Abstract
The specific binding of ligands by proteins and the coupling of this process to conformational changes is fundamental to protein function. We designed a fluorescence-based single-molecule assay and data analysis procedure that allows the simultaneous real-time observation of ligand binding and conformational changes in FeuA. The substrate-binding protein FeuA binds the ligand ferri-bacillibactin and delivers it to the ATP-binding cassette importer FeuBC, which is involved in bacterial iron uptake. The conformational dynamics of FeuA was assessed via Förster resonance energy transfer, whereas the presence of the ligand was probed by fluorophore quenching. We reveal that ligand binding shifts the conformational equilibrium of FeuA from an open to a closed conformation. Ligand binding occurs via an induced-fit mechanism, i.e., the ligand binds to the open state and subsequently triggers a rapid closing of the protein. However, FeuA also rarely samples the closed conformation without the involvement of the ligand. This shows that ligand interactions are not required for conformational changes in FeuA. However, ligand interactions accelerate the conformational change 10,000-fold and temporally stabilize the formed conformation 250-fold.
Significance
Ligand binding and the coupling of this process to conformational changes in proteins is fundamental to their function. We developed a single-molecule assay that allows the simultaneous observation of ligand binding and conformational changes in the protein FeuA. This allows for directly observing the ligand-binding process, ligand-driven conformational changes, and rare conformational transitions that are uncoupled from the ligand. These findings provide insights into the fundamental relation between ligand-protein interactions and protein conformational changes. Our findings are not only of interest to understand protein function, but the developed data analysis procedure allows the determination of (relative) distance changes in single-molecule Förster resonance energy transfer experiments in situations in which the donor and/or acceptor fluorophore are influenced by quenching.
Introduction
The non-covalent and specific interactions between ligands and proteins underlies almost all biological processes. The coupling of these binding events to conformational changes allows proteins to act as highly efficient enzymes, signal transducers, motors, switches, or pumps (1). Two basic models that describe the coupling between protein conformational changes and ligand binding are the induced-fit (2) and conformational selection mechanism (3). In the induced-fit mechanism, ligand interactions trigger a conformational change, whereas in the conformational selection mechanism, ligand interactions selectively stabilize a subset of conformations that already pre-exist in the unliganded protein (Fig. 1 A). Both mechanisms require intermediate states that are formed during the ligand-binding process. For example, when a protein switches between two conformational states, such as an open or a closed conformation (Fig. 1 A), an open-liganded state in the induced-fit mechanism and a closed-unliganded state in the conformational selection mechanism are essential intermediate states. However, the study of such transient and thermodynamically unstable states remains experimentally challenging.
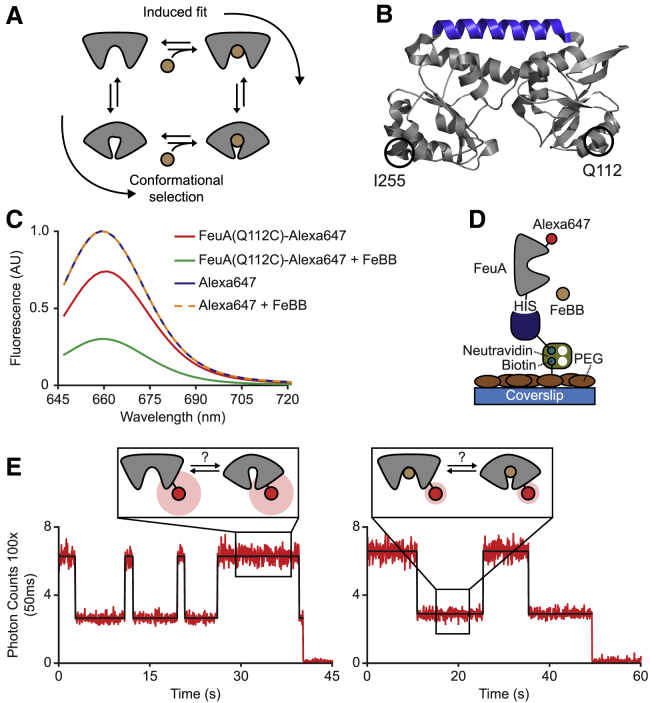
Figure 1.
Direct observation of ligand binding in FeuA using fluorescence quenching. (A) Ligand-binding mechanisms are shown. (B) X-ray crystal structure of the open-unliganded state of FeuA (Protein Data Bank [PDB]: 2WI8) is shown. Hinge region is indicated in blue. (C) Emission spectra of FeuA(Q112C) labeled with Alexa647 and free Alexa647 in the presence and absence of 5 μM FeBB are shown. (D) A schematic of surface-tethering FeuA proteins is given. (E) Fluorescence trajectories of FeuA(Q112C)-Alexa647 in the presence of 40 nM FeBB are shown. The fluorescent intensity (red) with the most probable state trajectory of the Poisson hidden Markov model (HMM) (black) is shown. The number of analyzed molecules is provided in Table S3. To see this figure in color, go online.
Primarily driven by high-resolution structural analysis of proteins adopting different conformations when free and in complex with ligand, an induced-fit mechanism was hypothesized for many proteins. However, development in single-molecule spectroscopy (4, 5, 6, 7), NMR (8, 9, 10), and other spectroscopic methods (11) revealed that proteins are highly dynamic and can undergo large conformational changes intrinsically, that is, in the absence of ligand. Examples are substrate-binding proteins of type I ABC importers (4), ABC exporters (7), adenylate kinase (9), RNase A (10), dihydrofolate reductase (12), ubiquitin (13), and DNA polymerase (14, 15). Because of the occurrence of intrinsic conformational changes, a conformational selection mechanism has been proposed to be the ligand-binding mechanism of many protein systems (3). However, the unambiguous determination of the binding mechanism requires simultaneous monitoring of ligand binding and the protein conformation. Moreover, the intrinsic conformational ensemble can, in principle, be investigated by studying the protein in the absence of ligand. However, trace contaminations of ligand can make this assessment experimentally difficult, especially under single-molecule conditions. To bypass these problems, we here established a fluorescence-based single-molecule assay and data analysis procedure that allows the simultaneous real-time observation of conformational changes and ligand binding in FeuA.
The substrate-binding protein FeuA is associated with the ATP-binding cassette (ABC) transporter FeuBC from Bacillus subtilis (16). This type II ABC importer is involved in the uptake of Fe3+ ions by an ATP-driven active transport of the siderophore bacillibactin (BB) in complex with Fe3+ (FeBB) (16). FeuA and other structurally related substrate-binding proteins (SBPs) (17) or domains (18) represent the primary receptors of bacterial ABC importers (19), tripartite ATP-independent periplasmic transporters (20), and others (17). These proteins capture the ligand from the external environment and deliver it to the membrane transporter for import into the cell. The structure of FeuA has a characteristic SBP fold (17) consisting of two subdomains connected by a hinge region, with ligand binding occurring between the subdomain interface (Fig. 1 B; (21)). Crystallography studies suggest that FeuA undergoes conformational changes involving a domain reorientation to engulf the ligand, leading to opening and closing of the protein (21). This apparently simple binary conformational switch, which is involved in molecular recognition, is investigated in this work to obtain insights into the coupling between ligand interactions and protein conformational changes.
Materials and Methods
Gene isolation, protein expression, and purification
The feuA gene (Uniprot: P40409) was isolated by PCR from the genome of B. subtilis subsp. subtilis str. 168. The primers (Table S1) were designed to exclude the signal peptide (amino acids 1–19), and cysteine 20 (which is probably post-translationally lipidated) with NdeI/HindIII restriction sites. The generated PCR fragment was A-tailed and ligated into the PGEM-T Easy Vector System (Promega, Madison, WI) (22). After removing the NdeI restriction site internal to the feuA gene by a silent mutation, the gene was subcloned in the pET20b vector (Merck, Darmstadt, Germany) using the NdeI/HindIII sites. Protein derivatives, including the cysteine and the silent mutation, were constructed using QuickChange mutagenesis (23). All sequences were checked for correctness by sequencing.
Cells harboring plasmids expressing the FeuAHis6 wild-type and derivatives were grown at 37°C until an optical density of 0.5 was reached. Protein expression was then induced by addition of 0.25 mM isopropyl β-D-1-thiogalactopyranoside. After 3 h of induction, cells were harvested. DNase 500 μg/mL (Merck) was added and passed twice through a French pressure cell at 1500 ψ. 2 mM phenylmethylsulfonyl fluoride was added to inhibit proteases. The soluble supernatant was isolated by centrifugation at 50,000 × g for 30 min at 4°C. The soluble material was purified and loaded on Ni2+-Sepharose resin (GE Healthcare, Chicago, IL) in 50 mM Tris-HCl (pH 8.0), 1 M KCl, 10% glycerol, 10 mM imidazole, 1 mM dithiothreitol (DTT). The immobilized proteins were washed (50 mM Tris-HCl (pH 8.0), 50 mM KCl, 10% glycerol, 10 mM imidazole, 1 mM DTT and subsequently with 50 mM Tris-HCl (pH 8.0), 1 M KCl, 10% glycerol, 30 mM imidazole, 1 mM DTT) and eluted (50 mM Tris-HCl (pH 8.0), 50 mM KCl, 10% glycerol, 300 mM imidazole, 1 mM DTT). Protein fractions were pooled (supplemented with 5 mM EDTA and 10 mM DTT), concentrated (10,000 MWCO Amicon; Merck), dialyzed against 100–1000 volumes of buffer (50 mM Tris-HCl, pH 8.0, 50 mM KCl, 50% glycerol, 10 mM DTT), aliquoted, and stored at −20°C.
Protein labeling
Labeling was performed with the maleimide dyes Alexa555 and Alexa647 (Thermo Fisher Scientific, Waltham, MA). The purified proteins were treated with 10 mM DTT for 30 min at 4°C to reduce oxidized cysteines. The protein sample was diluted to 1 mM DTT, immobilized on a Ni2+-Sepharose resin, and washed with 10 column volumes of buffer A (50 mM Tris-HCl (pH 7.4) and 50 mM KCl). The resin was incubated 2–8 h at 4°C with the dyes dissolved in buffer A. The molar dye concentration was 20 times higher than the protein concentration. Unbound dyes were removed by washing the column with 20 column volumes of buffer A and eluted with 400 mM imidazole. The labeled proteins were further purified by size-exclusion chromatography (Superdex 200; GE Healthcare) using buffer A. The sample composition was assessed by absorbance measurement at 280 nm (protein), 559 nm (Alexa555), and 645 nm (Alexa647) to determine labeling efficiency. For all samples, the labeling efficiency was >90%.
Ensemble fluorescence measurements
The fluorescence spectra were recorded on a scanning spectrofluorometer (Jasco FP-8300; Jasco, Tokyo, Japan). Emission spectra were recorded by excitation at 635 nm (5 nm bandwidth) in steps of 2 nm (2 nm emission bandwidth and 8 s integration time). Fluorescence anisotropy values were determined as described before (4). Fluorescence measurements were performed in buffer A at a concentration of 100–250 nM of labeled proteins and free fluorophores at room temperature.
Solution-based smFRET and ALEX
Solution-based single-molecule Förster resonance energy transfer (smFRET) and alternating laser excitation (ALEX) (24) experiments were performed using a home-built confocal microscope as described previously (4). In brief, two laser diodes (Coherent Obis; Coherent, Santa Clara, CA) with emission wavelengths of 532 and 637 nm were modulated in alternating periods of 50 μs. Average laser powers were 30 μW at 532 nm (∼30 kW/cm2) and 15 μW at 637 nm (∼15 kW/cm2). The fluorescence was collected by excitation at a depth of 20 μm and detected by two single-photon avalanche diodes (TAU-SPADs-100; Picoquant, Berlin, Germany). Measurements were carried out at 25–100 pM of labeled protein at room temperature in buffer A in the absence or presence of FeBB (EMC Biochemicals, Tübingen, Germany). Microscope no. 1.5H precision cover slides (VWR Marienfeld, Stockholm, Sweden) were coated with 1 mg/mL bovine serum albumin for 1 min, and unbound bovine serum albumin was subsequently removed by washing with buffer A.
Analysis of solution-based smFRET data
Photons were binned in 1 ms intervals, and only bins with a total of >200 photons considering all detection channels were analyzed. Three photon count rates were measured: N′DA (acceptor emission upon donor excitation), N′DD (donor emission upon donor excitation), and N′AA (acceptor emission upon acceptor excitation) (24). The background counts were estimated by excluding all time bins containing more than 20 counts and calculating the mean count rate over all remaining time bins. The leakage and direct excitation contributions were determined from the donor- and acceptor-only-labeled molecules as described by Lee et al. (25). Cross talk and background-correcting N′XY yields NXY. The proximity Förster resonance energy transfer (FRET) efficiency EPR is
| (1) |
and the stoichiometry S is
| (2) |
The donor- and acceptor-labeled protein (sub)populations within the EPR and S data set were clustered using a Gaussian mixture model, with one (apo) or two (holo) multivariate normal distributions. Molecules were assigned to the component yielding the highest posterior probability and are within 98% of the probability mass. For each cluster, the average NDA/NDD, NDD/NDA, and EPR were calculated. All postprocessing steps were programmed in MATLAB (MathWorks, Natick, MA).
Theory of interprobe distance ratio estimation
The distance between the donor and acceptor fluorophore r is related to the FRET efficiency E via
| (3) |
where R0 is the Förster radius and nDD and nDA are the background- and spectral-cross-talk-corrected donor and acceptor emission count rates when the donor is excited, respectively. γ = ϕAηA/ϕDηD depends on the donor and acceptor quantum yields—ϕD and ϕA, respectively—and the detection efficiencies of the donor and acceptor emission detection channels—ηD and ηA, respectively (25). Equation 3 can be rewritten as
| (4) |
Let r1 and r2 denote the (average) donor and acceptor fluorophore distance of two states, here denoted by state 1 and state 2. By using the definition of γ and noting that is proportional to ϕD, we find that the ratio between r1 and r2 satisfies
| (5) |
where ni ⋅ DA and ni ⋅ DD are the count rates nDA and nDD of state i (i = 1, 2), respectively, and ϕ1 ⋅ A and ϕ2 ⋅ A are the acceptor quantum yields of states 1 and 2, respectively. Let us now consider how the distance ratio (r1/r2)6 can be estimated from the data. We use the following notation: Ni ⋅ XY represents the measured count rate of ni ⋅ XY, that is, the background- and spectral-cross-talk-corrected count rate of Y emission (donor, acceptor) upon X excitation (donor, acceptor) when being in state i (i = 1, 2); and Ri and ri are the measured and true interprobe distances of state i (i = 1, 2), respectively. When we can assume that the relaxation times of the excited states of the fluorophores are short compared to the time between two consecutively detected photons, then there is no correlation between consecutive photons, and the distribution of Ni ⋅ XY can be approximated by a Poisson distribution (26). Then,
| (6) |
with
| (7) |
where k, p, and w denote the number of observations, is an unbiased and consistent estimator for (r1/r2)6. The sum in Eq. 7 extends over all observations, i.e., the total number of traces or time bins. When the acceptor quantum yield of both states is similar, we have ϕ1 ⋅ A = ϕ2 ⋅ A so that Eq. 5 becomes
| (8) |
An unbiased and consistent estimator for Eq. 8 is
| (9) |
In the presence of fluorophore quenching by the ligand, we use Eq. 6, and when no quenching occurs, we use Eq. 9 to infer the interprobe distance ratio. Details and a full derivation are provided in Appendix S1 in the Supporting Materials and Methods.
Scanning confocal microscopy
Confocal scanning microscopy was performed using a home-built confocal scanning microscope as described before (27, 28). In brief, scanning was performed using an XYZ-piezo stage (P-517-3CD with E-725.3CDA; Physik Instrumente, Karlsruhe, Germany). The detector signal was registered using a HydraHarp 400 ps event timer and a module for time-correlated single-photon counting (both Picoquant). Data were recorded with constant 532 nm excitation at intensities between 0.1 and 5 μW (∼25–1250 W/cm2) for smFRET measurements and 0.1 μW (∼25 W/cm2) when the protein was only labeled with Alex647 (640 nm excitation) or Alex555 (532 nm excitation). Scanning images of 10 × 10 μm were recorded with 50 nm step size and 1 ms integration time at each pixel. After each surface scan, the positions of labeled proteins were identified manually. Surface immobilization and a flow-cell arrangement was prepared as done previously (28, 29). Measurements were done at room temperature in buffer A with 1 mM 6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid and 10 mM cysteamine.
Analysis of fluorescence trajectories
Fluorescence trajectories were recorded in time bins of varying length as stated in the text or figure captions. We use the following notation: N′DY is the uncorrected count rate, N″DY is the background corrected count rate, and NDY is the background- and spectral-cross-talk-corrected count rate of Y emission (donor, acceptor) upon donor excitation. The apparent FRET efficiency is N′DA/(N′DD + N′DA). Only traces lasting longer than 20 time bins, having on average more than 10 photons per time bin that showed clear bleaching steps, were used for further analysis.
Equation 9 was used to estimate the interprobe distance ratio, with NDA/NDD = (N″DA − (l + dβγ)N″DD)/(1 + dβ)N″DD and NDD/NDA = (1 + dβ)N″DD/(N″DA − (l + dβγ)N″DD), where l, d, γ, and β are correction factors (25). Background was determined as the average count rate per channel when the fluorophores have bleached. The correction factors were determined using solution-based ALEX (25). In brief, the l and d factors were determined from the donor- and acceptor-only labeled FeuA molecules and the β and γ factors using the protein MalE (4) as reference standard. All correction factors were determined on the same microscope also used for the surface-based measurements (details of setup in Gouridis et al. (28)).
The state trajectories were modeled by a hidden Markov model (HMM) (30). For this, an implementation of HMM was programmed in MATLAB (MathWorks) as described previously (4). We assume that the FRET and acceptor-only and donor-only fluorescence trajectory can be considered as an HMM with only two states, having a one-dimensional Gaussian- or a Poisson-output distribution, respectively. The Gaussian distribution of state i (i = 1, 2) is defined by the average and variance. The Poisson distribution of state i (i = 1, 2) is defined by the average intensity of the acceptor or donor in state i. The likelihood function was maximized by using the Baum-Welch algorithm (31). The most probable state trajectory was found using the Viterbi algorithm (32). The individual lifetimes of state i were inferred from the most probable state trajectory and used to calculate the mean lifetime and the empirical cumulative distribution function of the lifetimes. Lifetimes that were only partially observed because of fluorophore bleaching were not included in the analysis. The relative population of state i was determined from total number of time bins the most probable state trajectories is in state i divided by the total number of time bins of the data set. The uncertainty of the relative population was determined from the SD of 105 bootstrapping steps on all traces. The quenching ratio for each molecule was obtained by taking the ratio of the intensity levels as obtained from the Poisson HMM.
Results
Direct observation of ligand binding and unbinding events
To investigate ligand binding by FeuA at the single-molecule level, we labeled FeuA with the Alexa647 fluorophore in one of its subdomains by introducing a single cysteine residue at a non-conserved position, which is solvent-exposed and distant from the binding pocket (Q112C; Fig. 1 B). First, we determined the emission spectra of FeuA-Alexa647 and free Alexa647 in the presence or absence of the ligand FeBB. We observed that the fluorescence intensity of FeuA-Alexa647 was quenched in the presence of 5 μM FeBB (Fig. 1 C). Because no quenching was observed for free Alexa647 (Fig. 1 C), we attribute this to binding of FeBB by FeuA.
To directly observe the binding and unbinding of ligand, the fluorophore emission of individual surface-tethered labeled proteins was measured over time by confocal scanning microscopy (Fig. 1 D). Representative fluorescence intensity trajectories of FeuA in the presence of 40 nM FeBB are shown in Fig. 1 E. All analyzed fluorescence trajectories show a single bleaching step, indicating that single molecules are examined (Fig. 1 E). Only in the presence of FeBB we observed stochastic switching between two intensity levels (Fig. 1 E), caused by fluorescence quenching of Alexa647 by FeBB. Thus, the intensity fluctuations can be interpreted as individual binding and unbinding events of FeBB to FeuA. To substantiate this claim, we determined the relative population of the lower intensity level to estimate the dissociation constant (KD) of FeBB binding by FeuA. From the analysis of 50 traces in the presence of 40 nM FeBB, we find that 66% of the time, FeuA is complexed with ligand. By using the Hill-Langmuir equation P = L/(L + KD), where P is the relative population of the ligand-bound state and L the ligand concentration, we obtain an estimated KD of 20 ± 3 nM (SD from bootstrapping; see also Fig. S1). This is in good agreement with the value obtained from intrinsic tryptophan fluorescence (KD = 27 ± 1 nM) (33). In summary, our assay can be used to directly probe the presence or absence of the ligand FeBB in FeuA. However, how the binding and unbinding events are coupled to the conformational changes in FeuA remains unclear.
Conformational states of FeuA
We used FRET to investigate whether ligand binding alters the FeuA conformation in solution and at room temperature. In our assay, each of the two subdomains was stochastically labeled with either a donor (Alexa555) or an acceptor fluorophore (Alexa647). Surface-exposed and non-conserved residues, showing large distance changes according to the crystal structures of the open and closed states (21), were chosen as cysteine positions for fluorophore labeling (Q112C/I255C; Fig. 1 B). The relationship between FRET efficiency and interprobe distance requires free fluorophore rotation, which was verified by steady-state anisotropy measurements (Table S2).
We used confocal microscopy with ALEX (24) to explore the conformational states of individual, freely diffusing proteins (Fig. 2 A). During the diffusional transit through the excitation volume of a confocal microscope, the labeled protein generates a short fluorescent burst, allowing for the determination of the apparent FRET efficiency and the stoichiometry S (see Materials and Methods for details). To retrieve interprobe distances, the apparent FRET efficiency was corrected for background and spectral cross talk to obtain the proximity ratio EPR. In our assays, changes in the apparent FRET efficiency and EPR can originate from interprobe distance changes, but also because of fluorophore quenching caused by FeBB binding (Fig. 2 B). Finally, S relates the total fluorescence recorded after donor excitation in the green and red detection channels to the total fluorescence after direct donor and acceptor excitation in each detection channel.
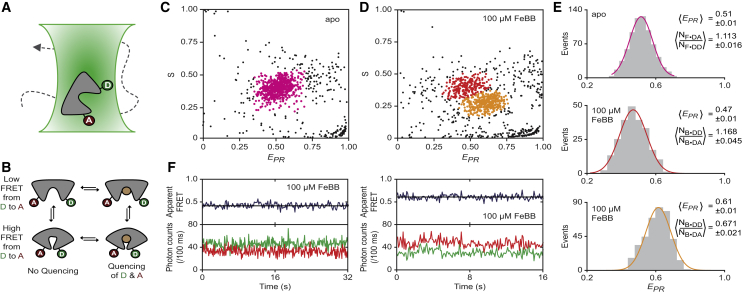
Figure 2.
Conformational changes of FeuA in solution. (A) A cartoon view of labeled proteins diffusing through the excitation volume of the confocal microscope is shown. (B) Quenching and FRET depend on the protein conformation and the ligand FeBB. (C and D) EPR and S of FeuA(Q112C/I255C) in the absence (C) and in the presence of 100 μM FeBB (D) are shown. The two donor- and acceptor-labeled protein populations in (D) are indicated in red and orange and the single population in (C) in pink. (E) EPR histogram of the selected molecules in (C) and (D). Color coding in (E) corresponds to the selected molecules in (C) and (D) with the same color. Solid line is a Gaussian fit. A 95% confidence interval for the average EPR and count rate ratios are shown. (F) Representative fluorescence trajectories of surface-immobilized FeuA(Q112C/I255C) in the presence of 100 μM FeBB are shown. Top panel shows calculated apparent FRET efficiency (blue) from the donor (green) and acceptor (red) photon counts as shown in the bottom panels. Black line indicates the average efficiency. The number of analyzed molecules is provided in Table S3. To see this figure in color, go online.
The EPR and S values of many individual proteins were acquired in the absence and presence of saturating concentrations of FeBB (100 μM) (Fig. 2, C and D). By separating donor-acceptor-labeled proteins from the donor- and acceptor-only-labeled proteins based on the S range, an EPR histogram was constructed (Fig. 2 E). The EPR histogram of ligand-free FeuA is unimodal and well fitted by a single Gaussian distribution. In the presence of 100 μM FeBB, two populations of donor-acceptor-labeled proteins are observed and are centered around different EPR and S values (Fig. 2, D and E). FRET analysis of surface-tethered proteins in the presence of 100 μM FeBB (83 traces) reveals that FeuA does not switch between these FRET states, i.e., fluorescence trajectories are obtained in either FRET state, with no switching between them (Fig. 2 F). The cysteine positions in the crystal structure have distinct distances to the ligand-binding site. Therefore, the two FRET states most likely arise because of the different donor- and acceptor-labeling positions. This is expected from stochastic labeling of two different cysteine positions, causing differences in fluorophore quenching by FeBB and thus differences in EPR and S values. Indeed, analysis of individual acceptor- and donor-only-labeled proteins shows that quenching is position- and fluorophore-dependent (Fig. 3; Table S3).
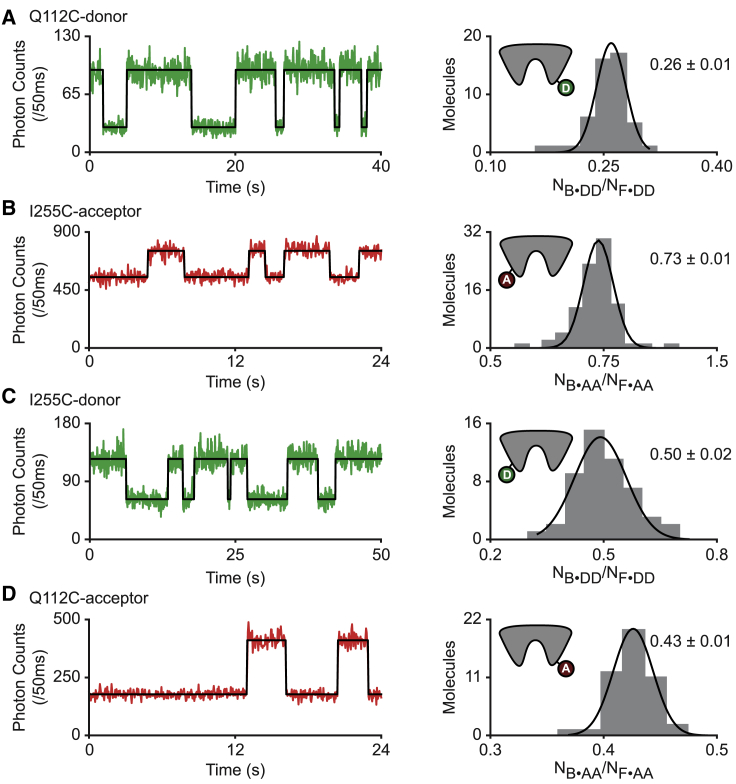
Figure 3.
Fluorescence quenching in FeuA depends on the labeling position and the fluorophore. (A–D) Fluorescence trajectories (left) of single-fluorophore-labeled FeuA in the presence of 25 nM FeBB (A and C) or 40 nM (B and D) and corresponding histogram of the count rate ratios NB⋅DD/NF⋅DD or NB⋅AA/NF⋅AA of all molecules (right) are shown. In the fluorescence trajectories, donor (green) and acceptor (red) count rate with the most probable state trajectory of the HMM (black) are shown. In the histogram, bars are the data and solid line a Gaussian fit. A 95% confidence interval for the average of the brightness ratios is indicated. The number of analyzed molecules is provided in Table S3. To see this figure in color, go online.
To correct the FRET efficiencies for fluorophore quenching, we related the populations in Fig. 2 D to their corresponding labeling positions. The low S value of the high FRET population in Fig. 2 D (orange population) implies that the quenching of the donor is more prominent than that of the acceptor. To quantify the quenching, we constructed and studied all four single-cysteine FeuA variants that were used in our FRET assays (Fig. 3). The quenching behavior of the high FRET population in Fig. 2 D (orange population) was observed to occur when Q112C was labeled with a donor and I255C with an acceptor (Fig. 3, A and B). The largely unaltered S value of the low FRET population in Fig. 2 D (red population) suggests that the donor and acceptor quenching is similar and was observed to occur when the labeling position is reversed, i.e., Q112C is labeled with an acceptor and I255C with a donor (Fig. 3, C and D).
To evaluate whether ligand binding causes conformational changes in FeuA, we developed an analysis scheme that 1) takes into account the influence of donor and acceptor quenching and 2) considers FRET between the donor and acceptor fluorophores. We show in the Materials and Methods and Appendix S1 in the Supporting Materials and Methods that
| (10) |
is an unbiased and consistent estimator for (rB/rF)6, where rB and rF are the true and unknown interprobe distances of the ligand-bound (B) and ligand-free (F) proteins, respectively, and RB and RF are the measured distances of rB and rF, respectively. In Eq. 10, Ni ⋅ XY denotes the measured count rate of Y emission (donor, acceptor) upon X excitation (donor, acceptor) when being in state i (bound, free), and denotes the average. Noteworthy, the distance ratio is independent of donor quenching, i.e., of NB ⋅ DD/NF ⋅ DD.
The average ratios NB ⋅ DD/NB ⋅ DA and NF ⋅ DA/NF ⋅ DD were obtained from the selected molecules in Fig. 2, C and D and noting that EPR = Ni ⋅ DA/( Ni ⋅ DA + Ni ⋅ DD) (Fig. 2 E). The average ratio NB ⋅ AA/NF ⋅ AA was obtained from individual acceptor-only-labeled proteins and by determining the ratio between the acceptor fluorescence intensity of the free or ligand-bound states (Fig. 3, B and D). All values used for the interprobe distance ratio estimation are summarized in Table 1. Finally, by using Eq. 10, we find that the interprobe distance ratio rB/rF when Q112C is labeled with the donor and I255C with the acceptor is estimated to be 0.90 ± 0.01 (95% confidence interval) and remains unaltered (0.91 ± 0.01) when the labeling position is reversed (Table 1). This is in agreement with the value obtained from the crystal structures (21), because the Cα-Cα distance between residues Q112 and I255 in the ligand-bound structure is reduced to 0.86 of the distance in the ligand-free structure.
Table 1.
Interprobe Distance Ratio Estimation
| Assumed labeling position | |||||
|---|---|---|---|---|---|
| Q112C donor and I255C acceptor | 0.733 ± 0.010 | 0.671 ± 0.021 | 1.113 ± 0.016 | 0.547 ± 0.020 | 0.904 ± 0.006 |
| Q112C acceptor and I255C donor | 0.432 ± 0.008 | 1.168 ± 0.045 | 1.113 ± 0.016 | 0.562 ± 0.025 | 0.908 ± 0.007 |
Error denotes a 95% confidence interval.
In summary, the analysis reveals that FeBB induces a conformational change in FeuA in solution and at room temperature. Moreover, the reduced interprobe distance in the presence of ligand is consistent with the view that the conformational transition from the open to the closed state in FeuA and related SBPs (4) is triggered by ligand interactions. However, the precise mechanism of ligand binding and whether there are short-lived intermediate states or fast conformational transitions cannot be concluded from these measurements.
Rare intrinsic conformational transitions in FeuA
To directly observe how binding and unbinding of the ligand FeBB are coupled to conformational changes in FeuA and to obtain insights into conformational dynamics, individual proteins were studied for extended times by investigating surface-tethered FeuA molecules with smFRET. It is worthwhile to note that in our surface-based smFRET assays we do not determine absolute distances, but only monitor relative interprobe distance changes by determining the instrument-dependent apparent FRET efficiency.
With this, we investigated the dynamics of ligand-free FeuA and addressed whether the protein can also close intrinsically, that is, when the ligand is absent. Compared to the solution-based smFRET experiments, examining individual surface-tethered proteins greatly increases the sensitivity to detect rare events. To investigate a truly ligand-free protein, a high concentration of unlabeled FeuA protein (∼20 μM) was added to scavenge any potential ligand contamination that could otherwise cause ligand-induced closing events.
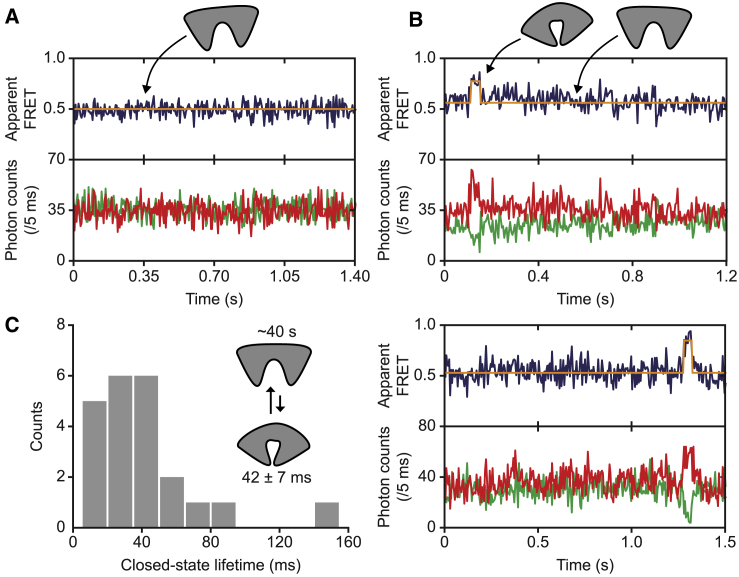
Consistent with the solution-based smFRET measurements (Fig. 2 C), FeuA is in a single FRET state (the open conformation) in the majority of the fluorescence-trajectories (437 traces out of 459; Fig. 4 A). In these traces, FeuA shows no detectable changes in the apparent FRET efficiency (or fluorescence intensity) related to conformational changes. However, in a small number of traces (22 traces out of 459), we observed rare transitions to a high FRET state, suggesting that FeuA can intrinsically close (Fig. 4 B). Indeed, by using Eq. 9 (see Materials and Methods), the average interprobe distance ratio between this high and low FRET state is 0.88 ± 0.02 (95% confidence interval). This shows that the interprobe distance is reduced and inferred to represent closure of the protein. Importantly, the absence of additional quenching effects provides direct evidence that the conformational change occurs independently of FeBB. Despite the low number of closing transitions that could be detected in the 459 traces (22 events), approximate estimates for the kinetics can be made. The ligand-free closed state has an average lifetime of 42 ± 7 ms (mean ± SEM; Fig. 4 C), and from the total observation time of ∼15 min, we estimate that this state is formed on average only once every ∼40 s (15 min/22 transitions). In summary, ligand-free FeuA is predominantly in an open conformation and can extremely rarely close, leading to the formation of a short-lived closed conformation.
Figure 4.
Ligand-free conformational dynamics of FeuA. (A and B) Fluorescence trajectories of surface-immobilized FeuA in the absence of ligand are shown. The top panel shows apparent FRET efficiency (blue), which was derived from the donor (green) and acceptor (red) photon count rates shown in the bottom panel. The number of analyzed molecules is provided in Table S3. (C) A histogram of the lifetime of the ligand-free closed state is shown. The cartoon depicts the ligand-free open and closed states with their estimated lifetimes. To see this figure in color, go online.
Ligand-bound FeuA is in the closed conformation
We then investigated the conformational dynamics of the ligand-bound protein. In our assay, the total fluorescence intensity reports on the presence of the ligand, whereas additional apparent FRET efficiency changes are indicative of protein conformational changes. However, in the 173 fluorescence-trajectories that were recorded in the presence of ∼KD concentrations of FeBB, we could not observe any FRET changes within the period a ligand was bound by FeuA (Fig. 5 A; Document S1. Supporting Materials and Methods, Fig. S1, and Tables S1–S3, Document S2. Article plus Supporting Material). The average apparent FRET efficiency of the initial and final 200 ms of the ligand-binding events were not significantly different from the period in between that (p = 0.28, one-way analysis of variance; Fig. 5 B). This suggests that once the ligand is bound, FeuA remains closed, and other conformational transitions, such as the formation of an open-liganded state, do not occur (or occur on timescales faster than the time resolution of 200 ms; see below).
Figure 5.
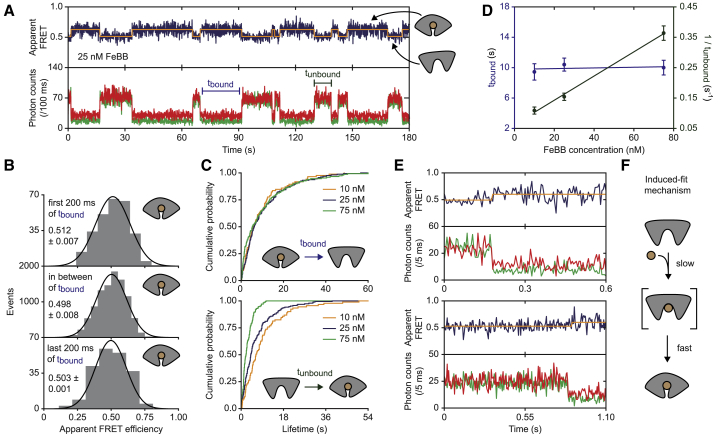
FeuA binds ligand via an induced-fit mechanism. (A) The fluorescence trajectory of FeuA(Q112C/I255C) is shown. In all fluorescence trajectories presented in the figure, the top panel shows apparent FRET efficiency (blue) and donor (green) and acceptor (red) photon counts are shown in the bottom panel. Orange lines are the most probable state trajectory of the HMM. (B) An apparent FRET efficiency histogram of the first and last 200 ms of the binding event and the period between that is given. Mean ± SEM is indicated. (C) Cumulative distribution function of the time FeuA has FeBB bound (tbound; top) or is free (tunbound; bottom) is shown at different FeBB concentrations. (D) tbound (purple) and the rate of binding (1/tunbound; green) as a function of FeBB concentration are shown. Data are mean ± SEM and the solid line is a linear fit. From the fit, binding and unbinding rates of ∼107 M−1 s1 and 0.10 s−1 are obtained. (E) Fluorescence trajectories of FeuA showing a single binding event are given. (F) A schematic of the induced-fit mechanism is shown. Number of analyzed molecules is provided in Table S3. To see this figure in color, go online.
With this in mind, we determined the lifetime of the ligand-bound state at varying FeBB concentrations (173 traces total). We observed that the lifetime of this state was largely concentration independent (p = 0.80, one-way analysis of variance) having an average lifetime of 10.0 ± 0.6 s (mean ± SEM; Fig. 5, C and D). Interestingly, the ligand-bound closed conformation is 250-fold longer-lived than the intrinsic closed state (10.0 ± 0.6 s vs. 42 ± 7 ms).
The ligand is bound via the induced-fit mechanism
Next, we investigated which conformational state binds the ligand (intrinsically closed or the open state), and thus, whether the ligand binds via a conformational selection or induced-fit mechanism (Fig. 1 A). We expect that when the intrinsic closed conformation binds the ligand, the ligand-binding frequency would be limited by the intrinsic closing frequency (∼1.5 min−1). So, we determined the lifetimes of the ligand-free states at varying FeBB concentrations (Fig. 5, C and D; Fig. S1 A). We observed that the binding frequency increases with FeBB concentration and is already 6.5 ± 0.6 min−1 (mean ± SEM) for the lowest concentration used (10 nM FeBB) (Fig. 5 D). Thus, ligand binding occurs at a faster rate than the intrinsic closing transitions. These data are consistent with an induced-fit mechanism. In addition, in 60 traces that were recorded with a higher time resolution of 5 ms, we observed that FeuA is in the open state before the ligand binds (Fig. 5 E). The average apparent FRET efficiency of the 10 ms period before the ligand binds (0.511 ± 0.014, mean ± SEM) and the period before that (0.516 ± 0.001, mean ± SEM), when the protein is in the open conformation, are not significantly different (p = 0.70, two-tailed unpaired t-test). This demonstrates that the open conformation binds the ligand and that ligand binding occurs via the induced-fit mechanism (Fig. 5 F).
The open-liganded state is extremely short-lived
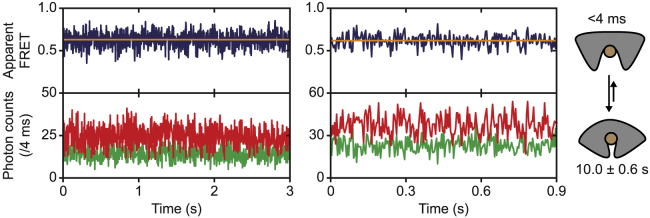
An essential intermediate state of the induced-fit mechanism is the open-liganded state (Fig. 1 A). Based on our data, we can already conclude that the open-liganded state has to be shorter-lived than 200 ms (see Ligand-Bound FeuA Is in the Closed Conformation). To further investigate the lifetime of this state, we increased the excitation intensity to obtain a time resolution of 4 ms. To probe the open-liganded state, we used saturated concentrations of FeBB. Under these conditions, the ligand-free open conformation is expected to be absent, and any detectable open state would consequently correspond to the open-liganded form. However, by examining 94 individual FeuA molecules with a total observation time of 104 s, we could not detect any opening transitions (Fig. 6). All these traces show FRET fluctuations, but those could not be separated from noise or did not originate from a clear anticorrelated donor and acceptor fluorescence change, as expected for real changes in FRET efficiency.
Figure 6.
Ligand-bound conformational dynamics of FeuA. Fluorescence trajectories of surface-immobilized FeuA(Q112C/I255C) in the presence of 100 μM FeBB are shown. Time resolution is 4 ms. The top panel shows calculated apparent FRET efficiency (blue) from the donor (green) and acceptor (red) photon counts as presented in the bottom panel. The orange lines indicate the average apparent FRET efficiency value. The cartoon depicts the ligand-bound open and closed states with the estimated lifetimes indicated. The number of analyzed molecules is provided in Table S3. To see this figure in color, go online.
Although we could not directly observe the open-liganded state, we can provide an upper bound for its lifetime. Based on the time resolution of the measurement, we conclude that the open-liganded state should be shorter-lived than 4 ms. Thus, when FeuA is in complex with FeBB, closing is accelerated more than 10,000-fold compared to when the ligand is absent (from <4 ms to 40 s). This suggests a mechanism in which the ligand drastically accelerates the conformational change that is already present in the ligand-free protein.
The energy landscape of FeuA
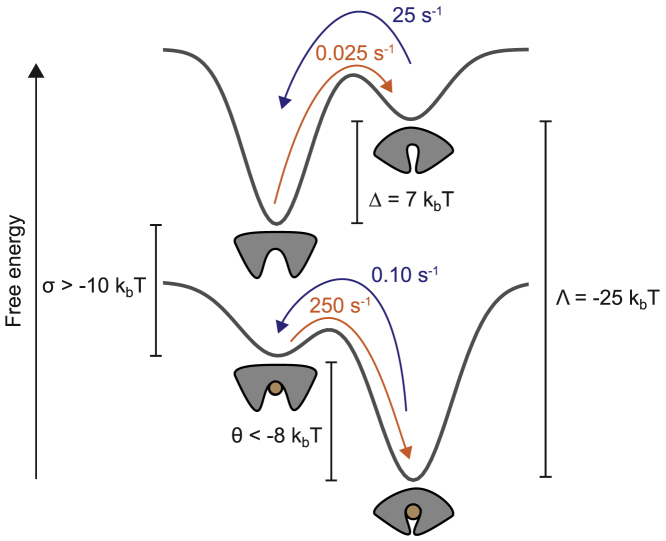
So far, we focused on the dynamic aspects of the molecular recognition process, that ultimately originates from the precise architecture of the energy landscape of the protein. Next, we used our single-molecule results to determine the thermodynamic properties of FeuA (Fig. 7).
Figure 7.
The conformational landscape of FeuA. A schematic representation of the thermodynamics of ligand binding and the conformational states of FeuA is given. Details on the determination of the energy values and rates can be found in the Results. To see this figure in color, go online.
The binding process can most easily be treated within the context of Gibbs ensembles. The grand partition function Ω(T, μ) of a single protein-ligand system, as shown in Fig. 7, is
| (11) |
where β is (kBT)−1, kB is the Boltzmann constant, T is the absolute temperature, Gi is the free energy of state i (O, open-unliganded; C, closed-unliganded; OL, open-liganded; CL, closed-liganded), and μ is the chemical potential. We assume that the ligand solution can be treated as ideal, so μ = μ0 + kBTlnL, where L is the ligand concentration (relative to 1 Molar) and μ0 is the standard chemical potential (μ = μ0 when L = 1).
The probability that the protein is in the intrinsic closed conformation is
| (12) |
where Δ = GC − GO is the ligand-free protein conformational free energy. From the fraction of time spend in the intrinsic closed conformation in the absence of ligand (Fig. 4), we find that P(C;L = 0) is 10−3, so Δ = 7 kBT.
In the presence of ligand, the fraction of proteins occupied by a ligand is
| (13) |
By treating μ as an ideal ligand solution (), we find that Eq. 13 is equal to
| (14) |
Equation 14 can be expressed as the Hill-Langmuir equation,
| (15) |
where KD is the dissociation constant as determined in our study (Fig. S1 B). By making the approximation that (see below), we find that KD is equal to
| (16) |
where Λ = (GCL − μ0) − GC is the protein-ligand interaction free energy of the closed conformation. We find that FeuA binds FeBB with a KD of 15–20 nM (Fig. S1 B), so Λ = −25 kBT.
When the protein is saturated with ligand, the probability of being in the closed conformation is
| (17) |
where θ = GCL − GOL is the ligand-bound protein conformational free energy and P(OL;L = ∞) = 1 − P(CL;L = ∞). We could not directly observe the OL state in the smFRET measurements (Fig. 6), but we can calculate an upper bound for P(OL;L = ∞). An estimator for P(OL;L = ∞) is
| (18) |
where τOL and τCL are the lifetimes of the OL and CL states, respectively. By using τOL < 4 ms and τCL = 10.0 s, we find that P(OL;L = ∞) < 410−4 so that θ < −8 kBT. Finally, we find that σ = (GOL − μ0) − GO = Δ + Λ − θ > −10 kBT, where σ is the protein-ligand interaction free energy of the open conformation.
Taken together, in the absence of ligand, the closed conformation is thermodynamically unstable and requires an energy input of Δ = 7 kBT for its formation. The situation is completely reversed when FeuA has a ligand bound: the open conformation is thermodynamically unstable and requires an energy input of more than −θ = 8 kBT for its formation. Furthermore, from the fact that Λ − σ = (GCL − GC) − (GOL − GO) < −15 kBT, we conclude that the protein-ligand interactions are at least 15 kBT stronger in the closed conformation compared to the open conformation.
Discussion
Modulation of the protein conformational landscape by ligand interactions is fundamental to the function and regulation of proteins. Here, we used a single-molecule approach to investigate the ligand-binding process and to understand how this is coupled to the conformational dynamics of the protein. For this, we analyzed the protein conformation and changes thereof via FRET and visualized the presence or absence of a ligand molecule via fluorophore quenching.
From the analysis of FeuA, combined with recent work on other proteins (4, 9, 10, 12, 34), a picture emerges in which ligand-bound protein conformations are also part of the conformational ensemble of the unliganded protein. It appears that ligand binding only alters the free energies of the equilibrium states and the barriers between them. For FeuA, the equilibrium lies toward the open conformation in the absence of ligand, with the closed conformation being 7 kBT higher in free energy. On the other hand, when FeuA has a ligand bound, the closed conformation is more than 8 kBT lower in free energy than the open conformation. Because of these large free energy differences, it is clear that in FeuA, there is a strong, but not an absolute, coupling between ligand binding and protein conformational changes. This shows that nature has fine-tuned the conformational landscape to approximate the behavior of FeuA by a simple binary switch that is regulated by ligand interactions.
Two basic mechanisms that connect the open and closed conformations with the unliganded and liganded states are the induced-fit and the conformational selection mechanism (Fig. 1 A). In the induced-fit mechanism, the ligand binds to the open conformation and triggers closing, whereas in the conformational selection mechanism, the ligand is directly bound by the closed conformation (Fig. 1 A). We demonstrate that ligand binding occurs via the induced-fit mechanism, by showing that the ligand binds to the open conformation and triggers closing (Fig. 5). We could argue that the conformational landscape provides the required directionality for the induced-fit mechanism. If ligand binding was to occur via the conformational selection mechanism, a substantial amount of thermal energy (Δ = 7 kBT) would be required to form the ligand-competent, intrinsic closed conformation, making the process highly inefficient (Fig. 7). The induced-fit mechanism would be more efficient because no thermodynamically unfavorable intermediate states need to be formed during the binding process (σ > −10 kBT, θ < −8 kBT; Fig. 7).
By using sensitive single-molecule methods, we observed that ligand-free FeuA can sample a temporally and thermodynamically unstable state that is different from the open conformation (Fig. 4). We provide direct evidence that the conformational change occurs independently of the ligand FeBB, because the FRET change occurs without any additional quenching effects that occurs when the ligand binds. Further investigations are required to characterize this conformation on a molecular level, but, based on the reduced interprobe distance relative to the open state, we infer that this rare protein state represents a closed conformation. We also note that we cannot exclude the existence of other conformations that exchange with the open conformation on the nanosecond-to-microsecond timescale. Such conformations could be other short-lived conformations such as a semi-closed state, as has been observed for the SBP maltose-binding protein (8). In this case, the apparent open state would be a temporal average of two states. For such scenarios, the ligand-binding mechanism could be more complex and might involve ligand binding to a short-lived conformation instead of the open conformation. To further elucidate this, methods with high(er) temporal resolution such as NMR (8), pulsed interleaved excitation spectroscopy (35), or multiparameter fluorescence detection (36) would be required.
To date, intrinsic closing has only been reported for SBPs of type I ABC importers (4, 28, 37, 38). ABC importers that employ SBPs can be subdivided as type I and II based on structural and mechanistic distinctions (19). FeuA belongs to the type II FeuBC importer, and, based on our data, we conclude that intrinsic closing occurs in SBPs of both type I and type II importers. Thus, some SBPs (4, 28, 37, 38), including FeuA, can close spontaneously and be triggered by ligands. In FeuA, ligand interactions drastically accelerate the closing transition more than 10,000-fold compared to the intrinsic closing rate (<4 ms vs. 40 s; Figs. 4 and 6). We speculate that once the open-liganded state is formed, direct ligand interactions pull the domains together, resulting in an acceleration of the closing transition.
The ligand does not only accelerate closing; it also temporally stabilizes this state by a factor of 250 (Figs. 4 and 5 D). Some insights into this temporal stabilization can be obtained from the crystal structures of FeuA (21). In the holo crystal structure, the ligand is engulfed by the protein, making favorable interactions with the residues of the binding site. Hence, a substantial input of (thermal) energy would be required to break these interactions and cross the energetic barrier to open the protein. In contrast, in the intrinsic closed conformation, these interactions are not present, allowing a more rapid crossing of the energy barrier.
Taken together, ligand interactions are not necessary for a conformational change in FeuA, however, these interactions accelerate the conformational change (10,000-fold) and temporally stabilize the formed conformation (250-fold). Both effects shift the conformational equilibrium towards the closed state. This shift may have been driven by mechanistic determinants to couple ligand-induced conformational changes in FeuA with transport in FeuBC. Ligand binding by FeuA and related SBPs via an induced-fit mechanism would allow the ABC transporter to discriminate between the ligand-free and ligand-bound states (39). The ligand-bound FeuA protein can be used to sense the presence of the correct ligand and initiate the transport process. Some SBPs have additional roles, as they are known to interact with chemoreceptors (40). Switching between two conformations allow the SBPs to transduce a signal, which is allosterically regulated by the ligand. Furthermore, we speculate that a wasteful conversion of chemical energy is prevented by the transient nature and high free energy of the intrinsic closed conformation, because any thermally driven mimic of the ligand-bound FeuA complex might be able to initiate the translocation cycle and consume the energy of ATP. In addition, a competition between the ligand-free and ligand-bound closed conformations to interact with the membrane-embedded transporter would inhibit substrate import (28).
As a final comment, we note that our data analysis approach to derive the interprobe distance ratio of two (conformational) states with altered quantum yield of donor and acceptor dyes could also be applied to situations in which FRET is changed because of protein-induced fluorescence enhancement (PIFE) (41, 42). The approach suggested here is particularly attractive for PIFE because the interprobe distance ratio is independent of the donor quantum yield, and thus Cy3, which is the most popular dye for PIFE, could be used in a straightforward fashion without additional knowledge of its quantum yield (changes).
Conclusions
We designed a single-molecule assay and data analysis procedure to probe the FeuA conformational changes via FRET and the presence of the ligand FeBB via fluorophore quenching. We show that FeuA exists in an open and a closed conformation in solution and at room temperature. In the absence of ligand, FeuA is (predominately) in the open state, and ligand shifts the equilibrium toward the closed state. Ligand binding occurs via the induced-fit mechanism, that is, a ligand binds to the open state and subsequently triggers a rapid closing of FeuA in less than 4 ms. Unbinding of the ligand also occurs almost simultaneously with the opening of the protein. However, FeuA also rarely samples the closed conformation without the involvement of the ligand, and thus, ligand interactions are not necessary for closing. However, such interactions accelerate the closing transition 10,000-fold and decrease the opening rate 250-fold in FeuA.
Author Contributions
M.d.B., G.G., and T.C. conceived and designed the study. T.C. supervised the project. G.G. and Y.A.M. performed the molecular biology. M.d.B. performed the measurements, analyzed the data, developed the theoretical work, and wrote the initial manuscript. M.d.B. and T.C. finalized the manuscript with input of all authors. All authors contributed to the discussion and interpretation of the results.
Acknowledgments
This work was financed by an Nederlandse Organisatie voor Wetenschappelijk Onderzoek Veni grant (722.012.012 to G.G.) and an European Research Council Starting Grant (No. 638536 – SM-IMPORT to T.C.). G.G. also acknowledges an European Molecular Biology Organization fellowship (long-term fellowship ALF 47–2012 to G.G.) and financial support by the Zernike Institute for Advanced Materials. G.G. is a Rega Foundation postdoctoral fellow. Y.A.M. was supported by the Indonesia Endowment Fund for Education (Layanan Beasiswa dan Pendanan PhD fellowship to Y.A.M.). T.C. was further supported by the Center of Nanoscience Munich, Deutsche Forschungsgemeinschaft within GRK2062 (project C03) and SFB863 (project A13), LMUexcellent, and the Center for Integrated Protein Science Munich.
Editor: Dimitrios Stamou.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.08.005.
Supporting Citations
Reference (43) appears in the Supporting Material.
Supporting Material
References
- 1.Alberts B., Johnson A., Walter P. Garland Science; New York: 2015. Molecular Biology of the Cell. [Google Scholar]
- 2.Koshland D.E. Application of a theory of enzyme specificity to protein synthesis. Proc. Natl. Acad. Sci. USA. 1958;44:98–104. doi: 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boehr D.D., Nussinov R., Wright P.E. The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 2009;5:789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Boer M., Gouridis G., Cordes T. Conformational and dynamic plasticity in substrate-binding proteins underlies selective transport in ABC importers. eLife. 2019;8:e44652. doi: 10.7554/eLife.44652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Husada F., Gouridis G., Cordes T. Watching conformational dynamics of ABC transporters with single-molecule tools. Biochem. Soc. Trans. 2015;43:1041–1047. doi: 10.1042/BST20150140. [DOI] [PubMed] [Google Scholar]
- 6.Kim E., Lee S., Kim H.S. A single-molecule dissection of ligand binding to a protein with intrinsic dynamics. Nat. Chem. Biol. 2013;9:313–318. doi: 10.1038/nchembio.1213. [DOI] [PubMed] [Google Scholar]
- 7.Husada F., Bountra K., Cordes T. Conformational dynamics of the ABC transporter McjD seen by single-molecule FRET. EMBO J. 2018;37:e100056. doi: 10.15252/embj.2018100056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang C., Schwieters C.D., Clore G.M. Open-to-closed transition in apo maltose-binding protein observed by paramagnetic NMR. Nature. 2007;449:1078–1082. doi: 10.1038/nature06232. [DOI] [PubMed] [Google Scholar]
- 9.Henzler-Wildman K.A., Thai V., Kern D. Intrinsic motions along an enzymatic reaction trajectory. Nature. 2007;450:838–844. doi: 10.1038/nature06410. [DOI] [PubMed] [Google Scholar]
- 10.Beach H., Cole R., Loria J.P. Conservation of mus-ms enzyme motions in the apo- and substrate-mimicked state. J. Am. Chem. Soc. 2005;127:9167–9176. doi: 10.1021/ja0514949. [DOI] [PubMed] [Google Scholar]
- 11.Busenlehner L.S., Armstrong R.N. Insights into enzyme structure and dynamics elucidated by amide H/D exchange mass spectrometry. Arch. Biochem. Biophys. 2005;433:34–46. doi: 10.1016/j.abb.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 12.Boehr D.D., McElheny D., Wright P.E. The dynamic energy landscape of dihydrofolate reductase catalysis. Science. 2006;313:1638–1642. doi: 10.1126/science.1130258. [DOI] [PubMed] [Google Scholar]
- 13.Lange O.F., Lakomek N.A., de Groot B.L. Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution. Science. 2008;320:1471–1475. doi: 10.1126/science.1157092. [DOI] [PubMed] [Google Scholar]
- 14.Santoso Y., Joyce C.M., Kapanidis A.N. Conformational transitions in DNA polymerase I revealed by single-molecule FRET. Proc. Natl. Acad. Sci. USA. 2010;107:715–720. doi: 10.1073/pnas.0910909107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rothwell P.J., Allen W.J., Seidel C.A. dNTP-dependent conformational transitions in the fingers subdomain of Klentaq1 DNA polymerase: insights into the role of the “nucleotide-binding” state. J. Biol. Chem. 2013;288:13575–13591. doi: 10.1074/jbc.M112.432690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miethke M., Klotz O., Marahiel M.A. Ferri-bacillibactin uptake and hydrolysis in Bacillus subtilis. Mol. Microbiol. 2006;61:1413–1427. doi: 10.1111/j.1365-2958.2006.05321.x. [DOI] [PubMed] [Google Scholar]
- 17.Berntsson R.P., Smits S.H., Poolman B. A structural classification of substrate-binding proteins. FEBS Lett. 2010;584:2606–2617. doi: 10.1016/j.febslet.2010.04.043. [DOI] [PubMed] [Google Scholar]
- 18.Schuurman-Wolters G.K., de Boer M., Poolman B. Protein linkers provide limits on the domain interactions in the ABC importer GlnPQ and determine the rate of transport. J. Mol. Biol. 2018;430:1249–1262. doi: 10.1016/j.jmb.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 19.Swier L.J.Y.M., Slotboom D.J., Poolman B. ABC transporters: a structure-function perspective. In: George A.M., editor. ABC Transporters - 40 Years on. Springer International Publishing; 2016. pp. 3–36. [Google Scholar]
- 20.Rosa L.T., Bianconi M.E., Kelly D.J. Tripartite ATP-independent periplasmic (TRAP) transporters and tripartite tricarboxylate transporters (TTT): from uptake to pathogenicity. Front. Cell. Infect. Microbiol. 2018;8:33. doi: 10.3389/fcimb.2018.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peuckert F., Miethke M., Marahiel M.A. Structural basis and stereochemistry of triscatecholate siderophore binding by FeuA. Angew. Chem. Int.Engl. 2009;48:7924–7927. doi: 10.1002/anie.200902495. [DOI] [PubMed] [Google Scholar]
- 22.Zhou M.Y., Clark S.E., Gomez-Sanchez C.E. Universal cloning method by TA strategy. Biotechniques. 1995;19:34–35. [PubMed] [Google Scholar]
- 23.Bok J.W., Keller N.P. Fast and easy method for construction of plasmid vectors using modified quick-change mutagenesis. Methods Mol. Biol. 2012;944:163–174. doi: 10.1007/978-1-62703-122-6_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kapanidis A.N., Lee N.K., Weiss S. Fluorescence-aided molecule sorting: analysis of structure and interactions by alternating-laser excitation of single molecules. Proc. Natl. Acad. Sci. USA. 2004;101:8936–8941. doi: 10.1073/pnas.0401690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee N.K., Kapanidis A.N., Weiss S. Accurate FRET measurements within single diffusing biomolecules using alternating-laser excitation. Biophys. J. 2005;88:2939–2953. doi: 10.1529/biophysj.104.054114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gopich I., Szabo A. Theory of photon statistics in single-molecule Förster resonance energy transfer. J. Chem. Phys. 2005;122:14707. doi: 10.1063/1.1812746. [DOI] [PubMed] [Google Scholar]
- 27.van der Velde J.H., Ploetz E., Cordes T. Mechanism of intramolecular photostabilization in self-healing cyanine fluorophores. Chemphyschem. 2013;14:4084–4093. doi: 10.1002/cphc.201300785. [DOI] [PubMed] [Google Scholar]
- 28.Gouridis G., Schuurman-Wolters G.K., Poolman B. Conformational dynamics in substrate-binding domains influences transport in the ABC importer GlnPQ. Nat. Struct. Mol. Biol. 2015;22:57–64. doi: 10.1038/nsmb.2929. [DOI] [PubMed] [Google Scholar]
- 29.Roy R., Hohng S., Ha T. A practical guide to single-molecule FRET. Nat. Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rabiner L.R. A tutorial on hidden markov models and selected applications in speech recognition. In: Waibel A., Lee K., editors. Readings in Speech Recognition. Morgan Kaufmann; 1990. pp. 267–296. [Google Scholar]
- 31.Baum L.E., Petrie T. Statistical inference for probabilistic functions of finite state Markov chains. Ann. Math. Stat. 1966;37:1554–1563. [Google Scholar]
- 32.Viterbi A.J. Error bounds for convolutional codes and an asymptotically optimum decoding algorithm. IEEE Trans. Inf. Theory. 1967;IT-13:260–269. [Google Scholar]
- 33.Miethke M., Skerra A. Neutrophil gelatinase-associated lipocalin expresses antimicrobial activity by interfering with L-norepinephrine-mediated bacterial iron acquisition. Antimicrob. Agents Chemother. 2010;54:1580–1589. doi: 10.1128/AAC.01158-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gouridis G., Hetzert B., Tampé R. ABCE1 controls ribosome recycling by an asymmetric dynamic conformational equilibrium. Cell Rep. 2019;28:723–734.e6. doi: 10.1016/j.celrep.2019.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Müller B.K., Zaychikov E., Lamb D.C. Pulsed interleaved excitation. Biophys. J. 2005;89:3508–3522. doi: 10.1529/biophysj.105.064766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sisamakis E., Valeri A., Seidel C.A. Accurate single-molecule FRET studies using multiparameter fluorescence detection. Methods Enzymol. 2010;475:455–514. doi: 10.1016/S0076-6879(10)75018-7. [DOI] [PubMed] [Google Scholar]
- 37.Flocco M.M., Mowbray S.L. The 1.9 A x-ray structure of a closed unliganded form of the periplasmic glucose/galactose receptor from Salmonella typhimurium. J. Biol. Chem. 1994;269:8931–8936. [PubMed] [Google Scholar]
- 38.Oswald C., Smits S.H., Bremer E. Crystal structures of the choline/acetylcholine substrate-binding protein ChoX from Sinorhizobium meliloti in the liganded and unliganded-closed states. J. Biol. Chem. 2008;283:32848–32859. doi: 10.1074/jbc.M806021200. [DOI] [PubMed] [Google Scholar]
- 39.Doeven M.K., van den Bogaart G., Poolman B. Probing receptor-translocator interactions in the oligopeptide ABC transporter by fluorescence correlation spectroscopy. Biophys. J. 2008;94:3956–3965. doi: 10.1529/biophysj.107.120964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y., Gardina P.J., Manson M.D. Model of maltose-binding protein/chemoreceptor complex supports intrasubunit signaling mechanism. Proc. Natl. Acad. Sci. USA. 1999;96:939–944. doi: 10.1073/pnas.96.3.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gidi Y., Götte M., Cosa G. Conformational changes spanning angstroms to nanometers via a combined protein-induced fluorescence enhancement-Förster resonance energy transfer method. J. Phys. Chem. B. 2017;121:2039–2048. doi: 10.1021/acs.jpcb.6b11495. [DOI] [PubMed] [Google Scholar]
- 42.Ploetz E., Lerner E., Cordes T. Förster resonance energy transfer and protein-induced fluorescence enhancement as synergetic multi-scale molecular rulers. Sci. Rep. 2016;6:33257. doi: 10.1038/srep33257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gopich I.V., Szabo A. Theory of the energy transfer efficiency and fluorescence lifetime distribution in single-molecule FRET. Proc. Natl. Acad. Sci. USA. 2012;109:7747–7752. doi: 10.1073/pnas.1205120109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.