Abstract
Sticholysins are pore-forming toxins of biomedical interest and represent a prototype of proteins acting through the formation of protein-lipid or toroidal pores. Peptides spanning the N-terminus of sticholysins can mimic their permeabilizing activity and, together with the full-length toxins, have been used as a tool to understand the mechanism of pore formation in membranes. However, the lytic mechanism of these peptides and the lipid shape modulating their activity are not completely clear. In this article, we combine molecular dynamics simulations and experimental biophysical tools to dissect different aspects of the pore-forming mechanism of StII1–30, a peptide derived from the N-terminus of sticholysin II (StII). With this combined approach, membrane curvature induction and flip-flop movement of the lipids were identified as two important membrane remodeling steps mediated by StII1–30. Pore formation by this peptide was enhanced by the presence of the negatively curved lipid phosphatidylethanolamine in membranes. This lipid emerged not only as a facilitator of membrane interactions but also as a structural element of the StII1–30 pore that is recruited to the ring upon its assembly. Collectively, these, to our knowledge, new findings support a toroidal model for the architecture of the pore formed by StII1–30 and provide new molecular insight into the role of phosphatidylethanolamine as a membrane component that can easily integrate into the ring of toroidal pores, thus probably aiding in their stabilization. This study contributes to a better understanding of the molecular mechanism underlying the permeabilizing activity of StII1–30 and peptides or proteins acting via a toroidal pore mechanism and offers an informative framework for the optimization of the biomedical application of this and similar molecules.
Significance
We provide evidence about the ability of sticholysin II (StII)1–30 to form toroidal pores. Because of pore assembly, StII1–30 induces membrane curvature and facilitates flip-flop movement of the lipids. The negatively curved lipid phosphatidylethanolamine relocates from the membrane into the pore ring, becoming a structural element of the lipidic pore formed by StII1–30. This peptide emerged as a, to our knowledge, new tool, together with the full-length toxin, to understand the mechanism of toroidal pore formation in membranes. This study provides new molecular insight into the role of curved lipids as cofactors of toroidal pores, which could aid in their stabilization by easily integrating into the ring. This framework could underpin strategies for the rational use of peptides or proteins acting via toroidal pores.
Introduction
The permeabilization of lipid membranes by pore-forming toxins (PFTs) has received special attention because of their role as potential virulence factors. These proteins bind and oligomerize in membranes, leading to membrane permeabilization and cell death (1, 2, 3). Therefore, they are also considered useful tools to study the basic molecular mechanisms of protein insertion into membranes. These toxins show a dynamic interplay with lipid membranes because they require a large conformational change for membrane insertion. In turn, they strongly modify the membrane structure upon interaction with the bilayer (4, 5). PFTs are usually classified according to the structure of the membrane-integrated domain as α- or β-PFTs (6). These lytic fragments commonly show structural homology with natural membranolytic peptides such as antimicrobial peptides (AMPs), typically molecules in which the pore-forming domain covers the entire length of the polypeptide (7). Based on this homology, synthetic peptides have been used as a valuable tool to study the contribution of lytic fragments in the activity of some pore-forming proteins such as the proapoptotic protein Bax (8) and actinoporins (9, 10, 11, 12, 13).
Actinoporins are a unique class of highly hemolytic eukaryotic α-PFTs, exclusively found in sea anemones, whose putative receptor is sphingomyelin (SM) (14, 15, 16, 17). Lipid mixing from different phases (18) and the induction of membrane curvature and lipid flip-flop (15, 19) are some membrane remodeling effects of these toxins, which are closely related to their pore-forming function. Indeed, these effects on membrane structure are commonly associated with their ability to form toroidal pores (15, 19). It has been widely accepted that the N-terminal α-helix of actinoporins is the protein fragment that builds the pore, which is also lined by phospholipids headgroups (15, 19, 20). In this regard, actinoporins represent a prototype of proteins acting through the formation of protein-lipid pores. Together with other virulence factors such as the cholesterol-dependent cytolysins, the apoptotic proteins Bax and Bak, and a number of AMPs such as melittin and magainins, they have been widely used as a tool to understand the mechanism of toroidal pore formation in membranes (4, 5, 21, 22, 23, 24).
Sticholysins I and II (StI and StII) are actinoporins produced by the Caribbean Sea anemone Stichodactyla helianthus (25) that form pores of around 2 nm diameter in natural and mimetic membrane systems (26). These toxins are of current interest as active components of a vaccine platform (27, 28) and of immunotoxin constructs that are active against tumor cells (29, 30). In line with the toroidal pore model, the activity of sticholysins is enhanced in the presence of lipids favoring the nonlamellar phase such as cardiolipin, phosphatidic acid, or phosphatidylethanolamine (PE) (15). These lipids, which are strong inducers of the negative curvature in the bilayer, seem to help in the formation of small toroidal pores by stabilizing the curvature along its edges (31, 32). Moreover, pore formation by sticholysins increases the transbilayer movement of lipid molecules, which should result from the fusion of the inner and outer leaflet of the membrane at the pore edge during toroidal pore formation (15).
Peptides reproducing the N-terminus of StI (StI1–31) and StII (StII1–30) (Fig. 1 A) can mimic the permeabilizing ability of these toxins in red blood cells and liposomes (10, 12, 13). In particular, StII1–30 is the most active peptide (12, 13, 33); it forms pores in human red blood cells of a similar size to those formed by the full-length toxin (9). StII1–30 has emerged as a good candidate to replace the whole toxins in some biomedical applications such as an active component of a vaccinal platform to promote the delivery of different molecules to the cytosol and enhance the immune response in anticancer therapies (27, 28). Previously, we have analyzed the conformational properties of StI1–31 and StII1–30 in solution and in membrane mimetic systems and their ability to permeabilize lipid vesicles (9, 12, 13, 33, 34). In solution, StII1–30 acquires an α-helical fold in which several monomers are associated together as oligomeric structures. This complex of associated α-helices can be stabilized in solution, without requiring membrane assistance (34). Based on these findings, we previously suggested a model in which StII1–30 membrane permeabilizing activity is favored by peptide preassociation in solution. However, the lytic mechanism of StII1–30 is not completely clear. Although it is known that negatively charged lipids enhance the activity of StII1–30 by facilitating electrostatic interactions with the membrane (12), the interplay between lipid shape and StII1–30-induced lipid rearrangements has not yet been explored (35). Moreover, no current high-resolution information exists about the structure of the pores that this peptide forms in lipid membranes.
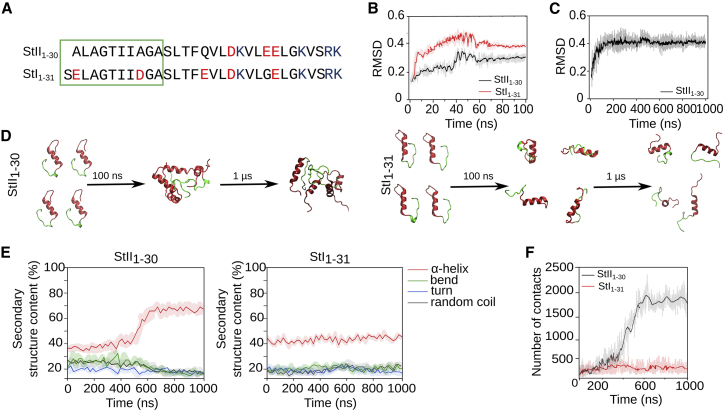
Figure 1.
StII1–30 has a high propensity to associate in solution. (A) Sequences of StII1–30 and StI1–31 are shown. Acidic amino acid residues are in red, and basic amino acid residues are in blue. The 1–10 (from StII1–30) and 1–11 (from StI1–31) hydrophobic regions are highlighted in green. (B) Root mean-square deviation calculated during equilibration of monomeric molecules of StII1–30 and StI1–31 in solution is shown. (C) Root mean-square deviation calculated for the association of StII1–30 in solution is shown. (D) Snapshots of the MD obtained with StII1–30 (left) and StI1–31 (right) at time 0, 100, and 1 μs of simulation in solution are shown. Each peptide is represented in two colors: the hydrophobic sequence 1–10 (from StII1–30) or 1–11 (from StI1–31) are in green, and the amphipathic helices 11–30 (from StII1–30) or 12–31 (from StI1–31) are in red. (E) The percentage of secondary structure of StII1–30 (left) and StI1–31 (right) obtained during the simulation time is shown. (F) The number of contacts among peptide molecules of StII1–30 or StI1–31 over the simulation time is shown. In (E) and (F), four peptide molecules were placed in the simulation box to allow their interaction. Dark lines in the plots represent the main values, and shadow lines represent the SD among three replicates. To see this figure in color, go online.
In this study, we characterized in atomistic detail the mechanism of membrane perturbation leading to pore formation by StII1–30. Using molecular dynamics (MD) simulations, we modeled membrane reshaping and pore formation in systems, including phosphatidylcholine (PC), SM, and PE. From our simulations, we calculated several structural properties of the pore including size, peptide topology, and lipid segregation into the pore. We predicted the ability of the peptide to induce membrane curvature and flip-flop movement of the lipids and the key role of PE in pore formation. In vitro, we focused on the effect of PE on StII1–30 binding to lipid monolayers and liposomes and on its permeabilizing activity in vesicles. Moreover, we also assessed the capability of StII1–30 to induce membrane curvature and lipid flip-flop. Our data identified PE as a critical membrane component that favors StII1–30 activity by modulating membrane properties required for more efficient binding and pore formation and also as a lipid constituent of the pore. Membrane curvature and lipid flip-flop induction emerged as key steps in the mechanism of membrane permeabilization by this peptide. This work provides the evidence supporting toroidal pore formation in the lytic action of StII1–30 and highlights the structural role of lipid shape in the pore formed by this peptide.
Materials and Methods
Chemicals and reagents
The following lipids were used:1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC), porcine SM, 1-palmitoyl-2-oleoyl-glycero-3-phosphoethanolamine (POPE), 1,2-dipalmitoleoyl-PE (DiPoPE), and pyrene-labeled PC (pyPC); they were purchased from Avanti Polar Lipids (Alabaster, AL) with 99% purity and used without further purification. The peptides were produced by solid-phase synthesis and further purified to >96% purity by reversed-phase high-performance liquid chromatography by GL Biochem (Shanghai, China). Peptide concentration was evaluated by a Micro BCA protein assay kit provided by Pierce (Illinois).
MD simulations
A detailed description of the systems created for MD simulations in solution and membranes is included in Supporting Materials and Methods. Membrane-peptide systems were generated using CHARMM-GUI and additional in-house-developed software (36). Systems were solvated using the TIP3 water model, and sodium and chloride ions were added to neutralize the system and to reach 150 mM final concentration. Each system was initially energy minimized for 50,000 steps. After minimizing its energy, the system was equilibrated for 150 ps before a 4-ns MD simulation at constant pressure (P = 1 atm, semiisotropic coupling) and temperature (T = 310 K). Periodic boundary conditions were applied in all three dimensions. Berendsen coupling protocols were used for the first 150 ps of the equilibration simulations (37, 38), and Nose-Hoover (39) and Parrinello-Rahman protocols were used for temperature and pressure coupling in the 4 ns of equilibration and the production simulation (40). Long-range electrostatic interactions were computed using the particle mesh Ewald method (41, 42). Lennard-Jones energies were truncated at 1.2 nm. Bond lengths were constrained with LINCS (43). After this initial setup, each system was simulated for 500 ns, at 310 K, with a time step of 2 fs. Three simulations were performed for each system, starting from different initial Maxwell distributions of random velocities. Frames were saved every 400 ps. Simulations were carried out using GROMACS version 5.0.2 (44). The force field employed was CHARMM 36 (45).
Analysis of the MD simulations
An overview of the analysis of the systems is provided in Fig. S1.
Lipid-peptide interactions and bilayer alterations
Lipid-peptide contacts were defined when the phosphodiester group of a given lipid (POPC, SM, or POPE) was found within 0.5 nm from the center of mass of each individual peptide. For a given frame, the total number of contacts was calculated as the sum of all the contacts established between phosphate groups of an individual lipid molecule and the amino acid residues of the peptides, following the procedure described by (46). The calculation was performed using the g_select tool implemented in GROMACS. Membrane thickness fluctuation in the process of pore formation was calculated in the region defined by the pore center using a cutoff of 1.5 nm, as described in (47). Because the shape of the pore is nonuniform across the membrane, the central region of the pore was defined as ±0.7 nm on each side of the center of the bilayer. The thickness of the membrane around the pore region was calculated using frames every 100 ns as the average phosphate-phosphate distance of opposite monolayers at each time point. Fluctuations in the thickness were calculated with respect to the initial simulation time.
Pore size
The presence of a transmembrane pore was determined following a clustering method as described in (47). Here, only the atomic coordinates of the phosphate atoms of POPC, POPE, and SM were analyzed. For each lipid, the phosphate atoms of each monolayer were considered as two independent clusters, and pores were identified when the phosphate groups of the lipids formed only one cluster. For the cluster analysis, a minimal distance of 1.0 nm between phosphate groups that form the upper and lower monolayer was established as a cutoff for pore definition. To estimate the size of the pore, the diameter of the area fitting the phosphate groups at the lipid-water interface was calculated in both the inner and outer monolayers, as described in (47). To validate these values, the diameter of the pore was also estimated by measuring the maximum distance between two water molecules located at the middle xy plane within the pore, as a function of the simulation time. The middle xy plane of the membrane was defined as the mean z coordinate (z′ = 0) between the lower and the upper leaflet.
Peptide structure and orientation
The helicity of the peptides was calculated using the DSSP criteria (48). The total number of contacts between peptide molecules was calculated within a 0.4 nm cutoff, using the gmx_mindist tool included in GROMACS. The variation of the tilt angle was calculated over the simulation time for each peptide, using the z axis perpendicular to the bilayer as a reference. Peptide orientation was estimated based on the distance of the z coordinate of amino acid residues located in three different regions of each peptide molecule (N-terminus: Ala1; center of mass: Gln15; and C-terminus: Lys30). The distances between the peptides and the center of the bilayer (z′ = 0) were averaged during the last 100 ns of simulation.
Lipid contribution to the pore
Mass density profiles of lipid phosphates and peptides were calculated using the tool gmx_density included in the GROMACS simulation package. Lipid density profiles were estimated as described in (49). In brief, the plane of the bilayer (xy) was divided into 10,000 grid cells of ∼0.2 × 0.2 nm, and the number of phosphate groups of each lipid type in each grid was calculated. Lipid densities were averaged over the last 100 ns of simulation, corresponding to the time period during which the number of each lipid type surrounding the peptides within 0.5 nm had equilibrated and the pore reached its maximum size. For each lipid type, lipid density values were normalized by considering the average density in the whole system.
The depletion-enrichment (DE) index of the lipids in the local environment of the pore was calculated considering a cutoff distance of 1.5 nm in the xy plane from a line going through the center of the pore. The cutoff distance was chosen considering that the final diameter of the pore was reached after 300 and 400 ns of simulation time in POPC:SM:POPE (50:40:10) and POPC:SM (50:50), respectively. The enrichment value of a lipid X (POPC, SM, or POPE) in the pore is defined as the ratio of its local concentration to its bulk concentration following the equations (50, 51) as follows:
| (1) |
in which the lipid distribution (LD) was calculated as follows:
| (2) |
and the lipid bulk (LB) was calculated as follows:
| (3) |
where the total lipids are the sum of all of them in each system.
Flip-flop movement of lipids
To analyze flip-flop movement of the lipids between the two leaflets of the membrane, we consider the phosphate group of each lipid type, located in the pore region (1.5 nm in the xy plane from a line going through the center of the pore). We defined the middle plane of the membrane (z′ = 0) as the mean z coordinate between the lower and the upper leaflet. The upper leaflet is defined as z′ > 0 and the lower leaflet as z′ < 0. Considering this, the z coordinate of the lipid phosphodiester group was calculated as a function of the simulation time.
Binding to lipid monolayers
Surface pressure (π) measurements were carried out as previously reported (17, 52, 53) with a μThrough-S system (Kibron, Helsinki, Finland) at room temperature under constant stirring. The aqueous phase consisted of Tris-buffered saline (TBS: 145 mM NaCl, 10 mM Tris-HCl (pH 7.4)). The lipid mixture (POPC:SM (50:50) or POPC:SM:POPE (50:45:10)) was predissolved in chloroform:methanol (2:1, v:v) and was gently spread over the surface. The desired initial surface pressure (πo) was attained by changing the amount of lipid applied to the air-water interface. StII1–30 was injected into the subphase, and the increment in surface pressure (Δπ) was recorded as a function of the elapsed time until a stable signal was obtained.
Binding to liposomes
Binding of StII1–30 to small unilamellar vesicles (SUVs) was followed by the increase in Phe or Trp fluorescence. Because StII1–30 does not contain any Trp in its primary sequence, the Trp-analog peptide StII1–30L2W was employed for Trp measurements (33). Fluorescence measurements were carried out at room temperature in an RF-5301PC spectrofluorophotometer (Shimadzu, Tokyo, Japan) using quartz cuvettes with excitation and emission slits of 5 and 10 nm, respectively. Peptide samples were excited at λexc 240 (Phe) or 280 (Trp) nm, and the emission spectra were recorded from λ 300 to 440 nm, respectively. The spectral correction was made by subtracting spectra measured under identical conditions but without peptide.
Differential scanning calorimetry
The effect of StII1–30 on the lamellar (L) to inverted hexagonal (HII) transition temperature of DiPoPE was assessed by differential scanning calorimetry (DSC). Lipid films were made by dissolving appropriate amounts of lipid in chloroform/methanol 2:1 (v/v) followed by solvent evaporation under a stream of nitrogen to deposit the lipid as a film on the walls of a tube. The tube was placed in a vacuum chamber for at least 2 h to remove possible traces of solvent. Films were hydrated with 20 mM PIPES buffer (1 mM EDTA, 150 mM NaCl, 0.002% NaN3 (pH 7.4)) in the presence or not of an appropriate amount of peptide and vortexed extensively to make multilamellar vesicles. Calorimetric scans were carried out on a MicroCal VP-DSC differential scanning calorimeter (Massachusetts). The reference and the sample solutions were degassed at room temperature before scanning. The scan rate was 1° Celsius ⋅ minute−1, with a delay of 10 min between sequential scans in a series to allow thermal equilibration. The scans corresponding to lipid transitions were recorded in the presence and in the absence of peptide using PIPES buffer as the reference. The temperature of transition (Tm) was calculated as the maximum of heat capacity versus temperature.
Vesicle permeabilization
Large unilamellar vesicles (LUVs) permeabilization was determined by measuring the fluorescence (λexc = 490 nm and λem = 520 nm) of released carboxyfluorescein (CF). Black plastic 96-well microplates (SPL Life Sciences, Seoul, South Korea) were pretreated with 0.1 mg/mL Prionex (Pentapharm, Basel, Switzerland), which strongly reduces unspecific binding of protein and vesicles to plastic. Each well was filled with the TBS plus 10 μM of lipids. Finally, the peptide was added in a total volume of 200 μL at the concentration reported in the text. After mixing vesicles and peptides, the release of CF produced an increase in fluorescence, F (because of the dequenching of the dye into the external medium), which was resolved in time. Spontaneous leakage of the dye was negligible under these conditions. Maximal release was always obtained by adding Triton X-100 (Sigma, EUA) at 1 mM and provided the fluorescence value f(total). The fraction of fluorophore release (F in percent) was calculated as follows:
| (4) |
where f(t) is the fluorescence at time t, f(total) is the maximum release upon addition of Triton X-100, and f(0) is the fluorescence before addition of the peptide.
Measurement of flip-flop movement of the lipids
The effect of StII1–30 on the transbilayer movement of lipids was assessed by fluorescence spectroscopy, as described before (54). For this, LUVs were prepared in TBS buffer. Briefly, the asymmetrically labeled liposomes were prepared by incubating the liposome suspension (20 μM final lipid concentration) with pyrene-labeled PC (pyPC) suspension (1 μM) for 20 min at 37°C. Changes in the fluorescence spectrum of PyPC upon peptide addition was followed in a spectrofluorometer (Hitachi F-4500, Tokyo, Japan) using an λexc of 344 nm and measuring the fluorescence intensity of monomers (IM) and excimers (IE) at λ 395 and 465 nm, respectively. The time course of the transbilayer redistribution of pyPC was followed through the calculation of the IM/IE ratio.
Results
Preassociated in solution StII1–30 forms pores in lipid membranes
Previous structural studies based on low-resolution circular dichroism measurements of StII1–30 have shown that this peptide is able to adopt α-helical secondary structure, self-associating in solution, without the assistance of the membrane (9, 34). Therefore, we performed atomistic MD simulations to obtain this type of associated structures of StII1–30 in solution. The homologous StI-derived peptide (StI1–31) that does not self-associate was used as a control (10, 34) (Fig. 1 A). Because it is known from previous circular dichroism studies that StII1–30 and StI1–31 have the potential to adopt similar structures to those found in the context of the protein (34), we began the simulations from the structure that these segments adopt in their respective parental sticholysins. The peptides were further equilibrated in solution for 100 ns (Fig. 1 B). StII1–30 and StI1–31 were stable after 100 ns, maintaining a predominantly α-helical conformation (Fig. S2 A). Therefore, four molecules of each peptide were further placed in a water box and simulated for 1 μs. During this time, both peptides remained mostly folded as α-helices (Fig. 1, C–E). Individual StII1–30 molecules interacted with each other at 100 ns, forming associated structures that remained stable after 1 μs simulation (Fig. 1, C, D, and F). For StII1–30, the content of α-helical structure increased because of its self-association (Fig. 1 E). Conversely, StI1–31 did not self-associate and remained unchanged during 1 μs of simulation (Fig. 1, D–F).
To model pore formation by StII1–30 in membranes, a series of atomistic MD simulations on lipid bilayers was performed in membranes containing POPC and SM with and without POPE. SM was selected because this lipid has been proposed as the natural receptor of actinoporins (55), whereas POPE was selected because of its capacity to form nonlamellar structures favoring negative curvature in the membrane in the absence of electrostatic interactions (56). Nonlamellar structures are essential for toroidal pore formation, in which the membrane adopts both a positive curvature in the direction parallel to the pore axis and a negative curvature in the direction perpendicular to the pore axis (32, 57).
In our simulations, two configurations were modeled (Fig. S1), each of them containing four molecules of StII1–30: 1) nonassociated StII1–30 and 2) preassociated StII1–30. Preassociated StII1–30 was obtained after 1 μs simulation (Fig. 1). Each system was simulated in three replicas. Fig. 2, A and B display representative snapshots of one replica of the simulations with preassociated StII1–30 in POPC:SM (50:50) or POPC:SM:POPE (50:40:10) membranes, respectively. Statistical analysis of the replicates is summarized in Table S1. The binding to the membrane took place only when StII1–30 began from a preassociated conformation in solution (Fig. 2 C). Upon membrane binding, the secondary structure and interpeptide molecule interactions of associated StII1–30 remained stable over the simulation time (Fig. S2, B and C). The interactions of hydrophobic amino acid residues of StII1–30 with the membrane increased over time, whereas those with the polar ones remained stable (Fig. S2 D). A detailed analysis of the first hydrophobic (1, 2, 3, 4, 5, 6, 7, 8, 9, 10) and the amphipathic (11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30) regions of StII1–30 showed that the apolar face of the amphipathic helices stablished stronger interactions with the membrane in both systems (Fig. S2 E). Because of membrane insertion, lipid contacts with those apolar amino acid residues located in the first hydrophobic segment increased over time. As a result, self-associated StII1–30 underwent some structural reorganization mostly involving the hydrophobic segments 1–10 (Fig. S2 F; Table S2). The increase in the average number of contacts of the peptides with the polar head of lipids in the membrane promoted an increase in membrane thickness (Fig. 2 D), most likely as a consequence of their recruitment to peptide-enriched regions.
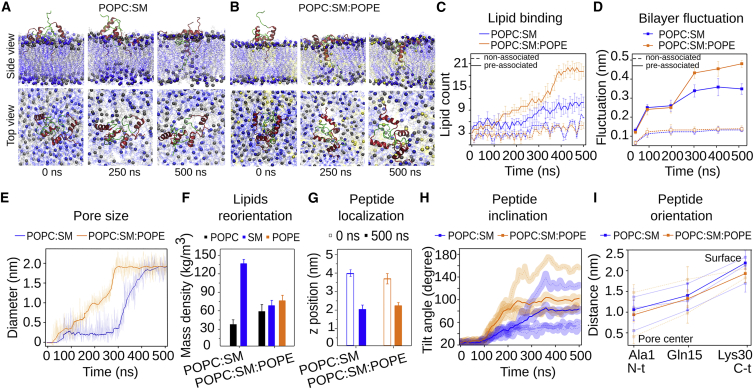
Figure 2.
POPE favors interaction with lipids and pore formation by preassociated StII1–30. Snapshots of the sequence of events in the simulations in (A) POPC:SM (50:50) and (B) POPC:SM:POPE (50:40:10) are shown. Phospholipid headgroups are represented in gray (POPC), blue (SM), or yellow (PE), and peptides are represented in two colors: the 1–10 segment is represented in green and the 11–30 in red. Water was removed for clarity. (C) The number of polar heads of the phospholipids that are found within 0.5 nm of the four molecules of StII1–30 is shown. (D) Shown are the kinetics of membrane thickness fluctuation around the pore region, measured as the distance between two different clusters of phosphate groups located at each monolayer. (E) An increase in size kinetics of the pore formed by preassociated StII1–30 in membranes is shown. The pore size was defined as the maximum distance between two water molecules located at the middle xy plane within the pore. (F) Reorientation of lipids in the membrane due to pore formation is shown. Mass density in the bilayer center (z axis position zero) was calculated after pore formation. This region was devoid of the headgroup of lipids at the beginning of the simulation. (G) Peptide localization in the membrane is shown. The z axis corresponding to the maximum of peptide mass density was calculated at the beginning and at the end of the simulation. (H) Kinetics of insertion into the bilayers of preassociated StII1–30. The tilt angle was calculated for each peptide molecule using the z axis perpendicular to the bilayer as a reference. (I) The orientation of preassociated StII1–30 molecules averaged over the last 100 ns of the simulation is shown. In (C)–(G), values correspond to the mean and the SD considering the three replicates of the system are shown. In (H) and (I), light lines correspond to the mean values of the four peptides of each system, and dark lines are the average of the three replicas. To see this figure in color, go online.
In both membrane systems, binding of self-associated StII1–30 promoted pore formation (Fig. 2, A and B) and the resulting water flux through the membrane (Fig. 2 E; Figs. S3 and S4). The final diameter of the pore was 2.0 nm in both membrane systems (Figs. 2 E and S4; Table S1). This size was enough to allow for the passage of water molecules through the membrane (Fig. S3 A). The pore size was similar to the one previously estimated experimentally with the use of osmoprotectants of different sizes (9). In the binary mixture, pore opening was a one-step process that began after 300 ns with a rapid increase in pore size (Fig. 2 E). In contrast, in POPE-containing membranes, this was a multistep process as reflected by the bimodal shape of the time course of the increase in the pore size. Initially, a slow rise in the pore diameter until ∼1.0 nm occurred, followed by a rapid increase to 2.0 nm in diameter. This result suggests that POPE could facilitate the process of pore opening by StII1–30 in membranes.
StII1–30-induced pores have a toroidal shape
In our MD simulations, StII1–30 induced membrane bending in both POPC:SM and POPC:SM:POPE systems as a consequence of pore formation (Figs. 2, A and B and S3, A and B). Moreover, our simulations showed that StII1–30 forms pores with a toroidal shaped structure, wherein the headgroups of the lipids contribute to the pore architecture (18). Strikingly, peptide molecules were located on one edge of the pore (Fig. 2, A and B; Fig. S3 B), which is enough to maintain the open configuration of toroidal pores, as previously reported for AMPs and for Bax (58, 59, 60, 61).
The imaging of toroidal pore structures has been difficult because of the intrinsic structural flexibility of peptide and lipid molecules (4, 5). Difficulties increase in the case of small-sized pores, such as those formed by AMPs, actinoporins, and sticholysin-derived peptides (9, 26, 31, 32). We took advantage of our MD simulations to gain insight into the structural dynamics and configuration of the pore formed by StII1–30. In the process of toroidal pore formation, lipid molecules reoriented, resulting in phosphate groups of opposite monolayers, which were in contact because of membrane bending in the membrane region containing the pore (Fig. 2 F; Fig. S3 B). StII1–30 initially appeared located parallel to the membrane surface, in contact with the headgroup of the phospholipids but partially inserted into the membrane in the pore configuration, without spanning the membrane (Fig. 2 G; Fig. S3 B). In the pore, StII1–30 tilted in an oblique orientation with respect to the bilayer normal (Fig. 2 H; Table S1). Molecules of StII1–30 remained at the pore edge, lying on the surface of the curved region of the membrane that directly forms part of the pore (Fig. S3 B). StII1–30 was oriented with its hydrophobic N-terminus buried into the pore section, and the C-terminus localized at the water-membrane interface (Fig. 2 I; Table S1). These results are also in line with the toroidal pore model in which lipid molecules form part of the ring and peptide molecules do not need to completely penetrate the membrane but rather destabilize its structure by inducing local membrane curvature.
PE has a dual role in enhancing membrane association and functioning as a structural component of the pore formed by StII1–30
The comparison of the simulations in the membranes with and without POPE allowed us to gain insight into the role of this lipid in the mechanism of the pore formation of StII1–30. We found that POPE enhanced both StII1–30 association to the membrane and pore formation (Fig. 2; Table S1). Although the final pore size reached upon equilibrium was similar in all the membrane systems (Ø ≈ 2 nm), the kinetics of pore opening were faster in POPC:SM:POPE membranes. These results indicate that PE plays a key role in the mechanism of the action of StII1–30 at favoring the initial binding to the membrane and enhancing the kinetics of pore opening.
To glean further insight into the mechanism of how PE facilitates pore formation by StII1–30, we compared the distribution of the different membrane lipids in the pore (Fig. 3). Fig. 3 A shows the distribution of individual lipids in POPC:SM and POPC:SM:POPE membranes containing StII1–30 pores. Because this peptide induces membrane curvature as a result of toroidal pore formation, lipids could be integrated in the pore region according to their geometric properties. Whereas POPC and SM are lipids with the propensity to form bilayer structures, POPE has a high tendency to adopt negative curvature because of its cone shape (32, 62) (Fig. 3 B). The latter class of lipids facilitates toroidal pore formation wherein the membrane bends to adopt a positive curvature in the direction parallel to the pore axis, and a negative curvature is defined at the pore boundaries (Fig. 3 C; (23, 32, 56, 63)).
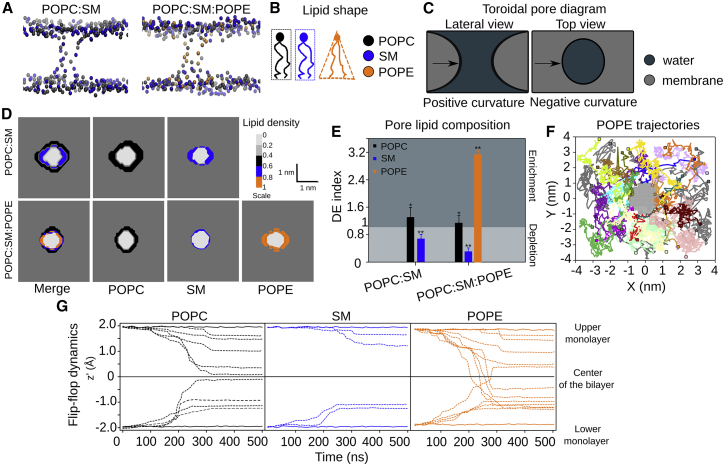
Figure 3.
StII1–30 pores are enriched in POPE. (A) Representation of the lipid composition of the toroidal pores induced by preassociated StII1–30 after 500 ns simulation is shown. (B) The shape of the lipids determines the propensity of the membrane to adopt planar or curved geometry. (C) The contribution of membrane curvature to the toroidal pore structure is shown: positive curvature in the direction of the membrane bending and negative curvature around the ring structure. In relatively small pores, negative curvature has a dominant contribution. (D) The profile density of the lipids around the pore is shown. (E) The depletion-enrichment (DE) index of the lipids during pore formation is shown. The index was obtained from the last 100 ns of simulation out of 500-ns-long simulation. Bars represent the average obtained from three replicas, and error bars represent their SD. Values greater than 1 indicate enrichment, and values less than 1 indicate depletion. Differences with p values ≥0.05 are indicated with ∗ and <0.05 with ∗∗. The statistical significance was calculated by a single sample t-test, according to (46). (F) A top-down view of the pore in the POPC:SM:POPE membrane is shown in which trajectories of individual POPE molecules during the 500 ns of simulation are shown. Each molecule is represented with different colors, initial position (boxes), and final position (stars). (G) Dynamics of the flip-flop movement of lipids in POPC:SM:POPE membrane are shown, represented as the mean z coordinate profile for the headgroup beads, calculated over windows of 10 ns. Individual headgroups (dashed line) are shown, with an average of the headgroups located at the interface of the upper and lower monolayer (solid line). To see this figure in color, go online.
To analyze the lipid composition of the pores formed by StII1–30 in the different membrane systems, we calculated their lipid density profiles (Fig. 3 D). In all the studied compositions, a specific lipid clustering was observed in the membrane region surrounding the pore (Fig. 3 D). The DE index allowed us to characterize the distinctive nature of the lipid environment surrounding the pore. This parameter quantifies the distribution of the individual lipids (POPC, SM, or POPE) in the pore relative to the bulk of lipids in the membrane (Fig. 3 E). DE values greater than 1 indicate enrichment and those less than 1 show depletion. This analysis revealed a slight enrichment of POPC in the pore formed by StII1–30 in POPC:SM membranes. In POPC:SM:POPE membranes, pores presented a significant enrichment of POPE, although they contained just 10% of this lipid. In fact, POPE was mostly confined to the pore in this system. Fig. S5 A shows the DE values obtained in the bulk membrane and the pore region over the simulation time. According to this parameter, POPE was initially distributed along the bilayer, but it was freely diffused to the pore region during 500 ns of simulation. This was also evidenced by the trajectories of individual POPE molecules during the simulation (Fig. 3 F). In contrast to POPC and SM, POPE did not show a preferential interaction with peptide molecules at the beginning of the simulation but only after pore opening (>200 ns) (Fig. S5 B). This result indicates that the relocation of POPE into the pore is not the result of a higher affinity for the peptide backbone but rather due to its intrinsic shape.
One relevant feature of toroidal pores is that membrane bending and curvature induction facilitate the flip-flop movement of the lipids through the pore ring (64, 65). To compare the contribution of each individual lipid to the flip-flop movement in the process of pore formation, we calculated the z coordinates of those lipids involved in the pore structure (within a 1.5-nm cutoff from the center of the pore, considering that the final size of the pore is 2 nm). Fig. 3 G shows the position of the lipids forming the pore as a function of the simulation time for POPC:SM:POPE membranes. A coordinate of z = 0 indicates the middle of the membrane, whereas +2 and −2 nm are the two interfaces with water on either side of the membrane. POPE and POPC showed more flip-flop events than SM. Altogether, our analysis suggests that in contrast to the full-length toxin, SM is not a critical component of the pore formed by StII1–30. Moreover, in the membrane with three components, the pore structure is mainly formed by POPE and POPC molecules. Among these lipids, POPE seems to play a critical role at facilitating pore formation because of its ability to form nonlamellar structures, which could assist membrane curvature induction during the assembly of toroidal pore structures and the resulting flip-flop movement of lipids.
StII1–30 activity in membrane systems is enhanced by PE
From our simulations, StII1–30 was predicted as being able to form toroidal pores in membranes, and PE emerged as a critical component of the membrane that could facilitate the activity of the peptide and as a structural element of the pore. This lipid has been classically considered an inductor of negative curvature in the membrane because its tail has a larger area than its head (Fig. 3 B; (62)). To have additional experimental data that validate our in silico predictions, we studied the pore-forming mechanism of StII1–30 with experimental biophysical tools. We assessed the StII1–30 binding to lipid monolayers and liposome vesicles and evaluated its permeabilizing activity in the later system. Membrane systems were composed of POPC:SM (50:50) or POPC:SM:POPE (50:40:10) to evaluate the contribution of PE to peptide membranotropic activity.
The increase in surface pressure (Δπ) by the association of peptides with previously formed lipid monolayers at the air-water interface can be employed to characterize their ability to interact with organized lipids (17, 52, 66). This method has the advantage of not requiring fluorescent amino acids or probes to detect lipid-peptide interactions, thus complementing fluorescence-based lipid binding assays (67). Fig. 4 A shows the kinetics of surface pressure (π) increase upon StII1–30 binding to a lipid monolayer. The injection of the peptide into the subphase of the monolayer triggered an increase in π (Δπ), which stabilized after ∼100 s (Fig. 4 A). As expected, Δπ decreased with the increase of initial surface pressure (π0) because of a tighter packing of the lipids (Fig. 4 B). A suitable parameter for the characterization of lipid-peptide interaction is the critical pressure (πc), obtained by extrapolating the Δπ at equilibrium as a function of π0 to zero (68, 69) (Table S3). This parameter corresponds to the minimum pressure that must be applied to avoid incorporation of the peptide into a monolayer and directly correlates with its affinity for the monolayer. StII1–30 attained the highest πc values in the PE-containing membrane, indicating that it probably penetrates more deeply into this lipid monolayer when compared with POPC:SM monolayers (Table S3).
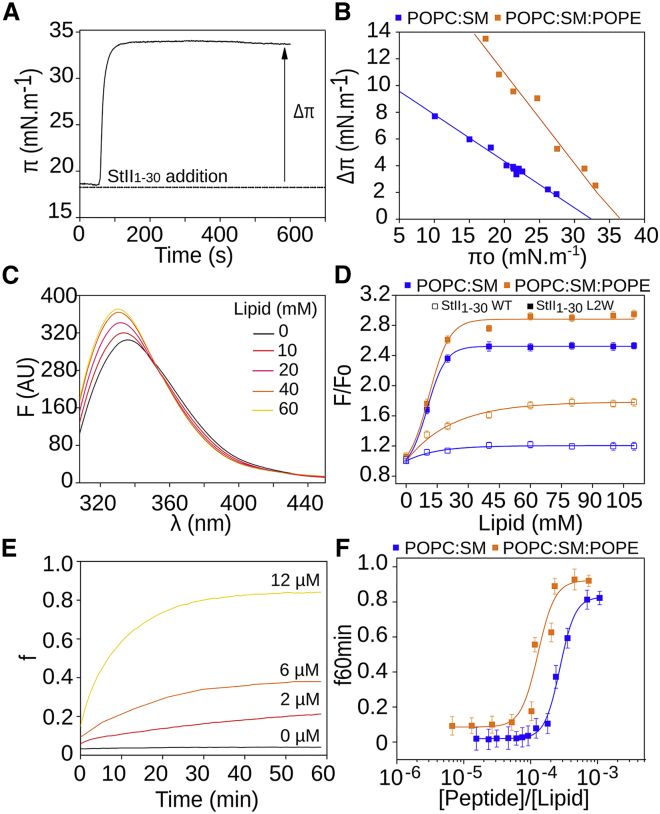
Figure 4.
POPE favors binding to lipids and membrane permeabilization by StII1–30. (A) Representative kinetics of StII1–30 binding to a lipid monolayer are shown. (B) Critical pressure induced by StII1–30 on monolayers of different lipid composition is shown. Lines represent the best linear fit of the Δπ as a function of the initial monolayer pressure (πo); r2 > 0.99. The peptide concentration is 1 μM. (C) Representative fluorescence spectra of StII1–30L2W in the presence of increasing amount of SUVs are shown. (D) Increase in the fluorescence intensity of StII1–30 or StII1–30L2W as a function of lipid concentration is shown. F0 and F stand for the fluorescence intensities (at λmax) from the spectra in the absence and the presence of SUVs, respectively. λexc is StII1–30_(Phe14) 251 nm, λexc is StII1–30L2W_(Trp2) 280 nm, and peptide concentration is 20 μM. (E) Representative kinetics of liposomes permeabilization induced by different concentrations of StII1–30 in LUVs are shown. (F) Release of CF from LUVs promoted by different concentrations of StII1–30 after 60 min of assay is shown. The lipid concentration is 10 μM, and the lipid composition is POPC:SM (50:50) or POPC:SM:POPE (50:40:10). Values in (E) and (F) are shown as the mean values and the SD among three replicates. To see this figure in color, go online.
The binding of StII1–30 to liposomes was measured by following the changes in the Phe (native StII1–30) or the Trp (StII1–30L2W) fluorescence emission spectra elicited by mixing with lipid vesicles (70). Fig. 4 C shows the typical spectra obtained upon StII1–30L2W titration with SUVs. The addition of increasing amounts of liposomes to the peptide solution triggered a progressive increase in fluorescence emission intensity as well as a blue shift in the position of maximum emission (λmax), indicating a change in the Trp local environment (Fig. 4 C). The magnitude of the change determined at high lipid concentration is measured by the parameter [F/F0]max. Vesicles containing PE showed higher values of [F/F0]max, suggesting that they were better membrane targets for StII1–30 (Fig. 4 D; Table S3). To characterize the affinity of StII1–30 to lipid vesicles, the L50 was determined. This parameter represents the lipid concentration at which half of the peptide molecules are bound to the liposomes (Table S3). The lowest L50 also showed that StII1–30 had more affinity for PE-containing membranes.
The permeabilization of lipid vesicles has provided direct evidence for the pore-forming activity of StII1–30 (12, 13, 18). Here, we examined whether PE modulated StII1–30 activity. Fig. 4 E shows the time course of StII1–30-elicited activity in liposomes. The addition of StII1–30 to a solution containing CF-loaded LUVs promoted the release of the dye at different rates and to different fractions in a concentration-dependent manner (Fig. 4 F). This peptide had higher activity in POPC:SM:POPE than in POPC:SM vesicles, which was reflected in the C50 parameter, which is directly related to the peptide concentration required to achieve the permeabilization of 50% of the vesicles (Table S3). Combined, our experimental results qualitatively support the MD simulations in that PE favors StII1–30 pore formation, most likely by promoting membrane binding and acting as an important element of the pore because of its intrinsic capability to form nonlamellar structures.
StII1–30 activity has typical functional features of toroidal pores
The toroidal pore model postulates the bending of the inner and outer monolayers of the membrane, forming half a torus. Therefore, in this pore configuration, the membrane adopts both a positive curvature in the direction of membrane bending and a negative curvature in the direction of the ring structure (Fig. 3 C; (31, 71)). Moreover, each pore behaves as a point of fusion between opposite monolayers, favoring the otherwise restricted transbilayer or flip-flop movement of lipids (62, 72). Our simulations predicted that StII1–30 forms toroidal pores, a process that is accompanied by the induction of membrane curvature and lipid flip-flop movement (Fig. 2). Therefore, we next sought to validate these predictions with biophysical experimental tools.
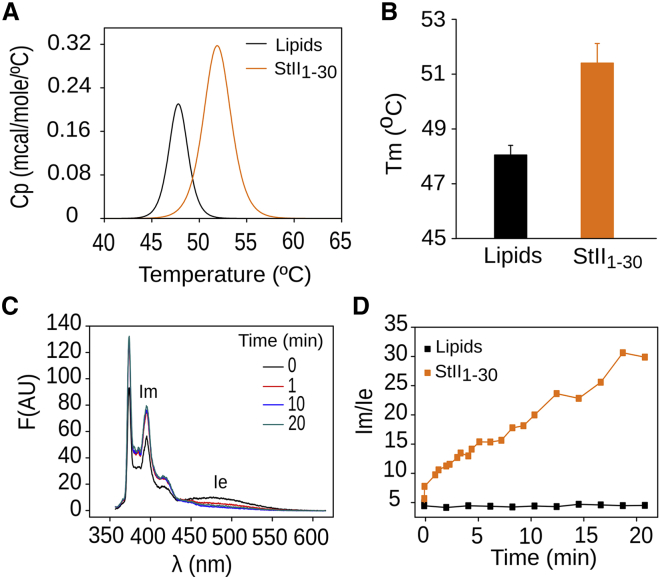
We evaluated the ability of StII1–30 to promote an increase in the L/HII phase transition temperature of DiPoPE, an approach that has been previously used to study the mechanism of toroidal pore formation of some AMPs (73, 74). Representative scans of DiPoPE in the presence or absence of StII1–30 were obtained by DSC (Fig. 5 A). Pure DiPoPE exhibited a thermotropic transition around 43°C that was shifted to a higher temperature when StII1–30 (52°C) was added to the liposomes (Fig. 5 B). An increase in Tm value for the L/HII phase transition of the DiPoPE can be interpreted as evidence of the capability of StII1–30 to induce positive curvature in the membrane. By stabilizing positive membrane curvature, StII1–30 promotes membrane stress in the opposite direction regarding curvature. Therefore, StII1–30 pores would essentially need to flip inside out to form the HII phase, a process that requires more energy, as evidenced by the higher Tm.
Figure 5.
StII1–30 stabilizes positive curvature and induces flip-flop movement of the lipids in the bilayer. (A) A DSC heating thermogram of multilamellar vesicles of DiPoPE in the presence or the absence of StII1–30 is shown. (B) The effect of StII1–30 on the bilayer (L) to hexagonal phase (HII) transition temperature of DiPoPE is shown. The lipid concentration is 10 mM. The peptide:lipid ratio is 1:100. (C) PC-pyrene spectra obtained after different time of addition of StII1–30 are shown. (D) The kinetics of transbilayer redistribution of pyPC are shown. It was followed by measuring the fluorescence intensity of pyrene monomers (IM) and excimers (IE) at λ 395 and 465 nm, respectively, and calculating the IE/IM ratio. The peptide concentration is 10 μM, the lipid concentration is 20 μM, and the lipid composition is POPC:SM:POPE (50:40:10). To see this figure in color, go online.
Finally, we investigated if the formation of StII1–30 pores promotes flip-flop movement of the lipids, as previously demonstrated for sticholysins (54) and for their shorter versions (75). We measured the transbilayer movement of a pyrene-labeled analog of PC (pyPC) in POPC:SM:POPE lipid vesicles. The shape of the emission spectra of the pyrene depends on the contribution of two populations made up by monomers and dimeric excimers. The extent of excimer formation depends on the collision frequency of pyrene-labeled analogs. Toroidal pore formation should facilitate flip-flop movement of the lipids, promoting the redistribution of initially asymmetric pyPC molecules in both leaflets (76) and the change of their fluorescent properties (54). The addition of StII1–30 induced a decrease of the excimer fluorescence intensities (Fig. 5 C) and the increase of the IM/IE ratio over time (Fig. 5 D). IM/IE did not change in the absence of the peptide, indicating the stability of the liposome system. These experiments allowed us to conclude that similar to what has been described for sticholysins (54), StII1–30 is able to induce flip-flop movement of the lipids. Given the ability of this peptide to induce membrane curvature and form pores in mimetic (12) and cellular membranes (9) and our structural data based on MD simulations, this evidence supports a mechanism in which StII1–30 forms toroidal structures involving membrane bending and lipid diffusion through the pore.
Discussion
Sticholysins are typical eukaryotic α-PFTs that form pores upon insertion of their N-terminal lytic fragment into the membrane (77). They act through the formation of protein-lipid pores and have been extensively used as a tool to investigate the mechanism of toroidal pore formation in membranes (4, 5, 21, 22). Synthetic peptides corresponding to the N-terminus of sticholysins are good models for the study of the structure and function of these toxins (9, 10, 12, 13). StII1–30 is the most active peptide derived from the N-terminus of actinoporins (13) and therefore may be a promising candidate for biomedical applications. Here, we combined MD simulations and biophysical tools to get further insight into the pore-forming mechanism of StII1–30. Specifically, we investigated the role of membrane curvature and lipid flip-flop on the mechanism of pore formation by this peptide. The combination of computational and experimental biophysical tools allowed us to gain information about the structure of the pore formed by StII1–30 in lipid membranes and the role of curved lipids in the assembly of toroidal pores.
We have previously proposed that the higher permeabilizing activity of StII1–30 could be ascribed to its higher propensity to fold and preassociate as α-helical structures in solution, without the assistance of the membrane (34). The results of this work support this hypothesis. In our model, we propose that preassociation of StII1–30 in solution might assist irreversible binding to lipids by increasing the local membrane-bound peptide concentration. The asymmetric attack of associated StII1–30 to one leaflet of the bilayer and its high local concentration at the membrane would facilitate bilayer destabilization and pore opening because of the increase of membrane tension, as proposed for other membranotropic peptides such as those derived from Bax (8, 78, 79).
MD simulations were used to obtain information about the interplay between StII1–30 and lipids during pore formation, with molecular details. In all the membrane systems containing preassociated StII1–30, we observed the same sequence of events, including 1) binding to the membrane interface, 2) enhancement of membrane thickness, 3) curvature induction due to membrane bending, and 4) pore formation. StII1–30 formed pores of ∼2 nm diameter size, which mirrors our previous experimental data in human red blood cells (9). Altogether, our predictions based on MD simulations and our results obtained here and in previous works with biophysical tools (33) support the toroidal pore model for the mechanism of action of StII1–30. A similar mechanism was also suggested for the full-length sticholysins (15, 80). In our simulations, StII1–30 molecules appear accommodated in the membrane in an oblique configuration with the N-terminus nearer the core of the ring and the C-terminus lying in the membrane interface, in agreement with previous data obtained in liposomes (13, 33). A configuration in which StII1–30 is located at the pore surface without spanning the membrane bilayer and only in one edge of the ring supports the toroidal pore model. In this model, one section of the pore can remain protein free, as observed for other proteins such as the cholesterol-dependent cytolysins and Bax or some AMPs (21, 22, 81).
We predicted and validated experimentally that StII1–30 induces positive curvature and flip-flop movement of the lipids as a consequence of pore formation. A similar ability has supported the mechanism of toroidal pore formation for the full-length protein and a shorter derived form (15, 54, 80). In the context of toroidal lipid pores, curvature induction and membrane bending represent critical steps for pore opening (4). In this case, the role of the peptides or lytic fragments is to help to reduce the stress caused by membrane distortion and stabilize curvature formation (71). Mechanistically, curvature induction could be explained based on the hydrophobic effect. Initial membrane disruption due to peptide binding can promote temporary exposure of the hydrophobic acyl chains of the lipids to the aqueous environment. To avoid the high energy cost of this process, lipids bend and form a highly curved, nonbilayer structure at the pore edge connecting the opposite monolayers of the membrane and forming a continuous surface. Afterward, lipids can easily exchange monolayers by simple diffusion over the pore edges. As a result, in the framework of this model, not only peptide molecules but also membrane mechanical properties play an important role in pore assembly (24, 47, 63, 65, 71).
Both computational and experimental approaches showed that membrane binding and pore formation by StII1–30 was enhanced by PE. The presence of this lipid in the membrane bilayer seems to facilitate pore formation by acting at two different stages of the process. On the one hand, PE could be considered a modulator of the membrane properties that are required for efficient pore formation because of its capability to form nonlamellar lipid phases (31, 82, 83). This capability seems to favor early stages of the membrane destabilization process by enhancing peptide insertion into the bilayer and membrane remodeling events, which are fundamental to accomplish stable pore opening. Given that the generation of a pore is energetically expensive, the presence of such nonlamellar lipids should contribute to pore stability (84). On the other hand, PE appears as a cofactor of the pore induced by StII1–30 because membrane regions comprising the pore assembly are especially enriched in this lipid.
There remains controversy regarding the effect of lipids that induce positive or negative curvature on the mechanism of pore formation of small AMPs and pore-forming proteins. PE and cardiolipin, which induce negative curvature, facilitate pore formation by sticholysins, Bax, and their derived peptides (8, 15, 78, 85). An opposite effect on pore formation has been found for magainins and melittin, whose activity is affected by PE but enhanced in the presence of the positively curved lipid lysophosphatidylcholine (31, 86, 87). Depending on peptide concentration, melittin pores can be promoted by positive or by negative curvature (88). In general, lipid shape and membrane curvature seem to modulate peptide partition, orientation, and penetration depth together with the local tilting of lipids, which are essential processes for the assembly of toroidal pores (88, 89, 90). Given the different effect of curved lipids on the activity of proteins/peptides acting via toroidal pores, it is plausible that some pore properties such as size and peptide or lipid orientation could have an impact on the ability of negative and positive curved lipids to relocate into these pores (71, 91). It is well known that the pore size strongly influences the balance of the contribution between positive and negative curvature in toroidal pores (57). Positive curvature is induced in the direction of the membrane bending and is modulated by the membrane thickness, whereas negative curvature is stabilized in the direction of the ring, parallel to the membrane (57). Knowing that curvature is inversely proportional to the radius, the smaller the pore size is, the higher the negative curvature along the pore edges. As a consequence, in the case of very small pores, like those formed by actinoporins (15, 26) or StII1–30 (9), negative curvature-inducing lipids (i.e., PE) would easily accommodate in the ring sections of the pore and play an additional role at stabilizing its edges.
Conclusions
We propose a mechanistic framework that explains in detail the lytic activity of StII1–30 and provide evidence for a toroidal model for the structure of the pore it forms. Induction of membrane curvature and flip-flop movement of the lipids were identified as two important membrane remodeling steps related to toroidal pore formation by StII1–30. We highlight the key role of PE and in general curvature-inducing lipids as cofactors of toroidal pores that can facilitate the adoption of the open state by easily accommodating in the ring. Understanding these aspects of the mode of action of StII1–30 could underpin strategies for the rational use of this and other peptides or proteins acting via a similar mechanism of toroidal pore formation as biotechnological tools in different lipid formulations.
Author Contributions
H.M.-G. performed and analyzed the MD simulations, the fluorescence assays, and the flip-flop experiments. P.A.V. and M.E.V.-T. performed some of the analysis from the MD. U.R. performed and analyzed the calorimetry and the monolayer experiments. P.A.V. and D.P.T. contributed to the MD simulation design. R.F.E. and R.M.E. supervised the calorimetry experiments and provided resources. C.A. and M.E.L. contributed to the design of the biophysics experiments and the conception of the project. H.M.-G., D.P.T., and U.R. wrote the manuscript with input from other authors. D.P.T. and U.R. designed and led the project.
Acknowledgments
H.M.-G. is an Alberta Innovates Health Solutions Scholar and Vanier Canada Research scholarship recipient. M.E.V.-T. is an Eyes High Doctoral Recruitment Scholarship (University of Calgary) recipient. D.P.T. holds the Alberta Innovates Technology Futures Strategic Chair in (Bio)Molecular Simulation. The authors thank Dr. Valentina Corradi for constant discussion and Dr. Katia Cosentino, Dr. Rodrigo Cuevas, and Dr. Lohans Pedrera for carefully reading the manuscript, Dr. Kathleen McWilliams for English corrections, and Orlando Quintero Tesoro for cover art design.
Computational resources were provided by Compute Canada, funded by the Canada Foundation for Innovation and partners. Additional support came from the Canada Research Chairs Program (D.P.T.) and the Natural Sciences and Engineering Research Council (D.P.T.). U.R. was a grantee from the International Foundation for Science (4616) and from the Emerging Leaders in America Program of the Canadian government.
Editor: Kalina Hristova.
Footnotes
Pedro A. Valiente’s present address is Donnelly Centre for Cellular and Biomolecular Research, University of Toronto, Toronto, Ontario, Canada.
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.09.018.
Contributor Information
D. Peter Tieleman, Email: tieleman@ucalgary.ca.
Uris Ros, Email: uris.ros@ifib.uni-tuebingen.de.
Supporting Material
References
- 1.Parker M.W., Feil S.C. Pore-forming protein toxins: from structure to function. Prog. Biophys. Mol. Biol. 2005;88:91–142. doi: 10.1016/j.pbiomolbio.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 2.Iacovache I., Bischofberger M., van der Goot F.G. Structure and assembly of pore-forming proteins. Curr. Opin. Struct. Biol. 2010;20:241–246. doi: 10.1016/j.sbi.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 3.Dal Peraro M., van der Goot F.G. Pore-forming toxins: ancient, but never really out of fashion. Nat. Rev. Microbiol. 2016;14:77–92. doi: 10.1038/nrmicro.2015.3. [DOI] [PubMed] [Google Scholar]
- 4.Ros U., García-Sáez A.J. More than a pore: the interplay of pore-forming proteins and lipid membranes. J. Membr. Biol. 2015;248:545–561. doi: 10.1007/s00232-015-9820-y. [DOI] [PubMed] [Google Scholar]
- 5.Cosentino K., Ros U., García-Sáez A.J. Assembling the puzzle: oligomerization of α-pore forming proteins in membranes. Biochim. Biophys. Acta. 2016;1858:457–466. doi: 10.1016/j.bbamem.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gouaux E. Channel-forming toxins: tales of transformation. Curr. Opin. Struct. Biol. 1997;7:566–573. doi: 10.1016/s0959-440x(97)80123-6. [DOI] [PubMed] [Google Scholar]
- 7.Iacovache I., van der Goot F.G., Pernot L. Pore formation: an ancient yet complex form of attack. Biochim. Biophys. Acta. 2008;1778:1611–1623. doi: 10.1016/j.bbamem.2008.01.026. [DOI] [PubMed] [Google Scholar]
- 8.García-Sáez A.J., Coraiola M., Salgado J. Peptides derived from apoptotic Bax and Bid reproduce the poration activity of the parent full-length proteins. Biophys. J. 2005;88:3976–3990. doi: 10.1529/biophysj.104.058008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casallanovo F., de Oliveira F.J., Schreier S. Model peptides mimic the structure and function of the N-terminus of the pore-forming toxin sticholysin II. Biopolymers. 2006;84:169–180. doi: 10.1002/bip.20374. [DOI] [PubMed] [Google Scholar]
- 10.Cilli E.M., Pigossi F.T., Schreier S. Correlations between differences in amino-terminal sequences and different hemolytic activity of sticholysins. Toxicon. 2007;50:1201–1204. doi: 10.1016/j.toxicon.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 11.Drechsler A., Potrich C., Norton R.S. Structure and activity of the N-terminal region of the eukaryotic cytolysin equinatoxin II. Biochemistry. 2006;45:1818–1828. doi: 10.1021/bi052166o. [DOI] [PubMed] [Google Scholar]
- 12.Ros U., Pedrera L., Ávarez C. The membranotropic activity of N-terminal peptides from the pore-forming proteins sticholysin I and II is modulated by hydrophobic and electrostatic interactions as well as lipid composition. J. Biosci. 2011;36:781–791. doi: 10.1007/s12038-011-9156-4. [DOI] [PubMed] [Google Scholar]
- 13.Ros U., Rodríguez-Vera W., Alvarez C. Differences in activity of actinoporins are related with the hydrophobicity of their N-terminus. Biochimie. 2015;116:70–78. doi: 10.1016/j.biochi.2015.06.024. [DOI] [PubMed] [Google Scholar]
- 14.Kem W. Sea anemone toxins: structure and action. In: Hessinger D.A., Lenhoff H.M., editors. The Biology of Nematocysts. Academic Press; 1988. pp. 375–405. [Google Scholar]
- 15.Valcarcel C.A., Dalla Serra M., Menestrina G. Effects of lipid composition on membrane permeabilization by sticholysin I and II, two cytolysins of the sea anemone Stichodactyla helianthus. Biophys. J. 2001;80:2761–2774. doi: 10.1016/S0006-3495(01)76244-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderluh G., Macek P. Cytolytic peptide and protein toxins from sea anemones (Anthozoa: Actiniaria) Toxicon. 2002;40:111–124. doi: 10.1016/s0041-0101(01)00191-x. [DOI] [PubMed] [Google Scholar]
- 17.Pedrera L., Fanani M.L., Alvarez C. Sticholysin I-membrane interaction: an interplay between the presence of sphingomyelin and membrane fluidity. Biochim. Biophys. Acta. 2014;1838:1752–1759. doi: 10.1016/j.bbamem.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 18.Ros U., Edwards M.A., Epand R.M. The sticholysin family of pore-forming toxins induces the mixing of lipids in membrane domains. Biochim. Biophys. Acta. 2013;1828:2757–2762. doi: 10.1016/j.bbamem.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 19.Anderluh G., Dalla Serra M., Menestrina G. Pore formation by equinatoxin II, a eukaryotic protein toxin, occurs by induction of nonlamellar lipid structures. J. Biol. Chem. 2003;278:45216–45223. doi: 10.1074/jbc.M305916200. [DOI] [PubMed] [Google Scholar]
- 20.Tanaka K., Caaveiro J.M., Tsumoto K. Structural basis for self-assembly of a cytolytic pore lined by protein and lipid. Nat. Commun. 2015;6:6337. doi: 10.1038/ncomms7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilbert R.J., Dalla Serra M., Anderluh G. Membrane pore formation at protein-lipid interfaces. Trends Biochem. Sci. 2014;39:510–516. doi: 10.1016/j.tibs.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 22.Gilbert R.J. Protein-lipid interactions and non-lamellar lipidic structures in membrane pore formation and membrane fusion. Biochim. Biophys. Acta. 2016;1858:487–499. doi: 10.1016/j.bbamem.2015.11.026. [DOI] [PubMed] [Google Scholar]
- 23.Bechinger B. Rationalizing the membrane interactions of cationic amphipathic antimicrobial peptides by their molecular shape. Curr. Opin. Colloid Interface Sci. 2009;14:349–355. [Google Scholar]
- 24.Bechinger B., Gorr S.U. Antimicrobial peptides: mechanisms of action and resistance. J. Dent. Res. 2017;96:254–260. doi: 10.1177/0022034516679973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lanio M.E., Morera V., de los Angeles Chavez M. Purification and characterization of two hemolysins from Stichodactyla helianthus. Toxicon. 2001;39:187–194. doi: 10.1016/s0041-0101(00)00106-9. [DOI] [PubMed] [Google Scholar]
- 26.Tejuca M., Dalla Serra M., Menestrina G. Sizing the radius of the pore formed in erythrocytes and lipid vesicles by the toxin sticholysin I from the sea anemone Stichodactyla helianthus. J. Membr. Biol. 2001;183:125–135. doi: 10.1007/s00232-001-0060-y. [DOI] [PubMed] [Google Scholar]
- 27.Laborde R.J., Sanchez-Ferras O., Lanio M.E. Novel adjuvant based on the pore-forming protein sticholysin II encapsulated into liposomes effectively enhances the antigen-specific CTL-mediated immune response. J. Immunol. 2017;198:2772–2784. doi: 10.4049/jimmunol.1600310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cruz-Leal Y., Grubaugh D., Lanio M.E. The vacuolar pathway in macrophages plays a major role in antigen cross-presentation induced by the pore-forming protein sticholysin II encapsulated into liposomes. Front. Immunol. 2018;9:2473. doi: 10.3389/fimmu.2018.02473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tejuca M., Anderluh G., Dalla Serra M. Sea anemone cytolysins as toxic components of immunotoxins. Toxicon. 2009;54:1206–1214. doi: 10.1016/j.toxicon.2009.02.025. [DOI] [PubMed] [Google Scholar]
- 30.Pentón D., Pérez-Barzaga V., Tejuca M. Validation of a mutant of the pore-forming toxin sticholysin-I for the construction of proteinase-activated immunotoxins. Protein Eng. Des. Sel. 2011;24:485–493. doi: 10.1093/protein/gzr002. [DOI] [PubMed] [Google Scholar]
- 31.Matsuzaki K., Sugishita K., Epand R.M. Relationship of membrane curvature to the formation of pores by magainin 2. Biochemistry. 1998;37:11856–11863. doi: 10.1021/bi980539y. [DOI] [PubMed] [Google Scholar]
- 32.Epand R.M. Lipid polymorphism and protein-lipid interactions. Biochim. Biophys. Acta. 1998;1376:353–368. doi: 10.1016/s0304-4157(98)00015-x. [DOI] [PubMed] [Google Scholar]
- 33.Ros U., Souto A.L., Alvarez C. Functional and topological studies with Trp-containing analogs of the peptide StII(1–30) derived from the N-terminus of the pore forming toxin sticholysin II: contribution to understand its orientation in membrane. Biopolymers. 2013;100:337–346. doi: 10.1002/bip.22211. [DOI] [PubMed] [Google Scholar]
- 34.Ros U., Carretero G.P.B., Alvarez C. Self-association and folding in membrane determine the mode of action of peptides from the lytic segment of sticholysins. Biochimie. 2019;156:109–117. doi: 10.1016/j.biochi.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 35.Corradi V., Sejdiu B.I., Tieleman D.P. Emerging diversity in lipid-protein interactions. Chem. Rev. 2019;119:5775–5848. doi: 10.1021/acs.chemrev.8b00451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee J., Cheng X., Im W. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 2016;12:405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berendsen H.J. Molecular dynamics simulations: the limits and beyond. In: Deuflhard P., Hermans J., Leimkuhler B., Mark A.E., Reich S., Skeel R.D., editors. Computational Molecular Dynamics: Challenges, Methods, Ideas. Springer; 1999. pp. 3–36. [Google Scholar]
- 38.Berendsen H.J., Postma J.P.M., Haak J. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 39.Hoover W.G. Canonical dynamics: equilibrium phase-space distributions. Phys. Rev. A Gen. Phys. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 40.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 41.Darden T., York D., Pedersen L. Particle mesh Ewald: an N-log (N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 42.Essmann U., Perera L., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 43.Hess B., Kutzner C., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 44.Abraham M.J., Murtola T., Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- 45.Huang J., MacKerell A.D., Jr. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 2013;34:2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barreto-Ojeda E., Corradi V., Tieleman D.P. Coarse-grained molecular dynamics simulations reveal lipid access pathways in P-glycoprotein. J. Gen. Physiol. 2018;150:417–429. doi: 10.1085/jgp.201711907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sengupta D., Leontiadou H., Marrink S.J. Toroidal pores formed by antimicrobial peptides show significant disorder. Biochim. Biophys. Acta. 2008;1778:2308–2317. doi: 10.1016/j.bbamem.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 48.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 49.Castillo N., Monticelli L., Tieleman D.P. Free energy of WALP23 dimer association in DMPC, DPPC, and DOPC bilayers. Chem. Phys. Lipids. 2013;169:95–105. doi: 10.1016/j.chemphyslip.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 50.Gu R.X., Baoukina S., Tieleman D.P. Cholesterol flip-flop in heterogeneous membranes. J. Chem. Theory Comput. 2019;15:2064–2070. doi: 10.1021/acs.jctc.8b00933. [DOI] [PubMed] [Google Scholar]
- 51.Corradi V., Mendez-Villuendas E., Tieleman D.P. Lipid-protein interactions are unique fingerprints for membrane proteins. ACS Cent. Sci. 2018;4:709–717. doi: 10.1021/acscentsci.8b00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pedrera L., Gomide A.B., Alvarez C. The presence of sterols favors sticholysin I-membrane association and pore formation regardless of their ability to form laterally segregated domains. Langmuir. 2015;31:9911–9923. doi: 10.1021/acs.langmuir.5b01687. [DOI] [PubMed] [Google Scholar]
- 53.Martinez D., Otero A., Lissi E. Effect of sphingomyelin and cholesterol on the interaction of St II with lipidic interfaces. Toxicon. 2007;49:68–81. doi: 10.1016/j.toxicon.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 54.Antonini V., Pérez-Barzaga V., Tejuca M. Functional characterization of sticholysin I and W111C mutant reveals the sequence of the actinoporin’s pore assembly. PLoS One. 2014;9:e110824. doi: 10.1371/journal.pone.0110824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bakrac B., Gutiérrez-Aguirre I., Anderluh G. Molecular determinants of sphingomyelin specificity of a eukaryotic pore-forming toxin. J. Biol. Chem. 2008;283:18665–18677. doi: 10.1074/jbc.M708747200. [DOI] [PubMed] [Google Scholar]
- 56.Epand R.M., Epand R.F. Lipid domains in bacterial membranes and the action of antimicrobial agents. Biochim. Biophys. Acta. 2009;1788:289–294. doi: 10.1016/j.bbamem.2008.08.023. [DOI] [PubMed] [Google Scholar]
- 57.Epand R.M., Vogel H.J. Diversity of antimicrobial peptides and their mechanisms of action. Biochim. Biophys. Acta. 1999;1462:11–28. doi: 10.1016/s0005-2736(99)00198-4. [DOI] [PubMed] [Google Scholar]
- 58.Xu X.P., Zhai D., Hanein D. Three-dimensional structure of Bax-mediated pores in membrane bilayers. Cell Death Dis. 2013;4:e683. doi: 10.1038/cddis.2013.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marquette A., Bechinger B. Biophysical investigations elucidating the mechanisms of action of antimicrobial peptides and their synergism. Biomolecules. 2018;8:E18. doi: 10.3390/biom8020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harmouche N., Aisenbrey C., Bechinger B. Solution and solid-state nuclear magnetic resonance structural investigations of the antimicrobial designer peptide GL13K in membranes. Biochemistry. 2017;56:4269–4278. doi: 10.1021/acs.biochem.7b00526. [DOI] [PubMed] [Google Scholar]
- 61.Bechinger B. The SMART model: Soft Membranes Adapt and Respond, also Transiently, in the presence of antimicrobial peptides. J. Pept. Sci. 2015;21:346–355. doi: 10.1002/psc.2729. [DOI] [PubMed] [Google Scholar]
- 62.Epand R.M. Membrane lipid polymorphism - relationship to bilayer properties and protein function. In: Dopico A.M., editor. Methods in Molecular Biology. Methods in Membrane Lipids. Humana Press; 2007. pp. 15–26. [DOI] [PubMed] [Google Scholar]
- 63.Aisenbrey C., Marquette A., Bechinger B. The mechanisms of action of cationic antimicrobial peptides refined by novel concepts from biophysical investigations. Adv. Exp. Med. Biol. 2019;1117:33–64. doi: 10.1007/978-981-13-3588-4_4. [DOI] [PubMed] [Google Scholar]
- 64.Matsuzaki K., Murase O., Miyajima K. An antimicrobial peptide, magainin 2, induced rapid flip-flop of phospholipids coupled with pore formation and peptide translocation. Biochemistry. 1996;35:11361–11368. doi: 10.1021/bi960016v. [DOI] [PubMed] [Google Scholar]
- 65.Mihajlovic M., Lazaridis T. Antimicrobial peptides in toroidal and cylindrical pores. Biochim. Biophys. Acta. 2010;1798:1485–1493. doi: 10.1016/j.bbamem.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alvarez C., Ros U., Lanio M.E. Biophysical and biochemical strategies to understand membrane binding and pore formation by sticholysins, pore-forming proteins from a sea anemone. Biophys. Rev. 2017;9:529–544. doi: 10.1007/s12551-017-0316-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maget-Dana R. The monolayer technique: a potent tool for studying the interfacial properties of antimicrobial and membrane-lytic peptides and their interactions with lipid membranes. Biochim. Biophys. Acta. 1999;1462:109–140. doi: 10.1016/s0005-2736(99)00203-5. [DOI] [PubMed] [Google Scholar]
- 68.Mesa-Galloso H., Delgado-Magnero K.H., Valiente P.A. Disrupting a key hydrophobic pair in the oligomerization interface of the actinoporins impairs their pore-forming activity. Protein Sci. 2017;26:550–565. doi: 10.1002/pro.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Valle A., Lopez-Castilla A., Schreier S. Cys mutants in functional regions of Sticholysin I clarify the participation of these residues in pore formation. Toxicon. 2011;58:8–17. doi: 10.1016/j.toxicon.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 70.Lakowicz J.R. Springer; New York: 2006. Principles of Fluorescence Spectroscopy. [Google Scholar]
- 71.Woo S.Y., Lee H. Effect of lipid shape on toroidal pore formation and peptide orientation in lipid bilayers. Phys. Chem. Chem. Phys. 2017;19:21340–21349. doi: 10.1039/c7cp02708g. [DOI] [PubMed] [Google Scholar]
- 72.Tieleman D.P., Marrink S.J. Lipids out of equilibrium: energetics of desorption and pore mediated flip-flop. J. Am. Chem. Soc. 2006;128:12462–12467. doi: 10.1021/ja0624321. [DOI] [PubMed] [Google Scholar]
- 73.Gottler L.M., Ramamoorthy A. Structure, membrane orientation, mechanism, and function of pexiganan--a highly potent antimicrobial peptide designed from magainin. Biochim. Biophys. Acta. 2009;1788:1680–1686. doi: 10.1016/j.bbamem.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Siegel D.P., Epand R.M. Effect of influenza hemagglutinin fusion peptide on lamellar/inverted phase transitions in dipalmitoleoylphosphatidylethanolamine: implications for membrane fusion mechanisms. Biochim. Biophys. Acta. 2000;1468:87–98. doi: 10.1016/s0005-2736(00)00246-7. [DOI] [PubMed] [Google Scholar]
- 75.Carretero G.P.B., Vicente E.F., Schreier S. Dissecting the mechanism of action of actinoporins. Role of the N-terminal amphipathic α-helix in membrane binding and pore activity of sticholysins I and II. PLoS One. 2018;13:e0202981. doi: 10.1371/journal.pone.0202981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Müller P., Schiller S., Herrmann A. Continuous measurement of rapid transbilayer movement of a pyrene-labeled phospholipid analogue. Chem. Phys. Lipids. 2000;106:89–99. doi: 10.1016/s0009-3084(00)00134-1. [DOI] [PubMed] [Google Scholar]
- 77.Álvarez C., Mancheño J.M., Lanio M.E. Sticholysins, two pore-forming toxins produced by the Caribbean Sea anemone Stichodactyla helianthus: their interaction with membranes. Toxicon. 2009;54:1135–1147. doi: 10.1016/j.toxicon.2009.02.022. [DOI] [PubMed] [Google Scholar]
- 78.García-Sáez A.J., Coraiola M., Salgado J. Peptides corresponding to helices 5 and 6 of Bax can independently form large lipid pores. FEBS J. 2006;273:971–981. doi: 10.1111/j.1742-4658.2006.05123.x. [DOI] [PubMed] [Google Scholar]
- 79.García-Sáez A.J., Chiantia S., Schwille P. Pore formation by a Bax-derived peptide: effect on the line tension of the membrane probed by AFM. Biophys. J. 2007;93:103–112. doi: 10.1529/biophysj.106.100370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Álvarez C., Casallanovo F., Schreier S. Binding of sea anemone pore-forming toxins sticholysins I and II to interfaces--modulation of conformation and activity, and lipid-protein interaction. Chem. Phys. Lipids. 2003;122:97–105. doi: 10.1016/s0009-3084(02)00181-0. [DOI] [PubMed] [Google Scholar]
- 81.Bechinger B., Lohner K. Detergent-like actions of linear amphipathic cationic antimicrobial peptides. Biochim. Biophys. Acta. 2006;1758:1529–1539. doi: 10.1016/j.bbamem.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 82.Matsuzaki K. Magainins as paradigm for the mode of action of pore forming polypeptides. Biochim. Biophys. Acta. 1998;1376:391–400. doi: 10.1016/s0304-4157(98)00014-8. [DOI] [PubMed] [Google Scholar]
- 83.Koller D., Lohner K. The role of spontaneous lipid curvature in the interaction of interfacially active peptides with membranes. Biochim. Biophys. Acta. 2014;1838:2250–2259. doi: 10.1016/j.bbamem.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 84.Lee M.T., Chen F.Y., Huang H.W. Energetics of pore formation induced by membrane active peptides. Biochemistry. 2004;43:3590–3599. doi: 10.1021/bi036153r. [DOI] [PubMed] [Google Scholar]
- 85.Basañez G., Sharpe J.C., Zimmerberg J. Bax-type apoptotic proteins porate pure lipid bilayers through a mechanism sensitive to intrinsic monolayer curvature. J. Biol. Chem. 2002;277:49360–49365. doi: 10.1074/jbc.M206069200. [DOI] [PubMed] [Google Scholar]
- 86.van den Bogaart G., Mika J.T., Poolman B. The lipid dependence of melittin action investigated by dual-color fluorescence burst analysis. Biophys. J. 2007;93:154–163. doi: 10.1529/biophysj.107.106005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee M.T., Hung W.C., Huang H.W. Many-body effect of antimicrobial peptides: on the correlation between lipid’s spontaneous curvature and pore formation. Biophys. J. 2005;89:4006–4016. doi: 10.1529/biophysj.105.068080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shi Y., Wan M., Fang W. Peptide-lipid interaction sites affect vesicles’ responses to antimicrobial peptides. Biophys. J. 2018;115:1518–1529. doi: 10.1016/j.bpj.2018.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Matsuzaki K. Membrane permeabilization mechanisms. Adv. Exp. Med. Biol. 2019;1117:9–16. doi: 10.1007/978-981-13-3588-4_2. [DOI] [PubMed] [Google Scholar]
- 90.Mescola A., Marín-Medina N., Alessandrini A. Magainin-H2 effects on the permeabilization and mechanical properties of giant unilamellar vesicles. J. Colloid Interface Sci. 2019;553:247–258. doi: 10.1016/j.jcis.2019.06.028. [DOI] [PubMed] [Google Scholar]
- 91.Paterson D.J., Tassieri M., Cooper J.M. Lipid topology and electrostatic interactions underpin lytic activity of linear cationic antimicrobial peptides in membranes. Proc. Natl. Acad. Sci. USA. 2017;114:E8324–E8332. doi: 10.1073/pnas.1704489114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.