Abstract
Systemic delivery of antisense oligonucleotides (AO) for DMD exon skipping has proven effective for reframing DMD mRNA, rescuing dystrophin expression, and slowing disease progression in animal models. In humans with Duchenne muscular dystrophy treated with AOs, low levels of dystrophin have been induced, and modest slowing of disease progression has been observed, highlighting the need for improved efficiency of human skipping drugs. Here, we demonstrate that dantrolene and Rycals S107 and ARM210 potentiate AO-mediated exon skipping of exon 44 or exon 45 in patient-derived myotube cultures with appropriate mutations. Further, dantrolene is shown to boost AO-mediated exon skipping in patient-derived, induced cardiomyocyte cultures. Our findings further validate the ryanodine receptors (RyR) as the likely target responsible for exon skip boosting and demonstrate potential applicability beyond exon 51 skipping. These data provide preclinical support of dantrolene trial as an adjuvant to AO-mediated exon-skipping therapy in humans and identify a novel Rycal, ARM210, for development as a potential exon-skipping booster. Further, they highlight the value of mutation-specific DMD culture models for basic discovery, preclinical drug screening and translation of personalized genetic medicines.
Keywords: Duchenne muscular dystrophy, combination therapy, dantrolene, dystrophin, exon skipping, armgo, ARM210, muscle, therapy
Introduction
Duchenne muscular dystrophy (DMD) is a lethal genetic disease of childhood, affecting 1/5,000 male births1 and is caused by mutations in the DMD gene. The DMD gene encodes the dystrophin protein, essential for the health of skeletal and heart muscle. Dystrophin is integral to the function of dystrophin-associated glycoprotein complex (DGC), which provides structural support and membrane stability during muscle contraction by linking the actin cytoskeleton to the extracellular matrix.2 Additionally, dystrophin controls asymmetric division in muscle stem cells, impacting stem cell renewal and myogenic fate determination during muscle repair.3,4 Without functional dystrophin, defects in sarcolemmal membrane stabilization lead to progressive muscle damage, while stem cell defects limit the efficiency of regeneration. Progressive muscle weakness leads to loss of virtually all muscle function, respiratory and cardiac failure, and premature death between ages 20 and 30.5
Most DMD mutations result from large frameshifting deletions (∼65%),6 leading to complete absence of dystrophin protein expression and DMD. A milder phenotype, referred to as Becker muscular dystrophy (BMD), typically results from in-frame deletions that lead to the production of a partially functional dystrophin protein with an internal deletion. “Exon skipping” is a therapeutic strategy that uses antisense oligonucleotides (AOs) to bypass one or more exons in the DMD pre-mRNA in order to restore reading frame and rescue expression of a partially functional dystrophin protein. Exon-skipping AOs have successfully restored the mRNA reading frame and rescued dystrophin expression in DMD and animal models of DMD.7, 8, 9, 10, 11, 12, 13, 14 Initial human exon-skipping studies focused on the targeted skipping of exons 51, 53, 45, 44, and 8, since these are predicted to rescue the most commonly occurring human DMD mutations and because mild phenotypes are associated with some naturally occurring in-frame deletions in these regions.13,15,16
Eteplirsen, a morpholino AO targeting exon 51 for skipping, recently received FDA accelerated approval based on the statistically significant induction of dystrophin expression as a surrogate biomarker.17,18 However, only low levels of dystrophin induction (< 1% of typical expression in skeletal muscle) have been observed so far in treated patients. While there is evidence that this low level of dystrophin induction slowed progression of disease, as measured by 6-min walk distance (6MWD), age at loss of ambulation (LOA), and forced vital capacity (FVC), pulmonary function changes relative to external DMD controls,18 it is likely that higher levels of dystrophin induction would be more therapeutically relevant. Thus, identification of agents capable of improving the efficacy of AO-mediated exon skipping are of great interest and hold the potential to further increase dystrophin levels in skeletal muscles and have a concomitant greater functional benefit.
Dantrolene is an FDA-approved drug primarily used for the treatment of malignant hyperthermia, neuroleptic malignant syndrome, and spasticity.19 We previously published that dantrolene boosts DMD AO-mediated exon 51 skipping in human DMD patient-derived fibroblasts, which were directly reprogrammed to myotubes through myogenic differentiation 1 (Myod1) induction (induced directly reprogrammed myotubes, termed iDRM).20 Further, systemic codelivery of dantrolene was able to increase AO-mediated exon 23 skipping and dystrophin protein rescue in mdx mice.20,21 However, it remains unclear whether dantrolene is able to increase the amount of dystrophin produced when used in combination with AO targeting other exons and how it functions to promote skipping.
Dantrolene can directly bind the Ryanodine receptors (RyR), which controls Ca+-regulated muscle contraction.19 Since, like dantrolene, ryanodine and Rycals S107 also increased exon skipping, we hypothesize that dantrolene targets the RyR to potentiate exon skipping. Using cultured myotubes generated from DMD patient-derived inducible directly reprogrammable myotubes (iDRMs) and patient-derived induced pluripotent stem cells (iPSCs), we investigated exon 45 and exon 44 skipping and demonstrate that dantrolene as well as Rycals S107 and ARM210 (a proprietary Rycal, ARMGO Pharma) can each boost AO-directed skipping in culture. Finally, DMD patient-derived iPSCs were differentiated into cardiomyocytes, and dantrolene was shown to boost AO-mediated exon skipping of exon 45. These findings serve to further elucidate the applicability and mechanism of RyR modulators as boosters of AO-mediated exon skipping in DMD.
Results
Dantrolene Boosts AO-Mediated DMD Exon 45 and Exon 44 Skipping in Patient-Derived Myotube Cultures Amenable to Rescue by Reframing
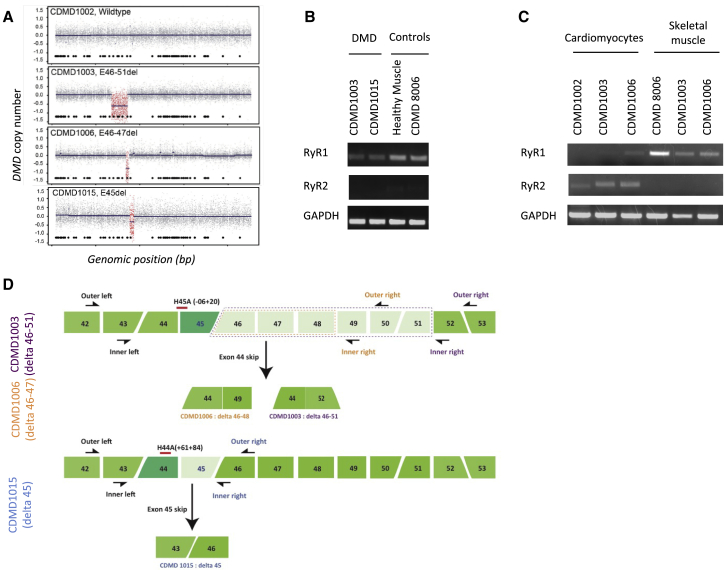
To determine if dantrolene increases AO-mediated exon skipping of clinically relevant DMD mutations, we created cell culture models from patients potentially amenable to exon 45 skipping (Center for Duchenne Muscular Dystrophy 10003-CDMD1003, deletion of exons 46–51; and CDMD1006, deletion of exons 46–47) or exon 44 skipping (CDMD1015, deletion of exon 45)22,23 (Figure 1). Comparative genomic hybridization confirmed the expected DMD mutations in primary fibroblasts (Figure 1A). RyR1 and RyR2 expression were determined by RT-PCR in DMD patient fibroblast directly reprogrammed to skeletal muscle myotubes (iDRMs) or first to iPSCs (Figures 1B and 1C) that are subsequently differentiated to cardiomyocytes. Consistent with their known expression patterns, DMD iDRM- and iPSC-derived myotubes express RyR1, but not RyR2, whereas iPSC-derived cardiomyocyte cultures predominantly express RyR2. The findings validate our reprogramming and differentiation schemes and demonstrate that RyRs, known dantrolene targets, are appropriately expressed in these model systems (Figures 1B and 1C). Figure 1D provides a schematic indicating the DMD mutations present in each cell model used in our studies and the primers used for detecting mRNA lacking the potentially skipped exon that would restore the reading frame if skipped (Figure 1D).
Figure 1.
Human DMD Patient and Normal Control-Cell-Based Skeletal Muscle and Cardiomyocyte Models Used in This Study
(A) DMD copy number variation confirms DMD mutations in primary DMD patient-derived fibroblasts. A custom CGH array was used to identify DMD gene mutations and breakpoint boundaries. Displayed are copy number data from individuals with a wild-type DMD gene (1002) or DMD patients with deletion spanning exons 46–51 (CDMD1003), 46–47 (CDMD1015), or exon 45 (CDMD1006). Probes within the deleted DMD region are highlighted in red. Tiled below are the genomic locations of the 79 DMD exons, with exon 1 beginning on the right and ending with exon 79 on the left. Patient fibroblasts were reprogrammed to iDRM or iPSC and subsequently differentiated into skeletal or cardiac muscle lineage. (B) DMD iDRM-derived myotubes express RyR1, but not RyR2. Patient-derived iDRMs were differentiated to myotubes through induced Myod activity and fusion conditions. Levels of RyR1 and Ryr2 expression were determined by RT-PCR in DMD iDRM CDMD1003- and CDMD1015-derived myotubes relative to healthy control muscle biopsy or primary myoblasts (CDMD8006). (C) iPSC-derived skeletal muscles predominantly express RyR1, whereas iPSC-derived cardiomyocytes predominantly express RyR2. DMD iPSC CDMD1003 and CDMD1006 or healthy control iPSC CDMD 1002 and 1006 were differentiated to myotubes or cardiomyocytes, and levels of RyR1 and RyR2 expression were determined using RT-PCR. CDMD8006 are healthy primary myoblasts. (D) Schematic representing three patient mutations and strategies for exon skipping to restore reading frame. Mutations for patients CDMD1003 and CDMD1006 are indicated by dashed boxes (purple and orange, respectively). The position of each primer pairs used for quantitating exon skipping are indicated outer primers for the first round of PCR and inner primers for the second round of PCR. Location and coordinates for AO H44A and H45A are also indicated.
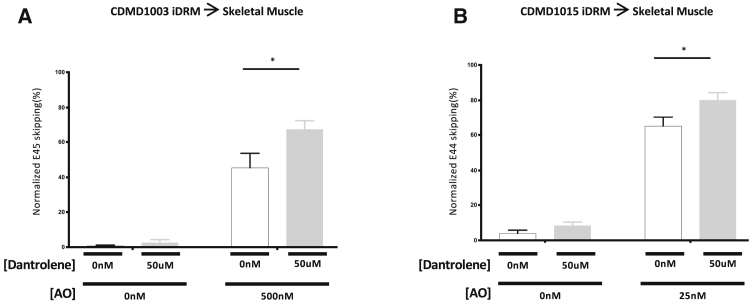
First, we tested the ability of dantrolene to boost exon 45 (iDRM CDMD1003; Figure 2A) or 44 (iDRM 1015; Figure 2B) skipping in iDRM-derived cultured myotubes. As shown in Figure 2, treatment with AO induces targeted exon (exon 45 or exon 44) exclusion and generation of a larger portion of skipped mRNA. Addition of dantrolene in combination with the appropriate AO increases the level of skipped mRNA relative to AO alone in both exon 44 and exon 45 skip-amenable DMD cell models (Figure 2).
Figure 2.
Dantrolene Boosts AO-Targeted Exon45 and Exon44 Skipping in Patient iDRM
(A and B) iDRM CDMD1003 (deletion of exons 46–51) (A) or CDMD1015 (deletion of exon 45) (B) were differentiated to myotubes and treated with AO targeting exon 45 skipping, H45A, with or without dantrolene, or dantrolene alone. Skipped and unskipped mRNA products were amplified using RT-PCR and quantified using an Agilent 2100 bioanalyzer. Data are expressed as normalized E45 or E44 skipping (skipped mRNA products over total mRNA products). Bars represent SEM. *p < 0.05; p values reflect a Student’s t test.
Rycals S107 and ARM210 Boost AO-Directed Exon 44 and 45 Skipping
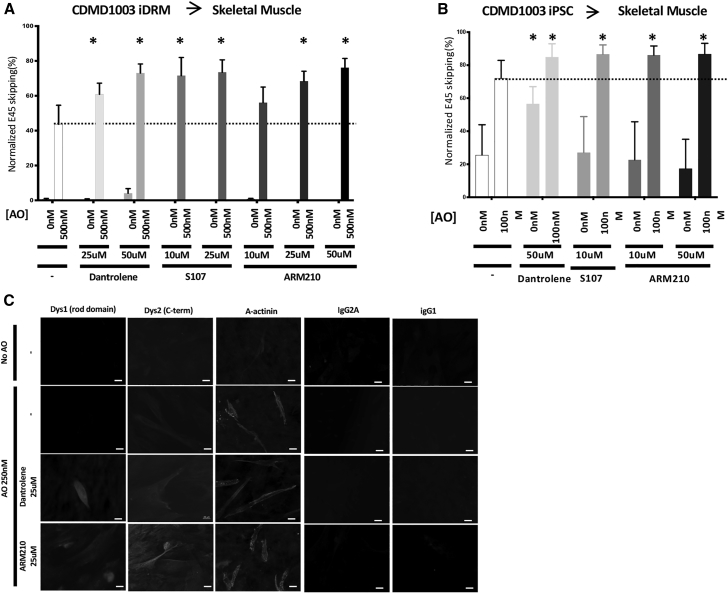
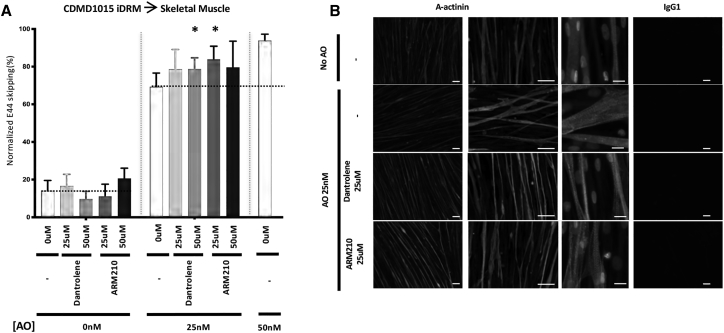
Next, we tested the Rycals S107 and ARM210 for their ability to boost AO-directed exon 45 or 44 skipping. We previously demonstrated that Rycal S107 could potentiate AO-directed exon skipping in iDRMs carrying a DMD mutation amenable to reframing by exon 51 skipping.20 ARM210 is a novel Rycal currently under development as a potential standalone treatment targeting calcium defects that may contribute to disease pathogenesis in DMD and which improves muscle strength and muscle histology in mdx mice.24 Here, we demonstrate in CDMD1003 iDRM- and iPSC-derived myotube cultures, treatment with AO alone induces targeted exon 45-skipped mRNA (Figures 3A and 3B). Treatments with AO/dantrolene, AO/Rycal S107, or AO/Rycal ARM210 combinations each boost exon 45 skipping relative to AO alone. Combination therapy similarly boosts dystrophin protein expression measure using in skeletal muscle cultures derived from CDMD1003 iPSC detected using immunofluorescence (Figure 3C). Similarly, dantrolene and Rycals S107 and ARM210 also boost AO-targeted exon 44 skipping in CDMD1015 iDRM patient-derived myotubes (Figure 4A). Immunofluorescence staining for alpha-actinin demonstrated striations in mature muscle cultures treated with AO alone or in combination with dantrolene or Rycal ARM210 (Figure 4B).
Figure 3.
Rycals ARM210 and S107 Boost AO-Directed Exon45 Skipping in Patient iDRM- and iPSC-Derived Myotubes
(A and B) iDRM CDMD1003 (deletion of exons 46–51) (A) or iPSC CDMD1003 (B) were differentiated to myotubes and treated with AO targeting exon 45 skipping, H45A, with or without dantrolene, Rycal S107, or Rycal ARM210 or treated with drugs alone. Skipped and unskipped mRNA products were amplified using RT-PCR and quantified using an Agilent 2100 bioanalyzer. Data are expressed as normalized E45 skipping (skipped mRNA products over total mRNA products). For each iDRM or iPSC, averages of two independent experiments performed in triplicates (iDRM) or duplicates (iPSC) are shown. Bars represent SEM. *p < 0.05; p values reflect a Student’s t test. Dystrophin immunostaining was performed using NCL-Dys1 (Dys1- rod domain) and NCL-Dys 2 (Dys 2- C-terminus) to measure the maturation of the myotubes. IgG control stainings were also performed. Scale bars are 20 μm (C).
Figure 4.
Rycal ARM210 and Dantrolene Boost AO-Mediated Exon44 RNA Skipping in Patient iDRM-Derived Myotubes
iDRM CDMD1015 (delta 45) were differentiated to myotubes and treated with AO targeting exon 44 skipping, H44A, with or without dantrolene or Rycal ARM210. Skipped and unskipped mRNA products were amplified using RT-PCR and quantified using an Agilent 2100 bioanalyzer (A). Data are expressed as normalized E45 or E44 skipping (mRNA skipped products over total mRNA products). Bars represent SEM. *p < 0.05; p values reflect a Student’s t test. Graphic represents two independent experiments run as triplicate. iDRM CDMD1015 (deletion of exon 45) were differentiated to myotubes and treated with AO targeting exon 44 skipping, H44A, with or without dantrolene or Rycal ARM210 and immunostaining for alpha-actinin (A-actinin) were performed to demonstrates maturation of the myotubes (B).
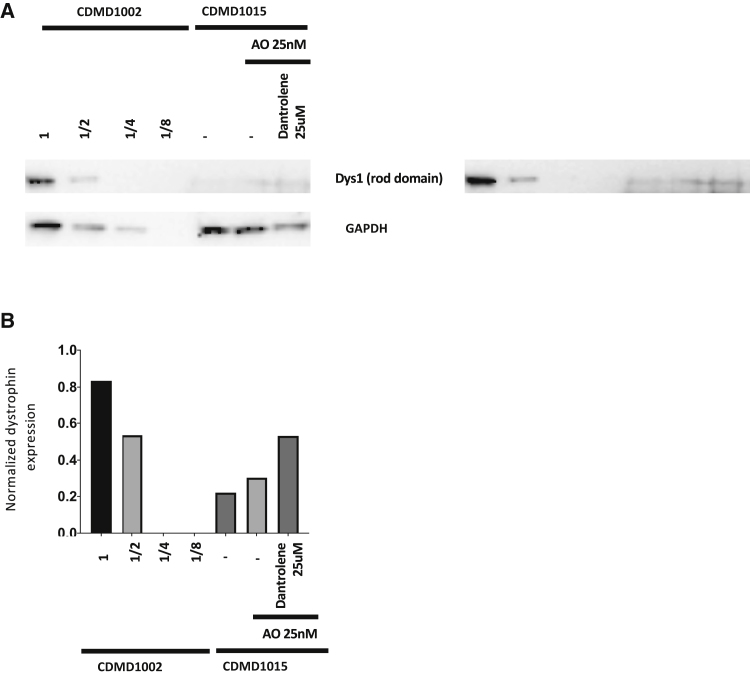
To determine if the enhanced mRNA skipping in CDMD1015 was translated into enhanced dystrophin protein rescue, we stained myotube cultures with antibody specific for dystrophin. After iDRM CDMD1015 differentiation, myotubes do not express dystrophin, consistent with the out-of-frame DMD mutation (Figure 5, upper panel). Low levels of dystrophin staining can be detected in some myotube cultures treated with AO alone (Figure 5, second panels from the top), and dystrophin levels are increased further when AO/dantrolene or AO/Rycal ARM210 are used in combination (Figure 5, lower two panels). Combination therapy also increased dystrophin protein expression as measured by western blotting (Figure 6). These findings demonstrate that observed increases in exon skipping translate into increased dystrophin protein rescue, consistent with our studies in mdx mice in vivo.20,21
Figure 5.
Rycal ARM210 and Dantrolene Boost AO-Mediated Exon44 Skipping in Patient iDRM-Derived Myotubes to Induce Dystrophin Protein Expression
Dystrophin immunostaining was performed using NCL-dys3 (Dys3, N-term), NCL-Dys1 (Dys1, rod domain), and NCL-Dys 2 (Dys 2, C-term) antibodies. IgG control staining were also performed. Scale bars represent 200 μm.
Figure 6.
Dantrolene Boosts the Induction of Dystrophin Protein Expression in iDRM-Derived Myotubes as Measured by Western Blotting
iDRM 1015 treated with AO H44A alone or in combination with dantrolene at 25 μM and immunoblotted using NCL-dys antibody to detect dystrophin protein. iDRM 1002 (WT) was used as a dystrophin standard control loaded in serial dilutions. GAPDH was used as loading control (A). ImageJ was used to quantitate the western blot results and are reported as values for Dystrophin divided by GAPDH values (B).
Dantrolene Boosts AO Exon Skipping in iPSC-Derived Cardiomyocytes
To assess the efficacy of AO/dantrolene combination treatment in DMD cardiac cell models, we differentiated CDMD1003 iPSC23 into cardiomyocytes in culture. As shown in Figure 7, treatment of CDMD1003 iPSC-derived cardiomyocytes with exon 45 skipping AO leads to induction of skipped DMD mRNA at 200 nM (Figures 7A and 7B). When AO is used in combination with dantrolene, increased exon skipping activity is detectable at both 50-nM and 250-nM AO concentrations (Figure 7A). These findings extend the observation that dantrolene can increase DMD exon skipping to in vitro differentiated human DMD cardiomyocytes.
Figure 7.
Rycal ARM210 and Dantrolene Boost AO-Directed Exon45 Skipping in iPSC-Derived Cardiomyocytes
iPSC CDMD1003 (deletion of exons 46–51) were differentiated to cardiomyocytes and treated with AO targeting exon 45 skipping, H45A, alone or in combination with dantrolene (A and B) or Rycal ARM210 (B). Skipped and unskipped mRNA products were amplified using RT-PCR and quantified using an Agilent 2100 bioanalyzer. Data are expressed as normalized E45 or E44 skipping (skipped mRNA products over total mRNA products). (B) To normalize across independent experiments, data are presented as fold change relative to AO treatment alone. Mean and SEM across two experiments are shown. iDRM CDMD1003 (deletion of exons 46–51) were differentiated to cardiomyocytes, replated as single cells, and treated with AO targeting exon 44 skipping, H44A, with or without dantrolene or Rycal ARM210.
Discussion
While DMD exon skipping AO have demonstrated substantial induction of dystrophin in animal models, only low levels of dystrophin rescue have been documented in human clinical trials.18 While even these low levels are plausibly slowing disease progression in skeletal muscle, greater dystrophin induction is predicted to yield better efficacy. Therefore, alternate chemistries, delivery mechanisms, or drug combinations that better promote exon skipping could be of significant value (for review see Barthélémy and Wein25).
We previously identified dantrolene as a skip booster in a drug screen aimed at identifying compounds with the capacity to augment AO-mediated DMD exon skipping in a reporter line.20 Dantrolene AO skip-boosting activity has now been validated in mdx mice over both long- and short-term systemic AO treatment. In these studies, exon 23 AO/dantrolene combination therapy demonstrated higher levels of exon 23-skipped mRNA and rescued dystrophin protein, improved muscle function, and diminished pathophysiology relative to those treated with AO or dantrolene alone.20,21
Dantrolene skip-boosting activity was further validated in patient iDRM-derived myotubes amenable to restoration of reading frame by exon 51 skipping, suggesting that boost activity of dantrolene may be relevant to human DMD exon-skipping therapies in clinical trial and being administered as a commercial product.20 Both 2-O-methyl and morpholino AO were boosted by dantrolene. However, we have yet to assess its activity in the context of alternate chemistries or AO targeting other DMD mutations currently in the therapeutic pipeline.26,27
Other compounds have been described with the ability to influence skipping in DMD models.28, 29, 30, 31 However, dantrolene is relatively unique, as it is a potentially practical therapy to augment exon skipping in humans, since it is an FDA-approved drug with a known safety profile, broad usage in pediatrics, and some previous experience in treatment of DMD.32 Our new data further demonstrate that dantrolene is able to augment exon skipping with AO targeting at least three different AO different exons.
Here, we demonstrate that dantrolene can increase AO-mediated skipping of exons 45 or 44, both of which are currently being targeted in preclinical or clinical studies. When combined with exon 45- or exon 44-skipping AO, dantrolene promotes exon skipping in relevant DMD patient iPSC-/iDRM-derived myotubes. Similar results are seen across cell models derived from two different exon 45 skip-amenable patients with distinct exon 45 mutations; deletion of exons 46–51 of DMD and deletion of exons 46–47 of DMD. Likewise, dantrolene boosts exon 44 skipping in patient-derived iDRM DMD myotube cultures carrying a mutation amenable to reframing by exon 44 skipping (deletion of exon 45 of DMD). While the degree of boost is modest, it is in keeping with the levels boosted in mdx mice that were previously shown to positively decrease disease pathology and in keeping with the literature indicating that even low levels of dystrophin can impact muscle function.18,21 Boost activity is seen in both patient iDRM- and iPSC-derived myotube models, adding confidence to our findings. Of note, differences in the robustness of the differentiation into myotubes from iPSCs or iDRMs provided some variation in AO dosing. iPSC-derived myotubes were more efficiently created in culture, and lower concentrations of AO (100 nM) were required to induce AO or AO/dantrolene exon skipping relative to iDRM (500 nM).
In skeletal muscle, dantrolene targets RyR1; a calcium channel involved in the regulation of intracellular calcium release from the sarcoplasmic reticulum during muscle contraction. In mdx mice, RyR1has been shown to be nitrosylated, leading to leaky channels,33 and similar results have been seen in muscle biopsies from DMD boys (A. Lacampagne, personal communication). For this reason, dantrolene has been tested in a small cohort of eight boys over 2 years in an attempt to control the leak and slow DMD progression.32 Consistent with studies in the mdx mouse, trends toward modest reduction of serum creatine kinase (CK) and increased muscle function did not reach statistical significance. Nonetheless, no adverse effects of dantrolene treatment were observed in this DMD cohort, aside from one patient in which dantrolene induced transient muscle weakness,32 which was reversed upon lowering the dantrolene dose by 25%. The patient received the lower dose for the study duration, highlighting the importance of monitoring potential acute weakness due to dantrolene treatment. While we saw no effect of dantrolene treatment alone on exon skipping or pathophysiology in mdx,20,21 it remains possible that in humans dantrolene may function alone to counter defective calcium regulation and downstream pathology in DMD, as suggested by others’ findings, as well as a skip booster when co-administered with AO, and both activities may slow disease progression.
Rycals regulate RyR activity by stabilizing calstabin:RyR binding and have been reported to normalize leaky RyR Ca+ flux observed in mdx mouse muscle.33 For this reason, Rycal ARM210 has been considered as a standalone therapy for DMD.24 We previously demonstrated that Rycal S107 and ryanodine, like dantrolene, also boost exon 51 AO skipping, leading us to hypothesize that the RyR is the relevant dantrolene target for promoting exon skipping.20 Here, we demonstrate that Rycals ARM210 and S107, like dantrolene, both boost exon 45 or exon 44 AO-targeted exon skipping, lending further support for skip regulation by a RyR-regulated pathway and identifying a novel Rycal with potential for boosting AO-mediated exon skipping in DMD. Because Rycals act to stabilize a closed conformation rather than block the active site, it has been suggested that they may not be limited by induced muscle weakness, which can be observed at high dantrolene doses in some patients.32, 33, 34 We have yet to determine the precise mechanism of RyR regulation of DMD exon skipping. However, previous studies have elucidated a role for tissue-specific calcium regulation of alternative RNA splicing, providing a plausible mechanistic hypothesis to test in future studies.35,36 Thus, it may be that leaky, nitrosylated RyR channels in DMD lead to high cytoplasmic calcium, and dantrolene and both Rycals lead to a reduction of the high calcium, leading to more efficient exon skipping.
Cardiac failure is a leading cause of death in DMD and thus is an important target for effective therapies.5,37 Studies in mouse and dog have indicated that AO exon-skipping treatments capable of rescuing dystrophin in skeletal muscle do not rescue its expression in cardiac tissue.14,38,39 The reason for this remains unclear, though published data support the suggestion that both AO delivery to the heart and limited skipping in cardiomyocytes limit the AO exon-skipping efficacy in cardiac tissue.40 In the experiments presented here, AO is artificially delivered to cardiomyocytes using oligofectamine, allowing us to examine the efficacy of AO/dantrolene boost activity in the context of cardiac muscle independently of delivery considerations. Prior studies described dantrolene as specifically targeting RyR1, expressed in skeletal, but not cardiac muscle.41 However, dantrolene has since been reported to also target RyR2,42 abundantly expressed in the heart and in our iPSC-derived cardiomyocytes, and is likely the relevant target for dantrolene boost. Notably, Rycals also will target both RyR1 and RyR2. It is widely suggested that effective DMD treatment will rely on a drug cocktail, targeting distinct aspects of DMD pathogenesis. Our findings highlight unexpected effects of drug combinations and encourage consideration of drug interactions and synergies in determining the ideal cocktail for particular DMD patients.
Materials and Methods
Collection, Isolation, and Propagation of Dermal Fibroblasts
Skin punches were obtained under institutional review board (IRB)-approved protocol (#11-001087) with informed patient consent, by coring a 3-mm diameter small piece of skin from the arm. The entire skin punch was placed in sterile DMEM (phenol red free; Thermo Fisher Scientific, Grand Island, NY, USA) + 1% penicillin/streptomycin (Thermo Fisher Scientific, Grand Island, NY, USA). Isolation of dermal fibroblasts was done as previously described (Barthelemy et al.43). Fibroblast growth media was changed every 3 days until 2–3 weeks, after which fibroblasts were typically sub-confluent and transferred for expansion into a T175 flask (Corning, Tewksbury, MA, USA). Each CDMD skin punch is assigned a unique four-digit identifier, beginning with CDMD1001 and subsequently increasing. Fibroblasts are considered as iDRMs once they are transduced with either LV-Myod-ER(T) or LV-Puro-Myod-ER(T).
Comparative Genomic Hybridization (CGH) Array Design and Analysis
A custom 14,022 probe oligonucleotide array was designed with probes tiling the DMD gene (Agilent, Santa Clara, CA, USA). Genomic DNA was labeled with Cy3, and normal male genomic DNA was labeled with Cy5 using a random priming kit (Agilent, Santa Clara, CA, USA), and labeled DNA was co-hybridized to the custom-designed array. Arrays were scanned with the DNA microarray scanner with Surescan high-resolution technology (Agilent, Santa Clara, CA, USA), and data were extracted with feature extraction software version 10.5.1.1. The values were extracted from the software and analyzed in R. Base-pair positions are reported relative to build HG18, and the log ratio of the Cy3/Cy5 (test/normal) intensity is plotted for all probes.
Myod1 Induction and Myotube Fusion of iDRM
iDRMs were seeded at 150,000 cells per well in fibroblast growth media (DMEM [+ phenol red, high glucose] [Thermo Fisher Scientific, Grand Island, NY, USA] + 15% fetal bovine serum [FBS] [Omega Scientific, Tarzana, CA, USA] + 1% nonessential amino acids [Thermo Fisher Scientific, Grand Island, NY, USA] + 1% penicillin/streptomycin [Thermo Fisher Scientific, Grand Island, NY, USA]) in 6-well plates (Corning, Tewksbury, MA, USA) precoated for 1 h with 2.5 mL of 5 μg/mL laminin in serum-free DMEM (BD Biosciences, San Jose, CA, USA). Myoblasts were seeded at 200,000 cells per well in skeletal muscle cell growth media (Promocell, Heidelberg, Germany) + 15% FBS + 1% penicillin/streptomycin in 6-well plates (Corning) precoated for 1 h with 0.1% porcine gelatin (Sigma, St. Louis, MO, USA). The following day, iDRMs were supplemented with 5 μM 4OH-tamoxifen (Sigma, St. Louis, MO, USA; dissolved in ethanol) in fibroblast growth media for 24 h. On day 3, cells were washed in 1× PBS (Thermo Fisher Scientific), and fusion media containing 1 μM 4OH-tamoxifen was added (1:1 Ham’s F-10:DMEM [phenol red free, high glucose], 2% horse serum [Thermo Fisher Scientific, Grand Island, NY, USA], 2% insulin-transferrin-selenium) (Thermo Fisher Scientific, Grand Island, NY, USA). Also, during the first week of differentiation, we added SB431542 at a final concentration of 5 μM. Cells were kept for 2 weeks in fusion conditions prior to treatment. Medium was changed every other day.
Collection of Primary Myoblasts and Differentiation
Primary myoblasts were obtained as previously described22 and induced to differentiate for 15 days prior to collection.
Human-Induced Pluripotent Stem Cell Culture and Differentiation
Human induced pluripotent stem cells (iPSCs) were reprogrammed as previously published.23 They were grown on human embryonic stem cells (hESC) qualified matrigel (Corning), fed daily with mTeSR1 medium (Stem Cell Technologies, Vancouver, BC, Canada), and passaged with 0.5 mM EDTA every 5–7 days. For differentiation of iPSC-cardiomyocytes, confluent iPSCs were enzymatically dissociated to form aggregates and differentiated into the cardiomyocyte lineage using the PSC cardiomyocyte differentiation kit (Thermo Fisher Scientific, Grand Island, NY, USA). The medium was changed every 2 days for 3 weeks. For immunohistochemistry, after transfection and drug treatments, cells were first split into single cells on matrigel-coated coverslips and grown for 1 day prior to imaging. For differentiation of hiPSC-derived skeletal muscle, cells were processed as previously published.23 In brief, cells were committed to the skeletal muscle lineage under the expression of a tamoxifen-inducible estrogen receptor responsive to tamoxifen (Myod-ERT). Following induction, the cells were differentiated in low-glucose DMEM with 5% horse serum and 1 μM tamoxifen for 14 days prior to treatment. Also, during the first 3 days of differentiation, we added SB431542 at a final concentration of 5 μM. Medium was changed daily.
AO Transfection
On day 5 of differentiation (iDRM, iPSC-derived skeletal muscle) or day 21 (iPSC-cardiomyocytes), cells were transfected with 0, 25, 50, or 500 nM of 2-O-methyl AO targeting exon 45 or exon 4413 (using oligofectamine; Thermo Fisher Scientific) transfection reagent according to our previously published protocol.43
RNA Isolation, PCR, and Quantification
On the day of harvesting, cell pellets were scrapped directly in trizol (Thermo Fisher Scientific) and total RNA isolated using the Purelink RNA mini-kit (Thermo Fisher Scientific). For exon-skipping analysis, 200–500 ng of total RNA was reverse transcribed with an exon 54 gene-specific primer [20]. For cell lines eligible for exon 45 skipping, a nested PCR was performed between DMD exons 43 and 52 using previously described primers for cell line CDMD1003 [20, 33] and between DMD exons 43 and 50 for cell line CDMD 1006 (Ex42-o, 5′-GTCCGTGAAGAAACGATGATG-3′ + Ex50-o, 5′-GTTTACCGCCTTCCACTCAG-3′ and Ex43-i, 5′-TCTCTCCCAGCTTGATTTCC-3′ + Ex49-i, 5′-CTGAGTGGCTGGTTTTTCCT-3′). For cell line CDMD1015, a nested PCR was performed between exons 42 and 46 (Ex42-o, 5′-CAATGCTCCTGACCTCTGTGC-3′ + Ex46-o, 5′-GCTCTTTTCCAGGTTCAAGTGG-3′ and Ex43-i, 5′-GTCTACAACAAAGCTCAGGTCG-3′ + Ex46-i, 5′-GCAATGTTATCTGCTTCCTCCAACC-3′). The amplified products were run on the Agilent 2100 bioanalyzer for exon-skipping quantification.
Compounds
All the drugs were prepared extemporaneously and re-suspended in water. We used a commercially available formulation of dantrolene (Revonto, Louisville, KY, USA) that is a formulation of dantrolene sodium (NDC: 27505-001-65) from DSM Pharmaceuticals. Rycal S107 and ARM210 were kindly provided by ARMGO Pharma.
Immunofluorescence
Cells were seeded at 40,000 cells per well on gelatin-coated 12-mm sterile glass coverslips in 24-well plates. Cells were grown according to the previously explained protocol. During all the procedures, cells were kept at room temperature unless specified otherwise. After myotube formation, cells were fixed with acetone.23 Primary antibodies were incubated overnight at 4°C: dystrophin NCL-1, NCL-2, or NCL-3 (Leica, Buffalo Grove, IL, USA) at a dilution of 1:100 and alpha actinin (Sigma, St. Louis, MO, USA) at a dilution of 1/250. Immunoglobulin G (IgG) controls were also used: IgG1, ref 554121 (BD Biosciences, San Jose, CA, USA), and IgG2A, ref 400202 (BioLegend, San Diego, CA, USA).
Secondary antibodies were used at a dilution of 1:500 (goat anti-mouse IgG [SA5-10173] and goat anti-rabbit [35553]) from Thermo Fisher Scientific for 1 h. Coverslips were mounted in ProLong gold antifade with DAPI (Thermo Fisher Scientific). Images for were acquired on a Zeiss AxioImager Z1 microscope and with the AxioVision rel. 4.6.3.0 acquisition software.
Western Blot
Total cell lysates were resolved on 4%–12% bolt protein gels by SDS-PAGE (Bolt Bis-Tris Plus gels, Thermo Fisher Scientific) and transferred to nitrocellulose membrane (Millipore). The membrane was blocked for 1 h in 3% nonfat dry milk in Tris-buffered saline (TBS) with 0.2% Tween 20 and incubated in primary antibodies overnight at 4°C. Incubation was performed with the following primary antibodies: dystrophin C-terminal (NCL-Dys1, 1:100, Leica Biosystems) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (MAB374, 1:50,000, Chemicon). Horseradish peroxidase-conjugated anti-rabbit IgG and anti-mouse IgG (GE Healthcare) secondary antibodies were used at 1:15,000 dilutions in 3% nonfat dry milk and incubated at room temperature (RT) for 2 h. Immunoblots were developed using enhanced chemiluminescence (SuperSignal West Pico Chemiluminescent Substrate; Thermo Fisher Scientific).
Author Contributions
F.B., C.H., and R.T.W. performed all experiments. E.D.D. provided technical support and help recruiting all the patients. F.B., M.C.M., E.E.M., and S.F.N. wrote the manuscript with critical input from all the other authors. F.B., M.C.M., and S.F.N. conceived the project and designed the experiments.
Conflict of Interests
S.F.N. and M.C.M., are inventors on a pending patent on identification of small molecules that enhance exon skipping filed by UCLA. M.C.M. and S.F.N. hold two patents for compounds related to enhancement of RyR targeted skipping.
Acknowledgments
F.B. was funded by an NIH NIAMS U54 AR052646 Wellstone Center of Excellence Training Fellowship. This work was funded from grants from the Department of Defense (award number W81XWH-15-1-0182), the UCLA Muscular Dystrophy Core Center (NIH grant 5P30AR057230), PPMD, and the Center for Duchenne Muscular Dystrophy at UCLA. Viral packaging was performed at the UCLA Vector Core. The UCLA Integrated Molecular Technologies Core is supported by CURE/P30 DK041301. This research was made possible by a grant from the California Institute for Regenerative Medicine (Grant Number CIRM TRX-05426). We would like to thank Dr. Elizabeth Gibbs for technical support.
References
- 1.Mendell J.R., Shilling C., Leslie N.D., Flanigan K.M., al-Dahhak R., Gastier-Foster J., Kneile K., Dunn D.M., Duval B., Aoyagi A. Evidence-based path to newborn screening for Duchenne muscular dystrophy. Ann. Neurol. 2012;71:304–313. doi: 10.1002/ana.23528. [DOI] [PubMed] [Google Scholar]
- 2.Tinsley J.M., Blake D.J., Roche A., Fairbrother U., Riss J., Byth B.C., Knight A.E., Kendrick-Jones J., Suthers G.K., Love D.R. Primary structure of dystrophin-related protein. Nature. 1992;360:591–593. doi: 10.1038/360591a0. [DOI] [PubMed] [Google Scholar]
- 3.Dumont N.A., Wang Y.X., von Maltzahn J., Pasut A., Bentzinger C.F., Brun C.E., Rudnicki M.A. Dystrophin expression in muscle stem cells regulates their polarity and asymmetric division. Nat. Med. 2015;21:1455–1463. doi: 10.1038/nm.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y.X., Feige P., Brun C.E., Hekmatnejad B., Dumont N.A., Renaud J.M., Faulkes S., Guindon D.E., Rudnicki M.A. EGFR-Aurka Signaling Rescues Polarity and Regeneration Defects in Dystrophin-Deficient Muscle Stem Cells by Increasing Asymmetric Divisions. Cell Stem Cell. 2019;24:419–432.e6. doi: 10.1016/j.stem.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bushby K., Finkel R., Birnkrant D.J., Case L.E., Clemens P.R., Cripe L., Kaul A., Kinnett K., McDonald C., Pandya S., DMD Care Considerations Working Group Diagnosis and management of Duchenne muscular dystrophy, part 2: implementation of multidisciplinary care. Lancet Neurol. 2010;9:177–189. doi: 10.1016/S1474-4422(09)70272-8. [DOI] [PubMed] [Google Scholar]
- 6.Bladen C.L., Salgado D., Monges S., Foncuberta M.E., Kekou K., Kosma K., Dawkins H., Lamont L., Roy A.J., Chamova T. The TREAT-NMD DMD Global Database: analysis of more than 7,000 Duchenne muscular dystrophy mutations. Hum. Mutat. 2015;36:395–402. doi: 10.1002/humu.22758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aoki Y., Nakamura A., Yokota T., Saito T., Okazawa H., Nagata T., Takeda S. In-frame dystrophin following exon 51-skipping improves muscle pathology and function in the exon 52-deficient mdx mouse. Mol. Ther. 2010;18:1995–2005. doi: 10.1038/mt.2010.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu Q.L., Rabinowitz A., Chen Y.C., Yokota T., Yin H., Alter J., Jadoon A., Bou-Gharios G., Partridge T. Systemic delivery of antisense oligoribonucleotide restores dystrophin expression in body-wide skeletal muscles. Proc. Natl. Acad. Sci. USA. 2005;102:198–203. doi: 10.1073/pnas.0406700102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mann C.J., Honeyman K., Cheng A.J., Ly T., Lloyd F., Fletcher S., Morgan J.E., Partridge T.A., Wilton S.D. Antisense-induced exon skipping and synthesis of dystrophin in the mdx mouse. Proc. Natl. Acad. Sci. USA. 2001;98:42–47. doi: 10.1073/pnas.011408598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McClorey G., Fall A.M., Moulton H.M., Iversen P.L., Rasko J.E., Ryan M., Fletcher S., Wilton S.D. Induced dystrophin exon skipping in human muscle explants. Neuromuscul. Disord. 2006;16:583–590. doi: 10.1016/j.nmd.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 11.McClorey G., Moulton H.M., Iversen P.L., Fletcher S., Wilton S.D. Antisense oligonucleotide-induced exon skipping restores dystrophin expression in vitro in a canine model of DMD. Gene Ther. 2006;13:1373–1381. doi: 10.1038/sj.gt.3302800. [DOI] [PubMed] [Google Scholar]
- 12.van Deutekom J.C., Bremmer-Bout M., Janson A.A., Ginjaar I.B., Baas F., den Dunnen J.T., van Ommen G.J. Antisense-induced exon skipping restores dystrophin expression in DMD patient derived muscle cells. Hum. Mol. Genet. 2001;10:1547–1554. doi: 10.1093/hmg/10.15.1547. [DOI] [PubMed] [Google Scholar]
- 13.Wilton S.D., Fall A.M., Harding P.L., McClorey G., Coleman C., Fletcher S. Antisense oligonucleotide-induced exon skipping across the human dystrophin gene transcript. Mol. Ther. 2007;15:1288–1296. doi: 10.1038/sj.mt.6300095. [DOI] [PubMed] [Google Scholar]
- 14.Yokota T., Lu Q.L., Partridge T., Kobayashi M., Nakamura A., Takeda S., Hoffman E. Efficacy of systemic morpholino exon-skipping in Duchenne dystrophy dogs. Ann. Neurol. 2009;65:667–676. doi: 10.1002/ana.21627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aartsma-Rus A., De Winter C.L., Janson A.A., Kaman W.E., Van Ommen G.J., Den Dunnen J.T., Van Deutekom J.C. Functional analysis of 114 exon-internal AONs for targeted DMD exon skipping: indication for steric hindrance of SR protein binding sites. Oligonucleotides. 2005;15:284–297. doi: 10.1089/oli.2005.15.284. [DOI] [PubMed] [Google Scholar]
- 16.Aartsma-Rus A., Straub V., Hemmings R., Haas M., Schlosser-Weber G., Stoyanova-Beninska V., Mercuri E., Muntoni F., Sepodes B., Vroom E., Balabanov P. Development of Exon Skipping Therapies for Duchenne Muscular Dystrophy: A Critical Review and a Perspective on the Outstanding Issues. Nucleic Acid Ther. 2017;27:251–259. doi: 10.1089/nat.2017.0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aartsma-Rus A., Krieg A.M. FDA Approves Eteplirsen for Duchenne Muscular Dystrophy: The Next Chapter in the Eteplirsen Saga. Nucleic Acid Ther. 2017;27:1–3. doi: 10.1089/nat.2016.0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mendell J.R., Goemans N., Lowes L.P., Alfano L.N., Berry K., Shao J., Kaye E.M., Mercuri E., Eteplirsen Study Group and Telethon Foundation DMD Italian Network Longitudinal effect of eteplirsen versus historical control on ambulation in Duchenne muscular dystrophy. Ann. Neurol. 2016;79:257–271. doi: 10.1002/ana.24555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krause T., Gerbershagen M.U., Fiege M., Weisshorn R., Wappler F. Dantrolene--a review of its pharmacology, therapeutic use and new developments. Anaesthesia. 2004;59:364–373. doi: 10.1111/j.1365-2044.2004.03658.x. [DOI] [PubMed] [Google Scholar]
- 20.Kendall G.C., Mokhonova E.I., Moran M., Sejbuk N.E., Wang D.W., Silva O., Wang R.T., Martinez L., Lu Q.L., Damoiseaux R. Dantrolene enhances antisense-mediated exon skipping in human and mouse models of Duchenne muscular dystrophy. Sci. Transl. Med. 2012;4:164ra160. doi: 10.1126/scitranslmed.3005054. [DOI] [PubMed] [Google Scholar]
- 21.Wang D.W., Mokhonova E.I., Kendall G.C., Becerra D., Naeini Y.B., Cantor R.M., Spencer M.J., Nelson S.F., Miceli M.C. Repurposing Dantrolene for Long-Term Combination Therapy to Potentiate Antisense-Mediated DMD Exon Skipping in the mdx Mouse. Mol. Ther. Nucleic Acids. 2018;11:180–191. doi: 10.1016/j.omtn.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang R.T., Barthelemy F., Martin A.S., Douine E.D., Eskin A., Lucas A., Lavigne J., Peay H., Khanlou N., Sweeney L. DMD genotype correlations from the Duchenne Registry: Endogenous exon skipping is a factor in prolonged ambulation for individuals with a defined mutation subtype. Hum. Mutat. 2018;39:1193–1202. doi: 10.1002/humu.23561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young C.S., Hicks M.R., Ermolova N.V., Nakano H., Jan M., Younesi S., Karumbayaram S., Kumagai-Cresse C., Wang D., Zack J.A. A Single CRISPR-Cas9 Deletion Strategy that Targets the Majority of DMD Patients Restores Dystrophin Function in hiPSC-Derived Muscle Cells. Cell Stem Cell. 2016;18:533–540. doi: 10.1016/j.stem.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Capogrosso R.F., Mantuano P., Uaesoontrachoon K., Cozzoli A., Giustino A., Dow T., Srinivassane S., Filipovic M., Bell C., Vandermeulen J. Ryanodine channel complex stabilizer compound S48168/ARM210 as a disease modifier in dystrophin-deficient mdx mice: proof-of-concept study and independent validation of efficacy. FASEB J. 2018;32:1025–1043. doi: 10.1096/fj.201700182RRR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barthélémy F., Wein N. Personalized gene and cell therapy for Duchenne Muscular Dystrophy. Neuromuscul. Disord. 2018;28:803–824. doi: 10.1016/j.nmd.2018.06.009. [DOI] [PubMed] [Google Scholar]
- 26.Aartsma-Rus A., Morgan J., Lonkar P., Neubert H., Owens J., Binks M., Montolio M., Phadke R., Datson N., Van Deutekom J., workshop participants Report of a TREAT-NMD/World Duchenne Organisation Meeting on Dystrophin Quantification Methodology. J. Neuromuscul. Dis. 2019;6:147–159. doi: 10.3233/JND-180357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watanabe N., Nagata T., Satou Y., Masuda S., Saito T., Kitagawa H., Komaki H., Takagaki K., Takeda S. NS-065/NCNP-01: An Antisense Oligonucleotide for Potential Treatment of Exon 53 Skipping in Duchenne Muscular Dystrophy. Mol. Ther. Nucleic Acids. 2018;13:442–449. doi: 10.1016/j.omtn.2018.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han G., Gu B., Cao L., Gao X., Wang Q., Seow Y., Zhang N., Wood M.J., Yin H. Hexose enhances oligonucleotide delivery and exon skipping in dystrophin-deficient mdx mice. Nat. Commun. 2016;7:10981. doi: 10.1038/ncomms10981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu Y., Wu B., Zillmer A., Lu P., Benrashid E., Wang M., Doran T., Shaban M., Wu X., Lu Q.L. Guanine analogues enhance antisense oligonucleotide-induced exon skipping in dystrophin gene in vitro and in vivo. Mol. Ther. 2010;18:812–818. doi: 10.1038/mt.2009.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martone J., Briganti F., Legnini I., Morlando M., Picillo E., Sthandier O., Politano L., Bozzoni I. The lack of the Celf2a splicing factor converts a Duchenne genotype into a Becker phenotype. Nat. Commun. 2016;7:10488. doi: 10.1038/ncomms10488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Leary D.A., Sharif O., Anderson P., Tu B., Welch G., Zhou Y., Caldwell J.S., Engels I.H., Brinker A. Identification of small molecule and genetic modulators of AON-induced dystrophin exon skipping by high-throughput screening. PLoS ONE. 2009;4:e8348. doi: 10.1371/journal.pone.0008348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bertorini T.E., Palmieri G.M., Griffin J., Igarashi M., Hinton A., Karas J.G. Effect of dantrolene in Duchenne muscular dystrophy. Muscle Nerve. 1991;14:503–507. doi: 10.1002/mus.880140603. [DOI] [PubMed] [Google Scholar]
- 33.Bellinger A.M., Reiken S., Carlson C., Mongillo M., Liu X., Rothman L., Matecki S., Lacampagne A., Marks A.R. Hypernitrosylated ryanodine receptor calcium release channels are leaky in dystrophic muscle. Nat. Med. 2009;15:325–330. doi: 10.1038/nm.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bellinger A.M., Reiken S., Dura M., Murphy P.W., Deng S.X., Landry D.W., Nieman D., Lehnart S.E., Samaru M., LaCampagne A., Marks A.R. Remodeling of ryanodine receptor complex causes “leaky” channels: a molecular mechanism for decreased exercise capacity. Proc. Natl. Acad. Sci. USA. 2008;105:2198–2202. doi: 10.1073/pnas.0711074105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee J.A., Tang Z.Z., Black D.L. An inducible change in Fox-1/A2BP1 splicing modulates the alternative splicing of downstream neuronal target exons. Genes Dev. 2009;23:2284–2293. doi: 10.1101/gad.1837009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee J.A., Xing Y., Nguyen D., Xie J., Lee C.J., Black D.L. Depolarization and CaM kinase IV modulate NMDA receptor splicing through two essential RNA elements. PLoS Biol. 2007;5:e40. doi: 10.1371/journal.pbio.0050040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simonds A.K. Respiratory complications of the muscular dystrophies. Semin. Respir. Crit. Care Med. 2002;23:231–238. doi: 10.1055/s-2002-33031. [DOI] [PubMed] [Google Scholar]
- 38.Alter J., Lou F., Rabinowitz A., Yin H., Rosenfeld J., Wilton S.D., Partridge T.A., Lu Q.L. Systemic delivery of morpholino oligonucleotide restores dystrophin expression bodywide and improves dystrophic pathology. Nat. Med. 2006;12:175–177. doi: 10.1038/nm1345. [DOI] [PubMed] [Google Scholar]
- 39.Yue Y., Ghosh A., Long C., Bostick B., Smith B.F., Kornegay J.N., Duan D. A single intravenous injection of adeno-associated virus serotype-9 leads to whole body skeletal muscle transduction in dogs. Mol. Ther. 2008;16:1944–1952. doi: 10.1038/mt.2008.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moulton H.M., Moulton J.D. Morpholinos and their peptide conjugates: therapeutic promise and challenge for Duchenne muscular dystrophy. Biochim. Biophys. Acta. 2010;1798:2296–2303. doi: 10.1016/j.bbamem.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 41.Zhao F., Li P., Chen S.R., Louis C.F., Fruen B.R. Dantrolene inhibition of ryanodine receptor Ca2+ release channels. Molecular mechanism and isoform selectivity. J. Biol. Chem. 2001;276:13810–13816. doi: 10.1074/jbc.M006104200. [DOI] [PubMed] [Google Scholar]
- 42.Oo Y.W., Gomez-Hurtado N., Walweel K., van Helden D.F., Imtiaz M.S., Knollmann B.C., Laver D.R. Essential Role of Calmodulin in RyR Inhibition by Dantrolene. Mol. Pharmacol. 2015;88:57–63. doi: 10.1124/mol.115.097691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barthelemy F., Wang D., Nelson S.F., Miceli M.C. Validation and Detection of Exon Skipping Boosters in DMD Patient Cell Models and mdx Mouse. Methods Mol. Biol. 2018;1828:309–326. doi: 10.1007/978-1-4939-8651-4_19. [DOI] [PubMed] [Google Scholar]