Abstract.
Aedes mosquitoes are the principal dengue vector in Taiwan, where the use of insecticides is a key element in the national control strategy. However, control efforts are constrained by the development of resistance to most insecticides, including pyrethroids. In this study, mutations in the voltage-gated sodium channel (VGSC) gene resulting in knockdown resistance (kdr) were examined in Aedes aegypti. Fragments of the VGSC gene were polymerase chain reaction (PCR)-amplified followed by restriction fragment length polymorphism analysis in samples from various settings in Southern Taiwan covering dry and wet seasons from 2013 to 2015. Three kdr mutations were identified: V1023G, D1794Y, and F1534C, with observed frequencies of 0.36, 0.55, and 0.33, respectively, in the dry season of 2013–2014. Exploring for temporal changes, the most important observation was the 1534C allele frequency increment in the following season to 0.60 (P < 0.05). This study suggests that continued insecticide pressure is driving the mutational changes, although the selection is ambiguous in the mosquito population.
The mosquito Aedes aegypti is the primary vector for several arboviral diseases, of which dengue is the most prominent in terms of geographic distribution and disease burden.1,2 In Taiwan, annual outbreaks have been reported over the past three decades3,4 with epidemic activity largely centered around Kaohsiung city. Two major outbreaks within the last 5 years included 15,732 (2014) and 43,784 (2015) confirmed cases.5 In Kaohsiung city, control efforts against Ae. aegypti are maintained throughout the year, however, with increased focus during rainy seasons in relation to intensified dengue activity, whereas in rural areas, such as Pingtung County, control efforts are mainly used in response to noted epidemic activity.3,4
Current control activities include environmental management and biological and chemical control targeting larval and adult stages of Ae. aegypti.4–6 One of the most used insecticide groups in control efforts are pyrethroids; however, pyrethroid resistance has been reported from Taiwan in recent years, suggesting reduced efficacy of ongoing control efforts targeting adult vectors.7,8
Various single nucleotide polymorphisms (SNPs) in the gene resulting in amino acid changes encoding the voltage-gated sodium channel (VGSC) have been associated with lowered sensitivity against pyrethroids also known as knockdown resistance (kdr).6,9 Knockdown resistance genes are known to be autosomal recessive, but several SNPs occurring together have been shown to result in increased levels of pyrethroid resistance.10 Several SNPs in the VGSC-coding gene resulting in amino acid changes of Ae. Aegypti have been observed in Asian countries, including S989P, V1023G, D1794Y, and the most frequently observed F1534C.9,10 In Taiwan, in particular, the coexistence of D1794Y and V1023G has previously been associated with pyrethroid resistance.11
To assess the prevalence and seasonal changes of SNPs in the VGSC gene as a proxy for pyrethroid resistance in Ae. aegypti populations of Southern Taiwan, Ae. aegypti larvae and pupae were collected in Kaohsiung city and the rural area of Pingtung County. Survey activities were completed at six randomly chosen study sites in urban Kaohsiung (three sites) and in rural Pingtung (three sites). One survey was carried out at each of the six study sites in the dry season from November 2013 to February 2014. For the wet season of 2014 (May–June), one survey was completed at two of the urban study sites, whereas for unforeseeable circumstances, the survey for the remaining four study sites was postponed to 2015 (May–July). The detailed sampling process was as previously described by Lin et al.4
A subset of 492 samples were available for this study; in the dry season (November 2013–February 2014) n = 180, 115 in the wet season of 2014 (May 2014–June 2014), and finally 197 Ae. aegypti in the wet season of 2015 (May 2015–July 2015). Genomic DNA was extracted by the Bender buffer method.12 Of the 492 Ae. aegypti collected, 488 (99.2%) samples were positive by species-specific PCR, as described by Higa et al.13 Three regions of the VGSC gene (GenBank, accession number: CM008045.1) containing known SNPs were targeted by PCR as described elsewhere.8,14,15 In brief, the three PCRs: 1534 (targeting F1534C), 1794 (targeting D1794Y), and 891623 (targeting S989P, V1016G/V1023G) were used with minor modifications: The annealing temperature was changed (to 54°C, 66°C, and 67°C for the 891623, 1794, and 1534 PCRs, respectively) and the extension time for 891623-PCR was prolonged to 90 seconds.
By PCR, 371 (76.0%), 363 (74.4%), and 341 (69.9%) were positive for the 1794, 1534, and 891623-PCRs, respectively. Overall, 69.3% (338/488) samples were positive in all the three PCRs. Restriction fragment length polymorphism analyses for identification of SNPs were designed by in silico digestion using NEBcutter V2.0 resulting in five enzymes (Table 1). All enzyme digestions were performed as single reactions according to the manufacturer’s protocol (New England Biolabs, Ipswich, MA), and fragments were visualized on a 1.5% agarose gel containing ethidium bromide.
Table 1.
Restriction enzyme and expected fragment sizes of the restriction fragment length polymorphism analysis
| Restriction enzyme | Target | Wild type | Mutant type | Amplicon size (bp) | Fragment, wild type (bp) | Fragment, mutation (bp) |
|---|---|---|---|---|---|---|
| HpyAV | D1794 | GAC | TAC | 754 | 483, 169, 102 | 588, 169 |
| FauI | 1534C | TTC | TGC | 635 | 476, 159 | 333, 159, 143 |
| HinfI | S989 | TCC | CCC | 1,020 | 481, 370, 169 | 539, 481 |
| RsaI | V1023 | GTA | GGA | 1,020 | 528, 332, 86, 74 | 528, 406, 86 |
| BslI | 1023G | GTA | GGA | 1,020 | 396, 336, 288 | 732, 288 |
Single-nucleotide polymorphisms were identified for VGSC gene positions 1794, 1534, and 1023, whereas 989 and 1016 displayed only wild types. None of the observed allele distributions were in Hardy–Weinberg equilibrium: V1023G (χ2 = 24, P < 0.05), F1534C (χ2 = 7.89, P = 0.02), and D1794Y (χ2 = 39.85, P < 0.05).
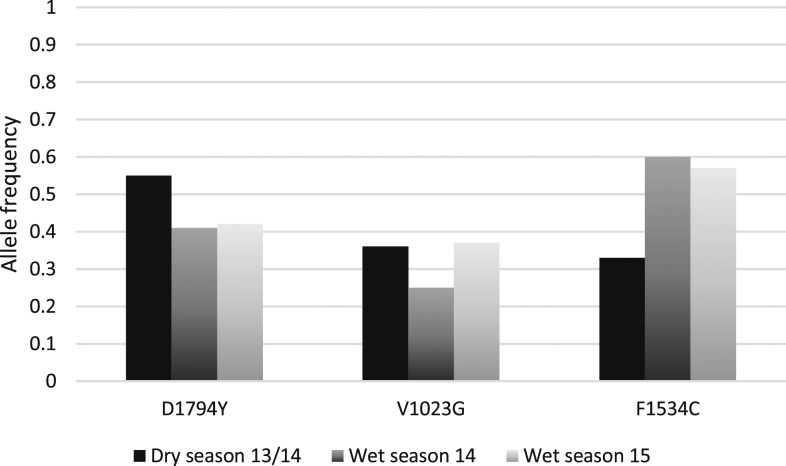
At position 1794, conclusive genotype results were attained for 265 of the 371 PCR-positive samples (71.4%). A significant decrease in 1794Y mutant allele frequency from the dry season of 2013–2014 at 0.55 to the wet season of 2014 at 0.41 was observed (χ2 = 6.15, P = 0.01). A similar low frequency was found in the wet season of 2015, at 0.42 (Figure 1).
Figure 1.
Mutant allele frequencies for 1794Y, 1023G and 1534C in Aedes aegypti collected in Kaohsiung City and Pingtung County from 2013 to 2015.
Of 341 samples, 250 (73.3%) gave conclusive results when genotyped for position 1023. Overall, the temporal distribution revealed a significant difference in frequency of the 1023G allele between the seasons (χ2 = 7.16, P = 0.02), with a mutant allele frequency of 0.36 in the dry season, and most notably, a significant increase in mutant alleles between the two wet seasons from 0.25 (2014) to 0.37 (2015) (χ2 = 6.58, P = 0.01) was observed. For digestion of PCR products covering position 1534, 255 of 363 (70.2%) samples gave conclusive results. The 1534C mutant allele frequency in the dry season was 0.33, followed by a significant increase in frequency to 0.60 for the wet season of 2014 (χ2 = 25.58, P < 0.05) and a similar frequency of 0.57 in the wet season of 2015 (Figure 1).
The extensive use of pyrethroid insecticides has resulted in kdr resistance in Aedes populations, generally associated with mutations in the VGSC gene.16 In this study, immature Ae. aegypti collected from Southern Taiwan was screened for kdr mutations resulting in amino acid changes in the VGSC. Relatively high mutant frequencies were observed for the first dry season at 0.55, 0.36, and 0.33 for 1794Y, 1023G, and 1534C, respectively. Allele frequencies for 1023G and 1794Y were also reported for Ae. aegypti populations sampled from January to July in 2008 in the Lingya district of Kaohsiung by Lin et al.,8 who observed somewhat similar mutant allele frequencies of 0.46 and 0.53 for 1794Y and 1023G, respectively, but did not examine the SNP 1534. Recently, a study by Chung et al.17 detected the mutation types S989P, V1023G, F1534C, and D1794Y from Ae. aegypti populations collected in Taiwan and furthermore explored by bioassays that these mutation types were associated with pyrethroid (cypermethrin) resistance. The most frequent mutation was V1016G (0.28) and the rarest is D1763Y (0.06); the proportions of S989P and F1534C were 0.18 and 0.22, respectively.17
In the present study, we observed the presence of the 1534C mutant allele, and the significant increase in prevalence observed from 2013 to 2014 strongly indicates that pyrethroids exerted pressure on local Ae. aegypti populations during the study period.
Most of the tested mosquitoes were heterozygotes for one or more kdr mutations (data not shown). Studies by Ishak et al.18 and Plernsub et al.10 looking at F/C1534 + V/G1023 and S/P989 + V/G1023 + F/C1534, respectively, have shown varying levels of resistance compared with mosquitos with homozygous mutations. Thus, an additive effect of mutations resulting in increased levels of resistance may also occur in these study sites as double and/or triple heterozygote mutants were frequently observed indicating an increasing complexity in the pattern of insecticide resistance in Ae. aegypti populations and possibly higher levels of pyrethroid resistance than in homozygote mutants only as discussed by Vera-Maloof et al.,19 where the combination of mutations is required for the resistance to occur.
The findings of this study suggest that the use of pyrethroid insecticides has led to kdr mutations and possible development of resistance in Ae. aegypti populations in Kaohsiung city and Pingtung County. The increase in mutant allele frequency as observed for position 1534 along with changes in double and triple mutants suggests that the potency of pyrethroids is compromised and that alternative control options for Ae. aegypti should be considered in Southern Taiwan.
Acknowledgments:
We thank the entomologists Y. H. Chang, C. T. Hsu, Y. T. Chang, L. H. Wang, H. T. Wang, S. E. Pan, M. Y. Chen, H. L. Keng, M. Y. Lu, C. C. Lin, I. Y. Chen, and T. T. Chang, and also Derua Yahya for providing us with mosquito DNA extraction protocols.
REFERENCES
- 1.Gloria-soria A, et al. 2016. Global genetic diversity of Aedes aegypti. Mol Ecol 25: 5377–5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kraemer MU, et al. 2015. The global distribution of the arbovirus vectors Aedes aegypti and Ae. albopictus. Elife 4: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin CH, Schiøler KL, Jepsen MR, Ho CK, Li SH, Konradsen F, 2012. Dengue outbreaks in high-income area, Kaohsiung city, Taiwan, 2003–2009. Emerg Infect Dis 18: 1603–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin CH, Schiøler KL, Ekstrøm CT, Konradsen F, 2018. Location, seasonal and functional characteristics of water holding containers with juvenile and pupal Aedes aegypti in southern Taiwan: a cross-sectional study using hurdle model analyses. PLoS Negl Trop Dis 12: e0006882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith LB, Kasai S, Scott JG, 2016. Pyrethroid resistance in Aedes aegypti and Aedes albopictus: important mosquito vectors of human diseases. Pestic Biochem Physiol 133: 1–12. [DOI] [PubMed] [Google Scholar]
- 6.Amelia-Yap ZH, Chen CD, Sofian-Azirun M, Low VL, 2018. Pyrethroid resistance in the dengue vector Aedes aegypti in southeast Asia: present situation and prospects for management. Parasit Vectors 11: 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsia W, Wu H, Yang Y, Lin C, 2011. Efficacy of commonly used insecticides to Aedes aegypti in southern Taiwan. Taiwan Epi Bul 27: 39–49. [Google Scholar]
- 8.Lin Y-H, Tsen WL, Tien NY, Luo YP, 2013. Biochemical and molecular analyses to determine pyrethroid resistance in Aedes aegypti. Pestic Biochem Physiol. 107: 266–276. [Google Scholar]
- 9.Moyes CL, et al. 2017. Contemporary status of insecticide resistance in the major Aedes vectors of arboviruses infecting humans. PLoS Negl Trop Dis 11: 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plernsub S, Saingamsook J, Yanola J, Lumjuan N, Tippawangkosol P, Sukontason K, Walton C, Somboon P, 2016. Additive effect of knockdown resistance mutations, S989P, V1016G and F1534C, in a heterozygous genotype conferring pyrethroid resistance in Aedes aegypti in Thailand. Parasit Vectors 9: 417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang C, Shen WK, Wang TT, Lin YH, Hsu EL, Dai SM, 2009. A novel amino acid substitution in a voltage-gated sodium channel is associated with knockdown resistance to permethrin in Aedes aegypti. Insect Biochem Mol Biol 39: 272–278. [DOI] [PubMed] [Google Scholar]
- 12.Collins FH, Mendez MA, Rasmussen MO, Mehaffey PC, Besansky NJ, Finnerty V, 1987. A ribosomal RNA gene probe differentiates member species of the Anopheles gambiae complex. Am J Trop Med Hyg 37: 37–41. [DOI] [PubMed] [Google Scholar]
- 13.Higa Y, Toma T, Tsuda Y, Miyagi I, 2010. A multiplex PCR-based molecular identification of five morphologically related, medically important subgenus stegomyia mosquitoes from the genus Aedes (Diptera: Culicidae) found in the Ryukyu archipelago, Japan. Jpn J Infect Dis 63: 312–316. [PubMed] [Google Scholar]
- 14.Li CX, et al. 2015. Relationship between insecticide resistance and kdr mutations in the dengue vector Aedes aegypti in southern China. Parasit Vectors 8: 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yanola J, Somboon P, Walton C, Nachaiwieng W, Somwang P, Prapanthadara L, 2011. High-throughput assays for detection of the F1534C mutation in the voltage-gated sodium channel gene in permethrin-resistant Aedes aegypti and the distribution of this mutation throughout Thailand. Trop Med Int Heal 16: 501–509. [DOI] [PubMed] [Google Scholar]
- 16.Faucon F, et al. 2015. Identifying genomic changes associated with insecticide resistance in the dengue mosquito Aedes aegypti by deep targeted sequencing. Genome Res 1347–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung HH, Cheng IC, Chen YC, Lin C, Tomita T, Teng HJ, 2019. Voltage-gated sodium channel intron polymorphism and four mutations comprise six haplotypes in an Aedes aegypti population in Taiwan. PLoS Negl Trop Dis 13: e0007291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishak IH, Jaal Z, Ranson H, Wondji CS, 2015. Contrasting patterns of insecticide resistance and knockdown resistance (kdr) in the dengue vectors Aedes aegypti and Aedes albopictus from Malaysia. Parasit Vectors 8: 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vera-Maloof FZ, Saavedra-Rodriguez K, Elizondo-Quiroga AE, Lozano-Fuentes S, Black WC, IV, 2015. Coevolution of the Ile1,016 and Cys1,534 mutations in the voltage gated sodium channel gene of Aedes aegypti in Mexico. PLoS Negl Trop Dis 9: e0004263. [DOI] [PMC free article] [PubMed] [Google Scholar]