation (see Table 2 and section 4.2). Protein, carbohydrate, DNA, RNA, and lipid composition were measured for cells growing on glucose in a chemostat at the reference dilution rate of 0.1 hr−1 used in S. cerevisiae model (Schulze, 1995). Reconstruction of biomass reaction to capture context-specific biomass composition such as variation in lipid fraction under different growth conditions (e.g., C/N ratio variation (Blazeck et al., 2014)) can be facilitated using the detailed formulation provided in Supplementary Materials 1. Further improvements include DNA base composition update based on GC content (Nakase and Komagatai, 1971), RNA bases composition informed from RNA-Seq data, and lipid’s acyl group composition measured by mass spectrometry (see section 4.2). Both biomass reactions for R. toruloides in carbon and nitrogen limited conditions imply a higher proportion for lipid than S. cerevisiae (in comparison to yeast 7.6’s biomass reaction) and a lower proportion of carbohydrate (and protein under carbon limitation). The DNA fraction for R. toruloides is also higher under both conditions compared to S. cerevisiae. Both the lipid and DNA fractions are higher while the RNA fraction is lower for cells growing under nitrogen limitation. Importantly, the combined coefficient-weighted molecular weights of all constituents were standardized to 1 g mmol−1 to ensure consistency of growth yield prediction (Chan et al., 2017). The biomass composition listed in Table 2, follows the core biomass definition (Feist et al., 2007; Feist and Palsson, 2010) consisting of growth-essential metabolite requirements. The inclusion of these metabolites in the biomass reaction was based on gene essentiality results (Coradetti et al., 2018) and experimental data from S. cerevisiae (Supplementary Materials 2). Compared to rhto-GEM model v. 1.1.1 (Tiukova et al., 2019), iRhto1108’s biomass reaction contains 23 additional constituents. In addition, iRhto1108 offers a nitrogen limited version (viz., conditions applicable for lipid production) underpinned by a significant biomass compositional difference. Visualizations of metabolic flux shift when switching between carbon and nitrogen limited biomass description are provided in Supplementary Materials 3.
Table 2.
Summary of iRhto1108’s biomass composition.
| Constituents | Composition (%) |
||||
|---|---|---|---|---|---|
| C-lim | N-lim | yeast7.6 | |||
| Proteina | 43.31 | 30.71 | 35.71 | ||
| l-Alanine | l-Arginine | l-Asparagine | |||
| l-Aspartate | l-Cysteine | l-Glutamine | |||
| l-Glutamate | Glycine | l-Histidine | |||
| l-Isoleucine | l-Leucine | l-Lysine | |||
| l-Methionine | l-Phenylalanine | l-Proline | |||
| l-Serine | l-Threonine | l-Tryptophan | |||
| l-Tyrosine | l-Valine | ||||
| Carbohydrate | 32.61 | 12.50 | 52.27 | ||
| 1,3-beta-d-Glucan | 1,6-beta-d-Glucan | Chitin | |||
| N-Glycanb | O-Glycanb | GPI-anchorb | |||
| Lipid | 12.33 | 44.84 | 0.74 | ||
| Episterol | Free fatty acids (7 species) c | Inositol-P-ceramide | |||
| Phosphatidylcholine | Phosphatidylethanolamine | Phosphatidylinositol | |||
| Phosphatidylserine | TAG | ||||
| RNAd | 6.73 | 4.69 | 5.85 | ||
| ATP | CTP | GTP | |||
| UTP | |||||
| DNAd | 1.12 | 3.36 | 0.34 | ||
| dATP | dCTP | dGTP | |||
| dTTP | |||||
| Cofactors and prosthetic groups | 0.06 | 0.06 | 0.03 | ||
| S-Adenosyl-L-methionine | Biotin | Coenzyme-A | |||
| FAD | Heme A | NAD | |||
| NADP | Riboflavin | Spermidine | |||
| Tetrahydrofolate | Thiamine diphosphate | ||||
| Inorganic ions | 3.85 | 3.85 | 5.06 | ||
| Calcium | Copper | Iron | |||
| Magnesium | Manganese | Phosphate | |||
| Potassium | Sulphate | Zinc | |||
Biomass constituents absent from yeast 7.6 are shown in boldface type. Different representations of yeast 7.6 biomass constituents are listed in the notes below. A detailed analysis of iRhto1108’s biomass composition is provided in section 4.2 and the full description in the Supplementary Materials 1.
Identical to those in yeast 7.6, amino acids in the biomass objective function are in charged-tRNA form.
The generic mannan (mannose-containing) metabolite in yeast 7.6 was replaced with three specific essential cell wall components (Orlean, 2012).
Seven free fatty acid species were abundant (>1% weight) in growth experiments detailed in section 4.7. These are palmitate (C16:0), stearate (C18:0), oleate (C18:1), linoleate (C18:2), linolenate (C18:3), docosanoate (C22:0), and tetracosanoate (C24:0).
Monophosphate ribonucleic and deoxyribonucleic acids in yeast 7.6 were replaced with the corresponding triphosphate ones.
In addition to the updated biomass composition derived for two separate growth conditions, we also revisited the ATP maintenance requirements (both growth and non-growth). Non-growth (NGAM) and growth associated maintenance (GAM) values were estimated by assessing the model’s optimal ATP production under glucose uptake restriction and growth yield requirement and experimentally recorded glucose uptake rates and growth rates. Correctly assessing ATP maintenance is important for properly quantifying energetic needs and growth yield (Feist et al., 2007). ATP maintenance requirements for iRhto1108 were calculated from available chemostat data for growth on glucose for both carbon and nitrogen limitation, respectively (Shen et al., 2013) (see section 4.4). An NGAM value of 1.01 mmol gDW−1 hr−1 for both conditions was recovered. In contrast, the growth associated maintenance (GAM) was condition-dependent with a value of 140.98 mmol gDW−1 under carbon limited and 154.94 mmol gDW−1 under nitrogen limited conditions. In yeast 7.6, NGAM is not modeled (though an earlier S. cerevisiae model (Mo et al., 2009) reported an NGAM value of 1 mmol gDW−1) and the GAM value is 59.28 mmol gDW−1. The GAM value quantifies growth-associated energy costs that are not captured in the biomass equation, alluding to higher energy demands for R. toruloides growth compared to S. cerevisiae. Under nitrogen limitation, GAM value is slightly higher (1.1-fold increase). Note that growth kinetics of R. toruloides under nitrogen limitation follows a different trend compared to carbon limitation (Shen et al., 2013). It appears that the assumption of constant GAM value across all growth rates may not hold under nitrogen limitation. However, higher ATP cost under nitrogen limitation is generally accepted (see Supplementary Materials 2). In rhto-GEM model v. 1.1.1 (Tiukova et al., 2019), a non-condition-specific GAM value of 132.7 mmol gDW−1 and NGAM value of 3 mmol gDW−1 hr−1 were reported. These values generally match the iRhto1108’s corresponding entries under carbon limitation.
2.3. Gene essentiality, growth viability, and phenotype predictions
iRhto1108 predictions were contrasted against gene essentiality and mutant auxotrophy data derived from the functional genomics study (Coradetti et al., 2018) and growth yield and viability data from multiple literature sources (see Supplementary Materials 2). To achieve an improved prediction-observation consistency, GPRs were curated to resolve incorrect gene essentiality predictions. The data collected and corresponding predictions are summarized in Table 3. Gene sequence disruptions with T-DNA insertions were carried out leading to evaluation of gene essentiality for 1079 of the 1108 genes in the model (Coradetti et al., 2018). Gene essentiality predictions are shown in Table 3. iRhto1108 achieved 84.5% accuracy for gene essentiality prediction (i.e., correct predictions of gene essentiality and non-essentiality over all predictions) which is similar to that of yeast 7.6 (i.e., 89.8%). The model is particularly adept at recovering mutant growth, measured by the sensitivity level of 94.3%. Positive mutant growth misses (NG-G) (i.e., 5.5%) by iRhto1108 were mainly due to differences in protein subunit assignments between S. cerevisiae and R. toruloides. For example, consistent with S. cerevisiae but in contrast to R. toruloides, GPI2 and GPI15 knockouts were predicted by iRhto1108 to be lethal since they are subunits of the enzyme catalyzing the first step of GPI anchor biosynthesis. We anticipate that inconsistencies similar to the GPI biosynthesis case could be resolved by elucidating the currently unknown gene-to-protein mapping structure. iRhto1108 relatively inaccurate predictions of negative mutant growth (G-NG) (i.e., specificity of 53.8%) matches the corresponding specificity for yeast 7.6 (i.e., 52.5%) as the GPR assignments were directly ported from yeast 7.6. For example, for l-methionine auxotrophic (i.e., rt6880 and rt3663) mutants upon knocking out cysteine biosynthesis via serine (Coradetti et al., 2018), iRhto1108 predicted growth without the need for l-methionine supplementation by allowing sulfur assimilation via the homoserine pathway. The lack of observed growth suggests the possible inactivity of the homoserine pathway (encoded by MET2, 6, and 17). The aforementioned case cannot be explained by the model using the information embedded in reaction stoichiometry and GPRs.
Table 3.
Summary of phenotype predictions by iRhto1108.
| Prediction | Statistics |
|---|---|
| Gene essentialitya | |
| G-G | 772 (72%) |
| G-NG | 120 (11%) |
| NG-G | 47 (4%) |
| NG-NG | 140 (13%) |
| Accuracy | 84.5% |
| Sensitivity | 94.3% |
| Specificity | 53.8% |
| Growth viability | |
| As carbon source | 13 |
| As nitrogen source (positive) | 5 |
| As nitrogen source (negative) | 1 |
| Mutant auxotrophy | |
| Arginine and/or methionine | 18/22 |
| Growth yield prediction | |
| d-Glucose | 4 validations |
| d-Xylose | 9 validations |
| Production yield prediction | |
| d-Arabitol | 6 validations |
Agreements or disagreements between model predictions and gene essentiality data are classified into four groups: G-G, G-NG, NG-G, and NG-NG; the first part of the group is in silico result, the second part is in vivo result, “G” stands for growth, and “NG” stands for non-growth. Accuracy = (G-G + NG-NG)/Total. Sensitivity = G-G/(G-G + NG-G). Specificity = NG-NG/(NG-NG + G-NG).
iRhto1108 was also tested in terms of its ability to predict growth on alternative substrates (see Table 3). The simulations were performed using the model with substrate uptake rate and secretion rate as inputs. R. toruloides can grow on d-glucose, d-xylose, acetate, glycerol, fructose, mannose, sucrose, cellobiose and fatty acids as carbon sources. Amino acids such as l-threonine, l-serine, l-proline, l-alanine, and l-arginine can support R. toruloides’ nitrogen needs. Growth experiments on cellobiose, mannose, sucrose, and amino acids were performed in this study whereas the other findings are collected from the literature (see Supplementary Materials 2). iRhto1108 predicts growth with these substrates upon activating the corresponding transporters. The activation of l-threonine transporter is seemingly in conflict with gene essentiality results since knockouts in de novo l-threonine biosynthesis are lethal in rich media (five steps from l-aspartate each encoded by HOM3, HOM2, THR1, THR4, and HOM6) (Coradetti et al., 2018). Further study on l-threonine biosynthesis pathway is necessary to explain why ex vivo l-threonine supplementation was insufficient to rescue those mutants. Other growth phenotypes such as mutant auxotrophy and quantitative growth yields are collected and used in model validations. Arginine and methionine auxotrophy predictions for 22 mutants are largely consistent with experimental findings (Coradetti et al., 2018) and explanations for inconsistencies (4 mutants) can be offered based on the model predictions such as the hypothesis of homoserine pathway being repressed under l-methionine abundance conditions, as mentioned above. Furthermore, iRhto1108 quantitative prediction of wild-type growth yield in exponential phase are close to the experimental numbers (see Supplementary Materials 2). Example visualizations of metabolic fluxes for growth maximization and triacylglycerol (TAG) overproduction are provided in Supplementary Materials 3.
2.4. Phenotypic change under nutrient starvation conditions
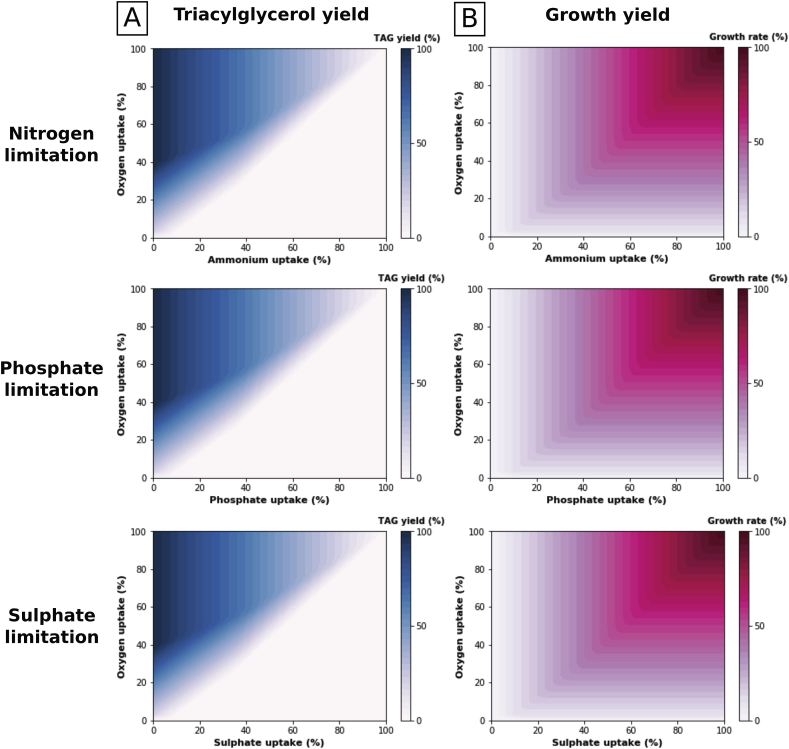
Nutrient starvation is a common strategy for enhancing lipid accumulation in oleaginous yeasts. It has been successfully implemented in R. toruloides (Shen et al., 2017), Y. lipolytica (Kerkhoven et al., 2016), and Lipomyces starkeyi (Liu et al., 2011). R. toruloides also over-accumulates lipid under phosphate (Wang et al., 2018) and sulphate (Wu et al., 2011) limitation. Triacylglycerol, the major compound in lipid accumulation, is stockpiled in lipid particles thus sequestering excess carbon substrate. We sought to recapitulate lipid accumulation in response to nutrient limitation conditions in iRhto1108 using two separate maximization problems. First, biomass production was prioritized by setting the flux balance analysis objective function to be maximization of growth yield given a nutrient-limited input. Second, lipid accumulation under low nitrogen was imposed to iRhto1108 by setting the model objective function to be maximization of TAG production. In addition, growth yield in the second problem was constrained to be equal to the maximal value found in the first problem. This optimization posture is hypothesized to be the regulatory outcome involving the TOR signaling pathway in R. toruloides NP11 (Zhu et al., 2012). Using these two maximization problems both growth and TAG yield were calculated under varying degrees of limitation for inorganic ions (i.e., ammonium, phosphate, and sulphate) and oxygen. Dissolved oxygen is an experimentally controllable variable that has been shown to affect lipid accumulation (Probst et al., 2016). In an experiment in which ammonium was limited for Y. lipolytica, reduced aeration rate was shown to enhance lipid production (Kavšček et al., 2015). Phenotype phase plane analysis (Edwards et al., 2002) has been used to examine growth and TAG yield with respect to these two sources of variation.
Under nutrient limitation, iRhto1108 predicts an increase in TAG yield and decrease in biomass yield which is in qualitative agreement with experimental observations for R. toruloides and other oleaginous yeast. The same trend is observed under nitrogen, phosphate, and sulphate limitation. This trend suggests that cell proliferation (growth maximization in the first simulation problem) is the primary cellular objective for R. toruloides in the absence of nutrient limitation whereas TAG storage (TAG production maximization in the second simulation problem) becomes the objective function for iRhto1108 in response to nutrient limitation conditions. The effect of oxygen limitation on TAG production in iRhto1108 follows a more complex trend depending on the degree of nutrient limitation. Moderate reduction in oxygen availability decreased growth yield with no effect on TAG yield. However, when oxygen level is reduced below a threshold (see the diagonal gradient region in Fig. 2, column A), TAG production is also compromised and growth yield is directly proportional to oxygen level (Fig. 2, column B). Note that reduction in TAG yield was also observed experimentally in Y. lipolytica under severe oxygen limitation with N2 aeration (Kavšček et al., 2015). This result is not surprising as oxygen is necessary to provide enough energy through oxidative phosphorylation for TAG synthesis. TAG biosynthesis requires one NAD(P)H for dihydroxyacetone reduction and an ATP plus two NADPH molecules for every two-carbon elongation step of the acyl groups. Acetyl-CoA biosynthesis required for fatty acid biosynthesis also consumes an ATP. Moderate reduction of oxygen availability did not improve TAG yield based on the phenotype phase plane analysis (Fig. 2). The result is in agreement with experimental results for R. toruloides (Bommareddy et al., 2015), but it is in contrast with results for Y. lipolytica (Kavšček et al., 2015). Overall, TAG accumulation appears to be insensitive to moderate oxygen availability reduction but becomes highly bottlenecked under very low oxygen availability. However, growth is affected by both oxygen and ammonium availability reaching a maximum when both are in excess.
Fig. 2.
Phenotype phase planes of TAG production (column A) and maximal growth yield (column B) in nutrient (i.e., ammonium, phosphate, sulphate) and oxygen limited conditions. Values on the figure are percentage of maximal allowed flux for nutrients uptake and maximal yield for TAG production and growth rate. Determined by the model, upper bounds of uptake values are minimal amounts required to sustain maximal growth (oxygen 12.78, ammonium 2.43, phosphate 0.20, and sulphate 0.03 mmol.gDW−1.hr−1). Maximal TAG production is 0.31 g/g glucose and maximal growth rate is 0.38 hr−1.
As described above, iRhto1108 captures changes in phenotype for R. toruloides under nutrient starvation. We next explored whether the corresponding flux changes for a new phenotype quantitatively match gene upregulation/downregulation data (Zhu et al., 2012) under the carbon and nitrogen limited version of the model (iRhto1108C and iRhto1108N, respectively). We used as a criterion of gene-reaction correlation that the change in gene expression level and associated reaction flux change is within a factor of two. In addition, genes with a p-value of less than 0.05 and a false discovery rate of less than 0.001 were excluded from the analysis as proposed in (Zhu et al., 2012). We identified 12 upregulated and 11 downregulated genes under nitrogen starvation that quantitatively matched changes in metabolic fluxes predicted by iRhto1108. Overall, we find very few genes (23 out of 1064) where the change in mRNA level quantitatively tracks the shift in model-predicted metabolic fluxes. Gene expression levels (taken from (Zhu et al., 2012)) and metabolic flux values are reported in Supplementary Materials 2. Increased energy demand under nitrogen limitation, was accompanied by upregulation for subunits of ATP synthase (ATP14, 16, and VMA9), NADH:ubiquinone oxidoreductase (complex I) (NDE2, rt0331, rt4846, rt1642, and rt2984), and ferrocytochrome-c:oxygen oxidoreductase (complex III) (rt2984). In addition, increased lipid fraction of biomass under nitrogen limitation, is consistent with upregulation of genes in sterol (ERG7, 12) and sphingolipid synthesis (rt3023). In contrast, a gene in phospholipid synthesis were downregulated (i.e., CHO1) which is consistent with the lower phospholipid fraction among lipid species under nitrogen limitation (Shen et al., 2017). The reduced carbohydrate fraction in biomass was accompanied by downregulations in cell wall biosynthesis (GSC2, ALG1, GFA1, GPI13, GPI14, SEC53, rt1388, CHS1/2, and MNT3). Overall, only a few genes (23 out of 1064) quantitatively tracked the corresponding flux changes. This finding re-emphasizes that flux distribution does not simply track changes in gene expression but rather is affected by many other factors such as transcriptional regulation, translation efficiency, substrate level regulation, and metabolite pools (Chubukov et al., 2013).
2.5. Predicting metabolic engineering strategies for enhanced triacylglycerol production
In this section, we explore the effectiveness of iRhto1108 to guide strain design by contrasting predictions for TAG overproduction with successful experimental interventions. A number of re-engineering strategies have been recently implemented in R. toruloides by mimicking effective interventions in Y. lipolytica (Blazeck et al., 2014). These interventions include upregulation of acetyl-CoA carboxylase, diacylglycerol transferase, malic enzyme, and stearoyl-CoA desaturase (Zhang et al., 2016b, 2016a). We applied the OptForce procedure (Ranganathan et al., 2010) using iRhto1108 as the metabolic map and contrasted with existing solutions. The goal was not necessarily to find new interventions but rather to assess whether iRhto1108 can indeed steer strain design algorithms towards promising designs. The OptForce procedure was applied for TAG overproduction using glucose as the carbon substrate (see section 4.6). 13C metabolic flux analysis (MFA) data for Y. lipolytica under nitrogen limitation (Wasylenko et al., 2015) was used as a stand-in to determine the reference flux distribution for wild-type strain as no such data is currently available for R. toruloides; Y. lipolytica is an oleaginous yeast with similar metabolic capability such as utilizing ATP citrate lyase for cytoplasmic acetyl-CoA production. Nitrogen availability level of 33% (of the maximum needed) was inferred from the MFA flux data by calculating the minimal amount of ammonium uptake. The model built for nitrogen limited conditions, iRhto1108N, was used throughout the OptForce simulation.
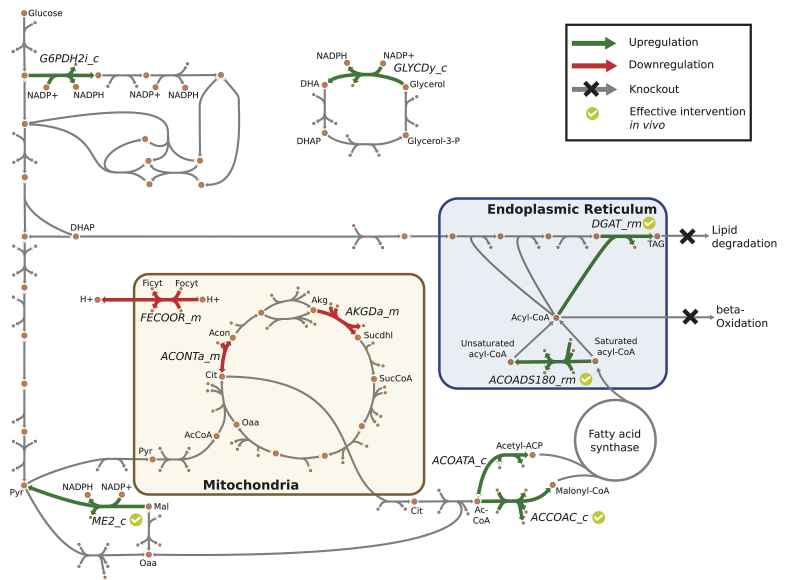
First, sets of candidates for overexpression (MUSTU), downregulation (MUSTL), and knockout (MUSTX) were determined by contrasting the flux ranges of wild-type and overproducing strain. We excluded in vivo essential reactions (Coradetti et al., 2018) from MUSTX and transport, exchange, and generalized reactions from all MUST sets. For a sequence of reactions in series only the first step was considered as a perturbation candidate. For example, among 31 reactions of the fatty acid synthase’s chain elongation, only the first step ACP S-acetyltransferase (ACOATA_c) was retained in MUSTU. For the MUSTL set, downregulations of biomass-coupled reactions (i.e., 119) were excluded from further analysis since those perturbations reduce cellular growth for production gain. No flux pairs that considered sums and differences were identified by the analysis (MUSTUU, MUSTLL and MUSTLU for overexpressed sum, downregulated sum, and overexpressed flux difference, respectively) (Ranganathan et al., 2010). This result is due to the linearity of the acyl-CoA (from acetyl-CoA) and TAG synthesis pathways (i.e., sequential attachment of acyl-CoA to dihydroxyacetone backbone) and the absence of converging paths towards TAG. Overall, few perturbations (i.e., 14 in MUST-single and none in MUST-pair) were suggested because under nitrogen limitation the reference wild-type fluxes already achieve a TAG production phenotype (though less than overproducing strain) and resemble the overproducing state. Surprisingly, we found that two key overproduction targets (Liang and Jiang, 2013), (i) ATP citrate lyase and (ii) cytoplasmic malic enzyme (ME2_c), were not included in the MUSTU set. ATP citrate lyase is the key enzyme in producing cytoplasmic acetyl-CoA. However, acetyl-CoA synthetase can functionally replace ATP citrate lyase thus both reactions form a MUSTUU pair. However, since acetyl-CoA synthetase can participate in a high-flux thermodynamically infeasible cycle that transports acetyl-CoA to mitochondria, hydrolyzes it to acetate, exports to cytoplasm, and re-synthesizes acetyl-CoA, the ranges of the flux sum of the pair for wild-type and overproducing strain overlapped. This discovered cycle was subsequently removed from the model by turning off the ethanol-induced acetyl-CoA transport via carnitine shuttle (Schmalix and Bandlow, 1993) and ATP citrate lyase and acetyl-CoA synthetase were added to the MUSTUU set. Malic enzyme can participate in a transhydrogenase cycle involving malate dehydrogenase and pyruvate carboxylase and produce cytoplasmic NADPH for acyl-CoA synthesis. The model contains three mechanisms for cytoplasmic NADPH production hence the malic enzyme contribution can only be detected by looking at non-overlapped flux triplets. The other two mechanisms are the oxidative pentose phosphate pathway (PPP) and glycerol dehydrogenase. All three NADPH production mechanisms were added to the MUSTU set and their overexpression levels were found by minimizing the reaction flux in the overproducing strain with the other two knocked out. Overall, there were ten candidates for overexpression (MUSTU), three candidates for downregulation (MUSTL), and no candidates for knockout (MUSTX). All reactions in MUST sets are provided in Supplementary Materials 2. OptForce was used to search for combinations of candidates in MUST sets that can lead to enhanced TAG overproduction and recorded these combinations into the FORCE set (Table 4). Without the lipid degradation knockout, no combinations could be identified since OptForce min-max objective function identified the worst-case scenario of TAG synthesis-degradation cycle. beta-Oxidation was not originally placed in the MUSTX set since both wild-type and overproducing strains can run (to some extend) TAG and fatty acid synthesis-degradation cycles. Beta-Oxidation knockout was manually added along with the removal of fatty acid secretion since this phenotype was not observed in both wild type and engineered strains (Zhang et al., 2016b), and OptForce was rerun (see Table 4).
Table 4.
Combinations of genetic perturbations suggested by OptForce procedure for triacylglycerol production under nitrogen limitation.
| Intervention a | WT | Mutant strains |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| k = 2 |
k = 3 |
k = 4 |
|||||||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | |||
| ↑ DGAT_rm | P (Rt) | X | X | X | X | X | X | X | |||||||||
| ↑ ACCOAC_c | P (Rt) | X | |||||||||||||||
| ↑ ACOATA_c | P (O) | X | X | X | |||||||||||||
| ↑ ACOADS180_rm | P (Rt) | X | X | X | X | ||||||||||||
| ↑ ME2_c | C (Rt) | X | X | ||||||||||||||
| ↑ G6PDH2i_c | C | X | X | X | |||||||||||||
| ↑ GLYCDy_c | C | X | X | X | |||||||||||||
| ↓ FECOOR_m | R | X | |||||||||||||||
| ↓ ACONTa_m | R | X | |||||||||||||||
| ↓ AKGDa_m | R | X | |||||||||||||||
| Δ beta-Oxidation b | D (O) | X | X | X | X | X | X | X | X | ||||||||
| Δ Lipid degradation c | D (O) | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | |
| TAG production flux (mmol/gDW/hr) | 0.185 | 0.567 | 0.570 | 0.569 | 0.569 | 0.568 | 0.572 | 0.572 | 0.565 | 0.565 | 0.566 | 0.569 | 0.569 | 0.568 | 0.568 | 0.567 | |
Reaction abbreviations are listed in the table and annotated in the text. Reactions are described in Supplementary Materials 1. These interventions are classified into types of strategies: P – “push-pull”, C – cofactor regeneration, R – respiratory disruption, D – degradation knockout. Successful implementations in R. toruloides (Rt) (Zhang et al., 2016b, 2016a) or other oleaginous yeast (O) (Blazeck et al., 2014; Tamano et al., 2013) are reported.
beta-Oxidation knockout was applied manually prior to finding FORCE set for strains #8-15 in order to block fatty acid degradation. The first step for beta-oxidation in peroxisome and mitochondria were knocked out, which are fatty acyl-CoA oxidase (encoded by POX1, rt4374, and rt4384) and dehydrogenase (encoded by rt5702, rt4202, and rt1644), respectively.
Lipid degradation knockout was applied to all designs to produce TAG, including TAG lipase (encoded by TGL2, TGL5, and ATG15) and monoacylglycerol lipase (YJU3).
OptForce identified a total of 15 sets of interventions (see Table 4 and Fig. 3). Several identified genetic interventions match successfully implemented strategies in R. toruloides (4 out of 12) (Zhang et al., 2016a, 2016b) and other oleaginous yeasts (3 out of 12) (Blazeck et al., 2014; Tamano et al., 2013). Identified TAG overproducing strategies can be divided into three group: upregulation of precursor production and TAG production (“push-pull” strategies) (Tai and Stephanopoulos, 2013), cofactor regeneration, and respiratory disruption (see Table 4). “Push-pull” interventions which directly increase the flux throughput present in all combinations. Cofactor regeneration and respiratory disruption interventions, which indirectly support TAG production, by themselves do not lead to robust TAG overproducing phenotypes. However, those interventions when applied in combination further increased TAG production for strains with “push-pull” interventions. When predicting the quantitative effectiveness of interventions, stoichiometric models cannot always capture the synergy of interventions strategies. For example, overexpression of diacylglycerol acyltransferase (DGAT_rm, DGA1) (Table 4) alone could achieve 89% of theoretical yield. However, both interventions, DGA1 and ACC1 (ACCOAC_c), were needed to derive a high-yield strain (Zhang et al., 2016b). Overall, a maximum of two interventions per FORCE set (excluding manually applied degradation knockout interventions) were suggested by OptForce (see Table 4). Supplying NADPH for acyl-CoA synthesis via overexpressing cytoplasmic malic enzyme (ME or rt4393) has shown improvement as a single intervention but decreased lipid yield in the triple overexpression of ACC1 DGA1 ME (Zhang et al., 2016a). Predictions with iRhto1108 showed a different trend, where single interventions had little effect thus double interventions following a “push-pull” strategy (e.g., DGA1) were required to improve yield. As discussed in (Zhang et al., 2016b), a hypothesis for the counterintuitive behavior under ME overexpression is that increasing the flux through malic enzyme might disrupt cellular balance affect lipid biosynthesis and the transhydrogenase cycle. Knocking out fatty acid and lipid degradation pathways is another strategy that was proven effective in Y. lipolytica (Blazeck et al., 2014) and subsequently tested in R. toruloides (Zhang et al., 2016a). For fatty acid degradation, the in vivo knockout targets have included genes involved in peroxisome biogenesis such as PEX10 (Blazeck et al., 2014), fatty acyl-CoA transporter (Ferreira et al., 2018), and beta-oxidation genes such as oxidase (Beopoulos et al., 2008) and (in Y. lipolytica) multifunctional enzyme - dehydrogenase and dehydratase (Blazeck et al., 2014). In the FBA simulation, we chose to disrupt beta-oxidation by blocking the first step of acyl-CoA oxidase in peroxisome (encoded by POX1, rt4374, and rt4384) and acyl-CoA dehydrogenase in mitochondria (encoded by rt5702, rt4202, and rt1644). For lipid degradation, TAG lipase genes have been selected for knockout in S. cerevisiae strain designs (Ferreira et al., 2018). In simulation, we chose to knock out TAG and monoacylglycerol lipase genes including TGL2, TGL5, YJU3, and ATG15. While TGL5 is a bifunctional TAG lipase and lysophosphatidate acyltransferase (for lipid biosynthesis), the latter function can be performed alternatively by SLC1. In addition, TGL5 knockout, in combination with TGL3 and TGL4 knockouts, was found to overall enhanced lipid accumulation in S. cerevisiae (Ferreira et al., 2018).
Fig. 3.
Visualization of triacylglycerol production pathway. Interventions identified by OptForce and implemented in vivo were annotated. Reaction abbreviations are listed in Table 4 and detailed in Supplementary Materials 1. Metabolite abbreviations: DHA – dihydroxyacetone, DHAP – DHA phosphate, Ficyt – ferricytochrome, Focyt – ferrocytochrome, Pyr – pyruvate, Mal – malate, AcCoa – acetyl-CoA, Oaa – oxaloacetate, Cit – citrate, Acon – aconitate, Akg – alpha-ketoglutarate, Sucdhl – S(8)-succinyldihydrolipoamide, SucCoA – succinyl-CoA, TAG – triacylglycerol.
We also compared the OptForce results with interventions that were implemented in other oleaginous organisms. As mentioned before, lipid and fatty acid degradation knockouts were effective in Y. lipolytica (Blazeck et al., 2014). Overexpression of fatty acid synthase (with ACOATA_c being the first step) was found to increase fatty acid and TAG production by more than two-fold in the oleaginous fungus Aspergillus oryzae (Tamano et al., 2013) but was otherwise ineffective in Y. lipolytica (Silverman et al., 2016) implying an organism-specific response. OptForce identified five new interventions that can increase TAG production when being applied in combination with a “push-pull” intervention. Two additional mechanisms for NADPH generation, glucose 6-phosphate dehydrogenase (G6PDH2i_c) in oxidative PPP or glycerol dehydrogenase (GLYCDy_c), are analogous to malic enzyme overexpression. Glycerol dehydrogenase upregulation is theoretically possible but introduces the toxic metabolite dihydroxyacetone (Molin et al., 2003). On the other hand, glucose 6-phosphate dehydrogenase and more broadly the oxidative PPP was found to be upregulated natively under nitrogen limitation in Y. lipolytica (two-fold flux increase in 13C-MFA study (Wasylenko et al., 2015)) and R. toruloides (inferred by mutant phenotypes (Zhang et al., 2016a)). Thus, OptForce correctly identified the importance of oxidative PPP upregulation though a directed intervention may not be necessary. Finally, downregulation perturbations were suggested for aconitase (ACONTa_m), oxoglutarate dehydrogenase (AKGDa_m), or ferrocytochrome-c:oxygen oxidoreductase (FECOOR_m). These downregulations decrease cellular respiration, repress growth, and indirectly allow more carbon to be used in TAG production. In detail, the downregulation of the citric acid cycle reduces the production of mitochondrial NADH which supports ATP production by the electron transport chain. Lower ATP production leads to less biomass formation, implying that the unused reduced cofactors are regenerated through the production of TAG. Growth repression can also be achieved using nutrient limitation and culture optimization (Blazeck et al., 2014) while allowing the cell to maintain its growth robustness trait. Overall, iRhto1108-driven strain redesign using OptForce identifies many “push-pull” strategies. Not all strategies are in agreement with the experimental findings but upon careful interpretation of the desired metabolic redirections, alternative ways of achieving the same goal can be designed that bypass the specified interventions.
3. Conclusion
In this work, we collect and organize functional genomics data (Coradetti et al., 2018) and prior knowledge into the genome-scale metabolic model iRhto1108. Essential cellular metabolism and growth capability of the model were validated extensively with experimental results, including gene essentiality (Coradetti et al., 2018) and growth data. iRhto1108 was also able to recapitulate experimentally-observed lipid accumulation phenotypes (Shen et al., 2017; Wang et al., 2018; Wu et al., 2011). We showed that iRhto1108 can comprehensively capture R. toruloides’s metabolism and provide meaningful predictions that were validated with experimental data including suggestion of genetic perturbations leading to triacylglycerol overproducing strains. We envision that in the future iRhto1108 will aid in exploring the metabolic potential of R. toruloides, following in the footsteps of the model organisms Saccharomyces cerevisiae (Borodina and Nielsen, 2014). Despite careful curation, a large number of blocked reactions (i.e., 677 out of 2204) remained in the model spanning multiple pathways. Most of them are transport reactions (i.e., 194 reactions) connecting the network. The rest participate in secondary metabolism and degradation of amino acids, fatty acids, and lipids. We chose to keep them in the hope that they would aid in gap-filling attempts in the future.
Because there is another R. toruloides model available (rhto-GEM v. 1.1.1) (Tiukova et al., 2019), we hereby provide our perspective on the contextual applicability of the two models for potential users to consider. First it is important to note that iRhto1108 and rhto-GEM were reconstructed for different R. toruloides strains (i.e., IFO0880 and NP11, respectively). To-date gene essentiality data is only available for strain IFO0880 and both models were benchmarked using this same dataset. iRhto1108 performed better in the following scores of accuracy (i.e., 84.5% vs. 73.0%), sensitivity (i.e., 94.3% vs. 86.6%), and specificity (i.e., 53.8% vs. 39.2%) (Tiukova et al., 2019). This may be due to the fact that the gene essentiality data were for exactly the same strain. In terms of substrate utilization, both models are comparable except that iRhto1108 can recapitulate growth on arabinose and cellobiose as carbon source and growth on proline as nitrogen source. In term of genome coverage, iRhto1108 contains slightly more genes than rhto-GEM (i.e., 1108 vs. 926 genes) or after the removal of blocked reactions (i.e., 806 vs. 624 genes). Lipid metabolism was also formulated at different levels of detail in the two models. For example, in iRhto1108, TAG was formulated as a single lipid species with the same acyl group in all three positions and the acyl group was composed of chain-length specific fatty acyl in the experimentally measured proportion. In rhto-GEM, TAG was formulated as 25 separate lipid species with a chain-specific acyl group of different length (i.e., SLIMEr formulation) (B. J. Sánchez et al., 2018b). Thus, rhto-GEM can accommodate high-resolution lipidomics data and track the relative abundances of lipid species of different acyl chain length variation. Due to the complete enumeration of lipid species, the numbers of reactions (i.e., 4930) and metabolites (i.e., 3374) in rhto-GEM are larger. Because lipid metabolism in iRhto1108 is described in a more economical fashion, iRhto1108 would be more tractable to use, unless a detailed lipidomics analysis is carried out. In addition, iRhto1108 uses BiGG ID format (King et al., 2016) to facilitate usability experience, especially in model analyses requiring manual inspection. Escher maps for metabolic network visualization (King et al., 2015a) are also provided for the same purpose.
Stoichiometric models can capture all known interconversion routes from substrates to biomass components and products and globally balance cofactor needs. However, they inherently cannot mechanistically link enzyme levels with metabolite concentrations and metabolic fluxes. To this end, kinetic models offer a promising formalism for integrating such heterogeneous datasets (A. Chowdhury et al., 2015a). Efforts towards this direction will require 13C-derived information on internal fluxes under a variety of genetic and environmental perturbations along with secreted products and biomass yield (Khodayari and Maranas, 2016). To this end, atom mapping models for all reactions in the R. toruloides model (Gopalakrishnan et al., 2015) will have to be constructed and robust methodologies for flux elucidation and kinetic model parameterization will have to be developed accounting for the multi-compartment nature of metabolism.
4. Methods
4.1. Draft reconstruction from existing fungal genome-scale reconstruction and model refinements
In general, the workflow used in this study followed an established protocol described in Mueller et al. (2013) for generating a metabolic model utilizing a previously built metabolic model for a closely related organism. This protocol provides a priority structure for assigning functions to genes with multiple annotations. The most recent genome sequence and gene annotations of R. toruloides was used for this reconstruction (Coradetti et al., 2018). The unannotated sequence of mitochondrial genome of R. toruloides was annotated using RAST (Brettin et al., 2015) and MAP (Huntemann et al., 2016) on the KBase platform (Arkin et al., 2018) (see Supplementary Materials 1). An initial draft reconstruction was assembled by mapping genes and reactions from the S. cerevisiae genome-scale model yeast 7.6 (Aung et al., 2013) with updated information from R. Chowdhury et al. (2015). Briefly, first, homologous genes were determined by a bidirectional protein BLAST procedure (Mueller et al., 2013) with an e-value cutoff of 10−5. The Boolean logic given by each gene-protein-reaction (GPR) association in yeast 7.6 was then evaluated using these bidirectional hits. A reaction was next added to the draft model only if its GPR satisfied can be satisfied with the present gene homologs necessary for a functional protein. This draft reconstruction was further extended with KBase’s “build fungal model” application (Arkin et al., 2018) which extracts homologous genes and associated reactions from a library of fungal genome-scale models using similar homologous genes identification schematics. We prioritized building the initial scaffold using yeast 7.6 model rather than KBase because the biochemical information in yeast 7.6 was experimentally verified whenever possible. Next, additional reactions and GPRs were manually added using the annotated genome and validated with NCBI’s Conserved Domain Database (Marchler-Bauer et al., 2015). Missing assignments of reaction compartments were resolved using the protein subcellular localization prediction software DeepLoc (Almagro Armenteros et al., 2017). Adjustments made to reactions reversibility and activation in the default model are commented in Supplementary Materials 1 whereas other adjustments made specifically to model simulations are stated in the main text.
In addition to the biologically relevant additions and curations made to iRhto1108 (see section 2.1, 2.2, 4.2), additional validations and refinements were performed to improve the model quality. Specifically, the GPRs of S. cerevisiae yeast 7.6 reactions that were recently updated in the yeast model repository (https://github.com/SysBioChalmers/yeast-GEM, version 8.3.3) were evaluated and modifications were made to the GPRs of 22 reactions. Furthermore, we ensured that every reaction is mass and charge balanced and as a result we updated 663 metabolite formulae (i.e., 33.5%) and 94 reaction stoichiometries (i.e., 4.4%) using standardized metabolite formulae from MetaCyc (Caspi et al., 2018) and ModelSEED (Henry et al., 2010) databases. Database verification for metabolite formulae was at 77.8% coverage and we manually assigned the formulae assignments for the remainders to ensure all reactions were mass and charge balance (excluding pseudo and exchange reactions). Further model curation involved identifying and fixing thermodynamically infeasible cycles. For instance, cycles that allowed the unbounded production of ATP were eliminated by blocking the reverse direction of the ATP hydrolysis reactions (Fritzemeier et al., 2017). iRhto1108 model structure was checked using the memote test suite (Lieven et al., 2018) with the model annotations standardized to the MIRIAM namespace (Juty et al., 2012), which is used by memote. The final version of the model passed all memote tests.
4.2. Generation of biomass reactions
Both experimental data from literature and those generated in this study (see section 4.2), alongside the original yeast 7.6 biomass reaction, were used to determine the metabolite coefficients in the biomass objective functions for carbon and nitrogen limitation conditions (Supplementary Materials 1). The macromolecular composition was measured for R. toruloides (see section 4.2). Other experimentally determined biomass specifications for R. toruloides were also incorporated, including genome GC content (Nakase and Komagatai, 1971), lipid composition in nitrogen limitation conditions (i.e., natural, phospho, and glycolipids composition) (Shen et al., 2017), relative abundances of RNA nucleotide (this study), and relative abundance of acyl groups and free fatty acids in lipid (this study). Experimentally determined specifications taken from S. cerevisiae used in this model were amino acid, inorganic compound (phosphate, sulphate, and metal ions), and cell wall compositions (Klis et al., 2014), as corresponding data for R. toruloides was not available. Additional data adopted from the yeast 7.6 model were lipid subspecies composition (e.g., phosphatidylinositol, phosphatidylcholine, phosphatidylethanolamine, and phosphatidylserine composition). These data from experiments on S. cerevisiae and yeast 7.6 were deemed acceptable as R. toruloides and S. cerevisiae are closely related. The list of biomass constituents was reviewed and validated with relevant literature and experimental gene essentiality results (see section 2.2). Without the measurement of the soluble metabolite pool, the coefficients of twelve cofactors and prosthetic groups were set to a small number of 10−4 so as to impose a biosynthesis requirement on the in silico model, resulting in 0.06% of the total biomass by weight. A similar measure was also adopted for the biomass reactions in other models such as those for S. cerevisiae models, including yeast 7.6 (Mo et al., 2009). Metabolite coefficients associated with growth-associated ATP maintenance were also updated (see section 4.4). Calculations and detailed listings of metabolites and coefficients in the biomass reaction are provided in the Supplementary Materials 1.
A total of 68 metabolites were included in the biomass component list for the R. toruloides model. The veracity of these inclusions was ascertained using data from single-gene knockout essentiality experiments in R. toruloides (Coradetti et al., 2018) and S. cerevisiae (see Supplementary Materials 2). Overlaps and differences between yeast 7.6 and iRhto1108’s list of biomass constituents are summarized in Table 1. Among the differences were eight nucleotide monophosphates which were replaced in iRhto1108 by the corresponding nucleotide triphosphates and pyrophosphate in order to directly account for DNA and RNA polymerization. The generic mannan metabolite was likewise substituted with N-glycan, O-glycan, and the glycosylphosphatidylinositol anchor (Goto, 2007; Orlean, 2012). Similarly, the “generic” free fatty acid designation in yeast 7.6 was replaced with seven distinct free fatty acid compounds found with abundances of >1% by weight in the measurement of saponified fatty acids using LC-MS (see section 4.7). In addition to the substitutions detailed above, nine cofactors and prosthetic groups suggested initially by Xavier et al. (2017) (Xavier et al., 2017) were added to the list of biomass constituents so as to improve iRhto1108’s gene essentiality predictions. Seven new inorganic ions were also included in the biomass reaction following the measurements for S. cerevisiae by Lange and Heijnen (2001) as these ions are known to be essential (Supplementary Materials 2). Although the biomass reaction in iRhto1108 is organism-specific, the list of biomass constituents is not unique to iRhto1108 and is applicable for the models of S. cerevisiae and possibly other closely related species such as Y. lipolytica. All newly added constituents in iRhto1108 were found to be essential for S. cerevisiae growth (see Supplementary Materials 2).
4.3. Modeling simulation
Flux balance analysis (FBA) was used throughout the process for model validation and prediction (Orth et al., 2010). Growth phenotypes were obtained using FBA with the objective of maximizing the biomass reaction (vbiom) whose flux is equivalent to the growth rate. In general, the substrate uptake rates such as glucose (vglc) were set to the experimentally determined values if available. Otherwise, carbon substrate (e.g., glucose, xylose, or glycerol) uptake rate for a simulation was set to 5 mmol gDW−1 hr−1 which was close to the physiological glucose uptake rate reported in Zhu et al. (2012). For examining the model’s ability to utilize amino acid as nitrogen source, a specific amino acid uptake rate was set to 0.25 mmol gDW−1 hr−1 (i.e., 5% of default substrate uptake rate of 5 mmol gDW−1 hr−1). All simulations were performed using the carbon limitation condition model iRhto1108C unless iRhto1108N was specified to be used (i.e., for the nitrogen limitation condition).
For gene essentiality and mutant auxotrophy predictions in rich media (Coradetti et al., 2018), supplementary compound uptake rates were set to 0.25 mmol gDW−1 hr−1 (i.e., 5% of default substrate uptake rate of 5 mmol gDW−1 hr−1). Rich media components were described in Coradetti et al. (2018) and are listed in Supplementary Materials 1. The undefined composition of yeast extract in Yeast-Peptone-Dextrose media was assumed to be that of YNB media plus 20 amino acids and d-glucose. The supplementary nutrients present in YNB included thiamine, riboflavin, nicotinate, pyridoxin, folate, (R)-pantothenate, 4-aminobenzoate, and myo-inositol. Oxygen and ammonium uptake rates were unconstrained in all simulations. Gene knockout was translated to the corresponding reaction(s) knockout by examining the Boolean gene-protein-reaction rules. A reaction was knocked out in the model by setting the corresponding upper and lower flux bounds to zero. A gene was determined to be essential if the knockout mutant’s maximal growth rate calculated by FBA was less than 0.0001 hr−1. The criteria for experimentally determined gene essentiality are described in Coradetti et al. (2018). The calculations were performed using the COBRApy package (version 0.13.4) (Ebrahim et al., 2013).
4.4. Determination of ATP maintenance requirements
Non-growth (NGAM) and growth associated ATP maintenance (GAM) values were determined using continuous chemostat data from Shen et al. (2013). A functional draft model utilizing a biomass reaction without a GAM demand was used to determine the biomass synthesis requirement excluding ATP maintenance. To calculate the ATP maintenance requirement per experimental data point, glucose uptake rate (vglc) and growth rate (vbiom) were set to the experimentally determined values. Next, ATP maintenance requirement was given by the ATP hydrolysis rate (vatpm) which is the maximal through the following reaction: ATP + H2O → ADP + H+ + HPO42−. An NGAM value of 1.01 mmol ATP gDW−1 hr−1 was found for no growth at vglc of 0.032 mmol gDW−1 hr−1, reported in Shen et al., 2013. GAM value was the slope of the line (found using linear regression) through all the maximal ATP maintenance rates constrained by the experimental vglc, vbiom, and NGAM (by setting the intercept to the NGAM value). For nitrogen limitation condition, because the lipid composition of the biomass varied with the growth rate (Shen et al., 2013), per chemostats data point, the coefficients of the biomass reaction were adjusted in order to account for the compositional change. Specifically, lipid composition was adjusted to the experimentally determined value and the other macromolecular compositions (such as protein, carbohydrate, RNA, and DNA) were adjusted while maintaining the original relative levels. For carbon limitation conditions, the compositional profile remained relatively constant across the growth rates (Shen et al., 2017, 2013) and thus no adjustments were made to the biomass reaction whilst determining the GAM value. Details of these simulations are provided in the Supplementary Materials 2.
4.5. Phenotype phase plane and gene-flux correlation analysis
Phenotype phase planes (Edwards et al., 2002) were used to calculate the maximal triacylglycerol yield under growth optimization priority under nutrient scarcity. Minimal oxygen, ammonium, phosphate, and sulphate uptake rates necessary for growth were found by minimizing the respective uptake rate subjected to maximal growth yield. The uptake rates were 12.8, 2.4, 0.20, and 0.03 mmol gDW−1 hr−1 for oxygen, ammonium, phosphate, and sulphate, respectively. Oxygen and ammonium (or phosphate or sulphate) uptake rates’ ranges were then discretized to 30 points between zero to the calculated minimal uptake value at maximal growth rate on the phenotype phase plane. For every point on the plane, a two-step procedure was applied. First, the growth rate was maximized subject to the limitation in oxygen and ammonium (or phosphate or sulphate) uptake rates. Second, triacylglycerol production was maximized subject to not only nutrient limitations but also first-priority growth optimization by constraining the biomass production to be at least the maximal amount determined in the previous step. These model-predicted phenotypes were used to construct the contour plots shown in Fig. 2. TAG’s molecular weight of 882.40 mg mmol−1 derived from lipid’s acyl group composition was used in calculating TAG yield (unit of g TAG/g Glucose). Next, to compare the in silico flux redistribution in iRhto1108 (constructed for strain IFO0880) to the experimentally-observed differential gene expression in strain NP11 (Zhu et al., 2012), we first established the mapping between NP11 genes and model reactions using bidirectional BLAST hit (Mueller et al., 2013). Once a mapping was established, the fold-change of the in silico metabolic flux can be calculated.
4.6. Identification of genetic perturbation for triacylglycerol overproduction
The OptForce procedure was used to identify sets of genetic perturbations that once implemented result in strain with overproducing phenotypes, hereby called overproducing strains. Detailed formulations and explanations were documented in Ranganathan et al. (2010) and additionally in (Chowdhury et al., 2015b). Throughout the simulation, diacylglycerol acyltransferase in lipid particle (DGAT_l) was turned off to allow flux to go through only endoplasmic reticulum version of the reaction (DGAT_rm). The first step in the OptForce procedure is to contrast flux ranges of a wild-type to those of an overproducing strain, one reaction at a time. Flux data obtained from 13C-MFA study of Y. lipolytica wild-type under nitrogen limitation (Wasylenko et al., 2015) was used to constrain wild-type fluxes in this study. Specifically, 13C-MFA flux data for wild-type strain (among four sets of MFA fluxes) without separating cytoplasmic and mitochondrial malic enzyme fluxes were used because the study concluded that the compartmentalized malic enzyme fluxes were indistinguishable (Wasylenko et al., 2015). In the simulation, 13C-MFA flux range for malic enzyme were constrained on the sum of three reactions, namely cytoplasmic (ME2_c), mitochondrial with NAD+ (ME1_m), and mitochondrial with NADP+ (ME2_m) malic enzyme. The flux basis for this study was the glucose uptake of 10 mmol gDW−1 hr−1. We assess metabolic fluxes for both wild-type and overproducing strains under nitrogen limited conditions. Nitrogen availability level of 33% (0.7 mmol gDW−1 hr−1 in the basis of glucose uptake) was used in all OptForce simulations and was calculated by minimizing ammonium uptake rate for the wild-type strain with respect to reaction fluxes constrained by 13C-MFA data. Reference fluxes for overproducing strain were constrained by forced production of TAG to 90% of theoretical yield, growth yield required at 10% of wild-type, and aforementioned nitrogen limitation. In this work, flux variability analysis (FVA) (Mahadevan and Schilling, 2003) were used on iRhto1108 to generate the flux ranges for both wild-type and overproducing strains. Reactions with non-overlapping flux ranges were potential candidates for genetic perturbation. Reactions with FVA ranges being lower, higher, or equal zero (only) in overproducing strain compared to wild-type strain were put in MUSTL, MUSTU, or MUSTX set, respectively. Value ranges of flux sums (e.g., v1 + v2) and differences (e.g., v1 – v2) were also evaluated using FVA. Reaction pairs with higher values of flux sums, lower values of flux sums, and higher value of flux differences were put in MUSTUU, MUSTLL, and MUSTLU set, respectively. Reaction in the pair are put in the respective MUSTU or MUSTL depending on their membership in MUST paired set. The combinations of overexpression (MUSTU), downregulations (MUSTL), or knockout (MUSTX) were determined using a bi-level mixed-integer linear programming optimization formulation and put in FORCE set (Ranganathan et al., 2010). Overexpression and downregulations were applied to the model by setting the reaction flux bound (lower and upper bound, respectively) to the level calculated in overproducing strain. Reaction knockout was applied by setting both the lower and upper bounds to zero. Because of the production minimization objective, OptForce could not suggest any overproducing strains due to lipid degradation pathways. To eliminate the TAG synthesis-degradation cycle that occurred under production minimization objective, TAG degradation and cycling pathways were knocked out in simulation, including neutral lipid lipase (6 reactions). In addition, fatty acid secretions are not observed in R. toruloides in wild-type or engineered strains and thus were knocked out (12 reactions). For strains #8-15, the fatty acid degradation pathway was also blocked by knocking out the first step, fatty acyl-CoA ABC transporter to peroxisome (12 reactions) and tetradecanoate diffusion. The list of these knocked-out reactions are provided in the Supplementary Materials 2. OptForce (Ranganathan et al., 2010) was performed using GAMS (version 24.8.5, GAMS Development Corporation) using IBM ILOG CPLEX solver (version 12.7.1.0) on the high-performance computing resource cluster of Pennsylvania State University’s Institute for CyberScience Advanced CyberInfrastructure (ICS-ACI).
4.7. Experimental determination of biomass composition
4.7.1. Strains and cultivation
Biomass composition was determined for R. toruloides IFO0880 cultivated in chemostat using two limiting nutrients (either glucose or nitrogen). An overnight stock was prepared in minimal medium that contains yeast nitrogen base without amino acid (YNB, Sigma Y0626) and 20 g/L glucose. It was then inoculated into 250 mL culture to grow in continuous mode in a 500-mL chemostat (Sixfors; Infors AG, Bottmingen, Switzerland). For the carbon limitation condition, YNB with 0.8 g/L glucose was used. For the nitrogen limitation condition, 20 g/L glucose and 0.05 g/L (NH4)2SO4 were supplemented to YNB without ammonium sulphate (Difco, 291940). For both conditions, the culture was stirred at 400 rpm for sufficient oxygen, and was kept at 0.1 h−1 growth rate. After the culture had reached steady state (pH, oxygen, and cell density), it was harvested for the biomass measurement.
4.7.2. Biomass component analysis
DNA was measured using diphenylamine reagent. 7.5 mL culture was pelleted and washed by 1 mL cold 1 mM HClO4. Serial dilutions of 1 mg/mL Calf thymus DNA (Sigma) were prepared for calibration. Samples were hydrolyzed in 500 μL 1.6 M HClO4 for 30 min at 70 °C, and then reacted with 1 ml diphenylamine reagent (0.5 g diphenylamine in 50 mL glacial acetate, 0.5 ml 98% H2SO4, and 0.125 ml 3.2% acetaldehyde water solution) at 50 °C for 3 h. After centrifugation, the supernatant was taken for OD600 measurement.
RNA was measured by 260 nm absorption. Basically, 2.5 mL culture was pelleted and washed, and digested with 300 μL 0.3M KOH at 37 °C for 60 min. DNA and protein were then precipitated by 100 μL 3M HClO4. Supernatant was taken and precipitate was washed with 600 μL 0.5M HClO4. Absorption of combined supernatant at 260 nm, 900 nm, and 977 nm was measured. RNA concentration can be calculated as 5.6*(A260-A260blank)/(A977-A900) in μg/mL. RNA composition was calculated from the relative abundances of bases in RNA-Seq data. Extracted RNA was sequenced on a HiSeq2500 (Illumina). Fastq files were generated and demultiplexed with the bcl2fastq v1.8.4 Conversion Software Reads were trimmed by Trimmomatic (Bolger et al., 2014) and analyzed using FastQC (Andrews, 2010). STAR version 2.5.4a (Dobin et al., 2013) and featureCounts from the Subread package, version 1.5.2 (Liao et al., 2014) were used to map the reads to the R. toruloides strain IFO0880 reference genome (Coradetti et al., 2018) and obtain relative counts.
Protein was measured using the Biuret method. Briefly, 2.5 mL culture was pelleted, washed and boiled in 100 μL 3M NaOH at 98 °C for 5 min. After cool down, the mixture was reacted with 100 μL CuCO4 for 5 min at room temperature. After centrifugation, supernatant was taken for determination of 555 nm absorption, which was calibrated by serial dilution of Bovine serum albumin (BSA) solutions (Thermo).
Lipid was determined by measuring saponified fatty acids using LC-MS. Briefly, cell pellet from 2.5 mL culture was extracted and saponified in 1 mL 0.3M KOH–MeOH solution at 80 °C for 60 min, then neutralized by 100 μL formic acid, and then extracted by 1 mL hexane 20 nM. 20 nM isotope-labeled fatty acid standards (U–13C–C16:0, U–13C–C18:1, U–13C–C18:2; Cambridge Isotope) were added before saponification as internal standards. Extracted fatty acids were dried under N2 and redissolved in 200 μL acetonitrile:methanol (1:1), and then analyzed by reversed-phase C8 column chromatography coupled to negative-ion mode, full-scan high-resolution LC-MS (Exactive, Thermo).
Carbohydrate was determined by hydrolyzing cell pellet from 2.5 mL culture in 100 μL 2M HCl at 80 °C for 1 h. 0.4 mg U–13C-glucose was added as internal control before hydrolyzation. The lysate was neutralized by 100 μL 2M NH4HCO3, diluted in 1.8 mL 80% MeOH and centrifuged. The supernatant was taken and analyzed by negative-ion mode LC-MS equipped with hydrophilic interaction liquid chromatography (Q Exactive Plus, Thermo). Mass peaks equivalent to C6H12O6 were selected for quantification.
4.8. Growth experiments in batch culture
4.8.1. Strain, media, and culture conditions
YPD medium (10 g/L yeast extract, 20 g/L peptone, and 20 g/L glucose) was used for routine growth of R. toruloides IFO0880. Growth rates of R. toruloides IFO0880 was tested in minimal medium (MM) using different C/N ratios (20 g/L d-glucose, 1.7 g/L yeast nitrogen base without amino acids and ammonium sulphate, 0.4–7.1 g/L NH4Cl, C/N = 5:1–90:1, 25 mM Na2HPO4, 150 mM KH2PO4, pH 5.6). Stationary phase R. toruloides IFO0880 seed cultures were obtained by inoculating single colonies from a YPD agar plate into 25 mL YPD liquid medium in 125 mL baffled flask. For growth, the seed cultures were then used to inoculate into 50 mL minimal medium in 250 mL baffled flask with a starting OD600 of 1. The cells were then grown at 30 °C and 250 rpm. All experimental conditions were performed with four replicates. Growth viability on amino acids was tested in minimal media with different amino acids as nitrogen source (11.9 g/L alanine, 14 g/L serine, 5.8 g/L arginine, 9.7 g/L lysine, 15.3 g/L proline, 15.9 g/L threonine and 7.1 g/L NH4Cl as control). The growth viability of cellobiose, sucrose, and mannose were tested in modified media (70 g/L of carbon source, 10 g/L yeast extract, 1.7 g/L yeast nitrogen base without amino acids and ammonium sulphate, 8 g/L (NH4)2SO4, and 1 g/L MgSO4, pH 5.6).
4.8.2. Dry cell weight measurement
Cell growth was measured by the absorbance at 600 nm using cell density meter. Dry cell weights were determined as follows. The 1–5 ml of culture samples were collected into pre-weighed tubes and centrifuged at 16,000×g for 5 min. Supernatant was discarded, and pellets were then washed twice with 50 mM phosphate buffered saline. Washed pellets were dried till constant weight at 65 °C for 24–48 h. The tubes were then weighed.
4.9. Data and materials
The models in MS-Excel spreadsheets format and the formulation of biomass reaction is available in Supplementary Materials 1. Gene essentiality predictions, phenotype predictions, and OptForce strain design suggestions are all provided in the Supplementary Materials 2. Additionally, the following files are available from this study: (i) a visual metabolic network map reconstructed using the Escher toolbox (King et al., 2015a) and (ii) memote’s report on model standardization, annotation, and network consistency (Lieven et al., 2018). The models (available formats include MS-Excel spreadsheets, JSON, SBML (version 3, level 1), MATLAB, and YAML) and the aforementioned data were provided in a memote-created repository published on GitHub (https://github.com/maranasgroup/iRhto_memote) (Lieven et al., 2018). The models were also imported to KBase Narrative (see GitHub repository).
Funding
This work was supported by the U. S. Department of Energy Office of Science, Office of Biological and Environmental Research (Award Number DE-SC0018260).
Acknowledgements
We would like to thank Lin Wang (Pennsylvania State University) for the advice on OptForce simulation, Debolina Sarkar (Pennsylvania State University) for the critical review of the manuscript, and Jose Juan Almagro Armenteros (Technical University of Denmark) for running DeepLoc to predict protein subcellular localization. We thank the Pennsylvania State University’s Institute for CyberScience Advanced CyberInfrastructure (ICS-ACI) for providing high-performance computing resources.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mec.2019.e00101.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Adrio J.L. Oleaginous yeasts: promising platforms for the production of oleochemicals and biofuels. Biotechnol. Bioeng. 2017;114:1915–1920. doi: 10.1002/bit.26337. [DOI] [PubMed] [Google Scholar]
- Almagro Armenteros J.J., Sønderby C.K., Sønderby S.K., Nielsen H., Winther O. DeepLoc: prediction of protein subcellular localization using deep learning. Bioinformatics. 2017;33:3387–3395. doi: 10.1093/bioinformatics/btx431. [DOI] [PubMed] [Google Scholar]
- Andrews S. 2010. FastQC: a Quality Control Tool for High Throughput Sequence Data.http://www.bioinformatics.babraham.ac.uk/projects/fastqc [WWW Document]. URL. [Google Scholar]
- Arkin A.P., Cottingham R.W., Henry C.S., Harris N.L., Stevens R.L., Maslov S., Dehal P., Ware D., Perez F., Canon S., Sneddon M.W., Henderson M.L., Riehl W.J., Murphy-Olson D., Chan S.Y., Kamimura R.T., Kumari S., Drake M.M., Brettin T.S., Glass E.M., Chivian D., Gunter D., Weston D.J., Allen B.H., Baumohl J., Best A.A., Bowen B., Brenner S.E., Bun C.C., Chandonia J.-M., Chia J.-M., Colasanti R., Conrad N., Davis J.J., Davison B.H., DeJongh M., Devoid S., Dietrich E., Dubchak I., Edirisinghe J.N., Fang G., Faria J.P., Frybarger P.M., Gerlach W., Gerstein M., Greiner A., Gurtowski J., Haun H.L., He F., Jain R., Joachimiak M.P., Keegan K.P., Kondo S., Kumar V., Land M.L., Meyer F., Mills M., Novichkov P.S., Oh T., Olsen G.J., Olson R., Parrello B., Pasternak S., Pearson E., Poon S.S., Price G.A., Ramakrishnan S., Ranjan P., Ronald P.C., Schatz M.C., Seaver S.M.D., Shukla M., Sutormin R.A., Syed M.H., Thomason J., Tintle N.L., Wang D., Xia F., Yoo H., Yoo S., Yu D. KBase: the United States department of energy systems biology knowledgebase. Nat. Biotechnol. 2018;36:566–569. doi: 10.1038/nbt.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung H.W., Henry S.A., Walker L.P. Revising the representation of fatty acid, glycerolipid, and glycerophospholipid metabolism in the consensus model of yeast metabolism. Ind. Biotechnol. 2013;9:215–228. doi: 10.1089/ind.2013.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beopoulos A., Mrozova Z., Thevenieau F., Le Dall M.-T., Hapala I., Papanikolaou S., Chardot T., Nicaud J.-M. Control of lipid accumulation in the yeast Yarrowia lipolytica. Appl. Environ. Microbiol. 2008;74:7779–7789. doi: 10.1128/AEM.01412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beopoulos A., Nicaud J.-M., Gaillardin C. An overview of lipid metabolism in yeasts and its impact on biotechnological processes. Appl. Microbiol. Biotechnol. 2011;90:1193–1206. doi: 10.1007/s00253-011-3212-8. [DOI] [PubMed] [Google Scholar]
- Blazeck J., Hill A., Liu L., Knight R., Miller J., Pan A., Otoupal P., Alper H.S. Harnessing Yarrowia lipolytica lipogenesis to create a platform for lipid and biofuel production. Nat. Commun. 2014;5:3131. doi: 10.1038/ncomms4131. [DOI] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommareddy R.R., Sabra W., Maheshwari G., Zeng A.-P. Metabolic network analysis and experimental study of lipid production in Rhodosporidium toruloides grown on single and mixed substrates. Microb. Cell Factories. 2015;14:36. doi: 10.1186/s12934-015-0217-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodina I., Nielsen J. Advances in metabolic engineering of yeast Saccharomyces cerevisiae for production of chemicals. Biotechnol. J. 2014;9:609–620. doi: 10.1002/biot.201300445. [DOI] [PubMed] [Google Scholar]
- Brettin T., Davis J.J., Disz T., Edwards R.A., Gerdes S., Olsen G.J., Olson R., Overbeek R., Parrello B., Pusch G.D., Shukla M., Thomason J.A., Stevens R., Vonstein V., Wattam A.R., Xia F. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015;5:8365. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzzini P., Innocenti M., Turchetti B., Libkind D., van Broock M., Mulinacci N. Carotenoid profiles of yeasts belonging to the genera Rhodotorula , Rhodosporidium , sporobolomyces , and sporidiobolus. Can. J. Microbiol. 2007;53:1024–1031. doi: 10.1139/W07-068. [DOI] [PubMed] [Google Scholar]
- Caspi R., Billington R., Fulcher C.A., Keseler I.M., Kothari A., Krummenacker M., Latendresse M., Midford P.E., Ong Q., Ong W.K., Paley S., Subhraveti P., Karp P.D. The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res. 2018;46:D633–D639. doi: 10.1093/nar/gkx935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castañeda M.T., Nuñez S., Garelli F., Voget C., De Battista H. Comprehensive analysis of a metabolic model for lipid production in Rhodosporidium toruloides. J. Biotechnol. 2018;280:11–18. doi: 10.1016/J.JBIOTEC.2018.05.010. [DOI] [PubMed] [Google Scholar]
- Chan S.H.J., Cai J., Wang L., Simons-Senftle M.N., Maranas C.D. Standardizing biomass reactions and ensuring complete mass balance in genome-scale metabolic models. Bioinformatics. 2017;33:3603–3609. doi: 10.1093/bioinformatics/btx453. [DOI] [PubMed] [Google Scholar]
- Chowdhury A., Khodayari A., Maranas C.D. Improving prediction fidelity of cellular metabolism with kinetic descriptions. Curr. Opin. Biotechnol. 2015;36:57–64. doi: 10.1016/J.COPBIO.2015.08.011. [DOI] [PubMed] [Google Scholar]
- Chowdhury A., Zomorrodi A.R., Maranas C.D. Bilevel optimization techniques in computational strain design. Comput. Chem. Eng. 2015;72:363–372. doi: 10.1016/J.COMPCHEMENG.2014.06.007. [DOI] [Google Scholar]
- Chowdhury R., Chowdhury A., Maranas C. Using gene essentiality and synthetic lethality information to correct yeast and CHO cell genome-scale models. Metabolites. 2015;5:536–570. doi: 10.3390/metabo5040536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubukov V., Uhr M., Le Chat L., Kleijn R.J., Jules M., Link H., Aymerich S., Stelling J., Sauer U. Transcriptional regulation is insufficient to explain substrate-induced flux changes in Bacillus subtilis. Mol. Syst. Biol. 2013;9:709. doi: 10.1038/msb.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coradetti S.T., Pinel D., Geiselman G.M., Ito M., Mondo S.J., Reilly M.C., Cheng Y.-F., Bauer S., Grigoriev I.V., Gladden J.M., Simmons B.A., Brem R.B., Arkin A.P., Skerker J.M. Functional genomics of lipid metabolism in the oleaginous yeast Rhodosporidium toruloides. Elife. 2018;7 doi: 10.7554/eLife.32110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahim A., Lerman J.A., Palsson B.O., Hyduke D.R. COBRApy: constraints-based reconstruction and analysis for Python. BMC Syst. Biol. 2013;7:74. doi: 10.1186/1752-0509-7-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards J.S., Ramakrishna R., Palsson B.O. Characterizing the metabolic phenotype: a phenotype phase plane analysis. Biotechnol. Bioeng. 2002;77:27–36. doi: 10.1002/bit.10047. [DOI] [PubMed] [Google Scholar]
- Evans C.T., Ratledge C. Possible regulatory roles of ATP:citrate lyase, malic enzyme, and AMP deaminase in lipid accumulation by Rhodosporidium toruloides CBS 14. Can. J. Microbiol. 1985;31:1000–1005. doi: 10.1139/m85-189. [DOI] [Google Scholar]
- Feist A.M., Henry C.S., Reed J.L., Krummenacker M., Joyce A.R., Karp P.D., Broadbelt L.J., Hatzimanikatis V., Palsson B.Ø. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol. Syst. Biol. 2007;3:121. doi: 10.1038/msb4100155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feist A.M., Palsson B.O. The biomass objective function. Curr. Opin. Microbiol. 2010;13:344–349. doi: 10.1016/J.MIB.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feist A.M., Zielinski D.C., Orth J.D., Schellenberger J., Markus J., Palsson B.Ø. Model-driven evalution of the production potential for growth coupled products of Escherichia coli. Metab. Eng. 2010;12:173–186. doi: 10.1016/j.ymben.2009.10.003.Model-driven. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira R., Teixeira P.G., Gossing M., David F., Siewers V., Nielsen J. Metabolic engineering of Saccharomyces cerevisiae for overproduction of triacylglycerols. Metabol. Eng. Commun. 2018;6:22–27. doi: 10.1016/J.METENO.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzemeier C.J., Hartleb D., Szappanos B., Papp B., Lercher M.J. Erroneous energy-generating cycles in published genome scale metabolic networks: identification and removal. PLoS Comput. Biol. 2017;13 doi: 10.1371/journal.pcbi.1005494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garay L.A., Sitepu I.R., Cajka T., Chandra I., Shi S., Lin T., German J.B., Fiehn O., Boundy-Mills K.L. Eighteen new oleaginous yeast species. J. Ind. Microbiol. Biotechnol. 2016;43:887–900. doi: 10.1007/s10295-016-1765-3. [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan S., Maranas C., Gopalakrishnan S., Maranas C.D. Achieving metabolic flux analysis for S. cerevisiae at a genome-scale: challenges, requirements, and considerations. Metabolites. 2015;5:521–535. doi: 10.3390/metabo5030521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto M. Protein O -glycosylation in fungi: diverse structures and multiple functions. Biosci. Biotechnol. Biochem. 2007;71:1415–1427. doi: 10.1271/bbb.70080. [DOI] [PubMed] [Google Scholar]
- Henry C.S., DeJongh M., Best A.A., Frybarger P.M., Linsay B., Stevens R.L. High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat. Biotechnol. 2010;28:977–982. doi: 10.1038/nbt.1672. [DOI] [PubMed] [Google Scholar]
- Hiltunen J.K., Mursula A.M., Rottensteiner H., Wierenga R.K., Kastaniotis A.J., Gurvitz A. The biochemistry of peroxisomal β-oxidation in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2003;27:35–64. doi: 10.1016/S0168-6445(03)00017-2. [DOI] [PubMed] [Google Scholar]
- Hsieh H.-C., Kuan I.-C., Lee S.-L., Tien G.-Y., Wang Y.-J., Yu C.-Y. Stabilization of d-amino acid oxidase from Rhodosporidium toruloides by immobilization onto magnetic nanoparticles. Biotechnol. Lett. 2009;31:557–563. doi: 10.1007/s10529-008-9894-z. [DOI] [PubMed] [Google Scholar]
- Hu C., Zhao X., Zhao J., Wu S., Zhao Z.K. Effects of biomass hydrolysis by-products on oleaginous yeast Rhodosporidium toruloides. Bioresour. Technol. 2009;100:4843–4847. doi: 10.1016/J.BIORTECH.2009.04.041. [DOI] [PubMed] [Google Scholar]
- Huntemann M., Ivanova N.N., Mavromatis K., Tripp H.J., Paez-Espino D., Tennessen K., Palaniappan K., Szeto E., Pillay M., Chen I.-M.A., Pati A., Nielsen T., Markowitz V.M., Kyrpides N.C. The standard operating procedure of the DOE-JGI Metagenome Annotation Pipeline (MAP v.4) Stand. Genomic Sci. 2016;11:17. doi: 10.1186/s40793-016-0138-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagtap S.S., Rao C.V. Production of d-arabitol from d-xylose by the oleaginous yeast Rhodosporidium toruloides IFO0880. Appl. Microbiol. Biotechnol. 2018;102:143–151. doi: 10.1007/s00253-017-8581-1. [DOI] [PubMed] [Google Scholar]
- Juty N., Le Novere N., Laibe C. Identifiers.org and MIRIAM Registry: community resources to provide persistent identification. Nucleic Acids Res. 2012;40:D580–D586. doi: 10.1093/nar/gkr1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavšček M., Bhutada G., Madl T., Natter K. Optimization of lipid production with a genome-scale model of Yarrowia lipolytica. BMC Syst. Biol. 2015;9:72. doi: 10.1186/s12918-015-0217-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkhoven E.J., Pomraning K.R., Baker S.E., Nielsen J. Regulation of amino-acid metabolism controls flux to lipid accumulation in Yarrowia lipolytica. NPJ Syst. Biol. Appl. 2016;2:16005. doi: 10.1038/npjsba.2016.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher S.J. Diversity and origin of alternative NADH:ubiquinone oxidoreductases. Biochim. Biophys. Acta Bioenerg. 2000;1459:274–283. doi: 10.1016/S0005-2728(00)00162-6. [DOI] [PubMed] [Google Scholar]
- Khodayari A., Maranas C.D. A genome-scale Escherichia coli kinetic metabolic model k-ecoli457 satisfying flux data for multiple mutant strains. Nat. Commun. 2016;7:13806. doi: 10.1038/ncomms13806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King Z.A., Dräger A., Ebrahim A., Sonnenschein N., Lewis N.E., Palsson B.O. Escher: a web application for building, sharing, and embedding data-rich visualizations of biological pathways. PLoS Comput. Biol. 2015;11 doi: 10.1371/journal.pcbi.1004321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King Z.A., Lloyd C.J., Feist A.M., Palsson B.O. Next-generation genome-scale models for metabolic engineering. Curr. Opin. Biotechnol. 2015;35:23–29. doi: 10.1016/J.COPBIO.2014.12.016. [DOI] [PubMed] [Google Scholar]
- King Z.A., Lu J., Ager A.D., Miller P., Federowicz S., Lerman J.A., Ebrahim A., Palsson B.O., Lewis N.E. BiGG Models: a platform for integrating, standardizing and sharing genome-scale models. Nucleic Acids Res. 2016;44 doi: 10.1093/nar/gkv1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klis F.M., de Koster C.G., Brul S. Cell wall-related bionumbers and bioestimates of Saccharomyces cerevisiae and Candida albicans. Eukaryot. Cell. 2014;13:2–9. doi: 10.1128/EC.00250-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V., Fedorova N.D., Jackson J.D., Jacobs A.R., Krylov D.M., Makarova K.S., Mazumder R., Mekhedov S.L., Nikolskaya A.N., Rao B., Rogozin I.B., Smirnov S., Sorokin A.V., Sverdlov A.V., Vasudevan S., Wolf Y.I., Yin J.J., Natale D.A. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004;5:R7. doi: 10.1186/gb-2004-5-2-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kot A.M., Błażejak S., Gientka I., Kieliszek M., Bryś J. Torulene and torularhodin: “new” fungal carotenoids for industry? Microb. Cell Factories. 2018;17:49. doi: 10.1186/s12934-018-0893-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange H.C., Heijnen J.J. Statistical reconciliation of the elemental and molecular biomass composition of Saccharomyces cerevisiae. Biotechnol. Bioeng. 2001;75:334–344. doi: 10.1002/bit.10054. [DOI] [PubMed] [Google Scholar]
- Lee J.J., Chen L., Cao B., Chen W.N. Engineering Rhodosporidium toruloides with a membrane transporter facilitates production and separation of carotenoids and lipids in a bi-phasic culture. Appl. Microbiol. Biotechnol. 2016;100:869–877. doi: 10.1007/s00253-015-7102-3. [DOI] [PubMed] [Google Scholar]
- Lee S.Y., Kim H.U. Systems strategies for developing industrial microbial strains. Nat. Biotechnol. 2015;33:1061–1072. doi: 10.1038/nbt.3365. [DOI] [PubMed] [Google Scholar]
- Li Y., Zhao Z. (Kent), Bai F. High-density cultivation of oleaginous yeast Rhodosporidium toruloides Y4 in fed-batch culture. Enzym. Microb. Technol. 2007;41:312–317. doi: 10.1016/J.ENZMICTEC.2007.02.008. [DOI] [Google Scholar]
- Liang M.-H., Jiang J.-G. Advancing oleaginous microorganisms to produce lipid via metabolic engineering technology. Prog. Lipid Res. 2013;52:395–408. doi: 10.1016/J.PLIPRES.2013.05.002. [DOI] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Lieven C., Beber M.E., Olivier B.G., Bergmann F.T., Ataman M., Babaei P., Bartell J.A., Blank L.M., Chauhan S., Correia K., Diener C., Dräger A., Ebert B.E., Edirisinghe J.N., Faria J.P., Feist A., Fengos G., Fleming R.M.T., Garcia-Jimenez B., Hatzimanikatis V., Helvoirt W. van, Henry C., Hermjakob H., Herrgard M.J., Kim H.U., King Z., Koehorst J.J., Klamt S., Klipp E., Lakshmanan M., Novere N. Le, Lee D.-Y., Lee S.Y., Lee S., Lewis N.E., Ma H., Machado D., Mahadevan R., Maia P., Mardinoglu A., Medlock G.L., Monk J., Nielsen J., Nielsen L.K., Nogales J., Nookaew I., Resendis O., Palsson B., Papin J.A., Patil K.R., Poolman M., Price N.D., Richelle A., Rocha I., Sanchez B., Schaap P., Sheriff R.S.M., Shoaie S., Sonnenschein N., Teusink B., Vilaca P., Vik J.O., Wodke J.A., Xavier J.C., Yuan Q., Zakhartsev M., Zhang C. 2018. Memote: A Community-Driven Effort towards a Standardized Genome-Scale Metabolic Model Test Suite. bioRxiv 350991. [DOI] [Google Scholar]
- Liu H., Zhao X., Wang F., Jiang X., Zhang S., Ye M., Zhao Z.K., Zou H. The proteome analysis of oleaginous yeast Lipomyces starkeyi. FEMS Yeast Res. 2011;11:42–51. doi: 10.1111/j.1567-1364.2010.00687.x. [DOI] [PubMed] [Google Scholar]
- Lopes H., Rocha I. Genome-scale modeling of yeast: chronology, applications and critical perspectives. FEMS Yeast Res. 2017;17 doi: 10.1093/femsyr/fox050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan R., Schilling C.H. The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab. Eng. 2003;5:264–276. doi: 10.1016/J.YMBEN.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Maia P., Rocha M., Rocha I. In silico constraint-based strain optimization methods: the quest for optimal cell factories. Microbiol. Mol. Biol. Rev. 2016;80:45–67. doi: 10.1128/MMBR.00014-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., Derbyshire M.K., Gonzales N.R., Lu S., Chitsaz F., Geer L.Y., Geer R.C., He J., Gwadz M., Hurwitz D.I., Lanczycki C.J., Lu F., Marchler G.H., Song J.S., Thanki N., Wang Z., Yamashita R.A., Zhang D., Zheng C., Bryant S.H. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015;43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo M.L., Palsson B.Ø., Herrgård M.J. Connecting extracellular metabolomic measurements to intracellular flux states in yeast. BMC Syst. Biol. 2009;3:37. doi: 10.1186/1752-0509-3-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molin M., Norbeck J., Blomberg A. Dihydroxyacetone kinases in Saccharomyces cerevisiae are involved in detoxification of dihydroxyacetone. J. Biol. Chem. 2003;278:1415–1423. doi: 10.1074/jbc.M203030200. [DOI] [PubMed] [Google Scholar]
- Monk J., Nogales J., Palsson B.O. Optimizing genome-scale network reconstructions. Nat. Biotechnol. 2014;32:447–452. doi: 10.1038/nbt.2870. [DOI] [PubMed] [Google Scholar]
- Mueller T.J., Berla B.M., Pakrasi H.B., Maranas C.D. Rapid construction of metabolic models for a family of Cyanobacteria using a multiple source annotation workflow. BMC Syst. Biol. 2013;7:142. doi: 10.1186/1752-0509-7-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakase T., Komagatai K. DNA base composition of some species of yeasts and yeast-like fungi. J. Gen. Appl. Microbiol. 1971;17:363–369. [Google Scholar]
- O’Brien E.J., Monk J.M., Palsson B.O. Using genome-scale models to predict biological capabilities. Cell. 2015;161:971–987. doi: 10.1016/j.cell.2015.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlean P. Architecture and biosynthesis of the Saccharomyces cerevisiae cell wall. Genetics. 2012;192:775–818. doi: 10.1534/genetics.112.144485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth J.D., Thiele I., Palsson B.Ø. What is flux balance analysis? Nat. Biotechnol. 2010;28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Österlund T., Nookaew I., Nielsen J. Fifteen years of large scale metabolic modeling of yeast: developments and impacts. Biotechnol. Adv. 2012;30:979–988. doi: 10.1016/J.BIOTECHADV.2011.07.021. [DOI] [PubMed] [Google Scholar]
- Park Y.-K., Nicaud J.-M., Ledesma-Amaro R. The engineering potential of Rhodosporidium toruloides as a workhorse for biotechnological applications. Trends Biotechnol. 2018;36:304–317. doi: 10.1016/J.TIBTECH.2017.10.013. [DOI] [PubMed] [Google Scholar]
- Probst K.V., Schulte L.R., Durrett T.P., Rezac M.E., Vadlani P.V. Oleaginous yeast: a value-added platform for renewable oils. Crit. Rev. Biotechnol. 2016;36:942–955. doi: 10.3109/07388551.2015.1064855. [DOI] [PubMed] [Google Scholar]
- Ranganathan S., Suthers P.F., Maranas C.D. OptForce: an optimization procedure for identifying all genetic manipulations leading to targeted overproductions. PLoS Comput. Biol. 2010;6 doi: 10.1371/journal.pcbi.1000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez B., Li F., Lu H., Kerkhoven E., Nielsen J. 2018. SysBioChalmers/yeast-GEM: Yeast 8.3.3. [DOI] [Google Scholar]
- Sánchez B.J., Li F., Kerkhoven E.J., Nielsen J. 2018. SLIMEr: Probing Flexibility of Lipid Metabolism in Yeast with an Improved Constraint-Based Modeling Framework. bioRxiv 324863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberger J., Lewis N.E., Palsson B.Ø. Elimination of thermodynamically infeasible loops in steady-state metabolic models. Biophys. J. 2011;100:544–553. doi: 10.1016/J.BPJ.2010.12.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmalix W., Bandlow W. The ethanol-inducible YAT1 gene from yeast encodes a presumptive mitochondrial outer carnitine acetyltransferase. J. Biol. Chem. 1993;268:27428–27439. [PubMed] [Google Scholar]
- Schulze U. Technical University of Denmark; 1995. Anaerobic Physiology of Saccharomyces cerevisiae. [Google Scholar]
- Shen H., Gong Z., Yang X., Jin G., Bai F., Zhao Z.K. Kinetics of continuous cultivation of the oleaginous yeast Rhodosporidium toruloides. J. Biotechnol. 2013;168:85–89. doi: 10.1016/J.JBIOTEC.2013.08.010. [DOI] [PubMed] [Google Scholar]
- Shen H., Zhang X., Gong Z., Wang Y., Yu X., Yang X., Zhao Z.K. Compositional profiles of Rhodosporidium toruloides cells under nutrient limitation. Appl. Microbiol. Biotechnol. 2017;101:3801–3809. doi: 10.1007/s00253-017-8157-0. [DOI] [PubMed] [Google Scholar]
- Shi S., Zhao H. Metabolic engineering of oleaginous yeasts for production of fuels and chemicals. Front. Microbiol. 2017;8:2185. doi: 10.3389/fmicb.2017.02185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman A.M., Qiao K., Xu P., Stephanopoulos G. Functional overexpression and characterization of lipogenesis-related genes in the oleaginous yeast Yarrowia lipolytica. Appl. Microbiol. Biotechnol. 2016;100:3781–3798. doi: 10.1007/s00253-016-7376-0. [DOI] [PubMed] [Google Scholar]
- Srinivasan S., Cluett W.R., Mahadevan R. Constructing kinetic models of metabolism at genome-scales: a review. Biotechnol. J. 2015;10:1345–1359. doi: 10.1002/biot.201400522. [DOI] [PubMed] [Google Scholar]
- Starmer W.T., Fell J.W., Catranis C.M., Aberdeen V., Ma L.-J., Zhou S., Rogers S.O. Yeasts in the genus Rhodotorula recovered from the Greenland ice sheet. In: Rogers S.O., Castello J.D., editors. Life in Ancient Ice. Princeton University Press; 2005. pp. 181–195. [Google Scholar]
- Tai M., Stephanopoulos G. Engineering the push and pull of lipid biosynthesis in oleaginous yeast Yarrowia lipolytica for biofuel production. Metab. Eng. 2013;15:1–9. doi: 10.1016/J.YMBEN.2012.08.007. [DOI] [PubMed] [Google Scholar]
- Tamano K., Bruno K.S., Karagiosis S.A., Culley D.E., Deng S., Collett J.R., Umemura M., Koike H., Baker S.E., Machida M. Increased production of fatty acids and triglycerides in Aspergillus oryzae by enhancing expressions of fatty acid synthesis-related genes. Appl. Microbiol. Biotechnol. 2013;97:269–281. doi: 10.1007/s00253-012-4193-y. [DOI] [PubMed] [Google Scholar]
- Tchakouteu S.S., Kopsahelis N., Chatzifragkou A., Kalantzi O., Stoforos N.G., Koutinas A.A., Aggelis G., Papanikolaou S. Rhodosporidium toruloides cultivated in NaCl-enriched glucose-based media: adaptation dynamics and lipid production. Eng. Life Sci. 2017;17:237–248. doi: 10.1002/elsc.201500125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele I., Palsson B. A protocol for generating a high quality genome-scale metabolic reconstruction. Nat. Protoc. 2010;5:93–121. doi: 10.1038/nprot.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiukova I.A., Prigent S., Sandgren M., Nielsen J., Kerkhoven E.J. 2019. Genome-scale Model of Rhodotorula Toruloides Metabolism. bioRxiv 528489. [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhang S., Zhu Z., Shen H., Lin X., Jin X., Jiao X., Zhao Z.K. Systems analysis of phosphate-limitation-induced lipid accumulation by the oleaginous yeast Rhodosporidium toruloides. Biotechnol. Biofuels. 2018;11:148. doi: 10.1186/s13068-018-1134-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylenko T.M., Ahn W.S., Stephanopoulos G. The oxidative pentose phosphate pathway is the primary source of NADPH for lipid overproduction from glucose in Yarrowia lipolytica. Metab. Eng. 2015;30:27–39. doi: 10.1016/J.YMBEN.2015.02.007. [DOI] [PubMed] [Google Scholar]
- Wei S., Jian X., Chen J., Zhang C., Hua Q. Reconstruction of genome-scale metabolic model of Yarrowia lipolytica and its application in overproduction of triacylglycerol. Bioresour. Bioprocess. 2017;4:51. doi: 10.1186/s40643-017-0180-6. [DOI] [Google Scholar]
- Wiebe M.G., Koivuranta K., Penttilä M., Ruohonen L. Lipid production in batch and fed-batch cultures of Rhodosporidium toruloides from 5 and 6 carbon carbohydrates. BMC Biotechnol. 2012;12:26. doi: 10.1186/1472-6750-12-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Zhao X., Shen H., Wang Q., Zhao Z.K. Microbial lipid production by Rhodosporidium toruloides under sulfate-limited conditions. Bioresour. Technol. 2011;102:1803–1807. doi: 10.1016/J.BIORTECH.2010.09.033. [DOI] [PubMed] [Google Scholar]
- Xavier J.C., Patil K.R., Rocha I. Integration of biomass formulations of genome-scale metabolic models with experimental data reveals universally essential cofactors in prokaryotes. Metab. Eng. 2017;39:200–208. doi: 10.1016/J.YMBEN.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S.-J., Chi Z.-M., Zhang Y., Li Y.-F., Liu G.-L., Jiang H., Hu Z., Chi Z.-M. Fatty acids from oleaginous yeasts and yeast-like fungi and their potential applications. Crit. Rev. Biotechnol. 2018;38:1–12. doi: 10.1080/07388551.2018.1428167. [DOI] [PubMed] [Google Scholar]
- Yang Z., Huang J., Geng J., Nair U., Klionsky D.J. Atg22 recycles amino acids to link the degradative and recycling functions of autophagy. Mol. Biol. Cell. 2006;17:5094–5104. doi: 10.1091/mbc.e06-06-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A.-Q., Pratomo Juwono N.K., Leong S.S.J., Chang M.W. Production of fatty acid-derived valuable chemicals in synthetic microbes. Front. Bioeng. Biotechnol. 2014;2:78. doi: 10.3389/fbioe.2014.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Ito M., Skerker J.M., Arkin A.P., Rao C.V. Metabolic engineering of the oleaginous yeast Rhodosporidium toruloides IFO0880 for lipid overproduction during high-density fermentation. Appl. Microbiol. Biotechnol. 2016;100:9393–9405. doi: 10.1007/s00253-016-7815-y. [DOI] [PubMed] [Google Scholar]
- Zhang S., Skerker J.M., Rutter C.D., Maurer M.J., Arkin A.P., Rao C.V. Engineering Rhodosporidium toruloides for increased lipid production. Biotechnol. Bioeng. 2016;113:1056–1066. doi: 10.1002/bit.25864. [DOI] [PubMed] [Google Scholar]
- Zhu Z., Zhang S., Liu H., Shen H., Lin X., Yang F., Zhou Y.J., Jin G., Ye M., Zou H., Zhao Z.K. A multi-omic map of the lipid-producing yeast Rhodosporidium toruloides. Nat. Commun. 2012;3:1112. doi: 10.1038/ncomms2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.