Abstract
Adeno-associated virus (AAV) vectors are clinically proven gene delivery vehicles that are attracting an increasing amount of attention. Non-genome-containing empty AAV capsids are by-products during AAV production that have been reported to potentially impact AAV product safety and efficacy. Therefore, the presence and amount of empty AAV capsids need to be characterized during process development. Multiple methods have been reported to characterize empty AAV capsid levels, including transmission electron microscopy (TEM), analytical ultracentrifugation (AUC), charge detection mass spectrometry (CDMS), UV spectrophotometry, and measuring capsid and genome copies by ELISA and qPCR. However, these methods may lack adequate accuracy and precision or be challenging to transfer to a quality control (QC) lab due to the difficulty of implementation. In this study, we used AAV serotype 6.2 (AAV6.2) as an example to show the development of a QC-friendly anion exchange chromatography (AEX) assay for the determination of empty and full capsid percentages. The reported assay requires several microliters of material with a minimum titer of 5 × 1011 vg/mL, and it can detect the presence of as low as 2.9% empty capsids in AAV6.2 samples. Additionally, the method is easy to deploy, can be automated, and has been successfully implemented to support testing of various in-process and release samples.
Keywords: AAV, AAV6.2, Chromatography, Anion exchange chromatography (AEC), Empty capsids, AUC, High-throughput
Introduction
Adeno-associated virus (AAV) is a non-enveloped parvovirus with a diameter of approximately 25 nm.1 The AAV capsid is composed of 60 copies of capsid proteins adopting a T = 1 icosahedron shape.2 Packed into this near-spherical capsid is a linear single-stranded DNA genome, approximately 4.7 kb long. The viral genome contains two 145-nt-long inverted terminal repeats (ITRs), one at each end of the DNA strand. In wild-type AAVs, the ITRs flank three viral genes that encode replication, capsid, and assembly-activating proteins. In recombinant AAVs (rAAVs), the viral genome is replaced by a transgene while the ITR is retained for proper viral packaging and transfection.3
Recombinant AAV vectors are becoming a powerful tool of choice for in vivo gene delivery due to their unique features, including a lack of pathogenicity and the ability to transduce both nondividing and dividing cells with stable long-term gene expression in a wide variety of tissues.4,5 To date, there have been three rAAV-based gene therapies approved for commercial use: Glybera approved by the European Medicines Agency (EMA) for rare familial lipoprotein lipase deficiency; Luxturna approved by the US Food and Drug Administration (FDA) for rare retina disease; and, very recently, Zolgensma approved by the FDA for spinal muscular atrophy. Additionally, hundreds of clinical trials are currently ongoing using rAAVs.6,7
Scalable manufacturing of rAAVs has been demonstrated using both mammalian and insect cells.1,6,8 Independent of the manufacturing approaches, one characteristic of AAV production is the formation of empty capsids, which contain no vector genome. The level of empty capsids can vary from 10% to 90%, as visualized by transmission electron microscopy (TEM).9 The effects of empty capsids on therapeutic outcomes are not yet fully understood. Gao et al.10 reported that empty capsids reduced transduction efficiency and increased hepatic transaminase levels in mouse models, which indicated exacerbated side effects. On the other hand, there is accumulated evidence suggesting that empty capsids can serve as decoys to overcome AAV clearance and improve gene transfer efficiency.9,11 Whether an impurity or a decoy, empty capsids need to be closely monitored to ensure consistent product quality from batch to batch.12,13
Multiple methods have been described to quantify empty capsids in AAV samples. TEM has been widely employed to visualize AAV particles after negative staining.14 The empty and genome-containing capsids (full capsids) can be differentiated using electron micrographs and counted for quantification. Exploiting the difference in protein and DNA UV spectra, Sommer et al.15 measured the absorbance at 260 and 280 nm to calculate the ratio and absolute concentration of capsid and vector genome. A sample denaturing step was needed to minimize the interference from Rayleigh scattering during UV spectroscopy experiments. Burnham et al.16 applied analytical ultracentrifugation (AUC) to characterize AAV subspecies packaged with none, fragmented, or intact genomes. These species have different buoyant densities, and they can be resolved by a sedimentation velocity experiment coupled with sophisticated modeling analysis. Pierson et al.17 applied charge detection mass spectrometry (CDMS) to determine molecular weight distribution of AAV particles by concurrently measuring the mass charge ratio and the charge of individual ions. Additionally, capsid and genome copies can be determined by ELISA and qPCR,18 respectively, to estimate the ratio of empty and full capsids. All these methods are fundamentally different and provide complementary insights into AAV sample quality. Nevertheless, a robust, versatile, high-throughput, and sample-sparing method that requires minimal expertise is highly desirable to support the ever-growing activities in AAV process development. A chromatography-based method could potentially address these requirements.
Anion exchange chromatography (AEX) has been used for purification and enrichment of full AAV particles of serotypes 1, 2, and 8.19, 20, 21 However, the application of AEX for accurate quantification of empty capsid level in AAV has not been thoroughly evaluated in the literature. In this study, we used AAV serotype 6.2 (AAV6.2) as a model for the development of an AEX assay determining empty and full capsid percentages. The effects of mobile phase solvents on capsid stability and empty and full (E/F) capsid separation are studied in detail. The relative response of E/F AAV capsids by different detection techniques and the impact on quantification results are discussed along with the method performance and applications. Finally, a comparison of the AEX and AUC methods is illustrated using an affinity-purified AAV6.2 sample.
Results
Initial Column Screening and On-Column Stability of AAV Capsids
Ion exchange chromatography has long been used for downstream purification of AAVs.22, 23, 24 Enrichment of genome-containing full AAV particles after AEX purification has been reported,20,25 yet chromatograms of E/F separation have been shown for only the AAV1, 2, and 8 serotypes.19, 20, 21 Different quaternary ammonium-based strong anion exchange columns have been used for these separations successfully, including POROS 50 HQ for AAV1 and 2 and CIM-QA for AAV8. Qu et al.20 attempted to separate AAV6 empty and full capsids using the AAV2 method, without much success.
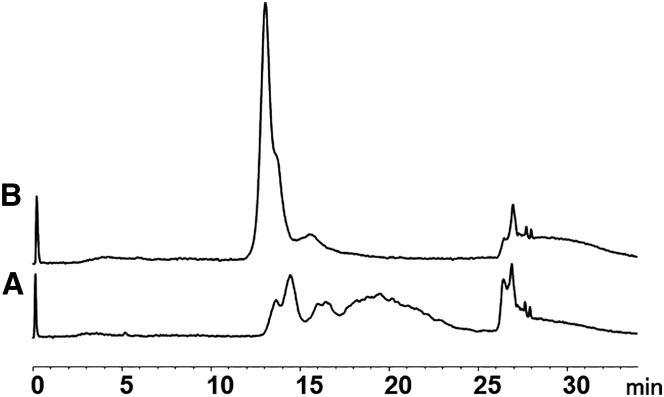
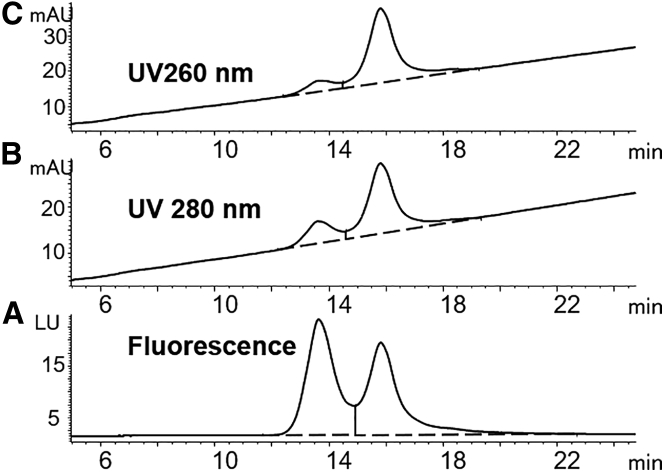
We started our method development for AAV6.2 E/F separation using the POROS 50 HQ and CIMac AAV full/empty column, which is a variation of the CIM-QA column advertised for AAV E/F separations. POROS resins are based on polystyrene-divinylbenzene beads with large pores, ranging in size from 0.05 to 1 mm. The CIMac AAV full/empty column is a monolithic column composed of polymethacrylate, containing interconnected, convective channels with a diameter of 1.3 mm. The monolithic structure allows high flow rates with low pressure drops, enabling increased sample throughput compared to traditional, bead-based resins. During our initial screenings on both columns, AAV6.2 resolved as multiple chromatographic peaks under certain conditions, especially when ionic strength was low (see Figure 1A as an example when the CIMac AAV full/empty column was used). More than five peaks were observed. The first two peaks (13–15 min) were assigned as empty and full AAV capsids, respectively, based on their UV profiles (i.e., A260/A280 ratio and maximum absorbance wavelength). The last set of peaks (17–25 min) was extremely broad, suggesting potential undesired nonspecific interactions or on-column degradation.
Figure 1.
Comparison of AAV6.2 Separations Using Mobile Phases
(A and B) Comparison of AAV6.2 separations using mobile phases without (A) and with (B) 2 mM MgCl2. Column, CIMac AAV full/empty; mobile phase condition, 50 mM Tris (pH 9.0) with 0–300 mM NaCl in 25 min. Fluorescence detection was used.
The structural integrity of AAV capsids under chromatographic conditions is a prerequisite for a separation method of empty and full capsids. The on-column stability of AAV capsids has not been reported previously as a concern. Divalent cations are known to stabilize non-enveloped viruses under thermal stress,26 and Urabe et al.19 included MgCl2 in the sample dilution buffer prior to loading AAV1 on preparative-scale columns. In our case, the presence of 2 mM MgCl2 resulted in the disappearance of the broad peaks and a dramatic increase in the intensity of the first two peaks (Figure 1B). Using a higher concentration of MgCl2 did not provide further improvement. Rather, the additional MgCl2 reduced the retention time and resolution between empty and full AAV particles. Calcium chloride was found to impair the separation more than MgCl2 did. Other agents, such as glycerol and Pluronic, were also evaluated, but no stabilizing effects were observed.
In the presence of MgCl2, a partial separation was observed on CIMac AAV full/empty column, which was selected for method optimization.
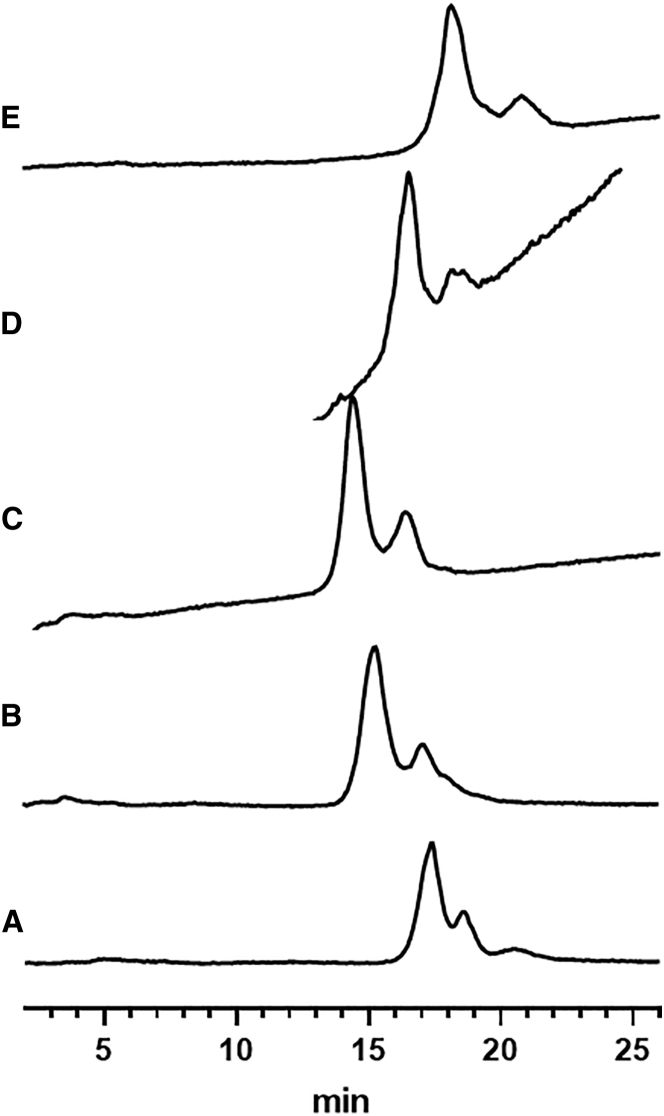
Salt Selection
Salts can potentially impact non-covalent interactions among capsids and interactions between AAV and column packing materials. For example, when studying poliovirus recovery from membrane filters, Farrah et al.27 observed increased hydrophobic adsorption to filters when antichaotropic salts were used. Urabe et al.19 reported improved E/F separation of AAV1 by using antichaotropes, such as NH4+ and (CH3)4N+, as compared to NaCl. Following a similar approach, we studied the salt effects on the separation of AAV6.2 by evaluating quaternary alkyl ammonium salts (QAASs) with different alkyl chains, including methyl, ethyl, propyl, and butyl. As shown in Figure 2, QAASs produced better separation than NaCl and the separation was greater when QAASs with a longer alkyl chain were used. Additionally, AAV stability gradually improved too, as evidenced by the disappearance of the last small peak observed under the NaCl condition, which was likely an on-column degradation product and was not observed in the final method. This result is consistent with the hypothesis that antichaotropes enhance the stabilizing hydrophobic interactions between viral capsid proteins. Other antichaotropes, including ammonium sulfate and sodium citrate, were also tested, but the separation was inferior to that obtained using QAASs. Furthermore, the recovery of AAV6.2 was negatively impacted when using sodium citrate as the elution salt.
Figure 2.
Comparison of AAV6.2 Separations Using Different Types of Salt
(A–E) Mobile phase conditions, 20 mM Tris (pH 9.0) and 2 mM MgCl2 with different salt gradients in 25 min. (A) 0–200 mM NaCl, (B) 0–300 mM (CH3)4NCl, (C) 0–300 mM (C2H5)4NCl, (D) 0–300 mM (C3H8)4NCl, and (E) 0–300 mM (C4H11)4NCl. Slightly different salt gradients were selected to produce similar retention times. Fluorescence detection was used.
Both tetraethyl and tetrapropyl ammonium salts showed strong UV absorbance in the range from 260 to 280 nm (data not shown). Tetrapropyl ammonium salt also caused a significant baseline drift of the fluorescence signal (Figure 2D). Tetrabutyl ammonium salt produced broader and less symmetric peaks, presumably due to an increased interaction between AAV and column packing materials. As a result, tetramethyl ammonium chloride was selected for the E/F separation.
pH and Buffering Agents
The isoelectric point (pI) of empty and full AAV capsids is around 6.3 and 5.9, respectively.7,28 Therefore, AEX separation of AAV6.2 was evaluated in the pH range from 7.5 to 9.0. At higher pH, AAV was more negatively charged, which resulted in longer retention time and better separation (Figure S1).
Different agents that provide buffering effects near pH 9 were evaluated for their impact on AAV6.2 E/F separation. The impact of buffering agents on the resolution between empty and full AAV capsids was found to be small. Bis-tris propane (BTP) was selected for the final method, because it produced a slightly better separation and did not interfere with either UV or fluorescence detection.
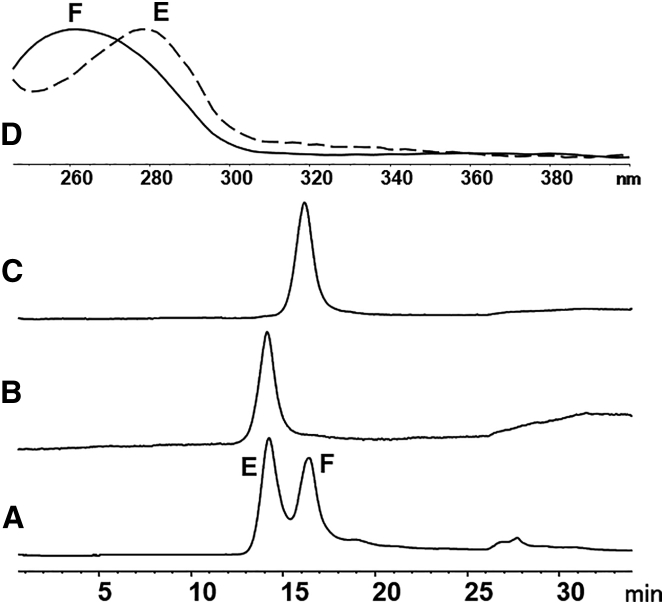
Confirmation of E/F Separation
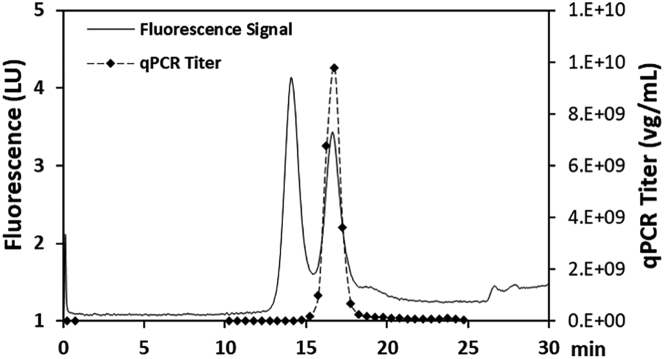
The assignment of the two peaks resolved by AEX as empty and full capsids was confirmed based on the following three observations. First, the retention times of the resolved peaks were identical to empty and full AAV6.2 standards, as shown in Figures 3A–3C. Second, the two peaks had distinct UV profiles that corresponded to empty and full AAV particles, respectively (Figure 3D). Finally, a representative AEX separation using the newly developed method was fractionated and analyzed by qPCR measurement of the genome copy number. The genomic DNA was only detected in the second chromatographic peak (Figure 4), while capsid was detected in both peaks by the capsid ELISA method (data not shown). These observations collectively confirmed that the first peak corresponded to empty capsid and the second peak corresponded to full AAV particles, as shown in Figures 3 and 4.
Figure 3.
Identification of AEX Peaks Using Purified Standards and UV Absorbance Spectra
(A–C) An affinity-purified AAV6.2 sample (A), empty AAV6.2 standard (B), and full AAV6.2 standard (C). (D) The UV spectra of the first peak (E, dashed line) and the second peak (F, solid line) from the sample shown in (A). Fluorescence detection was used.
Figure 4.
Overlay of Fluorescence Signal-Based Chromatography Profile (Solid Line) and qPCR Titer of Multiple Fractions over the Course of the Separation (Diamond Symbols, Dashed Line)
Reporting Empty and Full AAV Capsid Percentages
UV absorbance is the most commonly used detection method in chromatography. Empty and full AAV capsids have different UV responses because the viral genome contributes to UV absorption. The relative UV response factors of full to empty AAV6.2 particles are calculated to be 11.5 and 4.9 at 260 and 280 nm, respectively, using the published method.15,16 As a result, the full AAV particle concentration is overestimated by more than 10 times when monitored at 260 nm. Burnham et al.16 had similar observations when analyzing AUC data acquired at 260 nm. They found that the interference signal, which was based on refractive index, had much less response bias from empty and full AAV capsids. However, a higher sample concentration was required to compensate for lower sensitivity in interference AUC experiments.
We evaluated monitoring AAV E/F separation using fluorescence detection, a more sensitive technique than UV.29 Figure 5 compares the UV and fluorescence signals of a representative separation. The fluorescence signal had less baseline drift, improved signal-to-noise ratio, and less response bias between empty and full AAV particles, benefiting from minimal DNA intrinsic fluorescence. The quantitative calculation of protein and single-stranded DNA fluorescence is not straightforward. Empirically, we found that a relative response factor of 1.3 between full and empty capsids applied for AAV6.2 with different viral genomes. As shown in Table 1, after considering the relative response factors, both UV and fluorescence signals produced consistent quantification of empty and full AAV particle content. Fluorescence is the preferred detection method, mainly due to its higher sensitivity and significantly reduced response bias for empty and full capids.
Figure 5.
Separation of Empty and Full AAV6.2 Capsids Monitored by Different Detectors
(A–C) Fluorescence detection with 280-nm excitation and 340-nm emission (A), UV detection at 280 nm (B), and UV detection at 260 nm (C).
Table 1.
Quantification Results of Empty and Full AAV6.2 Capsid Content Using AEX Coupled with Different Detection Methods
| Detection | Area Percent E/F | Response Factor E/F | Percent E/F |
|---|---|---|---|
| UV 260 nm | 17.4/82.6 | 1/11.5 | 70.8/29.2 |
| UV 280 nm | 32.5/67.5 | 1/4.9 | 70.3/29.7 |
| Fluorescence | 65.2/34.8 | 1/1.3 | 70.9/29.1 |
Method Performance
Performance of the developed AEX method was evaluated for repeatability, intermediate precision, linearity, accuracy, and LOD/LOQ. The method qualification results for empty capsid percentages are presented in Table 2. The repeatability (n = 6) and intermediate precision (n = 6, different analyst, column, instrument, and day) are 1.1% and 2.0%, respectively, when using UV detection at 280 nm, and 0.5% and 1.5%, respectively, when using fluorescence detection. Excellent loading linearity (R2 = 0.9998) was observed. The LOD and LOQ were determined to be 1.6 × 1010 and 5.0 × 1010 vg/mL, respectively.
Table 2.
Summary of AEX Method Performance Evaluation Results for the Quantification of Empty Capsid Percentages
| Precision | UV280 nm |
Fluorescence |
||
|---|---|---|---|---|
| Mean | % CV | Mean | % CV | |
| Precision repeatability (6 injections) | 71.2 | 1.1 | 71.7 | 0.5 |
| Intermediate precision (3 injections/lab, 2 labs) | 72.6 | 2.0 | 72.2 | 1.5 |
| Loading Linearity (Fluorescence Detection) | ||||
| Linearity curve | y = 30.496 x + 0.6689 | |||
| R2 | 0.9998 | |||
| Linear rangea | 8.3 × 1010–8.3 × 1011 | |||
| LOD/LOQ | 1.6 × 1010/5.0 × 1010 | |||
| Empty % Linearity (Fluorescence Detection) | ||||
| Linearity curve | y = 1.0152 x – 0.0147 | |||
| R2 | 0.9983 | |||
| Linear range | 0–100 | |||
| LOD/LOQ (at nominal load of 5.0 × 1011) | 2.9%/8.8% | |||
| Accuracy (Fluorescence Detection, 3 Injections) | Recovery, % | % CV | ||
| Low (8.8%) | 90 | 4.3 | ||
| Middle (24.5%) | 95 | 1.3 | ||
| High (75.4%) | 99 | 0.4 | ||
The upper loading range can be extended by using AAV standards with higher titer.
Purified empty and full AAV capsids were mixed in defined ratios and measured by the developed AEX method. Chromatograms that cover the whole range are overlaid in Figures S2A–S2D. Excellent linearity (R2 = 0.9983) was obtained in the range of 0%–100% empty capsid (Figure S2E). At the loading of 5 × 1011, the LOD and LOQ of empty capsid were estimated to be 2.9% and 8.8%, respectively. The accuracy of the method was determined to be ≥90% at low, middle, and high levels of empty capsids (Table 2). These preliminary results demonstrated that the reported method can potentially be fully qualified to be a release assay for AAV6.2 empty capsid and full particle determination.
Discussion
Method Applications
The method reported in this study has been successfully applied to rAAVs produced by different upstream approaches, including transient transfection and helper virus-based expression. In-process samples from both column and density gradient purifications can also be analyzed as is. No sample preparation is needed even for samples in UV-absorbing matrices, such as iodixanol.
Comparing with Other Techniques
Existing methods for the quantification of empty and full AAV particles have limitations that prevent their broader use for lot release or characterization. For example, TEM provides more qualitative information and is not an accurate quantitative method.16 Capsid and genome quantification by ELISA/qPCR or UV absorbance suffer from poor accuracy. TEM and AUC experiments are low throughput and often require a dedicated facility and/or specially trained analysts. Among the existing methods, AUC is the most widely adopted method for characterization and quantitative measurement of empty and full AAV capsid percentages.
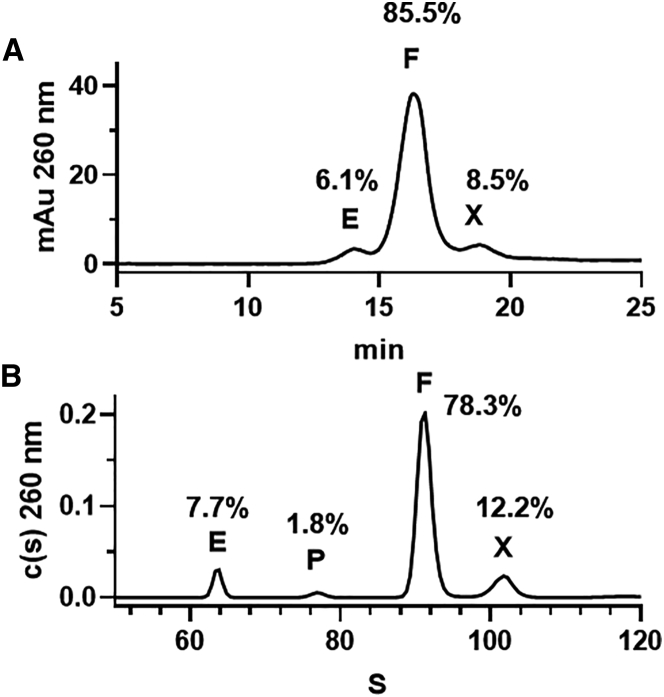
AEX can address these existing limitations. AEX was compared to sedimentation velocity AUC, the gold standard method. Whereas AUC showed a clearly better resolution, the empty and full AAV capsids were well resolved with a similar distribution by both methods (Figure 6). The percent area of empty capsid at 260 nm was determined to be 6.1% and 7.7% by AEX and AUC, respectively. Additionally, both methods detected an extra species after the full AAV particle peak, although the area percent reported was slightly different (8.5% versus 12.2%). Notably, AUC alone was able to resolve an additional species with 1.8% peak area and sedimentation coefficient of 78S, corresponding to AAV packed with fragmented genomes.16 In comparison, this partially filled AAV capsid could not be separated from full AAV capsids by AEX. The identification and characterization of these extra species are beyond the scope of the current study.
Figure 6.
Characterization of AAV empty and full capsids by AEX and AUC
Comparison of the AEX chromatogram (A) and AUC sedimentation coefficient distribution (B) of an affinity-purified AAV6.2 sample. E, empty capsid; F, full capsid; P, capsid with fragmented genome; X, unknown species. The peak percents labeled on the plots are area percents determined at UV 260 nm, without response factor corrections.
In summary, we have presented an AEX method for empty and full AAV6.2 capsid quantification. The method produces similar quantification results to the commonly used AUC method. Moreover, the method is executed on the quality control (QC)-friendly high-performance liquid chromatography (HPLC) platform, and it has demonstrated good linearity, accuracy, and intermediate precision. Finally, the AEX method uses only several microliters of material, requires no sample preparation or special expertise to operate, and can be automated for high-throughput HPLC analysis of in-process and release samples.
Materials and Methods
Production and Purification of AAV Vector and Empty AAV Capsid
AAV vectors were produced either via transient transfection or helper virus-based production. For the production of AAV vectors by transient transfection, HEK293 cells were co-transfected using polyethyleneimine and a pre-defined mixture of the three plasmids (ITR vector with gene of interest, AAV rep/cap, and Ad helper plasmid), and AAV vectors were harvested at 48–72 h post-transfection. For the production of AAV vectors by helper virus-based production system, BHK cells were co-infected with two rHSVs (rHSV-GOI and rHSV-rep/cap), and AAV vectors were harvested at 48 h post-infection. Empty capsids were generated by not including the GOI vector.
For both AAV production methods, AAV in cells and culture media were harvested and pooled for purification; purification was performed by affinity resin AVB Sepharose High Performance (GE Healthcare, Marlborough, MA, USA). Before loading to the column, raw AAV pool was incubated with 0.5% Triton X-100 and 50 U/mL benzonase at 37°C for 1 h and clarified by centrifugation at 10,000 × g for 15 min, followed by sequential filtrations through 1.2- and 0.45-mm filters. AAV was eluted from column with low-pH buffer, concentrated, and buffer-exchanged using Vivaspin 20, 50 kDa MWCO concentrator (GE Healthcare).
Isolation of AAV6.2 Full Capsid Particles from Affinity-Purified Product by Iodixanol Density Gradient
AAV6.2 full capsid was purified by iodixanol density gradient purification according to the method described by Crosson et al.,30 with the following modifications: 10 mL affinity-purified AAV6.2 was dispensed into a 32-mL OptiSeal centrifuge tube (Beckman Instruments, Palo Alto, CA, USA). Iodixanol (OptiPrep; Sigma-Aldrich, St. Louis, MO) solutions with increasing density fractions were laid under the sample in the following order: 5.5 mL of 15%, 5.5 mL of 25%, 5 mL of 40%, and 5 mL of 60%. The tubes were centrifuged in a 70Ti rotor (Beckman Instruments) at 69,000 rpm for 1 h at 18°C. Gradients were fractionated through an 18G needle inserted horizontally into the tube approximately 4 mm below the interface of the 40% and 60% step gradients. Fractions were analyzed by SDS-PAGE, qPCR, and AEX to determine purity, titer, and E/F contents.
Optimized AEX Method
The AEX separation method of AAV6.2 was developed on an Agilent 1260 HPLC system (Santa Clara, CA, USA) equipped with a quaternary pump, an autosampler, a photodiode array detector with 60-mm Max-Light flow cell, a fluorescence detector, and a fraction collector. The separation was accomplished on a CIMac AAV full/empty-0.1-mL column (BIA Separations, Ajdovščina, Slovenia) using a salt gradient, delivered from four channels of the quaternary pump. The mobile phases were A, water; B, 1 M tetramethylammonium chloride; C, 20 mM MgCl2; and D, 250 mM Bis-tris propane (pH 9.0). A linear gradient was set as 62%A/10%C/28%D at t0, 37%A/25%B/10%C/28%D at 25 min, and 72%B/28%D at 30 min. The column was equilibrated at the initial condition for 6 min between injections. The separation was monitored by photodiode array detector at 260 and 280 nm and fluorescence detector with 280-nm excitation and 340-nm emission.
AAV Vector Genome Copy Number Titration by qPCR
The AAV vector genome concentration was determined by qPCR on the Applied Biosystems 7500 Fast Real-Time PCR System. Briefly, fractions from HPLC were diluted 10-fold and digested with DNase I followed by proteinase K treatment. The treated samples were further diluted 1,000-fold and added to the TaqMan Fast Universal PCR Master mix (Applied Biosystems) along with the vector-specific primer and probe for qPCR run. A linearized vector plasmid was serially diluted and used as the standard in the qPCR run to determine the vector genome copy number in the HPLC fractions.
AAV Vector Capsid Titration by ELISA
Capsid concentration was measured by a sandwich ELISA (Progen Biotechnik). Pre-diluted AAV samples were captured on a microplate that was coated with a monoclonal antibody against a conformational epitope on assembled AAV6 capsids. Captured AAV6.2 capsids were bound to a biotin-conjugated anti-AAV6.2 antibody. Streptavidin peroxidase conjugate and substrate solution were then added to generate a color reaction for the quantitation of the bound AAV capsids. The capsid concentration was calculated against the titration curve of the kit control sample.
Sedimentation Velocity AUC
Sedimentation velocity AUC analysis was performed using Optima AUC (Beckman Coulter, Indianapolis, IN, USA). 400 μL sample was loaded into the sample sector of a two-sector velocity cell, and 400 μL buffer was loaded into the corresponding reference sector. The sample was placed in the eight-hole rotor and allowed to equilibrate in the instrument until a temperature of 20°C and a full vacuum were maintained for 1 h. Sedimentation velocity centrifugation was performed at 20,000 rpm and 20°C. Absorbance (260 and 280 nm) and Raleigh interference optics were used to simultaneously record the radial concentration as a function of time until the lightest sedimenting component cleared the optical window. Data were processed using the SEDFIT31 software following a previously published approach.16
Author Contributions
Conceptualization, C.W., S.H.R.M., and D.L.; Methodology, C.W.; Investigation, C.W., S.H.R.M., Z.C., J.D., X.Z., and C.G.; Writing – Original Draft, C.W.; Writing – Review & Editing, C.W., Z.C., T.L., A.E.S., and D.L.; Resources, G.X., T.L., L.C., and C.G.; Supervision, A.E.S. and D.L.
Acknowledgments
C.G. would like to thank Dr. Damien Marsic and Dr. Changshou Gao for their support of the work. The authors would like to thank Dr. Xiaoyu Chen and Dr. Carrie Sowers for their critical review of the manuscript. This research was developed with funding from the Defense Advanced Research Projects Agency under HR011-18-3-001. The views, opinions, and/or findings expressed are those of the authors and should not be interpreted as representing the official views or policies of the Department of Defense or the U.S. government.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtm.2019.09.006.
Contributor Information
Chunlei Wang, Email: chunlei.wang1@astrazeneca.com.
Dengfeng Liu, Email: liud@medimmune.com.
Supplemental Information
References
- 1.Samulski R.J., Muzyczka N. AAV-mediated gene therapy for research and therapeutic purposes. Annu. Rev. Virol. 2014;1:427–451. doi: 10.1146/annurev-virology-031413-085355. [DOI] [PubMed] [Google Scholar]
- 2.Xie Q., Bu W., Bhatia S., Hare J., Somasundaram T., Azzi A., Chapman M.S. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc. Natl. Acad. Sci. USA. 2002;99:10405–10410. doi: 10.1073/pnas.162250899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao L., During M., Xiao W. Replication competent helper functions for recombinant AAV vector generation. Gene Ther. 2002;9:1199–1206. doi: 10.1038/sj.gt.3301710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daya S., Berns K.I. Gene therapy using adeno-associated virus vectors. Clin. Microbiol. Rev. 2008;21:583–593. doi: 10.1128/CMR.00008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asokan A., Schaffer D.V., Samulski R.J. The AAV vector toolkit: poised at the clinical crossroads. Mol. Ther. 2012;20:699–708. doi: 10.1038/mt.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grieger J.C., Samulski R.J. Chapter 12: adeno-associated virus vectorology, manufacturing, and clinical applications. In: Friedmann T., editor. Volume 507. Academic Press; 2012. pp. 229–254. (Methods in Enzymology). [DOI] [PubMed] [Google Scholar]
- 7.Qu W., Wang M., Wu Y., Xu R. Scalable downstream strategies for purification of recombinant adeno- associated virus vectors in light of the properties. Curr. Pharm. Biotechnol. 2015;16:684–695. doi: 10.2174/1389201016666150505122228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naso M.F., Tomkowicz B., Perry W.L., 3rd, Strohl W.R. Adeno-associated virus (aav) as a vector for gene therapy. BioDrugs. 2017;31:317–334. doi: 10.1007/s40259-017-0234-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flotte T.R. Empty adeno-associated virus capsids: contaminant or natural decoy? Hum. Gene Ther. 2017;28:147–148. doi: 10.1089/hum.2017.29039.trf. [DOI] [PubMed] [Google Scholar]
- 10.Gao K., Li M., Zhong L., Su Q., Li J., Li S., He R., Zhang Y., Hendricks G., Wang J., Gao G. Empty virions in aav8 vector preparations reduce transduction efficiency and may cause total viral particle dose-limiting side-effects. Mol. Ther. Methods Clin. Dev. 2014;1:20139. doi: 10.1038/mtm.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wright J.F. AAV empty capsids: for better or for worse? Mol. Ther. 2014;22:1–2. doi: 10.1038/mt.2013.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wright J.F. Product-related impurities in clinical-grade recombinant AAV vectors: Characterization and risk assessment. Biomedicines. 2014;2:80–97. doi: 10.3390/biomedicines2010080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wright J.F. Manufacturing and characterizing AAV-based vectors for use in clinical studies. Gene Ther. 2008;15:840–848. doi: 10.1038/gt.2008.65. [DOI] [PubMed] [Google Scholar]
- 14.Horowitz E.D., Rahman K.S., Bower B.D., Dismuke D.J., Falvo M.R., Griffith J.D., Harvey S.C., Asokan A. Biophysical and ultrastructural characterization of adeno-associated virus capsid uncoating and genome release. J. Virol. 2013;87:2994–3002. doi: 10.1128/JVI.03017-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sommer J.M., Smith P.H., Parthasarathy S., Isaacs J., Vijay S., Kieran J., Powell S.K., McClelland A., Wright J.F. Quantification of adeno-associated virus particles and empty capsids by optical density measurement. Mol. Ther. 2003;7:122–128. doi: 10.1016/s1525-0016(02)00019-9. [DOI] [PubMed] [Google Scholar]
- 16.Burnham B., Nass S., Kong E., Mattingly M., Woodcock D., Song A., Wadsworth S., Cheng S.H., Scaria A., O’Riordan C.R. Analytical ultracentrifugation as an approach to characterize recombinant adeno-associated viral vectors. Hum. Gene Ther. Methods. 2015;26:228–242. doi: 10.1089/hgtb.2015.048. [DOI] [PubMed] [Google Scholar]
- 17.Pierson E.E., Keifer D.Z., Asokan A., Jarrold M.F. Resolving adeno-associated viral particle diversity with charge detection mass spectrometry. Anal. Chem. 2016;88:6718–6725. doi: 10.1021/acs.analchem.6b00883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lock M., McGorray S., Auricchio A., Ayuso E., Beecham E.J., Blouin-Tavel V., Bosch F., Bose M., Byrne B.J., Caton T. Characterization of a recombinant adeno-associated virus type 2 Reference Standard Material. Hum. Gene Ther. 2010;21:1273–1285. doi: 10.1089/hum.2009.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Urabe M., Xin K.-Q., Obara Y., Nakakura T., Mizukami H., Kume A., Okuda K., Ozawa K. Removal of empty capsids from type 1 adeno-associated virus vector stocks by anion-exchange chromatography potentiates transgene expression. Mol. Ther. 2006;13:823–828. doi: 10.1016/j.ymthe.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 20.Qu G., Bahr-Davidson J., Prado J., Tai A., Cataniag F., McDonnell J., Zhou J., Hauck B., Luna J., Sommer J.M. Separation of adeno-associated virus type 2 empty particles from genome containing vectors by anion-exchange column chromatography. J. Virol. Methods. 2007;140:183–192. doi: 10.1016/j.jviromet.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 21.Lock M., Alvira M.R., Wilson J.M. Analysis of particle content of recombinant adeno-associated virus serotype 8 vectors by ion-exchange chromatography. Hum. Gene Ther. Methods. 2012;23:56–64. doi: 10.1089/hgtb.2011.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burova E., Ioffe E. Chromatographic purification of recombinant adenoviral and adeno-associated viral vectors: methods and implications. Gene Ther. 2005;12(Suppl 1):S5–S17. doi: 10.1038/sj.gt.3302611. [DOI] [PubMed] [Google Scholar]
- 23.Okada T., Nonaka-Sarukawa M., Uchibori R., Kinoshita K., Hayashita-Kinoh H., Nitahara-Kasahara Y., Takeda S., Ozawa K. Scalable purification of adeno-associated virus serotype 1 (AAV1) and AAV8 vectors, using dual ion-exchange adsorptive membranes. Hum. Gene Ther. 2009;20:1013–1021. doi: 10.1089/hum.2009.006. [DOI] [PubMed] [Google Scholar]
- 24.Brument N., Morenweiser R., Blouin V., Toublanc E., Raimbaud I., Chérel Y., Folliot S., Gaden F., Boulanger P., Kroner-Lux G. A versatile and scalable two-step ion-exchange chromatography process for the purification of recombinant adeno-associated virus serotypes-2 and -5. Mol. Ther. 2002;6:678–686. doi: 10.1006/mthe.2002.0719. [DOI] [PubMed] [Google Scholar]
- 25.Davidoff A.M., Ng C.Y., Sleep S., Gray J., Azam S., Zhao Y., McIntosh J.H., Karimipoor M., Nathwani A.C. Purification of recombinant adeno-associated virus type 8 vectors by ion exchange chromatography generates clinical grade vector stock. J. Virol. Methods. 2004;121:209–215. doi: 10.1016/j.jviromet.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 26.Wallis C., Melnick J.L. Stabilization of poliovirus by cations. Tex. Rep. Biol. Med. 1961;19:683–700. [PubMed] [Google Scholar]
- 27.Farrah S.R., Shah D.O., Ingram L.O. Effects of chaotropic and antichaotropic agents on elution of poliovirus adsorbed on membrane filters. Proc. Natl. Acad. Sci. USA. 1981;78:1229–1232. doi: 10.1073/pnas.78.2.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venkatakrishnan B., Yarbrough J., Domsic J., Bennett A., Bothner B., Kozyreva O.G., Samulski R.J., Muzyczka N., McKenna R., Agbandje-McKenna M. Structure and dynamics of adeno-associated virus serotype 1 VP1-unique N-terminal domain and its role in capsid trafficking. J. Virol. 2013;87:4974–4984. doi: 10.1128/JVI.02524-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chan K.C., Veenstra T.D., Issaq H.J. Comparison of fluorescence, laser-induced fluorescence, and ultraviolet absorbance detection for measuring HPLC fractionated protein/peptide mixtures. Anal. Chem. 2011;83:2394–2396. doi: 10.1021/ac1032462. [DOI] [PubMed] [Google Scholar]
- 30.Crosson S.M., Dib P., Smith J.K., Zolotukhin S. Helper-free production of laboratory grade aav and purification by iodixanol density gradient centrifugation. Mol. Ther. Methods Clin. Dev. 2018;10:1–7. doi: 10.1016/j.omtm.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.