ABSTRACT
BACKGROUND:
The periodontal tissues are continuously exposed to specific bacterial components that have the ability to alter many local functions. Normal endogenous infections in healthy mouths cause disease when their numbers increase significantly.
OBJECTIVE:
Determine the percentage of different periodontal pathogenic bacteria and their association with periodontal status.
DESIGN:
Cross-sectional, analytical.
SETTINGS:
School children of both genders in Saudi Arabia.
PATIENTS AND METHODS:
Clinical examination consisted of measurement of the gingival and periodontal supporting tissue including attachment loss, probing pocket depth and furcation involvement following the National Health and Nutrition Examination Survey (NHANES) and taking samples of the subgingival bacterial flora.
MAIN OUTCOME MEASURES:
The percentage of periodontal pathogenic bacteria and its association with periodontal status in Saudi
Arabia.
SAMPLE SIZE:
Bacterial samples were collected from 277 subjects.
RESULTS:
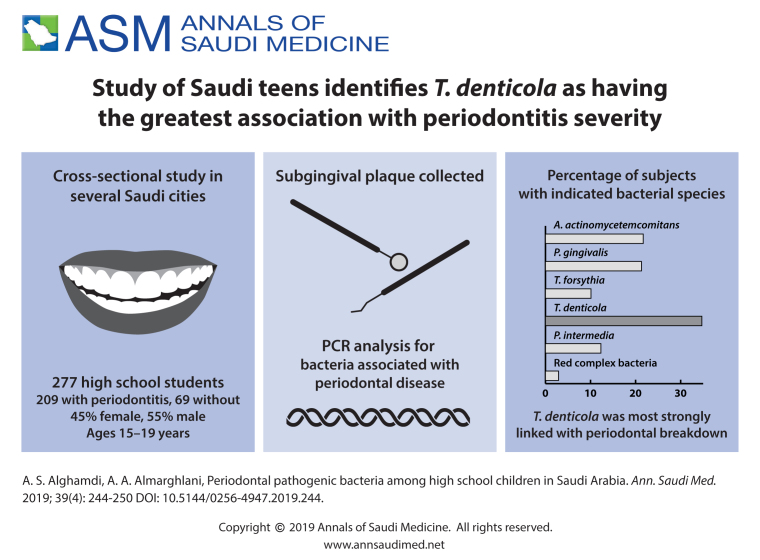
Aggregatibacter actinomycetemcomitans was present in 21.7% of the subjects, Porphyromonas gingivalis in 21.3%; Tannerella forsythia in 10.1%; Treponema denticola in 34.7% and Prevotella inter-media in 12.3%. The red complex bacteria were found in 2.9% of the subjects.
CONCLUSIONS:
The percentages of bacteria varied but only T denticola was significantly associated with periodontal breakdown. In addition, the presence of more than 2 of the 5 species tested were significantly associated with tissue damage.
LIMITATIONS:
Cannot be generalized to all of Saudi Arabia. Larger controlled studies are needed.
CONFLICT OF INTEREST:
None.
INTRODUCTION
Periodontal diseases are abundant among children, adolescents, and adults.1 They involve any inherited or acquired disorders of the tooth supporting structures (gingiva, cementum, periodontal ligament, and alveolar bone).2 The periodontal tissues are constantly exposed to certain bacterial components that have the capability to modulate local functions.3 Natural inflammatory processes protect the host and limit the pathogenic effect of biofilms, thus regulating some tissue destruction as a collateral effect of the defense.4 Host-bacterial interaction theory is accepted as the current etiological basis for the disease.5 According to the American Academy of Periodontology, “A bacterial infection alone is insufficient to result in periodontal disease. The host response plays a critical role in the tissue destruction seen in periodontitis and everyone is not equally susceptible to periodontal disease”.6 The concept of periodontal disease pathogenesis was introduced by Page in 1990,7 who summarized the multifactorial nature of the disease, stating that while bacteria are crucial in causing the disease, they alone are insufficient. Host and environmental factors strongly influence the onset, severity of the disease and the rate of progression as well as the response to treatment.8
Of the bacterial species currently known to inhabit the oral cavity, many possess the ability to destroy periodontal structures even when present in small quantities.9 As part of their pathogenic potential, these bacterial species are able to colonize the subgingival area, produce virulence factors that directly (enzymes and toxins) or indirectly (antigens and activators) lead to initiation of a destructive inflammatory reaction in the individual and injury of periodontal tissues.10,11 The host immune response is activated through microbial infection which leads to production of specific antibodies, cytokines and other humoral factors to defend host tissues from inflammatory agents, microbial penetration and injury. Subgingival plaque present in deep periodontal pockets is dominated by gram-negative anaerobic rods and spirochetes.12 Strong evidence has implicated Porphyromonas gingivalis13 and Aggregatibacter actinomycetemcomitans14 in the pathogenesis of adult periodontitis. In addition, Tannerella forsythia,15 Prevotella intermedia,16 Peptostreptococcus micros,17 and Fusobacterium nucleatum18 have been strongly associated with the initiation, severity and progression of periodontal diseases. There is strong evidence that these bacteria are associated and/or responsible for periodontal destruction.19
Motile rods, spirochetes and P intermedia are elevated proportionately in teenaged children with gingivitis and linked with clinical features such as bleeding on probing and the gingival index.20 In a group of healthy children, 2 to 18 years of age, investigators have found that 60% and 75% of the children had detectable levels of P gingivalis, A actinomycetemcomitans and T forsythia in their plaque. These bacterial species, which cause gingivitis in children, are endemic in healthy mouths, and cause disease when their numbers increase significantly.21 The current study aimed to examine the percentage of different periodontal pathogenic bacteria and their association with periodontal status based on cross-sectionally collected dental plaque samples from high school children in Saudi Arabia.
PATIENTS AND METHODS
The current study was part of an ongoing larger national cross-sectional descriptive study that is appraising the pattern of gingival and periodontal diseases among high school children in Saudi Arabia. In the large study, we examined a random sample of 2435 school children grades 10 to 12 (15–18 years old) of both genders in September 2012 until January 2016 (41 months) The study focused mainly on larger cities but will also include some smaller cities in more rural areas. The selected cities were: Riyadh, Jeddah, Dammam, Abha and Tabuk. A multistage clustered sampling design was followed to guarantee an adequate representation of all children in the country within the specified school grades.
Two hundred and seventy-seven subjects were included in the current study from a total sample of 2435 children. They were divided into children diagnosed with periodontitis (209 subjects) and periodontally healthy control (69 subjects) from the total sample of 2435 children. The study protocol was approved by the Institutional Review Board, Faculty of Dentistry, King Abdulaziz University (no. 073-09-12). Informed consent was provided by the parents of children to be included in the study. No children were admitted to the study without their parents' approval. Subject name, gender, age, address, and contact information were recorded. At an examination visit, the examiners reviewed the medical history with the subjects and recorded the information. A dental history questionnaire was completed by each subject and revised with the examiner. All examinations were performed on permanent teeth only; primary teeth (if present) and partially erupted teeth were excluded from the examination.
The periodontal examination consisted of measurement of the gingival and periodontal supporting tissue including gingivitis, attachment loss, probing pocket depth and furcation involvement; and the assessment of gingival bleeding, and dental plaque. Measurements were based on criteria set by the US National Health and Nutrition Examination Survey (NHANES) IV where Random Half-Mouth was used.22
In periodontally healthy children (69 subjects), samples were collected from two randomly selected subgingival sites. In children with periodontal diseases, samples were collected from the two deepest pockets. For sample collection, the tooth was dried and supragingival plaque was removed from sampling sites; a subgingival plaque sample was collected using a sterile curette in a single vertical stroke. Each sample was immediately placed in a sterile centrifuge tube containing 0.5 mL ethylenediamine tetraacetic acid (EDTA) buffer. The samples were sent to King Abdulaziz University Hospital microbiology lab for analysis. An improved multiplex nested PCR technique was used to allow sensitive detection of all five periodontal pathogens.23,24
The data obtained was analyzed using IBM SPSS version 22. A simple descriptive statistic was used to define the characteristics of the study variables as counts and percentages for the categorical and nominal variables while continuous variables are presented by mean and standard deviation. To establish a relationship between categorical variables, we used the chi-square test. When comparing two group means and more than two groups, an independent t test or oneway ANOVA were used. The conventional P value of <.05 was the criteria to reject the null hypothesis.
RESULTS
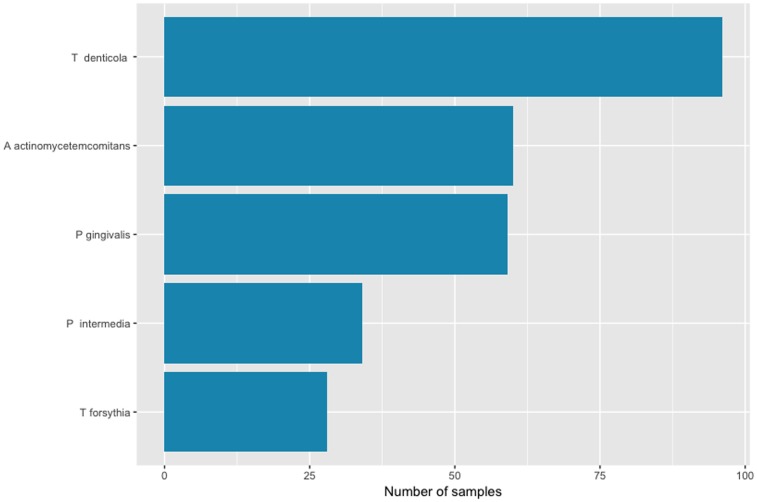
The 277 subjects ranged in age from 15 to 19 years with a mean (standard deviation) of 17.3 (1.0) years. Two hundred sixty-one (94%) were Saudi, the remainder were other Arab nationalities. More than half of the subjects were males (54.6%). Bacterial samples were obtained from the 277 subjects. A actinomycetemcomitans was present in 21.7%, P gingivalis in 21.3%; T forsythia in 10.1%; Treponema denticola in 34.7%; and P intermedia in 12.3% (Figure 1). The red complex bacteria were found in 2.9% of the subjects.25 About 57.4% of the participants had at least one type of bacteria, whereas 2 or more bacteria were found in 19.1%. Only 9.0% of the participants had 3 or more bacteria, and 4 or more bacteria were found in only 1.8%.
Figure 1.
Number of samples by type of bacteria.
Age had a significant relationship with A actinomycetemcomitans (Table 1). Table 2 shows characteristics of samples relative to the presence of P gingivalis. Pocket depth ≥4 mm had a significant relationship with the presence of T denticola (Table 3). None of the characteristics of the sample relative to T forsythia and P intermedia were significantly related to these bacteria. None of the characteristics of the sample relative to the presence of up to two bacteria showed a significant association. The mean PD had a significant relationship with the presence of 3 or more and 4 or more bacteria. The Gingival Index, mean PD and percentage of PD ?4mm had a significant relationship with the presence of 4 or more bacteria. None of the variables showed a significant association with the red complex bacteria.
Table 1.
Characteristics of the study samples relative to the presence of A actinomycetemcomitans.
| Variables | A actinomycetemcomitans | P value | |
|---|---|---|---|
| Yes | No | ||
| Total (n) | 60 (21.7) | 217 (78.3) | N/A |
| Age (years) | 17.0 (0.9) | 17.3 (1.0) | .032 |
| Gender | |||
| Male | 16 (15.8) | 85 (84.2) | .075 |
| Female | 44 (25.0) | 132 (75.0) | |
| Nationality | |||
| Non-Saudi | 12 (19.0) | 51 (81.0) | .567 |
| Saudi | 48 (22.4) | 166 (77.6) | |
| Smoker | |||
| Yes | 2 (11.8) | 15 (88.2) | .307 |
| No | 58 (22.3) | 202 (77.7) | |
| Plaque Index | 1.36 (0.8) | 1.52 (0.8) | .161 |
| Gingival Index | 1.30 (0.8) | 1.22 (0.8) | .505 |
| PD (mm) | 0.65 (0.2) | 0.68 (0.2) | .370 |
| PD ≥4 mm | 7.45 (9.6) | 7.46 (9.2) | .996 |
| CAL (mm) | 0.17 (0.3) | 0.18 (0.3) | .921 |
| CAL ≥1m | 9.36 (20.4) | 10.42 (23.4) | .758 |
Data are number (percentage) or mean (standard deviation). PD: Pocket depth, CAL: Clinical attachment loss Note: Numbers do not add up in some cells due to missing data
Table 2.
Characteristics of the study samples relative to the presence of P gingivalis.
| Variables | P gingivalis | P value | |
|---|---|---|---|
| Yes | No | ||
| Total (n) | 59 (21.3) | 218 (78.7) | N/A |
| Age (years) | 17.35 (0.9) | 17.25 (1.0) | .440 |
| Gender | |||
| Male | 21 (20.8) | 80 (79.2) | .876 |
| Female | 38 (21.6) | 138 (48.4) | |
| Nationality | |||
| Non-Saudi | 14 (22.2) | 49 (77.8) | .839 |
| Saudi | 45 (21.0) | 169 (79.0) | |
| Smoker | |||
| Yes | 3 (17.6) | 14 (82.4) | .704 |
| No | 56 (21.5) | 204 (78.5) | |
| Plaque Index | 1.49 (0.9) | 1.49 (0.8) | .997 |
| Gingival Index | 1.29 (0.8) | 1.22 (0.8) | .592 |
| Plaque diameter (mm) | 0.69 (0.2) | 0.67 (0.2) | .627 |
| PD ≥4mm | 8.78 (9.6) | 7.10 (9.2) | .219 |
| CAL (mm) | 0.17 (0.2) | 0.18 (0.3) | .832 |
| CAL ≥1m | 9.73 (20.3) | 10.31 (23.4) | .867 |
Data are number (percentage) or mean (standard deviation). Note: Numbers do not add up in some cells due to missing data.
Table 3.
Characteristics of the study samples relative to the presence of T denticola.
| Variables | T denticola | P value | |
|---|---|---|---|
| Yes | No | ||
| Total n | 96 (34.7) | 181 (65.3) | N/A |
| Age (years) | 17.18 (1.0) | 17.29 (0.9) | .406 |
| Gender | |||
| Male | 34 (33.7) | 67 (66.3) | .792 |
| Female | 62 (35.2) | 114 (64.8) | |
| Nationality | |||
| Non-Saudi | 21 (33.3) | 42 (66.7) | .802 |
| Saudi | 75 (35.0) | 139 (65.0) | |
| Smoker | |||
| Yes | 8 (47.1) | 9 (52.9) | .267 |
| No | 88 (33.8) | 172 (66.2) | |
| Plaque Index | 1.48 (0.8) | 1.50 (0.8) | .856 |
| Gingival Index | 1.27 (0.8) | 1.22 (0.8) | .598 |
| PD (mm) | 0.69 (0.2) | 0.67 (0.2) | .477 |
| PD ≥4 mm | 9.67 (11.5) | 6.28 (7.7) | .004 |
| CAL (mm) | 0.21 (0.3) | 0.16 (0.3) | .158 |
| CAL ≥1m | 12.18 (23.9) | 9.08 (22.0) | .303 |
Data are number (percentage) or mean (standard deviation). Numbers of subjects. Note: Numbers do not add up in some cells due to missing data.
DISCUSSION
We found A actinomycetemcomitans in 21.7% of the population and its presence was significantly associated with younger age. Lopez reported the prevalence of A actinomycetemcomitans to be between 6.25% and 12.5% in a population of adults.26 A actinomycetemcomitans was also found in very young children (6–36 months) with a percentage of up to 30%. A actinomycetemcomitans is a gram-negative small non-motile rod that is strongly associated with destructive and aggressive, i.e. grade C, forms of periodontal disease.27
P gingivalis and T forsythia were found in 21.3% and 10.1% of the population, respectively. A study showed that the percentage of P gingivalis ranged from 3% for Caucasians to 17% for Indian subjects.28 The latter population seems to be closer to the population in this study. The presence of these species was not associated with any of the characteristics examined in this population, which might indicate that they may be part of the normal flora in some individuals.
P gingivalis is a gram-negative black-pigmented anaerobic rod that is readily found in subgingival biofilms. It is widely recognized as a contributor to the development of periodontal infections and implicated as a major pathogen in adult periodontitis.29–32 P gingivalis showed the highest load while A actinomycetemcomitans had the lowest load in an adult Italian population.33 T forsythia (previously Bacteroides forsythus) is another gram negative non-motile rod which is found in periodontal sites with active destruction as well as with disease recurrence.34
The highest prevalence found in this study was that of T denticola, which was detectable in 34.7% of the samples. The presence of this microorganism was significantly associated with higher percentage of PD ≥4 mm. T denticola is also part of the red complex, which is a major contributor to common adult forms of periodontitis.34 It is a gram-negative motile anaerobe related to periodontal lesions. More commonly found in patients with severe periodontitis, rather than in patients with healthy periodontium or gingivitis. In both healthy and HIV-infected individuals in an adult Serbian population, T forsythia was the most and T denticola was the least frequent bacteria, which is opposite to our results. This can lead to primary conclusion that T denticola is more associated with younger individuals but more studies are needed.35,36
Table 4.
Characteristics of the study samples relative to the presence of 3 or more bacteria.
| Variables | 3 or more bacteria | P value | |
|---|---|---|---|
| Yes | No | ||
| Total (n) | 25 (9.0) | 252 (91.0) | N/A |
| Age (years) | 16.95 (1.1) | 17.28 (0.9) | .151 |
| Gender | |||
| Male | 9 (8.9) | 92 (91.1) | .960 |
| Female | 16 (9.1) | 160 (90.9) | |
| Nationality | |||
| Non-Saudi | 5 (7.9) | 58 (92.1) | .732 |
| Saudi | 20 (9.3) | 194 (90.7) | |
| Smoker | |||
| Yes | 1 (5.9) | 16 (94.1) | .641 |
| No | 24 (9.2) | 236 (90.8) | |
| Plaque Index | 1.26 (0.9) | 1.51 (0.8 | .143 |
| Gingival Index | 1.42 (0.9) | 1.22 (0.8 | .257 |
| PD (mm) | 0.77 (0.2) | 0.67 (0.2) | .021 |
| PD ≥4 mm | 11.61 (11.4) | 7.04 (9.0) | .062 |
| CAL (mm) | 0.25 (0.4) | 0.17 (0.3) | .275 |
| CAL ≥1m | 14.99 (27.7) | 9.67 (22.1) | .276 |
Note: Numbers do not add up in some cells due to missing data.
Table 5.
Characteristics of the study samples relative to the presence of 4 or more bacteria.
| Variables | 4 or more bacteria | P value | |
|---|---|---|---|
| Yes | No | ||
| Total (n) | 5 (1.8) | 272 (98.2) | N/A |
| Age (years) | 17.00 (0.7) | 17.26 (1.0) | .556 |
| Gender | |||
| Male | 0 | 101 (100.0) | .087 |
| Female | 5 (2.8) | 171 (97.2) | |
| Nationality | |||
| Non-Saudi | 0 | 63 (100.0) | .221 |
| Saudi | 5 (2.3) | 209 (97.7) | |
| Smoker | |||
| Yes | 0 (0.0) | 17 (100.0) | .564 |
| No | 5 (1.9) | 255 (98.1) | |
| Plaque Index | 1.78 (0.3) | 1.48 (0.8) | .417 |
| Gingival Index | 2.00 (0.3) | 1.22 (0.8) | .003a |
| PD (mm) | 0.42 (0.2) | 0.68 (0.2) | .008b |
| PD ≥4 mm | 3.46 (1.4) | 7.53 (9.4) | <.001a |
| CAL (mm) | 0.12 (0.3) | 0.18 (0.3) | .675 |
| CAL ≥1m | 1.90 (4.2) | 10.35 (22.9) | .411 |
Welch's t test <.05 level.
Independent t test Note: Numbers do not add up in some cells due to missing data.
Another bacteria strongly associated with periodontal disease which was checked for in this sample is P intermedia.16,34,37 The percentage of subjects with detectable P intermedia was 12.3%, but it was not associated with any other characteristics. Ellwood and coworkers reported a prevalence of 2% for subjects who were positive for P intermedia.28 In a study comparing Italian and Dutch groups, all bacterial loads differed significantly.38 T denticola and P gingivalis were significantly more prevalent in the Italian group when compared to Dutch group. This can suggest the importance of ethnicity and background in the bacterial profile related to periodontal disease.
More than half of the sample (57%) had 1 or more bacterial species detectable, while 19% had 2 or more bacterial species present. Neither was significantly related to any of the characteristics examined. On the other hand, 9% of the sample was positive for 3 or more bacterial species and this was positively associated with higher mean PD. Furthermore, 1.8% had 4 or more of the examined pathogenic bacteria. This was associated with higher gingival index, percentage of PD ≥4 mm and mean PD. Red complex bacteria was found in 2.9% of the sample, although not significantly related to any characteristic. However, the probability of having bacteria belonging to the red complex is 25% to 48% in the presence of bleeding on probing and with a pocket depth >6 mm by 20% to 37% in adult Italian population.39 This suggests that when more pathogenic bacteria are present there is an increased chance of periodontal breakdown. This appears to confirm the non-specific theory of bacterial colonization, which suggests that different combinations of indigenous bacteria, rather than just a single species, can produce the pathogenic potential necessary to cause progression from gingivitis to destructive periodontitis.40
The use of microbiological assays for detecting specific microbes as an adjunct in the diagnosis and management of periodontal disease has been strongly recommended. It is clear that plaque control and root planing is very important in the management of most forms of periodontal disease; however, identifying and monitoring of specific microorganisms represents a new method to the management of periodontal disease and provides a rationale for treatment of the specific infections of the periodontium. Future technologies may provide more efficient and improved tests.41
Within the limitations of the study, we can conclude that all bacterial species tested were present with variable prevalence, but only A actinomycetemcomitans and T denticola were significantly associated with periodontal breakdown. In addition, the presence of more than 2 of the 5 species tested was significantly associated with tissue damage. This study should be followed by further studies with larger sample sizes covering large and small cities in Saudi Arabia. Also, studies using real-time quantitative PCR for detecting whether the level of bacteria has an effect on the parameters studied can be compared to the results obtained in this study.42
Funding Statement
This study was funded by King Abdulaziz City for Science and Technology, Riyadh, Kingdom of Saudi Arabia (grant no. AT-32-9).
REFERENCES
- 1.Albandar JM, Tinoco EM. Global epidemiology of periodontal diseases in children and young persons. Periodontol 2000. 2002;29:153–76. Epub 2002/07/10. doi: [pii]. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 2.Papapanou PN SM, et al. Periodontitis: Consensus report of workgroup 2 of the 2017 World Workshop on the Classification of Periodontal and Peri?Implant Diseases and Conditions. J Clin Periodontol. 2018;45(Suppl 20):S162–S70. [DOI] [PubMed] [Google Scholar]
- 3.G H. Periodontitis: from microbial immune subversion to systemic inflammation. Nat Rev Immunol. 2015;15(1):30–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cekici AKA, Hasturk H, Van Dyke T. Inflammatory and immune pathways in the pathogenesis of periodontal disease. Periodontol 2000. 2014; Feb (64(1)):57–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finlay BB, McFadden G. Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell. 2006;124(4):767–82. Epub 2006/02/25. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 6.Silva NAL, Bravo D, et al. Host response mechanisms in periodontal diseases. J Appl Oral Sci. 2015;23(3):329–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Page RC. Host response tests for diagnosing periodontal diseases. J Periodontol. 1992;63(4 Suppl):356–66. Epub 1992/04/01. doi: 10.1902/jop.1992.63.4s.356 [doi]. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 8.Van Dyke TE, Sheilesh D. Risk factors for periodontitis. J Int Acad Periodontol. 2005;7(1):3–7. Epub 2005/03/02. PubMed PMID: . [PMC free article] [PubMed] [Google Scholar]
- 9.Paster BJ, Olsen I, Aas JA, Dewhirst FE. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol 2000. 2006;42:80–7. Epub 2006/08/26. doi: [pii] 10.1111/j.1600-0757.2006.00174.x [doi]. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 10.Brochut PF, Marin I, Baehni P, Mombelli A. Predictive value of clinical and microbiological parameters for the treatment outcome of scaling and root planing. J Clin Periodontol. 2005;32(7):695–701. Epub 2005/06/22. doi: [pii] 10.1111/j.1600-051X.2005.00730.x [doi]. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 11.Newman MG, Takei HH, Klokkevold PR, Carranza FA. Carranza's Clinical Periodontology. 10th ed. Philadephia: WB Saunders Co; 2006. [Google Scholar]
- 12.Haffajee AD, Socransky SS. Microbial etiological agents of destructive periodontal diseases. Periodontol 2000. 1994;5:78–111. Epub 1994/06/01. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 13.Kou Y, Inaba H, Kato T, Tagashira M, Honma D, Kanda T, et al. Inflammatory responses of gingival epithelial cells stimulated with Porphyromonas gingivalis vesicles are inhibited by hop-associated polyphenols. J Periodontol. 2008;79(1):174–80. Epub 2008/01/02. doi: 10.1902/jop.2008.070364 [doi]. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 14.Dogan B, Kipalev AS, Okte E, Sultan N, Asikainen SE. Consistent intrafamilial transmission of Actinobacillus actinomycetemcomitans despite clonal diversity. J Periodontol. 2008;79(2):307–15. Epub 2008/02/07. doi: 10.1902/jop.2008.070270 [doi]. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 15.Moore WE, Moore LV. The bacteria of periodontal diseases. Periodontol 2000. 1994;5:66–77. Epub 1994/06/01. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 16.Lovegrove JM. Dental plaque revisited: bacteria associated with periodontal disease. J N Z Soc Periodontol. 2004(87):7–21. Epub 2004/05/18. PubMed PMID: . [PubMed]
- 17.Tanabe S, Bodet C, Grenier D. Peptostreptococcus micros cell wall elicits a pro-inflammatory response in human macrophages. J Endotoxin Res. 2007;13(4):219–26. Epub 2007/10/25. doi: 13/4/219 [pii] 10.1177/0968051907081869 [doi]. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 18.Saito Y, Fujii R, Nakagawa KI, Kuramitsu HK, Okuda K, Ishihara K. Stimulation of Fusobacterium nucleatum biofilm formation by Porphyromonas gingivalis. Oral Microbiol Immunol. 2008;23(1):1–6. Epub 2008/01/05. doi: [pii] 10.1111/j.1399-302X.2007.00380.x [doi]. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 19.Colombo AMC, Hartenbach F, do Souto R, Silva-Boghossian C. Periodontal-disease-associated biofilm: A reservoir for pathogens of medical importance. Microbial Pathogenesis. 2016;94:27–34. [DOI] [PubMed] [Google Scholar]
- 20.Gafan GP, Lucas VS, Roberts GJ, Petrie A, Wilson M, Spratt DA. Prevalence of periodontal pathogens in dental plaque of children. J Clin Microbiol. 2004;42(9):4141–6. Epub 2004/09/15. doi: 10.1128/JCM.42.9.4141-4146.2004 [doi] 42/9/4141 [pii]. PubMed PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanner AC, Milgrom PM, Kent R Jr., Mokeem SA, Page RC, Riedy CA, et al. The microbiota of young children from tooth and tongue samples. J Dent Res. 2002;81(1):53–7. Epub 2002/02/05. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 22.Kingman A, Susin C, Albandar JM. Effect of partial recording protocols on severity estimates of periodontal disease. J Clin Periodontol. 2008;35(8):659–67. Epub 2008/06/03. doi: [pii] 10.1111/j.1600-051X.2008.01243.x [doi]. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 23.Cortelli JRFC, Costa FO, Cortelli SC, Kajiya M, Howell SC, Kawai T. Detection of periodontal pathogens in newborns and children with mixed dentition. Eur J Clin Microbiol Infect Dis. 2012;Jun;31(6):1041–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haffajee AD, Socransky SS. Microbial etiological agents of destructive periodontal diseases. Periodontol 2000. 1994;5:78–111. Epub 1994/06/01. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 25.Tran SDRJ. Improved multiplex PCR using conserved and species-specific 16S rRNA gene primers for simultaneous detection of Actinobacillus actinomycetemcomitans, Bacteroides forsythus, and Porphyromonas gingivalis. J Clin Microbiol. 1999;Nov;37(11):3504–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopez NJ. Occurrence of Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, and Prevotella intermedia in progressive adult periodontitis. J Periodontol. 2000;71(6):948–54. Epub 2000/07/29. doi: 10.1902/jop.2000.71.6.948 [doi]. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 27.Parameter on aggressive periodontitis. American Academy of Periodontology. J Periodontol. 2000;71(5 Suppl):867–9. Epub 2000/06/30. doi: 10.1902/jop.2000.71.5-S.867 [doi]. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 28.Ellwood R, Worthington HV, Cullinan MP, Hamlet S, Clerehugh V, Davies R. Prevalence of suspected periodontal pathogens identified using ELISA in adolescents of differing ethnic origins. J Clin Periodontol. 1997;24(3):141–5. Epub 1997/03/01. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 29.Brown LJ, Loe H. Prevalence, extent, severity and progression of periodontal disease. Periodontol 2000. 1993;2:57–71. Epub 1993/06/01. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 30.Mucci LA, Bjorkman L, Douglass CW, Pedersen NL. Environmental and heritable factors in the etiology of oral diseases–a population-based study of Swedish twins. J Dent Res. 2005;84(9):800–5. Epub 2005/08/20. doi: 84/9/800 [pii]. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 31.Cugini C, Klepac-Ceraj V, Rackaityte E, Riggs JE, Davey ME. Porphyromonas gingivalis: keeping the pathos out of the biont. J Oral Microbiol. 2013;5. Epub 2013/04/09. doi: 10.3402/jom.v5i0.19804 [doi] 19804 [pii]. PubMed PMID: . [DOI] [PMC free article] [PubMed]
- 32.How KYSK, Chan KG. Porphyromonas gingivalis: An Overview of Periodontopathic Pathogen below the Gum Line. Front Micro-biol. 2016;7:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Checchi VPG. Microbiological Response to Periodontal Therapy: A Retrospective Study. Open Dent J. 2018(12):837–45. [DOI] [PMC free article] [PubMed]
- 34.Ahmed A. N RR, Victor D. Identification of Tannerella Forsythia and Treponema Denticola in Down Syndrome Subjects and Healthy Subjects with Periodontal Disease-A PCR Study. Biomed Pharmacol J. 2018;11(1). [Google Scholar]
- 35.Kannosh I SD, Toljic B, Radunovic M, Pucar A, Matic Petrovic S, Grubisa I, Lazarevic M, Brkic Z, Knezevic Vukcevic J, Milasin J. The presence of periopathogenic bacteria in subgingival and atherosclerotic plaques – An age related comparative analysis. J Infect Dev Ctries. 2018(12):1088–95. [DOI] [PubMed] [Google Scholar]
- 36.Tolji? BTA, Petrovi? SM, Kannosh IY, Dragovi? G, Jevtovi? D, De Luka SR, Risti?-Djurovi? JL, Mila?in J. The new Microbiologica. 2018;41(1):61–6. [PubMed] [Google Scholar]
- 37.Lazar VDL, Curutiu C, Gheorghe I, Holban A, Popa M, Chifiriuc C. Impact of Dental Plaque Biofilms in Periodontal Disease: Management and Future Therapy, Periodontitis, Pachiappan Arjunan. IntechOpen. 2017.
- 38.Montevecchi MAF, Checchi V, Gatto MR, Checchi L. Microbiological Distribution of Six Periodontal Pathogens Between Untreated Italian and Dutch Periodontal Patients. Oral Health Prev Dent. 2016;14(4):329–37. [DOI] [PubMed] [Google Scholar]
- 39.Checchi L. GMR, Checchi V, Carinci F. Bacteria prevalence in a large Italian population sample: a clinical and microbiological study. J Biol Regul Homeost Agents. 2016. Apr-Jun (30(2 Suppl 1)):199–208. [PubMed] [Google Scholar]
- 40.Theilade E. The non-specific theory in microbial etiology of inflammatory periodontal diseases. J Clin Periodontol. 1986;13(10):905–11. Epub 1986/11/01. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 41.Genco RJZJ, Christersson LA. Use and interpretation of microbiological assays in periodontal diseases. Oral Microbiol Immunol. 1986;Nov;1(1):73–81. [DOI] [PubMed] [Google Scholar]
- 42.Schulz AMK, Schonian G, Fleischer B, Drosten C. Detection, differentiation, and quantitation of pathogenic Leishmania organisms by a fluorescence resonance energy transfer-based real-time PCR assay. J Clin Microbiol. 2003;41 (4):1529–35. [DOI] [PMC free article] [PubMed] [Google Scholar]