Abstract
Ciliates are a powerful model organism for the study of basal bodies and motile cilia. These single-celled protists contain hundreds of cilia organized in an array making them an ideal system for both light and electron microscopy studies. Isolation and subsequent proteomic analysis of both cilia and basal bodies have been carried out to great success in ciliates. These studies reveal that ciliates share remarkable protein conservation with metazoans and have identified a number of essential basal body/ciliary proteins. Ciliates also boast a genetic and molecular toolbox that allows for facile manipulation of ciliary genes. Reverse genetics studies in ciliates have expanded our understanding of how cilia are positioned within an array, assembled, stabilized, and function at a molecular level. The advantages of cilia number coupled with a robust genetic and molecular toolbox have established ciliates as an ideal system for motile cilia and basal body research and prove a promising system for future research.
Keywords: motile cilia, basal body, ciliate, doublet microtubules, ciliary array
Introduction
Ciliates are single-celled protozoans grouped under the phylum Ciliophora (Cavalier-Smith, 1993). Species of ciliates are morphologically diverse but, as their name suggests, ciliates are united in that they contain many motile cilia. Paramecium tetaurelia/caudatum and Tetrahymena thermophila are the two ciliate species that have had the biggest impact on our understanding of both motile cilia and their major nucleating/anchoring structure, the basal body.
Both Tetrahymena and Paramecium are genetically tractable systems. They have completely sequenced genomes (Aury et al., 2006; Eisen et al., 2006) and their genetics, though unconventional, lend themselves well to both forward and reverse genetic approaches, including genomic knockouts, knock-ins, and gene fusions (Bruns and Cassidy-Hanley, 2000; Hai et al., 2000; Kung et al., 2000; Yu and Gorovsky, 2000; Shang et al., 2002b; Beisson et al., 2010). Importantly, ciliate cilia are structurally and molecularly conserved with higher eukaryotes (Carvalho-Santos et al., 2011). Their cilia are arranged in an array that is comparable to human multi-ciliated cells, and their cilia beat in a biphasic whip-like motion that is consistent with their human counterparts (Wood et al., 2007; Funfak et al., 2015). Perhaps the most important advantage that ciliates have for cilia and basal body research is their large quantity of cilia per cell. A typical vertebrate multi-ciliated epithelial cell contains 200–300 cilia per cell, however, Tetrahymena and Paramecium cells contain upward of 750 and 4000 cilia per cell, respectively (Figure 1 and Supplementary Table S1; Pearson and Winey, 2009; Tilley et al., 2015). The large quantity of cilia has established ciliates as an ideal system for both light and electron microscopy as well as proteomics. In the following sections of this review we will specifically focus on the joint contributions of Tetrahymena and Paramecium to our understanding of basal body and motile cilia composition, assembly, structure, and function.
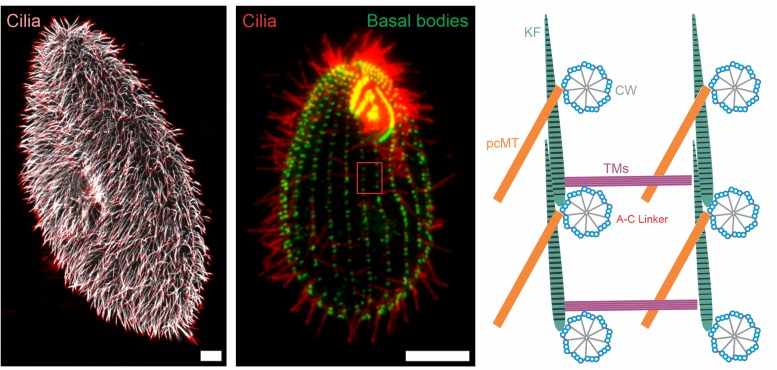
FIGURE 1.
Immunofluorescence images and schematic representation of ciliate cilia and accessory structures. Immunofluorescence images of Paramecium tetaurelia (left) and Tetrahymena thermophila (middle) cells. The Paramecium tetaurelia cell (left) is stained for cilia (α-GT335, white; α-TAP952, red) and the Tetrahymena thermophila cell (middle) is stained for basal bodies (α-centrin, green) and cilia (α-GT335, red). Scale bar = 10 μm. The red box in the Tetrahymena image (middle) is represented schematically in the right image showing a top down view of basal body organization with associated accessory structures. CW, cartwheel; pcMT, post ciliary microtubules; TM, transverse microtubules; KF, Kinetodesmal Fiber.
Basal Bodies
Basal bodies nucleate and anchor motile cilia in the cell, and as such, they are essential for the function of motile cilia. The basal bodies of ciliates provide a number of advantages for researchers. Ciliate basal bodies are strikingly similar to those of other eukaryotes in both structure and molecular composition (Carvalho-Santos et al., 2011). They are assembled in a stepwise process that is analogous to that of centrioles; however, ciliate basal bodies never function as centrioles (Dippell, 1968; Allen, 1969). This is experimentally advantageous because basal body defects are never conflated with mitotic defects. Additionally, like the multi-ciliated cells found in higher eukaryotes such as humans and Xenopus laevis, ciliate basal bodies are positioned and oriented into an array via a comprehensive network of accessory structures (Figure 1 and Supplementary Table S1) (Bayless et al., 2015; Tassin et al., 2015; Zhang and Mitchell, 2015; Vertii et al., 2016). Methods have also been developed to isolate and purify basal bodies, leaving researchers with pure fractions to perform biochemical and proteomic studies (Argetsinger, 1965; Tiedtke, 1985).
Identification of Basal Body Components
A proteome of Tetrahymena basal bodies was completed in 2007 (Kilburn et al., 2007). The Tetrahymena proteome displays a significant amount of overlap with two other basal body proteomes from human centrioles and Chlamydomonas basal bodies, a finding that highlights the high molecular conservation between phylogenetically distant species (Andersen et al., 2003; Keller et al., 2005). Importantly, the Tetrahymena proteome advanced the field by localizing identified proteins through GFP tagging and immuno-EM mapping (Kilburn et al., 2007). This localization data has served as an important first step in the functional characterization of many components, including the core basal body components Bld10, Poc1, and Sas6 (Culver et al., 2009; Pearson et al., 2009; Bayless et al., 2012; Meehl et al., 2016).
Basal Body Assembly and Maintenance
Basal body assembly begins with the formation of a radially symmetric cartwheel followed by the addition and elongation of triplet microtubules to the ends of each cartwheel spoke. Adjacent triplet microtubules are connected by A-C linkers then the entire basal body is capped at its distal end in a region called the transition zone, where the triplet microtubule of the basal body give way to the doublet microtubules of the ciliary axoneme (Figure 1; Carvalho-Santos et al., 2010). The first structural and temporal dissection of early basal body assembly was completed in ciliates in the late 1960’s (Dippell, 1968; Allen, 1969). These comprehensive cytological studies used electron microscopy to identify the early stages of basal body assembly, including the temporal ordering of assembly of the cartwheel and the microtubule walls (Dippell, 1968; Allen, 1969). In addition, these studies clearly define the major structural domains of the basal body like the cartwheel, A-C linkers, and the transition zone. Several of these structures are well defined in recent electron tomography studies (Höög et al., 2012; Li et al., 2012; Meehl et al., 2016; Greenan et al., 2018).
At the heart of basal body structure is the triplet microtubules. Triplet microtubule assembly and stability rely on a number of non-canonical tubulins, and ciliates have been impactful in their study and classification. The essential role of γ-tubulin in the earliest stages of basal body microtubule nucleation was first described in ciliates (Ruiz et al., 1999; Shang et al., 2002a). δ-tubulin has been shown in Paramecium to be necessary for the assembly of the basal body C-tubule (Garreau de Loubresse et al., 2001). Rarer tubulin isoforms ε- and ζ-tubulin are also found in ciliates and are necessary for basal body duplication and stabilization (Dupuis-Williams et al., 2002; Ruiz et al., 2004; Ross et al., 2013). Tetrahymena has also been used to study how ciliary beating affects the distribution of proteins and tubulin post-translational modifications at the triplet microtubules (Bayless et al., 2016). From this work both tubulin glutamylation and the triplet microtubule localizing protein Fop1 were shown to associate asymmetrically to the region of the basal body that experiences the most compressive force from ciliary beating (Bayless et al., 2016).
In many eukaryotic systems, basal body assembly is initiated by PLK4 (Habedanck et al., 2005). Ciliates have no clear PLK4 homolog, though they do contain many other key proteins involved in early assembly (Carvalho-Santos et al., 2010). Functional studies on the major constituent of the cartwheel, Sas-6, revealed that a fully formed cartwheel is necessary for basal body assembly (Culver et al., 2009). Attachment of the cartwheel to the triplet microtubules is facilitated by Bld10, a protein first identified and characterized in Chlamydomonas, though work in ciliates has established that Bld10 is essential for assembly and stability of basal bodies (Matsuura et al., 2004; Hiraki et al., 2007; Jerka-Dziadosz et al., 2010; Bayless et al., 2012). It is not only the link between cartwheels and the triplet microtubules that is necessary for stabilizing the basal body, the A-C linker between adjacent triplet microtubules is necessary as well. Tetrahymena Poc1 is necessary for establishing the A-C linker between adjacent triplet microtubules (Meehl et al., 2016). When Poc1 is lost from Tetrahymena cells, basal bodies fall apart under conditions where cilia-generated force is increased, highlighting the role of structural connections in maintaining the overall stability of the basal body (Pearson et al., 2009; Meehl et al., 2016).
Work in ciliates has established that Centrin 2 is necessary for basal body elongation and establishment of the transition zone, while Centrin 3 is necessary for anchoring the basal body in place (Guerra et al., 2003; Stemm-Wolf et al., 2005; Vonderfecht et al., 2011, 2012; Aubusson-Fleury et al., 2012; Jerka-Dziadosz et al., 2013). Taken together, these works represent the large impact ciliates have had on our understanding of basal body assembly and stability.
Basal Body Placement in an Array
The complex organization of basal body arrays have been well studied in ciliates. In both Paramecium and Tetrahymena, basal bodies are arranged into rows, called kinety, that extend along the anterior-posterior axis of the cell (Figure 1; Lynn, 1981; Iftode and Fleury-Aubusson, 2003). The positioning of basal bodies along a kinetid is complex and utilizes the coordination of at least three accessory structures: transverse microtubules, post-ciliary microtubules, and the striated rootlet (kinetodesmal fiber). The two microtubule based structures appear to be specific to ciliates with the transverse microtubules reaching across kinety and the post-ciliary microtubules reaching behind the basal body within a kinety (Figure 1; Lynn, 1981; Iftode and Fleury-Aubusson, 2003). The striated rootlet forges an attachment from the basal body to the plasma membrane and anchors it into place. Striated rootlets are found in most systems that contain multi-ciliated cells, thus highlighting the importance of this cortical connection (Steinman, 1968; Anderson, 1972; Sandoz et al., 1988). More recent work utilized automated image analysis to reveal the three dimensional arrangement of the Tetrahymena ciliary array (Galati et al., 2016). In a landmark study that tested the theory of structural inheritance, Janine Beisson and Tracy Sonneborn generated inverted kinety of basal bodies in Paramecium and found that all accessory structures built subsequently became inverted to match the inverted positioning of basal bodies (Beisson and Sonneborn, 1965). This experiment demonstrated that not all information is encoded by DNA, but some is, in fact, epigenetic. A result that was later replicated in Tetrahymena (Ng and Frankel, 1977). Furthermore, the kinetodesmal fiber alters its length in response to altered cilia-generated forces, suggesting that structural inheritance is tunable and able to adapt to environmental cues (Galati et al., 2016).
The molecular composition of basal body accessory structures is not well-defined but ciliates have contributed greatly to what we do know. The Tetrahymena striated fiber is made up, at least in part, by the DisAp protein (Galati et al., 2014). Orientation of basal bodies is disrupted by loss of Paramecium proteins Meckelin and Centrin 3, while ODF-1 and VFL3 are necessary for the docking of Paramecium basal bodies to the cell cortex (Jerka-Dziadosz et al., 2013; Picariello et al., 2014; Bengueddach et al., 2017). These studies highlight the power of ciliate reverse genetics approaches. Recent work has also made connections between the tubulin post-translational modification glycylation and attachment of Tetrahymena basal bodies at the cell cortex (Junker et al., 2019). The assembly, maintenance, and precise positioning of basal bodies remains an essential foundation for the building of a functional cilium and ciliates have, and will remain, at the forefront of basal body research.
Motile Cilia
Motile cilia and flagella function to move extra-cellular fluid. Structurally, the ciliate motile cilium is conventional consisting of nine set of doublet microtubules arranged radially around a central pair of single microtubules. These cilia beat in a biphasic, whip-like pattern that is consistent with the motile cilia of human cells (Supplementary Table S1; Tuxhorn et al., 1998; Bayless et al., 2015; Funfak et al., 2015). Experimentally, genetic techniques such as protein tagging allow for easy visualization of cilia and a simple calcium shock is sufficient to shear cilia from the cell allowing for easy isolation for biochemical assays (Rosenbaum and Carlson, 1969; Adoutte et al., 1980). As described below, research using ciliates has done much to illuminate the molecular composition, structure, assembly, and function of motile cilia.
Identification of Motile Cilia Components
In many of the same ways that ciliates have been impactful for identifying the molecular components of basal bodies, ciliates have also been used successfully in the identification of motile cilia molecular components. The significant overlap between the vertebrate and protist cilia proteome was made clear in a mass spectrometry screen using Tetrahymena cilia dubbed the “ciliome”(Smith et al., 2005). Isolation and purification of the Paramecium ciliary membrane also yielded a proteome of the ciliary membrane, the first of its kind (Yano et al., 2013). Outside of proteomics, whole genome expression profiling has been used to identify the genes that are upregulated during ciliogenesis, which offers a strong starting point for novel discovery and characterization of ciliogenesis proteins (Arnaiz et al., 2010).
Motile Cilia Structure
As with basal bodies, electron microscopy has been employed to investigate the structural makeup of ciliate ciliary axonemes. Detailed cytological analysis of the transition zone between basal bodies and ciliary axonemes was carried out in both Paramecium and Tetrahymena (Dippell, 1968; Allen, 1969; Hufnagel, 1969; Dute and Kung, 1978). At the distal end of the axoneme, the termination of the central pair of microtubules and the ciliary tip were first described in detail in Tetrahymena axonemes (Sale and Satir, 1977). Tetrahymena axonemes were also used in fine structural work detailing the radial spoke and axonemal dynein complexes that are necessary for ciliary movement (Goodenough and Heuser, 1985).
Perhaps the biggest impact that ciliates have made in the area of cilia structure has come from the use of single particle cryo-electron microscopy (cryo-EM) and cryo-electron tomography (cryo-ET) to investigate the structure of the ciliary doublet microtubules. Doublet microtubules are exceptionally stable making them an ideal substrate for cryo-EM and cryo-ET. Early studies used sea urchin sperm flagella for cryo-EM/ET, more recently Chlamydomonas flagella and Tetrahymena axonemes have become the model systems of choice (Pigino et al., 2012). Currently, single particle cryo-EM of Tetrahymena axonemes has achieved the best resolution of doublet microtubules to date at below a nanometer resolution (Ichikawa et al., 2017). There are two areas where the cryo-EM and cryo-ET structural work from Tetrahymena axonemes has been influential: our understanding of the machinery outside of the doublet microtubules and our understanding of the interior of the doublet microtubules. Radial spoke proteins and dynein arms make motile cilia move. The structure of these large complexes has been elucidated by structural work in Chlamydomonas but also with the help of Tetrahymena (Pigino et al., 2011). The lumen of the doublet microtubules has also been shown to have complex networks of proteins in them termed microtubule inner proteins or MIPs (Figure 2; Ichikawa et al., 2017; Ichikawa and Bui, 2018; Stoddard et al., 2018). These MIPs are thought to stabilize the doublet microtubule to allow for the repeated bending without break. The structural studies of doublet microtubules in ciliates have shown us just how complex the axoneme is, highlighting the importance of a precise assembly process.
FIGURE 2.
Structure of axoneme doublet microtubules. Cross-section of ciliary axoneme doublet microtubules. A- and B-tubules are shown. Tubulin protofilaments are colored gray and numbered. Microtubule inner proteins (MIPs) and filamentous MIPs (fMIPs) are distinguished by color as denoted in the table to the right. Adapted from Ichikawa and Bui (2018) and Stoddard et al. (2018).
Motile Cilia Assembly
Motile cilia assembly occurs through a concerted process called intraflagellar transport (IFT; Nakayama and Katoh, 2018). The advantage that ciliates have for studying ciliary assembly is two-fold. First, ciliates have a large number of cilia. Second, cilia assembly can be induced and followed in real time (Jiang et al., 2015). The first step in assembly of a cilium is to establish a ciliary cap. In Tetrahymena this is a multistep process that involves the confluence of three diverse structures (Seixas et al., 2017). After establishment of a ciliary cap, IFT is utilized to assemble the axoneme. In Paramecium, IFT46 is necessary for the trafficking of other IFT components between the cytoplasm and the ciliary axoneme, thus making it an upstream regulator of IFT (Shi et al., 2018). In Chlamydomonas and Paramecium axonemes, failure of IFT to traffic axonemal dynein into the cilium results in short cilia that are non-motile (Fassad et al., 2018). Furthermore, IFT trains have been shown to queue up at the base of Chlamydomonas and Tetrahymena axonemes while waiting their turn during assembly (Wingfield et al., 2017). Interestingly, the Tetrahymena LF4/MOK kinase family member, LF4a, regulates ciliary length by limiting the rate of IFT (Jiang et al., 2019). Besides IFT related mechanisms, ciliates have highlighted many other proteins necessary for assembly of the ciliary axoneme. Tetrahymena Dyf-1 is necessary for ciliogenesis as is PHLP2, both of which help with the assembly of axoneme microtubules (Dave et al., 2009; Bregier et al., 2013). The axoneme microtubules also display careful coordination of their length control. In Tetrahymena axonemes, FAP256/CEP104 promotes A-tubule elongations while CHE-12/Crescerin and ARMC9 specifically regulate the length of the B-tubule (Louka et al., 2018). Overall, work from ciliates has helped us understand the complexities of ciliary assembly, in particular the nuances of IFT.
Motile Cilia Function
Ciliary beating is a complex process that requires many molecular interactions. During beating, the ciliary axoneme must maintain a regular waveform all while remaining in concert with its neighboring cilia (Wan, 2018). One of the most fundamental questions about motile cilia was answered with the help of ciliates: How do cilia move? In the 1960’s Ian Gibbons identified and purified axonemal dynein from Tetrahymena axonemes (Gibbons, 1963; Gibbons and Rowe, 1965). This was the first microtubule molecular motor identified and its discovery represents a major milestone in cell biology. Follow-up studies using both Paramecium and Tetrahymena cilia identified how the sliding force between doublet microtubules is translated into the bending force that is seen during beating and the requirement of calcium and calcium signaling for propagation of ciliary beating (Bancroft, 1906; Satir, 1965, 1968; Mogami and Takahashi, 1983; Funfak et al., 2015; Yano et al., 2015). These works represent major advances in our understanding of how cilia beating is facilitated.
At the molecular level, the ease of reverse genetic approaches in ciliates allows for directed investigation of ciliary proteins. The conserved protofilament ribbon protein, Rib72, is not only a MIP, but it is necessary for the localization of a majority of MIPs in the A-tubule of Tetrahymena ciliary axonemes (Stoddard et al., 2018). When A-tubule MIPs are lost from axonemes, the structural integrity of the axoneme is compromised resulting in cilia that bend abnormally as they move through the typically rigid power stroke (Stoddard et al., 2018). As a result, their waveform and coordination with neighboring cilia is disrupted suggesting that MIPs play an important role in the structural support for the ciliary axoneme (Figure 2). Tetrahymena have also been used to functionally characterize ciliary components of radial spokes and the dynein arms, and both are necessary for proper ciliary beating (Urbanska et al., 2015, 2018).
A final and important area of research that ciliates have contributed greatly to is the study of tubulin post-translational modifications. Acetylation of tubulin is a common post-translational modifications found in stable populations of microtubules. The acetyltransferase MEC-17 is responsible for K40 tubulin acetylation and was first identified in Tetrahymena (Akella et al., 2010). Identification and characterization of the TTLL family of tubulin modifying enzymes has also been carried out in Tetrahymena (Janke et al., 2005). From these studies we find that tubulin glutamylation and glycylation affect the stability and waveform of ciliary beating in Tetrahymena axonemes (Wloga et al., 2008, 2009, 2010; Suryavanshi et al., 2010; Junker et al., 2019). Overall, ciliates have been utilized to demonstrate how motile cilia beat and the molecular players that affect ciliary structure and function.
Perspectives and Future Outlook
The strengths of ciliate research on cilia lies in their favorable genetics and the abundance of cilia in each cell. The simplicity of basal bodies and cilia isolation, combined with the ease of functional characterization of specific ciliary components, ensures that ciliates will be a viable option for basal body and motile cilia research. The resolution achieved by single particle cryo-EM and cryo-ET of Tetrahymena axonemes is exciting and provides a promising platform for future structural work. Ciliates are also poised to play a large part in understanding the biomechanics of ciliary beating. High speed imaging of ciliary beating is possible in ciliates and its use, coupled with genetic study of specific proteins, will be important for understanding how the whip-like ciliary beat stroke is performed and maintained (Funfak et al., 2015; Stoddard et al., 2018). Cilia disassembly is not well understood and the ease of inducing deciliation in ciliates could prove powerful for future research. Further, ciliates are positioned well for the study of human ciliary disorders. Genetic variants of Primary Ciliary Dyskinesia (PCD) have been modeled in Paramecium offering unparalleled insight into the pathology of this devastating disorder (Fassad et al., 2018). Given advances that have already been made, and blossoming avenues ripe for study, there is reason to believe that the future of ciliate research is bright and the best is yet to come.
Author Contributions
BB and MW conceived and wrote the manuscript. FN generated the figures and legends.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Sam Li and Amy Fabritius for their critical reading and insightful comments on the manuscript. The authors would also like to thank Anne-Marie Tassin and Pierrick le Borgne for the Paramecium image used in Figure 1.
Footnotes
Funding. This work was supported by funding from the National Institutes of Health (GM127571-02 to MW), Santa Clara University through a Dean’s Startup Grant from the College of Arts & Sciences (BB), and the Clare Boothe Luce Scholars Program (FN).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2019.00265/full#supplementary-material
A comparison of common model organisms used in motile cilia research.
References
- Adoutte A., Ramanathan R., Lewis R. M., Dute R. R., Ling K. Y., Kung C., et al. (1980). Biochemical studies of the excitable membrane of Paramecium tetraurelia. III. Proteins of cilia and ciliary membranes. J. Cell Biol. 84 717–738. 10.1083/jcb.84.3.717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akella J. S., Wloga D., Kim J., Starostina N. G., Lyons-Abbott S., Morrissette N. S., et al. (2010). MEC-17 is an alpha-tubulin acetyltransferase. Nature 467 218–222. 10.1038/nature09324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen R. D. (1969). The morphogenesis of basal bodies and accessory structures of the cortex of the ciliated protozoan Tetrahymena pyriformis. J. Cell Biol. 40 716–733. 10.1083/jcb.40.3.716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen J. S., Wilkinson C. J., Mayor T., Mortensen P., Nigg E. A., Mann M. (2003). Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426 570–574. 10.1038/nature02166 [DOI] [PubMed] [Google Scholar]
- Anderson R. G. (1972). The three-dimensional structure of the basal body from the rhesus monkey oviduct. J. Cell Biol. 54 246–265. 10.1083/jcb.54.2.246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argetsinger J. (1965). The isolation of ciliary basal bodies (Kinetosomes) from Tetrahymena pyriformis. J. Cell Biol. 24 154–157. 10.1083/jcb.24.1.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaiz O., Goût J.-F., Bétermier M., Bouhouche K., Cohen J., Duret L., et al. (2010). Gene expression in a paleopolyploid: a transcriptome resource for the ciliate Paramecium tetraurelia. BMC Genomics 11:547. 10.1186/1471-2164-11-547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubusson-Fleury A., Lemullois M., de Loubresse N. G., Laligné C., Cohen J., Rosnet O., et al. (2012). The conserved centrosomal protein FOR20 is required for assembly of the transition zone and basal body docking at the cell surface. J. Cell Sci. 125 4395–4404. 10.1242/jcs.108639 [DOI] [PubMed] [Google Scholar]
- Aury J.-M., Jaillon O., Duret L., Noel B., Jubin C., Porcel B. M., et al. (2006). Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature 444 171–178. [DOI] [PubMed] [Google Scholar]
- Bancroft F. W. (1906). On the influence of the relative concentration of calcium ions on the reversal of the polar effects of the galvanic current in Paramecium. J. Physiol. 34 444–463. 10.1113/jphysiol.1906.sp001167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayless B. A., Galati D. F., Junker A. D., Backer C. B., Gaertig J., Pearson C. G. (2016). Asymmetrically localized proteins stabilize basal bodies against ciliary beating forces. J. Cell Biol. 215 457–466. 10.1083/jcb.201604135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayless B. A., Galati D. F., Pearson C. G. (2015). Tetrahymena basal bodies. Cilia 5 1. 10.1186/s13630-016-0022-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayless B. A., Giddings T. H., Winey M., Pearson C. G. (2012). Bld10/Cep135 stabilizes basal bodies to resist cilia-generated forces. Mol. Biol. Cell 23 4820–4832. 10.1091/mbc.E12-08-0577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisson J., Bétermier M., Bré M.-H., Cohen J., Duharcourt S., Duret L., et al. (2010). Silencing specific Paramecium tetraurelia genes by feeding double-stranded RNA. Cold Spring Harb. Protoc. 2010:5363. [DOI] [PubMed] [Google Scholar]
- Beisson J., Sonneborn T. M. (1965). Cytoplasmic inheritance of the organization of the cell cortex in Paramecium aurelia. Proc. Natl. Acad. Sci. U.S.A. 53 275–282. 10.1073/pnas.53.2.275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengueddach H., Lemullois M., Aubusson-Fleury A., Koll F. (2017). Basal body positioning and anchoring in the multiciliated cell Paramecium tetraurelia: roles of OFD1 and VFL3. Cilia 6:6. 10.1186/s13630-017-0050-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregier C., Krzemień-Ojak L., Włoga D., Jerka-Dziadosz M., Joachimiak E., Batko K., et al. (2013). PHLP2 is essential and plays a role in ciliogenesis and microtubule assembly in Tetrahymena thermophila. J. Cell. Physiol. 228 2175–2189. 10.1002/jcp.24384 [DOI] [PubMed] [Google Scholar]
- Bruns P. J., Cassidy-Hanley D. (2000). Biolistic transformation of macro- and micronuclei. Methods Cell Biol. 62 501–512. 10.1016/s0091-679x(08)61553-8 [DOI] [PubMed] [Google Scholar]
- Carvalho-Santos Z., Azimzadeh J., Pereira-Leal J. B., Bettencourt-Dias M. (2011). Evolution: tracing the origins of centrioles, cilia, and flagella. J. Cell Biol. 194 165–175. 10.1083/jcb.201011152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho-Santos Z., Machado P., Branco P., Tavares-Cadete F., Rodrigues-Martins A., Pereira-Leal J. B., et al. (2010). Stepwise evolution of the centriole-assembly pathway. J. Cell Sci. 123 1414–1426. 10.1242/jcs.064931 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. (1993). Kingdom protozoa and its 18 phyla. Microbiol. Rev. 57 953–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver B. P., Meehl J. B., Giddings T. H., Winey M. (2009). The two SAS-6 homologs in Tetrahymena thermophila have distinct functions in basal body assembly. Mol. Biol. Cell 20 1865–1877. 10.1091/mbc.E08-08-0838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave D., Wloga D., Sharma N., Gaertig J. (2009). DYF-1 Is required for assembly of the axoneme in Tetrahymena thermophila. Eukaryot. Cell 8 1397–1406. 10.1128/EC.00378-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dippell R. V. (1968). The development of basal bodies in Paramecium. Proc. Natl. Acad. Sci. U.S.A. 61 461–468. 10.1073/pnas.61.2.461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis-Williams P., Fleury-Aubusson A., de Loubresse N. G., Geoffroy H., Vayssié L., Galvani A., et al. (2002). Functional role of epsilon-tubulin in the assembly of the centriolar microtubule scaffold. J. Cell Biol. 158 1183–1193. 10.1083/jcb.200205028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dute R., Kung C. (1978). Ultrastructure of the proximal region of somatic cilia in Paramecium tetraurelia. J. Cell Biol. 78 451–464. 10.1083/jcb.78.2.451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen J. A., Coyne R. S., Wu M., Wu D., Thiagarajan M., Wortman J. R., et al. (2006). Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 4:e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassad M. R., Shoemark A., Le Borgne P., Koll F., Patel M., Dixon M., et al. (2018). C11orf70 mutations disrupting the intraflagellar transport-dependent assembly of multiple axonemal dyneins cause primary ciliary dyskinesia. Am. J. Hum. Genet. 102 956–972. 10.1016/j.ajhg.2018.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funfak A., Fisch C., Abdel Motaal H. T., Diener J., Combettes L., Baroud C. N., et al. (2015). Paramecium swimming and ciliary beating patterns: a study on four RNA interference mutations. Integr. Biol. 7 90–100. 10.1039/c4ib00181h [DOI] [PubMed] [Google Scholar]
- Galati D. F., Abuin D. S., Tauber G. A., Pham A. T., Pearson C. G. (2016). Automated image analysis reveals the dynamic 3-dimensional organization of multi-ciliary arrays. Biol. Open. 5 20–31. 10.1242/bio.014951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galati D. F., Bonney S., Kronenberg Z., Clarissa C., Yandell M., Elde N. C., et al. (2014). DisAp-dependent striated fiber elongation is required to organize ciliary arrays. J. Cell Biol. 207 705–715. 10.1083/jcb.201409123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garreau de Loubresse N., Ruiz F., Beisson J., Klotz C. (2001). Role of delta-tubulin and the C-tubule in assembly of Paramecium basal bodies. BMC Cell Biol. 2:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons I. R. (1963). Studies on the protein components of cilia from Tetrahymena pyriformis. Proc. Natl. Acad. Sci. U.S.A. 50 1002–1010. 10.1073/pnas.50.5.1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons I. R., Rowe A. J. (1965). Dynein: a protein with adenosine triphosphatase activity from cilia. Science 149 424–426. 10.1126/science.149.3682.424 [DOI] [PubMed] [Google Scholar]
- Goodenough U. W., Heuser J. E. (1985). Substructure of inner dynein arms, radial spokes, and the central pair/projection complex of cilia and flagella. J. Cell Biol. 100 2008–2018. 10.1083/jcb.100.6.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenan G. A., Keszthelyi B., Vale R. D., Agard D. A. (2018). Insights into centriole geometry revealed by cryotomography of doublet and triplet centrioles. eLife 7:e36851. 10.7554/eLife.36851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra C., Wada Y., Leick V., Bell A., Satir P. (2003). Cloning, localization, and axonemal function of Tetrahymena centrin. Mol. Biol. Cell 14 251–261. 10.1091/mbc.e02-05-0298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habedanck R., Stierhof Y.-D., Wilkinson C. J., Nigg E. A. (2005). The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 7 1140–1146. 10.1038/ncb1320 [DOI] [PubMed] [Google Scholar]
- Hai B., Gaertig J., Gorovsky M. A. (2000). Knockout heterokaryons enable facile mutagenic analysis of essential genes in Tetrahymena. Methods Cell Biol. 62 513–531. 10.1016/s0091-679x(08)61554-x [DOI] [PubMed] [Google Scholar]
- Hiraki M., Nakazawa Y., Kamiya R., Hirono M. (2007). Bld10p constitutes the cartwheel-spoke tip and stabilizes the 9-fold symmetry of the centriole. Curr. Biol. 17 1778–1783. 10.1016/j.cub.2007.09.021 [DOI] [PubMed] [Google Scholar]
- Höög J. L., Bouchet-Marquis C., McIntosh J. R., Hoenger A., Gull K. (2012). Cryo-electron tomography and 3-D analysis of the intact flagellum in Trypanosoma brucei. J. Struct. Biol. 178 189–198. 10.1016/j.jsb.2012.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufnagel L. A. (1969). Cortical ultrastructure of Paramecium aurelia. Studies on isolated pellicles. J. Cell Biol. 40 779–801. 10.1083/jcb.40.3.779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa M., Bui K. H. (2018). Microtubule inner proteins: a meshwork of luminal proteins stabilizing the doublet microtubule. Bioessays 40 15035. 10.1002/bies.201700209 [DOI] [PubMed] [Google Scholar]
- Ichikawa M., Liu D., Kastritis P. L., Basu K., Hsu T. C., Yang S., et al. (2017). Subnanometre-resolution structure of the doublet microtubule reveals new classes of microtubule-associated proteins. Nat. Commun. 8:15035. 10.1038/ncomms15035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iftode F., Fleury-Aubusson A. (2003). Structural inheritance in Paramecium: ultrastructural evidence for basal body and associated rootlets polarity transmission through binary fission. Biol. Cell 95 39–51. 10.1016/s0248-4900(03)00005-4 [DOI] [PubMed] [Google Scholar]
- Janke C., Rogowski K., Wloga D., Regnard C., Kajava A. V., Strub J.-M., et al. (2005). Tubulin polyglutamylase enzymes are members of the TTL domain protein family. Science 308 1758–1762. 10.1126/science.1113010 [DOI] [PubMed] [Google Scholar]
- Jerka-Dziadosz M., Gogendeau D., Klotz C., Cohen J., Beisson J., Koll F. (2010). Basal body duplication in Paramecium: the key role of Bld10 in assembly and stability of the cartwheel. Cytoskelet 67 161–171. 10.1002/cm.20433 [DOI] [PubMed] [Google Scholar]
- Jerka-Dziadosz M., Koll F., Włoga D., Gogendeau D., Garreau de Loubresse N., Ruiz F., et al. (2013). A Centrin3-dependent, transient, appendage of the mother basal body guides the positioning of the daughter basal body in Paramecium. Protist 164 352–368. 10.1016/j.protis.2012.11.003 [DOI] [PubMed] [Google Scholar]
- Jiang Y.-Y., Lechtreck K., Gaertig J. (2015). Total internal reflection fluorescence microscopy of intraflagellar transport in Tetrahymena thermophila. Methods Cell Biol. 127 445–456. 10.1016/bs.mcb.2015.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y.-Y., Maier W., Baumeister R., Minevich G., Joachimiak E., Wloga D., et al. (2019). LF4/MOK and a CDK-related kinase regulate the number and length of cilia in Tetrahymena. PLoS Genet. 15:e1008099. 10.1371/journal.pgen.1008099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker A. D., Soh A. W. J., O’Toole E. T., Meehl J. B., Guha M., Winey M., et al. (2019). Microtubule glycylation promotes basal body attachment to the cell cortex. J. Cell Sci. 132:jcs233726. 10.1242/jcs.233726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller L. C., Romijn E. P., Zamora I., Yates J. R., Marshall W. F. (2005). Proteomic analysis of isolated Chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr. Biol. 15 1090–1098. 10.1016/j.cub.2005.05.024 [DOI] [PubMed] [Google Scholar]
- Kilburn C. L., Pearson C. G., Romijn E. P., Meehl J. B., Giddings T. H., Culver B. P., et al. (2007). New Tetrahymena basal body protein components identify basal body domain structure. J. Cell Biol. 178 905–912. 10.1083/jcb.200703109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung C., Saimi Y., Haynes W. J., Ling K. Y., Kissmehl R. (2000). Recent advances in the molecular genetics of Paramecium. J. Eukaryot. Microbiol. 47 11–14. [DOI] [PubMed] [Google Scholar]
- Li S., Fernandez J.-J., Marshall W. F., Agard D. A. (2012). Three-dimensional structure of basal body triplet revealed by electron cryo-tomography. EMBO J. 31 552–562. 10.1038/emboj.2011.460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louka P., Vasudevan K. K., Guha M., Joachimiak E., Wloga D., Tomasi R. F.-X., et al. (2018). Proteins that control the geometry of microtubules at the ends of cilia. J. Cell Biol. 217 4298–4313. 10.1083/jcb.201804141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn D. H. (1981). The organization and evolution of microtubular organelles in Ciliated Protozoa. Biol. Rev. 56 243–292. 10.1111/j.1469-185x.1981.tb00350.x [DOI] [Google Scholar]
- Matsuura K., Lefebvre P. A., Kamiya R., Hirono M. (2004). Bld10p, a novel protein essential for basal body assembly in Chlamydomonas: localization to the cartwheel, the first ninefold symmetrical structure appearing during assembly. J. Cell Biol. 165 663–671. 10.1083/jcb.200402022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehl J. B., Bayless B. A., Giddings T. H., Pearson C. G., Winey M. (2016). Tetrahymena Poc1 ensures proper intertriplet microtubule linkages to maintain basal body integrity. Mol. Biol. Cell 27 2394–2403. 10.1091/mbc.E16-03-0165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogami Y., Takahashi K. (1983). Calcium and microtubule sliding in ciliary axonemes isolated from Paramecium caudatum. J. Cell Sci. 61 107–121. [DOI] [PubMed] [Google Scholar]
- Nakayama K., Katoh Y. (2018). Ciliary protein trafficking mediated by IFT and BBSome complexes with the aid of kinesin-2 and dynein-2 motors. J. Biochem. 163 155–164. 10.1093/jb/mvx087 [DOI] [PubMed] [Google Scholar]
- Ng S. F., Frankel J. (1977). 180 degrees rotation of ciliary rows and its morphogenetic implications in Tetrahymena pyriformis. Proc. Natl. Acad. Sci. U.S.A. 74 1115–1119. 10.1073/pnas.74.3.1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C. G., Osborn D. P. S., Giddings T. H., Beales P. L., Winey M. (2009). Basal body stability and ciliogenesis requires the conserved component Poc1. J. Cell Biol. 187 905–920. 10.1083/jcb.200908019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C. G., Winey M. (2009). Basal body assembly in ciliates: the power of numbers. Traffic 10 461–471. 10.1111/j.1600-0854.2009.00885.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picariello T., Valentine M. S., Yano J., Van Houten J. (2014). Reduction of meckelin leads to general loss of cilia, ciliary microtubule misalignment and distorted cell surface organization. Cilia 3:2. 10.1186/2046-2530-3-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigino G., Bui K. H., Maheshwari A., Lupetti P., Diener D., Ishikawa T. (2011). Cryoelectron tomography of radial spokes in cilia and flagella. J. Cell Biol. 195 673–687. 10.1083/jcb.201106125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigino G., Maheshwari A., Bui K. H., Shingyoji C., Kamimura S., Ishikawa T. (2012). Comparative structural analysis of eukaryotic flagella and cilia from Chlamydomonas, Tetrahymena, and sea urchins. J. Struct. Biol. 178 199–206. 10.1016/j.jsb.2012.02.012 [DOI] [PubMed] [Google Scholar]
- Rosenbaum J. L., Carlson K. (1969). Cilia regeneration in Tetrahymena and its inhibition by colchicine. J. Cell Biol. 40 415–425. 10.1083/jcb.40.2.415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross I., Clarissa C., Giddings T. H., Winey M. (2013). ε-tubulin is essential in Tetrahymena thermophila for the assembly and stability of basal bodies. J. Cell Sci. 126 3441–3451. 10.1242/jcs.128694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz F., Beisson J., Rossier J., Dupuis-Williams P. (1999). Basal body duplication in Paramecium requires gamma-tubulin. Curr. Biol. 9 43–46. 10.1016/s0960-9822(99)80045-1 [DOI] [PubMed] [Google Scholar]
- Ruiz F., Dupuis-Williams P., Klotz C., Forquignon F., Bergdoll M., Beisson J., et al. (2004). Genetic evidence for interaction between eta- and beta-tubulins. Eukaryot. Cell 3 212–220. 10.1128/ec.3.1.212-220.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale W. S., Satir P. (1977). The termination of the central microtubules from the cilia of Tetrahymena pyriformis. Cell Biol. Int. Rep. 1 45–49. 10.1016/0309-1651(77)90008-x [DOI] [PubMed] [Google Scholar]
- Sandoz D., Chailley B., Boisvieux-Ulrich E., Lemullois M., Laine M. C., Bautista-Harris G. (1988). Organization and functions of cytoskeleton in metazoan ciliated cells. Biol. Cell 63 183–193. 10.1016/0248-4900(88)90057-3 [DOI] [PubMed] [Google Scholar]
- Satir P. (1965). Studies on cilia: II. examination of the distal region of the ciliary shaft and the role of the filaments in motility. J. Cell Biol. 26 805–834. 10.1083/jcb.26.3.805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satir P. (1968). Studies on cilia. 3. Further studies on the cilium tip and a “sliding filament” model of ciliary motility. J. Cell Biol. 39 77–94. 10.1083/jcb.39.1.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seixas C., Gonçalves J., Melo L. V., Soares H. (2017). Tetrahymena cilia cap is built in a multi-step process: a study by atomic force microscopy. Protist 168 697–717. 10.1016/j.protis.2017.10.001 [DOI] [PubMed] [Google Scholar]
- Shang Y., Song X., Bowen J., Corstanje R., Gao Y., Gaertig J., et al. (2002b). A robust inducible-repressible promoter greatly facilitates gene knockouts, conditional expression, and overexpression of homologous and heterologous genes in Tetrahymena thermophila. Proc. Natl. Acad. Sci. U.S.A. 99 3734–3739. 10.1073/pnas.052016199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y., Li B., Gorovsky M. A. (2002a). Tetrahymena thermophila contains a conventional gamma-tubulin that is differentially required for the maintenance of different microtubule-organizing centers. J. Cell Biol. 158 1195–1206. 10.1083/jcb.200205101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L., Shi X., Shen Y. (2018). Intraflagellar transport 46 (IFT46) is essential for trafficking IFT proteins between cilia and cytoplasm in Paramecium. Sci. Rep. 8:9259. 10.1038/s41598-018-27050-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. C., Northey J. G. B., Garg J., Pearlman R. E., Siu K. W. M. (2005). Robust method for proteome analysis by MS/MS using an entire translated genome: demonstration on the ciliome of Tetrahymena thermophila. J. Proteome Res. 4 909–919. 10.1021/pr050013h [DOI] [PubMed] [Google Scholar]
- Steinman R. M. (1968). An electron microscopic study of ciliogenesis in developing epidermis and trachea in the embryo of Xenopus laevis. Am. J. Anat. 122 19–55. 10.1002/aja.1001220103 [DOI] [PubMed] [Google Scholar]
- Stemm-Wolf A. J., Morgan G., Giddings T. H., White E. A., Marchione R., McDonald H. B., et al. (2005). Basal body duplication and maintenance require one member of the Tetrahymena thermophila centrin gene family. Mol. Biol. Cell 16 3606–3619. 10.1091/mbc.e04-10-0919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard D., Zhao Y., Bayless B. A., Gui L., Louka P., Dave D., et al. (2018). Tetrahymena RIB72A and RIB72B are microtubule inner proteins in the ciliary doublet microtubules. Mol. Biol. Cell 29 2566–2577. 10.1091/mbc.E18-06-0405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryavanshi S., Eddé B., Fox L. A., Guerrero S., Hard R., Hennessey T., et al. (2010). Tubulin glutamylation regulates ciliary motility by altering inner dynein arm activity. Curr. Biol. CB 20 435–440. 10.1016/j.cub.2009.12.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassin A.-M., Lemullois M., Aubusson-Fleury A. (2015). Paramecium tetraurelia basal body structure. Cilia 5:6. 10.1186/s13630-016-0026-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedtke A. (1985). Isolation of pure pellicles containing intact basal bodies of Tetrahymena pyriformis. J. Cell Sci. 77 155–165. [DOI] [PubMed] [Google Scholar]
- Tilley A. E., Walters M. S., Shaykhiev R., Crystal R. G. (2015). Cilia dysfunction in lung disease. Annu. Rev. Physiol. 77 379–406. 10.1146/annurev-physiol-021014-071931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuxhorn J., Daise T., Dentler W. L. (1998). Regulation of flagellar length in Chlamydomonas. Cell Motil. Cytoskeleton 40 133–146. [DOI] [PubMed] [Google Scholar]
- Urbanska P., Joachimiak E., Bazan R., Fu G., Poprzeczko M., Fabczak H., et al. (2018). Ciliary proteins Fap43 and Fap44 interact with each other and are essential for proper cilia and flagella beating. Cell. Mol. Life Sci. 75 4479–4493. 10.1007/s00018-018-2819-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanska P., Song K., Joachimiak E., Krzemien-Ojak L., Koprowski P., Hennessey T., et al. (2015). The CSC proteins FAP61 and FAP251 build the basal substructures of radial spoke 3 in cilia. Mol. Biol. Cell 26 1463–1475. 10.1091/mbc.E14-11-1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertii A., Hung H.-F., Hehnly H., Doxsey S. (2016). Human basal body basics. Cilia 5:13. 10.1186/s13630-016-0030-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonderfecht T., Cookson M. W., Giddings T. H., Clarissa C., Winey M. (2012). The two human centrin homologues have similar but distinct functions at Tetrahymena basal bodies. Mol. Biol. Cell 23 4766–4777. 10.1091/mbc.E12-06-0454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonderfecht T., Stemm-Wolf A. J., Hendershott M., Giddings T. H., Meehl J. B., Winey M. (2011). The two domains of centrin have distinct basal body functions in Tetrahymena. Mol. Biol. Cell 22 2221–2234. 10.1091/mbc.E11-02-0151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan K. Y. (2018). Coordination of eukaryotic cilia and flagella. Essays Biochem. 62 829–838. 10.1042/EBC20180029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingfield J. L., Mengoni I., Bomberger H., Jiang Y.-Y., Walsh J. D., Brown J. M., et al. (2017). IFT trains in different stages of assembly queue at the ciliary base for consecutive release into the cilium. eLife 6:2017. 10.7554/eLife.26609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloga D., Dave D., Meagley J., Rogowski K., Jerka-Dziadosz M., Gaertig J. (2010). Hyperglutamylation of Tubulin can either stabilize or destabilize microtubules in the same cell. Eukaryot. Cell 9 184–193. 10.1128/EC.00176-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloga D., Rogowski K., Sharma N., Van Dijk J., Janke C., Eddé B., et al. (2008). Glutamylation on alpha-tubulin is not essential but affects the assembly and functions of a subset of microtubules in Tetrahymena thermophila. Eukaryot. Cell 7 1362–1372. 10.1128/EC.00084-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloga D., Webster D. M., Rogowski K., Bré M.-H., Levilliers N., Jerka-Dziadosz M., et al. (2009). TTLL3 Is a tubulin glycine ligase that regulates the assembly of cilia. Dev. Cell 16 867–876. 10.1016/j.devcel.2009.04.008 [DOI] [PubMed] [Google Scholar]
- Wood C. R., Hard R., Hennessey T. M. (2007). Targeted gene disruption of dynein heavy chain 7 of Tetrahymena thermophila results in altered ciliary waveform and reduced swim speed. J. Cell Sci. 120 3075–3085. 10.1242/jcs.007369 [DOI] [PubMed] [Google Scholar]
- Yano J., Rajendran A., Valentine M. S., Saha M., Ballif B. A., Van Houten J. L. (2013). Proteomic analysis of the cilia membrane of Paramecium tetraurelia. J. Proteomics 78 113–122. 10.1016/j.jprot.2012.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano J., Valentine M. S., Van Houten J. L. (2015). Novel insights into the development and function of cilia using the advantages of the paramecium cell and its many cilia. Cells 4 297–314. 10.3390/cells4030297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Gorovsky M. A. (2000). Protein tagging in Tetrahymena. Methods Cell Biol. 62 549–559. 10.1016/s0091-679x(08)61556-3 [DOI] [PubMed] [Google Scholar]
- Zhang S., Mitchell B. J. (2015). Basal bodies in Xenopus. Cilia 5:2. 10.1186/s13630-016-0024-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A comparison of common model organisms used in motile cilia research.