Abstract
Although our knowledge of human diseases has increased dramatically, the molecular basis, phenotypic traits, and therapeutic targets of most diseases still remain unclear. An increasing number of studies have observed that similar diseases often are caused by similar molecules, can be diagnosed by similar markers or phenotypes, or can be cured by similar drugs. Thus, the identification of diseases similar to known ones has attracted considerable attention worldwide. To this end, the associations between diseases at the molecular, phenotypic, and taxonomic levels were used to measure the pairwise similarity in diseases. The corresponding performance assessment strategies for these methods involving the terms “category-based,” “simulated-patient-based,” and “benchmark-data-based” were thus further emphasized. Then, frequently used methods were evaluated using a benchmark-data-based strategy. To facilitate the assessment of disease similarity scores, researchers have designed dozens of tools that implement these methods for calculating disease similarity. Currently, disease similarity has been advantageous in predicting noncoding RNA (ncRNA) function and therapeutic drugs for diseases. In this article, we review disease similarity methods, evaluation strategies, tools, and their applications in the biomedical community. We further evaluate the performance of these methods and discuss the current limitations and future trends for calculating disease similarity.
Keywords: disease similarity, phenotypic traits, molecular basis, ncRNA function, therapeutic drugs
Introduction
Human disease is one of the permanent aspects of the human condition, similar to birth, aging, and death, from a philosophical point of view. The search for novel understanding of disease never stops. Although, currently, there has been great success with the development of biotechnology, the molecular basis of and therapeutic agents for most diseases remain unclear. Current studies have observed that similar diseases are often caused by similar molecules,1, 2, 3 can be diagnosed by similar markers or phenotypes,4, 5, 6 and are also cured by similar drugs.7, 8, 9, 10, 11 Based on this, novel functional molecules for a disease could, in theory, be revealed using prior knowledge of similar diseases.12, 13, 14, 15, 16, 17, 18 Thus, research on identifying the similarity between diseases has attracted increasing attention.
A pair of diseases with a high similarity score can be defined as being similar diseases. To measure disease similarity, prior knowledge of diseases plays a crucial role. The symptoms and signs accompanying diseases, also called phenotypes, are the intuitive characteristics of a disease.19,20 As early as 2004, Freudenberg and Propping21 used phenotypes sourced from the Online Mendelian Inheritance in Man (OMIM) website22 to calculate the similarity of OMIM diseases. With an ever-increasing number of phenotypes being observed by the biomedical community, abundant algorithms have been developed for measuring disease similarity at a phenotypic level.
Many studies have shown that the alterations of molecules can lead to the occurrence of diseases. Thus, the exploration of a common molecular basis is another way to measure disease similarity. With the development of next-generation sequencing technologies, a vast number of protein-coding genes (PCGs) and noncoding RNA (ncRNA) genes associated with diseases have been identified. For example, hemophilia A is an X-linked recessive bleeding disorder caused by a deficiency in the activity of coagulation factor VIII (F8), which can be affected by variations in the F8 genes.23,24 MicroRNA (miRNA)-155 is an endogenous ncRNA that regulates several mRNAs to cause B cell lymphomas.25,26 Based on the molecular basis of diseases, a large number of methods27, 28, 29, 30, 31, 32, 33 have been designed for calculating disease similarity, using this as a metric.
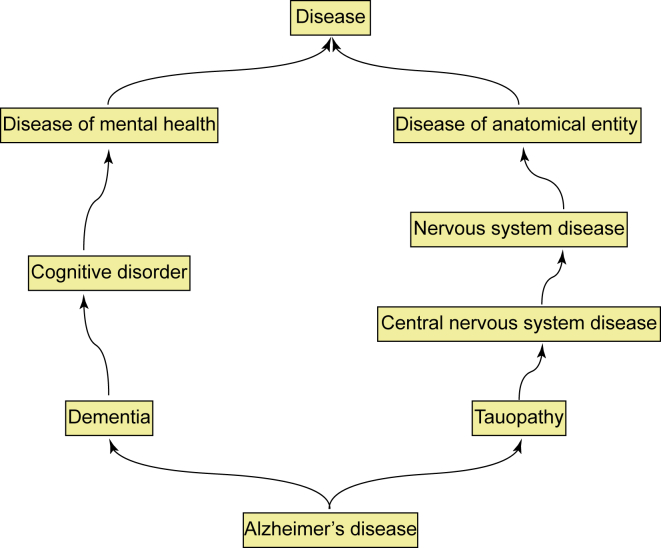
Recently, disease taxonomy has begun to play an important role in measuring disease similarity. One of the typical taxonomic classifiers for diseases is Disease Ontology (DO).34 In this, each disease term represents a disease with different names, and two terms can be linked on the basis of a set of inclusive relationships. For example, “Alzheimer’s disease” can be linked to “tauopathy.” All of the disease terms and the set of inclusion relationships forms the disease hierarchy and directed acyclic graph (DAG) of DO (Figure 1), where a node represents a disease term, and an edge is a set of inclusive relationships between the two terms. The common ancestors of two disease terms based on the DAG have often been utilized to calculate the similarity of two terms.35
Figure 1.
Sub-graph of the DO Hierarchy for Alzheimer’s Disease
Arrows represent an “IS_A” relationship for DO. For example, “Alzheimer’s disease” is linked to “Dementia” by an “IS_A” relationship. All of the terms that can be linked by “IS_A” relationships in the graph from “Alzheimer’s disease” are the ancestors of “Alzheimer’s disease.” All of the terms that can link to “Disease” by “IS_A” relationships are the descendants of “Disease.”
Currently, dozens of methods have been designed for calculating disease similarity based on prior disease knowledge at the phenotypic, molecular, and hierarchical levels. In this article, we review the main topics of investigation in disease similarity, including the proper selection of proper data, the design and implementation of methods, the evaluation of a method’s performance, and even the application of existing methods for predicting molecular factors of diseases.
Data Sources
Three types of data sources, including disease vocabularies, disease annotations, and gene functional annotations, are widely utilized for calculating disease similarity (Table 1). Here, we list and introduce these main data sources.
Table 1.
Summary of Data Sources
| Category and Name | Creation Date | Initiator | PMID |
|---|---|---|---|
| Disease Vocabulary | |||
| OMIM | 1960s | McKusick36 | 17357067 |
| MeSH | 1960s | Winifred Sewell38 | 14119288 |
| UMLS | 1980s | Olivier Bodenreider41 | 14681409 |
| SNOMED CT | 2001 | Wang et al.46 | 11825284 |
| DO | 2003 | Schriml et al.34 | 22080554 |
| MEDIC | 2012 | Davis et al.39 | 22434833 |
| Disease Annotations | |||
| GeneRIF | 2007 | 17990498 | |
| CTD | 2003 | 27651457 | |
| GAD | 2004 | Becker et al.48 | 15118671 |
| miR2Disease | 2008 | Jiang et al.54 | 18927107 |
| HPO | 2008 | Robinson et al.5 | 18950739 |
| SpliceDisease | 2011 | 22139928 | |
| lncRNADisease | 2012 | 23175614 | |
| HMDD v2.0 | 2013 | 24194601 | |
| SIDD | 2013 | Cheng et al.62 | 24146757 |
| OAHG | 2016 | Cheng et al.61 | 27703231 |
| Gene Functional Annotations | |||
| GOA | 2003 | Camon et al.63 | 12654719 |
| HumanNet | 2011 | Lee et al.66 | 21536720 |
Disease Vocabularies
Disease vocabularies document disease terms for distinguishing between different diseases. Each disease term in a vocabulary contains a unique identifier, preferred disease name, synonyms, abbreviations, and the definition of a disease. Parts of these vocabularies even provide a hierarchy of disease terms based on a set of inclusive relationships.
OMIM
The OMIM22,36 is a comprehensive, authoritative compendium of genetic diseases, which is freely available and updated daily. It was initiated in the early 1960s by Dr. Victor A. McKusick and has been developed for online usage by the NCBI since 1985.
MeSH
The Medical Subject Headings (MeSH)37,38 provides hierarchically organized terminology for indexing and cataloging biomedical information for PubMed. MeSH divides all biomedical terms into 16 categories, in which C and F03 contain disease names, containing more than 4,600 disease terms. In addition to the terms in these categories, MeSH also contains supplementary term records, which document thousands of disease terms.
MEDIC
The “merged disease vocabulary” (MEDIC)39 was established by the Comparative Toxicogenomics Database (CTD)40 biocurators and is composed of more than 10,000 unique diseases. To take advantage of the familiarity and immediate genetic data offered by OMIM terms, as well as the navigation utility and PubMed indexing feature of MeSH terms, MEDIC integrates OMIM terms with MeSH terms and hierarchical relationships.
UMLS
The Unified Medical Language System (UMLS)41 is a repository of biomedical vocabularies developed by the U.S. National Library of Medicine (NLM). The UMLS integrates over 2 million names for some 900,000 concepts from more than 60 families of biomedical vocabularies, as well as 12 million relations between these concepts. Vocabularies integrated in the UMLS Metathesaurus include MeSH, OMIM, Gene Ontology (GO),42 and so forth.
DO
The Disease Ontology (DO) database34 was developed to create a single structure for the classification of diseases that unifies the representation of disease between varied vocabularies into a relational ontology. DO terms can be linked in a hierarchy by a type of semantic association called an “IS_A” relationship43 (Figure 1). The initial builds of DO in 2003 and 2004 used the International Classification of Diseases (ICD-9)44 as the foundational vocabulary. Recent revisions have improved this with the reorganization of DO based on UMLS disease terms in conjunction with term mappings to Systematized Nomenclature of Medicine--Clinical Terms (SNOMED CT)45,46 and ICD-9. The current version of DO is organized into eight main classes to represent cellular proliferation, mental health, anatomical entity, infectious, and agent, etc.
Disease Annotations
The molecular basis and phenotypic characterization of a disease are two main aspects of prior knowledge often used for measuring disease similarity. Resources collecting these sources of prior knowledge are called disease annotations.
Disease Annotations of PCGs
Disease-related PCGs are mainly documented in the OMIM, Gene Reference into Function (GeneRIF),47 Genetic Association Database (GAD),48 SpliceDisease,49 and CTD databases. OMIM was intended for use primarily by physicians and other professionals concerned with genetic disorders. GeneRIF provides functional annotations of genes from the NCBI and allows scientists to add a short functional summary of NCBI genes that is limited to 425 characters. The GAD emphasizes genetic association data from complex diseases and disorders. SpliceDisease provides detailed descriptions of the relationships between gene variations, splicing defects, and diseases. The CTD documents the interactions between chemicals and gene products, as well as their relationships to diseases. The relationships between genes and diseases in the CTD often comes in the form of information about RNA splicing, SNPs, and so on.
Disease Annotations of miRNAs
miRNAs are a class of endogenous single-stranded small ncRNAs that play a crucial role in various human diseases by negatively regulating the expression of PCGs.50, 51, 52, 53 Two manually curated data sources of disease-miRNA relationships include miR2Disease54 and the Human miRNA Disease Database (HMDD) v2.0.55 Both of these two resources document miRNA deregulation in various human diseases.
Disease Annotations of lncRNAs
Long ncRNAs (lncRNAs) are mRNA-like transcripts that are longer than 200 nt and have little or no protein-coding capacity.56,57 According to the theory of competing endogenous RNA (ceRNA),58 they can affect the expression of PCGs through competitively binding with miRNAs. Thus, it becomes important to understand the role of lncRNAs in diseases.59 The LncRNADisease database has a manually accumulated set of relationships between lncRNAs and diseases.60
Disease Annotations of Phenotypes
Phenotypes are documented in the Clinical Synopsis section of the textual descriptions of each OMIM disease. Robinson et al.5 extracted all of the phenotypes from this text and constructed a human phenotype ontology (HPO) to annotate human diseases.
Integrated Resources of Disease Annotations
In previous efforts, we developed two integrated resources for disease annotations. integrated resource for annotating human genes with multi-level ontologies (OAHG)61 focused on the disease annotations of PCGs, miRNAs, and lncRNAs; and a semantically integrated database towards a global view of human disease (SIDD)62 documented disease-related molecular, phenotypic, and environmental features. The data sources integrated by OAHG involved OMIM, HMDD, and LncRNADisease. SIDD integrated up to 18 different data sources, including OMIM, GAD, CTD, LncRNADisease, and HPO.
Gene Functional Annotations
Similar molecular foundations of diseases may be influenced not only by common genes but also by different genes with common functions. Recently, associations between genes from gene functional annotation resources have been introduced for calculating disease similarity. Here, we list resources for the identification of gene functional annotations.
GOA
Disease-related PCGs can possess similar molecular functions (MFs), and may be involved in similar biological processes (BPs). This type of functional association of genes often exposes the similarity of different diseases. The GO annotation (GOA)63 of PCGs provides assignments of MF and BP terms of GO to gene products, in a project run by the European Bioinformatics Institute (EBI).
HumanNet
In addition to the GOA of PCGs, functional relationships between disease-related genes can also be reflected by protein-protein interactions,64 mRNA co-expression,65 and so forth. By integrating all of this data, HumanNet provides a more comprehensive relative score of pairwise PCG relationship.66
Disease Similarity Measures
The similarity between diseases can be reflected by their common phenotypic characteristic, molecular basis, and hierarchy structures. Therefore, we have classified the disease similarity methods into phenotype-based, molecule-based, hierarchy-based, and hybrid methods (Table 2).
Table 2.
Summary of Disease Similarity Methods
| Author(s) | Molecule Based | Phenotype Based | Hierarchy Based | Vocabulary | PMID (or Reference Number) | Year |
|---|---|---|---|---|---|---|
| Freudenberg and Propping21 | √ | OMIM | 12385992 | 2002 | ||
| van Driel et al.67 | √ | OMIM | 16493445 | 2006 | ||
| Köhler et al.68 | √ | OMIM | 19800049 | 2009 | ||
| Zhang et al.69 | √ | OMIM | 20659468 | 2010 | ||
| Zhou et al.72 | √ | MeSH | 24967666 | 2014 | ||
| Chen et al.73 | √ | UMLS | 25277758 | 2015 | ||
| Hoehndorf et al.119 | √ | DO | 26051359 | 2015 | ||
| Deng et al.120 | √ | OMIM | 25664462 | 2015 | ||
| Mabotuwana et al.92 | √ | SNOMED CT | 23850839 | 2013 | ||
| Mathur et al.99 | √ | DO | 21347137 | 2010 | ||
| Suthram et al.78 | √ | UMLS | 20140234 | 2010 | ||
| Gottlieb et al.8 | √ | UMLS | 21654673 | 2011 | ||
| Hamaneh and Yu82 | √ | OMIM/MeSH | 25360770 | 2014 | ||
| Kim et al.83 | √ | PharmGKB | 26212477 | 2015 | ||
| Wang et al.35 | √ | DO/MeSH | 17344234 | 2007 | ||
| Resnik27 | √ | √ | DO | 27 | 1995 | |
| Lin126 | √ | √ | DO | 28 | 1998 | |
| Schlicker et al.98 | √ | √ | 16776819 | 2006 | ||
| Mathur et al. | √ | √ | DO | 22166490 | 2012 | |
| Cheng et al.91 | √ | √ | DO | 24932637 | 2014 |
Phenotype-Based Methods
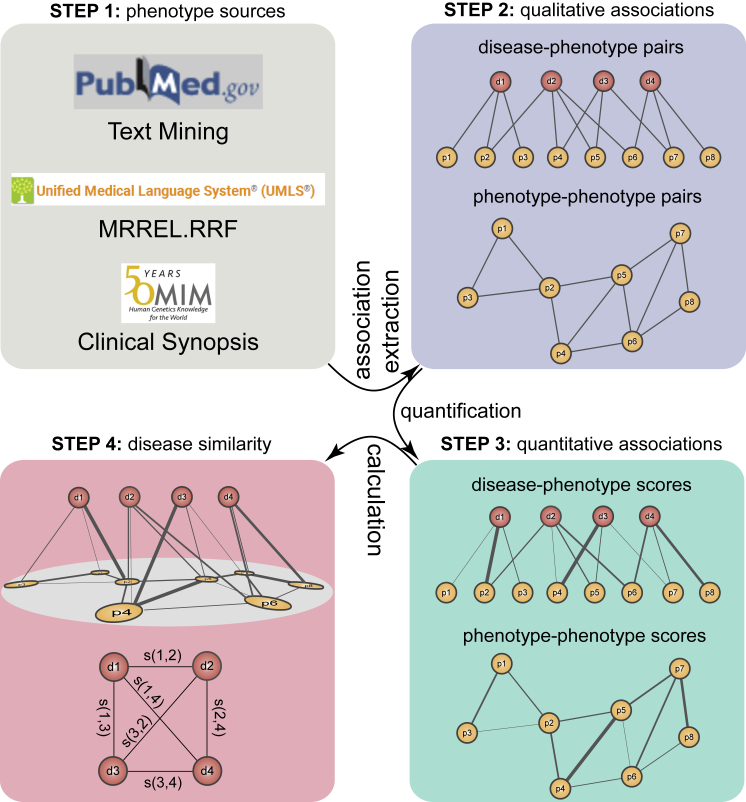
Figure 2 shows the schematic process of phenotype-based methods. First, qualitative associations between phenotypes and diseases are extracted from phenotype data sources. Then, each pair of qualitative associations is quantified as a disease-phenotype score or phenotype-phenotype score. Finally, these scores are utilized for calculating disease similarity.
Figure 2.
Schematic of the Process of Phenotype-Based Methods
Freudenberg’s Method
OMIM diseases were originally attributed manually by Freudenberg and Propping21 according to their phenotypic appearance, using the indices “periodicity,” “etiology,” “tissue,” “age of onset,” and “mode of inheritance.” The index “periodicity” is a Boolean variable, indicating an episodic occurrence of a disease in contrast to a linear progression. The index “etiology” is based on clinical signs and laboratory or pathological findings related to a disease. The index “tissue” is compiled as the anatomic location of phenotype. The index “inheritance” indicates whether a disease is inherited in an autosomal-dominant, autosomal-recessive, X chromosome, mitochondrial, or complex manner. The index “age of onset” refers to the age of a patient when symptoms are generally first noticed. Then, the similarity of diseases d1 and d2 is defined as the following:
| (Equation 1) |
where wi represents the contribution of a single index to the total similarity score, and sim(d1.indexi, d2.indexi) indicates the similarity between the ith indexes of d1 and d2.
van Driel’s Method
van Driel et al.67 calculated the similarity between over 5,000 diseases based on phenotypic features of OMIM records. For each OMIM disease, its phenotypic descriptions were extracted from “TX” and “CS” fields. Then, the OMIM diseases and phenotypic descriptions were mapped to the anatomy (category A) and the disease (category C) sections of MeSH to establish disease-term associations. Each disease-term association was then defined as a vector with three features as follows:
| (Equation 2) |
| (Equation 3) |
and
| (Equation 4) |
where t1 and d1 represent a phenotype term and a disease, respectively. In Equations 2 and 4, counted(t1,d1) means the occurrence number of t1 in the OMIM records of d1. In Equation 3, N is the total number of records analyzed, and n1 is the number of records that contain the term t1. In Equation 4, descendant(t1) is the number of descendant terms in the hierarchy of MeSH, and descendant(t1,d1) is the number of descendant terms in the OMIM records of d1. The similarity between diseases d1 and d2 is then defined as Equation 5 below:
| (Equation 5) |
where t1,i and t2,i mean the ith term vector of d1 and d2, respectively; and m is the total number of phenotypic terms.
Freudenberg’s Method
Phenotypic terms of the “CS” field of OMIM records were also manually extracted to construct an HPO by Freudenberg.68 Then, the similarity of pairwise phenotypic terms was calculated based on Resnik’s method27 as follows:
| (Equation 6) |
where a is the ancestor of phenotypes p1 and p2, N is the total number of genes associated with the phenotypes, and n(a) is the number of genes associated with a. Then, the similarity of pairwise diseases d1 and d2 is defined as follows:
| (Equation 7) |
and
| (Equation 8) |
where n and m represent the number of phenotypes associated with d1 and d2, respectively.
Zhang’s Method
Zhang et al.69 extracted phenotypic terms from the “TX” and “CS” fields of OMIM’s disease records using a MetaMap transfer tool.70 As a result, each disease could be represented as a set of phenotypes. Then the weights of phenotypic terms for diseases were calculated based on a term frequency-inverse document frequency (TF-IDF) weighting scheme.71 Subsequently, each disease was represented as a weighted vector of these phenotypic terms. Finally, the similarity of pairwise diseases was defined as the cosine of their corresponding phenotypic vectors.
Zhou’s Method
Zhou et al.65,72 define a disease as a set of symptoms, which were extracted from PubMed. Each disease was described as a weighted vector of phenotypic terms. Here the weight was calculated by a TF-IDF weighting scheme. The similarity of a pairwise disease was then defined as the cosine of their vectors.
Chen’s Method
Chen et al.73 extracted the disease-phenotype relationships from the UMLS file MRREL.RRF where disease-phenotype relationships were documented based on OMIM, Ultrasound Structured Attribute Reporting,74 and Minimal Standard Digestive Endoscopy Terminology.75 This group then used the information content (IC) to weight each phenotype concept as follows:
| (Equation 9) |
where N is the total number of diseases, and n1 is the number of diseases associated with a phenotype p1. Then they modeled the phenotype similarity of pairwise diseases by the cosine of their feature vectors.
Molecule-Based Methods
The schematic process of molecule-based methods is analogous to that of the previously stated phenotype-based methods. Here, genes are the mainly disease-related molecules. Phenotypic-based methods always utilized the semantics associations between phenotypes. In comparison, genes can be associated in more ways, such as in terms of protein-protein interactions (PPIs), co-expression, and so forth.
Mathur’s Method
SwissProt76 documents proteins that have been manually annotated with diseases, which were mapped to DO terms using MetaMap by Mathur and Dinakarpandian.77 Then, the similarity of diseases d1 and d2 was calculated based on their corresponding genes as follows:
| (Equation 10) |
where G1 and G2 are gene sets of diseases d1 and d2, respectively, |.| is the number of terms in the specified set, and N is the total number of genes.
Suthram’s Method
Suthram et al.78 compared diseases using an integrated analysis of disease-related mRNA expression data and the human protein interaction network.78 First, they identified conserved functional modules of genes using PathBLAST79 based on PPI data from the Human Protein Reference Database (HPRD).80 Next, they normalized the gene expression data in each microarray sample using a Z-score transformation and computed the activity level of each gene in a disease. Then, the module response score for each module in a disease was assigned to be the mean of the gene activity score of its component genes. Finally, they calculated the partial correlation coefficient between diseases based on the corresponding module response score and defined it as the disease similarity.
Gottlieb’s Method
Gottlieb et al.8 presented four algorithms for calculating disease similarity using the genetic signatures of diseases from gene expression experiments,8 which involved signature-based, signature sequence-based, signature PPI-based, and signature GO-based methods. The signature-based method utilized a Jaccard index between every pair of disease signatures to calculate disease similarity as follows:
| (Equation 11) |
where G1 and G2 are the signatures of diseases d1 and d2, respectively, and |.| is the number of terms in the specified set.
The signature PPI-based method calculated the distances between each pair of disease signatures based on their corresponding proteins using an all-pairs shortest paths algorithm on the human PPI network. Distances were transformed into similarity values using the following formula:
| (Equation 12) |
where P1 and P2 are the corresponding proteins of diseases d1 and d2, respectively, and D(P1, P2) is the shortest path between these proteins in the PPI network. A is a parameter chosen to be 0.9 × e by Perlman et al.81
The signature sequence-based method calculated the Smith-Waterman sequence alignment score between disease signatures and then divided the score by the geometric mean of the scores from aligning each sequence against itself. In addition, the signature GO-based method calculated the similarity between each pair of disease signatures based on their corresponding GO terms.
Hamaneh’s Method
Hamaneh and Yu82 devised a network-based measure to calculate disease similarity. First, they assigned weights to all proteins by using information flow from a disease to the human PPI network and back. As a result, each disease was represented as a weighted vector whose dimension is the number of proteins in the network. Then, the similarity of two diseases was defined as the cosine of the angle between their corresponding vectors.
Kim’s Method
Kim et al.83 extracted disease-gene pairs and disease-drug pairs from the literature and used the frequencies of co-occurrence relationships as features to calculate disease similarity.83 In this work, disease names, gene symbols, and drug names were from the Pharmacogenomics Knowledgebase (PharmGKB).84 This assumes that G1 and G2 are genes that occurred in the same sentence as diseases d1 and d2, respectively. D1 and D2 are drugs that occurred in the same sentence as diseases d1 and d2, respectively. The similarity of d1 and d2, therefore, can be defined as the following:
| (Equation 13) |
| (Equation 14) |
and
| (Equation 15) |
where N and M are the total number of genes and drugs, respectively.
Hierarchy-Based Methods
Hierarchy-based approaches are based only on the hierarchical structure of disease-related ontologies. In the previously mentioned studies, multiple methods have been presented for calculating the similarity of ontology terms using shared path and distance based on hierarchical structures85, 86, 87, 88, 89. However, currently only Wang’s method is widely utilized for calculating disease similarity.
Wang’s Method
Assuming that D1 is the set including d1 and all of its ancestor terms in an ontology-based “IS_A” relationship, the hierarchical contribution of the terms d to d1 is represented as follows:
| (Equation 16) |
where w is a hierarchical contribution factor for hierarchical association. According to Wang et al.35,90 and Cheng et al.,91 w is defined as 0.5 for an “IS_A” relationship of DO.34 Then, the value of the summation of all of the hierarchical contributions of D1 to d1 is SV(d1), which is defined as follows:
| (Equation 17) |
Assuming that D2 is the set including d2 and all of its ancestor terms, the similarity between d1 and d2 is defined by Wang’s method as follows:
| (Equation 18) |
Mabotuwana et al.’s Method
Mabotuwana et al.92 defined similarity of pairwise terms as inversely proportional to the distance between terms, as follows:
| (Equation 19) |
where d is the number of nodes in the shortest path between two diseases based on the DAG of ontology.
Hybrid Methods
Molecular and hierarchical associations between diseases have been combined as hybrid methods for calculating disease similarity. These methods often utilize disease-related genes to define the IC of diseases93, 94, 95 as follows:
| (Equation 20) |
where N denotes the total number of genes, and nd represents the number of genes of d. Here, disease-related genes are often based on OMIM,36 CTD,40 SIDD,62 OAHG,61 and so on.
Resnik’s Method
Early in 1995, Resnik27 presented a method for calculating the similarity between ontology terms. In 2002, this method was introduced for calculating the similarity between GO terms.96 In 2011, Li et al.97 utilized this method for calculating the similarity between DO terms. According to Resnik’s method, the similarity of pairwise diseases d1 and d227 equals the IC of the most informative common ancestor (MICA) of these two diseases as follows:
| (Equation 21) |
Lin’s Method
Concerned that the similarity between ontology terms should also be decided by the IC of the two terms, Lin28 improved Resnik’s method in 1998. According to Lin’s method28, the similarity of pairwise diseases d1 and d2 can be reflected by both the MICA of the disease pair and the IC of each disease as follows:
| (Equation 22) |
Schlicker’s Method
Schlicker et al.98 improved Resnik’s method from the same perspective as Lin, and they defined disease similarity as follows:
| (Equation 23) |
In this equation, ancestors(d1, d2) represents the common ancestor of diseases d1 and d2.
Mathur’s Method
In 2012, Mathur et al.99 designed a new method named PSB for calculating the similarity between DO terms. According to this method, the significance of related BPs terms from GO42 should be computed for each disease using a hypergeometric test.99 Assuming that d1 and d2 can be associated with m and n BP terms, respectively, the similarity of d1 and d2 is defined as follows:
| (Equation 24) |
where represents the similarity between two BPs p1i and p2j as follows:
| (Equation 25) |
Here, ICGO and ICDO represent the IC based on GO and DO, respectively. n(p1∩p2) and n(p1∪p2) denote the number of common genes of p1 and p2 and the number of total genes of p1 and p2, respectively.
Cheng’s Method
In addition to related BP, genes can be associated by PPI, co-expression, and so forth. Therefore, Cheng et al.91 presented the SemFunSim method to improve Mathur’s method by incorporating the gene functional network from HumanNet,66 which reflects the comprehensive gene associations from PPI, co-expression, BP, and so on. This assumes that G1 and G2 represent related gene sets of d1 and d2, respectively. Then, the similarity between t1 and t2 by Cheng et al.’s91 method is described by the following:
| (Equation 26) |
where |GMICA| represents the number of genes of MICA for t1 and t2 and m and n denote the number of genes in G1 and G2, respectively. Sim(g1i, g2j) is the functional similarity score between genes g1i and g2j from HumanNet.66
Performance Evaluation
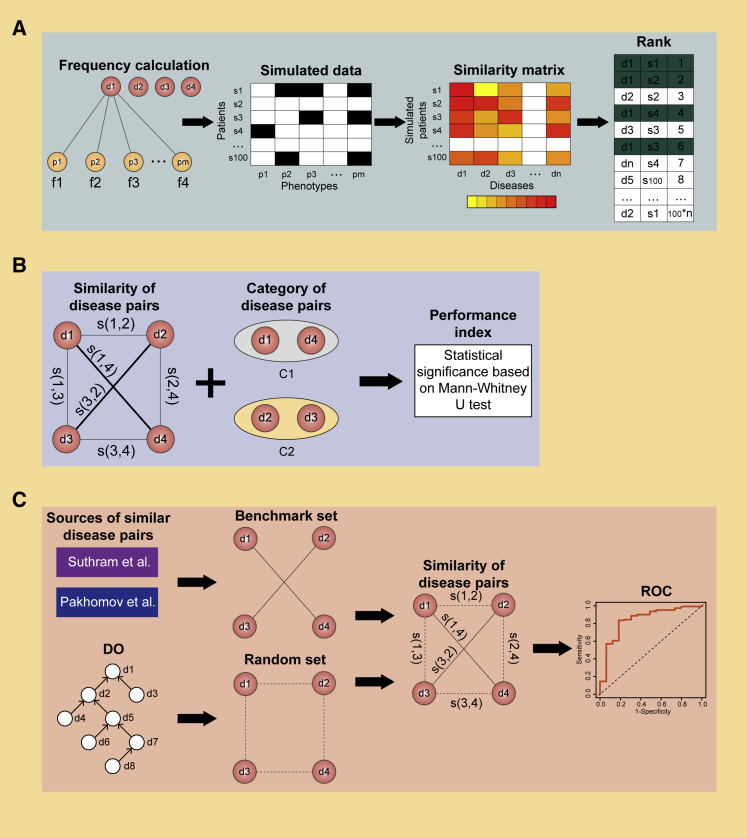
The performance of a disease similarity method can be affected by the quality of the prior knowledge it is based on. Most of the methods that utilize a manually curated dataset is high reliability. Some of the methods mentioned here use data from the literature extracted using text-mining tools. Data obtained in an unsupervised way should always be evaluated. In Mathur’s method,77 disease-related genes were mined from literature using MetaMap.70 The recall and precision were calculated based on a benchmark dataset from Monttaz et al.,100 which contained 200 records that were manually annotated by experts. The identified similarity pairs of diseases should always be then evaluated to measure the performance of the method used. Three types of classical evaluation strategies are introduced here (Figure 3).
Figure 3.
Schematic of the Process of Performance Evaluation
(A) Performance evaluation of a simulated patient-based method. (B) Performance evaluation of a term-category-based method. (C) Performance evaluation of a benchmark-data-based method.
Simulated-Patient-Based Strategy
In consideration of the difficulty in obtaining phenotypic information about a large number of patients, Sebastian et al.68 presented a simulated-patient-based method to evaluate their phenotype-based disease similarity method. We used 44 complex dysmorphology syndromes for which adequate frequency phenotypes were available, and then 100 virtual patients for each disease were generated on the basis of the frequency of phenotypes among persons diagnosed with a certain disease. For example, to generate patients with phenotypes A and B, in which A occurs in 40% and B occurs in 60% of patients, a random number generator was utilized to generate two random numbers uniformly distributed between 0 and 100. Subsequently, the similarity of the simulated patient to each of the OMIM diseases was calculated and then ranked. The average rank of all of the patients was returned to assess the performance of the original method.
Term-Category-Based Strategy
Sun et al.101 utilized information on disease-related molecules to design a disease similarity measurement method. Their results were evaluated using the disease classification terminologies found in the ICD-9. Their assumption was that two similar diseases should be subjected to the same categories in the ICD-9. Therefore, the correlation between the similarity of diseases and their classifications can reflect the performance of this method. Since similarity scores are not normally distributed, they used a nonparametric test—the Mann-Whitney U test102—to assess the statistical significance of the disease similarity.
Benchmark Data-Based Strategy
In the previous study, Cheng et al.91 constructed a benchmark set containing 70 pairs of similar diseases, which were manually integrated from two datasets. One dataset was adapted from Suthram et al.78 from the literature. The other dataset was curated by medical residents.103
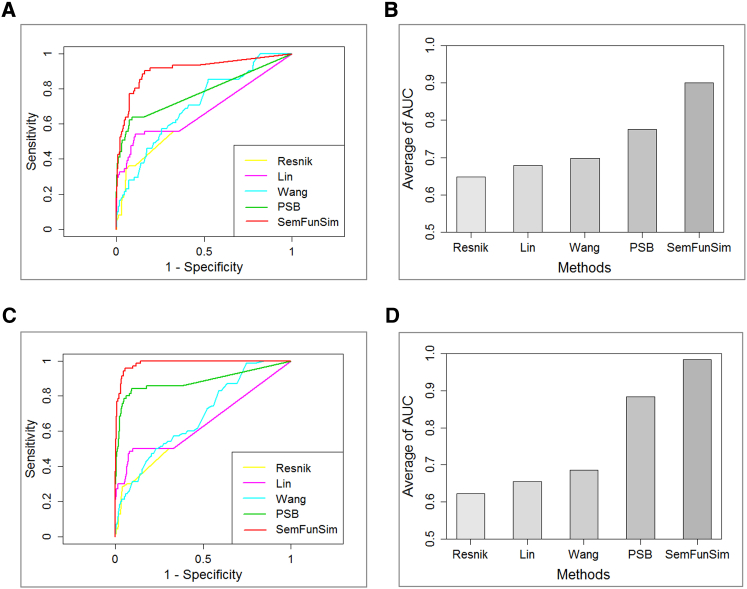
Here, we have evaluated the performance of Wang’s, Resnik’s, and Lin’s methods, PSB, and the SemFunSim using benchmark data. First, disease pairs of our benchmark dataset were deemed as positive groups, and 10-fold more disease pairs were randomly generated as a negative group. Next, the similarity of disease pairs of these two groups was calculated based on the aforementioned listed methods. Then, the area under receiver operating characteristic (ROC) curves (AUCs) was obtained. This process was iterated 100 times using different negative groups each time, and the average AUC reflects the respective performance of these methods.
Figure 4A shows the AUC of one of 100 iterations using disease-related genes from GeneRIF, while Figure 4B shows the average AUC of 100 iterations using disease-related genes from GeneRIF. The average AUC for Resnik’s, Lin’s, and Wang’s methods, PSB, and the SemFunSim were 0.6484, 0.6791, 0.6978, 0.7759, and 0.9008, respectively. Figures 4C and 4D show the results using disease-related genes from SIDD. The calculated average AUC for Resnik’s, Lin’s, and Wang’s methods, PSB, and the SemFunSim were 0.6209, 0.6351, 0.6849, 0.8843, and 0.9849, respectively.
Figure 4.
Performance Evaluation Using a Benchmark-Data-Based Strategy
(A) ROC curve for one of the 100 iterations using disease-related genes from GeneRIF. (B) The average AUC from 100 iterations using disease-related genes from GeneRIF. (C) ROC curve for one of the 100 iterations using disease-related genes from SIDD. (D) The average AUC from 100 iterations using disease-related genes from SIDD.
The performance of these methods are subject to the prior knowledge they used. Wang’s method only used the entire structure of the ontology; therefore, its performance is limited by the comprehensive of the ontology. Although Resnik’s and Lin’s methods incorporated the structure of ontology and ontology annotation, they do not utilize all the “IS_A” relationships of ontology. Thus, the performance of these three methods is not very good. In comparison with Resnik’s and Lin’s methods, PSB introduced GOA for associating disease-related genes. Thus, its performance improved a lot. Since disease-related genes could be associated in terms of PPIs, co-expression, and so on, the performance of PSB is improved much more by the SemFunSim method.
Applications
Disease similarity can be determined at the molecular, phenotypic, and hierarchical levels. Conversely, similar diseases reflect the correlations of their inducing molecules, phenotypes, and classifications. Therefore, disease similarity has been widely applied in the functional prediction of molecules, clinical diagnosis, and the establishment of disease associations.
The Functional Prediction of Molecules
This is based on the observation that genes causing similar diseases tend to lie close to one another in a network of PPI.104,105 Vanunu et al.104 constructed a comprehensive network using gene-disease association, disease similarity, and PPI data to predict disease-related PCGs using a random walk method.106
In comparison with PCGs, it is not easy to determine the function of ncRNAs due to limited knowledge with regard to their impact on proteins from wet lab experiments with these ncRNAs. Fortunately, disease similarity has been useful for this in previous investigations.90,107, 108, 109, 110 Based on prior knowledge of the associations between ncRNAs and diseases, functional similarity of ncRNAs can be calculated based on the similarities of their related diseases to construct a network in which an ncRNA is represented as a node and the similarity of pairwise ncRNAs is represented as edges.90 Just such a network was then utilized for predicting novel ncRNA-disease associations by the random walk with restart (RWR) method.106,108,109
Recently, disease similarity has been utilized for mining potential therapeutic drugs for diseases. Based on the observation that similar diseases can often be treated with similar drugs, Cheng et al.91,111 prioritized potential drugs for a disease based on their results with similar diseases. Gottlieb et al.8 combined disease similarity and drug similarity to predict novel drug indications.
Clinical Diagnosis
The diagnosis process can be a challenging undertaking, given the large number of hereditary disorders and the range of partially overlapping clinical features associated with them. To resolve this problem, Robinson et al.5,68 established an HPO to calculate the disease similarity and diagnose diseases according to clinical phenotype. According to Equations 6, 7, and 8, disease similarity can be calculated based on their phenotype sets. For an individual patient, the similarity between OMIM diseases and clinical features could also be calculated based on this method. The similarity score in this case then reflects the probability of a potential disease in the patient.
Construction of Qualitative Associations of Diseases
In 2006, Goh et al.112 utilized the common genetic origin of diseases to construct a human disease network (HDN) from the molecular level based on OMIM. This was an early study that established a qualitative association between diseases from a quantitative perspective. A portion of each disease stems not as the consequence of the single genetic defects but, rather, the breakdown in molecular interaction networks. Thus, their associations cannot be reflected by this network. Therefore, the network was extended based on PPIs, metabolic networks, and different pathways.113, 114, 115
Recently, Zhou et al.72 established an HDN at the phenotypic level, where the link weight between two diseases quantified the disease similarity. Here, the symptoms of diseases were extracted from literature in PubMed. Each disease was described as a vector of phenotypes. Then, the similarity between diseases was defined as the cosine similarity of their vectors.
Tools for Calculating Disease Similarity
Inspired by the wide recent application of machine learning methods in bioinformatics,116, 117, 118 various algorithms have been implemented for calculating disease similarity using R and web-based programs67,68,90,97,111,119, 120, 121, 122, 123, 124 (Table 3). These tools play important roles in disease diagnosis, the prediction of drugs, and so forth. Here, we introduce four frequently used tools in detail.
Table 3.
Summary of Disease Similarity Tools
| Author(s) | Name | Type | Web Site | Vocabulary | PMID | Year |
|---|---|---|---|---|---|---|
| van Driel et al.67 | MimMiner | webpage | OMIM | 16493445 | 2006 | |
| Robinson et al.5 | Phenomizer | webpage | http://compbio.charite.de/phenomizer/ | OMIM | 19800049 | 2009 |
| Wang et al.90 | MISIM | webpage | MeSH | 20439255 | 2010 | |
| Li et al.97 | DOSim | R package | DO | 21714896 | 2011 | |
| Hoehndorf et al.119 | NA | webpage | http://aber-owl.net/aber-owl/diseasephenotypes/ | OMIM | 26051359 | 2015 |
| Hamaneh and Yu123 | DeCoaD | webpage | https://www.ncbi.nlm.nih.gov/CBBresearch/Yu/mn/DeCoaD/ | DO | 26047952 | 2015 |
| Deng et al.120 | HPOSim | R package | https://sourceforge.net/p/hposim/summary/ | OMIM | 25664462 | 2015 |
| Yu et al.121 | DOSE | R package | http://www.bioconductor.org/packages/release/bioc/html/DOSE.html | DO | 25677125 | 2015 |
| Cheng et al.111 | DisSim | webpage | http://bio-annotation.cn/DisSim | DO | 27457921 | 2016 |
| Cheng et al.122 | DisSetSim | webpage | http://bio-annotation.cn/DisSetSim/ | DO | 29297411 | 2017 |
| Cheng et al.124 | DincRNA | webpage | http://bio-annotation.cn:18080/DincRNAClient/#/Home | DO | 29365045 | 2018 |
MimMiner
van Driel et al.67 designed a phenotype-based method and implemented it as a tool—namely, MimMiner—for calculating the similarity of OMIM diseases. This tool provides interfaces to query the similar diseases related to an input diseases and is widely used in bioinformatics community. It should be noted that this tool needs to be updated due to the rapid increase in the size of the OMIM disease database.
Phenomizer
Phenomizer is an online tool that can be helpful in the diagnosis processes and is based on disease similarity.68 Currently, thousands of genetic disorders characterized by specific combinations of phenotypic features are documented in OMIM. The diagnosis process based on phenotypes is difficult without computer-based tools. Phenomizer allows an automatic correlation between phenotypic abnormalities and hereditary disorders found in OMIM. The p values are generated to evaluate the statistical significance of those correlation scores given by Phenomizer. This tool is also useful for suggesting additional possible phenotypic alterations for further evaluation in a patient of interest.
DOSim
DOSim is an R package used for computing the similarity between DO terms97 based on Wang’s method35 and nine hybrid methods involving Resnik’s method, Lin’s method, and so forth.93, 94, 95,98,125, 126, 127. This tool also implements utilities to calculate the similarity of genes based on their inducing diseases and conduct DO enrichment analysis.
DisSim
DisSim111 is an online system for exploring similar diseases in DO. It provides both the similarity of pairwise diseases and the significance of their similarity score. In addition, the system integrates therapeutic drugs for known diseases to predict potential drugs for other human diseases based on the observation that similar diseases can be treated with similar drugs.78
Discussion
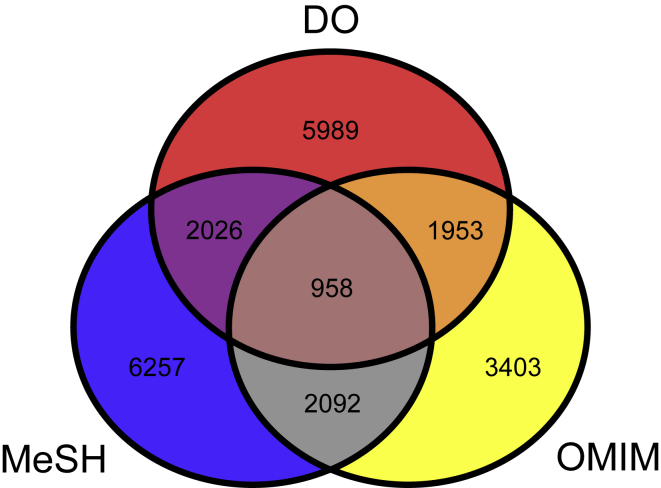
Most disease similarity methods depend on disease vocabularies and their annotations. Phenotype-based methods extract disease annotations of phenotypes from PubMed and OMIM. Disease names from these data sources are from MeSH and OMIM. Hierarchy-based methods utilize the structure of ontology from MeSH and DO. Current molecule-based methods mainly used the DO annotations of genes. In summary, DO, MeSH, and OMIM contain the most frequently used vocabularies for calculating disease similarity. However, not all disease terms are contained in any one of these vocabularies. For comparison, OMIM documents more specific disease terms, such as TYPE III SYNDACTYLY (OMIM: 186100). MeSH and DO involve classification of diseases, such as cancer (DOID: 162). Figure 5 shows the number of disease terms distributed across the different vocabularies. In total, 958 common disease terms are documented in DO, MeSH, and OMIM, which covers 8.8%, 8.5%, and 11.4% of DO, MeSH, and OMIM terms, respectively. Although OMIM and MeSH terms have been integrated into MEDIC, MEDIC lacks many DO terms and disease classifications. Therefore, combining all of the disease terms of DO, MeSH, and OMIM is critical for calculating disease similarity using the same vocabulary. In addition, a unified disease annotation database based on this integrated vocabulary is indispensable for improving the universality of similarity determining algorithms. In our previous studies, we provided a global view of human diseases by annotating disease-related molecule and phenotype features with DO.62,111 However, the absence of disease terms in DO limits its application.
Figure 5.
Distribution of Disease Terms in DO, MeSH, and OMIM
Disease-related ontologies only contain “IS_A” relationships, which limits the performance of hierarchy-based methods. For example, Wang’s method could be applied to multiple term associations of ontology, such as “IS_A,” “PART_OF,” “LOCATE_IN,” and so on. The performance evaluation results in Figure 4 shows that Wang et al.’s method could be improved, which may be achieved with the occurrence of more types of disease associations than the “IS_A” relationship.
Data quality and the quantity of disease annotations of phenotypes and molecules are crucial for the performance of molecule-based, phenotype-based, and hybrid-based methods. OMIM documents close but few disease-gene associations. Contrary to this, GeneRIF and SIDD retain loose but abundant associations. All of these datasets were combined together without distinction for calculating disease similarity in most cases. These methods could be improved by ranking all of the associations. For example, we can improve the disease annotations by adding the evidence for each disease-gene association such as that found in the GOA database.128
In general, newer methods should consider more types of prior knowledge, leading to better performance. Wang’s method,35 which is a hierarchy-based method, was presented in 2007. The SemFunSim method was presented in 2014, and it incorporates the hierarchical structure of DO, disease annotations of genes, and gene associations. The evaluation results in Figure 4 show that SemFunSim achieves a higher AUC than Wang’s method. Although hybrid methods integrate more types of prior knowledge of diseases, molecular and phenotypic associations of diseases were ignored. Therefore, it is possible that the performance of disease similarity methods could be further improved by fusing more disease knowledge types.
Although comprehensive knowledge benefits the calculative precision of disease similarity, these methods based on a single type of prior knowledge can also very valuable for biological applications. Diseases are often caused by the molecular mechanism and could be reflected by diverse phenotypes. Disease phenotypes can be detected from clinical diagnosis, while causal molecules are identified from wet labs. Gaps in phenotypic and molecular levels exist for understanding diseases. Here, disease similarity based on different types of knowledge could bridge the gap.
The purpose of calculating disease similarity is to identify similar diseases. However, it is not easy to determine similar diseases directly from most of the presented methods and tools. One feasible strategy for this purpose is provided here by DisSim,111 which provides the p values for each similarity score. According to current methods, the similarity of pairwise diseases can be obtained, which are then normalized to Z scores. Then, the one-side p values are calculated as a significance score for each similarity score. Another way to provide p values for similarity scores would be a permutation test.
Disease similarity plays important roles in mining the novel molecular features of diseases, clinical diagnosis, and so on. The exploration of the function of ncRNAs is a long-term challenge, as these RNAs do not produce proteins. Currently, disease similarity has been successful in predicting the function of ncRNAs, especially in prioritizing miRNA-disease14,129, 130, 131, 132, 133 and lncRNA-disease pairs.90,108 In the future, these methods can be used for comprehending the function of other types of ncRNAs, such as circular RNA (circRNAs).134 In a previous study, disease similarity was utilized for diagnosis based on phenotypes.68 This may also be helpful for molecular diagnosis. Alterations in the presence of metabolites are easily determined in the clinical, meaning metabolite-disease pairs can be prioritized based on disease similarity methods. Therefore, it is theoretically possible to predict potential diseases based on abnormalities in metabolite levels.
Author Contributions
L.C., J.H., S.L., and Q.J. conceived and designed the experiments. L.C., H.Z., P.W., W.Z., M.L., and T.L. analyzed data. L.C. wrote the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We thank LetPub (https://www.letpub.com) for its linguistic assistance during the preparation of the manuscript. This work was supported by the National Natural Science Foundation of China (grant nos. 61871160 and 61502125); the Heilongjiang Postdoctoral Fund (grant nos. LBH-TZ20 and LBH-Z15179); and the China Postdoctoral Science Foundation (grant nos. 2018T110315 and 2016M590291).
Contributor Information
Junwei Han, Email: hanjunwei1981@163.com.
Shulin Liu, Email: slliu@hrbmu.edu.cn.
Qinghua Jiang, Email: qhjiang@hit.edu.cn.
References
- 1.Aerts S., Lambrechts D., Maity S., Van Loo P., Coessens B., De Smet F., Tranchevent L.C., De Moor B., Marynen P., Hassan B. Gene prioritization through genomic data fusion. Nat. Biotechnol. 2006;24:537–544. doi: 10.1038/nbt1203. [DOI] [PubMed] [Google Scholar]
- 2.Franke L., van Bakel H., Fokkens L., de Jong E.D., Egmont-Petersen M., Wijmenga C. Reconstruction of a functional human gene network, with an application for prioritizing positional candidate genes. Am. J. Hum. Genet. 2006;78:1011–1025. doi: 10.1086/504300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chavali S., Barrenas F., Kanduri K., Benson M. Network properties of human disease genes with pleiotropic effects. BMC Syst. Biol. 2010;4:78. doi: 10.1186/1752-0509-4-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robinson P.N., Mundlos S. The human phenotype ontology. Clin. Genet. 2010;77:525–534. doi: 10.1111/j.1399-0004.2010.01436.x. [DOI] [PubMed] [Google Scholar]
- 5.Robinson P.N., Köhler S., Bauer S., Seelow D., Horn D., Mundlos S. The Human Phenotype Ontology: a tool for annotating and analyzing human hereditary disease. Am. J. Hum. Genet. 2008;83:610–615. doi: 10.1016/j.ajhg.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang W., Wan S., Yang Z., Teschendorff A.E., Zou Q. Tumor origin detection with tissue-specific miRNA and DNA methylation markers. Bioinformatics. 2018;34:398–406. doi: 10.1093/bioinformatics/btx622. [DOI] [PubMed] [Google Scholar]
- 7.Yu L., Ma X., Zhang L., Zhang J., Gao L. Prediction of new drug indications based on clinical data and network modularity. Sci. Rep. 2016;6:32530. doi: 10.1038/srep32530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gottlieb A., Stein G.Y., Ruppin E., Sharan R. PREDICT: a method for inferring novel drug indications with application to personalized medicine. Mol. Syst. Biol. 2011;7:496. doi: 10.1038/msb.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo H., Wang J., Li M., Luo J., Peng X., Wu F.X., Pan Y. Drug repositioning based on comprehensive similarity measures and Bi-Random walk algorithm. Bioinformatics. 2016;32:2664–2671. doi: 10.1093/bioinformatics/btw228. [DOI] [PubMed] [Google Scholar]
- 10.Yu L., Su R., Wang B., Zhang L., Zou Y., Zhang J., Gao L. Prediction of novel drugs for hepatocellular carcinoma based on multi-source random walk. IEEE/ACM Trans. Comput. Biol. Bioinformatics. 2017;14:966–977. doi: 10.1109/TCBB.2016.2550453. [DOI] [PubMed] [Google Scholar]
- 11.Yu L., Wang B., Ma X., Gao L. The extraction of drug-disease correlations based on module distance in incomplete human interactome. BMC Syst. Biol. 2016;10(Suppl 4):111. doi: 10.1186/s12918-016-0364-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X., Huang L. LRSSLMDA: Laplacian Regularized Sparse Subspace Learning for MiRNA-Disease Association prediction. PLoS Comput. Biol. 2017;13:e1005912. doi: 10.1371/journal.pcbi.1005912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen W., Feng P., Ding H., Lin H. Classifying included and excluded exons in exon skipping event using histone modifications. Front. Genet. 2018;9:433. doi: 10.3389/fgene.2018.00433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai H.Y., Feng C.Q., Zhang Z.Y., Tang H., Chen W., Lin H. A brief survey of machine learning application in cancerlectin identification. Curr. Gene Ther. 2018;18:257–267. doi: 10.2174/1566523218666180913112751. [DOI] [PubMed] [Google Scholar]
- 15.Chen X., Yan G.Y. Novel human lncRNA-disease association inference based on lncRNA expression profiles. Bioinformatics. 2013;29:2617–2624. doi: 10.1093/bioinformatics/btt426. [DOI] [PubMed] [Google Scholar]
- 16.Jiang L., Xiao Y., Ding Y., Tang J., Guo F. Discovering cancer subtypes via an accurate fusion strategy on multiple profile data. Front. Genet. 2019;10:20. doi: 10.3389/fgene.2019.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu L., Huang J., Ma Z., Zhang J., Zou Y., Gao L. Inferring drug-disease associations based on known protein complexes. BMC Med. Genomics. 2015;8(Suppl 2):S2. doi: 10.1186/1755-8794-8-S2-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L., Ping P.Y., Kuang L.N., Ye S.T., Lqbal F.M.B., Pei T.R. A novel approach based on bipartite network to predict human microbe-disease associations. Curr. Bioinform. 2018;13:141–148. [Google Scholar]
- 19.Albuisson J., Isidor B., Giraud M., Pichon O., Marsaud T., David A., Le Caignec C., Bezieau S. Identification of two novel mutations in Shh long-range regulator associated with familial pre-axial polydactyly. Clin. Genet. 2011;79:371–377. doi: 10.1111/j.1399-0004.2010.01465.x. [DOI] [PubMed] [Google Scholar]
- 20.Gurnett C.A., Bowcock A.M., Dietz F.R., Morcuende J.A., Murray J.C., Dobbs M.B. Two novel point mutations in the long-range SHH enhancer in three families with triphalangeal thumb and preaxial polydactyly. Am. J. Med. Genet. A. 2007;143A:27–32. doi: 10.1002/ajmg.a.31563. [DOI] [PubMed] [Google Scholar]
- 21.Freudenberg J., Propping P. A similarity-based method for genome-wide prediction of disease-relevant human genes. Bioinformatics. 2002;18(Suppl 2):S110–S115. doi: 10.1093/bioinformatics/18.suppl_2.s110. [DOI] [PubMed] [Google Scholar]
- 22.Amberger J., Bocchini C., Hamosh A. A new face and new challenges for Online Mendelian Inheritance in Man (OMIM®) Hum. Mutat. 2011;32:564–567. doi: 10.1002/humu.21466. [DOI] [PubMed] [Google Scholar]
- 23.Mannucci P.M., Tuddenham E.G. The hemophilias--from royal genes to gene therapy. N. Engl. J. Med. 2001;344:1773–1779. doi: 10.1056/NEJM200106073442307. [DOI] [PubMed] [Google Scholar]
- 24.Mazurier C., Parquet-Gernez A., Gaucher C., Lavergne J.M., Goudemand J. Factor VIII deficiency not induced by FVIII gene mutation in a female first cousin of two brothers with haemophilia A. Br. J. Haematol. 2002;119:390–392. doi: 10.1046/j.1365-2141.2002.03819.x. [DOI] [PubMed] [Google Scholar]
- 25.Kluiver J., Poppema S., de Jong D., Blokzijl T., Harms G., Jacobs S., Kroesen B.J., van den Berg A. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J. Pathol. 2005;207:243–249. doi: 10.1002/path.1825. [DOI] [PubMed] [Google Scholar]
- 26.Eis P.S., Tam W., Sun L., Chadburn A., Li Z., Gomez M.F., Lund E., Dahlberg J.E. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc. Natl. Acad. Sci. USA. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Resnik P. Using information content to evaluate semantic similarity in a taxonomy. arXiv. 1995 https://arxiv.org/abs/cmp-lg/9511007v1 arXiv:cmp-lg/9511007v1. [Google Scholar]
- 28.Lin D. An information-theoretic definition of similarity. ICML’98: Proceedings of the 15th International Conference on Machine Learning. 1998;98:296–304. [Google Scholar]
- 29.Jiang L., Xiao Y., Ding Y., Tang J., Guo F. FKL-Spa-LapRLS: an accurate method for identifying human microRNA-disease association. BMC Genomics. 2018;19(Suppl 10):911. doi: 10.1186/s12864-018-5273-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang L., Ding Y., Tang J., Guo F. MDA-SKF: similarity kernel fusion for accurately discovering miRNA-disease association. Front. Genet. 2018;9:618. doi: 10.3389/fgene.2018.00618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu L., Zhao J., Gao L. Drug repositioning based on triangularly balanced structure for tissue-specific diseases in incomplete interactome. Artif. Intell. Med. 2017;77:53–63. doi: 10.1016/j.artmed.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 32.Chen X., Wang L., Qu J., Guan N.N., Li J.Q. Predicting miRNA-disease association based on inductive matrix completion. Bioinformatics. 2018;34:4256–4265. doi: 10.1093/bioinformatics/bty503. [DOI] [PubMed] [Google Scholar]
- 33.Chen X., Sun Y.Z., Guan N.N., Qu J., Huang Z.A., Zhu Z.X., Li J.Q. Computational models for lncRNA function prediction and functional similarity calculation. Brief. Funct. Genomics. 2019;18:58–82. doi: 10.1093/bfgp/ely031. [DOI] [PubMed] [Google Scholar]
- 34.Schriml L.M., Arze C., Nadendla S., Chang Y.W., Mazaitis M., Felix V., Feng G., Kibbe W.A. Disease Ontology: a backbone for disease semantic integration. Nucleic Acids Res. 2012;40:D940–D946. doi: 10.1093/nar/gkr972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J.Z., Du Z., Payattakool R., Yu P.S., Chen C.F. A new method to measure the semantic similarity of GO terms. Bioinformatics. 2007;23:1274–1281. doi: 10.1093/bioinformatics/btm087. [DOI] [PubMed] [Google Scholar]
- 36.McKusick V.A. Mendelian Inheritance in Man and its online version, OMIM. Am. J. Hum. Genet. 2007;80:588–604. doi: 10.1086/514346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lowe H.J., Barnett G.O. Understanding and using the medical subject headings (MeSH) vocabulary to perform literature searches. JAMA. 1994;271:1103–1108. [PubMed] [Google Scholar]
- 38.Sewell W. Medical subject headings in MEDLARS. Bull. Med. Libr. Assoc. 1964;52:164–170. [PMC free article] [PubMed] [Google Scholar]
- 39.Davis A.P., Wiegers T.C., Rosenstein M.C., Mattingly C.J. MEDIC: a practical disease vocabulary used at the Comparative Toxicogenomics Database. Database (Oxford) 2012;2012:bar065. doi: 10.1093/database/bar065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis A.P., Grondin C.J., Johnson R.J., Sciaky D., King B.L., McMorran R., Wiegers J., Wiegers T.C., Mattingly C.J. The Comparative Toxicogenomics Database: update 2017. Nucleic Acids Res. 2017;45(D1):D972–D978. doi: 10.1093/nar/gkw838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32:D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith B., Ceusters W., Klagges B., Köhler J., Kumar A., Lomax J., Mungall C., Neuhaus F., Rector A.L., Rosse C. Relations in biomedical ontologies. Genome Biol. 2005;6:R46. doi: 10.1186/gb-2005-6-5-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deyo R.A., Cherkin D.C., Ciol M.A. Adapting a clinical comorbidity index for use with ICD-9-CM administrative databases. J. Clin. Epidemiol. 1992;45:613–619. doi: 10.1016/0895-4356(92)90133-8. [DOI] [PubMed] [Google Scholar]
- 45.Donnelly K. SNOMED-CT: The advanced terminology and coding system for eHealth. Stud. Health Technol. Inform. 2006;121:279–290. [PubMed] [Google Scholar]
- 46.Wang A.Y., Barrett J.W., Bentley T., Markwell D., Price C., Spackman K.A., Stearns M.Q. Mapping between SNOMED RT and Clinical Terms version 3: a key component of the SNOMED CT development process. Proc. AMIA Symp. 2001;2001:741–745. [PMC free article] [PubMed] [Google Scholar]
- 47.Mitchell J.A., Aronson A.R., Mork J.G., Folk L.C., Humphrey S.M., Ward J.M. Gene indexing: characterization and analysis of NLM’s GeneRIFs. AMIA Annu. Symp. Proc. 2003;2003:460–464. [PMC free article] [PubMed] [Google Scholar]
- 48.Becker K.G., Barnes K.C., Bright T.J., Wang S.A. The genetic association database. Nat. Genet. 2004;36:431–432. doi: 10.1038/ng0504-431. [DOI] [PubMed] [Google Scholar]
- 49.Wang J., Zhang J., Li K., Zhao W., Cui Q. SpliceDisease database: linking RNA splicing and disease. Nucleic Acids Res. 2012;40:D1055–D1059. doi: 10.1093/nar/gkr1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 51.Chen Y., Yang X., Xu Y., Cao J., Chen L. Genomic analysis of drug resistant small cell lung cancer cell lines by combining mRNA and miRNA expression profiling. Oncol. Lett. 2017;13:4077–4084. doi: 10.3892/ol.2017.5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen X., Xie D., Zhao Q., You Z.H. MicroRNAs and complex diseases: from experimental results to computational models. Brief. Bioinform. 2019;20:515–539. doi: 10.1093/bib/bbx130. [DOI] [PubMed] [Google Scholar]
- 53.Chen X., Yin J., Qu J., Huang L. MDHGI: matrix decomposition and heterogeneous graph inference for miRNA-disease association prediction. PLoS Comput. Biol. 2018;14:e1006418. doi: 10.1371/journal.pcbi.1006418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang Q., Wang Y., Hao Y., Juan L., Teng M., Zhang X., Li M., Wang G., Liu Y. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009;37:D98–D104. doi: 10.1093/nar/gkn714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Y., Qiu C., Tu J., Geng B., Yang J., Jiang T., Cui Q. HMDD v2.0: a database for experimentally supported human microRNA and disease associations. Nucleic Acids Res. 2014;42:D1070–D1074. doi: 10.1093/nar/gkt1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mercer T.R., Dinger M.E., Mattick J.S. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 57.Cheng L., Wang P., Tian R., Wang S., Guo Q., Luo M., Zhou W., Liu G., Jiang H., Jiang Q. LncRNA2Target v2.0: a comprehensive database for target genes of lncRNAs in human and mouse. Nucleic Acids Res. 2019;47(D1):D140–D144. doi: 10.1093/nar/gky1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vučićević D., Schrewe H., Orom U.A. Molecular mechanisms of long ncRNAs in neurological disorders. Front. Genet. 2014;5:48. doi: 10.3389/fgene.2014.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen G., Wang Z., Wang D., Qiu C., Liu M., Chen X., Zhang Q., Yan G., Cui Q. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013;41:D983–D986. doi: 10.1093/nar/gks1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cheng L., Sun J., Xu W., Dong L., Hu Y., Zhou M. OAHG: an integrated resource for annotating human genes with multi-level ontologies. Sci. Rep. 2016;6:34820. doi: 10.1038/srep34820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cheng L., Wang G., Li J., Zhang T., Xu P., Wang Y. SIDD: a semantically integrated database towards a global view of human disease. PLoS ONE. 2013;8:e75504. doi: 10.1371/journal.pone.0075504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Camon E., Magrane M., Barrell D., Lee V., Dimmer E., Maslen J., Binns D., Harte N., Lopez R., Apweiler R. The Gene Ontology Annotation (GOA) database: sharing knowledge in UniProt with Gene Ontology. Nucleic Acids Res. 2004;32:D262–D266. doi: 10.1093/nar/gkh021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ortutay C., Vihinen M. Identification of candidate disease genes by integrating Gene Ontologies and protein-interaction networks: case study of primary immunodeficiencies. Nucleic Acids Res. 2009;37:622–628. doi: 10.1093/nar/gkn982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stuart J.M., Segal E., Koller D., Kim S.K. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 66.Lee I., Blom U.M., Wang P.I., Shim J.E., Marcotte E.M. Prioritizing candidate disease genes by network-based boosting of genome-wide association data. Genome Res. 2011;21:1109–1121. doi: 10.1101/gr.118992.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Driel M.A., Bruggeman J., Vriend G., Brunner H.G., Leunissen J.A. A text-mining analysis of the human phenome. Eur. J. Hum. Genet. 2006;14:535–542. doi: 10.1038/sj.ejhg.5201585. [DOI] [PubMed] [Google Scholar]
- 68.Köhler S., Schulz M.H., Krawitz P., Bauer S., Dölken S., Ott C.E., Mundlos C., Horn D., Mundlos S., Robinson P.N. Clinical diagnostics in human genetics with semantic similarity searches in ontologies. Am. J. Hum. Genet. 2009;85:457–464. doi: 10.1016/j.ajhg.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang S., Wu C., Li X., Chen X., Jiang W., Gong B.S., Li J., Yan Y.Q. From phenotype to gene: detecting disease-specific gene functional modules via a text-based human disease phenotype network construction. FEBS Lett. 2010;584:3635–3643. doi: 10.1016/j.febslet.2010.07.038. [DOI] [PubMed] [Google Scholar]
- 70.Aronson A.R. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc. AMIA Symp. 2001;2001:17–21. [PMC free article] [PubMed] [Google Scholar]
- 71.Wilbur W.J., Yang Y. An analysis of statistical term strength and its use in the indexing and retrieval of molecular biology texts. Comput. Biol. Med. 1996;26:209–222. doi: 10.1016/0010-4825(95)00055-0. [DOI] [PubMed] [Google Scholar]
- 72.Zhou X., Menche J., Barabási A.L., Sharma A. Human symptoms-disease network. Nat. Commun. 2014;5:4212. doi: 10.1038/ncomms5212. [DOI] [PubMed] [Google Scholar]
- 73.Chen Y., Zhang X., Zhang G.Q., Xu R. Comparative analysis of a novel disease phenotype network based on clinical manifestations. J. Biomed. Inform. 2015;53:113–120. doi: 10.1016/j.jbi.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bell D.S., Greenes R.A., Doubilet P. Form-based clinical input from a structured vocabulary: initial application in ultrasound reporting. Proc. Annu. Symp. Comput. Appl. Med. Care. 1992;1992:789–790. [PMC free article] [PubMed] [Google Scholar]
- 75.Tringali M., Hole W.T., Srinivasan S. Integration of a standard gastrointestinal endoscopy terminology in the UMLS Metathesaurus. Proc. AMIA Symp. 2002;2002:801–805. [PMC free article] [PubMed] [Google Scholar]
- 76.UniProt Consortium The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mathur S., Dinakarpandian D. Automated ontological gene annotation for computing disease similarity. Summit Transl. Bioinform. 2010;2010:12–16. [PMC free article] [PubMed] [Google Scholar]
- 78.Suthram S., Dudley J.T., Chiang A.P., Chen R., Hastie T.J., Butte A.J. Network-based elucidation of human disease similarities reveals common functional modules enriched for pluripotent drug targets. PLoS Comput. Biol. 2010;6:e1000662. doi: 10.1371/journal.pcbi.1000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sharan R., Suthram S., Kelley R.M., Kuhn T., McCuine S., Uetz P., Sittler T., Karp R.M., Ideker T. Conserved patterns of protein interaction in multiple species. Proc. Natl. Acad. Sci. USA. 2005;102:1974–1979. doi: 10.1073/pnas.0409522102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Keshava Prasad T.S., Goel R., Kandasamy K., Keerthikumar S., Kumar S., Mathivanan S., Telikicherla D., Raju R., Shafreen B., Venugopal A. Human Protein Reference Database—2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Perlman L., Gottlieb A., Atias N., Ruppin E., Sharan R. Combining drug and gene similarity measures for drug-target elucidation. J. Comput. Biol. 2011;18:133–145. doi: 10.1089/cmb.2010.0213. [DOI] [PubMed] [Google Scholar]
- 82.Hamaneh M.B., Yu Y.K. Relating diseases by integrating gene associations and information flow through protein interaction network. PLoS ONE. 2014;9:e110936. doi: 10.1371/journal.pone.0110936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim H., Yoon Y., Ahn J., Park S. A literature-driven method to calculate similarities among diseases. Comput. Methods Programs Biomed. 2015;122:108–122. doi: 10.1016/j.cmpb.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 84.Thorn C.F., Sharma M.R., Altman R.B., Klein T.E. PharmGKB summary: pazopanib pathway, pharmacokinetics. Pharmacogenet. Genomics. 2017;27:307–312. doi: 10.1097/FPC.0000000000000292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.del Pozo A., Pazos F., Valencia A. Defining functional distances over gene ontology. BMC Bioinformatics. 2008;9:50. doi: 10.1186/1471-2105-9-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu X., Zhu L., Guo J., Zhang D.Y., Lin K. Prediction of yeast protein-protein interaction network: insights from the Gene Ontology and annotations. Nucleic Acids Res. 2006;34:2137–2150. doi: 10.1093/nar/gkl219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wu H., Su Z., Mao F., Olman V., Xu Y. Prediction of functional modules based on comparative genome analysis and Gene Ontology application. Nucleic Acids Res. 2005;33:2822–2837. doi: 10.1093/nar/gki573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yu H., Gao L., Tu K., Guo Z. Broadly predicting specific gene functions with expression similarity and taxonomy similarity. Gene. 2005;352:75–81. doi: 10.1016/j.gene.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 89.Cheng J., Cline M., Martin J., Finkelstein D., Awad T., Kulp D., Siani-Rose M.A. A knowledge-based clustering algorithm driven by Gene Ontology. J. Biopharm. Stat. 2004;14:687–700. doi: 10.1081/bip-200025659. [DOI] [PubMed] [Google Scholar]
- 90.Wang D., Wang J., Lu M., Song F., Cui Q. Inferring the human microRNA functional similarity and functional network based on microRNA-associated diseases. Bioinformatics. 2010;26:1644–1650. doi: 10.1093/bioinformatics/btq241. [DOI] [PubMed] [Google Scholar]
- 91.Cheng L., Li J., Ju P., Peng J., Wang Y. SemFunSim: a new method for measuring disease similarity by integrating semantic and gene functional association. PLoS ONE. 2014;9:e99415. doi: 10.1371/journal.pone.0099415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mabotuwana T., Lee M.C., Cohen-Solal E.V. An ontology-based similarity measure for biomedical data—application to radiology reports. J. Biomed. Inform. 2013;46:857–868. doi: 10.1016/j.jbi.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 93.Jiang J.J., Conrath D.W. Semantic similarity based on corpus statistics and lexical taxonomy. arXiv. 1997 https://arxiv.org/abs/cmp-lg/9709008 arXiv:cmp-lg/9709008. [Google Scholar]
- 94.Pesquita C., Faria D., Bastos H., Falco A., Couto F.M. Evaluating GO-based semantic similarity measures. Ismb/eccb Sig. Meet. Program Mater. Iscb. 2007;37:37–40. [Google Scholar]
- 95.Li B., Wang J.Z., Feltus F.A., Zhou J., Luo F. Effectively integrating information content and structural relationship to improve the GO-based similarity measure between proteins. arXiv. 2010 https://arxiv.org/abs/1001.0958 arXiv:1001.0958. [Google Scholar]
- 96.Lord P.W., Stevens R.D., Brass A., Goble C.A. Investigating semantic similarity measures across the Gene Ontology: the relationship between sequence and annotation. Bioinformatics. 2003;19:1275–1283. doi: 10.1093/bioinformatics/btg153. [DOI] [PubMed] [Google Scholar]
- 97.Li J., Gong B., Chen X., Liu T., Wu C., Zhang F., Li C., Li X., Rao S., Li X. DOSim: an R package for similarity between diseases based on Disease Ontology. BMC Bioinformatics. 2011;12:266. doi: 10.1186/1471-2105-12-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schlicker A., Domingues F.S., Rahnenführer J., Lengauer T. A new measure for functional similarity of gene products based on Gene Ontology. BMC Bioinformatics. 2006;7:302. doi: 10.1186/1471-2105-7-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mathur S., Dinakarpandian D. Finding disease similarity based on implicit semantic similarity. J. Biomed. Inform. 2012;45:363–371. doi: 10.1016/j.jbi.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 100.Mottaz A., Yip Y.L., Ruch P., Veuthey A.L. Mapping proteins to disease terminologies: from UniProt to MeSH. BMC Bioinformatics. 2008;9(Suppl 5):S3. doi: 10.1186/1471-2105-9-S5-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sun K., Gonçalves J.P., Larminie C., Przulj N. Predicting disease associations via biological network analysis. BMC Bioinformatics. 2014;15:304. doi: 10.1186/1471-2105-15-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nachar N. The Mann-Whitney U: a test for assessing whether two independent samples come from the same distribution. Tutor. Quant. Methods Psychol. 2008;4:13–20. [Google Scholar]
- 103.Pakhomov S., McInnes B., Adam T., Liu Y., Pedersen T., Melton G.B. Semantic similarity and relatedness between clinical terms: an experimental study. AMIA Annu. Symp. Proc. 2010;2010:572–576. [PMC free article] [PubMed] [Google Scholar]
- 104.Vanunu O., Magger O., Ruppin E., Shlomi T., Sharan R. Associating genes and protein complexes with disease via network propagation. PLoS Comput. Biol. 2010;6:e1000641. doi: 10.1371/journal.pcbi.1000641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ganegoda G.U., Sheng Y., Wang J. ProSim: a method for prioritizing disease genes based on protein proximity and disease similarity. BioMed Res. Int. 2015;2015:213750. doi: 10.1155/2015/213750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Köhler S., Bauer S., Horn D., Robinson P.N. Walking the interactome for prioritization of candidate disease genes. Am. J. Hum. Genet. 2008;82:949–958. doi: 10.1016/j.ajhg.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hu Y., Zhou M., Shi H., Ju H., Jiang Q., Cheng L. InfDisSim: a novel method for measuring disease similarity based on information flow. In: Tian T., Jiang Q., Liu Y., Burrage K., Song J., Wang Y., Hu X., Morishita S., Zhu Q., Wang G., editors. Proceedings of the 2016 IEEE International Conference on Bioinformatics and Biomedicine. BIBM; 2016. pp. 20–26. [Google Scholar]
- 108.Sun J., Shi H., Wang Z., Zhang C., Liu L., Wang L., He W., Hao D., Liu S., Zhou M. Inferring novel lncRNA-disease associations based on a random walk model of a lncRNA functional similarity network. Mol. Biosyst. 2014;10:2074–2081. doi: 10.1039/c3mb70608g. [DOI] [PubMed] [Google Scholar]
- 109.Chen X., Yan C.C., Luo C., Ji W., Zhang Y., Dai Q. Constructing lncRNA functional similarity network based on lncRNA-disease associations and disease semantic similarity. Sci. Rep. 2015;5:11338. doi: 10.1038/srep11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yu L., Zhao J., Gao L. Predicting potential drugs for breast cancer based on miRNA and tissue specificity. Int. J. Biol. Sci. 2018;14:971–982. doi: 10.7150/ijbs.23350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cheng L., Jiang Y., Wang Z., Shi H., Sun J., Yang H., Zhang S., Hu Y., Zhou M. DisSim: an online system for exploring significant similar diseases and exhibiting potential therapeutic drugs. Sci. Rep. 2016;6:30024. doi: 10.1038/srep30024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Goh K.I., Cusick M.E., Valle D., Childs B., Vidal M., Barabási A.L. The human disease network. Proc. Natl. Acad. Sci. USA. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lee D.S., Park J., Kay K.A., Christakis N.A., Oltvai Z.N., Barabási A.L. The implications of human metabolic network topology for disease comorbidity. Proc. Natl. Acad. Sci. USA. 2008;105:9880–9885. doi: 10.1073/pnas.0802208105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li Y., Agarwal P. A pathway-based view of human diseases and disease relationships. PLoS ONE. 2009;4:e4346. doi: 10.1371/journal.pone.0004346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang X., Zhang R., Jiang Y., Sun P., Tang G., Wang X., Lv H., Li X. The expanded human disease network combining protein-protein interaction information. Eur. J. Hum. Genet. 2011;19:783–788. doi: 10.1038/ejhg.2011.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chen W., Yang H., Feng P., Ding H., Lin H. iDNA4mC: identifying DNA N4-methylcytosine sites based on nucleotide chemical properties. Bioinformatics. 2017;33:3518–3523. doi: 10.1093/bioinformatics/btx479. [DOI] [PubMed] [Google Scholar]
- 117.Dao F.Y., Lv H., Wang F., Feng C.-Q., Ding H., Chen W., Lin H. Identify origin of replication in Saccharomyces cerevisiae using two-step feature selection technique. Bioinformatics. 2018;35:2075–2083. doi: 10.1093/bioinformatics/bty943. [DOI] [PubMed] [Google Scholar]
- 118.Feng C.Q., Zhang Z.Y., Zhu X.J., Lin Y., Chen W., Tang H., Lin H. iTerm-PseKNC: a sequence-based tool for predicting bacterial transcriptional terminators. Bioinformatics. 2019;35:1469–1477. doi: 10.1093/bioinformatics/bty827. [DOI] [PubMed] [Google Scholar]
- 119.Hoehndorf R., Schofield P.N., Gkoutos G.V. Analysis of the human diseasome using phenotype similarity between common, genetic, and infectious diseases. Sci. Rep. 2015;5:10888. doi: 10.1038/srep10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Deng Y., Gao L., Wang B., Guo X. HPOSim: an R package for phenotypic similarity measure and enrichment analysis based on the human phenotype ontology. PLoS ONE. 2015;10:e0115692. doi: 10.1371/journal.pone.0115692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yu G., Wang L.G., Yan G.R., He Q.Y. DOSE: an R/Bioconductor package for disease ontology semantic and enrichment analysis. Bioinformatics. 2015;31:608–609. doi: 10.1093/bioinformatics/btu684. [DOI] [PubMed] [Google Scholar]
- 122.Hu Y., Zhao L., Liu Z., Ju H., Shi H., Xu P., Wang Y., Cheng L. DisSetSim: an online system for calculating similarity between disease sets. J. Biomed. Semantics. 2017;8(Suppl. 1):28. doi: 10.1186/s13326-017-0140-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hamaneh M.B., Yu Y.K. DeCoaD: determining correlations among diseases using protein interaction networks. BMC Res. Notes. 2015;8:226. doi: 10.1186/s13104-015-1211-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cheng L., Hu Y., Sun J., Zhou M., Jiang Q. DincRNA: a comprehensive web-based bioinformatics toolkit for exploring disease associations and ncRNA function. Bioinformatics. 2018;34:1953–1956. doi: 10.1093/bioinformatics/bty002. [DOI] [PubMed] [Google Scholar]
- 125.Resnik P. Vol. 1. Morgan Kaufmann Publishers; 1995. pp. 448–453. (Using information content to evaluate semantic similarity in a taxonomy. Proceedings of the 14th International Joint Conference on Artificial Intelligence). [Google Scholar]
- 126.Lin D. Vol. 1. Morgan Kaufmann Publishers; 1998. pp. 296–304. (An information-theoretic definition of similarity. Proceedings of the 15th International Conference on Machine Learning). [Google Scholar]
- 127.Couto F.M., Silva M.J., Coutinho P. Semantic similarity over the gene ontology: family correlation and selecting disjunctive ancestors. CIKM ’05 Proceedings of the 14th ACM International Conference on Information and Knowledge Management. 2005:343–344. [Google Scholar]
- 128.Li Y., Yu H. Vol. 2014. Oxford; 2014. p. bau113. (A robust data-driven approach for gene ontology annotation. Database). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zou Q., Li J., Song L., Zeng X., Wang G. Similarity computation strategies in the microRNA-disease network: a survey. Brief. Funct. Genomics. 2016;15:55–64. doi: 10.1093/bfgp/elv024. [DOI] [PubMed] [Google Scholar]
- 130.Liu Y., Zeng X., He Z., Zou Q. Inferring microRNA-disease associations by random walk on a heterogeneous network with multiple data sources. IEEE/ACM Trans. Comput. Biol. Bioinformatics. 2017;14:905–915. doi: 10.1109/TCBB.2016.2550432. [DOI] [PubMed] [Google Scholar]
- 131.Chen X., Huang L., Xie D., Zhao Q. EGBMMDA: Extreme Gradient Boosting Machine for MiRNA-Disease Association prediction. Cell Death Dis. 2018;9:3. doi: 10.1038/s41419-017-0003-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chen X., Xie D., Wang L., Zhao Q., You Z.H., Liu H. BNPMDA: Bipartite Network Projection for MiRNA-Disease Association prediction. Bioinformatics. 2018;34:3178–3186. doi: 10.1093/bioinformatics/bty333. [DOI] [PubMed] [Google Scholar]
- 133.Chen X., Yan C.C., Zhang X., You Z.H. Long non-coding RNAs and complex diseases: from experimental results to computational models. Brief. Bioinform. 2017;18:558–576. doi: 10.1093/bib/bbw060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zeng X., Lin W., Guo M., Zou Q. A comprehensive overview and evaluation of circular RNA detection tools. PLoS Comput. Biol. 2017;13:e1005420. doi: 10.1371/journal.pcbi.1005420. [DOI] [PMC free article] [PubMed] [Google Scholar]