Abstract
Genetic deficiency of the nuclease DCLRE1C/Artemis causes radiosensitive severe combined immunodeficiency (RS-SCID) with lack of peripheral T and B cells and increased sensitivity to ionizing radiations. Gene therapy based on transplanting autologous gene-modified hematopoietic stem cells could significantly improve the health of patients with RS-SCID by correcting their immune system. A lentiviral vector expressing physiological levels of human ARTEMIS mRNA from an EF1a promoter without post-transcriptional regulation was developed as a safe clinically applicable candidate for RS-SCID gene therapy. The vector was purified in GMP-comparable conditions and was not toxic in vitro or in vivo. Long-term engraftment of vector-transduced hematopoietic cells was achieved in irradiated Artemis-deficient mice following primary and secondary transplantation (6 months each). Vector-treated mice displayed T and B lymphopoiesis and polyclonal T cells, had structured lymphoid tissues, and produced immunoglobulins. Benign signs of inflammation were noted following secondary transplants, likely a feature of the model. There was no evidence of transgene toxicity and no induction of hematopoietic malignancy. In vitro, the vector had low genotoxic potential on murine hematopoietic progenitor cells using an immortalization assay. Altogether, these preclinical data show safety and efficacy, and support further development of the vector for the gene therapy of RS-SCID.
Keywords: gene therapy, SCID, primary immunodeficiency, Artemis, T cell, B cell, mouse, preclinical study
Introduction
Among severe combined immunodeficiency (SCID), rare cases of radiosensitive SCID (RS-SCID)1 are caused by loss-of-function mutations in the DNA cross-link repair enzyme 1C (DCLRE1C) gene, also known as the ARTEMIS gene (OMIM: 602450).2 Such cases of T-B-NK+ SCID present early in life with severe infections, diarrhea, and failure to thrive. Patients demonstrate increased radiosensitivity and an autosomal recessive pattern of inheritance. Artemis is a ubiquitously found nuclear protein, which is a member of the metallo-beta-lactamase superfamily of nucleases used to repair DNA double-strand breaks (DSBs).3 By digesting DNA overhangs and DNA hairpins, Artemis is a key factor for the non-homologous end joining (NHEJ) mechanism that is used to repair DSBs during all phases of the cell cycle and used to rearrange the T cell receptor (TCR) and immunoglobulin (Ig) genes during T and B lymphoid development.4 Artemis also controls DNA replication efficiency by breaking DNA in stalled replication forks, thereby limiting chromosomal instability.5 Conventional treatment of RS-SCID is based on the transplantation of allogeneic hematopoietic stem cells (HSCs) to restore T and B lymphopoiesis and to provide adaptive immune function in treated patients. Allogeneic HSC transplant from matched family donors provides a survival probability greater than 85% in RS-SCID patients and about 65% for haplo-identical transplants.6 However, compared with other SCIDs, the allo-HSC-transplanted RS-SCID patients have high rates of complications including late complications such as growth delay and nutritional problems.6, 7 Autologous gene therapy based on the transplantation of gene-corrected autologous HSC is an option for RS-SCID patients without human leukocyte antigen (HLA)-compatible donors, although late complications concerning non-hematopoietic tissues are not expected to be avoided by gene therapy.
Several groups including ourselves have already shown that recombinant HIV-1-derived lentiviral vectors (LVs) can be used to express either murine or human Artemis proteins in hematopoietic cells to restore B and T lymphopoiesis in murine models of RS-SCID or in xeno-transplant models engrafted with transduced patient cells.8, 9, 10, 11, 12 High levels of Artemis expression appear to be toxic in vitro and in vivo.11, 13, 14 Loss of cell viability, perturbed cell cycle, increased DNA damage, and apoptosis were reported in cultured murine or human cell lines transduced with LV that strongly express either human or murine Artemis.13 The in vitro effects were vector dose dependent and correlated to the strength of the promoter used in the Artemis transfer cassette. The strong elongation factor-1α (EF1a) promoter caused greater in vitro toxicity than the weaker human phosphoglycerate kinase (PGK) promoter, which is about 25% less strong in a luciferase reporter assay.11 Artemis-deficient HSC transduced by a LV with a murine PGK promoter driving expression of murine Artemis successfully engrafted in vivo and reconstituted B and T lymphocyte compartments in Artemis-deficient mice,8 whereas HSC transduced with a LV using stronger cytomegalovirus (CMV) or EF1a promoters failed to support lymphoid reconstitution after transplantation in RAG-1-deficient animals.10
Thus, a safe and clinically applicable Artemis LV should express transgene levels that are non-toxic and corrective. One option is to use the ARTEMIS gene promoter consisting of a 1-kb sequence upstream of the translational start site,15 which provides a much weaker (about 80% less) expression than the EF1a promoter in a luciferase reporter assay.11 LVs expressing ARTEMIS from its own promoter were not toxic in vitro or in vivo, enabling lymphoid reconstitution in Artemis-deficient mice.11, 12 The strategy that we chose to develop is to use the EF1a promoter, which has been used in several human gene therapy trials for SCIDs, but to omit the woodchuck post-transcriptional regulatory element (WPRE) in the expression cassette. WPRE can increase transgene levels by at least 2-fold in other systems.16 All of the previously tested Artemis expression cassettes that were found to cause toxicity included a WPRE.10, 11, 13 We found that expressing the ARTEMIS cDNA under control of EF1a promoter and without WPRE (EF1a-hArtemis LV) provided physiologically relevant levels of Artemis expression that permitted safe long-term engraftment and lymphoid correction in Artemis-deficient mice. Gene therapy-treated Artemis-deficient mice were analyzed for a period of up to 1 year through primary and secondary transplants. In parallel, the vector was also tested in an in vitro immortalization (IVIM) assay using murine hematopoietic progenitor cells to estimate the vector genotoxicity as previously reported.17 Results in mice suggest that the EF1a-hArtemis LV would be a safe and efficient vector for the gene therapy of RS-SCID.
Results
A Self-inactivating LV for RS-SCID Gene Therapy
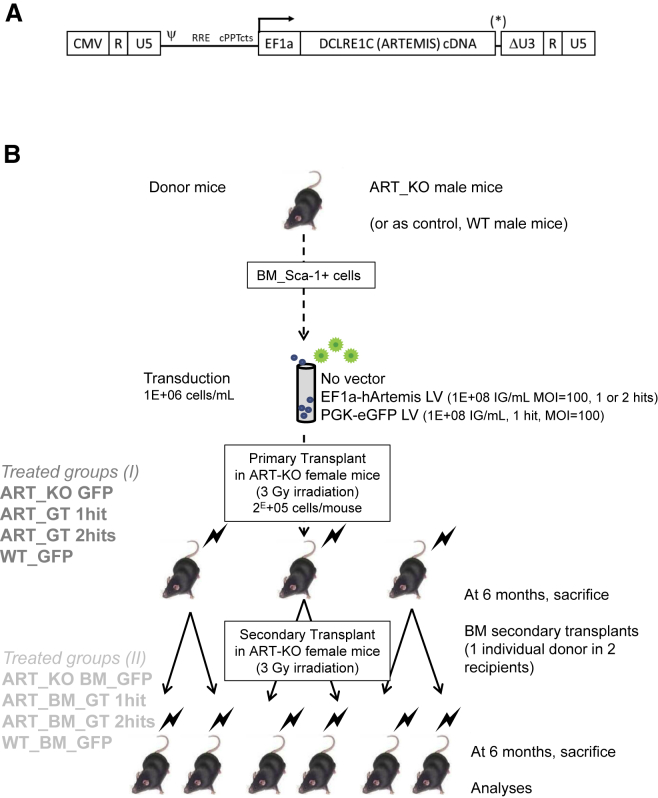
The vector tested in the present study is an advanced self-inactivated HIV-1-derived LV (pCCL backbone) expressing the native ARTEMIS cDNA under control of the short intronless EF1a promoter, without WPRE (Figure 1A). The WPRE was omitted because Artemis overexpression reportedly prevents engraftment of transduced murine hematopoietic cells.11, 13 Indeed, adding a WPRE (Figure 1A, position *) strongly enhanced Artemis mRNA expression by about 5-fold per copy of integrated vector when tested in human fibroblasts derived from a RS-SCID subject, compared with the vector lacking this element (Figure S1A). These differential effects were confirmed at the protein level (Figure S1B) and prompted us not to choose this vector. To estimate the strength of the vector expressing ARTEMIS and lacking WPRE, we transduced Artemis-deficient mouse cells with this vector and compared expression against levels found in normal mice using a standardized qRT-PCR system calibrated on a plasmid containing both human ARTEMIS and murine Artemis genes sequences. This method showed that two copies of the EF1a-hArtemis vector per cell were needed to obtain transgene mRNA levels equivalent to those found in normal murine hematopoietic cells (Figure S1C). Based on these results and data from the literature, it was reasonable to expect that the EF1a-hArtemis LV could function in a physiologically relevant and safe manner to correct the T and B cell defects of Artemis-deficient mice.
Figure 1.
Artemis LV and Schematic Design of Mouse Transplant Experiments
(A) EF1a-hArtemis LV construct. A self-inactivated HIV-1-derived LV (pCCL backbone) expressing human ARTEMIS cDNA under control of the EF1α promoter. (B) Purified bone marrow (BM) Sca-1+ cells from ART_KO or WT mice were transduced by EF1a-hArtemis or PGK-eGFP LV at the concentration of 1E+8 IG/mL for one or two hits of infection, and 2E+5 cells/mice were transplanted into irradiated primary recipient ART_KO mice. Two groups of 10 mice each received cells transduced with one or two hits of EF1a-hArtemis LV (groups ART_GT 1hit and ART_GT 2hits). A control treatment group of 10 mice received GFP-transduced ART_KO Sca-1+ BM cells from which no functional correction was expected (group ART_KO GFP). A positive control treatment group of eight mice received GFP-transduced WT cells that were expected to restore immune development (group WT_GFP). Primary transplanted mice were followed up for up to 6 months at which point the BM cells from each of the surviving animals were transplanted into two secondary recipients (irradiated ART_KO female mice) for a longitudinal follow-up of 6 months (68 mice total). Longitudinal follow-up of the mice included regular observations, weighing, blood counts, measures of engraftment and transduction, and autopsy and histology at sacrifice. In addition to the treated mice, primary and secondary transplant experiments also included control non-transplanted groups with irradiated and non-irradiated ART-KO mice providing background results (groups ART_KO and ART_KO irr) and non-irradiated WT mice for normal comparison (group WT).
Biosafety Study Design
A formal preclinical study was designed to evaluate the efficiency and safety of the EF1a-hArtemis LV in primary and in secondary transplanted Artemis-deficient mice as outlined in Figure 1B. The design of long-term observations, 1 year in total, was previously used in preclinical protocols of gene therapy aiming to reveal vector-induced insertional mutagenesis or transgene toxicity.8, 18, 19 Engraftment was measured by the levels of male donor cells into female recipient mice as previously reported.20 The protocol also measured transduction and transgene expression levels, functional correction, health status, hematological parameters and occurrence of tumors. For these studies, we generated a quality-controlled batch of EF1a-hArtemis LV, which was produced and purified in conditions similar to other clinical-grade LV vectors produced at Genethon.21 The purified EF1a-hArtemis LV was sterile, had low levels of DNA and protein contaminants, and lacked replicative particles (Table S1). A PGK-eGFP LV that was also prepared and purified in the same conditions was used as a transduction efficiency control.
Sca-1+ hematopoietic progenitor cells were used for the transplant experiments outlined in Figure 1B, because this population enriched in progenitor cells is known to engraft and to restore T and B lymphoid development and function following transduction with a murine Artemis transgene.8 In two separate experiments, Sca-1+ bone marrow (BM) progenitor cells obtained from male Artemis-deficient (ART_KO) mice were efficiently transduced with the EF1a-hArtemis LV (Table S2A), and these cells were transplanted in female ART_KO mice. Efficient transduction was confirmed with the PGK-eGFP LV, which provided a similar range of VCN values as the EF1a-hArtemis vector and showed 65%–70% GFP-positive cells in the bulk cultures (Table S2A). One or two cycles of infection (hits) provided on average 1.4 vector copy numbers per cell (VCNs) in cells after bulk culture. The effects of repeated infection with the Artemis vector were more evident on more primitive cell transduction providing, respectively, between 1 and 2.2 VCNs in clonogenic hematopoietic progenitor cells, and Artemis transgene expression levels that were between 32% and 47% of normal C57BL/6 wild-type (WT) cell levels (Table S2A). Individually picked colonies confirmed that repeated transduction increased the frequency of transduced hematopoietic progenitor cells and increased their transduction levels because 91% positive colonies with an average of 2.3 VCNs were obtained after two hits versus 86% positive colonies with 1.4 VCNs after one hit of vector (Tables S2A and S2B). None of the vectors tested altered the hematopoietic differentiation and clonogenic potential of the cells as measured by colony-forming unit (CFU-C) cell counts (data not shown).
Primary transplanted mice were followed up for up to 6 months, at which point the BM cells from each of the surviving animals were transplanted into two secondary recipients for a longitudinal additional follow-up of 6 months. In the EF1a-hArtemis-LV-treated groups, between 0.6 and 10 (average 4E+6) male donor BM cells (BMCs) were secondarily transplanted, corresponding to the infusion of an average of 7E+3 transduced hematopoietic progenitor cells per mouse, containing 2.3 ± 1.2 VCNs per cell (Table S3).
In Vivo Engraftment of Transduced Cells and Correction of the Artemis Phenotype
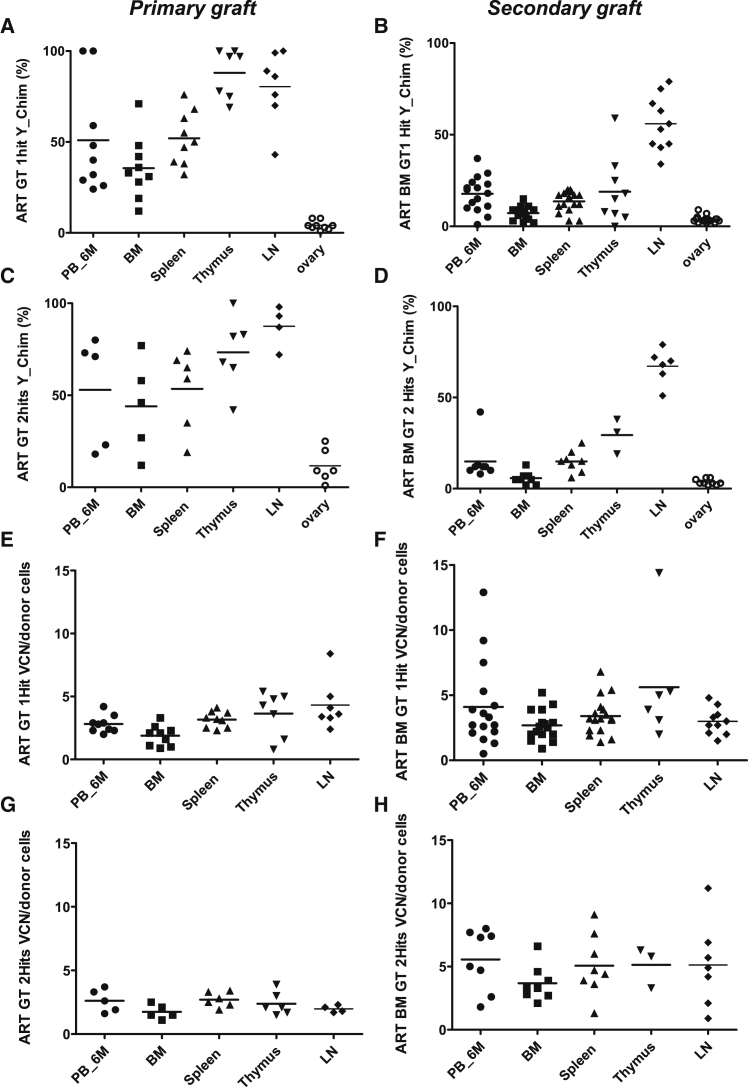
The engraftment of gene-modified donor cells was observed following primary or secondary transplants in groups treated with one or two hits of the EF1a-hArtemis LV (Figures 2A–2H). Donor chimerism (Figures 2A–2D) and vector integration (Figures 2E–2H) were detected in various hematopoietic and lymphoid organs, but not in non-hematopoietic tissues such as ovaries, as measured by qPCR. Transduction levels in vivo ranged between two and four copies per cell, according to the organs tested. Levels of engraftment with transduced donor cells were higher in primary transplant (ranging from 50% to 80% on average) than in secondary transplants (from 10%–65% on average depending on the organs). Lower levels of engraftment in secondary transplants, particularly in the BM, were not specific for the EF1a-hArtemis LV because this also occurred with the PGK-eGFP LV (Figures S2A–2H). The markedly higher engraftment level in lymph nodes, and to a lesser extent in thymus, compared with BM could be a feature of the model, which is using sublethal irradiation conditioning that limits myelo-erythroid engraftment in BM. Transplanting whole BMCs in secondary grafts provided a relatively low number of hematopoietic progenitor cells (between 0.2 and 1.4E+4 donor colony forming cells (CFCs) per mouse; Table S3), which limits engraftment in BM. Alternatively, the BMCs infused for the secondary grafts could have contained immature or mature T cells that engrafted preferentially in the secondary hosts. Indeed, after secondary transplant, lymph nodes contained greater than 50% of donor-derived transduced cells in groups treated with the EF1a-hArtemis LV, indicating that transduced lymphoid cells can persist for long periods of time (Figure 2).
Figure 2.
Engraftment and Transduction Levels in Primary and Secondary Grafts
Primary (A, C, E, and G) and secondary (B, D, F, and H) grafts show the percentage of donor cells measured by Y chimerism in different tissues and the level of transduction measured in by VCN, respectively, in the ART GT 1Hit group (A, B, E, and F) and ART GT2Hits group (C, D, G, and H). VCN was determined by qPCR and normalized for the percentage of engraftment. Each data point is from a single mouse. Horizontal bars represent mean values.
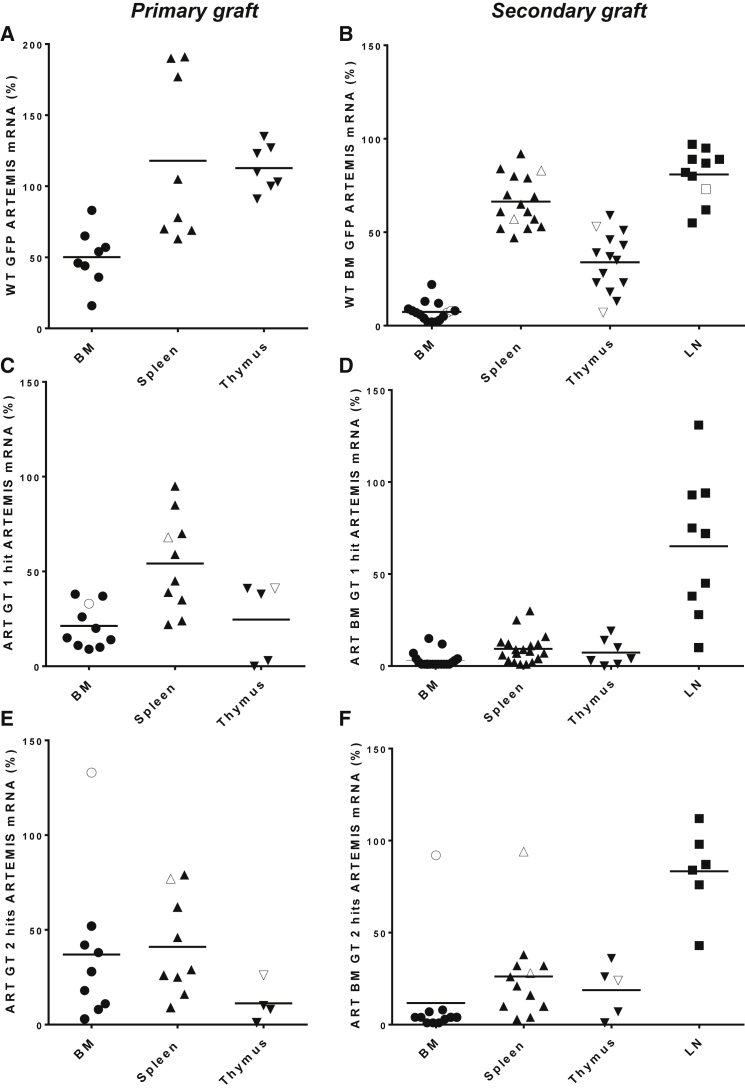
Transgene expression was detected at various levels in lymphoid and hematopoietic organs following primary and secondary transplantation (Figures 3A–3F). In primary grafts, for instance, 54% ± 25% and 41% ± 23% relative expression of ARTEMIS mRNA was measured in the spleen of mice treated, respectively, with one or two hits of EF1a-hArtemis LV, compared with 0% mRNA in the negative control groups and 117% ± 58% mRNA in mice treated by WT cells (Figures 3A, 3C, and 3E). In the BM, about half as much ARTEMIS mRNA was provided by the EF1a-hArtemis LV transduction compared with WT cells (50% ± 20% in WT_GFP mice versus 21% ± 13% in ART-GT one-hit mice and 25% ± 18% in ART-GT two-hit mice). In secondary grafts, ARTEMIS mRNA expression was generally lower than in primary grafts but still detectable in BM, spleen, thymus, and lymph nodes in EF1a-hArtemis LV-treated mice (Figures 3B, 3D, and 3F). In the spleen, 9.4% ± 8%, 19% ± 12%, and 66% ± 13% of relative mRNA are found, respectively, in mice treated with one or two hits of EF1a-hArtemis LV and with WT cells. Following secondary transplants, the lymph nodes were highly reconstituted, expressing similar levels of ARTEMIS mRNA in mice treated with one or two hits of EF1a-hArtemis LV or with WT cells (65% ± 36% in ART-GT one-hit mice and 83% ± 21% in ART-GT two-hit mice versus 81% ± 13% in WT-GFP mice). Some mice developed tumors from donor-derived transduced cells (as discussed below), and values of mRNA for these mice are represented by open symbols in Figure 3. Outlier mRNA values corresponding to tumor samples were not included in the mRNA level group comparisons.
Figure 3.
ARTEMIS mRNA Expression after Primary and Secondary Engraftment
Primary (A, C, and E) and secondary (B, D, and F) grafts show ARTEMIS mRNA levels in hematopoietic and lymphoid tissues in the WT GFP group (A and B), ART GT1hit group (C and D) and ART GT2Hits group (E and F). (A–F) Each data point is from a single mouse. Open symbols correspond to mice that developed tumors from donor-derived transduced cells. Horizontal bars represent mean values.
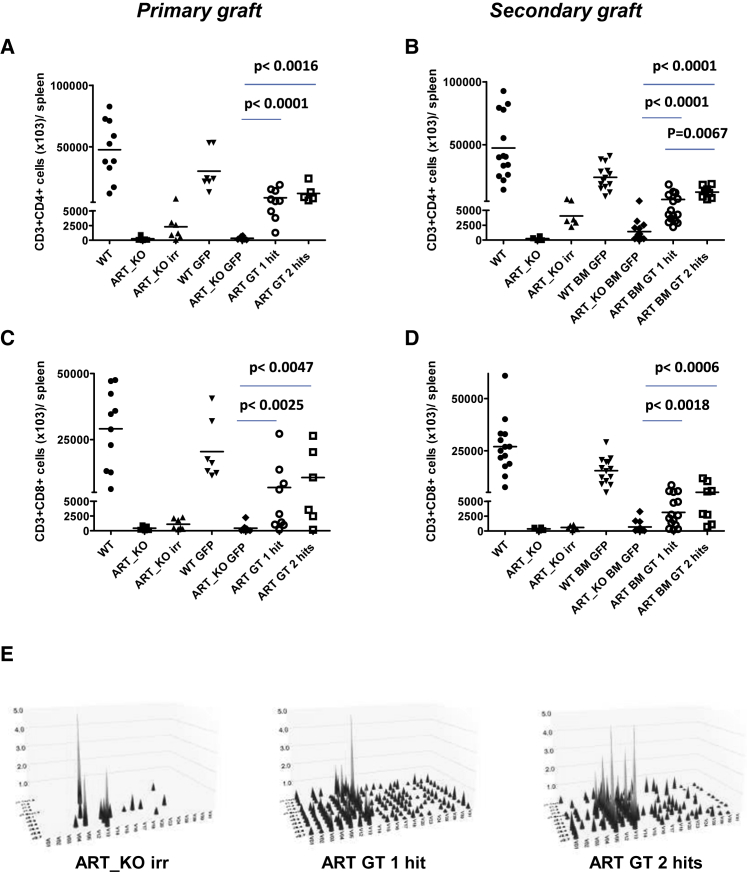
Gene therapy with the EF1a-hArtemis LV restored T cell development. The recovery of thymopoiesis was evident in ARTEMIS gene therapy-treated mice, particularly in primary transplants. The number of CD4+CD8+ double-positive thymocytes per thymus × E+03 was 11,383 ± 13,201 and 7,471 ± 8,656 in mice treated with one and two hits of Artemis vector, respectively, as compared with 464 ± 575 cells in GFP-treated Artemis-deficient negative controls, with these values excluding mice with thymomas. Thymic recovery was also evident by strong immunohistological marking of CD3 throughout the thymus in ARTEMIS gene therapy-treated mice, whereas the staining was very low to absent in Artemis-deficient controls (Figure S3). Correction of T cell development was also shown by the presence of statistically higher numbers of CD3+CD4+ and CD3+CD8+ T cells in lymphoid organs such as the spleen (Figures 4A–4D), whereas control ART-KO mice had very few T cells. The effects of one or two hits of transduction were not statistically different on T cell reconstitution, except that two hits provided higher levels of CD4+ T cells in secondary transplants (Figure 4B). TCR Vβ diversity was measured by PCR and confirmed that ART-KO mice used in these experiments and that have a severe block in T cell development have a very limited, oligoclonal, and skewed TCR repertoire with a representation of V12-J1.5, V13-J2.7, and V05-J2.5 TCR β chain usage.2, 22 After irradiation, TCR diversity was around 6% in the spleen (Figure 4E). Following gene therapy, the EF1a-hArtemis LV restored a polyclonal pool of CD3+ T lymphocytes with a T cell repertoire (TCR) β chain diversity of 54%–71% and 46%–78%, respectively, in groups treated with one and two hits of vector (Figure 4E). These values remained inferior to what was observed in mice treated with WT cells that had a TCRβ diversity of 85% in spleen (data not shown). The same V12-J1.5, V13-J2.7, and V05-J2.5 detected in ART-KO mice were still detected in mice treated by ARTEMIS gene therapy but no longer represented a majority of the Vβ usage. Increased numbers of TCRγδ+ T cells were also observed in EF1a-hArtemis LV-treated mice compared with negative controls (data not shown).
Figure 4.
T Lymphocyte Reconstitution in Spleen from Recipient Mice Treated by ARTEMIS Gene Therapy
Mice were analyzed after 6 months of transplantation for T phenotype in hematopoietic organs. Representative recovery of CD3+CD4+ (A and B) and CD3+CD8+ (C and D) cells in spleen of recipient female ART_KO mice from primary (A and C) and secondary graft (B and D). (A–D) Each data point is from a single mouse. Horizontal bars represent mean values. p values were calculated with a Mann Whitney test. (E) Analysis of T cell receptor Vβ (TRVB) repertoire in the spleens of primary grafts with a 3D representation. Each peak represents a rearrangement between a family of V genes and a J segment. The x axis represents TRBV, y axis represents TRBJ, and z axis represents the relative intensity of the signal.
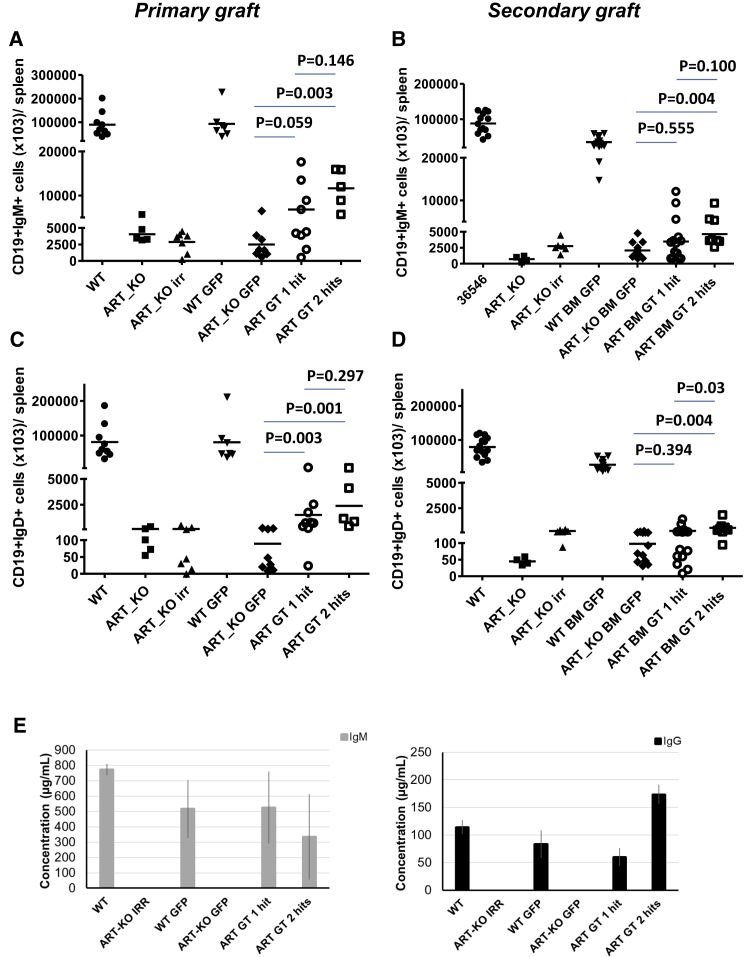
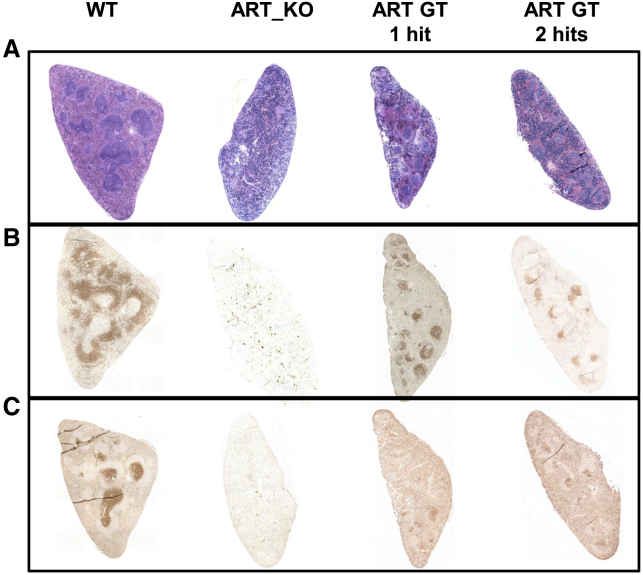
Gene therapy with the EF1a-hArtemis LV also significantly restored B lymphocyte development, although B cell recovery was not as high as in mice treated by WT cells (Figures 5A–5D). However, the presence of CD19+IgM+ and CD19+IgD+ cell populations was detectable over background in the blood (data not shown) and was significantly higher in the spleen of mice treated with two hits of the EF1a-hArtemis LV compared with GFP-treated mice in primary and secondary transplants (Figures 5A–5D). Circulating levels of IgM and IgG were detected in the sera of mice treated with the EF1a-hArtemis LV and were similar to the levels found in WT mice or mice treated with WT cells, whereas IgG and IgM were undetectable in negative control mice (Figure 5E). Furthermore, immunohistological staining of spleens in EF1a-hArtemis LV-treated mice was structured (Figure 6A) and showed the presence of B220+ cells organized in B cell follicles (Figure 6B) surrounding CD3+ T cell-dense areas indicative of germinal centers (Figure 6C). The splenic organ structuration of B cell and T cell areas was clearly induced by EF1a-hArtemis LV gene therapy, although it was not as marked as in mice treated with WT cells. Altogether the results show that EF1a-hArtemis LV enables efficient engraftment and secondary transplantation, and induces partial but effective recovery of Artemis mRNA expression capable of inducing T and B cell development with possible immune function as shown by Ig production.
Figure 5.
B Lymphocyte Reconstitution in Spleen and Sera from Recipient Mice Treated by Artemis Gene Therapy
As previously described, mice were analyzed also for B phenotype in hematopoietic organs after 6 months of transplantation. Representative recovery of CD19+IgM+ and CD19+IgD+ cells in spleen of recipient female ART_KO mice from primary (A and C) and secondary graft (B and D). (A–D) Each data point is from a single mouse. Horizontal bars repreesent mean values. p values were calculated with a Mann-Whitney test. (E) Evaluation of IgM and IgG in serum of mice from the primary graft. Data are represented as mean ± standard deviation.
Figure 6.
Histological Evaluation (×4) of Spleen from Recipient Mice Treated by Artemis Gene Therapy in Comparison with the WT or ART_KO Control Mice
Representative sections of spleen from the indicated groups (top) treated with H&E staining (A) or immunostaining for B220 (B) or immunostaining for CD3 (C).
Evaluation of Health in Artemis-Treated Mice
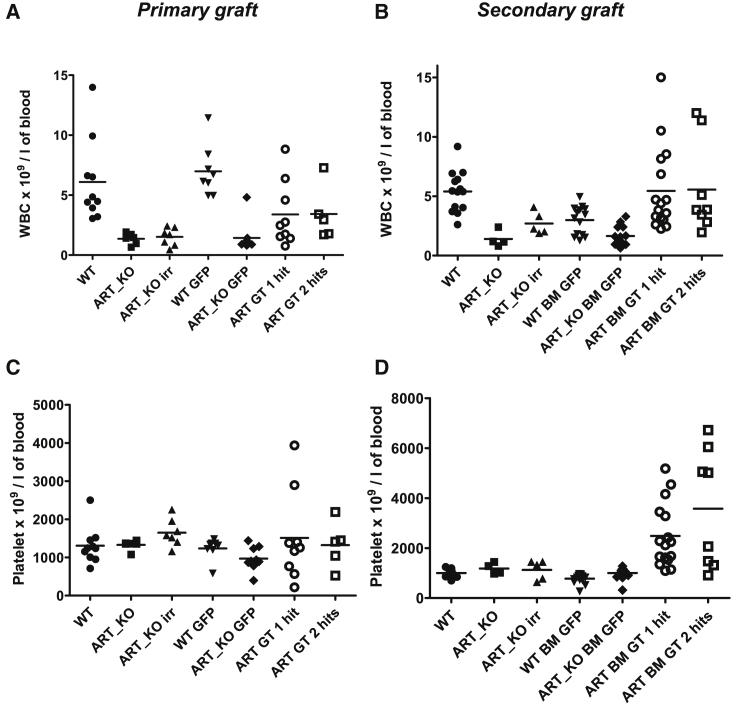
Blood counts, hemoglobin, and hematocrit were measured 3 and 6 months post-transplantation to assess the health of mice. Besides, mice were regularly observed for health status and weight. In the follow-up of primary and secondary transplants, there was no difference in any of the groups for red blood cell (RBC) counts (Figure S4), hemoglobin, and hematocrit parameters (data not shown). The levels of leukocytes (white blood cells [WBCs]), which are lower in Artemis-deficient mice compared with WT mice, were significantly increased by gene therapy (p < 0.001) after primary transplants (Figure 7A), although not as high as in mice treated with WT cells, in coherence with partial restoration of T and B cell levels (Figures 5 and 6). Platelets counts were not significantly different among groups in primary grafts (Figure 7C) but were clearly increased in the Artemis-treated mice after secondary transplants (Figure 7D). Such an increase coincided with higher levels of WBCs, although these were not higher than in WT mice (Figure 7B). These increased WBC counts were not due to the occurrence of malignancies, and mice with tumors will be detailed below. Such biological data suggested possible inflammation, which is known to increase WBC and platelet counts. Indeed, in secondary transplanted mice we detected low levels of circulating inflammatory cytokines such as interferon-γ (IFN-γ), interleukin-6 (IL-6), and tumor necrosis factor alpha (TNF-α), which appeared to be at higher levels than in Artemis-deficient non-treated controls, but some of these cytokines were found at the same levels in WT mice (Table S4). The sanitary status of the animals and of the facility was verified, and there were no detectable infectious agents detected at the time of the experiments. In the mice without tumors, the appearance and behavior of the animals were unremarkable. Some weight differences were noted among groups. Compared with WT mice, which weigh about 20 g between 6 and 8 weeks of age and display a moderate weight gain over time, Artemis-deficient mice were slightly heavier at start and continuously gained weight over time, even after irradiation, reaching body weights over 30 g after 6 months (data not shown). The transplantation of WT cells into irradiated ART_KO mice did not modify their increasing body weight curve, whereas EF1a-hArtemis LV-treated mice normalized their weight curve, which was the same, and not below, that of WT mice (data not shown). Analysis of individual mice showed no correlation between cytokine levels, body weight, or platelet counts (data not shown). We conclude that EF1a-hArtemis LV-treated mice do not suffer from acute hematopoietic or immune toxicity.
Figure 7.
Peripheral Blood Count Analysis before the Sacrifice of the Recipient Mice after Primary and Secondary Engraftment
Primary (A and C) and secondary (B and D) graft analysis of white blood cell (WBC) (A and B) and platelets (C and D) blood counts as determined with an automated cell counter. Each data point is a single mouse. Horizontal bars represent mean values.
No Evidence of Oncogenic Potential of the EF1-hArtemis LV
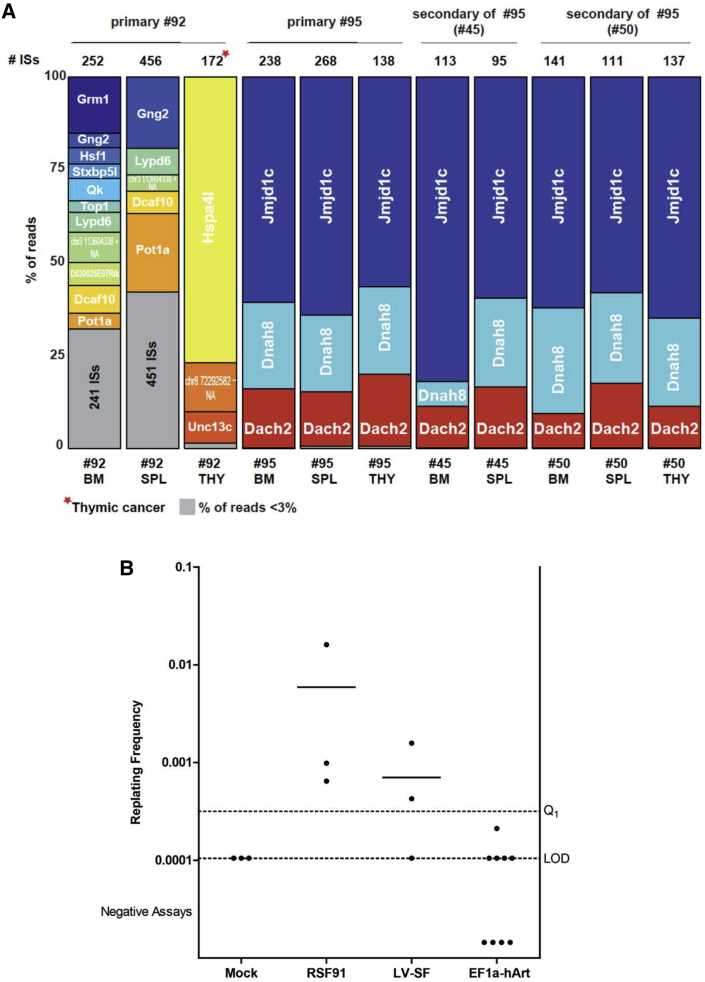
The ART-KO mice are tumor prone in the conditions used, but the protocol aimed to evaluate whether the Artemis vector caused an additional risk for oncogenicity. As expected, in the irradiated ART_KO group, the background level of tumors found within a 6-month period was 35%, which is a high level, consistent with the known role of Artemis in the control of chromosomal stability. In the EF1a-hArtemis LV-treated groups there were 20%–25% animals with tumors, which was not higher than any other group. All of the tumors identified in any of the groups were T cell lymphomas, variously expressing CD3, CD4, or CD8, which were either localized thymomas or disseminated in all lymphoid organs and sometimes in liver as mice displayed signs of splenomegaly and hepatomegaly (Table S5A). Molecular analyses could determine the origin of the tumors by detecting if they consisted of donor male cells or contained significant levels of vector copies. In total, 19 treated mice out of 88 (22%) were found to bear lymphomas regardless of groups of treatment, which was not higher than the background ART_KO irradiated control group (Table 1). Most tumors originated from host non-transduced cells as in the control group. Yet, six treated mice were found to have tumors originating from donor-derived transduced cells. Two of these tumors were transduced with the PGK-eGFP vector. Four mice had tumors with the EF1a-hArtemis LV, two of which arose in primary transplants (Table 1). One of these mice was unrecognized at the time and served as a BM donor, transmitting the tumor to the two mice generated by secondary transplants. The origin of the tumors with the EF1a-hArtemis LV was tracked by the analysis of vector insertion sites (ISs). One of the two primary graft tumors was a disseminated lymphoma that had three dominant vector insertions in Jmjd1c, Dnah8, and Dach2 genes, and these insertions were also identified in the secondary transplant tumors derived from this animal (Figure 8; Table 1; Table S5B). The other tumor was a localized thymoma that had two insertion sites in Hspa41 and Unc13c genes, and an intergenic IS mapped on the chr8 72,292,582. These insertions were not frequently represented in the other organs analyzed, confirming that the tumor was restricted to the thymus (Figure 8A). In the spleen and BM of this animal, a high number of diverse vector IS insertion sites was found as expected from the polyclonal population of cells and from the histological evaluation of these tissues that appeared to be normal (Table S5B). In addition, animals transplanted with the BM from this animal did not develop tumors. None of the gene insertions that were identified in tumor cells or in normal tissues analyzed in the whole study were found to be oncogenes or associated with known T cell lymphoma processes as determined by interrogation of the AmiGO2 MGI (Mouse Genome Informatics), and NCBI databases. Some of the gene vector insertions found in tumors (jmjd1c, hspa4l, dach2) or in non-tumoral tissue (qk) have previously been reported as sites of LV insertion in the murine genome.23, 24
Table 1.
Tumor Analysis
| Vector | Category | Group | Nb Mice with Tumor | Tumor Origin | Characteristics of Donor-Derived Transduced Tumors |
|---|---|---|---|---|---|
| None | controls | ART_KO irr | 7/20 (35%) | all tumors are from host, no vector | |
| PGK-eGFP LV | secondary transplants | WT BM GFP | 2/16 (12.5%) | 2 tumors: 2 donor-derived, transduced |
the 2 donor-derived tumors are localized thymomas the vector insertion sites were not defined |
| ART_KO BM GFP | 5/18 (28%) | 5 tumors: 5 host-derived, no vector |
|||
| EF1 hArtemis LV | primary transplants | ART GT 1 Hit | 5/20 (25%) | 1 tumor: 1 donor-derived, transduced |
thymoma, CD3+CD8+ cells vector insertions in Hspa4I; chr8_72292582; Unc13c genes |
| ART GT 2 Hits | 4 tumors: 1 donor-derived, transduced 2 host-derived, no vector 1 donor-derived, no vector |
Disseminated T cell lymphoma, CD3highCD4+CD8+ cells vector insertions in Jmjd1c; Dnah8; Dach2 genes gave rise to the two tumors in secondary transplants | |||
| secondary transplants | ART BM GT 1 Hit | 7/34 (20%) | 3 tumors: 3 host-derived, no vector |
||
| ART BM GT 2 Hits | 4 tumors: 2 donor-derived, transduced 2 host-derived |
the two donor-derived tumors are disseminated T cell lymphoma both carrying the vector insertions in Jmjd1c; Dnah8; Dach2 genes that match with the donor BM |
Figure 8.
Genotoxicity
Vector insertions in tumors or tissues (A) and IVIM assay (B). (A) Vector genomic insertions were analyzed in BM, spleen, and thymus of mice bearing tumors containing EF1a-hArtemis-LV-transduced donor cells. The read counts are indicated in each sample analyzed, and the percentage of reads for each IS is represented by individual colors, with gray representing IS with less than 3% of relative abundance. Samples are identified by mouse number and tissue (BM=bone marrow), SPL (spleen) and THY (thymus). (B) Insertional mutagenesis leads to clonal outgrowth after low-density seeding in the IVIM assay. Non-immortalized cells do not grow (negative, below limit of detection [LOD]). The number of wells containing insertion mutants is used to calculate the replating frequency (RF) according to Poisson statistics. Assays with an RF ≥ 3.17 × 10−4 (Q1 level) are counted as positive. Each dot represents one assay. In the three experiments, we obtained immortalized clones from the cells transduced by the positive control vectors RSF91 or LV-SF, whereas all plates containing EF1a-Artemis transduced cells were below the Q1 level. Horizontal bars represent mean values.
To complete the evaluation of the oncogenic potential of the EF1a-hArtemis LV, we performed an IVIM assay as described.17 The assay detects the induction of a survival advantage and transformation by insertional activation of cellular proto-oncogenes following transduction of murine hematopoietic cells with LTR-driven gamma-retroviral vectors such as pRSF91.GFPgPRE (RSF91) or SIN-LVs with strong internal promoters like RRL.PPT.SF.eGFP.pre* (LV-SF). In three independent IVIM assays, there was no detection of mutagenic potential from the EF1a-hArtemis LV (Figure 8B). Altogether, we conclude that there is no evidence of oncogenic potential from the EF1a-hArtemis LV and that, in some cases, transduction with the vector could be passively associated to, but not causative of, the oncogenic processes that can occur in Artemis-deficient cells.
Discussion
The present study is the first preclinical and comprehensive biosafety evaluation of a LV developed for the gene therapy of RS-SCID. This controlled study using a GMP-comparable vector with extensive survey of the health and biological parameters of treated mice provides additional information to previous reports showing that hArtemis-encoding LV can correct Artemis deficiency in mice.11, 12 It is shown that removing the WPRE decreased the level of expression to a safer level. Indeed, the EF1a-hArtemis LV expressed low but functional levels of Artemis that were not toxic in vitro or in vivo on murine hematopoietic cells, enabling long-term engraftment up to 1 year in secondary transplants in mice, and reconstituting T and B cell development without evidence of vector-induced toxicity on blood parameters, or of vector-induced mutagenesis. The lack of toxicity of the EF1a-hArtemis LV was also confirmed in vitro on human cells, causing no effect on cell growth, cell viability, and clonogenicity following transduction of umbilical cord blood CD34+ hematopoietic progenitor cells (data not shown). In contrast, the same vector with WPRE expressed greater levels of hArtemis, which, based on prior reports with a similar vector, would be expected to hamper engraftment of transduced cells. There is no direct molecular mechanism linking high Artemis expression with cellular or hematopoietic toxicity, and the toxicity of high Artemis levels is only inferred from correlative studies with different promoter systems.10, 11, 12 Artemis transgenic mice expressing up to 40 times more Artemis than WT mice do not display hematopoietic or lymphoid anomalies.22 In addition, hematopoietic progenitor cells of normal mice that were transduced to overexpress Artemis engrafted successfully (data not shown). Thus, mechanisms causing in vivo or in vitro toxicity observed with the strongly expressing Artemis vectors should be further studied to determine whether other parameters could be involved in the lack of engraftment that was observed. At the practical level, our study has established disease correction and no adverse events related to the vector. Such EF1a-hArtemis vector is a good candidate to further develop for clinical use in humans. Because it lacks unnecessary viral element (WPRE), it reduces the integration of foreign DNA sequences in the genome of patients. It is using a promoter that has already been used in successful lentiviral gene therapy trials in humans in other types of SCIDs,25 and it can be produced in GMP-comparable conditions demonstrating its ability to correct the disease phenotype in mice without acute toxicity.
Treatment of Artemis-deficient mice with the EF1a-hArtemis LV releases the T and B cell differentiation block caused by Artemis deficiency as reported in other studies using LV with murine8 or human Artemis10, 11, 12 transgenes. As a consequence, it also normalizes the natural killer (NK) cell counts, which were higher in non-treated Artemis-deficient mice (data not shown). Reconstitution of T cells, and particularly of peripheral T cells, was very effective at inducing high cell counts, thymopoiesis, and a polyclonal TCR repertoire, being equivalent to that obtained with LV using the Artemis promoter.11, 12 B cell levels were only partially corrected in BM or spleen. Yet, EF1a-hArtemis LV clearly restructured T and B cell zones in lymphoid tissues in treated animals including appearance of bronchus-associated lymphoid tissue (data not shown) and the production of Igs. The relatively low correction of B cell numbers by EF1a-hArtemis gene therapy is likely due to intrinsic limitations of the murine model. Indeed, differential T/B reconstitution and limited B cell recovery were also observed following BM transplant in Artemis-deficient mice.26 Poor B cell reconstitution following gene therapy was also seen with the Artemis promoter LV studies in mice in which about 1% donor-derived transduced B cells were recovered in blood compared with about 12% following infusion of WT cells.12 The EF1a-hArtemis LV achieved about 3%–4% donor B cells in blood versus 60% from WT cells. In BM, 3%–4% donor CD19+ B cells were obtained versus about 14% from WT cells, and these levels are similar to that reported with the Artemis promoter LV. Whereas B cell reconstitution is low in murine transplantation models, on the contrary, human HSC transplant studies using the NSG mouse model support efficient B cell differentiation and have shown that the ARTEMIS gene promoter enables the development of high levels of transduced B cells, comparable with WT cells.12 Thus, in spite of limitations of the models, we expect that the EF1a-hArtemis LV, which has a similar effect to the ARTEMIS promoter LV in mice, will also correct human B cells, but this will have to be demonstrated in future studies. We can speculate that gene therapy with the EF1a-hArtemis LV should improve the immune status of patients because the ARTEMIS promoter LV, which leads to a similar reconstitution in mice, permits functional recovery of immune responses as measured by antigen-specific T cell proliferation and antibody production.12
Adverse events could occur following gene therapy. There is a known association between Artemis deficiency and lymphomas, particularly aggressive B cell lymphomas in patients with hypomorphic Artemis mutations. Yet, lymphomas are not particularly increased in human RS-SCID patients treated by HSC transplant, so this suggests that the risk for adverse events such as lymphoma would not be significantly increased by gene therapy. Indeed, the risk for oncogenesis by insertional mutagenesis or by overt transgene toxicity did not manifest in our study. T cell lymphomas occurred in the mice and were primarily caused by the uncorrected Artemis-deficient background and not attributable to the effects of the vector. There was no evidence of in vitro transformation in the IVIM assay, although this assay essentially detects myeloid transformation by vectors containing strong promoters.17 Similar results were found in the IVIM assay with the ARTEMIS promoter LV.12 Inflammation is a recognized feature of Artemis deficiency in humans. Hypomorphic mutations and leaky phenotypes of Artemis deficiency can cause Omenn syndrome associated with inflammation.27 The production of circulating inflammatory cytokines such as IFNγ was recently reported in Artemis-deficient subjects and was linked to activation of neutrophils and NET formation following oxidative stress.28 In our study, we observed increased inflammatory cytokines in the Artemis vector-treated mice, but not in the Artemis-deficient non-treated controls or in mice treated with WT cells. The levels of cytokines observed in the vector-treated mice were modest and did not cause visible behavior change in the animals. Transgene toxicity can be excluded because it should have manifested earlier when high levels of ARTEMIS mRNA were observed in the primary transplants, and it did not. In addition, Artemis-transgenic mice that can express up to 40 times the normal levels of Artemis have no increased cytokine levels or inflammation.22 Also, transplanting HSC overexpressing ARTEMIS in C57BL/6 mice did not trigger inflammation (data not shown). One hypothesis is that the inflammation observed in the two-hit secondary transplanted animals was linked to a higher peripheral CD4+ T cell reconstitution in these animals because this was the only statistically significant difference compared with one hit (Figure 4B). These cells could have been activated. Although we failed to detect infectious pathogenic agents in the animal facility, other non-tested agents including endogenous retroviruses could have been involved. Alternatively, an imbalanced hematopoietic reconstitution may hinder immune homeostasis via elevated myeloid cells, which are prone to amplify inflammatory signals, and indeed the mice had increased platelet counts and high CD11b+ myeloid cell numbers in blood (data not shown). Inflammation may have been caused by insufficient correction in Artemis expression, as seen in patients with hypomorphic mutations.27 However, the levels of inflammation observed in mice seemed to be lower than reported in Artemis-deficient individuals,28 and this hypothesis is weakened by the fact that the observation was made only in the group treated by two hits of vector in which reconstitution was at least as high as in the group treated with only one hit. We conclude that the moderate and infra-clinical inflammation that was observed in mice treated with two hits of the EF1a-hArtemis LV after secondary transplant is more likely due to an effect of the model and by incomplete immune or hematopoietic homeostasis accompanied by T cell activation than to an effect of the vector. In RS-SCID patients treated by gene therapy, the immune follow-up is likely to rapidly identify the occurrence of inflammatory responses, should they occur.
Thus, altogether, the data presented here show that the EF1a-hArtemis LV permits a durable engraftment of hematopoietic progenitor cells in mice and reconstitutes their immune system without vector-related adverse events. These results support further development of this vector for future gene therapy in humans. Based on the results of this murine study, high levels of engraftment should be sought for optimal immune reconstitution; therefore, the infusion of a product containing a high frequency of transduced cells is recommended.
Materials and Methods
Animals
Artemis-deficient (ART_KO) mice with a Dclre1c exon 12-deleted allele on C57BL/6J background were reported previously8, 29 and were rederived by embryo transfer (Charles River) prior to the study to ensure a non-leaky phenotype and lack of infection. The rederived mice were bred and housed for experiments in a specific pathogen-exempt environment at Genethon Animal Facility, Evry, France (national agreement D 91-228-107) in agreement with local guidelines and national regulation. Animal health status monitoring of the facility was conducted several times during the study (Charles River). Experiments were performed in accordance to international ethics principles and the study was approved (CE10-005) by the local Animal Ethical Committee.
Vector
The vesicular stomatitis virus G glycoprotein (VSVG)-pseudotyped EF1a-hArtemis vector was produced by transient transfection of HEK293T cells in Cell Factory 10 stacks (CF10). In brief, 6 CF10 were transfected with pKLgagpol, pKRev, pKG, and pEF1a-hArtemis plasmids. After a medium change, Benzonase (1.13 U/mL) was added to the culture medium (DMEM supplemented with 10% fetal calf serum [FCS] and 5 mM MgCl2) to remove plasmid and cellular DNA. After two harvests, 12 L of supernatant was clarified, purified, and concentrated by anion exchange, tangential flow filtration, and gel filtration as previously reported.21 The final product was filtered through 0.22 μm, aliquoted, and transferred to <−70°C for storage. The lot was quality controlled before use and titered 2.3E+08 IG/mL (Infectious Genome/mL as defined on HCT116 cells). A PGK-eGPF LV encoding enhanced green fluorescent protein (eGFP) was produced an purified in the same conditions and titered 7.8E+08 IG/mL.
Preparation and Transduction of Progenitor Cells for the Primary Graft
BM cells were flushed from femurs and tibias of WT C57BL/6 mice and ART_KO male mice. The BM Sca-1+ cells were purified from WT or ART_KO BM, using magnetic bead cell sorting (Miltenyi Biotec). BMCs or BM_SCA+ cells were pre-activated overnight by culture in X-vivo 20 medium (Lonza, Walkersville, USA), 50 U/mL penicillin, 50 ng/mL streptomycin (PS) (Invitrogen, Cergy Pontoise, France), murine IL-3 (20 ng/mL), human thrombopoietin (TPO) (100 ng/mL), human Flt-3 ligand (100 ng/mL) (R&D Systems, Lille, France), and murine stem cell factor (100 ng/mL) (Abcys, Paris, France), at 37°C, 5% CO2. The transduction of ART_KO BMCs or ART_KO BM SCA+ cells (1E+6 cells/mL) was performed with 1E+8 infectious genomes (i.g.)/mL of EF1a-hArtemis or PGK-GFP-WPRE vector (MOI equivalent to 100) in the presence of protamine sulfate (6 μg/mL) and cytokines for 6 h. Fresh medium was added, and cells were washed. For the cells that received two hits of infection, a second exposure of vector was performed overnight in the presence of protamine sulfate (6 μg/mL) and cytokines. Fresh medium was added, and cells were washed. One part of cells was injected in the mice and the other part was expanded for 7 days in the presence of cytokines in a humidified atmosphere (37°C, 5% CO2 in air) and was plated in MethoCult medium (H3434; STEMCELL Technologies, Vancouver, CA, USA). After 1 week, CFU-E/BFU-E, CFU-GM, and CFU-GEMM were counted by microscopy, and individual colonies were collected and genomic DNA (gDNA) was extracted to measure VCN. Seven days post-infection, the efficiency of transduction was evaluated in the population of cells expanded in vitro from Sca1+ cells with cytokines and in individual CFCs by qRT-PCR to detect VCN. qRT-PCR was performed to measure ARTEMIS transcript levels in the population of cells expanded in vitro from Sca-1+ cells.
Transplantation of Progenitor Cells
One day post-infection, the transduced progenitor cells (2E+5 cells/mouse) were injected intravenously (retro-orbital) into irradiated ART_KO females. Recipient female ART_KO mice were irradiated in ventilated plexiglass containers using an X-ray source at a dosage rate of 0.84 Gy/min (Faxitron CP160). Total body irradiation was given as a single exposure of 3 Gy at 2–3 h before cell transplantation.
Preparation of BM Cells for the Secondary Graft
After isoflurane anesthesia, mice from primary graft were killed by cervical dislocation and their femur and tibias removed to flush BMCs. One part of cells was used for the secondary transplant, and the other part was used for different analyses (VCN, mRNA expression, immune-phenotype, and insertion site analyses for mice with tumors). For the secondary graft, BMCs (1E+7 cells) from each mouse from the primary graft were injected intravenously into two irradiated Artemis-deficient females. Recipient female ART_KO mice were irradiated as described above for primary transplants.
Evaluation of Transduction, Engraftment, and Transgene Expression in Treated Mice
Mice were regularly observed during the protocol and, if needed, were sacrificed before the end of the protocol and autopsied. At 3 and 6 months (at sacrifice) after transplantation, blood was sampled by retro-orbital puncture (200 μL of blood with citrate 3.8%: 1/10 vol). At the endpoint (6 months post-transplantation), remaining live mice were killed by cervical dislocation and following macroscopic analysis, organs were taken for histopathological analysis. BMCs were obtained after flushing. Cell suspensions from lymph nodes, spleen, and thymus were obtained by passing the disrupted tissue through a 40-μm filter in PBS buffer. The efficiency of transduction was evaluated by measuring VCN using qPCR. Male donor cell engraftment into female recipient mice was evaluated using qPCR of a male Y sequence calibrated on a series of genomic DNA samples prepared from varying proportions of male/female blood cells, as previously described.20
Genomic DNA was extracted from spleen, BM, thymus, and lymph node. For greater than 1E+6 cells, the “Wizard genomic DNA purification kit” (Promega Corporation, Madison, WI, USA) was used. When cell quantity was inferior to 1E+6 cells, Proteinase K extraction was performed. The “QIAamp DNA Blood Mini kit” was used for gDNA extraction from peripheral blood cells, and the “DNeasy Blood & Tissue Kit” (QIAGEN, Hilden, Germany) for gDNA extraction from ovary.
Total RNA from spleen, BM, thymus, and lymph node was extracted using the SV total RNA isolation system (Promega). One microgram of each total RNA sample was reverse transcribed into cDNA with the SuperScript II RT (GIBCO BRL) using random hexamer primers at 42°C for 50 min. The RT was inactivated by incubation for 15 min at 70°C.
All primers and probes used are listed in Table S6. Oligonucleotide primers and TaqMan probes (with a 5′ reporter dye [6-carboxyfluorescein] and a downstream 3′ quencher dye [6-carboxytetramethyl rhodamine]) were designed with Primer Express and Oligo 4.0 Primer Analysis software (Perkin Elmer Applied Biosystem). qPCR was performed as previously reported.24 All measures were performed at least in duplicate. Negative controls were included in each analysis. Data were edited using the Primer Express software. VCN was measured by PCR quantification of HIV-Psi sequence per cell in reference to the murine TTN gene. Results were expressed as copies of vector per cell and calculated from a standard curve based on dilutions of a plasmid containing both the HIV-Psi and mouse TTN sequences. Donor chimerism or Y-Chimerism was measured by PCR quantification of murine Y sequence per cell in reference to the murine Ttn gene. Results were expressed as percentages of male cells in the population of cells tested. Percentages of Y-chimerism were calculated by qPCR using a reference titration curve consisting of varying proportions of male and female blood cells. ARTEMIS mRNA was measured by qRT-PCR in reference to murine TfIId gene expression and this ratio was normalized to the ratio of murine Artemis/TfIId using a standard that consists of gDNA from murine WT cells spiked with a plasmid containing a copy each of murine and human Artemis transgene sequences. Using this standard, WT murine cells provided the 100% value.
Flow Cytometry
Antibodies used for the study are listed in Table S7. Stainings were done after Fc blocking. Cells were analyzed with the Beckman Coulter FC 500 using the Kaluza version 1.2 software. At 3 and 6 months post-transplantation, lysed peripheral blood cells were stained with the following multicolor antibody panels: CD3-allophycocyanin (APC), CD4-PECy7, CD8-ECD/IgM APC, CD19-PECy7, IgD-PE/CD11b APC, TCRab-PE-Cy7, and NK1.1 PE. Thymus cells were stained by CD3-APC, CD4 PE-Cy7, CD8-ECD and CD4-PECy7, CD8-ECD, CD44-PE, and CD25-APC. Lymph node cells were stained by CD3-APC, CD4 PE-Cy7, CD8-ECD/IgM APC, CD19-PECy7, and IgD-PE. Cells from spleen were stained with the same panel as lymph node and in addition by CD11b APC, TCRab-PE-Cy7, NK1.1 PE, and TCRgd-PE-CY7. Cells from BM were stained by CD3-APC, CD4 PE-Cy7, CD8-ECD; IgM APC, CD19-PECy7, IgD-PE; and CD11b APC, TCRab-PE-Cy7, NK1.1 phycoerythrin (PE).
Combinatorial Diversity Analysis (Multiplex PCR Assay)
Mouse TCR diversity was measured by ImmunID (ImmunID Technologies, Grenoble, France) using a patented multi-N-plex qPCR technology to detect and characterize V-J rearrangements of the T and B cell receptor at the genomic DNA level from cells of spleen or thymus. The assay allows the simultaneous detection of several V-J rearrangements in the same reaction and covers 100% of the possible combinatorial rearrangements. Results of immune diversity are expressed as a percentage. This percentage represents the number of detected V-J rearrangements compared with the maximum number of possible recombinations between V families and J segments.
Serum Analysis for Ig and Cytokine Quantification
Mouse IgM and IgG levels were measured by quantifying serial dilutions of serum samples with an ELISA kit (quantification ELISA kit; Bethyl, Montgomery, TX, USA), according to the manufacturer’s instructions.
Inflammatory cytokines were measured in selected mouse sera by Cytometric Bead Array mouse inflammation Kit (BD reference 552364) using 1:5 dilution of serum. The BD CBA Mouse Inflammation Kit can be used to quantitatively measure IL-6, IL-10, monocyte chemoattractant protein-1 (MCP-1), IFN-γ, TNF, and IL-12p70 (IL-12p70) protein levels in a single sample. The kit performance has been optimized for analysis of specific proteins in tissue culture supernatants, EDTA plasma, and serum samples. The kit limit of detection provided by the manufacturer is 5 pg/mL for IL-6, 17.5 pg/mL for IL-10, 52.7 pg/mL for MCP-1, 2.5 pg/mL for IFN-γ, 7.3 pg/mL for TNF, and 10.7 pg/mL for IL-12p70.
Histological Evaluation
Tissues removed from euthanized animals was fixed in 10% formalin and embedded in paraffin. Tissue sections (4 μm) were stained with H&E for histological examination. Immunostaining was performed for the spleen (CD3+, B220+) and thymus (CD3+). Analysis and interpretation of the results were verified by an independent outside contractor, Biodoxis.
Blood Counts
Blood samples were analyzed for standard hematological parameters (WBC counts, RBC, hemoglobin, hematocrit, and platelet counts) using an MS9.3 counter (Schloessing Melet, Cergy-Pontoise, France).
Vector Insertion Site Analysis
LV integration sites (IS) have been recovered, processed, and mapped as previously described.24 In brief, genomic DNA extracted from either BM, thymus, and spleen was digested with Tru9I restriction enzyme, and LTR vector-genome junctions were amplified by LM-PCR. Nested PCRs were performed with specific primers annealing to the linker and the 3′ vector LTR adapted to Illumina sequencing and containing a 4-bp sample-specific barcode for sample identification. Libraries were quantified and loaded on a 1% agarose gel, and amplicons ranging from 200 to 500 kb were manually extracted from the gel, purified, and processed with MiSeq Reagent Kit v3, barcoded with a 6-bp sample-specific tag, and sequenced to saturation on an Illumina MySeq machine at IGA Technology Services (Udine, Italy). Raw reads were bioinformatically trimmed to recover the murine genome sequences adjacent to the 3′ proviral LTR by the Skewer software (mismatch rate of 0.12, minimal length of 20 bp after trimming, and a minimal match length equal to the pre-trimming sequence), and mapped on the reference genome (murine GRCm38/mm10) with the Bowtie2 software (>95% identity). The genomic coordinates of the first nucleotide in the host genome adjacent to the viral LTR were indicated as IS. ISs identified by different reads as mapping in the same genomic position were collapsed, and the number of corresponding reads defined the read counts. Genes having the IS inside the gene (defined as the most upstream TSS and the most downstream end of gene, in case of multiple isoforms) were annotated as target genes.
In Vitro Immortalization Assay
The method has been previously reported.17 In brief, after transduction, WT lineage-negative cells were expanded as mass cultures for 2 weeks in Iscove's Modified Dulbecco's Medium (IMDM) containing 50 ng/mL murine stem cell factor (mSCF), 100 ng/mL hFlt-3 ligand, 100 ng/mL hIL-11, 10 ng/mL, mIL-3, 10% FCS, 1% penicillin/streptomycin, and 2 mM glutamine. After mass culture expansion, cells were seeded into 96-well plates at a density of 100 cells per well. Two weeks later, wells containing actively proliferating cells were counted microscopically and by absorbance measurement of a reduced formazan (MTT assay) in a microplate reader. The replating frequency (RF) of insertional mutants was calculated based on Poisson statistics using the L-Calc software (STEMCELL Technologies, Vancouver, BC, Canada). Plates with an RF ≥ 3.17 × 10−4 were defined as positive assays.
Statistical Analyses
The non-parametric Mann-Whitney test was used (GraphPad software) to compare groups.
Author Contributions
Conceptualization, A.F., J.-P.d.V., M.C., S.H.-B.-A., and A.G.; Methodology, S.C., F.M., A.S., and A.G.; Investigation, S.C., C.L.-P., V.P., M.R., G.C., and B.G.; Writing – Original Draft: S.C. and A.G.; Writing –Review & Editing, C.L.-P., M.C., V.P., J.-P.d.V., M.R., A.F., and S.H.-B.-A.; Funding Acquisition, M.C., A.S., F.M., S.H.-B.-A., and A.G.; Supervision, M.C. and A.G.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
The authors would like to thank Drs. Ariana Moiani and Guillaume Corre for help with the insertion site analyses; Peggy Sanatine from the Imaging/Flow cytometry platform of Genethon for help with the flow cytometry analyses; Céline Sagrère, Sandrine Ferrand, Armelle Viornery, Roseline Yao, Séverine Charles, Simon Jimenez, and Laetitia van Wittenberghe for help with bioexperimentation and histology; the Bioproduction team of Genethon for help with vector production; and Erika Cantelli for help with manuscript preparation. The work was supported in part by funds from INSERM/DGOS (to A.G. and S.H.-B.-A.), by the European Union’s FP7 research and innovation programme under grant agreement CELL-PID (No. 261387 to M.C. and A.S.), by the European Union's Horizon 2020 research and innovation prgramme SCIDNET (No. 666908 to FM and AG) by the French Muscular Dystrophy Association/Telethon (AFM/Telethon) (to F.M. and A.G.), and also by equipment funds from “Region Ile-de-France,” “Conseil General de l'Essonne,” Genopole, and Evry University (to F.M. and A.G.).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtm.2019.08.014.
Supplemental Information
References
- 1.Dvorak C.C., Haddad E., Buckley R.H., Cowan M.J., Logan B., Griffith L.M., Kohn D.B., Pai S.Y., Notarangelo L., Shearer W. The genetic landscape of severe combined immunodeficiency in the United States and Canada in the current era (2010-2018) J. Allergy Clin. Immunol. 2019;143:405–407. doi: 10.1016/j.jaci.2018.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moshous D., Callebaut I., de Chasseval R., Corneo B., Cavazzana-Calvo M., Le Deist F., Tezcan I., Sanal O., Bertrand Y., Philippe N. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell. 2001;105:177–186. doi: 10.1016/s0092-8674(01)00309-9. [DOI] [PubMed] [Google Scholar]
- 3.Poinsignon C., Moshous D., Callebaut I., de Chasseval R., Villey I., de Villartay J.P. The etalol-beta-lactamase/beta-CASP domain of Artemis constitutes the catalytic core for V(D)J recombination. J. Exp. Med. 2004;199:315–321. doi: 10.1084/jem.20031142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drouet J., Frit P., Delteil C., de Villartay J.P., Salles B., Calsou P. Interplay between Ku, Artemis, and the DNA-dependent protein kinase catalytic subunit at DNA ends. J. Biol. Chem. 2006;281:27784–27793. doi: 10.1074/jbc.M603047200. [DOI] [PubMed] [Google Scholar]
- 5.Bétous R., Goullet de Rugy T., Pelegrini A.L., Queille S., de Villartay J.P., Hoffmann J.S. DNA replication stress triggers rapid DNA replication fork breakage by Artemis and XPF. PloS Genet. 2018;14:e1007541. doi: 10.1371/journal.pgen.1007541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schuetz C., Neven B., Dvorak C.C., Leroy S., Ege M.J., Pannicke U., Schwarz K., Schulz A.S., Hoenig M., Sparber-Sauer M. SCID patients with ARTEMIS vs RAG deficiencies following HCT: increased risk of late toxicity in ARTEMIS-deficient SCID. Blood. 2014;123:281–289. doi: 10.1182/blood-2013-01-476432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neven B., Leroy S., Decaluwe H., Le Deist F., Picard C., Moshous D., Mahlaoui N., Debré M., Casanova J.L., Dal Cortivo L. Long-term outcome after hematopoietic stem cell transplantation of a single-center cohort of 90 patients with severe combined immunodeficiency. Blood. 2009;113:4114–4124. doi: 10.1182/blood-2008-09-177923. [DOI] [PubMed] [Google Scholar]
- 8.Benjelloun F., Garrigue A., Demerens-de Chappedelaine C., Soulas-Sprauel P., Malassis-Séris M., Stockholm D., Hauer J., Blondeau J., Rivière J., Lim A. Stable and functional lymphoid reconstitution in artemis-deficient mice following lentiviral artemis gene transfer into hematopoietic stem cells. Mol. Ther. 2008;16:1490–1499. doi: 10.1038/mt.2008.118. [DOI] [PubMed] [Google Scholar]
- 9.Lagresle-Peyrou C., Benjelloun F., Hue C., Andre-Schmutz I., Bonhomme D., Forveille M., Beldjord K., Hacein-Bey-Abina S., De Villartay J.P., Charneau P. Restoration of human B-cell differentiation into NOD-SCID mice engrafted with gene-corrected CD34+ cells isolated from Artemis or RAG1-deficient patients. Mol. Ther. 2008;16:396–403. doi: 10.1038/sj.mt.6300353. [DOI] [PubMed] [Google Scholar]
- 10.Mostoslavsky G., Fabian A.J., Rooney S., Alt F.W., Mulligan R.C. Complete correction of murine Artemis immunodeficiency by lentiviral vector-mediated gene transfer. Proc. Natl. Acad. Sci. USA. 2006;103:16406–16411. doi: 10.1073/pnas.0608130103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Multhaup M.M., Podetz-Pedersen K.M., Karlen A.D., Olson E.R., Gunther R., Somia N.V., Blazar B.R., Cowan M.J., McIvor R.S. Role of transgene regulation in ex vivo lentiviral correction of artemis deficiency. Hum. Gene Ther. 2015;26:232–243. doi: 10.1089/hum.2014.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Punwani D., Kawahara M., Yu J., Sanford U., Roy S., Patel K., Carbonaro D.A., Karlen A.D., Khan S., Cornetta K. Lentivirus Mediated Correction of Artemis-Deficient Severe Combined Immunodeficiency. Hum. Gene Ther. 2017;28:112–124. doi: 10.1089/hum.2016.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Multhaup M., Karlen A.D., Swanson D.L., Wilber A., Somia N.V., Cowan M.J., McIvor R.S. Cytotoxicity associated with artemis overexpression after lentiviral vector-mediated gene transfer. Hum. Gene Ther. 2010;21:865–875. doi: 10.1089/hum.2009.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ulus-Senguloglu G., Arlett C.F., Plowman P.N., Parnell J., Patel N., Bourton E.C., Parris C.N. Elevated expression of artemis in human fibroblast cells is associated with cellular radiosensitivity and increased apoptosis. Br. J. Cancer. 2012;107:1506–1513. doi: 10.1038/bjc.2012.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Multhaup M.M., Gurram S., Podetz-Pedersen K.M., Karlen A.D., Swanson D.L., Somia N.V., Hackett P.B., Cowan M.J., McIvor R.S. Characterization of the human artemis promoter by heterologous gene expression in vitro and in vivo. DNA Cell Biol. 2011;30:751–761. doi: 10.1089/dna.2011.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zanta-Boussif M.A., Charrier S., Brice-Ouzet A., Martin S., Opolon P., Thrasher A.J., Hope T.J., Galy A. Validation of a mutated PRE sequence allowing high and sustained transgene expression while abrogating WHV-X protein synthesis: application to the gene therapy of WAS. Gene Ther. 2009;16:605–619. doi: 10.1038/gt.2009.3. [DOI] [PubMed] [Google Scholar]
- 17.Modlich U., Navarro S., Zychlinski D., Maetzig T., Knoess S., Brugman M.H., Schambach A., Charrier S., Galy A., Thrasher A.J. Insertional transformation of hematopoietic cells by self-inactivating lentiviral and gammaretroviral vectors. Mol. Ther. 2009;17:1919–1928. doi: 10.1038/mt.2009.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marangoni F., Bosticardo M., Charrier S., Draghici E., Locci M., Scaramuzza S., Panaroni C., Ponzoni M., Sanvito F., Doglioni C. Evidence for long-term efficacy and safety of gene therapy for Wiskott-Aldrich syndrome in preclinical models. Mol. Ther. 2009;17:1073–1082. doi: 10.1038/mt.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou S., Ma Z., Lu T., Janke L., Gray J.T., Sorrentino B.P. Mouse transplant models for evaluating the oncogenic risk of a self-inactivating XSCID lentiviral vector. PloS ONE. 2013;8:e62333. doi: 10.1371/journal.pone.0062333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charrier S., Stockholm D., Seye K., Opolon P., Taveau M., Gross D.A., Bucher-Laurent S., Delenda C., Vainchenker W., Danos O., Galy A. A lentiviral vector encoding the human Wiskott-Aldrich syndrome protein corrects immune and cytoskeletal defects in WASP knockout mice. Gene Ther. 2005;12:597–606. doi: 10.1038/sj.gt.3302440. [DOI] [PubMed] [Google Scholar]
- 21.Merten O.W., Charrier S., Laroudie N., Fauchille S., Dugué C., Jenny C., Audit M., Zanta-Boussif M.A., Chautard H., Radrizzani M. Large-scale manufacture and characterization of a lentiviral vector produced for clinical ex vivo gene therapy application. Hum. Gene Ther. 2011;22:343–356. doi: 10.1089/hum.2010.060. [DOI] [PubMed] [Google Scholar]
- 22.Rivera-Munoz P., Abramowski V., Jacquot S., André P., Charrier S., Lipson-Ruffert K., Fischer A., Galy A., Cavazzana M., de Villartay J.P. Lymphopoiesis in transgenic mice over-expressing Artemis. Gene Ther. 2016;23:176–186. doi: 10.1038/gt.2015.95. [DOI] [PubMed] [Google Scholar]
- 23.Bartholomae C.C., Arens A., Balaggan K.S., Yáñez-Muñoz R.J., Montini E., Howe S.J., Paruzynski A., Korn B., Appelt J.U., Macneil A. Lentiviral vector integration profiles differ in rodent postmitotic tissues. Mol. Ther. 2011;19:703–710. doi: 10.1038/mt.2011.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poletti V., Charrier S., Corre G., Gjata B., Vignaud A., Zhang F., Rothe M., Schambach A., Gaspar H.B., Thrasher A.J., Mavilio F. Preclinical Development of a Lentiviral Vector for Gene Therapy of X-Linked Severe Combined Immunodeficiency. Mol. Ther. Methods Clin. Dev. 2018;9:257–269. doi: 10.1016/j.omtm.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Ravin S.S., Wu X., Moir S., Anaya-O’Brien S., Kwatemaa N., I P., Theobald N., Choi U., Su L., Marquesen M. Lentiviral hematopoietic stem cell gene therapy for X-linked severe combined immunodeficiency. Sci. Transl. Med. 2016;8:335ra57. doi: 10.1126/scitranslmed.aad8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li L., Salido E., Zhou Y., Bhattacharyya S., Yannone S.M., Dunn E., Meneses J., Feeney A.J., Cowan M.J. Targeted disruption of the Artemis murine counterpart results in SCID and defective V(D)J recombination that is partially corrected with bone marrow transplantation. J. Immunol. 2005;174:2420–2428. doi: 10.4049/jimmunol.174.4.2420. [DOI] [PubMed] [Google Scholar]
- 27.Villa A., Notarangelo L.D., Roifman C.M. Omenn syndrome: inflammation in leaky severe combined immunodeficiency. J. Allergy Clin. Immunol. 2008;122:1082–1086. doi: 10.1016/j.jaci.2008.09.037. [DOI] [PubMed] [Google Scholar]
- 28.Gul E., Sayar E.H., Gungor B., Eroglu F.K., Surucu N., Keles S., Guner S.N., Findik S., Alpdundar E., Ayanoglu I.C. Type I IFN-related NETosis in ataxia telangiectasia and Artemis deficiency. J. Allergy Clin. Immunol. 2018;142:246–257. doi: 10.1016/j.jaci.2017.10.030. [DOI] [PubMed] [Google Scholar]
- 29.Rivera-Munoz P., Soulas-Sprauel P., Le Guyader G., Abramowski V., Bruneau S., Fischer A., Pâques F., de Villartay J.P. Reduced immunoglobulin class switch recombination in the absence of Artemis. Blood. 2009;114:3601–3609. doi: 10.1182/blood-2008-11-188383. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.