Abstract
We evaluated an immunochromatographic strip for the detection of avian avulavirus 1 (Newcastle disease virus, NDV) based on a high-affinity monoclonal antibody (mAb) that specifically recognizes the hemagglutinin-neuraminidase (HN) protein. The anti-HN mAb was labeled with colloidal gold as the detector. A chicken anti-NDV polyclonal antibody and staphylococcal protein A (SPA) were blotted on the nitrocellulose membrane for the test and control lines, respectively. The strip specifically recognized the NDV antigen with no cross-reactivity to other viruses that were examined. Furthermore, it specifically recognized a variety of NDV isolates, including virulent and attenuated strains. These results were confirmed using hemagglutination (HA) and RT-PCR assays. The NDV detection strip detected 104.9 EID50 viruses/0.1 mL in the NDV-infected sample, which is comparable to the classical HA test (105.2 EID50/0.1 mL). Following experimental infection, NDV was detected using the detection strip in infected tissues as early as 36 h after experimental infection and prior to development of clinical signs and appearance of gross anatomic lesions. The diagnostic sensitivity and specificity of the NDV detection strip for NDV infection were 83.3% and 100%, respectively, as confirmed by RT-PCR.
Keywords: avian avulavirus 1, immunochromatographic strip, monoclonal antibodies, Newcastle disease virus
Newcastle disease (ND) is a highly contagious viral disease of poultry caused by virulent avian avulavirus 1 (avian paramyxovirus 1; Newcastle disease virus [NDV]; family Paramyxoviridae, genus Avulavirus).12 ND is listed by the World Health Organization for Animals (OIE), and considered the fourth most problematic disease of poultry.1,4,5 In China, intensive vaccination with lentogenic NDV strains is commonly used to control ND outbreaks. However, chronic NDV infections still occur frequently in vaccinated flocks accompanied by subclinical signs and atypical lesions, which makes ND more difficult to diagnose at an early stage of infection.
Virus isolation (VI) and reverse-transcription PCR (RT-PCR) remain the definitive assays for NDV detection.15 Meanwhile, a wide variety of detection methods, such as slide enzyme-linked immunosorbent assay (ELISA),2 multiplex RT-PCR,10 nested RT-PCR,3 real-time PCR,8,13,16 and microarray,11 have been developed successfully for NDV detection. However, those methods have limited application in many diagnostic laboratories because they are time-consuming and require special instruments and professional skills. We evaluated an immunochromatographic strip test for the detection of NDV in a clinical or field environment.
The NDV standard strain F48E8 was obtained from the China Institute of Veterinary Drug Control (Beijing, China). Field isolates were kindly provided by the China Animal Health and Epidemiology Center (Qingdao, China), South China Agricultural University (Guangzhou, China), and Xinxiang University (Xinxiang, China), respectively. The lentogenic strains La Sota and Clone 30 were obtained from Harbin Weike Biotechnology Development (Harbin, China), Harbin Pharmaceutical Group Bio-Vaccine (Harbin, China), Yangling Green Biological Engineering (Xianyang, China), Qianyuanhao Biological (Beijing, China), Zhejiang Nuobeiwei Biology Technology (Hangzhou, China), and Beijing Veterinary Biological Products Factory (Beijing, China), respectively; strains F and HB1 were from Guangxi Liyuan Biology (Nanning, China). The mesogenic strain Mukteswar was obtained from Guangxi Liyuan Biology, Qilu Animal Health Products (Jinan, China) and Jilin Zhengye Biological Products (Jilin, China). The avirulent strain V4HR was obtained from Jiujiang Bomeilai Biological Products (Jiujiang, China).
Six-wk-old female BALB/c mice were purchased from the Laboratory Animal Center, Zhengzhou University (Zhengzhou, China). Ten-d-old specific pathogen–free (SPF) chicken eggs and 15-d-old chickens were obtained from Beijing Merial Vital Laboratory Animal Technology (Beijing, China). One-d-old chickens were provided by Henan Agricultural University (Zhengzhou, China) and raised to 12 wk old without vaccination under strictly controlled conditions.
NDV-infected tissues were obtained from chickens that were naturally infected with NDV and exhibiting classical signs and lesions. Furthermore, certain samples were obtained from chickens experimentally infected or immunized with NDV standard strain F48E8 or the vaccine strain La Sota. Uninfected samples were obtained from 15-d-old SPF chickens. Infectious bursal disease virus (IBDV) vaccine (strain B87) was obtained from Harbin Weike Biotechnology Development, Marek’s disease virus (MDV) vaccine (strain Fc-126) and infectious laryngotracheitis virus (ILTV) vaccine (strain K317) from Harbin Pharmaceutical Group Bio-Vaccine, and infectious bronchitis virus (IBV) vaccine (strain W93) from Liaoning Yikang Biological (Liaoyang, China). Egg-drop syndrome virus (EDSV-76; strain AV-127) was obtained from the China Institute of Veterinary Drugs (Beijing, China). The avian influenza A virus (subtypes H5, H7, and H9) hemagglutination inhibition (HI) test antigens were obtained from Harbin Weike Biotechnology Development.
A set of mAbs specific for NDV were obtained by immunizing mice with the purified NDV strain F48E8 as described previously.9 Briefly, spleen cells isolated from immunized mice were fused with NS0 myeloma cells using polyethylene glycol 1500 (Roche Diagnostics, Mannheim, Germany).17 Hybridoma supernatants were screened for specific binding to NDV antigen by an immunoperoxidase monolayer assay.9 Fourteen hybridomas secreting antibodies to NDV were isolated and cloned by limiting dilution. The mAbs were then purified from ascitic fluid by a caprylic acid and ammonium sulfate precipitation method.14 A high-affinity mAb (4D2), which was shown to recognize specifically the HN protein of most NDV isolates, yielded high HI activity (Table 1). This mAb was then used as a conjugated antibody.
Table 1.
Anti–Newcastle disease virus (NDV) antibodies used in the NDV detection strip.
| Antibody | Subtype |
Concentration (mg/mL) | HI titer | Usage | |
|---|---|---|---|---|---|
| Heavy | Light | ||||
| mAb 4D2 | IgG2b | κ | 4.2 | 1:29 | Conjugate |
| Chicken pAb | NT | 4.5 | 1:211 | Test line | |
HI = hemagglutination inhibition; mAb = monoclonal antibody; NT = not tested; pAb = polyclonal antibody.
Twelve-wk-old chickens were immunized 4 times at 2-wk intervals with the NDV live vaccine (La Sota strain) and the inactivated vaccine (Harbin Weike Biotechnology Development). Chickens were bled 10 d after the last injection, and sera were collected; the antibody titer against NDV was 1:211 by an HI test (Table 1). The chicken anti-NDV polyclonal antibody (pAb) IgG was then purified by ammonium sulfate precipitation and used for the test line blotting.
The purified mAb 4D2 was labeled with colloidal gold (~30 nm diameter) as described previously.6 Conjugated mAb was then dispensed (XYZ 3000 dispenser; Bio-Dot, Irvine, CA) to fiberglass pads (G041 glass fiber conjugate pad sheets; Millipore, Billerica, MA) to generate conjugate pads. On a 2-cm nitrocellulose membrane (Hi-Flow Plus 135 2 mil backing master roll slit width 2.0 cm; Millipore), the chicken anti-NDV pAb (1 mg/mL) and staphylococcal protein A (SPA; 0.2 mg/mL; Sigma-Aldrich) solutions were dispensed as test and control lines at a speed of 1 μL/cm, respectively. The fiberglass sample pad, conjugate pad, blotted nitrocellulose membrane, and filter paper absorption pad were assembled on the support board (Shanghai Liangxin Technology, Shanghai, China) sequentially, with 1–2 mm overlapping each other and cut into 2.8-mm pieces (CM 4000 cutter; Bio-Dot), which was then placed into a cassette (Shanghai Liangxin Technology).
To detect NDV in tissues and vaccines, the sample solution (80–120 μL) was applied to the NDV detection strip and placed horizontally for 10–15 min. If NDV was present in the effluent, the colloidal gold–labeled mAb dissolved and recognized the NDV antigen; the NDV-mAb complexes were then captured by the chicken anti-NDV pAb immobilized on the test line, and the excess colored mAb was trapped by SPA on the control line, forming 2 red lines on the membrane, which was recorded as a positive result. In the absence of NDV, the immobilized pAb on the test line was unable to trap the colloidal gold conjugate, resulting in the single control line, which was considered to be a negative result (Fig. 1).
Figure 1.
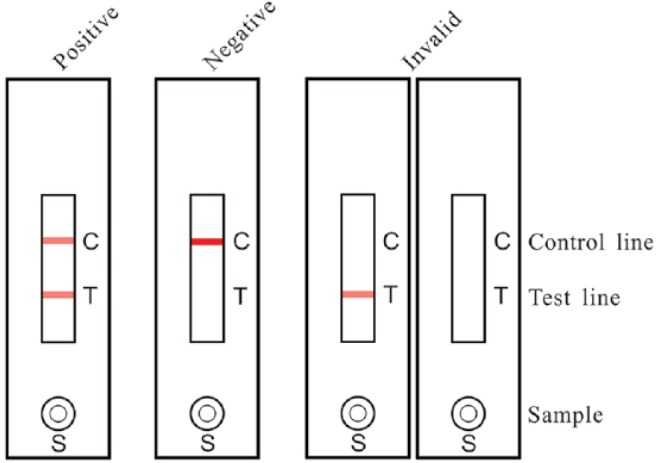
Detection of Newcastle disease virus (NDV) using the NDV immunochromatographic strip. The sample solution (80–120 μL) was applied to the NDV detection strip at S and placed horizontally for 10–15 min before observing the result. If both the test and control lines turned red, the sample was considered NDV positive. The sample was considered negative when only the control line (not the test line) was colored. No control band appearing on the membrane suggested that the testing procedure was improper or the strip invalid.
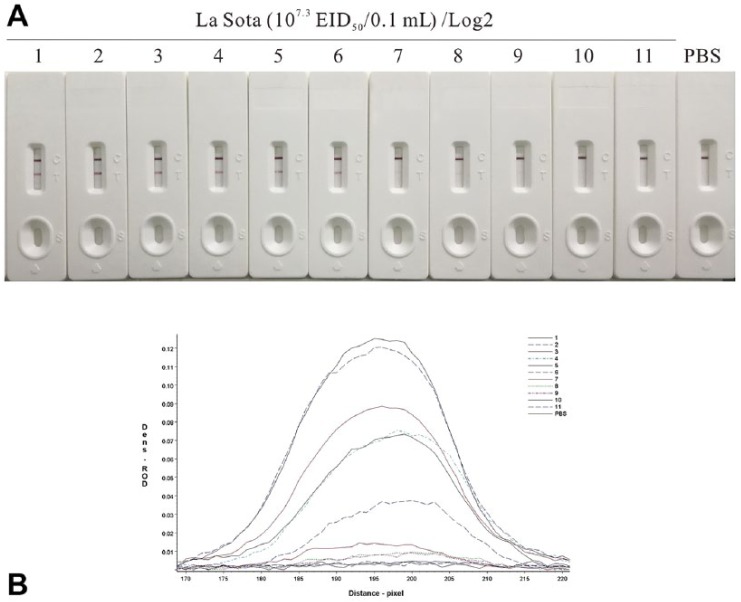
The La Sota sample containing 107.3 50% embryo infectious dose (EID50)/0.1 mL virus was 2-fold serially diluted and tested by the NDV detection strip. The HA test and RT-PCR were performed as references to test the sensitivity of the strip.15 The colored membranes were screened (TSR-3000 plate reader; Bio-Dot), and data acquisition was conducted (AIS software v.6.0; Bio-Dot). The NDV detection strip detected as little as 104.9 EID50/0.1 mL NDV in the sample, which was comparable to the HA test (105.2 EID50/0.1 mL; Fig. 2, Table 2). However, this test was less sensitive than RT-PCR (104.0 EID50/0.1 mL).
Figure 2.
Determination of the sensitivity of the Newcastle disease virus (NDV) immunochromatographic strip. A. The La Sota culture (107.3 EID50/0.1 mL) was 2-fold serially diluted from 1:21 to 1:211, and diluted samples were then applied to the NDV detection strip and placed horizontally for 10–15 min before observing the result. B. The colored membranes were screened under a TSR-3000 Reader, and relative optical density (ROD) values were analyzed by AIS software.
Table 2.
Sensitivity of the Newcastle disease virus detection strip compared with HA and RT-PCR assays.
| Sample dilution | Virus titer (EID50/0.1 mL) | Strip test |
HA test | RT-PCR | |
|---|---|---|---|---|---|
| ROD | Result | ||||
| 1:2 | 107.0 | 153.85 | + | + | + |
| 1:4 | 106.7 | 152.48 | + | + | + |
| 1:8 | 106.4 | 107.55 | + | + | + |
| 1:16 | 106.1 | 90.47 | + | + | + |
| 1:32 | 105.8 | 86.76 | + | + | + |
| 1:64 | 105.5 | 41.73 | + | + | + |
| 1:128 | 105.2 | 16.41 | + | + | + |
| 1:256 | 104.9 | 11.30 | + | − | + |
| 1:512 | 104.6 | 10.17 | − | − | + |
| 1:1,024 | 104.3 | 5.26 | − | − | + |
| 1:2,048 | 104.0 | 6.29 | − | − | + |
| PBS | 0 | 5.69 | − | − | − |
+ = positive; − = negative; HA = hemagglutination; PBS = phosphate-buffered saline: ROD = relative optical density; RT-PCR = reverse-transcription PCR.
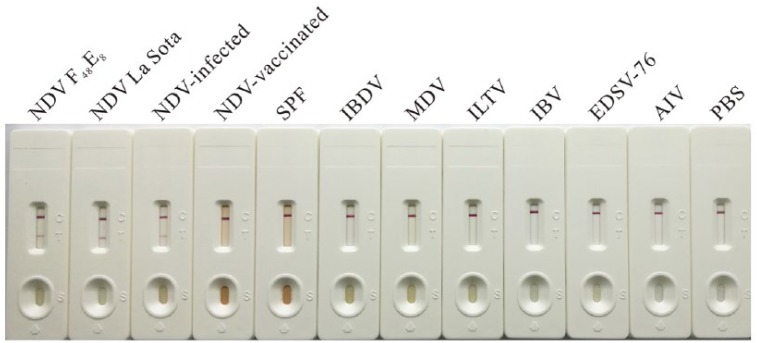
The NDV isolates and vaccine strains, as well as the infected specimens from various areas, were identified by RT-PCR in advance, and positive samples were selected to determine specificity of the NDV detection strip. Meanwhile, the NDV-vaccinated and normal tissues as well as the other virus samples, including AIV, IBDV, MDV, IBV, ILTV, and EDSV-76, were used as negative controls. The NDV detection strips detected NDV antigen in 12 NDV isolates, 18 vaccine strains including La Sota (6), Clone 30 (6), F (1), B1 (1) Mukteswar (3), and V4HR (1), and 25 infected specimens, which were identified as positive by the reference RT-PCR (Table 3). No positive band was observed on the strips that were tested in samples of the vaccinated (0 of 30) or normal (0 of 30) tissues, as well as other poultry viruses (e.g., IBDV, MDV, ILTV, IBV, EDSV-76, and AIV [H5, H7, H9 subtypes]; Fig. 3).
Table 3.
Specificity of the Newcastle disease virus (NDV) detection strip test.
| Sample | NDV strip test |
Positive rate (%) | ||
|---|---|---|---|---|
| Positive | Negative | Total | ||
| NDV strain | ||||
| Total isolates | 12 | 0 | 12 | 100 |
| La Sota | 6 | 0 | 6 | 100 |
| Clone 30 | 6 | 0 | 6 | 100 |
| F | 1 | 0 | 1 | 100 |
| HB1 | 1 | 0 | 1 | 100 |
| Mukteswar | 3 | 0 | 3 | 100 |
| V4HR | 1 | 0 | 1 | 100 |
| Chicken tissue | ||||
| NDV-infected | 25 | 0 | 25 | 100 |
| NDV-vaccinated | 0 | 30 | 30 | 0 |
| Related virus* | ||||
| SPF | 0 | 30 | 30 | 0 |
| IBDV | 0 | 1 | 1 | 0 |
| MDV | 0 | 1 | 1 | 0 |
| ILTV | 0 | 1 | 1 | 0 |
| IBV | 0 | 1 | 1 | 0 |
| EDSV-76 | 0 | 1 | 1 | 0 |
| AIV (H5/H7/H9) | 0 | 3 | 3 | 0 |
See text for definition of abbreviations.
Figure 3.
Determination of the specificity of the Newcastle disease virus (NDV) immunochromatographic strip. The diluted NDV strains F48E8 and La Sota, the tissues from NDV-infected, vaccinated, and SPF chickens. The related virus vaccines and detection antigens were applied to the NDV detection strips and placed horizontally for 10–15 min before observing the result.
Evaluation of assay repeatability within and between batches was performed as described previously.7 Fifteen NDV-infected tissues from experimentally NDV-infected chickens and 15 normal samples from SPF chickens were selected for the repeatability test, which were identified by RT-PCR. For intra-assay (within batch) repeatability, 3 replicates of each sample were assigned in the same batch strips; 3 replicates of each sample were run with strips from different batches for interassay repeatability. A similarly colored test line was generated by NDV-infected samples applied to NDV detection strips (n = 3) from the same batch, as well as those from 3 different batches, indicating good repeatability of the NDV detection strip.
To determine storage life, NDV detection strips were placed in a plastic bag sealed with desiccant. The strips were then examined for specificity and sensitivity by testing the NDV vaccine strain La Sota as well as NDV-infected, vaccinated, and normal tissues at 0, 3, 6, 9, 12, and 18 mo after storage. Valid results were obtained in tests of NDV detection strips sealed with desiccant in a plastic bag at room temperature for up to 18 mo (Table 4).
Table 4.
Sensitivity and specificity of the Newcastle disease virus (NDV) detection strip test at various storage times.
| Storage time (mo) | Sensitivity |
Specificity |
||
|---|---|---|---|---|
| La Sota (EID50/0.1 mL) |
NDV-infected chicken tissues | NDV-vaccinated chicken tissues | Normal tissues | |
| 0 | 104.9 | + | − | − |
| 3 | 104.9 | + | − | − |
| 6 | 104.9 | + | − | − |
| 9 | 104.9 | + | − | − |
| 12 | 104.9 | + | − | − |
| 18 | 105.2 | + | − | − |
+ = positive; − = negative.
Thirty-five, 15-d-old, SPF chickens were experimentally challenged with 106 EID50 of NDV standard strain F48E8 by nasal and eye-drop routes; 20 chickens were inoculated with the vaccine strain La Sota as controls. The F48E8-infected chickens showed obvious nervous signs of torticollis and opisthotonus at 36 h post-infection (hpi) followed by diarrhea and respiratory distress at 48 hpi and began to die at 56–72 hpi, whereas the La Sota–vaccinated chickens were healthy and without any clinical signs. To confirm NDV infection, 3–5 of the infected chickens, or chickens that died, and 4 of the vaccinated chickens were selected at different times post-inoculation (Table 5), the tissues (lung, spleen, cecal tonsil, intestine, and glandular stomach) were dissected from each chicken, and these samples were tested using the NDV detection strip and RT-PCR, respectively. NDV antigens were detected from lung (1 of 5) and spleen (1 of 5) at 24 hpi and from all samples (25 of 25) at 36 hpi by the NDV detection strips, with a total positive rate of 72.6% (127 of 175; Table 5). NDV RNA was detected from lung (1 of 5) at 12 hpi and from all tissue types (6 of 25) at 24 hpi, with a total positive rate of 75.4% (132 of 175). These results demonstrated that the strip test and RT-PCR could detect NDV in infected tissues at 36–120 hpi with 97.1% confidence (170 of 175). Conversely, neither NDV antigen nor RNA was detected from any samples (0 of 100) from La Sota–vaccinated chickens using the NDV detection strip or RT-PCR (Table 5). The experimental procedure was authorized and regulated by the Ethical and Animal Welfare Committee of Key Laboratory of Animal Immunology, Henan Academy of Agricultural Sciences, China.
Table 5.
Detection of Newcastle disease virus antigen in the infected or vaccinated chickens.
| Post-infection time | F48E8 infection |
La Sota vaccination |
||||
|---|---|---|---|---|---|---|
| Strip | RT-PCR | Coincidence (%) | Strip | RT-PCR | Coincidence (%) | |
| 12 h | 0/25* | 1/25 | 96 | NT | NT | NT |
| 24 h | 2/25 | 6/25 | 84 | 0/20 | 0/20 | 100 |
| 36 h | 25/25 | 25/25 | 100 | NT | NT | NT |
| 2 d | 30/30 | 30/30 | 100 | NT | NT | NT |
| 3 d | 30/30 | 30/30 | 100 | 0/20 | 0/20 | 100 |
| 4 d | 25/25 | 25/25 | 100 | NT | NT | NT |
| 5 d | 15/15 | 15/15 | 100 | 0/20 | 0/20 | 100 |
| 10 d | NT | NT | NT | 0/20 | 0/20 | 100 |
| 14 d | NT | NT | NT | 0/20 | 0/20 | 100 |
| Total | 127/175 | 132/175 | 97.1 | 0/100 | 0/100 | 100 |
NT = not tested; RT-PCR = reverse-transcription PCR.
Positive number/total sample number including lung, spleen, cecal tonsils, intestine, and glandular stomach.
To validate the NDV detection strips, 1,023 tissue samples including lung (213), spleen (219), cecal tonsils (202), intestine (184), and glandular stomach (205) were collected from 15 different chicken flocks in Henan province, China. NDV RNA was detected by RT-PCR in 6 of 1,023 tissues from diagnostic chicken submissions; 5 of these 6 samples were NDV antigen–positive as tested by the NDV detection strips (Table 6). Using RT-PCR as a reference, the diagnostic sensitivity (DSn), diagnostic specificity (DSp), and accuracy of the NDV detection strip were calculated as 83.3%, 100%, and 99.9% according to the formula: DSn = TP/(TP + FN); DSp = TN/(TN + FP), and accuracy = (TP + TN)/total number of samples tested, in which TP, FP, TN, and FN indicated true positive, false positive, true negative, and false negative, respectively.7
Table 6.
Comparison of the Newcastle disease virus (NDV) detection strip with reverse-transcription PCR (RT-PCR) for clinical samples.
| NDV detection strip | RT-PCR reference |
||
|---|---|---|---|
| Positive | Negative | Total | |
| Positive | 5 (TP) | 0 (FP) | 5 |
| Negative | 1 (FN) | 1,017 (TN) | 1,018 |
| Total | 6 | 1,017 | 1,023 |
FN = false negative; FP = false positive; TN = true negative; TP = true positive.
NDV antigen was detected by the NDV detection strip in lung and spleen of virulent F48E8 NDV–infected chickens as early as 24 hpi, which was earlier than the appearance of clinical signs and gross anatomic lesions; tests remained positive throughout the infection. Conversely, NDV antigens failed to be detected from chickens vaccinated with the La Sota strain. These results suggested that the viral load of NDV in chicken tissues has significant differences between virulent and attenuated strains. The virulent NDV strains multiply rapidly and accumulate in the respiratory system, leading to severe lesions, whereas low-virulence strains, such as La Sota, induced no obvious disease because of reduced viral replication. Our study clearly demonstrated that the NDV detection strip had similar sensitivity and specificity to HA and RT-PCR for the detection of NDV infection of chickens in the field.
Acknowledgments
We thank Dr. Clinton Jones of Oklahoma State University, Stillwater, OK for his review of the manuscript.
Footnotes
Declaration of conflicting interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
ORCID iD: Jun Luo  https://orcid.org/0000-0002-7351-8180
https://orcid.org/0000-0002-7351-8180
Funding: This work was supported by grants from the National Key Research and Development Program of China (2016YFD0500800), the Special Fund for Agro-scientific Research in the Public Interest (201303033), the Key Science and Technology Program of Henan Province, China (162102110051), and the Budget Special Program of Henan Province, China (20188119).
References
- 1. Brown VR, et al. A review of virulent Newcastle disease viruses in the United States and the role of wild birds in viral persistence and spread. Vet Res 2017;48:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Desingu PA, et al. Development of slide ELISA (SELISA) for detection of four poultry viral pathogens by direct heat fixation of viruses on glass slides. J Virol Methods 2014;209:76–81. [DOI] [PubMed] [Google Scholar]
- 3. Desingu PA, et al. A rapid method of accurate detection and differentiation of Newcastle disease virus pathotypes by demonstrating multiple bands in degenerate primer based nested RT-PCR. J Virol Methods 2015;212:47–52. [DOI] [PubMed] [Google Scholar]
- 4. Dimitrov KM, et al. Newcastle disease vaccines: a solved problem or a continuous challenge? Vet Microbiol 2017;206:126–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ganar K, et al. Newcastle disease virus: current status and our understanding. Virus Res 2014;184:71–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Herreara GA, et al. Colloidal gold labeling for diagnostic pathology. In: Hayat MA, ed. Colloidal Gold: Principles, Methods, and Applications. Vol. 3 San Diego, CA: Academic Press, 1990:321–345. [Google Scholar]
- 7. Jacobson RH. Validation of serological assays for diagnosis of infectious diseases. Rev Sci Tech 1998;17:469–526. [DOI] [PubMed] [Google Scholar]
- 8. Laamiri N, et al. A multiplex real-time RT-PCR for simultaneous detection of four most common avian respiratory viruses. Virology 2018;515:29-37. [DOI] [PubMed] [Google Scholar]
- 9. Li QM, et al. Production and characterization of neutralizing monoclonal antibodies against Newcastle disease virus. J Henan Agri Sci 2017;46:127–131. [in Chinese]. [Google Scholar]
- 10. Liu H, et al. Multiplex RT-PCR for rapid detection and differentiation of class I and class II Newcastle disease viruses. J Virol Methods 2011;171:149–155. [DOI] [PubMed] [Google Scholar]
- 11. Lung O, et al. Electronic microarray assays for avian influenza and Newcastle disease virus. J Virol Methods 2012;185:244–253. [DOI] [PubMed] [Google Scholar]
- 12. Miller PJ, et al. Newcastle disease. In: Swayne DE, et al., eds. Diseases of Poultry. 13th ed. Hoboken, NJ: Wiley-Blackwell, 2013:89–138. [Google Scholar]
- 13. Nidzworski D, et al. Detection and differentiation of virulent and avirulent strains of Newcastle disease virus by real-time PCR. J Virol Methods 2011;173:144–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Page M, et al. Purification of IgG by precipitation with sodium sulfate or ammonium sulfate. In: Walker JM, ed. The Protein Protocols Handbook. 2nd ed. Totowa, NJ: Humana Press, 2002:983–985. [Google Scholar]
- 15. World Organization for Animal Health (OIE). Newcastle disease. In: Manual of Diagnostic Tests and Vaccines for Terrestrial Animals: Mammals, Birds and Bees. Biological Standards Commission. Paris, France: OIE, 2012:555–574. [Google Scholar]
- 16. Yacoub A, et al. Development of a novel real-time PCR-based strategy for simple and rapid molecular pathotyping of Newcastle disease virus. Arch Virol 2012;157:833–844. [DOI] [PubMed] [Google Scholar]
- 17. Zola H. Monoclonal Antibodies: A Manual of Techniques. Boca Raton, FL: CRC Press, 1987. [Google Scholar]