Abstract
During metastasis, cancer cells navigate through a spatially heterogeneous extracellular matrix (ECM). Physical properties of ECM, including the degree of confinement, influence cell migration behavior. Here, utilizing in vitro three-dimensional collagen microtracks, we demonstrate that cell-ECM interactions, specifically the degree of spatial confinement, regulate migratory behavior. We found that cells migrate faster when they are fully confined, contacting all four walls (top, bottom, and two sides) of a collagen microtrack, compared with cells that are partially confined, contacting less than four walls. When fully confined, cells exhibit fewer but larger vinculin-containing adhesions and create greater strains in the surrounding matrix directed toward the cell body. In contrast, partially confined cells develop a more elongated morphology with smaller but significantly more vinculin-containing adhesions and displace the surrounding matrix less than fully confined cells. The resulting effect of increasing cell contractility via Rho activation is dependent on the number of walls with which the cell is in contact. Although matrix strains increase in both fully and partially confined cells, cells that are partially confined increase speed, whereas those in full confinement decrease speed. Together, these results suggest that the degree of cell-ECM contact during confined migration is a key determinant of speed, morphology, and cell-generated substrate strains during motility, and these factors may work in tandem to facilitate metastatic cell migration.
Significance
During metastatic migration, cells can use pores and interstitial spaces to move through tissue. Here, we show that their migratory behavior is dependent on the extent to which they interact with their surroundings. Increased wall contact on all sides of the cell results in faster migration, larger adhesions, and increased cell-induced matrix deformations. These results point toward novel microenvironment-specific behaviors in metastatic migration.
Introduction
Spatial heterogeneity is a hallmark of the tumor microenvironment and is particularly important in metastatic cancer cell migration. The stromal matrix is composed of complex, permeable structures in which pore sizes can range from 1 to 20 μm in diameter (1, 2), which cancer cells must navigate to metastasize. Moreover, metastatic cells are able to maneuver through the stroma, creating tunnel-like channels within the matrix using proteases (3). These channels, whether pre-existing or created by other cells, present migrating cells with varying degrees of confinement and mechanical cues that influence migration behavior and mechanisms used to move through these spatial restrictions. However, the mechanisms cells employ to navigate confinement within the primary tumor environment are still not well understood.
Several models have recreated these three-dimensional (3D) tracks in vitro (2, 4, 5, 6, 7, 8, 9). Polydimethylsiloxane (PDMS) is a widely used polymer that is molded to simulate 3D, single-cell microchannels with variable stiffness ranges, whereas polyacrylamide (PA) gels are similarly used for their versatility and tunability (8, 9, 10, 11, 12, 13). These models have revealed distinct behaviors of migratory cells, depending on the degree of confinement. When bound in channels molded from PA gels, cells in narrow channels moved significantly faster than those in wider channels, adopting a more amoeboid-like movement (8). On the other hand, decreased and biphasic behavior in cell speed has been observed for cells in decreasing-sized PDMS channels, suggesting that the mechanisms regulating confined migration are not yet fully understood (13, 14, 15).
In addition to having an effect on migration speed, it has been shown that confinement induces changes in the size and density of focal adhesions (FAs) in migrating cells (15, 16). It has been reported that cells exhibit a smaller or reduced presence of FAs, which are important for the transduction of contractile forces from the cell to its surrounding matrix in narrow confined PDMS channels (15, 16). Interestingly, these FAs vary in size and intensity, and little is known about their mechanism of formation and traction force transmission during confined migration (17).
Most studies on cell migration in confined channels have been performed using PDMS or PA gels with a comparatively higher stiffness than native tissue (8, 9, 18), making it difficult to quantify cellular traction stress as these materials can be impervious to deformation by cells. Moreover, these systems lack several other key features of the in vivo microenvironment, including similar adhesive site presentation and fiber structure. Therefore, in this study, we utilized our previously developed 3D collagen microtrack platform to study the effect of confinement on cell migration and cell-induced matrix deformations (as an indirect measure of traction stresses) (19, 20, 21). This system more closely mimics physiological conditions by using fibrous collagen, in which the channel dimensions and wall stiffness can be tuned while also permitting the quantification of matrix deformations due to cellular traction stresses. We introduced microbeads into the collagen surrounding the microtracks, enabling the quantification of cell-induced matrix strains and studied cell behavior in two types of confinement within the microtrack—full and partial confinement. We define cells as fully confined when they are in contact with all four walls—top, bottom, and two side walls—of the collagen track during migration, and we define cells as partially confined when they contact some fraction of, but not all, of the four walls. We then examined cell geometry, vinculin-containing FA area, density, and cell speed as well as cell-induced matrix deformations as a function of spatial confinement within the collagen microtracks. Our data indicate that fully confined migration utilizes different cellular mechanisms than either two-dimensional or even 3D migration, creating larger FA complexes and differential cell-induced matrix deformations. Our results indicate that the cell response to spatial confinement depends on the number of walls a cell contacts rather than simply the size of the channel. Together, these findings provide further insight into the mechanisms cancer cells utilize during confined migration in spatially complex 3D microenvironments.
Materials and Methods
Cell culture and reagents
Highly metastatic breast adenocarcinoma cells MDA-MB-231 (HTB-26; American Type Culture Collection, Manassas, VA) were maintained at 37°C and 5% CO2 in Dulbecco’s Modified Eagle’s Medium (Life Technologies, Grand Island, NY) supplemented with 10% fetal bovine serum (Atlanta Biologicals, Flowery Branch, GA) and 1% penicillin-streptomycin (Life Technologies). For a subset of experiments, MDA-MB-231 cells were incubated in complete medium supplemented with 0.125 μg/mL Rho Activator II (CN03-A; Cytoskeleton, Denver, CO).
DNA constructs and lentiviral transductions
The lentiviral vector, pRRL-vinculin-EGFP, was created by replacing venus from pRRL-vinculin-venus (plasmid number 111833; Addgene, Cambridge, MA) with enhanced green fluorescent protein (EGFP) from pHIV-EGFP (plasmid number 21373; Addgene; deposited by Bryan Welm and Zena Werb). EGFP was amplified by PCR using forward and reverse primers, ccgattagaattcatggtgagcaagggcgagga and ttacttgtacagctcgtccatgcctctagataatcgg, respectively. EGFP was then inserted using XbaI and EcoRI cloning sites. pRRL-vinculin-EGFP lentiviral particles were prepared by the transient transfection of HEK293T cells with the lentiviral expression vector and second-generation packaging constructs psPAX2 and pMD2.G using TransIT-LT1, according to manufacturer’s instructions (Mirus Bio, Madison, WI). Lentiviral particles were harvested from HEK293T media at 48 and 72 h post-transfection, concentrated 100-fold with Lenti-X Concentrator (Clontech Laboratories, Mountview, CA), and used for stable cell transduction in the presence of 8 μg/mL polybrene.
3D microtrack fabrication
Collagen microtracks were prepared as previously described (19, 22, 23). In brief, a silicon wafer mold was used to cast polydimethylsiloxane (PDMS; Dow Corning, Midland, MI) stamps by curing cross-linker and monomer (1:10 ratio) at 60°C for 2 h. Acid-extracted type I collagen stock solution (10 mg/mL) from rat tail tendons was diluted to 3 mg/mL concentration using ice-cold complete culture medium and neutralized with 1 N NaOH. PDMS stamps were lightly coated with neutralized collagen solution, inverted over a drop of neutralized collagen solution, and allowed to polymerize for at least 90 min at 37°C. PDMS stamps were removed, and MDA-MB-231 cells were seeded onto patterned collagen at a low density of 70,000 cells/mL to minimize cell-cell interactions. Patterned collagen matrices were closed from the top with a collagen-layered glass coverslip to form 3D collagen microtracks. The final length and depth of the collagen microtracks were 1000 and 15 μm, respectively. Straight longitudinal microtracks were created with widths of 5, 10, 15, and 20 μm and tapered microtracks with a 20 to 5 μm wide spatial gradient. Collagen microtracks were prepared on plastic-bottom six-well plates for phase contrast imaging and glass-bottom six-well plates with no. 1.5 cover glass (Cellvis, Mountain View, CA) for confocal imaging. For matrix displacement experiments, 0.5-μm diameter carboxyl-modified fluorescent polystyrene beads (Invitrogen, Grand Island, NY) were dispersed in neutralized collagen solution before polymerization.
Time-lapse and confocal microscopy
Time-lapse phase contrast imaging was performed on a Zeiss Axio Observer Z1 inverted microscope equipped with a Hamamatsu ORCA-ER camera using a 10×/0.3 NA objective and operated by AxioVision software. For cell speed measurements, MDA-MB-231 cells were imaged every 20 min for 12 h. For persistence measurements, cells were imaged every 5 min for 12 h. Before all time-lapse imaging, cells were incubated in the tracks in complete media for 4–6 h to allow for cell adhesion and spreading. Rho Activator II was added to complete media where indicated. Confocal reflectance was performed with a Zeiss LSM800 inverted confocal microscope equipped with a 40×/1.1 NA long working distance water-immersion objective. To observe vinculin, vinculin-EGFP-expressing MDA-MB-231 cells were imaged at 0.50 μm z axis intervals measuring only adhesions larger than 0.04 μm2 to exclude values below the optical resolution of microscope. Before imaging, cells were fixed with 3.2% paraformaldehyde for 15 min, followed by permeabilization with 1% Triton X-100 for 3 h, and washed with phosphate-buffered saline/0.2% tween three times for 15 min each to remove cytoplasmic background noise. To track fluorescent bead displacement, time-lapse confocal fluorescence imaging was performed every 20 min for 12 h at 0.50 μm z axis intervals using a 20×/0.8 NA objective. All live-cell imaging was performed in an environmental chamber maintained at 37°C and 5% CO2.
Migration analysis
In 3D collagen microtracks, cell migration speed was calculated by averaging the distance between cell centroids (from frame to frame in the time-lapse series) and dividing by the total time step. Cell centroid position was determined by outlining cells in ImageJ (version 1.49b; National Institutes of Health, Bethesda, MD). Speed measurements were taken over a minimum of 6 h. Cells that divided or interacted with other cells during migration were excluded from analysis.
Persistence was quantified using the same migration analysis to obtain the distance traveled between cell centroids for each time step. The sum of the distance traveled in each 5-min time step was calculated for 20-min intervals, and the total path distance was divided by the end-to-end path distance for the 20-min period to obtain persistence values from 0 to 1.
3D vinculin-containing adhesion structure analysis
Confocal z-stack images of vinculin-EGFP-expressing MDA-MB-231 cells were converted to maximum intensity z-projections in ImageJ and processed to subtract background and increase the visibility of adhesions. After thresholding the image, adhesion number was quantified with the ImageJ particle analyzer. Vinculin-adhesion area, cell area, and cell volume were calculated by counting pixel and voxel number, respectively (24). Cell area was measured from a maximum z-projection in the x-y plane as it is in this plane that the dimension of the microtrack was altered. Quantified cells were randomly selected to exclude selection bias.
Measurement of bead and matrix displacement
To quantify bead displacement, bead positions were recorded in cell-free collagen microtracks, and final cell-induced displacement was tracked using the Trackmate plugin in ImageJ (25). In Trackmate, the median filter was used to automatically detect fluorescent beads, and the track analysis function was utilized to quantify displacement of the beads in an unstressed matrix versus a stressed matrix and to obtain bead positions within the matrix. Cells were outlined using ImageJ and measured to obtain centroid position and aspect ratio (major axis of cell divided by minor axis). Distance from the bead position to the cell centroid was then quantified. Images of fluorescent beads in the collagen microtrack with the cell out of frame were overlaid with images of the cell in the frame to obtain qualitative analysis of bead displacement in the track.
Statistical analysis
Data in graphical form are presented as the mean ± SEM, min to max box-and-whisker plots, or histograms. Statistical analysis was conducted using GraphPad Prism 7.0. Normality in the spread of data for each experiment was tested using the D’Agostino-Pearson omnibus normality test. To evaluate statistical significance, analysis of variance with a Tukey’s honestly significant difference test and multiple comparisons was used to compare more than two groups, and two-tailed Student’s t-test was used to compare two groups. Statistical significance was considered with a p-value <0.05. All data are representative of a minimum of three independent replicate studies.
Results
Increased cell-matrix contact increases cell migration speed
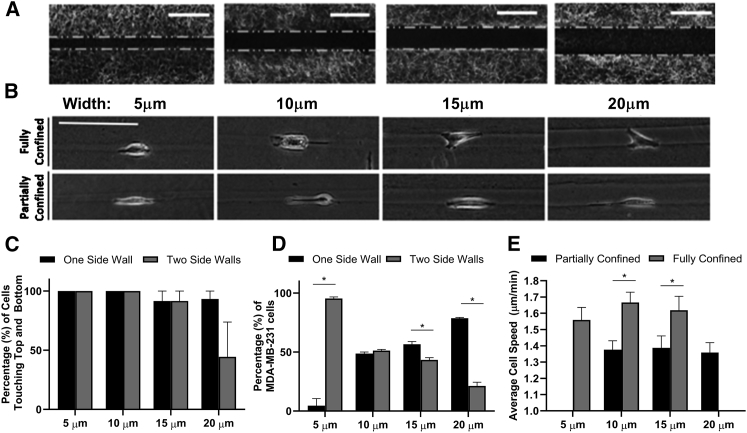
To recapitulate the range of confining pores in the extracellular microenvironment found in vivo (1, 26, 27) and examine the effects of spatial confinement on migration, we utilized microfabrication to create collagen microtracks. Straight, uniform width microtracks were created with widths of 5, 10, 15, or 20 μm and a height of 15 μm (Fig. 1 A). To differentiate cells based on their cell-matrix contact, we characterized cells as either fully confined—when they contacted all four walls of the microtrack (top, bottom, and two side walls)—or partially confined—when they contacted three walls (top, bottom, and one side wall) (Fig. 1 B). Using confocal microscopy, we verified cells were able to contact all side walls for all of the microtrack widths used; however, in 20-μm tracks, cells contacting both side walls did not consistently contact top and bottom walls as well (Fig. 1 C). Therefore, we categorized cells in 20-μm tracks as partially confined. Side wall contact was also highly dependent on microtrack width as cells predominately contacted both side walls and were fully confined in narrower tracks but only contacted a single side wall in wider tracks (Fig. 1 D).
Figure 1.
Cell migration speed depends on the extent of confinement. (A) Confocal reflectance images of 5, 10, 15, and 20 μm width collagen microtracks, scale bar = 25 μm. (B) Phase contrast images of 5, 10, 15, and 20 μm width microtracks with single MDA-MB-231 cells, scale bar = 50 μm. (C) Percentage of MDA-MB-231 cells that are able to reach both top and bottom walls and at least one side wall in each track size (n = 20–27 cells per condition). (D) Percentage of MDA-MB-231 cells that are either partially confined or fully confined by all four walls as a function of track width. (E) Average migration speed of MDA-MB-231 cells during migration in 5, 10, 15, and 20 μm width-sized microtracks; n = 35–50 cells per condition. ∗ indicates p < 0.05, with only significant differences denoted.
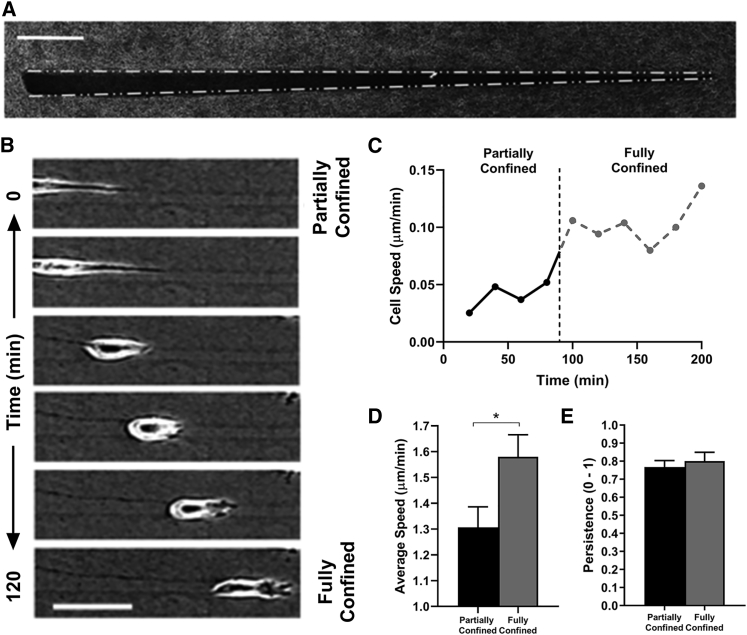
Spatial confinement and cell-matrix contact are important regulators of 3D cell migration (28, 29). Therefore, we next examined how track width and the degree of confinement affects cell speed during confined migration. Notably, cell migration speed significantly increased when cells were fully confined compared with when cells were partially confined, regardless of track width (Fig. 1 E). To determine if cells actively adjust their velocity to changes in spatial confinement and the number of walls contacted, we studied cell migration in a tapered collagen microtrack decreasing in width from 20 to 5 μm (Fig. 2 A). Indeed, as cells transition from a wider to narrower region of the tapered microtrack, where they were fully confined, migration speed increased (Fig. 2, B–D). Furthermore, we verified there was no difference in persistence between partially and fully confined cells, indicating no difference in directional migration (Fig. 2 E). Together, these results suggest that the degree of cell-matrix contact and spatial confinement plays an important role in regulating migration efficiency.
Figure 2.
Cells respond to increased matrix contact by increasing speed. (A) Confocal reflectance image of a tapered collagen microtrack with decreasing width ranging from 20 to 5 μm, scale bar = 25 μm. (B) Time-lapse phase contrast images (scale bar = 50 μm) and (C) migration speed of a single MDA-MB-231 cell when partially confined and fully confined by all four walls within the tapered microtrack; black dotted line indicates the switch in confinement. (D) Average migration speed of MDA-MB-231 cells when partially confined and fully confined by all four walls; n = 47 cells per condition. (E) Average persistence of cells in tracks was calculated using time-lapse images taken every 5 min rather than 20 min; n = 11 cells per condition. ∗ indicates p < 0.05, with only significant differences denoted.
Confinement induces changes in cell geometry but not volume or area
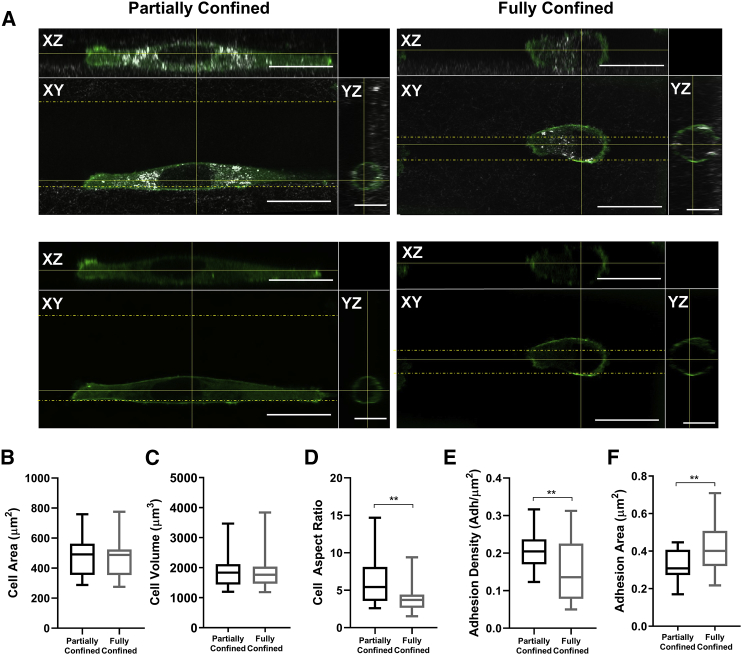
Cell geometry and spreading have both been reported to influence cell behavior during confined migration, in which an increase in cell size or aspect ratio has been linked to decreased migration efficiency (15, 30). To determine whether the degree of confinement produces morphological changes during migration, we examined the morphology of cells in full confinement using the 5-μm tracks and of cells in partial confinement using the 20-μm tracks (Fig. 3 A). Interestingly, both fully and partially confined cells were of a similar area when comparing maximum projections in the z-plane and similar volume when comparing 3D projections regardless of track size (Fig. 3, B and C). However, cells exhibited significant changes in aspect ratio dependent on the degree of confinement. Fully confined cells exhibited a decreased aspect ratio, whereas partially confined cells exhibited an increased aspect ratio (Fig. 3 D). Contact with four walls of a channel caused cells to exhibit a more rounded morphology, whereas cells that are not confined in channels tend to be more elongated, suggesting that cytoskeletal remodeling occurs based on matrix contact.
Figure 3.
Vinculin-containing FA size and density varies based on confinement. (A) Confocal reflectance (top) and fluorescent images (bottom) of single cells when partially confined and fully confined by all four walls within the microtracks (5-μm tracks for fully confined, 20-μm tracks for partially confined). fluorescence indicates vinculin-containing FA protein; XY, ZX, and YZ represent top, side, and front view of microtrack, respectively. Scale bars, 20 μm in the XY and XZ views. Scale bars, 10 μm in the YZ view. (B) Cell area, (C) cell volume, and (D) cell aspect ratio when partially confined and fully confined in microtracks; n = 20 cells per condition. (E) Density of vinculin-containing adhesions and (F) area of vinculin-containing adhesions are >0.04 μm2 in cells that are partially confined and fully confined; n = 20 cells per condition. ∗∗ indicates p < 0.01, with only significant differences denoted. To see this figure in color, go online.
Cells exhibit a decreased density of larger vinculin-containing adhesions when fully confined in tracks
Given that cells in full confinement are more rounded than those that are partially confined, which tend to be more elongated, we sought to investigate whether the extent of confinement alters FA area and/or density. FA size has been suggested to play a key role in translating structural and chemical cues in cells to cell shape (15, 31). Vinculin is one of the key structural proteins recruited to FA complexes and plays a well-established role in regulating cell polarity and directionality (23). To investigate the relationship between cell spreading, confinement, and cell-matrix adhesion, we seeded vinculin-EGFP-expressing cells within microtracks and quantified the density and area of vinculin adhesions (Fig. 3, E and F). To ensure that the number of adhesions on the surface of cells was not simply due to changes in surface area of the cell based on confinement, we measured adhesion density (adhesions per μm2 of cell surface) rather than simply the total number of adhesions. Although there was no statistical difference in cell area or volume in partial versus full confinement (Fig. 3, B and C), fully confined cells exhibited fewer total vinculin-containing adhesions per μm2 of the cell surface compared to partially confined cells (Fig. 3 E). Additionally, cells in full confinement exhibited significantly larger adhesions than partially confined cells (Fig. 3 F), suggesting a greater accumulation of vinculin in fully confined cells that supports stable cytoskeletal structure and attachment to the matrix (23). Taken together, these results indicate that spatial confinement induces changes in the number and size of vinculin-containing adhesions that mediate cell-matrix interactions.
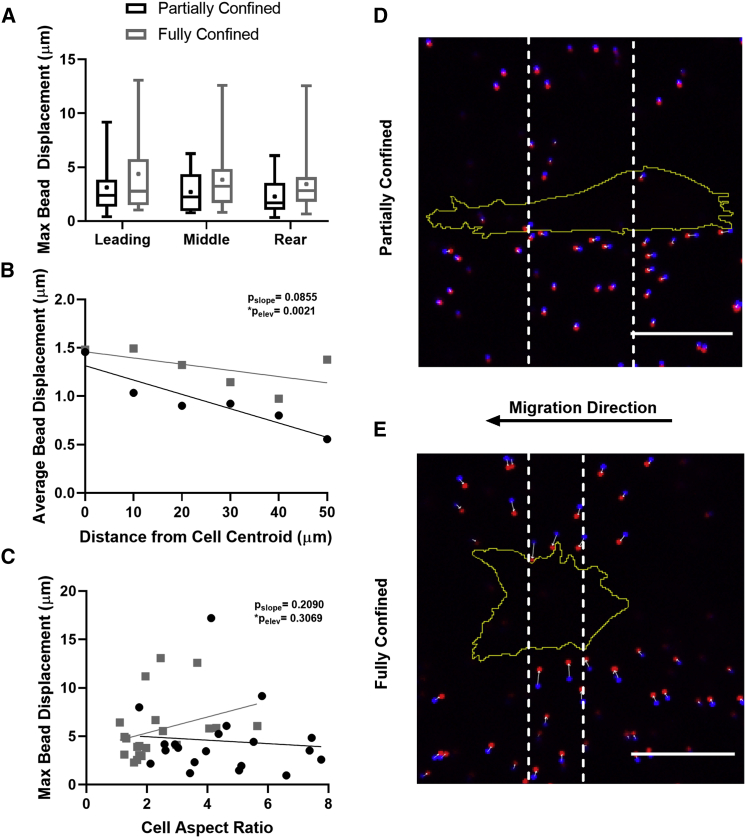
Magnitude and direction of strain varies based on confinement
FAs form foci at which forces are exerted by the cell on the substrate (32, 33, 34). These forces are transmitted from the cell to the surrounding matrix, facilitating cell motility (35) and causing strains within the matrix. Previous work has indicated that cell geometry may play an important role in regulating cellular strains and matrix deformation (36), and we therefore sought to determine whether the changes in FA area and density as well as changes in cell aspect ratio induced by confinement had an effect on the matrix strains created by the cell. Collagen matrix deformation was quantified by measuring cell-induced fluorescent bead displacement during fully versus partially confined migration (20, 21). Beads were first measured at an unstressed state, when the cell was not in frame, and then measured in a stressed state, when cells were interacting with the surrounding collagen matrix. Bead displacement was quantified as a function of the distance from the cell centroid to determine the distance the strains were measurably transmitted through the matrix (21). Bead displacement was also compared between the region at the leading edge of the cell, the middle region of the cell, and at the trailing edge of the cell to determine whether matrix deformations are a function of location relative to the cell body. Interestingly, there were no regional differences in bead displacement (Fig. 4 A). However, cells that were fully confined in the collagen microtracks displace beads to a greater extent compared to partially confined cells, suggesting that cells fully confined by the matrix may exert greater forces (Fig. 4 B). As expected, beads located closer to the cell body displaced more than those located further away, with no significant differences in the slopes of the partially and fully confined cells (Fig. 4 B).
Figure 4.
Strain from cell-generated matrix deformation is dependent on the extent of confinement. (A) Average of maximum bead displacements from when the cell is absent to when the cell is present in the leading, middle, and rear region of the cell, with black indicating partially confined cells, and gray indicating fully confined cells. (B) Average bead displacement as a function distance of the bead from the cell centroid with significance displayed and (C) maximum bead displacement as a function of cell aspect ratio was measured as the major axis divided by the minor axis of the cell, with circular points denote partially confined cells, square points denote fully confined cells. (D and E) Overlay images of beads when the cell is absent (blue position) and when the cell is present (red position) with cell position outlined in yellow, representative image of partially confined (D) and fully confined (E) cells with dashed white lines indicating leading, middle, and rear regions, both cells moving left, indicated by black arrow. The direction of bead displacement is indicated by white arrows. Scale bars, 20 μm; n = 20 cells per condition, and ∼100 beads per cell were tracked. To see this figure in color, go online.
To investigate whether a relationship exists between cell geometry and matrix strains because of confinement, we examined maximum bead displacement as a function of cell aspect ratio (Fig. 4 C). Interestingly, bead displacement did not significantly change with cell shape in both fully and partially confined cells, suggesting that strains are not correlated with or dictated by cell morphology. In observing bead displacements qualitatively, we found that cells pulled on the matrix during migration rather than pushing (Fig. 4, D and E). Notably, however, when confined, cells tend to pull the matrix inward toward the cell body (perpendicular to the direction of motion), whereas in partial confinement, cells pull the matrix parallel to their cell body. Altogether, these data indicate that the degree of confinement, not cell shape, determines the magnitude and direction of cell-induced matrix strains.
Response to changes in Rho activation are confinement dependent
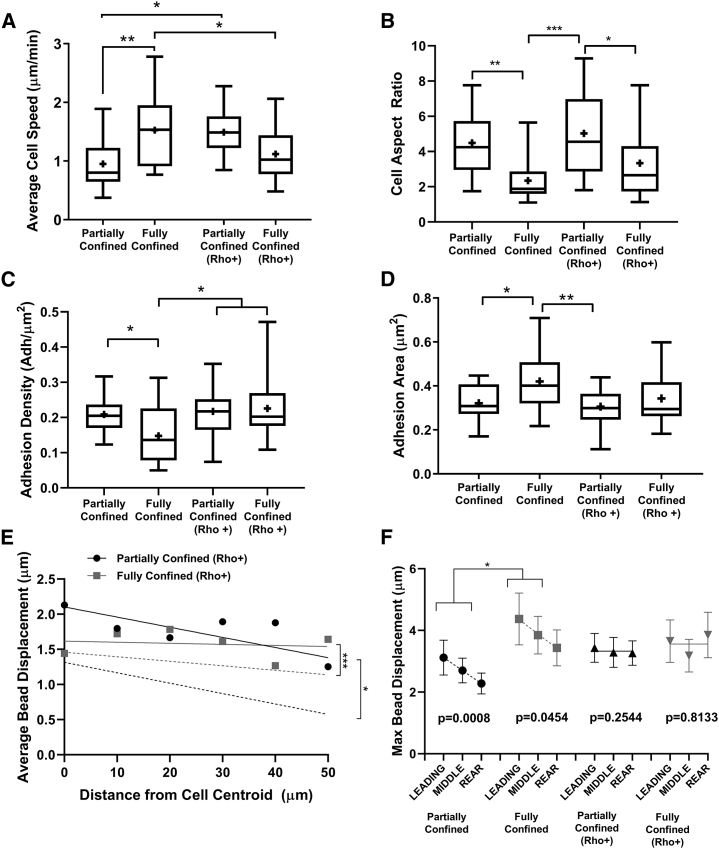
Our results suggest that the area and density of vinculin adhesions as well as matrix deformation are affected by the degree of confinement in 3D. We find that as cells change their shape in confinement, the subsequent changes in matrix contact alter adhesion formation and growth, as well as cell-induced matrix strains, and changes in cell speed. Given that contractility modulates the assembly of vinculin-containing FAs and directs traction force generation (37, 38, 39, 40), we investigated whether alterations to cell contractility would differentially modify migration behavior, depending on the extent of cellular confinement. We treated cells with Rho Activator II, a reagent that activates the Rho GTPases by targeting the Switch II region of these GTPases to increase cell contractility, and measured cell speed as a function of spatial confinement (41). With Rho activation, cell speed significantly increased when cells are partially confined and decreased when cells are fully confined compared to controls (Fig. 5 A). After Rho activation, cells still exhibited a significantly higher aspect ratio when partially confined compared to treated cells that were fully confined (Fig. 5 B), suggesting that cell shape is more dependent on confinement rather than contractility.
Figure 5.
Activation of Rho alters cell migration behavior in confinement. (A) Average cell migration speed; n = 15–45 cells per condition. (B) Cell aspect ratio was measured as major axis divided by minor axis of untreated and treated partially or fully confined cells; n = 20 cells per condition. (C) Adhesion density in either Rho-treated or untreated partially and fully confined cells was measured as number of adhesions per μm2, measuring only adhesions >0.04 μm2; n = 20 cells per condition. (D) Area of vinculin-containing adhesions in either Rho-treated or untreated partially and fully confined cells; n = 20 cells per condition. (E) Average bead displacement as a function of the distance from the bead position to the cell centroid with slopes, where black lines represent partially confined cells, and gray lines represent fully confined cells. Untreated counterparts have been transposed over treated cells as dotted lines for comparison with significance values displayed; n = 20 cells per condition, and ∼100 beads per cell were tracked. (F) Maximum displacement of Rho-treated cells in the leading, middle, and rear regions surrounding the cells, displaying the mean y-values of treated and untreated cells, fully or partially confined, fit to a linear curve with p-values indicating a non-zero slope displayed; n = 20 cells per condition, and ∼100 beads per cell were tracked. ∗ indicates p < 0.05, ∗∗ indicates p < 0.01, and ∗∗∗ indicates p < 0.001, with only significant differences denoted.
Because of previous work showing that a relationship exists between adhesion size and contractility (37), we investigated the effect of Rho activation on adhesion formation. Interestingly, adhesion density in fully confined cells treated with Rho activator increased significantly compared to untreated fully confined cells, though there was no longer a statistical difference in adhesion density between treated fully and partially confined cells (Fig. 5 C). The adhesion density increases in fully confined cells after Rho activation, but the adhesion density in partially confined cells remains the same. Although we had found that cells in full confinement contain larger adhesions than cells in partial confinement, when we examined adhesion area in cells treated with Rho activator, no statistical significance between the treated groups was observed (Fig. 5 D). In summary, Rho activation results in an increase in adhesion density in fully confined cells, with no significant difference in adhesion area in treated cells. These data indicate that changes in intracellular contractility differentially affect cells, depending on the extent of cell confinement.
After Rho activation, regardless of the degree of confinement, treated cells pulled the matrix significantly farther than their untreated counterparts (Fig. 5 E). However, when comparing treated partially confined cells to treated fully confined cells, there was no significant difference between the two, suggesting that cell-induced matrix strains are dependent on contractility rather than the degree of confinement. The direction of bead motions resembles the motions without Rho activator; in full confinement, cells pulled the beads perpendicular toward their cell body, whereas in partial confinement, bead motion was more parallel to the direction of motion. When comparing the displacement of beads as a function of distance from the cell centroid, there was no significant difference in the slopes of the treated fully and partially confined cells (Fig. 5 E). In comparing bead motions in treated cells, we found no regional differences in the maximum bead displacement at the leading, middle, and rear edges of the cell (Fig. 5 F). However, untreated cells in both full and partial confinement exhibit a linearly decreasing trend from the front to the rear edge of the cell. Notably, this decrease in bead displacement from the front to the rear of the cell is lost in both configurations when cells are treated with RhoA activator (Fig. 5 F). In both full and partial confinement, cells still pulled the matrix toward them as they migrated, rather than pushing the beads away. These data indicate that increasing contractility via Rho activation increases cell-induced matrix strains, regardless of the extent of confinement, but it ablates the front-to-rear asymmetry in force generation.
Discussion
During metastatic invasion, cancer cells must navigate a mechanically and spatially heterogeneous microenvironment where physical cues from the extracellular matrix determine requirements for migration. Confinement can modify intracellular signaling and change cancer cell migration behavior and molecular mechanisms of migration (8, 42). Notably, confined migration utilizes distinct mechanisms compared to two-dimensional and 3D migration (22, 23, 43) because confining microenvironments can provide strong guidance cues that facilitate more directed and efficient migration (19). However, the mechanisms employed by cells during confined migration are not yet fully understood. Here, we utilized collagen microtracks to recapitulate confining tracks experienced by cancer cells in vivo and investigated how the degree of cell-matrix contact during confined migration influences migration behavior.
Our data indicate that the degree of cell-matrix contact, rather than the width of the microtrack, alters vinculin-containing adhesion size as well as density, cell-generated matrix strain, and migration speed. When cells are fully confined and contacting all walls of the microtrack, they exhibit larger, though fewer, adhesions and greater matrix strains, and they migrate more quickly through the microtrack. The extent of strain in the matrix is determined by the contractile state of the cell (44), suggesting fully confined cells are more contractile than partially confined cells. The cells in full confinement, in addition to being more contractile, also migrate faster, suggesting that contractility contributes to migration. However, when contractility was increased pharmacologically through Rho activation, speed decreases in fully confined cells. In partially confined cells, however, contractility also increases upon Rho activation, and speed also increases. These data suggest that the role of contractility in driving migration is different, depending on the extent of confinement.
Cancer cells typically use one of two main modes of migration to navigate their surrounding environment, mesenchymal and amoeboid, and they are able to switch plastically from one mode to the next, though it is unclear what drives this transition (45). Mesenchymal migration is characterized by Rac1 activity, which drives actin polymerization at the leading edge of the cell to form protrusions, followed by adhesion to the surrounding substrate and retraction of the cell rear (46, 47). Conversely, amoeboid migration is driven by RhoA-induced contractility and is typically characterized by a rounder cell morphology and the movement of a cell through squeezing. In amoeboid migration, faster migration is dominated by squeezing and blebbing rather than adhesion (15, 48). Given this, it is possible that cells in full confinement adopt an intermediate migratory mode, exhibiting traits of both mesenchymal and amoeboid migration. The rounded morphology and faster migration speed observed in full confinement appears to suggest amoeboid migration, whereas the presence of large vinculin-adhesion sites is more characteristic of mesenchymal migration. Our data suggest that for fully confined cells, contractility, rather than actin polymerization, dominates cell motility. However, an adhesion gradient from the front of the cell to the rear still exists, which may provide the mechanical asymmetry required for cell migration.
Notably, cells in full confinement exert substantially greater forces normal to the cell body than cells in partial confinement that exert primarily tangential forces. The mechanism by which cells create these forces and the role of these forces in migration are not clear. In prior work, we showed that the actin cytoskeleton of fully confined cells is primarily cortical (19). Work from others has shown that the actin cortex stabilizes the cell during migration and generates normal forces on the surrounding matrix to balance tension and intracellular pressure (49, 50). Though the role of normal forces in confined cell migration (where cells move faster) is somewhat counterintuitive, it is possible that they provide support for cells adhering to the microtrack walls. It is possible that fully confined cells utilize these normal forces to facilitate migration using an internal pressure-based mechanism like the nuclear piston model, which suggests that cells employs hydrostatic pressure generated in the cytoplasm to drive the cell forward (51). In either case, the generation of directional normal forces in full confinement appears to confer a migration speed advantage that more closely resembles amoeboid migration. However, the cell also clearly retains mesenchymal migration traits due to the recruitment of the mechanical linker protein, vinculin, which is uniquely capable of stabilized cell-matrix junctions in response to directional forces (52), and is present in the form of large adhesions, which is more suggestive of mesenchymal migration. Though it is unclear whether the size of the adhesions or their density is more crucial to modulating confined cell migration, it is likely that the increased normal forces encourage greater vinculin recruitment to adhesion sites, resulting in larger, though fewer, vinculin-containing adhesions. Spatial organization of these adhesions may lend to the mechanical asymmetry of the cell, forming a gradient that is required to drive cell migration (53, 54, 55).
Pharmacologically activating Rho has opposing effects, depending on whether the cells are in full or partial confinement. Rho activation has been reported to have global effects across the cell by locally balancing forces in the activated region so that unbalanced forces at the cell periphery are then able to generate traction (56). Thus, during global RhoA activation in full confinement, it is likely that the mechanical asymmetry of the cell is ablated through either the loss of local RhoA activity or by altering transmission of cytoskeletal force through adhesions already existing across the cell. The global increase in contractility may also disrupt the cortical integrity of the actin core, thereby dysregulating the balance with cortical tension to effectively abrogate efficient cell migration (49). Though mechanical asymmetry is similarly disrupted in partially confined cells, there is an increase in migration speed, suggesting that these cells may utilize alternative signals, such as Rac1 activation, to enhance protrusion formation. Rac1 activity, which dominates during mesenchymal migration, utilizes adhesion and actin polymerization to facilitate protrusion formation (47). It is likely that a potential increase in lamellipodia activity during enhanced contractility may result in the faster cell migration observed in partially confined cells.
Though confined migration has been heavily studied, this complex, dynamic process is still not fully understood. We propose that cells migrate through confinement using a transitional mode of migration exhibiting characteristics of both mesenchymal and amoeboid migration, in which cell contractility, rather than actin polymerization, is the driving force behind cell motility, though adhesion formation is still utilized by the cell to form a mechanically asymmetric gradient and facilitate cell migration. Regardless, it is clear that the effects of cell contractility on adhesion and migration are confinement dependent.
Author Contributions
J.A.M., A.R.-Z., F.B., B.D.H., and C.A.R.-K. designed the research. B.D.H. supplied reagents. A.R.-Z., J.A.M., and J.A.V. performed experiments and analyzed data. A.R.-Z. and J.A.M. wrote the manuscript. C.A.R.-K., F.B., M.R.Z., and B.D.H. reviewed and edited the manuscript.
Acknowledgments
We acknowledge the use of equipment and resources at the Cornell NanoScale Science and Technology Facility, a member of the National Nanotechnology Coordinated Infrastructure that is supported by the National Science Foundation (grant ECCS-1542081). This work was supported by a National Science Foundation-National Institutes of Health Physical and Engineering Sciences in Oncology Award 1233827 to C.A.R.-K., a National Science Foundation award (1454257) and a National Institutes of Health Award (EB022166) to B.D.H., and National Science Foundation Graduate Fellowships under grant number DGE-1650441 to A.R.-Z., J.A.V., and M.R.Z. and grant number DGE-1937963 to J.A.M. This work was additionally supported by the National Institutes of Health (GM131178).
Editor: Margaret Gardel.
Footnotes
Jenna A. Mosier and Aniqua Rahman-Zaman contributed equally to this work.
References
- 1.Wolf K., Alexander S., Friedl P. Collagen-based cell migration models in vitro and in vivo. Semin. Cell Dev. Biol. 2009;20:931–941. doi: 10.1016/j.semcdb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paul C.D., Hung W.C., Konstantopoulos K. Engineered models of confined cell migration. Annu. Rev. Biomed. Eng. 2016;18:159–180. doi: 10.1146/annurev-bioeng-071114-040654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedl P., Alexander S. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell. 2011;147:992–1009. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 4.Paul C.D., Shea D.J., Konstantopoulos K. Interplay of the physical microenvironment, contact guidance, and intracellular signaling in cell decision making. FASEB J. 2016;30:2161–2170. doi: 10.1096/fj.201500199R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Irimia D., Charras G., Toner M. Polar stimulation and constrained cell migration in microfluidic channels. Lab Chip. 2007;7:1783–1790. doi: 10.1039/b710524j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faure-André G., Vargas P., Lennon-Duménil A.M. Regulation of dendritic cell migration by CD74, the MHC class II-associated invariant chain. Science. 2008;322:1705–1710. doi: 10.1126/science.1159894. [DOI] [PubMed] [Google Scholar]
- 7.Tong Z., Balzer E.M., Konstantopoulos K. Chemotaxis of cell populations through confined spaces at single-cell resolution. PLoS One. 2012;7:e29211. doi: 10.1371/journal.pone.0029211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pathak A., Kumar S. Independent regulation of tumor cell migration by matrix stiffness and confinement. Proc. Natl. Acad. Sci. USA. 2012;109:10334–10339. doi: 10.1073/pnas.1118073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiu D.T., Jeon N.L., Whitesides G.M. Patterned deposition of cells and proteins onto surfaces by using three-dimensional microfluidic systems. Proc. Natl. Acad. Sci. USA. 2000;97:2408–2413. doi: 10.1073/pnas.040562297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Irimia D., Toner M. Spontaneous migration of cancer cells under conditions of mechanical confinement. Integr. Biol. 2009;1:506–512. doi: 10.1039/b908595e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rolli C.G., Seufferlein T., Spatz J.P. Impact of tumor cell cytoskeleton organization on invasiveness and migration: a microchannel-based approach. PLoS One. 2010;5:e8726. doi: 10.1371/journal.pone.0008726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heuzé M.L., Collin O., Piel M. Cell migration in confinement: a micro-channel-based assay. Methods Mol. Biol. 2011;769:415–434. doi: 10.1007/978-1-61779-207-6_28. [DOI] [PubMed] [Google Scholar]
- 13.Lautscham L.A., Kämmerer C., Fabry B. Migration in confined 3D environments is determined by a combination of adhesiveness, nuclear volume, contractility, and cell stiffness. Biophys. J. 2015;109:900–913. doi: 10.1016/j.bpj.2015.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hung W.C., Chen S.H., Konstantopoulos K. Distinct signaling mechanisms regulate migration in unconfined versus confined spaces. J. Cell Biol. 2013;202:807–824. doi: 10.1083/jcb.201302132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y.J., Le Berre M., Piel M. Confinement and low adhesion induce fast amoeboid migration of slow mesenchymal cells. Cell. 2015;160:659–672. doi: 10.1016/j.cell.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 16.Balzer E.M., Tong Z., Konstantopoulos K. Physical confinement alters tumor cell adhesion and migration phenotypes. FASEB J. 2012;26:4045–4056. doi: 10.1096/fj.12-211441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beningo K.A., Dembo M., Wang Y.L. Nascent focal adhesions are responsible for the generation of strong propulsive forces in migrating fibroblasts. J. Cell Biol. 2001;153:881–888. doi: 10.1083/jcb.153.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mak M., Reinhart-King C.A., Erickson D. Microfabricated physical spatial gradients for investigating cell migration and invasion dynamics. PLoS One. 2011;6:e20825. doi: 10.1371/journal.pone.0020825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kraning-Rush C.M., Carey S.P., Reinhart-King C.A. Microfabricated collagen tracks facilitate single cell metastatic invasion in 3D. Integr. Biol. (Camb.) 2013;5:606–616. doi: 10.1039/c3ib20196a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bloom R.J., George J.P., Wirtz D. Mapping local matrix remodeling induced by a migrating tumor cell using three-dimensional multiple-particle tracking. Biophys. J. 2008;95:4077–4088. doi: 10.1529/biophysj.108.132738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gjorevski N., Nelson C.M. Mapping of mechanical strains and stresses around quiescent engineered three-dimensional epithelial tissues. Biophys. J. 2012;103:152–162. doi: 10.1016/j.bpj.2012.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carey S.P., Rahman A., Reinhart-King C.A. Comparative mechanisms of cancer cell migration through 3D matrix and physiological microtracks. Am. J. Physiol. Cell Physiol. 2015;308:C436–C447. doi: 10.1152/ajpcell.00225.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rahman A., Carey S.P., Reinhart-King C.A. Vinculin regulates directionality and cell polarity in 2D, 3D matrix and 3D microtrack migration. Mol. Biol. Cell. 2016;27:1431–1441. doi: 10.1091/mbc.E15-06-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo M., Pegoraro A.F., Weitz D.A. Cell volume change through water efflux impacts cell stiffness and stem cell fate. Proc. Natl. Acad. Sci. USA. 2017;114:E8618–E8627. doi: 10.1073/pnas.1705179114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tinevez J.Y., Perry N., Eliceiri K.W. TrackMate: an open and extensible platform for single-particle tracking. Methods. 2017;115:80–90. doi: 10.1016/j.ymeth.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 26.Weigelin B., Bakker G.J., Friedl P. Intravital third harmonic generation microscopy of collective melanoma cell invasion: principles of interface guidance and microvesicle dynamics. Intravital. 2012;1:32–43. doi: 10.4161/intv.21223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tadeo I., Álvaro T., Noguera R. Composition and Function of the Extracellular Matrix in the Human Body. 2016. Tumor microenvironment heterogeneity: a review of the biology masterpiece, evaluation systems and therapeutic implications. [Google Scholar]
- 28.Wolf K., Te Lindert M., Friedl P. Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J. Cell Biol. 2013;201:1069–1084. doi: 10.1083/jcb.201210152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaman M.H., Trapani L.M., Matsudaira P. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc. Natl. Acad. Sci. USA. 2006;103:10889–10894. doi: 10.1073/pnas.0604460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leal-Egaña A., Letort G., Théry M. The size-speed-force relationship governs migratory cell response to tumorigenic factors. Mol. Biol. Cell. 2017;28:1612–1621. doi: 10.1091/mbc.E16-10-0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rape A.D., Guo W.H., Wang Y.L. The regulation of traction force in relation to cell shape and focal adhesions. Biomaterials. 2011;32:2043–2051. doi: 10.1016/j.biomaterials.2010.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burridge K., Chrzanowska-Wodnicka M. Focal adhesions, contractility, and signaling. Annu. Rev. Cell Dev. Biol. 1996;12:463–518. doi: 10.1146/annurev.cellbio.12.1.463. [DOI] [PubMed] [Google Scholar]
- 33.Miyamoto S., Teramoto H., Yamada K.M. Integrin function: molecular hierarchies of cytoskeletal and signaling molecules. J. Cell Biol. 1995;131:791–805. doi: 10.1083/jcb.131.3.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schoenwaelder S.M., Burridge K. Bidirectional signaling between the cytoskeleton and integrins. Curr. Opin. Cell Biol. 1999;11:274–286. doi: 10.1016/s0955-0674(99)80037-4. [DOI] [PubMed] [Google Scholar]
- 35.Butler J.P., Tolić-Nørrelykke I.M., Fredberg J.J. Traction fields, moments, and strain energy that cells exert on their surroundings. Am. J. Physiol. Cell Physiol. 2002;282:C595–C605. doi: 10.1152/ajpcell.00270.2001. [DOI] [PubMed] [Google Scholar]
- 36.Oakes P.W., Banerjee S., Gardel M.L. Geometry regulates traction stresses in adherent cells. Biophys. J. 2014;107:825–833. doi: 10.1016/j.bpj.2014.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dumbauld D.W., Shin H., García A.J. Contractility modulates cell adhesion strengthening through focal adhesion kinase and assembly of vinculin-containing focal adhesions. J. Cell. Physiol. 2010;223:746–756. doi: 10.1002/jcp.22084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lemmon C.A., Chen C.S., Romer L.H. Cell traction forces direct fibronectin matrix assembly. Biophys. J. 2009;96:729–738. doi: 10.1016/j.bpj.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rothenberg K.E., Scott D.W., Hoffman B.D. Vinculin force-sensitive dynamics at focal adhesions enable effective directed cell migration. Biophys. J. 2018;114:1680–1694. doi: 10.1016/j.bpj.2018.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.LaCroix A.S., Lynch A.D., Hoffman B.D. Tunable molecular tension sensors reveal extension-based control of vinculin loading. eLife. 2018;7:e33927. doi: 10.7554/eLife.33927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmidt G., Sehr P., Aktories K. Gln 63 of Rho is deamidated by Escherichia coli cytotoxic necrotizing factor-1. Nature. 1997;387:725–729. doi: 10.1038/42735. [DOI] [PubMed] [Google Scholar]
- 42.Hung W.C., Yang J.R., Konstantopoulos K. Confinement sensing and signal optimization via piezo1/PKA and myosin II pathways. Cell Rep. 2016;15:1430–1441. doi: 10.1016/j.celrep.2016.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paul C.D., Mistriotis P., Konstantopoulos K. Cancer cell motility: lessons from migration in confined spaces. Nat. Rev. Cancer. 2017;17:131–140. doi: 10.1038/nrc.2016.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kraning-Rush C.M., Carey S.P., Reinhart-King C.A. The role of the cytoskeleton in cellular force generation in 2D and 3D environments. Phys. Biol. 2011;8:015009. doi: 10.1088/1478-3975/8/1/015009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Talkenberger K., Cavalcanti-Adam E.A., Deutsch A. Amoeboid-mesenchymal migration plasticity promotes invasion only in complex heterogeneous microenvironments. Sci. Rep. 2017;7:9237. doi: 10.1038/s41598-017-09300-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamazaki D., Kurisu S., Takenawa T. Involvement of Rac and Rho signaling in cancer cell motility in 3D substrates. Oncogene. 2009;28:1570–1583. doi: 10.1038/onc.2009.2. [DOI] [PubMed] [Google Scholar]
- 47.Huang B., Lu M., Ben-Jacob E. The three-way switch operation of Rac1/RhoA GTPase-based circuit controlling amoeboid-hybrid-mesenchymal transition. Sci. Rep. 2014;4:6449. doi: 10.1038/srep06449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sahai E., Marshall C.J. Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nat. Cell Biol. 2003;5:711–719. doi: 10.1038/ncb1019. [DOI] [PubMed] [Google Scholar]
- 49.Álvarez-González B., Meili R., Del Álamo J.C. Three-dimensional balance of cortical tension and axial contractility enables fast amoeboid migration. Biophys. J. 2015;108:821–832. doi: 10.1016/j.bpj.2014.11.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salbreux G., Charras G., Paluch E. Actin cortex mechanics and cellular morphogenesis. Trends Cell Biol. 2012;22:536–545. doi: 10.1016/j.tcb.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 51.Petrie R.J., Koo H., Yamada K.M. Generation of compartmentalized pressure by a nuclear piston governs cell motility in a 3D matrix. Science. 2014;345:1062–1065. doi: 10.1126/science.1256965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang D.L., Bax N.A., Dunn A.R. Vinculin forms a directionally asymmetric catch bond with F-actin. Science. 2017;357:703–706. doi: 10.1126/science.aan2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmidt C.E., Horwitz A.F., Sheetz M.P. Integrin-cytoskeletal interactions in migrating fibroblasts are dynamic, asymmetric, and regulated. J. Cell Biol. 1993;123:977–991. doi: 10.1083/jcb.123.4.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Broussard J.A., Webb D.J., Kaverina I. Asymmetric focal adhesion disassembly in motile cells. Curr. Opin. Cell Biol. 2008;20:85–90. doi: 10.1016/j.ceb.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 55.Gupton S.L., Waterman-Storer C.M. Spatiotemporal feedback between actomyosin and focal-adhesion systems optimizes rapid cell migration. Cell. 2006;125:1361–1374. doi: 10.1016/j.cell.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 56.Oakes P.W., Wagner E., Gardel M.L. Optogenetic control of RhoA reveals zyxin-mediated elasticity of stress fibres. Nat. Commun. 2017;8:15817. doi: 10.1038/ncomms15817. [DOI] [PMC free article] [PubMed] [Google Scholar]