Abstract
High-mannose-type glycans (HMGs) are aberrantly enriched on HIV envelope glycoproteins. However, there is currently no drug selectively targeting HIV-associated HMGs. Here, we describe a novel HMG-targeting “lectibody,” a recombinant Fc-fusion protein comprising human IgG1 Fc and a novel actinohivin lectin variant (Avaren) obtained by structure-guided modifications for improved overall surface charge properties (AvFc). AvFc was engineered and produced using a rapid and scalable plant-based transient overexpression system. The lectibody exhibited potent antiviral activity against HIV-1 groups M and O primary viruses, as well as HIV-2 and simian immunodeficiency virus (SIV) strains, without affecting normal human blood cells. Furthermore, the lectibody induced Fc-mediated cell killing activity against HIV-1-infected cells and selectively recognized SIVmac239-infected macaque mesenteric lymph node cells in vitro. AvFc showed an extended serum half-life in rats and rhesus macaques, while no discernible toxicity was observed upon repeated systemic dosing in mice. These results highlight AvFc’s potential as a biotherapeutic targeting HIV-associated HMGs of cell-free virions, as well as productively infected cells, providing a foundation for new anti-HIV strategies. Efficient and cost-effective bioproduction in greenhouse facilities may open unique possibilities for further development of AvFc.
Keywords: plant virus vector, protein engineering, Nicotiana benthamiana, lectin, Fc fusion, HIV, high-mannose-type glycan
High-mannose-type glycans are clustered on HIV envelope, but there is currently no drug targeting these glycans. Matoba and colleagues developed a novel lectin-Fc fusion protein, or “lectibody,” selectively recognizing this glycobiomarker, which exhibited potent HIV neutralization and Fc-mediated HIV+ cell-killing activities without affecting uninfected normal cells.
Introduction
In human and other eukaryotic cells, high-mannose-type glycans (HMGs) appear in the early-stage N-glycosylation of nascent polypeptides within the endoplasmic reticulum. These “immature” oligosaccharides are subsequently processed into hybrid and complex glycoforms during protein secretion through the Golgi apparatus before they reach the cell membrane. Hence, HMGs are rarely found in the extracellular environment under normal conditions.1, 2 However, it is widely known that the envelope (Env) glycoproteins of HIV are heavily glycosylated with HMGs. While these glycans generally serve as a shield to protect neutralization-sensitive, conserved Env regions from humoral immune attack, recent studies have revealed that a fraction of broadly neutralizing antibodies (bNAbs) can evolve to target this HMG shield in some HIV-infected individuals, providing implications for HIV vaccine development.3, 4, 5
Carbohydrate-binding proteins, or lectins, are widely found in animals, plants, and microorganisms, playing critical roles in diverse biological processes, including cellular signaling, microbe-host interactions, and host defense.6, 7 Lectins are utilized in many biological applications, including cell profiling, lectin microarrays, affinity chromatography using immobilized lectins, histochemistry and cytochemistry, medical diagnosis as biosensors, and environmental monitoring.8, 9 In addition, several lectins of diverse origins have shown immunomodulatory, anti-tumor, anti-microbial, and antiviral effects,10, 11, 12 highlighting their potential use as drugs. Notwithstanding therapeutic potential demonstrated in vitro, the development of lectin-based antiviral therapeutics remains an underdeveloped area due to toxicity and/or limited availability. Actinohivin (AH) is an actinomycete-derived lectin recognizing the terminal α1,2-linked mannose residues on HMGs.13, 14 AH was shown to neutralize HIV without inducing cytotoxicity or mitogenicity in human blood cells,15 suggesting that this lectin may provide a useful tool for developing a new HMG-targeting agent. A major drawback, however, is that AH is highly hydrophobic, prone to aggregate, and, thus, recalcitrant to efficient recombinant production, hampering its development as a drug and/or biological tool.15 Here, we engineered a new AH variant with improved biochemical and pharmaceutical properties, using a plant-virus-vector-based transient overexpression system as a rapid screening and scalable production platform. A soluble variant, termed Avaren (actinohivin variant expressed in Nicotiana), was obtained by structure-guided mutations and subsequently fused to the fragment crystallizable region (Fc) of the human immunoglobulin G1 (IgG1) subclass to create the “lectibody” AvFc. Our data show that AvFc is highly producible in a scalable plant-based system, has anti-HIV activity derived from Fc functions, and lacks significant toxicity, highlighting its potential utility in novel HMG-targeting anti-HIV strategies.
Results
Design of a Bioproducible AH Variant
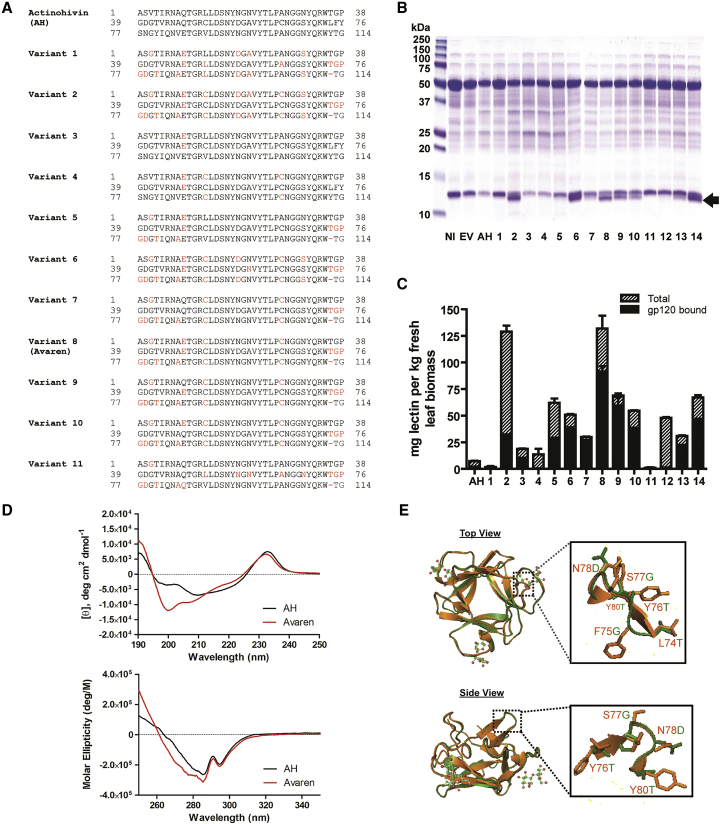
AH is composed of three near-homologous tandem repeats of a 38-amino-acid domain harboring a binding pocket for an HMG.13, 14 We hypothesized that the high aggregation propensity of AH could be improved by neutralizing its surface charge variation. Thus, we designed multiple AH variants by changing amino acids in one or two domains to corresponding residues in the other domain(s) so that all three domains are alike and have similar overall surface charge properties. For example, Gln9 (domain 1) and Gln47 (domain 2) were mutated to Glu to correspond to Glu85 in domain 3 (Figure 1A). To screen these variants, we used a plant-virus-vector-based transient overexpression system (magnICON) in N. benthamiana.16 Five days post-vector inoculation (dpi), several variants showed higher levels of accumulation in leaf tissue than that of AH (Figures 1B and 1C). These variants were readily extracted from leaf tissue in PBS, indicative of their improved solubility. Direct ELISA and HIV Env glycoprotein gp120-capture ELISA showed that variant 8 accumulated at the highest level, reaching over 100 mg kg−1 of leaf tissue, and retained gp120-binding capacity (Figure 1C) as well as anti-HIV activity (Figure S1). By contrast, AH recovered from leaf tissue by PBS extraction was only ∼1 mg kg−1 of leaf tissue (data not shown). Variant 8 was then named Avaren. In total, of the 114 amino acids in AH, 17 were modified to create Avaren (Figure 1A). Additionally, a disulfide bond was introduced at the rim of the sugar-binding pocket in domains 1 and 3, as originally found in domain 2 (confirmed by peptide mapping; Figure S2). Circular dichroism (CD) analysis revealed an increase in random coils in Avaren’s structure (Figure 1D, top panel). Nevertheless, the overall structure appeared to be similar to that of AH (Figure 1D, bottom panel). Homology modeling17 showed closely superimposed structures of AH and Avaren, albeit with a loss of a β strand in the hinge between domains 2 and 3 of Avaren (Figure 1E). The melting temperatures of Avaren and AH determined by CD were both 72.9°C, indicative of their similar structural stability. Collectively, it was concluded that Avaren has a better recombinant producibility in plants than AH while retaining overall conformational and functional properties.
Figure 1.
Molecular Engineering of Highly Bioproducible AH Variants
(A) The amino acid sequences of AH (UniProt: Q9KWN0) and AH variants. The three domains of mature AH (amino acids 1–38, 39–76, and 77–114) are aligned. Variant 8 was designated as Avaren (see Results). (B) SDS-PAGE was performed to analyze crude extracts of N. benthamiana leaves expressing AH or its variants and stained with Coomassie brilliant blue. At 5 dpi, leaf proteins were extracted with PBS (pH 7.2) containing 40 mM ascorbic acid using a 3:1 buffer-to-leaf ratio. NI, non-infiltrated leaf extract; EV, empty-vector-infiltrated leaf extract; AH, AH-expressing leaf extract. Lanes 1–14: leaf extracts of AH variants. (C) Quantification of AH variants in N. benthamiana leaf tissue using direct and gp120-capture ELISA for total protein (hatched bars) and gp120-binding protein (black bars) detected by a rabbit anti-AH antiserum. Variant 8 was designated as Avaren. (D) CD analysis. Far-UV CD (top) and near-UV CD (bottom) spectra of AH and Avaren. (E) Crystal structure of AH (PDB: 4G1R) (orange) superimposed with a homology model of Avaren (green), shown from the top and side views (PyMOL software). Homology modeling was performed with SWISS-MODEL, using AH as a template. Zoomed images of the surface-exposed loop between domains 2 and 3 (amino acids 74–80) are boxed.
Design, Production, and Binding Profiles of AvFc
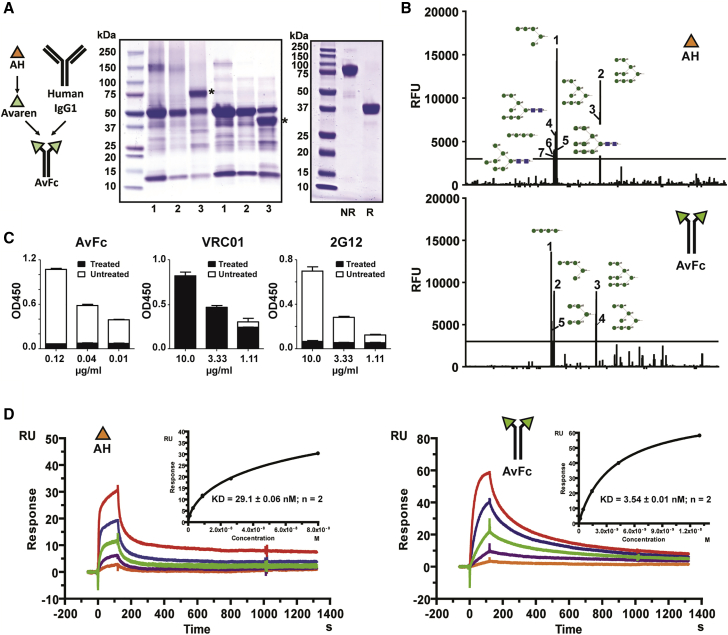
Avaren was translationally fused to the Fc region of a human IgG1 antibody (AvFc) in anticipation that such an antibody-like molecule (“lectibody”) may possess several advantages over the parent lectin, including higher HMG-binding avidity via dimerization, prolonged in vivo half-life, and Fc-mediated antiviral/anti-tumor functions such as antibody-dependent cell-mediated cytotoxicity (ADCC).18 Upon expression in N. benthamiana using a plant virus vector, AvFc accumulated to a significant portion of the total soluble leaf protein in 7 days and was efficiently purified to >95% homogeneity (Figure 2A). Approximately 1 g of purified AvFc was obtained from 10 kg plant biomass in a pilot-scale production facility. To determine AvFc’s sugar binding specificity, a glycan array analysis was performed. Among 610 mammalian glycans analyzed, both AH and AvFc showed high specificity to oligomannose glycans containing terminal α-1,2-linked mannose (Figure 2B). In a gp120-capture ELISA, the lectibody’s sugar-binding ability was abrogated upon gp120 treatment with a mannosidase, unlike the CD4 binding-site-specific bNAb VRC0119 but similarly to the Env-glycan-specific bNAb 2G1220, 21 (Figure 2C). These results demonstrate that AvFc retains the high HMG specificity of the original lectin AH. Notwithstanding, surface plasmon resonance (SPR) analysis revealed that the lectibody has increased affinity to recombinant HIV gp120 proteins by approximately 10-fold, compared to AH (Figure 2D; Figure S3).
Figure 2.
Design, Production, and HMG-Binding Profiles of AvFc
(A) Expression and purification of AvFc. Reducing SDS-PAGE and non-reducing SDS-PAGE were performed to analyze crude leaf extracts and purified AvFc, stained with Coomassie brilliant blue. Representative gel images are shown. Left: 1, non-infiltrated leaf extract; 2, empty-vector-infiltrated leaf extract; 3, AvFc-expressing leaf extract. Asterisks indicate AvFc. Right: purified AvFc under non-reducing and reducing conditions. NR, non-reducing conditions. R, reducing conditions. (B) Glycan array analysis. Sugar-binding profiles of AH (top) and AvFc (bottom) were analyzed in a mammalian glycan array with 610 glycans by the Consortium for Functional Glycomics. Glycans with a mean relative fluorescence unit exceeding 3,000 were ranked and indicated with schematic diagrams. Green circles indicate mannose; blue squares indicate N-acetylglucosamine. See Data Availability for a complete dataset. (C) Analysis of AvFc’s binding to gp120 treated with α-mannosidase. The gp120-binding ELISA was performed on AvFc, VRC01 (a HIV-1 CD4 binding-site-specific monoclonal antibody), and 2G12 (an HIV-1 envelope HMG-binding monoclonal antibody), using a recombinant HIV-1 gp120 or gp120 treated with α (1-2,3,6) mannosidase. The gp120-binding activities of AvFc and 2G12, but not of VRC01, were abolished by treating the Env protein with α-mannosidase, demonstrating AvFc’s specificity to the terminal mannose residues of HMGs. (D) SPR analysis of the binding affinities of AH and AvFc to gp120SF162. The binding kinetics and affinities of AH (left) and AvFc (right) to gp120SF162 were measured using a Biacore X100 2.0 instrument at ambient temperature. Representative sensorgrams are shown. A recombinant gp120SF162 was captured on a sensor chip to a surface density of about 50–100 RU. 3-fold serial dilutions of AvFc (1 μg/mL to 0.0123 μg/mL) or AH (1 μg/mL to 0.0123 μg/mL) were injected at a flow rate of 5 μL/min. KD was determined based on steady state (inset). Data indicate mean ± SEM from two independent analyses.
The Anti-HIV Activity of AvFc
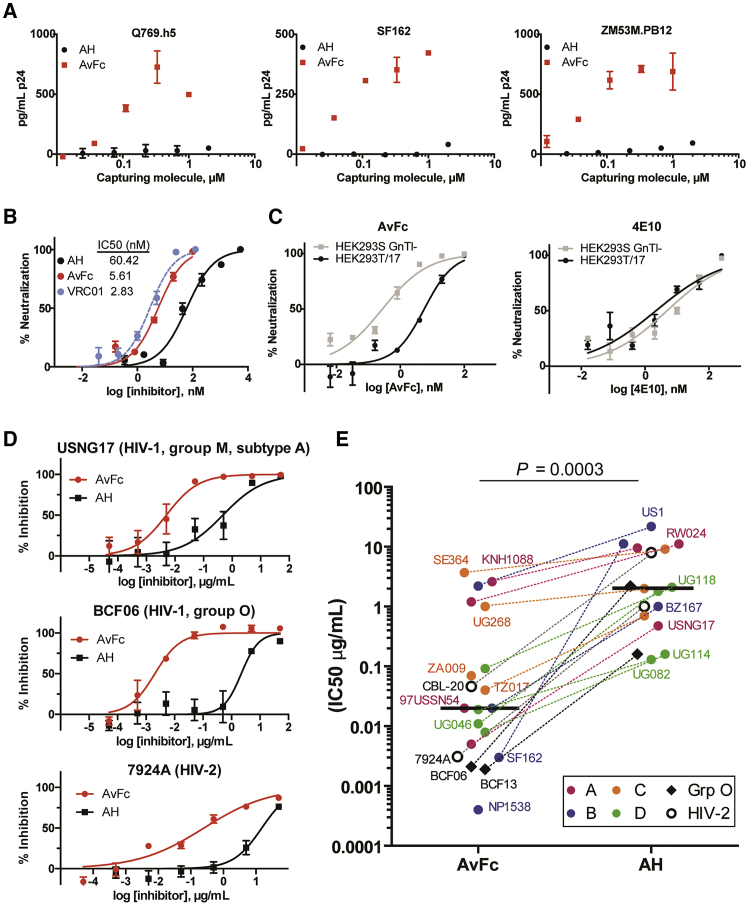
To investigate whether the increased gp120-binding affinity of AvFc translates into enhanced antiviral effects, we first performed a virus capture assay using Env-pseudotyped HIV-1 viruses. As shown in Figure 3A, AvFc caught the virions more effectively than AH for all three HIV-1 strains tested. In a pseudovirus neutralization assay using the tier-2 HIV-1 QH0692 clone,22 AvFc showed significantly more potent activity than AH, with a 50% inhibitory concentration (IC50) of 5.6 nM (versus 60.4 nM for AH), which is similar to that of VRC01 (Figure 3B). These results demonstrate that AvFc has significantly more potent HIV-1 neutralization activity than AH. Additionally, the lectibody showed a significantly stronger neutralization effect for the virus that was produced in the HEK293S GnTI− cell line (thus displaying higher amounts of Env HMGs23) than for the same virus produced in the standard HEK293T/17 cell line. This was in sharp contrast to the gp41-peptide-specific bNAb 4E10,24 whose neutralization activity appeared to be indifferent to Env HMG levels (Figure 3C). Thus, the results indicate that the HIV-1 neutralization activity of AvFc is proportional to the total amount of HMGs present on the viral surface. To further confirm AvFc’s anti-HIV activity, we tested its inhibitory activity against 20 different primary HIV strains that were produced in human peripheral blood mononuclear cells (PBMCs). Furthermore, we used human PBMCs from four different donors as target cells instead of a CD4-expressing cell line that was used in the pseudovirus assays. The lectibody, again, exhibited significantly stronger inhibitory activity than AH (p = 0.0003, Wilcoxon matched-pairs test), not only against HIV-1 Group M viruses but also against Group O and HIV-2 strains, with a median IC50 of 0.3 nM or 0.02 μg/mL for AvFc versus 156.3 nM or 2.00 μg/mL for AH (Figures 3D and 3E).
Figure 3.
AvFc’s Anti-HIV Activity
(A) Virus capture assay. Varying concentrations of AH or AvFc captured on a plate, to which Env-pseudotyped viruses (Q769.h5, Clade A; SF162, Clade B; or ZM53M.PB12, Clade C) were added and incubated for 1 h at 37°C. Captured virions were quantified by measuring the core HIV antigen p24. (B) The neutralizing activities of AH, AvFc, and the CD4-binding-site-specific bNAb VRC01 against Env-pseudotyped HIV-1 QH0692.42 virus in HOS-CD4-CCR5+ cells. Percent neutralization was calculated by dividing the luminescence of sample wells by that of virus-only control wells. Representative neutralization curves and the IC50s for AH, AvFc, and VRC01 (determined by nonlinear regression analysis; GraphPad Prism 5.0) are shown. (C) The impact of HIV-1 virion glycan structure on the antiviral activity. HIV-1 QH0692.42 pseudoviruses were produced in HEK293T/17 or HEK293S GnTI− cells, the latter of which give rise to more HMGs on Env. The neutralization curves of AvFc (leftpanel) or 4E10 (the anti-gp41-specific anti-HIV-1 bNAb; right panel) against these viruses were compared. (D and E) Human PBMC-based primary HIV inhibition assay. The viruses were produced in human PBMCs. Representative graphs in (D) show inhibition of group M, group O, and HIV-2 strains. IC50s of AvFc and AH are shown in (E), with horizontal bars representing median values (2.00 μg/mL and 0.02 μg/mL for AH and AvFc, respectively). Each assay was performed in quadruplicate using four different donor PBMCs as target cells. The Wilcoxon matched-pairs signed-rank test (GraphPad Prism 5) was used to analyze the differences.
Fc Functions of AvFc
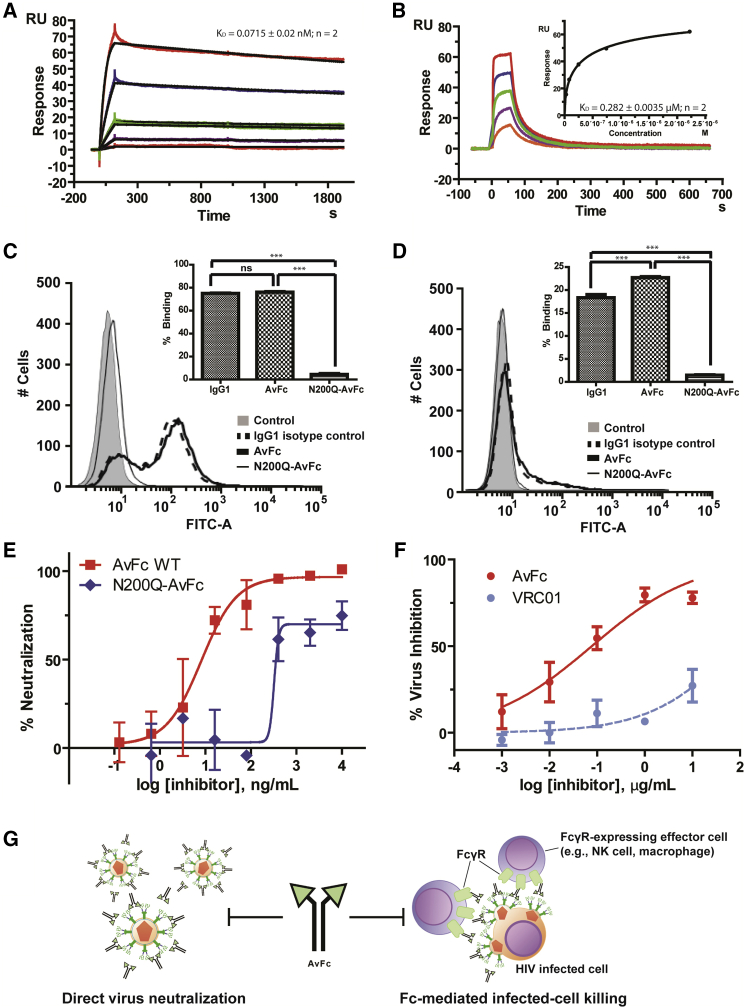
We next investigated the biochemical integrity of the Fc region of AvFc. SPR analysis showed that AvFc bound to two representative human activating Fcγ receptors, FcγRI and FcγRIIIa (Figures 4A and 4B), as well as the neonatal Fc receptor (FcRn; Figure S4), with KD values similar to those of a human IgG1κ isotype control (Table 1). AvFc’s capability to bind to the aforementioned FcγRs was confirmed by flow cytometry using FcγRI- and FcγRIIIa-expressing cells (Figures 4C and 4D). By contrast, AvFc with an Asn200 → Gln mutation (N200Q-AvFc), which eliminates N-glycosylation in the Fc region corresponding to Asn297 of human IgG1 and thereby significantly reduces affinity to these FcγRs,25, 26 showed dramatically reduced binding to these receptors (Figures 4C and 4D; Table 1). These data suggest that the Fc region of AvFc may be capable of exhibiting its functional property for Fc-mediated biological activities.
Figure 4.
Fc Functions of AvFc
(A and B) SPR analysis of AvFc binding to FcγRI (A) and FcγRIIIa (B). Analysis was done in two independent experiments, and average KD values are shown. (A) A representative FcγRI sensorgram. The raw (colored lines) and fitted (black lines) curves represent the concentrations of FcγRIA (1.2, 0.4, 0.13, 0.044, and 0.015 μg/mL, respectively, from top to bottom). KD was determined based on the 1:1 binding kinetics. (B) A representative FcγRIIIa sensorgram. The raw data curves (colored lines) represent the concentrations of FcγRIIIA (100, 33.3, 11.1, 3.7, 1.2, and 0.41 μg/mL, respectively, from top to bottom). The KD was determined based on steady state (inset). (C and D) Flow cytometry analysis of AvFc binding to FcγRI-expressing (C) and FcγRIIIa-expressing (D) TZM-bl cells. To eliminate background Avaren binding, samples were pre-incubated with yeast mannan. Representative flow histograms from triplicate analysis are shown. (E) HIV primary virus inhibition assay in PBMCs with aglycosylated (N200Q) AvFc. The assay was done in quadruplicate using four different PBMC preparations. (F) ADCVI activities of AvFc and VRC01. CD4+ lymphocytes were infected with HIV-192US657 at an MOI of approximately 0.05 for 72 h prior to the addition of AvFc (solid line) or VRC01 (broken line) and NK cells at an E:T ratio of 2:1. Data shown are representative of three independent triplicate experiments (expressed as mean ± SEM). (G) A schematic diagram showing AvFc’s anti-HIV mechanisms based on virion neutralization and Fc-mediated infected-cell killing.
Table 1.
Binding Affinity (KD Values) of AvFc, Human IgG1 Control, and N200Q-AvFc to Fc Receptors
| Protein | FcγRI (CD64) (nM) | FcγRIIIa (CD16) (μM) | FcRn, (nM) |
|---|---|---|---|
| AvFc | 0.07 ± 0.02 | 0.28 ± 0.00 | 60.01 ± 2.18 |
| Human IgG1 control | 0.11 ± 0.00 | 0.68 ± 0.01 | 18.00 ± 1.90 |
| N200Q-AvFc | 80.4 ± 19.6 | 1.70 ± 0.20 | NT |
KD values were determined using a Biacore X100 2.0 instrument. Data are mean ± SEM of two independent analyses. NT, not tested.
Because AvFc binds to FcγRI and FcγRIIIa, we next investigated whether AvFc’s anti-HIV activity could also be mediated by Fc functions. Indeed, in a human PBMC infection assay, N200Q-AvFc showed significantly reduced anti-HIV activity compared with AvFc (Figure 4E), likely reflecting the mutant’s significantly lower binding affinity to FcγRs (and, hence, lacking Fc-mediated antiviral effects). Furthermore, AvFc showed a dose-dependent inhibitory effect in an antibody-dependent cell-mediated virus inhibition (ADCVI) assay27 using primary human natural killer (NK) cells (Figure 4F). AvFc’s ADCVI activity was prominent at concentrations < 1 μg/mL and much stronger than that of VRC01, which had marginal effects at the same concentration range as shown in previous reports.28, 29 Collectively, these data indicate that, in addition to virion neutralization activity, AvFc can elicit Fc-mediated anti-HIV activity via binding to HMGs on viruses and infected cells (Figure 4G).
Safety Assessment of AvFc
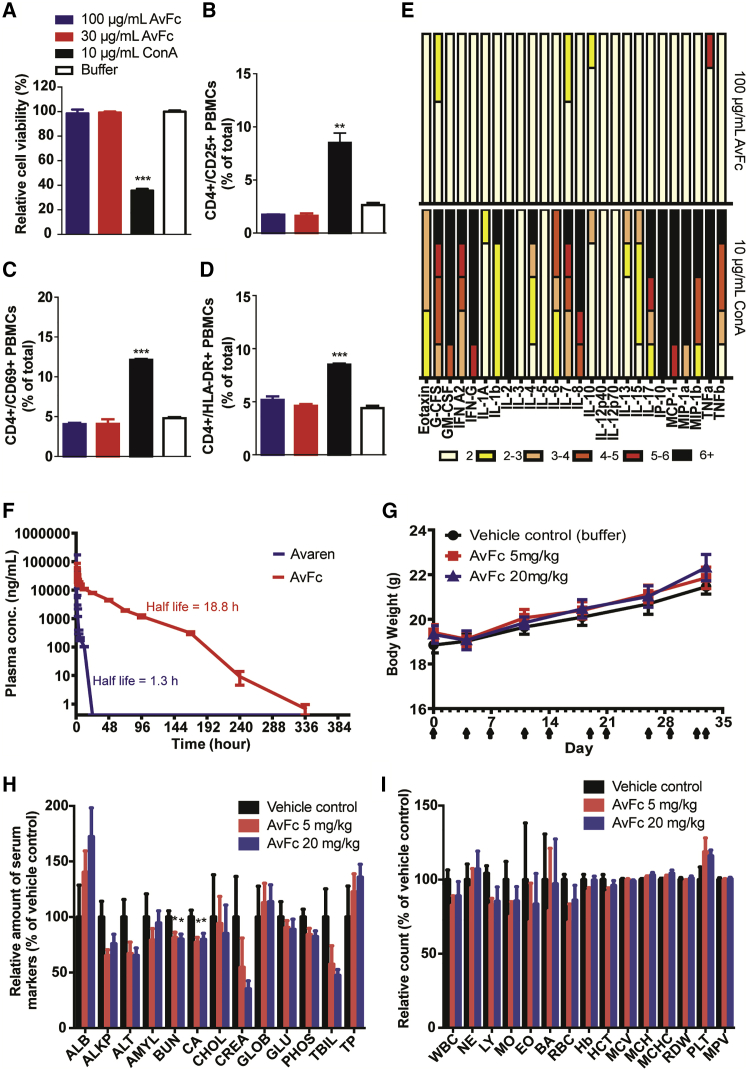
Given that AvFc may be developed as a novel anti-HIV agent, we next assessed its safety in in vitro and preclinical animal models. The lectibody showed no significant binding beyond that observed with VRC01 (Figure S5) or cytotoxicity (Figure 5A) at 100 μg/mL in human PBMCs, corresponding to 50,000 times the median HIV-neutralization IC50 (Figure 3E). This was in sharp contrast to concanavalin A (ConA), a well-known mannose-binding lectin15, which induced significant cytotoxicity at 10 μg/mL. Similarly, in flow cytometry analysis, AvFc showed no induction of the activation markers CD25, CD69, and HLA-DR in CD4 cells (Figures 5B–5D) or size and morphological changes in the PBMC population (Figure S6) that were clearly observed with ConA. Cytokine and chemokine release profiles in PMBCs stimulated with AvFc, analyzed using a multiplex bead array, ELISA, and qPCR, showed marginal impacts, if any (Figures 5E and S7; Table S1). These results suggest that AvFc has little mitogenicity or immunostimulatory activity that could pose an increased risk in HIV treatment. Next, rats were intravenously administered 4 mg kg−1 of AvFc or Avaren. AvFc was detectable in the blood circulation up to 10 days post-administration, whereas Avaren was cleared within 24 h (Figure 5F). The extended half-life of AvFc is likely attributed to neonatal Fc receptor affinity (Table 1; Figure S4). Despite its persistence in vivo, AvFc did not appear to induce any adverse effect; to probe its potential toxicity further, the lectibody was administered at 5 or 20 mg kg−1 twice a week over 5 weeks (10 doses total) to mice. No change was observed in behavior or body weight in all mice during the experiment (Figure 5G). An increase of liver and spleen weights was observed after 35 days for the high-dose AvFc treatment group treated twice a week (p = 0.01 and 0.0001, respectively; Figure S8A). This is likely due to an immune reaction to AvFc, as no distinct pathologies were observed in these tissues (Figure S8B); both Avaren and human Fc components are xenogeneic to mice. Serum chemistry showed a decrease in blood urea nitrogen and calcium levels for both doses of AvFc (p = 0.017 and 0.012, respectively; Figure 5H; Table S2). However, the values were still within or close to the normal physiological ranges. Additionally, although not statistically significant, a trend of decrease was noted in creatinine and total bilirubin levels in AvFc-dosed groups. There was no significant difference in any of the complete blood count parameters tested between AvFc-treated and vehicle control-treated mice (Figure 5I; Table S3). Together, these results showed an overall lack of major toxicity upon AvFc systemic administration.
Figure 5.
AvFc Lacks Major Toxicity in In Vitro and Animal Models
(A) Human PBMC viability assessed by flow cytometry after staining with propidium iodide (PI). (B–D) Analysis of PBMC activation. PBMCs were treated with a vehicle control, ConA, or AvFc and analyzed for CD25 (B), CD69 (C), and HLA-DR (D) after dual fluorescent staining. (E) Cytokine and chemokine secretion by PBMCs (from five different donors) stimulated with AvFc or ConA for 72 h, assessed by a multiplex bead array. Changes in expression levels are subdivided into 1- to 2-fold (white bars), 2- to 3-fold (yellow bars), 3- to 4-fold (apricot bars), 4- to 5-fold (red-orange bars), 5- to 6-fold (red bars), and >6-fold (black bars) increase. (F) Pharmacokinetic evaluation of Avaren and AvFc. Serum Avaren and AvFc concentrations were measured by specific immunoassays (see Materials and Methods) at different time points after single bolus dose administration via the tail vein. Data represent mean ± SEM obtained for each group (n = 4), and half-life values were derived using nonlinear regression (curve fitting) in the GraphPad Prism software. (G–I) Effects of AvFc repeated systemic dosing on body weight (G), blood chemistry (H), and complete blood count (I) in mice (n = 10). Blood chemistry and complete blood count data were analyzed 1 day after the last dose in the mouse repeated-dosing study. Values are relative to the vehicle control group (mean ± SEM). Actual measured values are shown in Tables S2 and S3. AvFc showed significantly lower BUN and Ca levels than the vehicle control (*p < 0.05; one-way ANOVA with Bonferroni’s posttests); however, these values remained within the normal range for mice (10–33 mg/dL for BUN and 8.0–15.5 mg/dL for Ca, according to the University of Louisville Pathology Laboratory).
Evaluation of AvFc toward Its Future Testing in Rhesus Macaque Simian Immunodeficiency Virus Infection Models
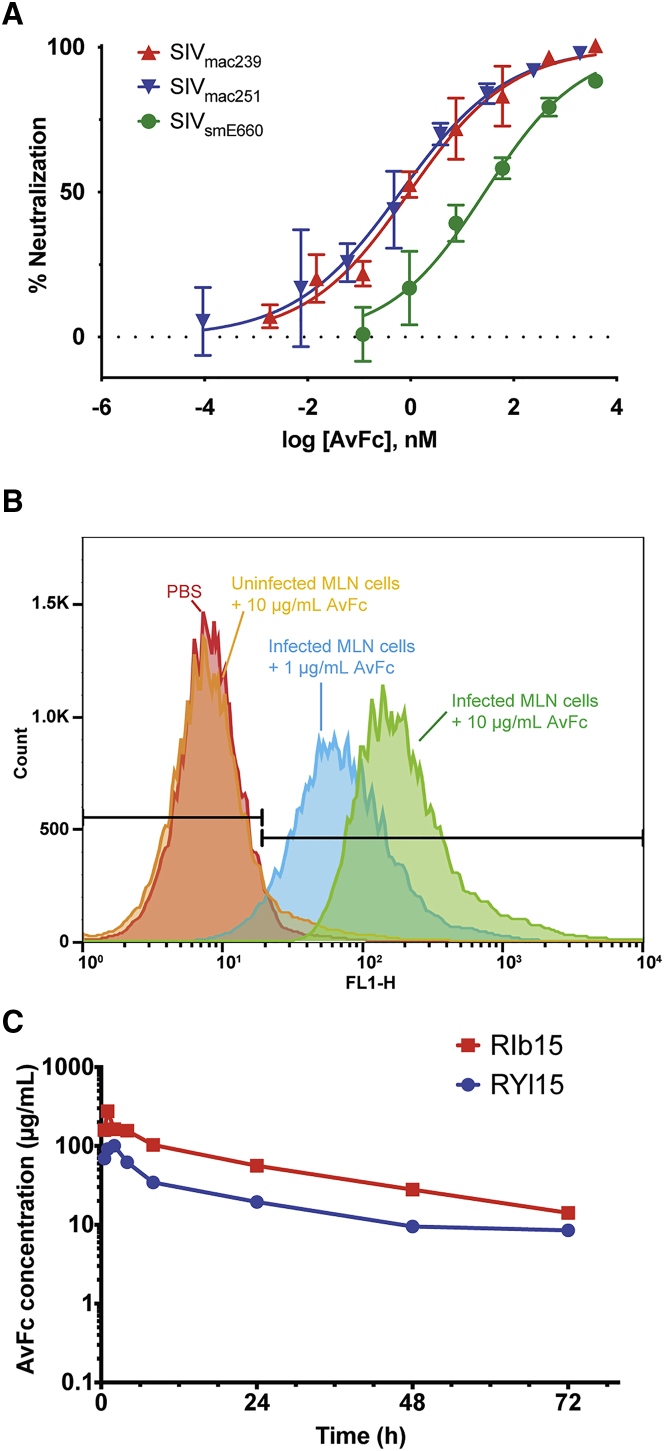
The aforementioned results justify the testing of AvFc’s therapeutic potential in non-human primate models of HIV infection. Toward this end, we first tested AvFc’s capacity to neutralize three simian immunodeficiency virus (SIV) strains, including SIVsmE660, SIVmac239, and SIVmac251, propagated in CEMx174 cells. The lectibody effectively neutralized these viruses, with IC50 values of 15.3, 6.6, and 3.8 nM, respectively (Figure 6A). Additionally, AvFc’s ability to recognize infected cells was examined by flow cytometry, using rhesus macaque mesenteric lymph node (MLN) cells that were infected by SIVmac239 in vitro. The results showed that AvFc at 1 and 10 μg/mL could dose-dependently recognize SIVmac239-infected rhesus MLN cells. Notably, the higher concentration of AvFc showed no binding to uninfected MLN cells, indicating the lectibody’s high selectively to infected cells (Figure 6B). Lastly, two adult female rhesus macaques were subcutaneously administered with a single bolus dose of 21 mg (3.6 and 3.7 mg/kg, respectively). The lectibody was well tolerated, exhibiting an average half-life of 27.7 h (Figure 6C). Flow cytometry analysis of macaque PBMCs isolated 0, 4, and 72 h after AvFc administration showed evidence of a selective and temporal redistribution of some cell subsets, including monocytes, dendritic cells, and NK cells, but no evidence of T cell activation (Figures S9–S16), consistent with data shown in Figure 5 for human PBMCs.
Figure 6.
SIV-Neutralizing Activity, SIV-Infected Cell Binding, and Macaque Subcutaneous Administration of AvFc
(A) Neutralization of SIV by AvFc. SIV strains smE660, mac239, and mac251 were propagated in CEMx174 cells. TZM-bl cells were infected with SIV in the presence of a vehicle control or serially diluted AvFc starting from 300 μg/mL (for mac239 and smE660) or 150 μg/mL (for mac251) at 37°C for 2 days. RLUs were converted to percent infection using the values from the no-drug control wells as 100% infection. AvFc neutralized SIVsmE660, SIVmac239, and SIVmac251 with IC50 values of 15.3, 6.6, and 1.8 nM respectively. Representative data from 3 independent experiments are indicated. (B) AvFc binding to rhesus macaque MLN cells. As shown, AvFc bound to SIVmac239-infected cells but not non-infected ones. (C) Pharmacokinetics of AvFc in rhesus macaques. A single bolus dose of 21 mg administered to each of two female macaques was well tolerated, with an average half-life of 27.7 h. Peak concentration occurred between 1 and 2 h post-injection.
Discussion
It is well known that HMGs accumulate on the surface of HIV.12 Nevertheless, currently there is no drug targeting this glycobiomarker. In this report, we have developed the lectibody AvFc, a recombinant HMG-specific lectin-Fc fusion protein. AvFc was created by a combined approach using structure-guided modifications and a scalable plant-virus-vector-based transient overexpression system. The lectibody inhibited a wide range of HIV strains and elicited Fc-mediated cytotoxicity to HIV-infected human PMBCs while not inducing detectable cell proliferation or causing noticeable toxicity upon systemic dosing at effective concentrations in animals. The present study illustrated that plant viral expression systems can provide a powerful tool for rapid screening and efficient production of novel recombinant proteins. The quantity of protein obtained and the ease with which multiple mutants can be made and tested using a transient plant expression system demonstrate the robustness and flexibility of this production platform, which can greatly facilitate drug discovery and production.
Our initial efforts focused on the engineering of AH mutants with improved solubility. Despite mutating as many as 15% of AH’s amino acids, Avaren was shown to retain overall structural integrity and HMG-binding activity (Figure 1). Thus, our approach of modifying AH’s surface charge property has met with success. Using the ExPASy ProtParam tool, it was determined that Avaren has a theoretical isoelectric point (pI) of 5.13 and a grand average of hydropathicity (GRAVY) value of −0.912, whereas AH has a theoretical pI of 8.59 and a GRAVY value of −0.778. Accordingly, Avaren has overall lower surface charge and hydrophobicity than AH. Raghunathan et al.30 have previously shown that protein surface charge engineering can modulate protein stability and aggregation properties. Thus, these findings suggest improved manufacturability of recombinant proteins. Other critical mutations responsible for Avaren’s high production yield appeared to be the modification of the surface-exposed loop between domains 2 and 3 and the C-terminal amino acids to mimic the more hydrophilic corresponding residues between domains 1 and 2. Additionally, a charged surface-exposed residue located at position 9 in each module (i.e., Gln9, Gln47, and Glu85 of AH) appeared to be a critical factor determining bioproducibility; Glu seemed to be more favorable than Gln (see variants 7–10). Furthermore, Avaren has two new disulfide bonds at the rim of the sugar-binding pocket in domains 1 and 3, as originally found in domain 2. These new disulfide bonds appear to have partly contributed to the high accumulation level of Avaren in leaf tissue (compare variants 5 and 8), as the cysteine-free variants, variants 1, 11, and 12, showed a poor level of tissue accumulation similar to AH (Figures 1B and 1C). It is possible that these disulfide bonds collectively facilitated proper tertiary structure formation in planta, leading to a high yield. Interestingly, variant 2 accumulated relatively well in leaf tissue yet lost the large part of gp120-binding ability (Figures 1B and 1C). This variant is similar to Avaren with 6 additional mutations, including Asn19 → Asp, Asn21 → Ala, Asn31 → Ser, Asn95 → Asp, Asn97 → Ala, and Asn107 → Ser. Combined with the results of variants 6, 13, and 14, we postulate that Asn21 and Asn97 may play a role in gp120 binding. These observations support the previously proposed notion that the open space between each sugar-binding pocket has Asn/Asp residues that help interact with HMGs.14
It is notable that Avaren dimerization via Fc fusion had virtually no impact on the sugar-binding profile of the original molecule AH, again highlighting successful molecular engineering in the present study. Our data indicated that AvFc had higher capacity to capture HIV virions than AH, which was also reflected in the lectibody’s increased functional affinity to gp120 in SPR analysis (Figures 2D, 3A, and S3). High avidity is a critical factor for lectins’ antiviral activity. For example, the dimeric lectin griffithsin significantly lost its anti-HIV activity when the protein was mutated into a monomeric form.31 Similarly, the monomer form of cyanovirin-N, which contains two sugar-binding sites, showed reduced anti-HIV activity when one of the sugar binding sites was mutated but restored activity upon dimerization.32 Thus, the enhanced antiviral activity of AvFc over that of AH can be partly explained by increased avidity to Env glycans. However, findings in human primary cell-based assays (Figures 3D, 3E, 4E, and 4F) suggest that a significant part of AvFc’s anti-HIV activity may be brought about by Fc functions. In particular, the strong ADCVI activity in Figure 3F indicates that AvFc can effectively recognize and aid in killing HIV-infected cells. This notion is further supported by our findings that AvFc showed no cytotoxicity or sugar-specific interaction in normal human PBMCs (Figures 5A and S5), while it efficiently bound to SIVmac239-infected, but not uninfected, macaque MLN cells (Figure 6B). These results highlight AvFc’s capacity to selectively target HIV-infected cells, which is an unprecedented feature for Env-glycan-targeting lectins described to date. The results further point to the possibility that AvFc could be used to detect and/or eliminate sites of residual viral replication in vivo, in addition to neutralizing free viruses in the circulation.
We analyzed a potential correlation between the predicted number of Env N-glycans and AvFc IC50s for HIV-1 strains tested in Figure 3E. While there is a trend of an inverse correlation (i.e., a lower IC50 with an increased number of glycans), it was not statistically significant (p = 0.4558, Pearson correlation coefficient; Figure S17). The slope of AvFc’s inhibitory curves appeared to be lower for HIV-2 than for HIV-1 strains (Figure 3D), which may be due to difference in their Env glycosylation patterns33 and indicative of distinct antiviral mechanisms.34 Additionally, among the three SIV strains tested in Figure 6A, the most AvFc-resistant strain, E660, has the least number of predicted Env N-glycans compared to mac239 and mac251 (21, 24, and 23, respectively; NetNGlyc 1.0 Server tool). We speculate that some glycans at specific locations on Env, rather than the total number of glycans, are critical to AvFc’s neutralization effects. Identification of such glycans is currently underway. There is some evidence to suggest that HIV can evolve to obtain resistance to antiviral lectins via deglycosylation.35, 36 However, deglycosylation of Env glycoproteins may result in the unmasking of critical epitopes, making the virus more sensitive to host immunoglobulins,12, 35, 37 and AvFc may still be able to recognize and kill infected cells expressing partially deglycosylated Env, since complete deglycosylation of Env is unlikely due to the critical roles of at least some N-glycans in Env assembly and viral infectivity.38, 39, 40 Additionally, the ability of AvFc to neutralize not only group M viruses but also group O, HIV-2, and SIV strains may be an advantage over (or a complement to) contemporary bNAbs, which may not recognize and neutralize beyond group M viruses. Because HMGs are widely found on the glycoproteins of enveloped viruses,12 there is a possibility that AvFc exhibits neutralization activity against other viruses as well. In our preliminary study, we found that AvFc holds nanomolar affinity to glycoproteins of hepatitis C virus, various influenza strains, severe acute respiratory syndrome coronavirus, and Middle East respiratory syndrome coronavirus (Figure S18; Table S4). Although these glycoproteins showed a lower affinity than HIV-1 gp120, the results suggest that AvFc could be used as a broad-spectrum antiviral agent, although binding to envelope glycoproteins alone does not guarantee antiviral activity. Studies are currently underway to address this possibility.
There are at least two major aspects that may need to be addressed further for the use of AvFc as an antiviral therapeutic agent. The first is the potential for immunogenicity to the molecule that could curtail its efficacy and/or safety in repeated treatments. The present animal studies were not suited to address the potential immunogenicity of AvFc, because the Fc domain of AvFc is of human origin, which itself would be immunogenic in mice and monkeys. Given that experimental animal models have a limited predictive value for the assessment of therapeutic protein immunogenicity in humans,41, 42 the use of AvFc variants with a species-specific Fc domain is, at least, essential to address the immunogenicity question in preclinical studies. Should this, indeed, be an issue, several potential remedies may be tested to minimize immunogenicity, including T cell epitope deletion, pegylation, and others, though these studies were beyond the scope of the present report. Second, AvFc showed a relatively short serum half-life of 27.7 h in rhesus macaques (Figure 6C). This may be, in part, due to slightly decreased affinity to FcRn compared to a human IgG1 isotype control, as shown in Table 1. A potential solution may be the modification of the Fc region to improve FcRn affinity, such as those described by Zalevsky et al.43 Additionally, studies are underway to determine the biodistribution profile and off-target binding potential of AvFc. While normal cells do not typically display high amounts of HMGs at their surface, these immature glycans have been found to be enriched in the plasma membranes of cancer cells44, 45, 46 and embryonic stem cells.47, 48 A future study should carefully evaluate what cell types other than HIV- or SIV-infected cells are recognized by AvFc.
In conclusion, AvFc is a novel biologic candidate targeting HIV-associated HMGs. The ability of AvFc to inhibit diverse HIV strains and recognize HIV- and SIV-infected primary cells, coupled with the apparent lack of toxicity upon systemic administration, warrants additional exploration of its therapeutic potential in non-human primate models of HIV infection. Additional engineering to improve pharmacological profile, such as deimmunization and Fc modifications for extended serum half-life and enhanced ADCC activity, may be necessary to further AvFc’s drug potential. In this regard, efficient, cost-effective, and rapid plant-based bioproduction in greenhouse facilities will facilitate such endeavors and may open unique possibilities for lectibody-based antiviral drug development.
Materials and Methods
Animal Housing and Care
Rodents: 8-week-old female Sprague-Dawley rats (Charles River Laboratories, Wilmington, MA, USA) weighing approximately 250 g (for the single-dose biodistribution study) and 6- to 8-week-old female BALB/c mice (Jackson Laboratory; for the repeated-dose toxicity study) were housed in a temperature- and humidity-controlled environment, with an alternating 12-h:12-h light/dark cycle and free access to standard diet and water. All experimental procedures were approved by the University of Louisville’s Institutional Animal Care and Use Committee. Rhesus macaques: two adult female rhesus macaques of Indian origin (RIb15 and RYl15), weighing 5.76 and 5.72 kg, respectively, were selected from the colonies of the New Iberia Research Center, University of Louisiana at Lafayette and housed there for the pharmacokinetics (PK) study. They were cared for in conformance to the guidelines of the Committee on the Care and Use of Laboratory Animals. The animals were fed a primate-specific diet (Purina) supplemented daily with fresh fruit and/or vegetables. All animal assessments were conducted under anesthesia. All experimental protocols and procedures were reviewed and approved by the University of Louisiana at Lafayette Animal Care and Use Committee.
Vector Construction of AH and AH Variants
A ‘‘deconstructed’’ tobamovirus replicon system16, 49 (magnICON; Icon Genetics) was used to express AH and AH variants in N. benthamiana. AH (GenBank: AB032371) and AH variant coding sequences (N. benthamiana codon optimized; GenBank: MK503330 for Avaren) were generated synthetically and sub-cloned into the magnICON vector pICH11599 via standard molecular biology or subcloning procedures using Nco I/Sac I restriction sites.
Viral-Vector-Based Overexpression of AH and AH Variants in Nicotiana benthamiana
Plant expression of AH and AH variants was performed using the magnICON system. The 3′ provector of AH and each AH variant was used with the 5′ provectors pICH20155 and pICH14011. The vectors were delivered into N. benthamiana leaves using the Agrobacterium vacuum infiltration method.15 After 5 days, leaf material was homogenized by a Precellys 24 homogenizer (Bertin Technologies, Rockville, MD, USA) in extraction buffer (PBS [pH 7.2], 40 mM ascorbic acid). The resulting extract was clarified by centrifugation at 10,000 × g for 5 min and analyzed for protein expression by SDS-PAGE and by direct and gp120-capture ELISA.15
CD Studies
AH and Avaren were buffer exchanged into a low-salt buffer (10 mM phosphate buffer [pH 7.0]) using 3.5 molecular weight cut-off (MWCO) dialysis cassettes. CD spectra were collected on a JASCO J-815 CD spectrometer (JASCO, Great Dunmow, Essex, UK) with a 1-nm bandwidth, using 1.0-mm and 10.0-mm pathlength cuvettes for far-UV and near-UV scans, respectively. The scans were performed at 20°C and corrected for the blank (low-salt buffer) control. Protein concentrations were determined by absorbance at 280 nm and converted to mean residue ellipticity (θ) or molar ellipticity.
Biomass Production and Agro-infiltration of AvFc
K1DFX-P2 N. benthamiana plants were grown for 25 days within a contained-growth room environment at Kentucky BioProcessing under artificial lighting. AvFc agrobacterium vectors were transiently delivered into the plants by vacuum infiltration (24” vacuum for 2 min). Post-infiltration, the plants were incubated for 7 days, and whole leaf tissues were harvested by mechanical cutting.
Extraction and Clarification of AvFc
The harvested biomass was mechanically disintegrated in a 2:1 buffer (20 mM sodium phosphate, 40 mM ascorbic acid [pH 7.0])-to-tissue ratio. The raw extract was adjusted to a pH of 7.0 ± 0.05 using sodium hydroxide. Celpure diatomaceous earth (DE; Advanced Minerals) was added at 33 g/L as a filter aid. The extract and DE slurry was then clarified using a plate-frame filter press and 0.3-μm cellulose pads (Ertel Alsop). The packed press was then washed with 10 vol of extraction buffer to improve recovery. The filter press filtrate and wash were combined to create the Protein A feed.
Purification of AvFc
AvFc from the clarified extract was captured on a MabSelect SuRe Protein A column (GE Healthcare), which was equilibrated with 5 column volumes (CVs) of 20 mM sodium phosphate (pH 7.0); the clarified extract was applied through a 1.2-μm glass fiber filter with a residence time of 2–10 min per CV. After feed application, the column was washed with equilibration buffer (5 mM sodium phosphate [pH 6.8]) to reach the UV baseline. The column was then eluted using a step to 2 M arginine (pH 3.0). The elution fraction was immediately neutralized to pH 6.8 ± 0.05 using 1 M Tris (pH 8.0) buffer, and sterile-filtered prior to the next chromatography step. AvFc was further purified using a CHT Type II 40-μm Column (Bio-Rad). The column was conditioned using 1 CV of neutralization buffer (250 mM sodium phosphate [pH 6.8]) and equilibrated using 10 CV of equilibration buffer. The protein A elution pool was diluted to ⩽10 mS/cm with water for irrigation (WFI) and loaded onto the column with a 2-min residence time. After feed application, the column was washed with 5 CV of equilibration buffer to reach the UV baseline and eluted using a 15-CV linear gradient to 5 mM sodium phosphate + 800 mM NaCl (pH 6.8). After collection of the elution peak, the CHT column was stripped using 250 mM sodium phosphate, 4 M NaCl (pH 6.8). The CHT elution pool was concentrated 10- to 20-fold using a 30-kDa tangential flow ultrafiltration cassette (Sartorius), followed by a 7× diafiltration against Dulbecco’s PBS (Invitrogen). The retentate was sterile filtered using a 0.2-μm cellulose acetate filter prior to storage at 2°C–8°C. N200Q-AvFc and AvFclec− were purified using the same procedure.
ELISA-Based Binding Analysis Using HIV-1 gp120 Treated with α-Mannosidase
Recombinant gp120 (SF162, HIV1/Clade B, Immune Technology, New York, NY, USA) was cleaved with α(1-2,3,6) mannosidase (PROzyme, Hayward, CA, USA). A volume of 1 μL gp120 was added to 10 μL 5× reaction buffer (supplied from PROzyme), 20 μL α(1-2,3,6) mannosidase, and 19 μL de-ionized water. A control untreated reaction was set up in the same manner replacing α(1-2,3,6) mannosidase with de-ionized water. The treated and untreated reactions were incubated for 24 h at 37°C. After 24 h, a gp120-capture ELISA (gp120 ELISA) was performed. ELISA plates were coated with treated or untreated reactions (diluted to 3 mL in 50 mM carbonate-bicarbonate [pH 9.6] coating buffer; final concentration of 330 ng/mL gp120) and blocked with blocking buffer (PBS [pH 7.2], 0.05% Tween 20, 5% [w/v] non-fat dry milk). 3-fold serially diluted antibodies (AvFc, VRC01, and 2G12) starting at 1 μg/mL were applied onto the plates and incubated for 1 h at 37°C. The gp120-bound antibodies were detected by HRP-conjugated mouse anti-human IgG (Fc) (SouthernBiotech, Birmingham, AL) at a 1:5,000 dilution. The tetramethylbenzidine (TMB) substrate (BioFX Laboratories, Owings Mills, MD, USA) was used for detection; absorbance at 450 nm was measured on a plate reader (Bio-Tek, Winooski, VT, USA). Graphs were plotted with the GraphPad Prism 5.0 software.
SPR (for gp120 Binding)
The binding affinity (KD) of AH or AvFc to gp120 was measured on a Biacore X100 2.0 instrument at ambient temperature. Recombinant His-tagged gp120 (Q769.h5, Immune Technology, #IT-001-0012p; SF162, Immune Technology, #IT-001-0028p-PBS; and ZM53M.PB12 Immune Technology, #IT-001-RC8p) proteins were captured on a sensor chip NTA following the manufacturer’s instructions to a surface density of about 30 RUs. A reference flow cell was utilized to correct response contributions such as bulk shifts that occur equally in the sample and reference flow cells. Serial dilutions of AH or AvFc were made in running buffer (HPS-P+ with 50 μM EDTA, GE Healthcare) and injected at a flow rate of 5 μL/min for a contact time of 120 s and a dissociation time of 1,200 s. A blank cycle (running buffer) was performed, and all sample injections were blank subtracted to correct the sensorgrams for drifts and other disturbances that affect the reference subtracted curve. Between sample injections, the system was washed with NTA wash buffer, with the surface regenerated with the NTA regeneration solution. A replicate of a non-zero concentration of AH or AvFc and the blank were injected in each experiment for double referencing, thus verifying the reliability of the immobilized chip throughout the experiment. The data were assessed by steady-state binding analysis.
Virus Capture Assay
3-fold serial dilutions of either AH, starting at 2.0 μM, and AvFc, starting at 1.0 μM, were coated on a 96-well plate in duplicate for each of three pseudoviruses for 1 h at 37°C. Wells were washed three times with PBS containing 0.05% Tween 20 (PBS-T) and then blocked with DMEM containing 20% fetal bovine serum (FBS) for 1 h at 37°C. Wells were washed three times with water followed by the addition of Q769.h5 (Clade A), SF162 (Clade B), or ZM53M.PB12 (Clade C) pseudoviruses for 1 h at 37°C. Following the capture of pseudoviruses, wells were washed three times with DMEM containing 10% FBS to wash away all unbound virions. Next, DMEM containing 1% Triton X-100 was added to each well to lyse the virions and release the core antigen p24 into the lysate. A p24-detection ELISA (Sino Biological, Beijing, China) was performed by transferring the lysate to an anti-p24-coated 96-well plate. The amount of pseudovirus captured by AH or AvFc was obtained based on a standard p24 starting at 10,000 pg/mL with 2-fold serial dilutions.
Env-Pseudotyped HIV-1 Neutralization Assay
The antiviral activity of AvFc was assessed based on reduction in luciferase reporter gene expression after infection of HOS-CD4+/CCR5+ cells with Env-pseudotyped viruses. The assay was performed in triplicate essentially as described elsewhere.50 Antiviral activity was reflected by IC50, which is the sample concentration yielding 50% of relative luminescence units (RLUs) compared with those of virus control after subtraction of background RLUs. Env-pseudotyped viruses were prepared by co-transfection of HEK293T/17 or HEK293S GnTI− cells (American Type Culture Collection [ATCC]) with various env-expressing plasmids and an env-deficient HIV-1 backbone vector (pNL4-3.Luc.R-.E-) and titrated in HOS-CD4+/CCR5+ cells to determine the optimal viral dilution yielding ∼150,000 RLUs. Samples and virus preparations were incubated for 1 h at 37°C, and 10,000 HOS cells per well were added for 72 h. Luciferase activity was measured using the Luciferase Assay System (Promega).
Primary HIV Inhibition Assay
Fresh PBMCs (ZenBio, Research Triangle Park, NC, USA) were stimulated with phytohemagglutinin (PHA) (Roche, Indianapolis, IN, USA) (5 μg/mL) for 24 h at 37°C in a humid environment containing 5% CO2 in 1:1 (v/v) RPMI 1640 (GIBCO/Invitrogen, Carlsbad, CA, USA) and AIM-V medium, supplemented with 10% FBS (Hyclone), 100 μg/mL streptomycin, 100 U/mL penicillin, L-glutamine, and N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES) (cRPMI). PHA blasts were then grown in cRPMI supplemented with 50 U/mL recombinant human IL-2 (ZeptoMetrix, Buffalo, NY, USA). Samples and virus solutions were incubated for 1 h at 37°C with 5% CO2 prior to the addition of 250,000 cells per well. After 72 h of culture, the media were changed for further 24 h of incubation. Upon addition of lysis buffer (1% Triton X-100), the plates were stored at −20°C. IC50s, expressed as 50% reduction of p24, were determined according to the manufacturer’s protocol (Sino Biological, Beijing, China). The assay was performed in quadruplicate for each virus strain.
SPR (for FcγR Binding)
The binding affinity (KD) of recombinant human Fc gamma receptors (FcγRs), IA or IIIA, to AvFc was measured on a Biacore X100 2.0 instrument at ambient temperature. AvFc was captured on a sensor chip via an anti-human IgG (Fc) antibody of IgG1 isotype, and varying concentrations of each recombinant FcγR were used as analytes. Briefly, monoclonal mouse anti-human IgG (Fc) antibody of IgG1 isotype (25 μg/mL) was immobilized on a CM5 sensor chip (GE Healthcare Biosciences) to 10,000 resonance units (RU) using the Human Antibody Capture Kit (GE Healthcare Biosciences). A reference flow cell was immobilized with the antibody to correct response contributions such as bulk shifts that occur equally in the sample and reference flow cells. AvFc was captured on the anti-human IgG (Fc) chip to a surface density of about 200 RU. In the FcγRI sensorgram in Figure 4A, the raw (colored lines) and fitted (black lines) curves represent FcγRI concentrations (1.2, 0.4, 0.13, 0.044, and 0.015 μg/mL from top to bottom). In the FcγRIIIa sensorgram in Figure 4B, the raw data curves (colored lines) represent FcγRIIIa concentrations (100, 33.3, 11.1, 3.7, 1.2, and 0.41 μg/mL from top to bottom). The analysis was performed in two independent experiments. The average equilibrium dissociation constant, KD (mean ± SEM), was determined based on 1:1 binding kinetics for FcγRI or on steady state for FcγRIIIA.
Flow Cytometry (for FcγR Binding)
FcγR binding by AvFc was demonstrated on a BD FACSAria flow cytometer using both FcγRI (CD64+) and FcγRIIIa (CD16A+) TZM-bl cells (NIH AIDS Reagent Program). Initially, background binding of Avaren to TZM-bl cells was blocked by incubating AvFc with mannan. Then, a 1:1 mixture of blocked AvFc and 1 × 106 FcγR-expressing cells was mixed for 1 h at room temperature, followed by three independent washes with Dulbecco’s PBS (DPBS). AvFc bound to FcγR was detected by incubating the cells with a goat F(ab’)2 anti-human IgG (Fc)-FITC (fluorescein isothiocyanate) secondary antibody. After 1 h, cells were again washed three times with DPBS. Cells were then fixed with 1% paraformaldehyde and analyzed on a flow cytometer. A non-FcγR-binding AvFc mutant was used as a negative control, and human IgGκ was utilized as a positive control. The analysis was performed in triplicate. Percent binding was taken as the percentage of the total viable cell population with a fluorescence intensity greater than a cell-only control.
ADCVI Assay
HIV-192US657 is an R5 primary isolate from the NIH AIDS Reagent Program. The virus was propagated by infecting PHA-stimulated PBMCs from a healthy donor by spinoculation (1,200 × g at room temperature for 2 h). The virus was passaged twice on PHA-stimulated PBMCs, and stocks were stored at −80°C until use. Human PBMCs obtained by Ficoll-Hypaque gradient centrifugation were allowed to adhere to polystyrene flasks for 1 h. Non-adherent cells were collected and stimulated for 24 h with 5 μg/mL PHA-L in cRPMI. CD4+ lymphocytes and NK effector cells were then magnetically separated from PBMCs with anti-CD4 and anti-CD56 monoclonal antibodies (mAbs), respectively (Miltenyi Biotech, Auburn, CA, USA). CD4+ lymphocytes were infected with HIV-192US657 at an MOI of approximately 0.05 for 48–72 h. CD4+ lymphocytes were initially propagated for 3 days in cRPMI supplemented with PHA-L (5 μg/mL) and IL-2 (20 U/mL), followed by maintenance with just IL-2. NK cells were propagated in cRPMI supplemented with IL-2 (20 U/mL). Next, 5 × 104 CD4+-infected lymphocytes and 1 × 105 NK cells, an effector/target cell (E:T) ratio of 2:1, were co-incubated with various concentrations of AvFc and VRC01, respectively, in 96-well plates for 72 h. Supernatants were sampled for p24 detection by ELISA (ZeptoMetrix; Buffalo, NY, USA). Control wells lacking the compound but containing NK cells (effector control, CD4+ lymphocytes + NK cells) and viral replication control wells lacking both the compound and NK cells (CD4+ lymphocytes only) were included in every 96-well plate. The assay was performed in triplicate. Virus inhibition was calculated as follows: % inhibition = {1 − ([p24t]/[p24ut])} × 100, where [p24t] is the p24 concentration in the supernatants of wells containing a given compound, and [p24ut] is the concentration of p24 from wells lacking the compound.
Human PBMC Viability Assay
Cryopreserved human PBMCs from three different donors were purchased from Precision Bioservices (Frederick, MD, USA). Cell viability of PBMCs from three different donors was assessed by flow cytometry after staining with propidium iodide, a dye excluded by live cells. Data were acquired on a BD FACSCalibur (BD Biosciences, San Jose, CA, USA), counting 10,000 events per sample; analyses were carried out with the CellQuest Pro software from BD Biosciences. ConA (10 μg/mL) and PBS were used as controls.
Evaluation of PBMC Activation Markers
Freshly thawed human PBMCs from three different donors (Precision Bioservices) were cultured for 3 days and analyzed by flow cytometry after dual fluorescent staining with mAbs purchased from BD PharMingen (San Diego, CA, USA). Briefly, cell cultures were transferred from plates to 5-mL round-bottom tubes and washed with 5% inactivated FBS in PBS (washing solution). After blocking for 10 min with purified rat anti-mouse CD16/CD32 (Mouse BD Fc Block), the cells were incubated in the dark with FITC-conjugated anti-CD4 mAb in combination with PE-conjugated anti-CD25, anti-CD69, or anti HLA-DR mAb for 30 min on ice. After a final washing step, data were acquired on a BD FACSCalibur (BD Biosciences, San Jose, CA, USA), counting 10,000 events per sample, and analyzed using the BD CellQuest Pro software from BD BioSciences. ConA (10 μg/mL) and PBS were used as controls.
Immunoassays for Cytokine Detection in PBMC Supernatants
PBMCs from 5 different blood donors (Precision Bioservices) were cultured in the presence of 30 or 100 μg/mL AvFc for 3 days, and the concentrations of interleukin (IL)-1α, IL-1b, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-8, IL-10, IL-12p40, IL-12p70, IL-13, IL-15, IL-17, eotaxin, granulocyte-macrophage colony-stimulating factor (GM-CSF or CSF2), granulocyte colony-stimulating factor (G-CSF or CSF3), interferon gamma (IFN-γ), IFN-α2, interferon-γ-inducible protein-10 (IP-10), monocyte chemoattractant protein-1(MCP-1), macrophage inflammatory protein (MIP)-1α, MIP-1β, tumor necrosis factor (TNF)-α, and TNF-b were evaluated in culture supernatants by the Luminex IS100 system (Millipore) using the MILLIPLEX Human Cytokine/Chemokine 26-Plex Kit according to the manufacturer’s instructions. Data were generated with the xPONENT software. Individual ELISAs were used to evaluate the amounts of IL-1β, IL-6, and IL-8 in cell-culture supernatants collected from PBMC cultures after 24 h of incubation in the presence of AvFc (30 or 100 μg/mL) or controls (10 μg/mL ConA or PBS). ELISA Ready-SET-Go! Kits were purchased from eBioscience and used in these experiments following the manufacturer’s instructions. The experiments were repeated for a total of at least two times.
Rat Treatments and Sample Collection
The animals were randomly divided into three groups (n = 4) to receive endotoxin free samples, including AvFc (4 mg/kg), Avaren (4 mg/kg), and an equivalent volume of the formulation buffer (30 mM histidine [pH 7.4], 250 mM sucrose), respectively, administered by tail vein injection. Following treatment, blood samples were collected by tail vein at various time points for serum preparation (centrifugation at 6,000 × g for 5 min). At 14 days post-treatment, the rats were sacrificed, and blood was collected via the posterior vena cava. At termination, potassium-EDTA anticoagulated whole blood was collected for complete blood count.
Mouse Treatments and Sample Collection
To evaluate the effects of repeated AvFc dosing, mice were randomly assigned to three groups (n = 10) and injected subcutaneously with buffer control, 5 mg/kg AvFc, or 20 mg/kg AvFc, twice a week for 5 weeks. Mouse weights were assessed before each injection. The animals were sacrificed on day 35, and blood was collected from the inferior vena cava. Kidneys, livers, spleens, hearts, and lungs were excised, weighed, fixed in 10% buffered formalin for 16 h, and placed in 70% ethanol until use.
Hematology Parameters and Serum Chemistry
Complete blood count was carried out in a blinded manner for mouse samples (n = 10) on a Hemavet 950 system (Drew Scientific) standardized for mouse blood. The following parameters were quantified in potassium-EDTA anticoagulated whole blood: red blood cell count (RBC; 104 cells per milliliter), total and differential leukocyte counts (neutrophils, lymphocytes, monocytes, eosinophils, and basophils were quantitated as 103 cells per milliliter or percent), hemoglobin concentration (HGB; grams per deciliter), hematocrit (HCT; percent), mean corpuscular volume (MCV; femtoliters), mean cell hemoglobin (MCH; picograms), mean cell hemoglobin concentration (MCHC; grams per deciliter), red cell distribution width (RDW; percent), platelet count (PLT; 104 cells per milliliter), and mean platelet volume (MPV; femtoliters). Levels of the following serum chemistry parameters were assessed, with the differences analyzed by two-way ANOVA: serum albumin (ALB), alkaline phosphatase (ALKP), amylase (AMY), alanine aminotransferase (ALT), blood urea nitrogen (BUN), calcium (CA), cholesterol (CHOL), creatinine (CREAT), globulin (GLOB), glucose (GLU), phosphorus (PHOS), total bilirubin (TBIL), and total protein (TP).
Immunoassays for Avaren and AvFc Quantitation in Serum Samples
Avaren and AvFc concentrations were quantitated in serum samples by ELISAs. For AvFc, 96-well plates were coated with 50 μL per well of 0.3 μg/mL gp120 (Protein Sciences, Meriden, CT, USA). After overnight incubation at 4°C, plates were blocked with 5% non-fat dry milk in PBS containing 0.05% Tween 20 (PBS-T). Then, samples diluted at 1:50–1:500 in blocking buffer were added and incubated at 37°C for 1 h. AvFc was detected by mouse anti-human IgG (Fc) conjugated to horseradish peroxidase (HRP) from SouthernBiotech (Birmingham, AL, USA). Plates were developed with SureBlue TMB Microwell Peroxidase Substrate, and reactions were stopped with 1 N H2SO4. Finally, absorbance at 450 nm and at 570 nm was measured on a Bio-Tek Synergy HT plate reader. Avaren detection was carried out by sandwich ELISA using mouse monoclonal anti-Avaren IgG and guinea pig anti-AH serum (produced in our laboratory) as capture and detection antibodies, respectively. Goat anti-guinea pig IgG conjugated to HRP (Santa Cruz Biotechnology, Dallas, TX, USA) was used as secondary antibody, and Avaren quantification was similar to what was previously described for AvFc. Serial dilutions of purified AvFc or Avaren were run in parallel to generate the respective standard curves.
SIV Neutralization Assay
The SIV neutralization assay was carried out in white opaque 96-well plates with TZM-bl cells as host cells. Three strains of SIV (smE660, mac239, and mac251) were propagated in CEMx174 cells for the experiment. Briefly, AvFc was prepared in 1:5 serial dilutions starting from 300 μg/mL (for mac239 and smE660) or 150 μg/mL (for mac251). In addition, wells were left to serve as controls, with some containing no drug, no drug or virus, or just growth medium. The drug and virus were then incubated together at 37°C for 1 h, with the amount of virus being diluted to give an appropriate amount of luciferase signal (at least 10× the background signal). Then, 10,000 TZM-bl cells were added to each well, and plates were further incubated at 37°C for 2 days, at which point cells were lysed and 100 μL luciferase assay reagent was added. Plates were immediately read on a luminescence-enabled plate reader. RLUs were converted to percent infection using the values from the no-drug control wells as 100% infection. These values were then plotted against drug concentration and fit by non-linear regression using the GraphPad Prism software.
Flow Cytometry Analysis of SIV-Infected MLN Cells
Cells, previously isolated from rhesus macaque MLNs and cryopreserved, were infected with SIVmac239 and cultured in RPMI 1640 supplemented with 10% FBS and 40 U/mL of human IL-2 (Tonbo Biosciences, #21-8029). Uninfected cells were also cultured as the control group. Cultures were monitored for p27 expression using a commercial ELISA kit (Sino Biological, KIT11695, Beijing, China). After infection was confirmed by a positive ELISA test, cells were harvested and blocked with a solution of PBS-T containing 3% BSA and human Fc block (BD #564219) for 30 min on ice. Cells were then washed and successively stained with 1 or 10 μg/mL AvFc and goat anti-human IgG-FITC secondary antibodies (Abcam #ab97224). Flow cytometry was performed on the BD FACScan system. Captured cell populations were gated for live lymphocytes, and unstained cells were used to determine the background fluorescence.
Pharmacokinetic Analysis of AvFc in Rhesus Macaques
Two adult female rhesus macaques (RIb15 and RYl15), weighing 5.76 and 5.72 kg, respectively, were administered a 21-mg bolus intravenous injection of AvFc equaling doses of 3.6 and 3.7 mg/kg, respectively. Blood was sampled at 0, 0.5, 1, 2, 4, 8, 24, 48, and 72 h and placed in tubes containing EDTA. Plasma was then separated by centrifugation and assayed for AvFc by a gp120 ELISA, using varying concentrations of purified AvFc spiked into neat plasma (0.064–1,000 ng/mL) as standards to determine the concentration of the drug. The assay lower limit of quantification was 1.93 ng/mL. The plasma concentrations of AvFc were then plotted against time, and PK parameters were estimated with PK Solver.51
Statistical Analyses
Group means and SEMs were derived from the values obtained in three individual replicates, and assays were performed at least twice independently unless otherwise noted. For all data, outliers were determined by statistical analysis using the Grubb’s test (p < 0.05) and excluded from further analysis. Statistical significance was analyzed by one-way ANOVA with Bonferroni’s post-tests or Wilcoxon matched-pairs signed rank test as indicated in the figure legends, using the GraphPad Prism 6 software. Differences were considered statistically significant if p < 0.05.
Data Availability
All data generated or analyzed during the present study are available from the corresponding author on reasonable request. The complete glycan array data (Figure 2B) are available at: http://www.functionalglycomics.org/glycomics/search/jsp/result.jsp?query=avaren&cat=all. The coding sequence of Avaren has been deposited to GenBank: MK503330.
Author Contributions
K.T.H., J.C.K., M.W.D., T.N.G., and N.M. wrote the manuscript. K.T.H., A.S.H., M.W.D., and S.D.H. produced the proteins. K.T.H. performed biochemical and biophysical experiments. A.S.H., T.N.G., M.W.D., and M.K.M. performed antiviral experiments. J.C.K. performed toxicological experiments. K.A.R., F.V., and C.V.H. provided technical support and helped analyze data for antiviral assays. K.T.H., J.C.K., and M.W.D. contributed equally to the work. N.M. conceived and designed experiments, secured funding, and supervised the study.
Conflicts of Interest
A.S.H. and N.M. are co-inventors on a patent related to the findings in this work (U.S. patent number 8802822).
Acknowledgments
We thank ICON Genetics GmbH, Halle (Saale), Germany, for providing the magnICON vector, Kentucky BioProcessing for pilot-scale AvFc production, Sudha Sankaran and Joshua Royal for technical assistance, and Leslie Barnes for N. benthamiana growth and maintenance. AH was kindly provided by Dr. Haruo Tanaka. We acknowledge the participation of the Protein-Glycan Interaction Resource of the CFG and the supporting grant R24 GM098791. This work was supported by an NIH grant (R21/R33 AI088585).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2019.07.021.
Supplemental Information
References
- 1.Chu C.S., Niñonuevo M.R., Clowers B.H., Perkins P.D., An H.J., Yin H., Killeen K., Miyamoto S., Grimm R., Lebrilla C.B. Profile of native N-linked glycan structures from human serum using high performance liquid chromatography on a microfluidic chip and time-of-flight mass spectrometry. Proteomics. 2009;9:1939–1951. doi: 10.1002/pmic.200800249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Molinari M. N-glycan structure dictates extension of protein folding or onset of disposal. Nat. Chem. Biol. 2007;3:313–320. doi: 10.1038/nchembio880. [DOI] [PubMed] [Google Scholar]
- 3.Doores K.J. The HIV glycan shield as a target for broadly neutralizing antibodies. FEBS J. 2015;282:4679–4691. doi: 10.1111/febs.13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behrens A.J., Crispin M. Structural principles controlling HIV envelope glycosylation. Curr. Opin. Struct. Biol. 2017;44:125–133. doi: 10.1016/j.sbi.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ward A.B., Wilson I.A. The HIV-1 envelope glycoprotein structure: nailing down a moving target. Immunol. Rev. 2017;275:21–32. doi: 10.1111/imr.12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharon N. Lectins: carbohydrate-specific reagents and biological recognition molecules. J. Biol. Chem. 2007;282:2753–2764. doi: 10.1074/jbc.X600004200. [DOI] [PubMed] [Google Scholar]
- 7.Ingale A.G., Hivrale A.U. Plant as a plenteous reserve of lectin. Plant Signal. Behav. 2013;8:e26595. doi: 10.4161/psb.26595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dan X., Liu W., Ng T.B. Development and applications of lectins as biological tools in biomedical research. Med. Res. Rev. 2016;36:221–247. doi: 10.1002/med.21363. [DOI] [PubMed] [Google Scholar]
- 9.Barouch D.H., Whitney J.B., Moldt B., Klein F., Oliveira T.Y., Liu J., Stephenson K.E., Chang H.W., Shekhar K., Gupta S. Therapeutic efficacy of potent neutralizing HIV-1-specific monoclonal antibodies in SHIV-infected rhesus monkeys. Nature. 2013;503:224–228. doi: 10.1038/nature12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oliveira C., Teixeira J.A., Domingues L. Recombinant lectins: an array of tailor-made glycan-interaction biosynthetic tools. Crit. Rev. Biotechnol. 2013;33:66–80. doi: 10.3109/07388551.2012.670614. [DOI] [PubMed] [Google Scholar]
- 11.Lam S.K., Ng T.B. Lectins: production and practical applications. Appl. Microbiol. Biotechnol. 2011;89:45–55. doi: 10.1007/s00253-010-2892-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balzarini J. Targeting the glycans of glycoproteins: a novel paradigm for antiviral therapy. Nat. Rev. Microbiol. 2007;5:583–597. doi: 10.1038/nrmicro1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang F., Hoque M.M., Jiang J., Suzuki K., Tsunoda M., Takeda Y., Ito Y., Kawai G., Tanaka H., Takénaka A. The characteristic structure of anti-HIV actinohivin in complex with three HMTG D1 chains of HIV-gp120. Chembiochem. 2014;15:2766–2773. doi: 10.1002/cbic.201402352. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki K., Tsunoda M., Hoque M.M., Zhang F., Jiang J., Zhang X., Ohbayashi N., Tanaka H., Takénaka A. Peculiarity in crystal packing of anti-HIV lectin actinohivin in complex with α(1-2)mannobiose. Acta Crystallogr. D Biol. Crystallogr. 2013;69:1818–1825. doi: 10.1107/S0907444913017812. [DOI] [PubMed] [Google Scholar]
- 15.Matoba N., Husk A.S., Barnett B.W., Pickel M.M., Arntzen C.J., Montefiori D.C., Takahashi A., Tanno K., Omura S., Cao H. HIV-1 neutralization profile and plant-based recombinant expression of actinohivin, an Env glycan-specific lectin devoid of T-cell mitogenic activity. PLoS ONE. 2010;5:e11143. doi: 10.1371/journal.pone.0011143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marillonnet S., Giritch A., Gils M., Kandzia R., Klimyuk V., Gleba Y. In planta engineering of viral RNA replicons: efficient assembly by recombination of DNA modules delivered by Agrobacterium. Proc. Natl. Acad. Sci. USA. 2004;101:6852–6857. doi: 10.1073/pnas.0400149101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arnold K., Bordoli L., Kopp J., Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 18.Czajkowsky D.M., Hu J., Shao Z., Pleass R.J. Fc-fusion proteins: new developments and future perspectives. EMBO Mol. Med. 2012;4:1015–1028. doi: 10.1002/emmm.201201379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu X., Yang Z.Y., Li Y., Hogerkorp C.M., Schief W.R., Seaman M.S., Zhou T., Schmidt S.D., Wu L., Xu L. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scanlan C.N., Pantophlet R., Wormald M.R., Ollmann Saphire E., Stanfield R., Wilson I.A., Katinger H., Dwek R.A., Rudd P.M., Burton D.R. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1-->2 mannose residues on the outer face of gp120. J. Virol. 2002;76:7306–7321. doi: 10.1128/JVI.76.14.7306-7321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scanlan C.N., Pantophlet R., Wormald M.R., Saphire E.O., Calarese D., Stanfield R., Wilson I.A., Katinger H., Dwek R.A., Burton D.R., Rudd P.M. The carbohydrate epitope of the neutralizing anti-HIV-1 antibody 2G12. Adv. Exp. Med. Biol. 2003;535:205–218. doi: 10.1007/978-1-4615-0065-0_13. [DOI] [PubMed] [Google Scholar]
- 22.Seaman M.S., Janes H., Hawkins N., Grandpre L.E., Devoy C., Giri A., Coffey R.T., Harris L., Wood B., Daniels M.G. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J. Virol. 2010;84:1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doores K.J., Bonomelli C., Harvey D.J., Vasiljevic S., Dwek R.A., Burton D.R., Crispin M., Scanlan C.N. Envelope glycans of immunodeficiency virions are almost entirely oligomannose antigens. Proc. Natl. Acad. Sci. USA. 2010;107:13800–13805. doi: 10.1073/pnas.1006498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zwick M.B., Jensen R., Church S., Wang M., Stiegler G., Kunert R., Katinger H., Burton D.R. Anti-human immunodeficiency virus type 1 (HIV-1) antibodies 2F5 and 4E10 require surprisingly few crucial residues in the membrane-proximal external region of glycoprotein gp41 to neutralize HIV-1. J. Virol. 2005;79:1252–1261. doi: 10.1128/JVI.79.2.1252-1261.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saxena A., Wu D. Advances in therapeutic Fc engineering – Modulation of IgG-associated effector functions and serum half-life. Front. Immunol. 2016;7:580. doi: 10.3389/fimmu.2016.00580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jennewein M.F., Alter G. The immunoregulatory roles of antibody glycosylation. Trends Immunol. 2017;38:358–372. doi: 10.1016/j.it.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 27.Forthal D.N., Landucci G., Cole K.S., Marthas M., Becerra J.C., Van Rompay K. Rhesus macaque polyclonal and monoclonal antibodies inhibit simian immunodeficiency virus in the presence of human or autologous rhesus effector cells. J. Virol. 2006;80:9217–9225. doi: 10.1128/JVI.02746-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smalls-Mantey A., Doria-Rose N., Klein R., Patamawenu A., Migueles S.A., Ko S.Y., Hallahan C.W., Wong H., Liu B., You L. Antibody-dependent cellular cytotoxicity against primary HIV-infected CD4+ T cells is directly associated with the magnitude of surface IgG binding. J. Virol. 2012;86:8672–8680. doi: 10.1128/JVI.00287-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosenberg Y., Sack M., Montefiori D., Forthal D., Mao L., Hernandez-Abanto S., Urban L., Landucci G., Fischer R., Jiang X. Rapid high-level production of functional HIV broadly neutralizing monoclonal antibodies in transient plant expression systems. PLoS ONE. 2013;8:e58724. doi: 10.1371/journal.pone.0058724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raghunathan G., Sokalingam S., Soundrarajan N., Madan B., Munussami G., Lee S.G. Modulation of protein stability and aggregation properties by surface charge engineering. Mol. Biosyst. 2013;9:2379–2389. doi: 10.1039/c3mb70068b. [DOI] [PubMed] [Google Scholar]
- 31.Moulaei T., Shenoy S.R., Giomarelli B., Thomas C., McMahon J.B., Dauter Z., O’Keefe B.R., Wlodawer A. Monomerization of viral entry inhibitor griffithsin elucidates the relationship between multivalent binding to carbohydrates and anti-HIV activity. Structure. 2010;18:1104–1115. doi: 10.1016/j.str.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matei E., Zheng A., Furey W., Rose J., Aiken C., Gronenborn A.M. Anti-HIV activity of defective cyanovirin-N mutants is restored by dimerization. J. Biol. Chem. 2010;285:13057–13065. doi: 10.1074/jbc.M109.094938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang M., Gaschen B., Blay W., Foley B., Haigwood N., Kuiken C., Korber B. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology. 2004;14:1229–1246. doi: 10.1093/glycob/cwh106. [DOI] [PubMed] [Google Scholar]
- 34.Webb N.E., Montefiori D.C., Lee B. Dose-response curve slope helps predict therapeutic potency and breadth of HIV broadly neutralizing antibodies. Nat. Commun. 2015;6:8443. doi: 10.1038/ncomms9443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu Q., Mahmood N., Shattock R.J. High-mannose-specific deglycosylation of HIV-1 gp120 induced by resistance to cyanovirin-N and the impact on antibody neutralization. Virology. 2007;368:145–154. doi: 10.1016/j.virol.2007.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alexandre K.B., Moore P.L., Nonyane M., Gray E.S., Ranchobe N., Chakauya E., McMahon J.B., O’Keefe B.R., Chikwamba R., Morris L. Mechanisms of HIV-1 subtype C resistance to GRFT, CV-N and SVN. Virology. 2013;446:66–76. doi: 10.1016/j.virol.2013.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou T., Doria-Rose N.A., Cheng C., Stewart-Jones G.B.E., Chuang G.Y., Chambers M., Druz A., Geng H., McKee K., Kwon Y.D. Quantification of the impact of the HIV-1-glycan shield on antibody elicitation. Cell Rep. 2017;19:719–732. doi: 10.1016/j.celrep.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kong L., Wilson I.A., Kwong P.D. Crystal structure of a fully glycosylated HIV-1 gp120 core reveals a stabilizing role for the glycan at Asn262. Proteins. 2015;83:590–596. doi: 10.1002/prot.24747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mathys L., Balzarini J. Several N-glycans on the HIV envelope glycoprotein gp120 preferentially locate near disulphide bridges and are required for efficient infectivity and virus transmission. PLoS ONE. 2015;10:e0130621. doi: 10.1371/journal.pone.0130621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mathys L., François K.O., Quandte M., Braakman I., Balzarini J. Deletion of the highly conserved N-glycan at Asn260 of HIV-1 gp120 affects folding and lysosomal degradation of gp120, and results in loss of viral infectivity. PLoS ONE. 2014;9:e101181. doi: 10.1371/journal.pone.0101181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bugelski P.J., Treacy G. Predictive power of preclinical studies in animals for the immunogenicity of recombinant therapeutic proteins in humans. Curr. Opin. Mol. Ther. 2004;6:10–16. [PubMed] [Google Scholar]
- 42.van Meer P.J., Kooijman M., Brinks V., Gispen-de Wied C.C., Silva-Lima B., Moors E.H., Schellekens H. Immunogenicity of mAbs in non-human primates during nonclinical safety assessment. MAbs. 2013;5:810–816. doi: 10.4161/mabs.25234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zalevsky J., Chamberlain A.K., Horton H.M., Karki S., Leung I.W., Sproule T.J., Lazar G.A., Roopenian D.C., Desjarlais J.R. Enhanced antibody half-life improves in vivo activity. Nat. Biotechnol. 2010;28:157–159. doi: 10.1038/nbt.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Christiansen M.N., Chik J., Lee L., Anugraham M., Abrahams J.L., Packer N.H. Cell surface protein glycosylation in cancer. Proteomics. 2014;14:525–546. doi: 10.1002/pmic.201300387. [DOI] [PubMed] [Google Scholar]
- 45.Newsom-Davis T.E., Wang D., Steinman L., Chen P.F., Wang L.X., Simon A.K., Screaton G.R. Enhanced immune recognition of cryptic glycan markers in human tumors. Cancer Res. 2009;69:2018–2025. doi: 10.1158/0008-5472.CAN-08-3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu X., Nie H., Zhang Y., Yao Y., Maitikabili A., Qu Y., Shi S., Chen C., Li Y. Cell surface-specific N-glycan profiling in breast cancer. PLoS ONE. 2013;8:e72704. doi: 10.1371/journal.pone.0072704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.An H.J., Gip P., Kim J., Wu S., Park K.W., McVaugh C.T., Schaffer D.V., Bertozzi C.R., Lebrilla C.B. Extensive determination of glycan heterogeneity reveals an unusual abundance of high mannose glycans in enriched plasma membranes of human embryonic stem cells. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.M111.010660. M111. 010660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Montacir H., Freyer N., Knöspel F., Urbaniak T., Dedova T., Berger M., Damm G., Tauber R., Zeilinger K., Blanchard V. The cell-surface N-glycome of human embryonic stem cells and differentiated hepatic cells thereof. ChemBioChem. 2017;18:1234–1241. doi: 10.1002/cbic.201700001. [DOI] [PubMed] [Google Scholar]
- 49.Gleba Y., Klimyuk V., Marillonnet S. Magnifection--a new platform for expressing recombinant vaccines in plants. Vaccine. 2005;23:2042–2048. doi: 10.1016/j.vaccine.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 50.Hamorsky K.T., Grooms-Williams T.W., Husk A.S., Bennett L.J., Palmer K.E., Matoba N. Efficient single tobamoviral vector-based bioproduction of broadly neutralizing anti-HIV-1 monoclonal antibody VRC01 in Nicotiana benthamiana plants and utility of VRC01 in combination microbicides. Antimicrob. Agents Chemother. 2013;57:2076–2086. doi: 10.1128/AAC.02588-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y., Huo M., Zhou J., Xie S. PKSolver: an add-in program for pharmacokinetic and pharmacodynamic data analysis in Microsoft Excel. Comput. Methods Programs Biomed. 2010;99:306–314. doi: 10.1016/j.cmpb.2010.01.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during the present study are available from the corresponding author on reasonable request. The complete glycan array data (Figure 2B) are available at: http://www.functionalglycomics.org/glycomics/search/jsp/result.jsp?query=avaren&cat=all. The coding sequence of Avaren has been deposited to GenBank: MK503330.