Abstract
The surface protective antigen (Spa) protein of Erysipelothrix rhusiopathiae is an important component in protecting pigs against swine erysipelas. The Spa protein has been antigenically divided into 3 types: SpaA, SpaB, and SpaC. Swine erysipelas vaccines are formulated with strains of serovar 1 and/or 2, both of which are SpaA-possessing serovars. The association of Spa type with E. rhusiopathiae serovar has been reported, and therefore, the determination of the Spa type and the serovar of clinical isolates are important to assess vaccine efficacy. An E. rhusiopathiae strain, designated Ireland, was isolated from a diseased pig and identified as serovar 6 by a conventional agar gel precipitation test. Sequence analysis of the chromosomal locus presumably defining the serovar antigenicity of E. rhusiopathiae revealed that the gene content and organization of the chromosomal regions of the Ireland strain were identical to those of the serovar 6 reference strain (Tuzok). Sequence analysis of the spa gene and dot blots using a SpaA-specific monoclonal antibody confirmed that, unlike the Tuzok strain possessing SpaB, the Ireland strain expressed SpaA, indicating that the Spa type is not associated with the serovar in this strain. Thus, further investigation into the association between Spa type and serovar of clinical swine isolates is warranted.
Keywords: Erysipelothrix rhusiopathiae, serovar 6, Spa type
Erysipelothrix rhusiopathiae is a gram-positive intracellular pathogen that causes a variety of diseases, called erysipelas, in many animals.20 E. rhusiopathiae is ubiquitous in nature and has been isolated from a wide range of hosts, including domesticated and wild animals, humans, poultry, wild birds, aquatic mammals, and on the exterior surfaces of fish and crustaceans.20
The genus Erysipelothrix comprises at least 4 species, E. rhusiopathiae, E. tonsillarum, E. inopinata, and E. larvae, and 3 unnamed species.17 Historically, Erysipelothrix species have been differentiated based on their serovar, which is determined by their cell wall peptidoglycan antigens.20 The 2 major Erysipelothrix species are E. rhusiopathiae (serovars 1a, 1b, 2, 4–6, 8, 9, 11, 12, 15–17, 19, 21, 23, and N; serovar N lacks serovar-specific antigens) and E. tonsillarum (serovars 3, 7, 10, 14, 20, 22, 24–26), with the latter species being commonly found in the tonsils of healthy animals.16 Among the Erysipelothrix serovars, 1a, 1b, and 2 are mainly associated with disease in pigs, chickens, and humans.3,12,18 In 2018, the chromosomal locus essential for the serovar-specific antigen and virulence of serovar 1 and 2 strains was identified.10 This genetic region is required to express and maintain the molecular integrity of the capsule, which is the most important virulence factor of E. rhusiopathiae.13 It was further revealed that the serovar N or untypeable strains, which are often isolated from diseased pigs, had mutation(s) in the serovar-defining genetic region and had lost the serovar-specific antigen, suggesting that most, if not all, of the serovar N or untypeable strains were originally clinically important serovars, namely, 1a, 1b, or 2.10,15
Swine erysipelas, which is characterized by acute septicemia, subacute urticaria, and chronic endocarditis and polyarthritis, causes great economic losses in the swine industry worldwide.20 Live attenuated vaccines and inactivated bacterins are available to control swine erysipelas.20 It has been reported that the Spa (surface protective antigen) protein, which is the most important vaccine antigen of E. rhusiopathiae,4,7,14 shows genetic and antigenic diversity and is divided into 3 types, SpaA, SpaB, and SpaC.19 The Spa type has been shown to be associated with the serovar of the strain. For example, E. rhusiopathiae serovars 1a, 1b, 2, 5, 8, 9, 12, 15–17, 23, and N possess SpaA, and E. rhusiopathiae serovars 4, 6, 11, 19, and 21 possess SpaB.19 Additionally, E. tonsillarum strains do not possess the Spa protein, and serovar 18, a yet unnamed species, possesses SpaC.19 It has been reported that the Spa type is more variable in fish and cetacean strains.5 It was also observed that a fish isolate of serovar 8, which was previously reported as a SpaA-possessing serovar, possessed SpaB.11 These findings suggest that the Spa type and E. rhusiopathiae serovar are not strictly associated in strains of aquatic origin. Swine erysipelas vaccines are produced from E. rhusiopathiae serovar 1 and/or 2 strains, and therefore, the current SpaA-based vaccines do not confer full protection against the strains possessing a different Spa type.19 In our study reported herein, we identified a serovar 6 swine isolate possessing the SpaA protein, suggesting that the SpaA type and serovar are also not strictly associated in certain strains of terrestrial origin.
We used the E. rhusiopathiae reference strains Fujisawa (serovar 1a) and Tuzok (serovar 6). As well, we used 3 isolates, one named Ireland, obtained from different urticarial skin lesions of a diseased pig, known to have been given an erysipelas vaccine. The bacteria were grown at 37°C for 16 h in brain-heart infusion broth (Becton, Dickinson, Baltimore, MD) supplemented with 0.1% Tween 80 and 0.3% Tris–HCl (pH 8.0). Serotyping was carried out by agar gel precipitation tests with autoclaved cell extracts and rabbit antisera raised against formalin-killed cells of the relevant reference strain as described previously.10
Genomic DNA of the E. rhusiopathiae strains was prepared as described previously,9 with the following modifications: after cell lysis using 10% sodium dodecyl sulfate, the samples were mixed with an equal volume of phenol–chloroform solution, centrifuged (21,880 × g, 15 min), and then DNA was recovered by ethanol precipitation. Based on the whole genome sequence of the Fujisawa strain, primers spa-F (5’-ACCGTTTATCGCGAGAGTCA-3’) and spa-R (5’-CCTCGCATTAAAGATGTTTC-3’) were designed to amplify a 2.3-kbp DNA fragment including the whole spa gene. PCR was performed in a 50-µL reaction mixture containing 50 ng of template DNA, 0.3 µM of each primer, 0.4 mM of each dNTP, PCR buffer, and 1.0 U of KOD FX DNA polymerase (Toyobo, Osaka, Japan). PCR amplification was performed (T-100 thermal cycler; Bio-Rad, Hercules, CA) with the following conditions: initial denaturation, 95°C for 2 min; and 3 steps of amplification (35 cycles), 98°C for 10 s, 60°C for 30 s, and 68°C for 2 min.
Sequencing of the chromosomal region defining serovar-specific antigenicity was performed as described previously.10 Briefly, the chromosomal region between ERH_1438 and ERH_1451 was divided into 2 genetic regions, and each region was PCR-amplified with the following primer sets: seq1F (5’-TGACGATTTCCTGGGCAATTCCCGCG-3’) and 6R (5’-TTCATGGCATGGTGGTGGCG-3’); 6F (5’-CAAGGCTTGCGCGTTTGGAC-3’) and seq3R (5’-TGGCATTTATCCTTAACGGC-3’). PCR was performed as described above with the following conditions: initial denaturation, 94°C for 2 min; and 3 amplification steps (35 cycles) consisting of 98°C for 10 s, 60°C for 30 s, and 68°C for 6 min. The PCR products were directly sequenced (BigDye Terminator v.3.1 cycle sequencing kit; ABI PRISM 3130xl genetic analyzer; Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions. Sequence identity was analyzed (Genetyx software; Genetyx, Tokyo, Japan).
A SpaA-specific monoclonal antibody (mAb; laboratory stock, unpublished) was used to detect SpaA expression in E. rhusiopathiae strains. Briefly, 3 µL of bacterial culture was spotted onto a nylon membrane (MagnaGraph; Funakoshi, Tokyo, Japan) and air-dried. The nylon membrane was blocked with 1% skim milk in phosphate-buffered saline containing 0.05% Tween 20 and incubated with the SpaA-specific mAb. The membrane was further treated with horseradish peroxidase conjugated with goat anti-mouse immunoglobulin antibody (IgG, IgM, and IgA; H+L; Zymed Laboratories, San Francisco, CA). The blots were developed with 3,3-diaminobenzidine tetrahydrochloride/hydrogen peroxide (WAKO, Tokyo, Japan).
We deposited the nucleotide sequences obtained in the DDBJ/GenBank/EMBL database under accessions LC425606 (spaA from the Ireland strain), LC425605 (spaB from the Tuzok strain), LC425604 (serovar-defining region of the Ireland strain), and LC425603 (serovar-defining region of the Tuzok strain). The whole genome sequence of the Fujisawa strain (accession AP012027) was retrieved from the database.
By conventional agar gel precipitation test, the 3 isolates from the diseased pig produced a precipitation line with serovar 6 antiserum (data not shown). One selected strain, designated Ireland, failed to produce a precipitation line with the other serovar-specific rabbit antisera raised against formalin-killed cells of all 26 Erysipelothrix reference strains (data not shown).
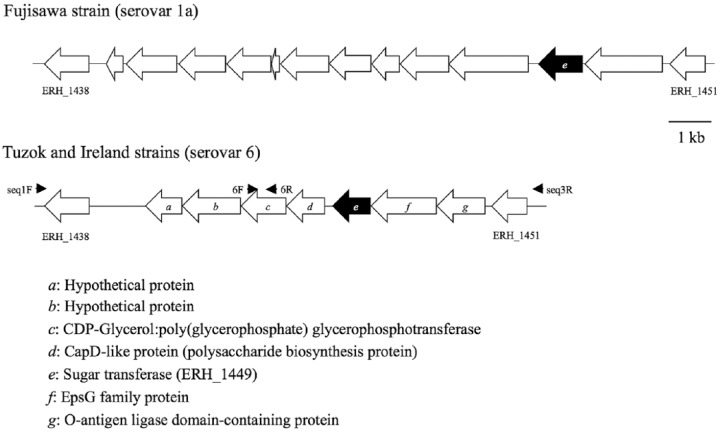
Sequences of the chromosomal region responsible for serovar antigenicity of the Fujisawa strain and the corresponding regions of Ireland and Tuzok were compared. In the serovar 6 Ireland and Tuzok strains, there were 7 common genes between ERH_1438 and ERH_1451. Among these common genes, the gene “e” was shared with strain Fujisawa (Fig. 1). The gene content and organization in this genomic region was identical between the two serovar 6 strains, with >99.4% amino acid sequence identity between each gene.
Figure 1.
Schematic representations of the chromosomal region defining the antigenicity of serovar 1a of Erysipelothrix rhusiopathiae and corresponding regions in serovar 6 strains. Identical genes are indicated by the same letter. Small arrows indicate the orientation and corresponding locations of the primers used for sequencing. Putative functions of the genes (a–g) are shown.
The spa genes of the Ireland strain and the serovar 6 reference strain Tuzok were amplified by PCR, sequenced, and compared to that of the Fujisawa strain. Sequence comparison revealed that the Ireland strain exhibited 99.2% and 61.2% amino acid sequence identity with the Fujisawa and Tuzok strains, respectively. The sequence of the N-terminal protective domain, which comprises amino acids 12–195 of the SpaA protein of the Fujisawa strain,14 was 99.5% identical to that of the Ireland strain. The Fujisawa and Ireland strains reacted with the SpaA-specific mAb in the immunoblot (data not shown), indicating that the Ireland strain expresses the SpaA protein on the cell surface.
A previous study6 observed incomplete protection of a commercial inactivated swine vaccine (serovar 2) against dolphin strains of unknown Spa type. It has also been reported that protection in mice vaccinated with a bacterin (serovar 2) was incomplete or nonexistent against non–spaA-type strains of aquatic origin (herring fish, beluga whale, and dolphin).5 Thus, disassociation of the Spa type and serovar is problematic if the Spa type of the vaccine and field isolates are different. The serovar 6 swine isolate analyzed in our study possessed the SpaA protein. Therefore, the pig, which had received an erysipelas vaccine, should have been protected against this strain. The reason for the lack of protection is unknown. It is possible that disease may have been the result of inappropriate vaccine handling or management and/or the effect of transportation stress.
Our study showed that disassociation of the Spa type and serovar of E. rhusiopathiae strains can occur in swine isolates. A serovar 11 strain isolated from a pig carcass at a slaughterhouse in the United States that possessed SpaA has been described.1 Although the most common serovars in diseased pigs are serovars 1 and 2, other serovars are frequently isolated from clinically affected pigs.8,18 In 2018, we suggested that serovar switching may occur in E. rhusiopathiae.10 This hypothesis is based on the following observations: 1) several Erysipelothrix serovars are shared by different Erysipelothrix species,17 2) some E. rhusiopathiae strains belonging to phylogenetically distinct clades displayed the same serovars,2 and 3) the G+C contents of the E. rhusiopathiae serovar-defining chromosomal region differ from those in other parts of the genome,10 suggesting that the serovar-defining region may consist of laterally transferred sequences. It is possible that less dominant serovar strains in clinical cases, including the serovar 6 swine strain analyzed in our study, may have previously been another serovar and then acquired foreign genes, resulting in a change of serovar. Importantly, our data suggest that some strains of clinically dominant serovars 1 and 2 may possess a heterologous Spa type. To determine if the vaccines provide complete cross-protection against field isolates, investigations of the Spa type and serovar of the field strains are required.
Footnotes
Declaration of conflicting interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
References
- 1. Bender JS, et al. Erysipelothrix spp. genotypes, serotypes, and surface protective antigen types associated with abattoir condemnations. J Vet Diagn Invest 2011;23:139–142. [DOI] [PubMed] [Google Scholar]
- 2. Forde T, et al. Genomic analysis of the multi-host pathogen Erysipelothrix rhusiopathiae reveals extensive recombination as well as the existence of three generalist clades with wide geographic distribution. BMC Genomics 2016;17:461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Harada K, et al. Serological and pathogenic characterization of Erysipelothrix rhusiopathiae isolates from two human cases of endocarditis in Japan. New Microbiol 2011;34:409–412. [PubMed] [Google Scholar]
- 4. Imada Y, et al. Truncated surface protective antigen (SpaA) of Erysipelothrix rhusiopathiae serotype 1a elicits protection against challenge with serotypes 1a and 2b in pigs. Infect Immun 1999;67:4376–4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ingebritson AL, et al. Erysipelothrix rhusiopathiae: association of Spa-type with serotype and role in protective immunity. Vaccine 2010;28:2490–2496. [DOI] [PubMed] [Google Scholar]
- 6. Lacave G, et al. Induction of cross-protection in mice against dolphin Erysipelothrix rhusiopathiae isolates with a swine commercial vaccine. Vet Microbiol 2001;80:247–253. [DOI] [PubMed] [Google Scholar]
- 7. Makino S, et al. Properties of repeat domain found in a novel protective antigen, SpaA, of Erysipelothrix rhusiopathiae. Microb Pathog 1998;25:101–109. [DOI] [PubMed] [Google Scholar]
- 8. McNeil M, et al. Serotypes and Spa types of Erysipelothrix rhusiopathiae isolates from British pigs (1987 to 2015). Vet J 2017;225:13–15. [DOI] [PubMed] [Google Scholar]
- 9. Ogawa Y, et al. The genome of Erysipelothrix rhusiopathiae, the causative agent of swine erysipelas, reveals new insights into the evolution of firmicutes and the organism’s intracellular adaptations. J Bacteriol 2011;193:2959–2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ogawa Y, et al. Identification of the chromosomal region essential for serovar-specific antigen and virulence of serovar 1 and 2 strains of Erysipelothrix rhusiopathiae. Infect Immun 2018;86:e00324-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shen HG, et al. Identification of surface protective antigen (spa) types in Erysipelothrix reference strains and diagnostic samples by spa multiplex real-time and conventional PCR assays. J Appl Microbiol 2010;109:1227–1233. [DOI] [PubMed] [Google Scholar]
- 12. Schmitt F, et al. Erysipelas in a free-range layer flock with conjunctival oedema as an unusual clinical sign. Berl Munch Tierarztl Wochenschr 2014;127:183–187. [PubMed] [Google Scholar]
- 13. Shimoji Y. Pathogenicity of Erysipelothrix rhusiopathiae: virulence factors and protective immunity. Microbes Infect 2000;2:965–972. [DOI] [PubMed] [Google Scholar]
- 14. Shimoji Y, et al. Immunological characterization of a protective antigen of Erysipelothrix rhusiopathiae: identification of the region responsible for protective immunity. Infect Immun 1999;67:1646–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shiraiwa K, et al. Identification of serovar 1a, 1b, 2, and 5 strains of Erysipelothrix rhusiopathiae by a conventional gel-based PCR. Vet Microbiol 2018;225:101–104. [DOI] [PubMed] [Google Scholar]
- 16. Takahashi T, et al. DNA relatedness among Erysipelothrix rhusiopathiae strains representing all twenty-three serovars and Erysipelothrix tonsillarum. Int J Syst Bacteriol 1992;42:469–473. [DOI] [PubMed] [Google Scholar]
- 17. Takahashi T, et al. A taxonomic study on Erysipelothrix by DNA-DNA hybridization experiments with numerous strains isolated from extensive origins. Microbiol Immunol 2008;52:469–478. [DOI] [PubMed] [Google Scholar]
- 18. Takahashi T, et al. Serovars of Erysipelothrix strains isolated from pigs affected with erysipelas in Japan. J Vet Med Sci 1996;58:587–589. [DOI] [PubMed] [Google Scholar]
- 19. To H, Nagai S. Genetic and antigenic diversity of the surface protective antigen proteins of Erysipelothrix rhusiopathiae. Clin Vaccine Immunol 2007;14:813–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wood RL, Henderson LM. Erysipelas. In: Straw BE, et al., eds. Diseases of Swine. 9th ed. Ames, IA: Blackwell, 2006:629–638. [Google Scholar]