Abstract
Transposable elements represent the largest components of many eukaryotic genomes and different genomes harbor different combinations of elements. Here, we discovered a novel DNA transposon in the genome of the clubmoss Selaginella lepidophylla. Further searching for related sequences to the conserved DDE region uncovered the presence of this superfamily of elements in fish, coral, sea anemone, and other animal species. However, this element appears restricted to Bryophytes and Lycophytes in plants. This transposon, named GingerRoot, is associated with a 6 bp (base pair) target site duplication, and 100–150 bp terminal inverted repeats. Analysis of transposase sequences identified the DDE motif, a catalytic domain, which shows similarity to the integrase of Gypsy-like long terminal repeat retrotransposons, the most abundant component in plant genomes. A total of 77 intact and several hundred truncated copies of GingerRoot elements were identified in S. lepidophylla. Like Gypsy retrotransposons, GingerRoots show a lack of insertion preference near genes, which contrasts to the compact genome size of about 100 Mb. Nevertheless, a considerable portion of GingerRoot elements was found to carry gene fragments, suggesting the capacity of duplicating gene sequences is unlikely attributed to the proximity to genes. Elements carrying gene fragments appear to be less methylated, more diverged, and more distal to genes than those without gene fragments, indicating they are preferentially retained in gene-poor regions. This study has identified a broadly dispersed, novel DNA transposon, and the first plant DNA transposon with an integrase-related transposase, suggesting the possibility of de novo formation of Gypsy-like elements in plants.
Keywords: GingerRoot, transposon, integrase, Selaginella lepidophylla, gene duplication
Introduction
Transposable elements (TEs), also called transposons, are DNA sequences that move within or among genomes. Transposons were first discovered by Barbara McClintock, who labeled them “controlling elements” (McClintock 1956). Transposons are widely dispersed and vary in structure, transposition mechanism, genomic location, and copy numbers in nearly all genomes studied to date. Due to these factors, they are associated with diverse evolutionary histories. Transposons mobilize in a genome through transposition, which entails either a DNA or RNA mechanism. This mechanism is used to organize TEs into two classes where Class I utilizes a transcribed RNA intermediate that is reverse transcribed into DNA before insertion into the genome (Lisch 2013). In plants, the most abundant Class I transposons are long terminal repeat (LTR) retrotransposons. In addition to reverse transcriptase, transposition of LTR elements requires integrase, which is responsible for the insertion of the elements into the genome. Class II DNA elements transpose via a DNA mechanism and are excised from their original locus and inserted at another site in the genome. Some DNA elements contain an open reading frame encoding a transposase (Kidwell and Lisch 2000). This enzyme recognizes the DNA element at the terminal inverted repeat (TIR), and excises and inserts the element at another locus. This endonuclease activity produces staggered cuts on the DNA strands which are repaired using the DNA repair mechanism (Craig 2002). The sequence duplications caused by staggered cuts followed by the repair are termed the target site duplication, or TSD, for their distinct pattern of nucleotides that are found flanking the terminal sequences of elements (Craig 2002).
In addition to the transposition mechanism, transposons are classified by their ability to transpose. A nonfunctional transposase or lack of transposase will prevent an element from mobilizing by itself (Craig 2002). These elements that do not encode the necessary transposase, in the case of DNA elements, or other proteins, such as reverse transcriptase or integrase for RNA elements, are termed nonautonomous as they are unable to self-catalyze transposition. Conversely, elements encoding functional proteins required for transposition are called autonomous elements. Transposases from DNA elements contain an open reading frame with several domains, of which the DDE domain is responsible for catalyzing the DNA strand cleavage (Kidwell and Lisch 2000). This DDE motif is critical for transposase function and can classify DNA elements into superfamilies (Yuan and Wessler 2011). In addition to DNA transposons, the integrase of many LTR retrotransposons and retroviruses also contains a DDE motif (Capy et al. 1997).
TEs vary in number and structure and are one of the primary drivers of genome size variation, along with polyploidy, by increasing in copy number. Many plant genomes comprised 104 copies of LTR retrotransposons (Bennetzen et al. 2005), whereas other genomes such as yeast have minimal transposons (Kidwell 2002). Differences in genomic regulation, such as histone marks, DNA methylation, and DNA maintenance mechanisms are thought to contribute to transposon abundance (Baniaga et al. 2016).
To date, seven superfamilies of DNA transposons have been identified in plants. Barbara McClintock discovered the first superfamily of DNA transposons, Ac/Ds (Ac/Ds/hAT/DTA), in 1950 (McClintock 1950). Like Ac/Ds, most other plant transposons were initially identified in early genetic and genomic studies focused on model plants such as maize, rice, and Arabidopsis. Those include En/Spm/CACTA/DTC, Mutator/MULE/DTM, PIF/Harbinger/Tourist/DTH, Tc1/Mariner/Stowaway/DTT, and Helitron/DHH (Wicker et al. 2007). Ten years ago, the Sola superfamily was identified in Hydra magnipapillata and other species including those of plants (Bao et al. 2009). With the exception of Helitron, all of plant DNA transposons are associated with TIR. Class II DNA transposons are generally found in lower copy numbers than Class I elements due to their transposition mechanism which does not necessarily increase copy number when active, leading to reduced proliferation in the genome compared with Class I RNA-based elements (Bennetzen 2000). In total, these differences contribute to distinct life histories and evolutionary strategies resulting in differences in element biology (Lisch and Slotkin 2011). Also, there are levels of evidence of genomic contraction or DNA removal for genome size control (Devos et al. 2002; Bennetzen et al. 2005; Brookfield and Johnson 2006; Hawkins et al. 2009). Because different genomes are associated with a variable level of DNA removal and epigenetic control, a unique combination of TEs is present in even closely related genomes.
The genus Selaginella contains over 600 species that have been dated back to a most recent common ancestor in the Pteridophytes, 400 Ma, which has since led to significant diversification (Banks 2009). In this study, we identified GingerRoot, a novel DNA transposon in the lycophyte Selaginellalepidophylla and related elements in additional species. As S. lepidophylla is phylogenetically basal to both Monilophytes and Spermatophytes, the identification of this new superfamily of transposons allows for comparative studies of repetitive sequences in vascular non-seed plants (Banks et al. 2011).
Materials and Methods
Element Identification
The GingerRoot elements in S. lepidophylla were identified in a genome-wide characterization of TEs in S. lepidophylla (VanBuren et al. 2018). Briefly, nonautonomous DNA transposons were identified by MITE-Hunter (Han and Wessler 2010). Candidate long terminal repeat retrotransposons (LTR-RTs) were identified using LTR_retriever (Ou and Jiang 2018). The remainders of repetitive sequences were collected using RepeatModeler (http://www.repeatmasker.org/RepeatModeler/; last accessed March 2019). The repeats collected by RepeatModeler were then categorized into two groups: sequences with and without identities. Those without identities were searched against the known transposase database and if they had a match, they were considered as the relevant transposons. For completely unknown repetitive sequences, manual curation was conducted to determine their identity and 5′ and 3′ boundaries. This was done in a stepwise process. First, the relevant sequences were initially used to search the genome and retrieve at least ten hits (BlastN, expect value <10−10) with the corresponding 100 bp (base pair) (or longer if necessary) of 5′ and 3′ flanking sequences. Second, recovered sequences were then aligned using DIALIGN2 (Morgenstern 1999), to determine the possible boundary between elements and their flanking sequences. In this case, a boundary was defined as the position to which sequence homology is conserved over more than half of the aligned sequences. Finally, sequences with defined boundaries were examined for the presence of TSD. To classify the relevant TEs, features in the terminal ends and TSD were used. Each transposon family is associated with distinct features in their terminal sequences and TSDs which can be identified (Wicker et al. 2007).
All GingerRoot-related sequences in the repeat library were manually curated and used to mask the genomic sequences of S. lepidophylla. The output of RepeatMasker was used to identify intact and truncated elements. Criteria for intact elements are: 1) TIRs should belong to the same element family; 2) TIRs should be in inverted orientation with terminal sequences outwards (as that in all DNA elements with TIR); 3) A 6–8 bp (mostly 6 bp) TSD was present immediately adjacent to the TIR; 4) The distance between the TIRs was not over 30 kb. TIRs that do not fall into the above criteria were considered to belong to truncated elements.
Distance between Elements and Adjacent Genes
The gene annotation information, including the transcripts and protein sequences and position of genes (gff files) in S. lepidophylla, was obtained from a previous study (VanBuren et al. 2018). If 50% or more of the transcript of a gene is masked by annotated TEs in S. lepidophylla or the protein sequence match a transposase (expect value <10−10), the gene is excluded from further consideration. Thereafter the distance between GingerRoot elements and the adjacent genes were calculated based on their positions. In each case, the nearest gene was not always present due to scaffold breaks before an annotated gene, whereby these cases were excluded from further analysis. In several cases the nearest annotated gene was marked unknown, these sequences were searched against the National Center for Biotechnology Information (NCBI) nucleotide database for the best matches. If the best hit was either unknown proteins or had similarity to a TE, these sequences were removed from further consideration.
Divergence Estimation of GingerRoot Elements in S. lepidophylla
The divergence of GingerRoots was performed by performing an All versus All BlastN search (Altschul et al. 1990). The resulting paired best match elements were used to compute their identity as a proxy for age as a molecular clock for the genome was unavailable. These values were then plotted in R using the beeswarm package.
DDE Motif Identification
The annotated protein sequences overlapping with intact GingerRoot elements were aligned using MUSCLE (Edgar 2004). The conserved regions were examined and putative DDE domains were deduced. The position of the DDE domain was further verified by alignment with similar proteins from other species, where the DDE residues were the only conserved residues. The amino acid sequences in the DDE regions were used for phylogenetic analysis.
Phylogeny
The amino acid sequences of the DDE motif of identified elements were aligned in MEGA (Tamura et al. 2011) using MUSCLE (Edgar 2004), together with published DDE sequences in (Yuan and Wessler 2011), using default settings. This alignment was then utilized in UGENE to create a Maximum likelihood phylogeny using JTT model + gamma + invariant distribution with 1,000 bootstraps (Okonechnikov et al. 2012). All the DDE motif sequences used in the phylogeny analysis are listed in supplementary file S1, Supplementary Material online.
Identification of Gene Fragments within GingerRoot Elements
To search for gene fragments within elements, the sequences of intact GingerRoot and Mutator-like elements (MULEs) were searched (BlastN, expect value <10−10) against the genomic sequence of genes from (VanBuren et al. 2018), with TEs removed as described above. If an element matches multiple genes, the one with the highest overall score would be considered as the parental gene. To test whether there is a bias toward a different portion of genes; the resulting fragments were then compared with the gene using a normalized length of the genomic sequences of the parental gene to create ten bins. The acquired fragments were called into each bin present. This was done for each acquisition identified. These calls were summed and plotted as frequency per bin. A χ2 goodness of fit test was done to test whether observed acquired fragments differed from a random event.
Identification of GingerRoot-Like Elements in Other Species Using Sequence Read Archive (SRA) Data Sets
The NCBI SRA database (Sayers et al. 2019) was searched using TBlastN, with a GingerRoot DDE sequence as the query. Default settings were used except for an expect value <0.1. The sequence used was from Sl-GR-005:
“KLSMYGIRIHGAIDASSHCVVYMVLAMDKRATTIYRAFSAATALFGRPRRVRSDCAVEHELVAQDMERHWPNAPKPPFITGSSTHNQKIEAFWRHLYEKVVWYYKETLWRMCDSG.” Matches were sorted by classification, copy number, and expect value.
Expression of GingerRoot Elements in S. lepidophylla
RNA-seq data set from S. lepidophylla was generated and processed in a previous study (VanBuren et al. 2018). The reads were pseudo-aligned and quantified for each TE sequence using Kallisto (Bray et al. 2016). These were then visualized using xQUARTZ tablet software. The resulting transcript per million data were converted to FPKM by using the formula, FPKM = (TPM_i × (sum_j FPKM_j)) × 10−6. The top ten elements with highest average FPKM values were then plotted using excel.
Methylation Level Quantification
The Bisulfite sequencing results were adapted from VanBuren et al. (2018), where the methylation level of each “C” in the three contexts (CpG, CHG, and CHH) was generated. We extracted data corresponding to GingerRoot, MULEs, parental genes, and gene fragments. The total average of methylation level was used for further analysis.
Results
A Novel Element Family from the Clubmoss S. lepidophylla
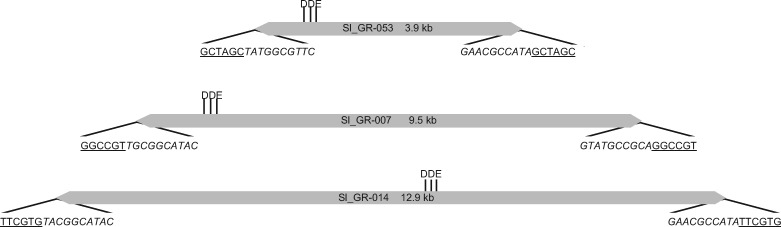
The genome of S. lepidophylla is one of the few sequenced genomes from non-seed land plants (VanBuren et al. 2018). Despite its small genome size (109 Mb), ∼25% of the genome is contributed by recognizable TEs (VanBuren et al. 2018). In this study, we further characterized the elements in this genome building from previous work (VanBuren et al. 2018). In addition to element families that were previously characterized in plants, a novel element family was identified, named GingerRoot. One prominent feature of the GingerRoot elements is the presence of a 6 bp TSD (fig. 1), which is relatively rare among known TEs. Those include some LTR elements (most of LTR elements are associated with a 5 bp TSD), Maverick/Polinton elements, and Ginger2/TDD DNA transposons (Marschalek et al. 1989; Kapitonov and Jurka 2006; Pritham et al. 2007; Wicker et al. 2007; Bao et al. 2010). In the S. lepidophylla genome, a total of 77 intact GingerRoots were identified. The majority (72 out of 77) of elements have terminal sequences of TA (5′-TA…TA-3′). The remaining five have an altered second nucleotide resulting in terminal sequences of 5′-TG…CA-3′ (fig. 1). Both types of motifs resemble those for LTR retrotransposons except TG…CA is more prevalent for LTR elements (Ou and Jiang 2018). The TIRs range in length from 100 to 150 bp and elements vary in size from 3,931 to 12,903 bp (supplementary table S1, Supplementary Material online). In addition, 287 GingerRoot elements with a single TIR sequence were identified in the S. lepidophylla genome (supplementary table S2, Supplementary Material online). These unpaired TIRs may have been derived from truncated elements (where one TIR sequence is deleted), elements interrupted by other elements, elements with mutated TSD, with mutated TIRs. In this case, it is possible either one TIR represents one element or two individual unpaired TIRs belong to a single element. As a result, the estimated total copy number of GingerRoot ranges from 221 to 364. Overall, the sequences from the intact and truncated elements account for 2.1 Mb, contributing to 1.7% of the total genome size.
Fig. 1.
—Element schematic showing GingerRoot elements in S. lepidophylla. DDE motif region is noted; TIRs are depicted as gray triangles; the flanking underlined sequences represent the TSD; and sequences in italics are the most terminal nucleotides of the TIR. Element sizes are not to scale.
Among the intact elements, the nucleotide level pairwise identity of the 77 elements ranges from 99% to outside methodological cutoffs. Because each individual insertion is derived from its immediate ancestral copy, an approximate distribution of elements over time can be estimated through the highest pairwise similarity of elements in the genome. Using an All versus All match the identity of the elements varied from 92.18% to 99.96% identical (supplementary fig. S1, Supplementary Material online). The distribution of elements shows the presence of many recent elements and few older elements with an average identity of 98.35%, following evolutionary dynamics that are seen in other transposons (Kronmiller and Wise 2008). Particularly, there are no two elements that are identical, suggesting a lack of current or extremely recent transposition activity.
GingerRoot Belongs to a New Superfamily of DNA Transposons
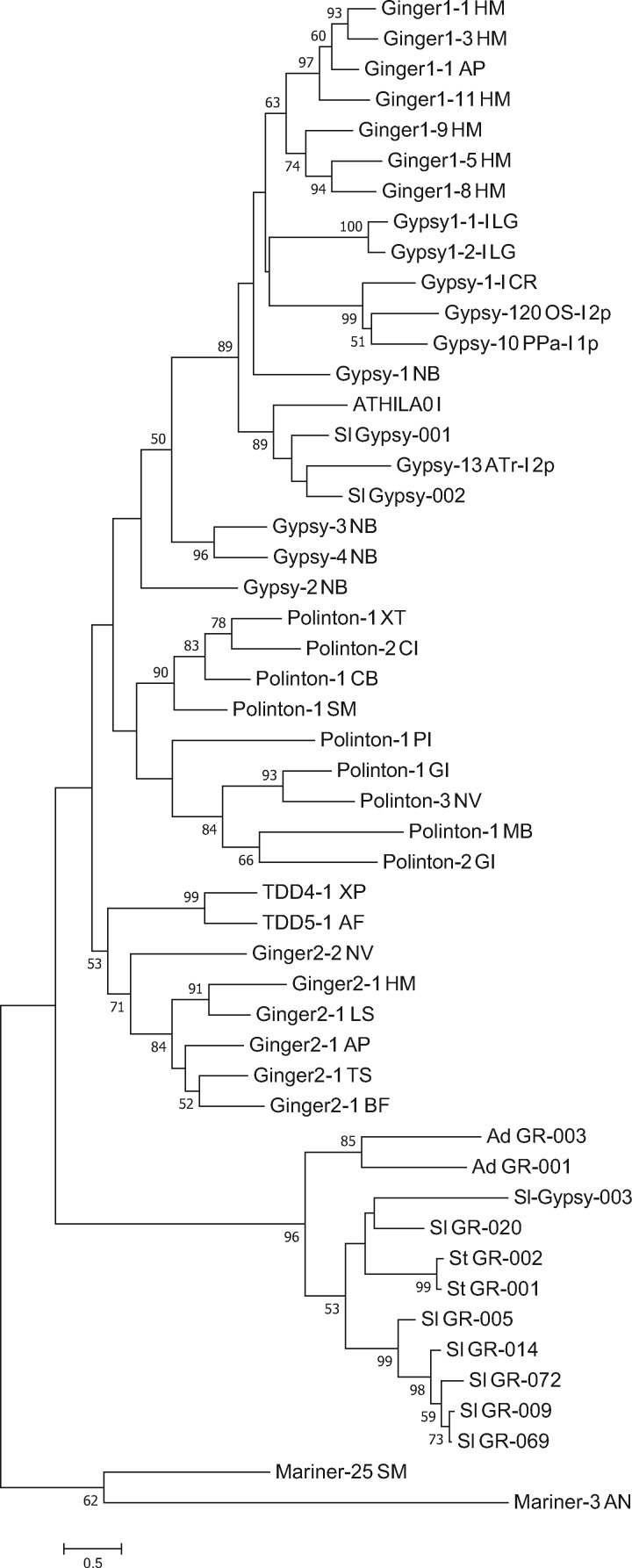
As aforementioned, autonomous DNA elements encode transposase proteins, containing the catalytic domain of three conserved amino acid residues, Aspartic acid (D), Aspartic acid (D), and Glutamic acid (E), referred to as the DDE motif (Henikoff 1992; Doak et al. 1994). After searching and extracting the DDE motif and flanking sequences, 54 out of the 77 intact elements were found to contain DDE motifs. These sequences were compared with a published DDE TE sequence data set (Yuan and Wessler 2011). Using the GingerRoot DDE motif as a query to search against elements from Repbase and elements described in Yuan and Wessler (2011), the best match retrieved was Ginger element Ginger2-1_HM (e value= 5 × 10−16) (Bao et al. 2010, 2015; Yuan and Wessler 2011). As the new elements identified in S. lepidophylla appeared to be closest to Ginger elements, they were named GingerRoot elements. Subsequently, the DDE motifs from GingerRoot were compared with a set of known TEs. As shown in figure 2, this phylogeny illustrates that GingerRoots from several species share a more recent common ancestor to one of the S. lepidophylla Gypsy-like LTR retrotransposons than other DNA elements including Ginger elements. In addition, GingerRoot elements form a single monophyletic clade, supporting a new superfamily classification for GingerRoot.
Fig. 2.
—The phylogeny of DDE motif of GingerRoots from S. lepidophylla and other organisms as well as other DDE motifs from Repbase using Maximum likelihood method. Model JTT + Gamma model + Invariant, of 1,000 bootstrap replicates. Numbers are % bootstrap support. AD, Acropora digitifera; AN, Aspergillus nidulans; AP, Acyrthosiphon pisum; AT, Amborella trichopoda; BF, Branchiostoma floridae; CB, Caenorhabditis briggsae; CI, Ciona intestinalis; CR, Chlamydomonas reinhardtii; DD, Dictyostelium discoideum; GR, GingerRoot; HM, Hydra magnipapillata; LG, Lottia gigantea; LS, Littorina saxatilis; MB, Monosiga brevicollis; NB, Nosema bombycis; NV, Nematostella vectensis; OS, Oryza sativa; PI, Phytophthora infestans; PP, Physcomitrella patens; RI, Rhizophagus intraradices; SL, Selaginella lepidophylla; SM, Schmidtea mediterranea; ST, Selaginella tamariscina; TS, Trichinella spiralis; XT, Xenopus tropicalis.
Because the transposase of both Ginger elements and Maverick/Polinton elements is similar to Gypsy-like integrase (Kapitonov and Jurka 2006; Pritham et al. 2007; Bao et al. 2010), the GingerRoot elements were further compared with these two superfamilies of elements. For Gypsy elements, we identified the YPYY, HHCC, and GPY motifs in Gypsy-like elements (Ebina et al. 2008), from S. lepidophylla for GingerRoot comparisons. These motifs flank the DDE domain in two identified Gypsy elements in S. lepidophylla and have been reported to be present in Ginger1 elements (Bao et al. 2010). However, none of these motifs were found flanking the DDEs identified in GingerRoots. Further using Polinton-related proteins from the Repbase database, a BLAST search and manual alignment of DDE flanking regions resulted in no apparent conserved motifs in GingerRoot elements, suggesting GingerRoot elements are not close relatives of Maverick/Polinton elements.
Gene Fragments inside GingerRoot
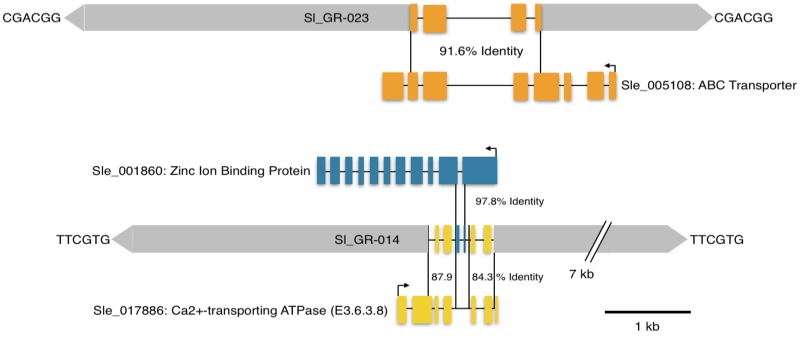
As many TEs are capable of duplicating genes or gene fragments (Cerbin and Jiang 2018), a query was made to see if the GingerRoot elements have the same ability. A search was performed using a gene data set from the S. lepidophylla genome excluding TE matches in S. lepidophylla as well as Repbase TEs. This approach identified 22 GingerRoot elements (out of 77, 28.6%) containing gene fragments (supplementary table S3, Supplementary Material online). These 22 GingerRoot elements contain fragments from a total of ten genes. Compared with their parental genes, these acquired regions contain both introns and exons (fig. 3), suggesting that the gene acquisition occurred at the DNA level, not the RNA or cDNA level. The size of the acquired gene sequences varies from 84 to 1,654 bp (supplementary table S3, Supplementary Material online). All of the acquired gene sequences represent gene fragments, not entire genes, and several GingerRoots have acquired fragments from two different genes. The sequence identity between the acquired region and their parental gene ranges from 79.2% to 97.8% (average of 88.7%), suggesting the acquisition/duplication activity has occurred over a wide range of time. GingerRoot elements with acquired gene fragments had significantly differing pairwise identities, suggesting distinct ages of the elements, compared with elements without gene fragments (97.35 vs. 98.65%, respectively), (t-test, P = 0.014) with a similar average length (8.57 vs. 8.88 kb, respectively) (t-test, P = 0.632). The GC content of the acquired regions is 48.3% GC, which is slightly lower than the overall gene GC content of 50.1%. GingerRoots upon acquiring a segment from a parental gene showed no preference for the 5′ or 3′ region (supplementary fig. S2, Supplementary Material online). Genes in monocots often demonstrate a negative GC-gradient, that is, the GC content at the 5′ end is higher than that at the 3′ end (Clément et al. 2015). Genes in S. lepidophylla vary dramatically in terms of GC content, ranging from 26% to 78%. However, GC gradient is largely absent from the genes in S. lepidophylla (supplementary fig. S3, Supplementary Material online). In addition, only 6 out of the 22 (27.3%) GingerRoot elements with gene fragments are associated with identifiable DDE motif, whereas 49 out of the 55 (89.1%) remaining GingerRoot elements are associated with DDE motif, suggesting that the acquisition of gene fragments is often accompanied by the loss of transposase coding regions.
Fig. 3.
—Acquired gene fragments in GingerRoot elements. Scale schematic of GingerRoots (gray) and the acquired gene fragments (orange, blue, and yellow). Acquired gene fragments are labeled by their annotation and gene identifier. Sequences flanking the elements are the TSD, and the numbers with percentages are the % identity of the acquired fragment to their parental genes.
Among the TIR elements, MULEs are well known for their capability to duplicate host genes (Cerbin and Jiang 2018). MULEs that carry genes or gene fragments are called Pack-MULEs. For comparison, one MULE family in S. lepidophylla was also searched for the presence of genic sequences within the elements. This family of MULEs totals 57 elements with 20 (35.1%) of them containing gene fragments and are therefore Pack-MULEs (supplementary table S4, Supplementary Material online). As a result, the fraction of elements with gene fragments in this family of MULEs is close to that in GingerRoot elements. The nucleotide sequence identity between acquired fragments and the parental genes range from 93.0% to 99.0%, suggesting the observed acquisition events for Pack-MULEs occurred in a much more narrow and recent time frame than that for GingerRoot elements. Like the GingerRoot elements, Pack-MULEs in S. lepidophylla do not demonstrate significant bias in terms of the position of acquired regions in parental genes and the GC content (average value 49.0%) of the acquired fragments (supplemental fig. S2, Supplementary Material online). In contrast to GingerRoot elements, Pack-MULEs and other MULEs have similar pairwise identities (table 1). However, Pack-MULEs are longer than other MULEs without gene fragments (8.81 vs. 5.43 kb, t-test, P = 0.003).
Table 1.
Comparison of GingerRoots and a Family of MULEs in S. lepidophylla
|
GingerRoot
|
MULE |
|||
|---|---|---|---|---|
| With Gene Fragments | Without Gene Fragments | With Gene Fragments (Pack-MULEs) | Without Gene Fragments | |
| Number of elements | 22 | 55 | 20 | 37 |
| Average size (kb) | 8.57 | 8.88 | 8.81 | 5.43 |
| Average pairwise identity (%) | 97.35 | 98.65 | 97.34 | 97.39 |
| Identity to parental gene (%) | 79.2–97.8 | N/A | 93.0–99.0 | N/A |
Target Specificity
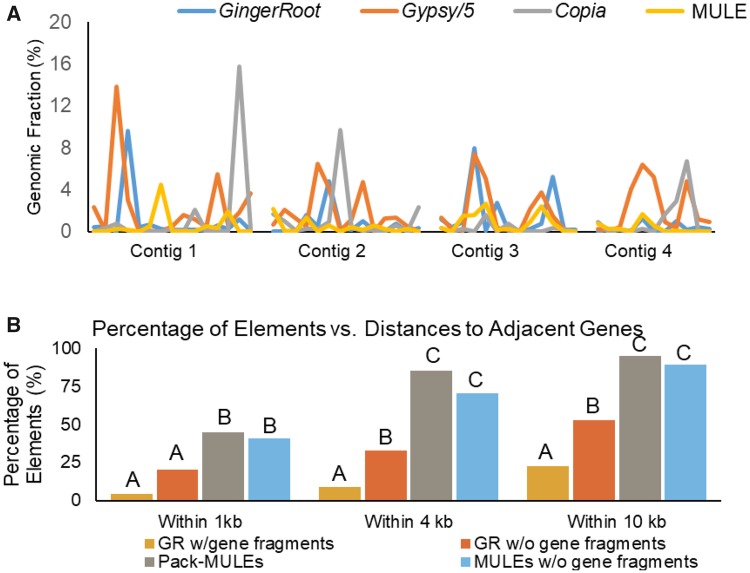
To compare the distribution of GingerRoot and other TEs in the genome, the distribution of each type of TEs was calculated in a 100 kb window. In figure 4A, the distribution of Gypsy, Copia-like retrotransposons, as well as that of GingerRoots and MULEs, was shown in the four longest contigs of the assembly. It appears that the GingerRoot elements (blue) are largely co-localized with Gypsy-like retrotransposons (orange), suggesting they have similar target specificity. In contrast, there is less overlap between GingerRoot elements and Copia-like elements (gray) or MULEs (yellow). In plants and fungi, many Gypsy-like LTR retrotransposons harbor a chromodomain at the C-terminus of the integrase. Those elements with chromodomains are more likely located in gene-poor heterochromatic regions than other LTR retrotransposons (Gao et al. 2008). When the C-terminus of transposase from the GingerRoot elements was examined, no previously described chromodomains were identified.
Fig. 4.
—The target specificity of GingerRoot elements in S. lepidophylla. (A) The distribution of GingerRoot, Gypsy, Copia elements, and MULEs in the four longest contigs of the S. lepidophylla assembly. The x axis represents the physical distance on the contig and the y axis represents the genome fraction of each type of elements. The bin size is 100 kb. Because Gypsy elements are much more abundant than other elements, its fraction value was divided by five to enable the visibility of other elements. (B) The distance of GingerRoot elements and MULEs to their adjacent genes. Distance categories, 0–1, 0–4, and 0–10 kb, on the x axis and percentage of elements in these categories on the y axis. Within distance categories difference letters represent significantly different percentages, χ2 comparison test: P < 0.05.
Because most of the previously characterized DNA transposons preferentially insert into genic regions, the nearest annotated genes were identified to elucidate the genomic context of GingerRoots and compared with that of MULEs. As shown in figure 4B, over 40% of MULEs are within 1 kb to a non-TE gene, and the majority (77%) are within 4 kb region of a gene. In contrast, only about 25% of the GingerRoot elements are within 4 kb of a gene, and over half of them located more than 10 kb away from a gene. The fraction of MULEs and GingerRoots is significantly different in all distance groups (χ2 comparison test; P < 0.01) (fig. 4B), showing GingerRoots are more distal to adjacent genes than MULEs in the genome. The average distance between GingerRoot elements and genes are farther apart than the average gene density of 4.0 kb/gene (VanBuren et al. 2018), supporting the preference for insertion in gene-poor regions of the genome.
GingerRoot elements carrying gene fragments are both older elements (see above) and farther away from genes than those without carrying gene fragments (fig. 4B). In contrast, Pack-MULEs seem to be more enriched in genic regions than their counterparts without carrying gene fragments, but the difference is not significant (fig. 4B).
GingerRoot is associated with a 6 bp TSD but there are a few exceptions including five out of the 77 elements having a 7 bp TSD and one element is associated with an 8 bp TSD. From the 72 intact elements with 6 bp TSD, the nucleotide preference was not apparent (supplementary fig. S4, Supplementary Material online). However, it was noticed that the TSD sequences were much more GC rich than the genomic average (61.2% vs. 49.6%, P < 10−5, χ2 test). As a result, the target specificity is not nucleotide specific but shows a GC-rich sequence preference.
Methylation Status of GingerRoots
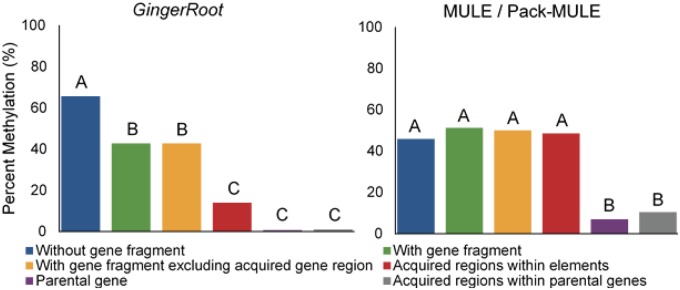
Using previously generated methylation data we interrogated CpG, CHG, and CHH methylation states (VanBuren et al. 2018). The level of methylation at different contexts (CpG, CHG, CHH) varies, yet the trend is similar among contexts, therefore we use the methylation at CpG context as an example (see supplementary fig. S5, Supplementary Material online for CHG and CHH). For GingerRoots without gene fragments, the average methylation level is 65.6%, which is similar to that for intact LTR retrotransposons (VanBuren et al. 2018). For GingerRoot elements with gene fragments, the average methylation level (42.8%) is significantly less than that for GingerRoots without gene fragments (Mann–Whitney test; P = 7.8 × 10−3). The acquired region is associated with an even lower methylation level (14.0%) than other regions in GingerRoots. However, the overall low methylation level of elements with gene fragments is not fully attributed to the acquired region, because the methylation level of non-acquired region in GingerRoots with gene fragments is still significantly lower than those without gene fragments (fig. 5). This suggests that the presence of gene fragments may result in reduced methylation levels in the surrounding regions in GingerRoots. Compared with the parental genes, the acquired regions in GingerRoots are more methylated but the difference is not significant due to wide variation among elements. In contrast to GingerRoot elements, the overall methylation level of MULEs is in between the GingerRoot elements with and without gene fragments. Surprisingly, there is no significant variation of methylation level among MULE elements with and without gene fragments (Mann–Whitney test; P = 0.333). Moreover, the methylation status of acquired regions inside the MULE elements is similar to the non-acquired regions of the elements and they are significantly more methylated than the corresponding regions inside the parental genes (fig. 5).
Fig. 5.
—CpG Methylation for GingerRoot and MULE/Pack-MULE elements. y Axis is the average % Methylation. x Axis is categories with and without a gene fragment, elements with gene fragments excluding the acquired gene region, parental genes, and the acquired regions within parental genes. Statistics are Mann–Whitney tests with different letters signifying significantly different means at P < 0.05.
Dearth of Expression of GingerRoot Elements in S. lepidophylla
To test whether the GingerRoot elements are expressed, we analyzed the RNA-seq reads over a time course experiment of dehydration to rehydration previously reported in VanBuren et al. (2018). RNA-seq reads were mapped to GingerRoots elements using Kallisto (Bray et al. 2016), adjusted for element length and excluding reads mapped to nested insertions, there were 10 elements that had reads mapping at greater than 1 × 10−4 FPKM, with average expression ranging from 7.35 × 10−2 to 5.09 × 10−4 FPKM, respectively (supplementary fig. S6, Supplementary Material online). As the FPKM values are quite low, these data do not show significant expression of GingerRoot elements, including the acquired gene fragments, under these conditions. No reads mapping to the GingerRoot DDE regions were found in this data set. In total under these experimental conditions, GingerRoot elements did not show evidence of reasonable transcriptional activity.
The Presence of GingerRoot in Other Organisms and Association with Low Abundance of LTR Elements in Animals
Utilizing the GingerRoot DDE motif, a search was conducted to find other related sequences in the NCBI Sequence Read Archive (SRA) database. This database contained both RNA (17) and DNA (46) data sets, and queried whether GingerRoot element DDEs are unique to S. lepidophylla or are more widely distributed. This search retrieved GingerRoot elements in a diverse set of taxonomic groups, including plants and animals (table 2, supplementary tables S5 and S6, Supplementary Material online). Noteworthy, was the absence of GingerRoot elements in plants outside of Lycophytes and Bryophytes. These data demonstrate that GingerRoot is not unique to S. lepidophylla as it is present in several plant clades such as Bryophytes and Lycophytes but is absent in more basal Charophytes as well as the Angiosperm and Gymnosperm clades.
Table 2.
Number of Matches of GingerRoot DDE Motif in SRA Search
| Number of Species | Read Number Range | Database Type | |
|---|---|---|---|
| Actinopterygii (ray-finned fish) | 29 | 1–32 | DNA |
| Invertebrates | 14 | 1–26 | DNA |
| Bryopsida | 18 | 2 to >100 | DNA/RNA |
| Lycopodiopsida | 3 | 4–210 | DNA |
We searched for homologous GingerRoot elements using two RNA-seq data sets collected from leaf tissue of the Bryophytes Meteoridium remotifolium and Antitrichia curtipendula (hanging moss) (Johnson et al. 2016). The other 21 species of pleurocarpous mosses used in this study (Johnson et al. 2016) did not return results in our search. To avoid redundancy, only the first 40 amino acids of the DDE motif was queried so that each element would not be represented by multiple nonoverlapping reads (read length = 100 bp). From both data sets, there are over ten unique reads with more than ten copies, suggesting multiple GingerRoot-like elements are expressed at a reasonable level from these two species (table 3).
Table 3.
RNA-Seq Reads Corresponding to DDE Motif in Meteoridium and A. curtipendula
| Organism | Meteoridium | A. curtipendula (Hanging Moss) |
|---|---|---|
| Accession number | SRR2518094 | SRR2518092 |
| Spots | 22.8 M | 21.9 M |
| Total reads mapped to the first 40 amino acids of DDE | 267 | 398 |
| Nonredundant reads (identical reads excluded) | 45 | 110 |
| Single copy reads | 25 | 76 |
| Reads with 2–9 copies (FPKM ∼1–4) | 7 | 19 |
| Reads with 12–86 copies (FPKM ∼5–39) | 13 | 15 |
In addition to plants, we queried the NCBI databases for the presence of GingerRoot in other non-plant organisms. In our search, we identified numerous homologous sequences from various other eukaryotic species. Predominantly matches were found in fish, invertebrate cnidarians, and others. Several identified include Acropora digitifera (coral spp.), Crassostrea gigas (pacific oyster), Bemisia tabaci (sweet potato whitefly), Oryzias latices (Japanese rice fish), and Danio rerio (zebra fish). These represent a diversity of the tree of life, and many of them are aquatic or marine organisms. To understand more about those genomes, we searched for publicly available data about the composition of TEs in the relevant genomes. Among the 16 animal genomes with data, the TE content ranges from 5% (Southern platyfish) to 51% (Zebra fish) (supplementary table S7, Supplementary Material online). Despite the dramatic variation of total TE content, the amount of LTR elements in those genomes is uniformly low, ranging from 0.01% to 4.56%, suggesting a competitive relationship between GingerRoot elements and LTR elements. This search shows GingerRoot elements are more widely distributed outside of Bryophytes. As shown in figure 2, the DDE motif of GingerRoot elements from S. lepidophylla and coral form a clade. In addition to the conservation of transposase, elements from coral seem to have similar termini as the GingerRoot elements in S. lepidophylla (supplementary fig. S7, Supplementary Material online), suggesting they indeed belong to the same superfamily.
Discussion
The First DNA Transposon Encoding Integrase-Related Transposase in Plant Genomes
TEs are of ancient origin. Many eukaryotic transposons can be traced back to insertion sequences in bacteria, which are DNA transposons (Feschotte and Pritham 2007). For example, the Tc1/Mariner-like DNA transposons are related to the IS630 family. Interestingly, the integrase of LTR retrotransposons is related to the transposase of IS3/IS481 family (Fayet et al. 1990; Capy et al. 1996; Bao et al. 2010). As a result, it was proposed that LTR retrotransposons arose through a fusion between a non-LTR retrotransposon and a DNA transposon (Eickbush and Malik 2002; Capy et al. 1997). Despite the abundance of LTR retrotransposons in most plant genomes and some of the animal genomes, few DNA transposons harboring integrase-related transposase have been reported in eukaryotic organisms. The Maverick/Polinton elements represent self-synthesizing transposons in that they contain DNA polymerase domain in addition to integrase-like proteins (Kapitonov and Jurka 2006; Pritham et al. 2007).
The Maverick/Polinton elements have 6 bp TSD yet the element termini (5′-AG…TC-3′) do not resemble that of LTR retrotransposons. Another integrase-related DNA transposons are the Ginger/TDD elements, which are associated with 4 (Ginger1) or 6 (Ginger2) bp TSD. It is worth mentioning that in this study, Ginger1 and Ginger2 form different clusters (fig. 2). Together with the different length of TSD, we consider Ginger1 and Ginger2 may represent two independent superfamilies of TEs. The termini of the Ginger elements are the same as that of LTR retrotransposons (5′-TG…CA-3′). In addition to the similar DDE motif in integrase of Gypsy-like LTR retrotransposons, Ginger1 elements have other signatures of Gypsy integrase motifs, such as the YPYY motif, H2C2 zinc finger domain, and the GPY motif. As a result, it was proposed Ginger1 could be derived from a Gypsy-like element, which represents “reverse evolution” (Bao et al. 2010). Nonetheless, Maverick/Polinton and Ginger elements have not been reported in plants.
The presence of GingerRoot elements in plants and the fact that it has amplified to a few hundred copies in S. lepidophylla genome indicate that DNA transposons bearing integrase-like transposase are capable of surviving in plant genomes. Unlike the Ginger1 elements, the GingerRoot elements do not harbor other signature proteins of Gypsy-like integrase, so they are unlikely derived from Gypsy-like elements. This is consistent with the fact that the termini of the majority of the GingerRoot elements are “5′-TA…TA-3′,” whereas that of the majority of Gypsy-like elements, are “5′-TG…CA-3′.” The GingerRoot elements are also distinct from Maverick/Polinton elements because of the lack of DNA polymerase proteins and the different element termini. Based on the above features and phylogeny analysis, it is likely that the transposase in GingerRoot elements and the integrase of Gypsy-like elements share common ancestors but have taken independent evolutionary paths. Particularly, some Gypsy-like LTR elements in S. lepidophylla are located on the same branch with GingerRoot elements, not with other Gypsy-like elements (fig. 2). This may indicate that Gypsy elements have multiple origins and the integrase for some of those in S. lepidophylla share a common origin with the GingerRoot transposase. More importantly, if LTR elements are indeed derived from the fusion of a non-LTR retrotransposon and a DNA transposon, the presence of integrase-related transposons in plants implies that novel Gypsy-like elements could have arisen in planta.
In plants, GingerRoot elements are only detected in Lycophytes and Bryophytes and are absent elsewhere (Monilophytes, Gymnosperms, and Angiosperms). In contrast, their distribution is much more widespread in animals, mostly aquatic. Apparently, all of those animal genomes are associated with low abundance of LTR retrotransposons (supplementary table S7, Supplementary Material online), so it is possible the reduced competition for target sites favor the survival of GingerRoot elements (also see following section). Alternatively, the genetic sequences of aquatic animals are more readily released to the environment and could be absorbed by other animals directly or indirectly through feeding or other activities. Such processes, combined with the lack of activity of LTR retrotransposons, may favor the horizontal transfer and widespread distribution of GingerRoots in aquatic animals.
Because Bryophytes resemble the initial plants migrating from an aquatic environment to a land environment, a parsimonious explanation for these observations would indicate that the eukaryotic GingerRoot or its ancestor arose in aquatic organisms, and were retained in the common ancestor of Lycophytes and Bryophytes but were lost in the most recent common ancestor of Monilophytes, Gymnosperms, and Angiosperms. Certainly, we cannot rule out the possibility that GingerRoot is present in some of these divisions (Monilophytes, Gymnosperms, and Angiosperms) yet the relevant plant genome sequences are not available. If such species do exist, the presence of GingerRoot in higher plants should still be very isolated. In plants, horizontal transfer of TEs occurs often, as it was estimated that there were 2 million horizontal transfer events of LTR retrotransposons in monocots and dicots species (El Baidouri et al. 2014). Given this fact, the very limited or isolated distribution of GingerRoot elements in plants suggests either the horizontal transfer events for this element are very rare, or there are some intrinsic genomic features of higher plants that make GingerRoot less competitive in the genome. The two possibilities are not mutually exclusive. Based on the expression pattern and pairwise similarity, the GingerRoot elements are unlikely active at the current time in S. lepidophylla. However, the fact that sequences corresponding to the DDE motif are detected from the RNA-seq libraries from multiple species indicates the possibility that this family of TEs is still actively amplifying in some of the species such as hanging moss (table 3).
The Target Selection of GingerRoot
Although TEs insert into many different locations in the genome, many elements demonstrate a certain level of target specificity (Craig 1997; Linheiro and Bergman 2012). Some transposons insert into a specific sequence, and this could be reflected by the TSD and the sequences flanking the TSD. For example, the Tc1/Mariner elements only insert into “TA” motifs. In addition to selection on the primary sequence, TEs also choose their targets at the higher structure or chromatin level. It is well known that Gypsy-like elements preferentially insert into heterochromatic, gene-poor regions. In contrast, the majority of DNA transposons are located in the euchromatic and gene-rich regions (Cresse et al. 1995; Zhao et al. 2016). GingerRoot does not have strict sequence specificity except it preferentially inserts into GC-rich sequences. Unlike most DNA transposons, GingerRoot tends to be located in gene-poor regions. The underlying mechanism for such distribution is unclear. However, because the transposase of GingerRoot is related to the integrase of Gypsy-like retrotransposons, it is not surprising that they have similar target specificity (fig. 4).
In well-characterized plant genomes such as Arabidopsis, rice, and maize, Gypsy-like retrotransposons are the sole dominant TEs in gene-poor regions. In addition to their target specificity, the dominance of Gypsy-like retrotransposons is likely due to the “copy and paste” transposition mechanism as well as the relative low selection pressure against insertion in the gene-poor regions. LTR elements have two identical or similar LTRs at the termini. When intra-element recombination occurs between the two LTRs, the internal region of the element, as well as one of the LTRs, is eliminated, leading to the formation of a solo LTR. Due to the loss of internal regions which encode transposition machinery, the formation of solo LTRs is one of the most effective factors that limit the further amplification of LTR elements. According to our previous study, the removal of LTR-RTs via formation of solo LTRs is more effective in S. lepidophylla than that in Arabidopsis and rice (VanBuren et al. 2018). It is possible the more frequent unequal homologous recombination in S. lepidophylla makes Gypsy-like elements less competitive so that GingerRoot elements are able to survive in the gene-poor region. One hypothesis is that in the ancestor of Monilophytes, Gymnosperms, and Angiosperms, the GingerRoot elements became extinct due to lower competitiveness with Gypsy-like elements arising from the less efficient formation of solo LTRs and the rapid amplification of Gypsy-like LTR elements. In the future, it would be intriguing to test the relationship between the abundance of GingerRoot elements and the recombination frequency of the relevant genomes. Taken together, the isolated distribution of GingerRoot in plant genomes might be attributed to the exceptional success of LTR elements in most plants.
Gene Acquisition by GingerRoot Elements
Many families of TEs are carrying sequences from regular genes in their internal regions. However, the frequency of such incidents varies dramatically. In general, DNA transposons seem to carry genes more frequently than retrotransposons. In maize, for example, retrotransposons contribute 75% of the maize genomic sequence, yet only 400 cases of gene capture events were detected. In contrast, DNA transposons only account for 9% of the genome, and they are associated with over 1,600 gene capture events (Schnable et al. 2009). It raises the question of whether the high frequency of gene capture with DNA transposons is due to their association with genes because it is physically convenient for them to duplicate gene sequences. Obviously, a considerable portion (29%) of the GingerRoot elements carries gene fragments, but the majority of them are not close to genes. Particularly, the elements carrying gene fragments are not closer to genes than those without gene fragments (fig. 4). As a result, the physical distance between the elements and the genes does not seem to be a critical factor for gene duplication. The mechanism for gene duplication by DNA transposons is still unclear. In maize, alternative transposition of Ac/Ds elements causes segmental duplication and generation of chimeric new genes (Zhang et al. 2013; Wang et al. 2015). However, it is unclear whether such activity would result in the integration of genic sequences into the elements which are competent for further transposition. For Pack-MULEs in rice, GC-rich, 5′ end gene sequences are preferred (Jiang et al. 2011; Ferguson et al. 2013). Yet, such a preference is not detected for either MULEs or GingerRoot elements in S. lepidophylla. This suggests that the preference of gene duplication demonstrated by Pack-MULEs in rice might be enabled by some host-specific factors. Although the mechanism of duplication of gene sequences by GingerRoot is unclear, it is evident that these elements are locating some gene fragments in gene-poor regions, resulting in the redistribution of genic sequences.
The Acquired Gene Fragments May Influence the Epigenetic Status and Retention of GingerRoot Elements
In rice, 40% of Pack-MULEs have evidence of expression and the epigenetic status (methylation and histone modification) of those expressed elements resemble that of protein coding genes (Zhao et al. 2018). In this study, we failed to detect a reasonable level of expression for any of the GingerRoot elements, so it is not surprising to observe that GingerRoot elements have much higher methylation level than the protein coding genes, which have very little body methylation in S. lepidophylla (VanBuren et al. 2018). Nevertheless, not all GingerRoot elements demonstrate similar levels of methylation. Despite their distance from genes, GingerRoot elements with gene fragments are associated with much lower methylation level than those elements without gene fragments, and the acquired regions are among the lowest methylated regions inside the elements. It could be because the acquired gene fragments are not as repetitive as the true transposon sequences and therefore are subject to a reduced level of silencing. However, even if we exclude the acquired fragments, GingerRoots carrying genes are still associated with a lower level of methylation, suggesting the acquired gene fragments influence the epigenetic status of the flanking regions.
Giving that gene fragments acquired by TEs might have the potential to evolve some function, it is surprising to observe that GingerRoot elements carrying gene fragments are more distal to genes than their counterparts without gene fragments (fig. 4). Considering the fact that GingerRoot elements carrying gene fragments are older than those without gene fragments, one possibility is that an island with a relatively low methylation level among the otherwise highly methylated heterochromatin is selectively retained because it is beneficial to the host. This could occur, for example, the presence of GingerRoot elements with gene fragments reduces the chance of further insertion of Gypsy-like elements residing in heterochromatic regions. In other words, the gene duplication by GingerRoots may prevent the further expansion of heterochromatin and influence the overall chromosomal structure. This is consistent with the fact that Pack-MULEs in rice are retained longer in pericentromeric regions than those in chromosomal arms (Zhao et al. 2018).
Compared with the differentiation (in terms of distribution and methylation) between GingerRoot elements with and without gene fragments, it is puzzling that Pack-MULEs and other MULEs have similar distribution, retention time, and methylation level in S. lepidophylla. The similar pairwise identity between Pack-MULEs and other MULEs implies that Pack-MULEs are not selectively retained. This is likely due to target specificity of MULEs, which prefer genic regions. On the one hand, the rapid removal of repetitive sequences in genic regions (compared with intergenic regions) in S. lepidophylla may prevent Pack-MULEs from evolving any function and selectively retained. This is consistent with the facts that the recognizable Pack-MULEs in S. lepidophylla harbor much more recent acquisitions than that of GingerRoots (table 1). On the other hand, the Pack-MULEs in S. lepidophylla are larger than other MULEs, so they could be subject to additional surveillance (Panda et al. 2016), which also hinders evolution of function. The average Pack-MULEs in rice are about 2 kb in length (Hanada et al. 2009), which is much smaller than Pack-MULEs in S. lepidophylla. Moreover, the similarity between Pack-MULEs and parental genes could be as low as 78% (Jiang et al. 2004), indicating Pack-MULEs in rice retain much longer than that in S. lepidophylla. As a result, Pack-MULEs in rice and in S. lepidophylla may have distinct evolutionary trajectory due to their different life span and element size.
In summary, GingerRoot represents a novel superfamily of DNA transposons. Unlike other plant DNA transposons, GingerRoot elements encode a transposase related to integrase and therefore resemble Gypsy-like retrotransposons in terms of target selection. Moreover, the length of TSD and element termini mimics that of LTR retrotransposons. Nevertheless, its element structure is similar to typical DNA transposons due to the presence of TIR, and it often carries gene fragments and therefore causes relocation of genic sequences. It is challenging for a DNA transposon to be located in the gene-poor regions to compete with Gypsy-like elements, and the high recombination rate in S. lepidophylla may prevent the excessive amplification of LTR retrotransposons so it favors the survival of GingerRoot. The unique features of GingerRoot may attribute to its orthogonal distribution in plants.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Dr Shujun Ou (Michigan State University) for assistance with the search for LTR elements in S. lepidophylla. This study was supported by National Science Foundation [MCB-1121650, IOS-1126998, IOS-1740874 to N.J.]; United States Department of Agriculture National Institute of Food and Agriculture and AgBioResearch at Michigan State University [Hatch grant MICL02408 to N.J.].
Literature Cited
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Baniaga AE, Arrigo N, Barker MS.. 2016. The small nuclear genomes of Selaginella are associated with a low rate of genome size evolution. Genome Biol Evol. 8(5):1516–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks JA. 2009. Selaginella and 400 million years of separation. Annu Rev Plant Biol. 60(1):223–238. [DOI] [PubMed] [Google Scholar]
- Banks JA, et al. 2011. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332(6032):960–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, Jurka MG, Kapitonov VV, Jurka J.. 2009. New superfamilies of eukaryotic DNA transposons and their internal divisions. Mol Biol Evol. 26(5):983–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, Kapitonov VV, Jurka J.. 2010. Ginger DNA transposons in eukaryotes and their evolutionary relationships with long terminal repeat retrotransposons. Mob DNA 1(1):3.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, Kojima KK, Kohany O.. 2015. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA 6:11.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL. 2000. Transposable element contributions to plant gene and genome evolution. Plant Mol Biol. 42(1):251–269. [PubMed] [Google Scholar]
- Bennetzen JL, Ma J, Devos KM.. 2005. Mechanisms of recent genome size variation in flowering plants. Ann Bot [Internet] 95(1):127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L.. 2016. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 34(5):525–527. [DOI] [PubMed] [Google Scholar]
- Brookfield JFY, Johnson LJ.. 2006. The evolution of mobile DNAs: when will transposons create phylogenies that look as if there is a master gene? Genetics 173(2):1115–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capy P, Langin T, Higuet D, Maurer P, Bazin C.. 1997. Do the integrases of LTR-retrotransposons and Class II element transposases have a common ancestor? Genetica 100(1–3):63–72. [PubMed] [Google Scholar]
- Capy P, Vitalis R, Langin T, Higuet D, Bazin C.. 1996. Relationships between transposable elements based upon the integrase-transposase domains: is there a common ancestor? J Mol Evol. 42(3):359–368. [DOI] [PubMed] [Google Scholar]
- Cerbin S, Jiang N.. 2018. Duplication of host genes by transposable elements. Curr Opin Genet Dev. 49:63–69. [DOI] [PubMed] [Google Scholar]
- Clément Y, Fustier M-A, Nabholz B, Glémin S.. 2015. The bimodal distribution of genic GC content is ancestral to monocot species. Genome Biol Evol. 7(1):336–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig NL. 2002. Mobile DNA: an Introduction. In: Craig NL, editor. Mobile DNA II. Washington DC: American Society of Microbiology. p. 3–11. [Google Scholar]
- Cresse AD, Hulbert SH, Brown WE, Lucas JR, Bennetzen JL.. 1995. MU1-related transposable elements of maize preferentially insert into low copy number DNA. Genetics 140(1):315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos KM, Brown JKM, Bennetzen JL.. 2002. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 12(7):1075–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doak TG, Doerder FP, Jahn CL, Herrick G.. 1994. A proposed superfamily of transposase genes: transposon-like elements in ciliated protozoa and a common “D35E” motif. Proc Natl Acad Sci U S A. 91(3):942–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebina H, Chatterjee AG, Judson RL, Levin HL.. 2008. The GP(Y/F) domain of TF1 integrase multimerizes when present in a fragment, and substitutions in this domain reduce enzymatic activity of the full-length protein. J Biol Chem. 283(23):15965–15974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickbush TH, Malik HS.. 2002. Origins and Evolution of Retrotransposons In: Craig NL, editor: Mobile DNA II. Washington DC: American Society of Microbiology p. 1111–1144. [Google Scholar]
- El Baidouri M, et al. 2014. Widespread and frequent horizontal transfers of transposable elements in plants. Genome Res. 24(5):831–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayet O, Ramond P, Polard P, Prère MF, Chandler M.. 1990. Functional similarities between retroviruses and the IS3 family of bacterial insertion sequences? Mol Microbiol. 4(10):1771–1777. [DOI] [PubMed] [Google Scholar]
- Ferguson AA, Zhao D, Jiang N.. 2013. Selective acquisition and retention of genomic sequences by Pack-Mutator-like elements based on guanine–cytosine content and the breadth of expression. Plant Physiol. 163(3):1419–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Pritham EJ.. 2007. DNA Transposons and the evolution of eukaryotic genomes. Annu Rev Genet. 41(1):331–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Hou Y, Ebina H, Levin HL, Voytas DF.. 2008. Chromodomains direct integration of retrotransposons to heterochromatin. Genome Res. 18(3):359–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Wessler SR.. 2010. MITE-Hunter: a program for discovering miniature inverted-repeat transposable elements from genomic sequences. Nucleic Acids Res. 38(22):e199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, et al. 2009. The functional role of Pack-MULEs in rice inferred from purifying selection and expression profile. Plant Cell 21(1):25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins JS, Proulx SR, Rapp RA, Wendel JF.. 2009. Rapid DNA loss as a counterbalance to genome expansion through retrotransposon proliferation in plants. Proc Natl Acad Sci U S A. 106(42):17811–17816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. 1992. Detection of Caenorhabditis transposon homologs in diverse organisms. New Biol. 4(4):382–388. [PubMed] [Google Scholar]
- Jiang N, Bao Z, Zhang X, Eddy SR, Wessler SR.. 2004. Pack-MULE transposable elements mediate gene evolution in plants. Nature 431(7008):569–573. [DOI] [PubMed] [Google Scholar]
- Jiang N, Ferguson AA, Slotkin RK, Lisch D.. 2011. Pack-Mutator-like transposable elements (Pack-MULEs) induce directional modification of genes through biased insertion and DNA acquisition. Proc Natl Acad Sci U S A. 108(4):1537–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MG, Malley C, Goffinet B, Shaw AJ, Wickett NJ.. 2016. A phylotranscriptomic analysis of gene family expansion and evolution in the largest order of pleurocarpous mosses (Hypnales, Bryophyta). Mol Phylogenet Evol. 98:29–40. [DOI] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J.. 2006. Self-synthesizing DNA transposons in eukaryotes. Proc Natl Acad Sci U S A. 103(12):4540–4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell MG. 2002. Transposable elements and the evolution of genome size in eukaryotes. Genetica 115(1):49–63. [DOI] [PubMed] [Google Scholar]
- Kidwell MG, Lisch DR.. 2000. Transposable elements and host genome evolution. Trends Ecol Evol. 15(3):95–99. [DOI] [PubMed] [Google Scholar]
- Kronmiller BA, Wise RP.. 2008. TEnest: automated chronological annotation and visualization of nested plant transposable elements. Plant Physiol. 146(1):45–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linheiro RS, Bergman CM.. 2012. Whole genome resequencing reveals natural target site preferences of transposable elements in Drosophila melanogaster. Stajich JE, editor. PLoS One 7(2):e30008.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch D. 2013. How important are transposons for plant evolution? Nat Rev Genet. 14(1):49–61. [DOI] [PubMed] [Google Scholar]
- Lisch D, Slotkin RK.. 2011. Strategies for silencing and escape. Int Rev Cell Mol Biol. 292:119–152. [DOI] [PubMed] [Google Scholar]
- Marschalek R, Brechner T, Amon-Böhm E, Dingermann T.. 1989. Transfer RNA genes: landmarks for integration of mobile genetic elements in Dictyostelium discoideum. Science 244(4911):1493–1496. [DOI] [PubMed] [Google Scholar]
- McClintock B. 1950. The origin and behavior of mutable loci in maize. Proc Natl Acad Sci U S A. 36(6):344–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. 1956. Controlling elements and the gene. Cold Spring Harb Symp Quant Biol. 21(0):197–216. [DOI] [PubMed] [Google Scholar]
- Morgenstern B. 1999. DIALIGN 2: improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics 15(3):211–218. [DOI] [PubMed] [Google Scholar]
- Okonechnikov K, Golosova O, Fursov M.. 2012. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28(8):1166–1167. [DOI] [PubMed] [Google Scholar]
- Ou S, Jiang N.. 2018. LTR_retriever: a highly accurate and sensitive program for identification of long terminal repeat retrotransposons. Plant Physiol. 176(2):1410–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda K, et al. 2016. Full-length autonomous transposable elements are preferentially targeted by expression-dependent forms of RNA-directed DNA methylation. Genome Biol. 17(1):170.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritham EJ, Putliwala T, Feschotte C.. 2007. Mavericks, a novel class of giant transposable elements widespread in eukaryotes and related to DNA viruses. Gene 390(1–2):3–17. [DOI] [PubMed] [Google Scholar]
- Sayers EW, et al. 2019. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 47(D1):D23–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, et al. 2009. The B73 maize genome: complexity, diversity, and dynamics. Science 326(5956):1112–1115. [DOI] [PubMed] [Google Scholar]
- Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28(10):2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBuren R, et al. 2018. Extreme haplotype variation in the desiccation-tolerant clubmoss Selaginella lepidophylla. Nat Commun. 9(1):13.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, et al. 2015. Alternative transposition generates new chimeric genes and segmental duplications at the maize p1 locus. Genetics 201(3):925–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, et al. 2007. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 8(12):973–982. [DOI] [PubMed] [Google Scholar]
- Yuan Y-W, Wessler SR.. 2011. The catalytic domain of all eukaryotic cut-and-paste transposase superfamilies. Proc Natl Acad Sci U S A. 108(19):7884–7889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zuo T, Peterson T.. 2013. Generation of tandem direct duplications by reversed-ends transposition of maize Ac elements. PLoS Genet. 9(8):e1003691.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D, Ferguson AA, Jiang N.. 2016. What makes up plant genomes: the vanishing line between transposable elements and genes. Biochim Biophys Acta 1859(2):366–380. [DOI] [PubMed] [Google Scholar]
- Zhao D, et al. 2018. The unique epigenetic features of Pack-MULEs and their impact on chromosomal base composition and expression spectrum. Nucleic Acids Res. 46(5):2380–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.