Here, we report 17 nearly complete genome sequences of enterovirus D68 (EV-D68) isolated from Kansas City, MO, in 2018. Phylogenetic analysis suggests that these strains belong to subclade B3, similar to the ones that caused the 2016 epidemics in the United States but different from the 2014 outbreak B1 strains.
ABSTRACT
Here, we report 17 nearly complete genome sequences of enterovirus D68 (EV-D68) isolated from Kansas City, MO, in 2018. Phylogenetic analysis suggests that these strains belong to subclade B3, similar to the ones that caused the 2016 epidemics in the United States but different from the 2014 outbreak B1 strains.
ANNOUNCEMENT
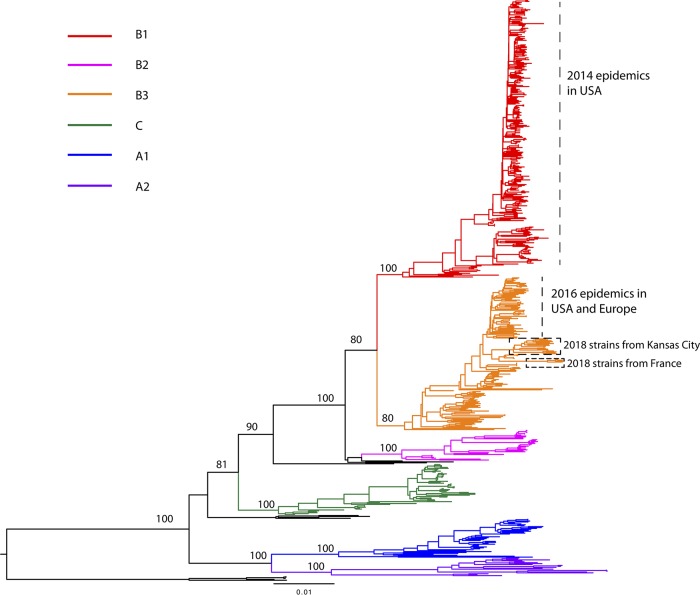
Enterovirus D68 (EV-D68) belongs to the genus Enterovirus in the family Picornaviridae, with a single positive-strand RNA genome of ∼7.5 kb in length coding for 4 structural proteins (VP1 to VP4) and 7 nonstructural proteins (2A to 2C and 3A to 3D) (1). EV-D68 was first isolated in California in 1962 from patients with bronchiolitis and pneumonia (2). Between 1962 and 2014, EV-D68 was sporadically detected in different parts of the world (3, 4). However, in August 2014, a new subclade of EV-D68, B1, caused a nationwide outbreak in the United States and an increased number of acute flaccid myelitis (AFM) cases (5, 6). In 2016, a different subclade of EV-D68 strains, B3, spread in some states, such as New York and Missouri, and caused local epidemics (Fig. 1) (7, 8). EV-D68 continues to spread sporadically and was reported in Europe, with more AFM cases involved, in 2018 (9, 10).
FIG 1.
We combined the 17 newly acquired complete EV-D68 genome sequences collected in 2018 in Kansas City with EV-D68 genome sequences available in GenBank (as of 31 January 2019) and generated a data set with a total of 628 complete genome sequences of EV-D68. Sequences were aligned using the MUSCLE program in MEGA6, with manual adjustments (15). The global phylogenetic tree of EV-D68 was inferred using the neighbor-joining method conducted in MEGA6, with the Kimura 2-parameter test and bootstrap test of 1,000 replicates (16). The tree is midpoint rooted and drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. Well-supported nodes by bootstrap values over 70% are shown next to the branches. Six major clades/subclades (B1, B2, B3, C, A1, and A2) are described in the tree. New 2018 sequences sampled from Kansas City are marked in the tree as well.
Previously, we had developed a high-throughput complete genome sequencing pipeline for EV-D68 to describe the early outbreak of EV-D68 in Kansas City, MO, in 2014 (11). Here, we report the genome sequence of EV-D68, which was obtained from 17 patients from Kansas City between August and October 2018. EV-D68 was confirmed by quantitative PCR (qPCR) (12). After RNA extraction, full-length cDNA was reverse transcribed with the first-strand synthesis SuperMix kit. Two overlapping amplicons (a small [S] 904-bp and large [L] 6.8-kbp amplicon) were generated using EV-D68-specific primers, as described before (11). Libraries were constructed with the NEBNext Ultra II FS DNA kit, samples were pooled, and sequencing was performed using an Illumina MiSeq instrument with 2 × 250-bp reads.
The median number of reads generated per sample was 376,488 (interquartile range [IQR], 326,864 to 421,360 reads). Sequencing reads were binned by barcode, adapters were trimmed with Cutadapt (v 1.18) (13), and low-quality bases were removed with Trimmomatic (v 0.36) (14). Reads were normalized using BBTools (v 38.34) (14) and initially assembled de novo using the SPAdes assembler (v 3.13.0) (15), using default parameters. BLASTN searches of the resulting contigs identified an EV-D68 isolate from 2016, with GenBank accession number KY385889 (enterovirus D68 isolate NY212_16, complete genome), as the closest match. Using this sequence as a reference, consensus sequences were produced for all 17 samples using CLC Genomics Workbench (v 11.0.1), with length and similarity fraction thresholds set at 0.9. The average depth of coverage varied between 224× and 4,228×. All 17 sequences are 7,331 bp in length, and their G+C contents vary between 41.6% and 41.9%. They were annotated using the VAPiD annotation pipeline (v 1.6.2) (16) and submitted to GenBank.
Although these 17 2018 EV-D68 isolates from Kansas City belonged to the B3 subclade, phylogenetic analysis shows that they did not cluster with the strains that caused the 2016 epidemics in the United States and Europe (∼98.21% similarity) or the strains reported in Europe (France) in 2018 (∼97.93% similarity), which also are in subclade B3 (Fig. 1). The similarity between the 2018 Kansas City EV-D68 strains and the 2014 outbreak B1 subclade was ∼94.83%. These are the first publicly available EV-D68 genome sequences collected in the United States during the 2018 epidemics. With the increasing number of AFM cases related to EV-D68 infection, our newly reported 2018 EV-D68 strains could help us understand the epidemiological dynamics and clinical implications of EV-D68 in the United States and globally.
Data availability.
All sequences were submitted to GenBank with accession numbers MK659588 to MK659604. All raw reads were submitted to SRA. The published SRA submission can be found at BioProject accession number PRJNA532464.
ACKNOWLEDGMENTS
This project was funded with startup funds from the Vanderbilt University Medical Center (VUMC) to S.R.D. S.R.D. is also supported by the NIH-funded Tennessee Center for AIDS Research (grant P30 AI110527), U19 AI 095227, U01s (U01AI132004, U01IP001083, and U01IP001063), and the Vanderbilt Institute for Clinical and Translational Research (grant support from the National Center for Advancing Translational Sciences under award number UL1TR000445). R.H.M. is funded through the Vanderbilt Training Program in a Big Biomedical Data Science training grant (5T32124123).
The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the VUMC or Children’s Mercy Hospitals and Clinics.
REFERENCES
- 1.Imamura T, Oshitani H. 2015. Global reemergence of enterovirus D68 as an important pathogen for acute respiratory infections. Rev Med Virol 25:102–114. doi: 10.1002/rmv.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schieble JH, Fox VL, Lennette EH. 1967. A probable new human picornavirus associated with respiratory diseases. Am J Epidemiol 85:297–310. doi: 10.1093/oxfordjournals.aje.a120693. [DOI] [PubMed] [Google Scholar]
- 3.Khetsuriani N, Lamonte-Fowlkes A, Oberst S, Pallansch MA, Centers for Disease Control and Prevention . 2006. Enterovirus surveillance–United States, 1970–2005. MMWR Surveill Summ 55:1–20. [PubMed] [Google Scholar]
- 4.Tokarz R, Firth C, Madhi SA, Howie SR, Wu W, Sall AA, Haq S, Briese T, Lipkin WI. 2012. Worldwide emergence of multiple clades of enterovirus 68. J Gen Virol 93:1952–1958. doi: 10.1099/vir.0.043935-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steele MT, Walsh I. 2015. Commentary. Severe respiratory illness associated with Enterovirus D68—Missouri and Illinois, 2014. Ann Emerg Med 65:335. doi: 10.1016/j.annemergmed.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 6.Greninger AL, Naccache SN, Messacar K, Clayton A, Yu G, Somasekar S, Federman S, Stryke D, Anderson C, Yagi S, Messenger S, Wadford D, Xia D, Watt JP, Van Haren K, Dominguez SR, Glaser C, Aldrovandi G, Chiu CY. 2015. A novel outbreak enterovirus D68 strain associated with acute flaccid myelitis cases in the USA (2012–14): a retrospective cohort study. Lancet Infect Dis 15:671–682. doi: 10.1016/S1473-3099(15)70093-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang W, Yin C, Zhuge J, Farooq T, Yoon EC, Nolan SM, Chen D, Fallon JT, Wang G. 2016. Complete genome sequences of nine Enterovirus D68 strains from patients of the Lower Hudson Valley, New York, 2016. Genome Announc 4:e01394-16. doi: 10.1128/genomeA.01394-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wylie KM, Wylie TN, Storch GA. 2017. Genome sequence of Enterovirus D68 from St. Louis, Missouri, USA, 2016. Genome Announc 5:e01630-16. doi: 10.1128/genomeA.01630-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bal A, Sabatier M, Wirth T, Coste-Burel M, Lazrek M, Stefic K, Brengel-Pesce K, Morfin F, Lina B, Schuffenecker I, Josset L. 2019. Emergence of enterovirus D68 clade D1, France, August to November 2018. Euro Surveill 24:1800699. doi: 10.2807/1560-7917.ES.2019.24.3.1800699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The United Kingdom Acute Flaccid Paralysis (AFP) Task Force. 2019. An increase in reports of acute flaccid paralysis (AFP) in the United Kingdom, 1 January 2018–21 January 2019: early findings. Euro Surveill 24:1900093. doi: 10.2807/1560-7917.ES.2019.24.6.1900093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan Y, Hassan F, Schuster JE, Simenauer A, Selvarangan R, Halpin RA, Lin X, Fedorova N, Stockwell TB, Lam TT, Chappell JD, Hartert TV, Holmes EC, Das SR. 2016. Molecular evolution and intraclade recombination of Enterovirus D68 during the 2014 outbreak in the United States. J Virol 90:1997–2007. doi: 10.1128/JVI.02418-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kujawski SA, Midgley CM, Rha B, Lively JY, Nix WA, Curns AT, Payne DC, Englund JA, Boom JA, Williams JV, Weinberg GA, Staat MA, Selvarangan R, Halasa NB, Klein EJ, Sahni LC, Michaels MG, Shelley L, McNeal M, Harrison CJ, Stewart LS, Lopez AS, Routh JA, Patel M, Oberste MS, Watson JT, Gerber SI. 2019. Enterovirus D68-associated acute respiratory illness—New Vaccine Surveillance Network, United States, July–October, 2017 and 2018. MMWR Morb Mortal Wkly Rep 68:277–280. doi: 10.15585/mmwr.mm6812a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 14.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. doi: 10.1007/bf01731581. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All sequences were submitted to GenBank with accession numbers MK659588 to MK659604. All raw reads were submitted to SRA. The published SRA submission can be found at BioProject accession number PRJNA532464.