Abstract
Background
For children with cancer, the clinical integration of precision medicine to enable predictive biomarker–based therapeutic stratification is urgently needed.
Methods
We have developed a hybrid-capture next-generation sequencing (NGS) panel, specifically designed to detect genetic alterations in paediatric solid tumours, which gives reliable results from as little as 50 ng of DNA extracted from formalin-fixed paraffin-embedded (FFPE) tissue. In this study, we offered an NGS panel, with clinical reporting via a molecular tumour board for children with solid tumours. Furthermore, for a cohort of 12 patients, we used a circulating tumour DNA (ctDNA)–specific panel to sequence ctDNA from matched plasma samples and compared plasma and tumour findings.
Results
A total of 255 samples were submitted from 223 patients for the NGS panel. Using FFPE tissue, 82% of all submitted samples passed quality control for clinical reporting. At least one genetic alteration was detected in 70% of sequenced samples. The overall detection rate of clinically actionable alterations, defined by modified OncoKB criteria, for all sequenced samples was 51%. A total of 8 patients were sequenced at different stages of treatment. In 6 of these, there were differences in the genetic alterations detected between time points. Sequencing of matched ctDNA in a cohort of extracranial paediatric solid tumours also identified a high detection rate of somatic alterations in plasma.
Conclusion
We demonstrate that tailored clinical molecular profiling of both tumour DNA and plasma-derived ctDNA is feasible for children with solid tumours. Furthermore, we show that a targeted NGS panel–based approach can identify actionable genetic alterations in a high proportion of patients.
Keywords: Paediatric oncology, Clinical targeted sequencing, Personalised medicine, Circulating tumour DNA
Highlights
-
•
The hybrid-capture panel gives reliable results from as little as 50 ng of DNA extracted from formalin-fixed paraffin-embedded tissue.
-
•
A tailored sequencing approach identifies ≥1 genetic alteration in 70% of samples and potentially actionable genetic alterations in 51% of patients.
-
•
In paired samples, from different treatment time points, there were differences in the genetic alterations detected.
1. Introduction
In adult malignancies, precision medicine initiatives enabling standardised, high-throughput molecular profiling and predictive biomarker–based stratification have been implemented to maximise clinical efficacy of targeted therapeutics [1], [2], [3], [4], [5], [6], [7]. Similar initiatives are urgently needed for childhood cancer, which remains the primary cause of death in children after infancy [8].
In children, comprehensive molecular profiling programmes have incorporated whole-exome sequencing (WES) and RNA sequencing (RNA-seq) and, in some cases, copy number analysis, whole-genome sequencing (WGS), microarray or methylation arrays. Such initiatives have detected potentially actionable findings in 46–60.9% of patients [9], [10], [11]. However, logistical and financial practicalities limit large-scale implementation of this approach in most health-care settings. Targeted next-generation sequencing (NGS) panels are typically more cost-effective and can be tailored to the study population and standardised according to regulatory requirements. Therefore, this may present a more suitable alternative for implementation into health-care systems.
Generic adult cancer gene panels have been used in children [12], [13]; however, the spectrum of mutations differs between adult and paediatric tumours. For example, recurrent H3 mutations are a hallmark of paediatric high-grade glioma [14], [15], and rearrangements upstream to the TERT promoter are frequent in neuroblastoma [16]. These differences necessitate a tailored approach to determine common and actionable events; hence, we have developed and clinically validated a paediatric-specific solid tumour NGS panel for use in precision medicine [17].
In children with relapsed/refractory cancer, access to adequate biopsy material remains challenging [18], [19]. Therefore, our strategy has been to optimise the paediatric panel for use on formalin-fixed paraffin-embedded (FFPE) tissue if frozen tissue is unavailable and, in parallel, begin evaluating more-easily accessible sources of tumour DNA, such as plasma.
Plasma-derived circulating tumour DNA (ctDNA) has been shown to be an alternative to repeat biopsy in common adult malignancies [20], [21], [22], [23]. ctDNA analysis is minimally invasive, amenable to serial sampling and may also give more representative information regarding tumour heterogeneity [24], [25]. Limited studies in children with cancer have detected somatic mutations in small volumes of plasma [26], [27], [28], [29], [30].
Here, we report the development of version 2 of our paediatric solid tumour–specific NGS panel and the national implementation of clinical NGS panel sequencing. We report on assay performance and the clinical relevance of the findings. In parallel, we evaluate the feasibility of performing targeted sequencing of ctDNA in a clinical laboratory setting using a ctDNA-specific NGS panel.
2. Materials and methods
2.1. Patients
A Royal Marsden Hospital (RM) pilot study for patients aged ≤24 years with solid tumours treated at our Children and Young People's Unit commenced in March 2016 and was subsequently expanded nationally for children aged ≤16 years. Ethical approval was obtained from the National Research Ethics Service (reference: 15/LO/07) and the Biological Studies Steering Group of the Children's Cancer and Leukaemia Group (reference: 2015 BS 09). Participants and/or guardians gave informed consent. Patients were eligible to enrol at any time including diagnosis and relapse/progression. Blood was taken for germline DNA analysis, and archival tissue was retrieved from the most recent surgery, or if indicated, a repeat biopsy could be requested at the treating clinician's discretion.
2.2. Sample preparation and sequencing
Sample preparation, DNA extraction, library preparation and sequencing were performed according to established protocols [17], [31]. Two different panels were used: version 1 (v1, 78 genes, 311 kb) and version 2 (v2, 91 genes, 473 kb) (Table S1). The custom hybridisation panel is capable of detecting single-nucleotide variants (SNVs), small insertions and deletions (indels), copy number variations (CNVs) and structural variants for which we capture the region where the breakpoint occurs, for instance, 50 kb upstream to the TERT promoter [16]. Sequencing output files were processed as previously reported [31]. Only somatic variants, detected after subtraction of germline findings, were reported.
Samples were analysed initially using MiSeq Reporter version 2.5 (http://emea.support.illumina.com/sequencing/sequencing_software/miseq_reporter/downloads.html). Analysis was later executed using an in-house developed pipeline Molecular Diagnostic Information Management System version 3.0 (MDIMSv3) using the following bioinformatic software and versions: demultiplexing was performed using bcl2fastq 2.17.1.14, reads were aligned using BWA 0.7.12, structural variants were identified using Manta 0.29.6, SNVs and indels were called with GATK 3.5.0 and variants were annotated with Oncotator version 1.5.1.0. CNVs were assessed as previously described [17].
2.3. Gene panel capture version 2, design and validation
Integral to the study design was the ability to update and adapt the regions included on the panel according to clinical need and target prioritisation. For v2, genes were ranked by consensus expert opinion according to set selection criteria (Table S1). The panel was validated using four cell blends (Tru-Q1-4 Horizon Discovery, Cambridge, United Kingdom [UK]) and 10 FFPE samples with known variants (SNVs = 554, indels = 79). Quality and coverage metrics were calculated across all the samples including (i) total reads, (ii) percentage of reads mapped to the reference sequence, (iii) percentage of duplicates, (iv) percentage of bases from unique reads deduplicated on target and (v) mean depth. Sensitivity, specificity and accuracy were determined by comparing the cell blends and FFPE samples with known variants and known true negatives.
2.4. Molecular tumour board
A monthly molecular tumour board (MTB) was established for discussion of findings, and the interpreted results were then reported to the treating clinician. The MTB core members included paediatric/adolescent oncologists, experts in early clinical trials, molecular pathologists, bioinformaticians and paediatric tumour biologists, from the RM, Great Ormond Street Hospital and The Institute of Cancer Research, London. OncoKB was used as a basis to define tiers of actionability [32]. In addition, COSMIC [33]-defined mutations/SNVs, genetic amplifications, gains or losses, for which a paediatric clinical trial was currently recruiting, were also considered, as well as alterations where compelling preclinical paediatric data existed for that target (Table S1). Heterozygous gene loss and missense mutations outside of defined hotspot regions were defined as not actionable.
2.5. ctDNA extraction and analysis
A total of 12 plasma samples were identified for sequencing where the corresponding tumour samples contained at least one genetic alteration present on the ctDNA panel. The plasma ctDNA sequencing results were not reported back to the MTB.
About 5 to 10 mL of blood was collected into cell-free DNA blood collection tubes (Streck, La Vista, United States of America) and centrifuged twice at 1600 g. ctDNA extraction and sequencing using a commercially available hybrid-capture panel (Avenio ctDNA expanded kit, Roche) was performed according to the manufacturer's instructions.
3. Results
3.1. Version 2 of the paediatric solid tumour panel
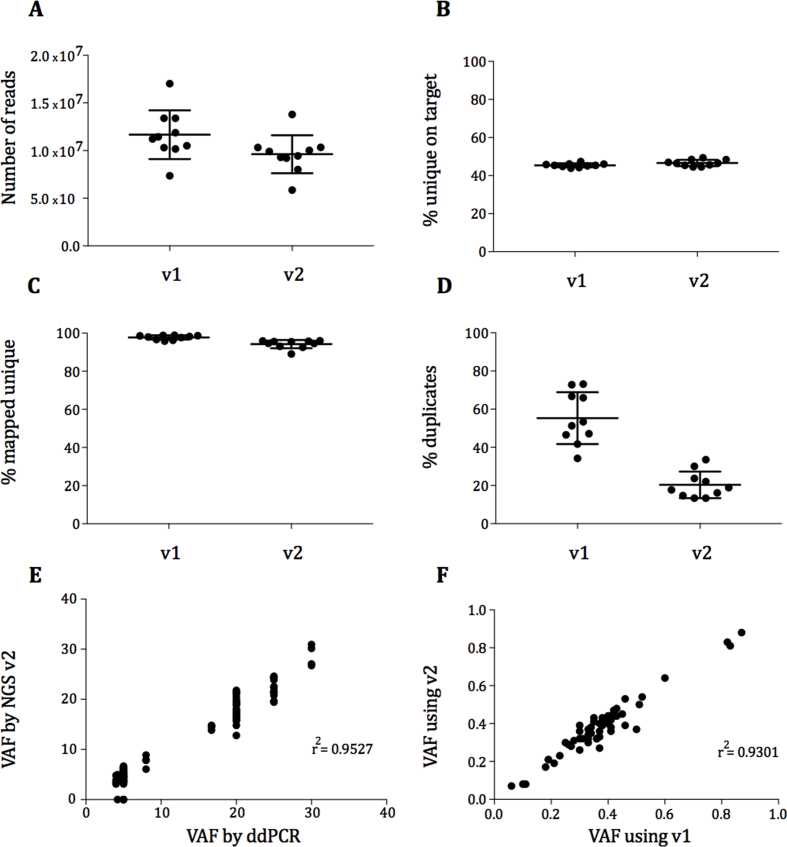
v1 of the panel was validated as previously reported [17]. v2 was also validated to Good Laboratory and Clinical Practice standards and performed well, comparable with v1, obtaining a similar number of reads and percentage of unique on-target reads (Figure S1A-C). The polymerase chain reaction (PCR) duplicate percentage was improved (v1 = 55.3% and v2 = 20.3%) (Figure S1D). The sensitivity for detection of SNVs was ≥99% and ≥90% for indels at ≥5% variant allele frequency (VAF) (Table S2). The specificity for SNVs was ≥98% at ≥5% allele frequency. The correlation (r2) of VAF for SNVs and indels between droplet digital polymerase chain reaction (ddPCR) and v2 was 0.9527 (Figure S1E) and between v1 and v2 was 0.9301 (Figure S1F).
3.2. Patient samples and overall performance
An overview of the study is given in Fig. 1. A total of 255 samples were submitted from 223 patients. Although patients were eligible to enrol at any time, 90% of evaluable patients had at least one episode of progression/relapse before study enrolment. FFPE tissue from the most recent surgery was requested for all but 3 patients where fresh frozen tissue was used.
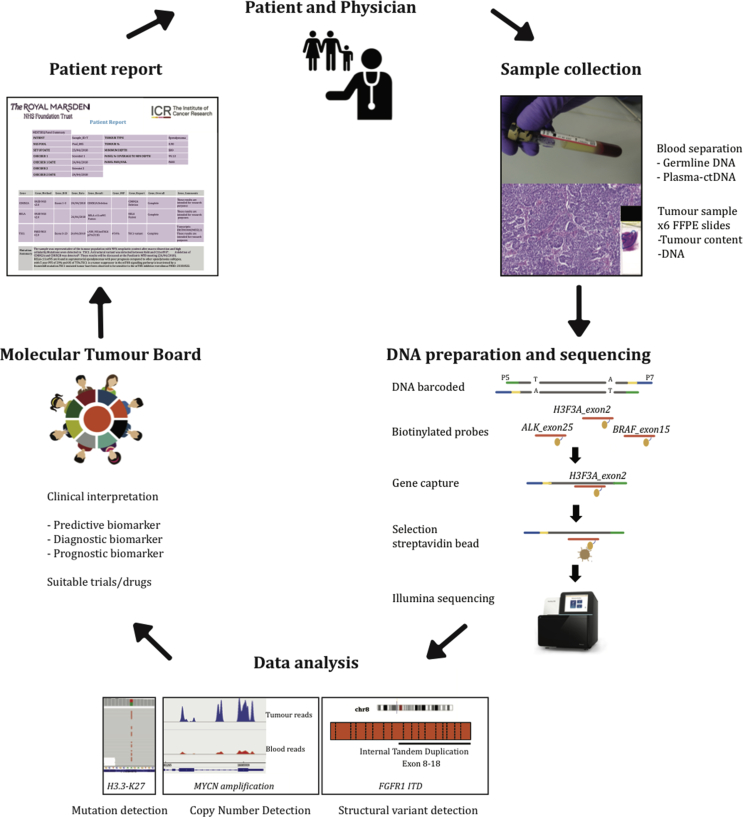
Fig. 1.
Study overview. After obtaining informed consent, tumour and blood samples were collected. DNA was extracted, and sequence libraries were prepared using the capture-based paediatric solid tumour panel. After sequencing, samples underwent an in-house data analysis pipeline that detects mutations, structural variants and copy number changes. Genomic alterations were manually reviewed by two independent scientists and then discussed in a molecular tumour board before a clinical report was issued. FFPE, formalin-fixed paraffin-embedded.
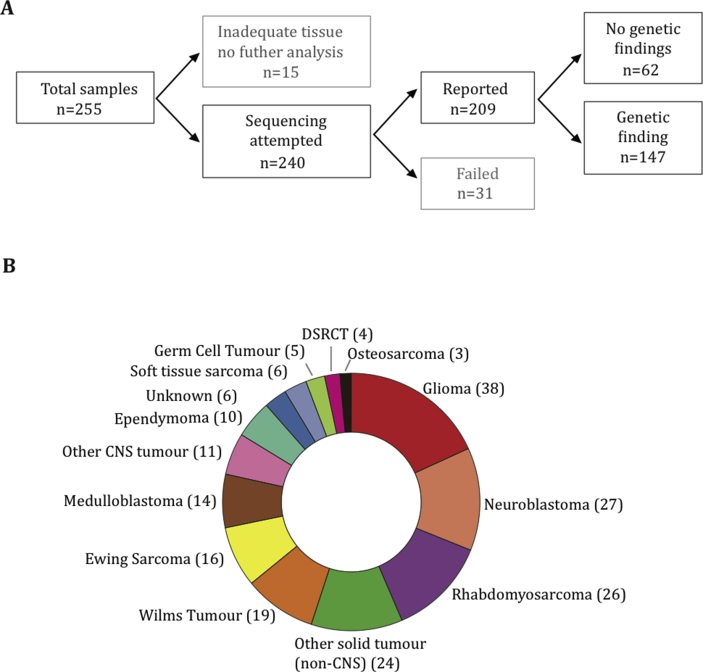
Adequate coverage for clinical reporting of results was obtained in 82% of submitted samples (Fig. 2A). Reasons for sample rejection or failure were as follows: tumour content less than 10%, DNA less than 20 ng and/or excessive DNA fragmentation. The median depth of coverage for all reported samples was 495 (interquartile range: 264–868). The most common cancers sequenced were glioma (38), neuroblastoma (27) and rhabdomyosarcoma (26) (Fig. 2B).
Fig. 2.
Tumour samples submitted for sequencing. Summary of sample flow and the total number of samples successfully sequenced (A). Distribution of tumour types among reported cases (B). DSRCT, desmoplastic small round cell tumour; CNS, central nervous system.
3.3. Genetic findings
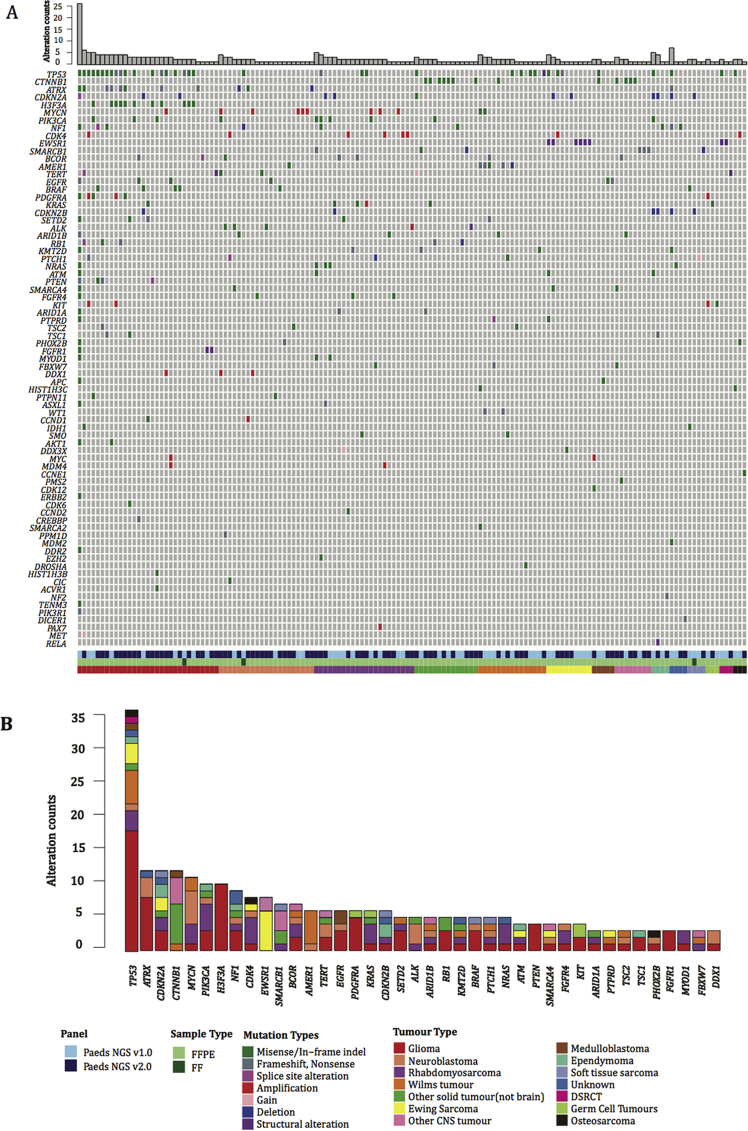
At least one genetic alteration was detected in 70% (145/209) of samples at an allele frequency ≥ 5%. The somatic genetic alterations detected, grouped according to underlying diagnosis, are summarised in Fig. 3, Table S3 and Fig S2. In keeping with other studies [34], the most frequently mutated gene was TP53 in 36/209 (17%); in addition high frequencies of alterations in genes known to be recurrently altered in paediatric malignancies such as ATRX, CDKN2A, CTNNB1 in 12/209 (5.7%), MYCN in 11/209 (5.2%) and H3F3A, PIK3CA in 10/209 (4.3%) were detected.
Fig. 3.
Overview of sequencing results. Oncoprint represents somatic mutations and gains, amplification and deletions detected in genes that are covered by the targeted panel. Samples are grouped in columns with genes displayed along rows. Samples are arranged according to the tumour type and genes sorted by frequency. Panel version, sample type, molecular annotations and diagnosis are provided as bars according to the included key (A). Bar plot of most recurrent altered genes, sorted by frequency and colour coded according to the tumour type (B). FFPE, formalin-fixed paraffin-embedded; DSRCT, desmoplastic small round cell tumour; CNS, central nervous system; FF, fresh frozen.
3.4. Clinical actionability
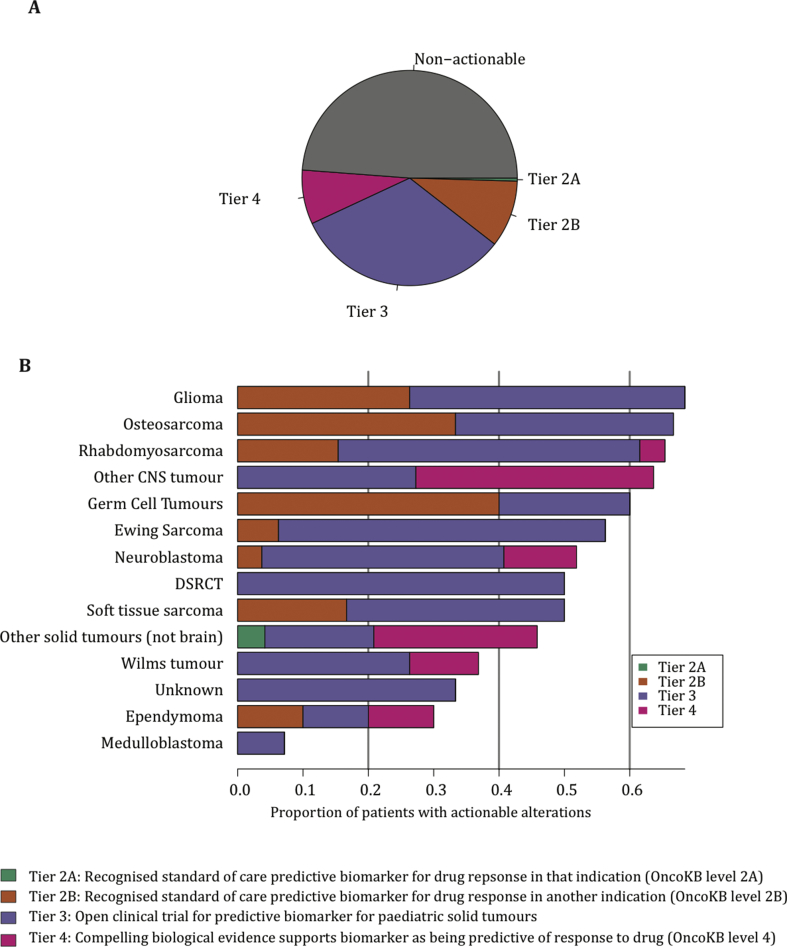
Potentially targetable alterations, defined by OncoKB tiers of actionability in addition to predictive biomarkers for currently recruiting paediatric clinical trials, were detected in 51% of sequenced samples (Fig. 4A). Of the 107 tumour samples classified as potentially actionable, 42 (39%) had greater than one actionable alteration detected. For each tumour sample, only the alteration for which there was the highest tier of evidence for actionability was included. Glioma was the tumour type with more defined actionable alterations found, followed by osteosarcoma and rhabdomyosarcoma (Fig. 4B). No tier 1 alterations (US Food and Drug Administration [FDA]–recognised biomarker predictive of response to an FDA-approved drug) were detected, indicative of the lack of regulatory approvals for paediatric indications. Only one patient had a tier 2A alteration: a patient with an inflammatory myofibroblastic tumour, harbouring an ALK:SQSTM1 translocation. The patient had a complete surgical resection and did not require systemic therapy.
Fig. 4.
Clinical actionability. Somatic alterations were defined according to OncoKB levels of evidence. Actionability tiers are described in the key. Distribution of actionability tiers for the entire sequenced cohort (A). Distribution of actionability tiers across common tumours, colour coded according to the tumour type (B). DSRCT, desmoplastic small round cell tumour; CNS, central nervous system.
As a feasibility study, follow-up data were not routinely collected for all patients. Of the 57 patients with a tier 2B or 3 alteration and available follow-up data, only four (7%) received targeted therapies:
Three patients with BRAFV600E mutations were treated with dabrafenib/trametinib combination therapy: patient 1 had a pleomorphic xanthoastrocytoma and was commenced on dabrafenib/trametinib after third disease progression. The patient remains on treatment with stable disease after 9 months. Patient 2 had glioblastoma multiforme and was commenced on dabrafenib/trametinib after disease progression. The patient had stable disease for 13 months before further progression. Patient 3 had multiply relapsed metastatic ameloblastic fibro-odontosarcoma [35]; by day 28 of treatment, there had been a partial response but asymptomatic cardiac toxicity, required discontinuation of both drugs. On normalisation of the shortening and ejection fractions, the patient was recommenced on single-agent dabrafenib and had sustained partial response for 15 further months. A patient with multiply relapsed metastatic germinoma and PDGFRA/KIT amplification was given dasatinib, but progressed on treatment.
One patient with high-grade glioma (patient ID 045-T) had a total of 49 somatic mutations (Table S3) (in ∼0.18 Mb) consistent with a hypermutator phenotype, associated with mismatch repair deficiency and predictive of potential sensitivity to immune checkpoint blockade [36]. However, the patient was not fit for clinical trial enrolment by the time the sequencing results were available.
Other patients had findings that informed prognosis: a mutation in CTNNB1 was found in a patient originally diagnosed with supratentorial primitive neuroectodermal tumour (PNET), biologically more in keeping with a WNT-activated medulloblastoma. Other examples included an MYOD1 mutation in a patient with embryonal rhabdomyosarcoma, associated with distinct clinical features and poor prognosis [37], and a RELA-c11orf95 fusion in a patient with supratentorial ependymoma, associated with high-risk disease [38].
3.5. Analysis of paired samples
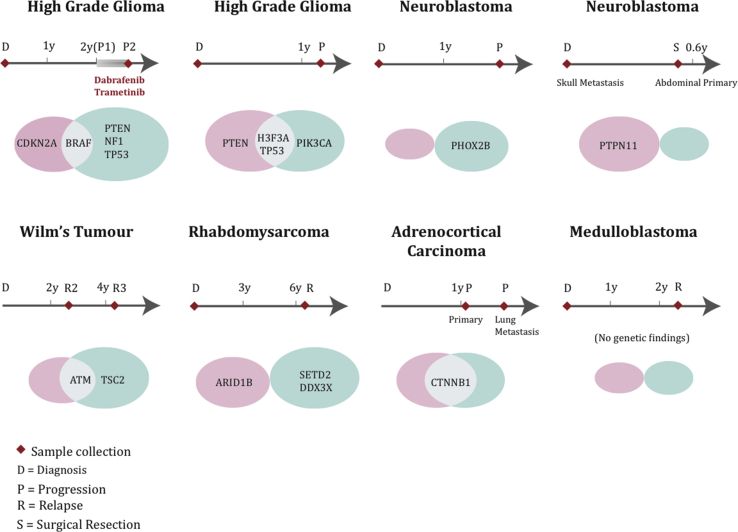
For eight patients, paired samples were sequenced at different stages of treatment (Fig. 5). In six of these, there were differences between the variants detected at different time points. Mutations in PTEN, NF1 and TP53 were observed in a patient with high-grade glioma (patient 2) after dabrafenib/trametinib treatment but not in the pre-treatment sample. The patient subsequently received everolimus but progressed after 3 months on treatment. The acquisition of NF1 mutations as a resistance mechanism after BRAF inhibition is consistent with findings in BRAFV600E-mutant melanoma [39], [40]. in another child with glioma sequenced at diagnosis and progression, the tumour harboured shared alterations in H3F3A and TP53, whereas PTEN was only present at diagnosis and PIK3CA at progression. In a patient with Wilms tumour, a potentially targetable TSC2 mutation was found in the 3rd relapse sample, which was not present in the previous sample.
Fig. 5.
Comparison of results from paired samples, sequenced at different time points. Venn diagrams compare the genetic findings in eight patients. Shared alterations are illustrated by the intersection of the two ovals. Alterations detected at only the 1st time point are represented in the pink oval, and alterations identified at the 2nd time point only are represented in the green oval. The size of the oval represents the number of variants identified in each patient.
3.6. ctDNA analysis
ctDNA was sequenced in a cohort of 12 patients with extracranial tumours, in whom the tumour panel had detected a genetic alteration that was also covered by a commercially available ctDNA sequencing panel. In 3 patients, in whom ctDNA and FFPE were sequenced from the same time point, there was a direct concordance between findings. However, in 5 patients, from whom plasma was collected after at least one subsequent relapse, variants were detected in the plasma that were not detected in FFPE samples (Table 1). For example, in a patient with neuroblastoma, an ALK F1174L mutation was detected in both tumour and plasma; however, an additional ALK hotspot mutation was also detected in the plasma that was not present in the tumour sample. In addition, of note, in 2 cases, variants detected in plasma at relapse were only identified at very low levels in diagnostic tumour samples, below the predefined limit of detection for clinical reporting.
Table 1.
Results of ctDNA panel sequencing of matched plasma samples and comparison with tumour panel sequencing for genes covered by both panels, ordered by the time elapsed between samples.
| Diagnosis | Days between samples | Treatment position with FFPE sample | Treatment position with blood sample | Isolated ctDNA (ng) | Gene | Amino acid change | AF FFPE DNA | AF ctDNA | Sequencing depth ctDNA | Sequencing depth tumour |
|---|---|---|---|---|---|---|---|---|---|---|
| Neuroblastoma | 5 | 5th relapse | 5th relapse | 18.54 | TP53 | C135F | 74.0% | 20.30% | 13348 | 393 |
| Wilms tumour | 19 | Post induction | Post induction | 32.22 | TP53 | G245D | 77.0% | 7.44% | 5498 | 402 |
| Ewing sarcoma | 84 | 2nd relapse | 2nd relapse | 50 | TP53 | C176Y | 87.0% | 49.90% | 3453 | 70 |
| Neuroblastoma | 214 | Diagnosis | 2nd relapse | 7.5 | ALK | R1275Q | N/D | 3.11%c | 2954 | 528 |
| ALK | F1174L | 17.0% | 3.88% | 2242 | 354 | |||||
| APC | R499* | 0.24%a | 0.31% | 2580 | 412 | |||||
| Ewing sarcoma | 315 | Diagnosis | Relapse | 34.02 | TP53 | R273C | 48.0%b | N/D | 3557 | 314 |
| TP53 | R337C | N/D | 31.40%c | 5237 | 391 | |||||
| CDKN2A | R80* | 3.0%a | 25.53% | 2064 | 899 | |||||
| ACC | 427 | 3rd progression | VGPR to 4th-line therapy | 51.96 | CTNNB1 | S33Pro | 33.00%b | N/D | 5194 | 777 |
| RMS | 444 | Diagnosis | 2nd relapse | 18.6 | TP53 | V173M | F | 11.43% | 2782 | 17 |
| PIK3CA | E542K | 15.0%b | N/D | 2166 | 167 | |||||
| PIK3CA | E545K | 17.0% | 0.56%a | 2065 | 180 | |||||
| Osteosarcoma | 514 | Diagnosis | 2nd relapse | 33.96 | TP53 | R248T | 78.0% | 11.08% | 6334 | 91 |
| TP53 | Y220C | N/D | 0.29%c | 5510 | 542 | |||||
| Neuroblastoma | 738 | Post induction | 1st relapse | 168.6 | TP53 | R249S | N/D | 0.05%c | 14825 | 193 |
| ALK | D1091N | 8.0% | 0.03% | 22632 | 308 | |||||
| RMS | 954 | Diagnosis | 2nd relapse | 29.52 | KRAS | G12C | 92.0% | 0.09%a | 3233 | 1453 |
| Wilms tumour | 1211 | Diagnosis | 3rd relapse | 50.76 | TP53 | R273C | 100.0% | 23.96% | 3961 | 74 |
| Wilms tumour | 1322 | Post induction | 3rd relapse | 19.86 | TP53 | R181C | 86.0% | 3.72% | 2525 | 141 |
| TP53 | C176Y | N/D | 3.03%c | 2439 | 174 |
FFPE, formalin-fixed paraffin-embedded; ctDNA, circulating tumour DNA; RMS, rhabdomyosarcoma; ACC, adrenocortical carcinoma; VGPR, very good partial response, postinduction, surgical resection after routine induction chemotherapy, AF, allele fraction; F, failed coverage; N/D, not detected.
Below limit of detection.
Detected in tumour only.
Detected in plasma only.
4. Discussion
Comprehensive molecular profiling strategies have been shown to be feasible in children with cancer [9], [10], [11] and show encouraging results. However, wide-scale implementation is impractical in most health-care settings, and even if resources were unlimited, it is also restricted by the availability of biopsy material. We show that using as little as 50 ng of DNA, this assay is an accurate, reproducible and practical platform for molecular stratification and identification of actionable targets, required to accelerate precision medicine clinical trials in childhood tumours.
We are aware that although capture-based panel sequencing is an excellent tool, it has limitations. With our targeted panel approach, only a small portion of the genome is sequenced, and therefore, it is not always possible to distinguish between focal gains or deletions and larger chromosomal gains or losses. Therefore, in version 3 of the panel, we are incorporating a new assay to determine this, which includes probes located across the chromosomes. In addition, as novel gene discoveries and/or targeted inhibitors become available, a wider approach is required for certain indications including a more extensive method for detection of structural variants/translocations. Capture NGS panels are able to detect translocations in DNA with the ability to determine the single-nucleotide breakpoint, so long as those breakpoints occur in or close to a targeted region. We used MANTA to detect spanning pair reads and split reads, thereby identifying fusion gene partners. However, detection of fusion genes is inevitably restricted. We are therefore currently validating a panel using anchored multiplex PCR-based enrichment to detect fusions from RNA, removing the need to sequence long and complex intronic regions. Furthermore, methylation profiling is particularly relevant for precise diagnostic classification of central nervous system (CNS) tumours, many of which harbour few if any recurrent somatic alterations.
Therefore, in the Stratified Medicine Paediatrics (SMPaeds) national molecular profiling study for children with relapsed and refractory cancers, we will retain the practical advantages of panel sequencing and run this alongside other more comprehensive profiling modalities including WES, RNA-seq, low-coverage WGS and methylation to support biomarker-driven clinical trials in the UK, such as eSMART [41]. Furthermore, where sufficient tissue is available, concurrent analysis via the National Health Service England WGS programme will be compared with SMPaeds genomic and clinical data. This approach will provide an unbiased assessment of the clinical utility and cost-effectiveness of multiple different modalities to enable formal recommendations for implementation into routine molecular diagnostics.
Despite the high detection rate of potentially actionable alterations, few patients received treatment with targeted agents. The reasons for this were multifactorial and include the following: lack of available clinical trials, difficulties accessing novel drugs on a compassionate-use basis and/or clinical deterioration of the patient. In addition, although many patients had relapsed/refractory disease, a considerable proportion of patients were still on either first-line therapy or proven standard relapse therapies at the time of sequencing. A number of patients were also enrolled in available phase I/II trials that did not require biomarker screening.
This was a pilot study, requiring retrieval of archival tissue, batching of samples for sequencing and infrequent MTBs. However, for the prospective SMPaeds study, which mandates biopsy at relapse for molecular preselection for clinical trials, samples will be processed in a clinically relevant time frame, which after clinical feedback is currently 3–4 weeks, with the final goal of returning data in two weeks. For children with primary solid tumours (who are not enrolled in SMPaeds), as a result of this study, NGS panel sequencing on the paediatric solid tumour panel v2 is now offered in the UK as part of routine National Health Service diagnostic testing with a turnaround time of 4 weeks from sample dispatch to reporting. Owing to ethical and consent constraints, we were not permitted to report germline findings in the present study. However, given the obvious clinical importance of predisposing mutations in paediatric cancer, we have now obtained suitable consents to report germline mutations via an accredited genetics clinic at Great Ormond Street Hospital.
The sequencing of paired tumour samples at different times demonstrates the importance of tumour heterogeneity and evolution, adding to the mounting literature in support of the clinical importance of biopsy at relapse for children with cancer [19], [42]. Notably, many tumour mutations emerging at the time of relapse (PTEN, NF1, PIK3CA and TSC2) are recognised predictive biomarkers of a targeted therapeutic response.
Although sequencing tissue samples of patients is crucial, liquid biopsies offer the possibility of a non-invasive source for tumour genotyping and disease monitoring. Our preliminary findings from a small number of children demonstrate that high-depth sequencing of ctDNA can identify actionable somatic variants. We also identified some discrepancies between tumour and plasma, most likely a reflection of tumour heterogeneity and evolution. However, large-scale validation studies comparing tumour and serial ctDNA findings in children with cancer are needed to define the clinical utility of ctDNA analysis, for which a bespoke ctDNA panel for paediatric solid tumours is currently being developed to be incorporated as part of the diagnostics pipeline.
In summary, we demonstrate the value of targeted gene sequencing as a practical and cost-effective clinical tool to enable improved diagnosis, prognostication and therapeutic stratification for children with cancer.
Acknowledgements
This work was supported by Christopher's Smile, the National Institute of Health Research (NIHR) Royal Marsden Biomedical Research Centre (BRC), Children With Cancer UK (CWC UK) Cancer Research UK (CRUK), Abbie's Fund, the Rosetree Trust and the KiCa Fund, managed by the King Baudouin Foundation. Roche provided support for Panel development. T.S.J. is funded by The Brain Tumour Charity, CWC UK, GOSH Children's Charity (GOSH CC), CRUK, the Olivia Hodson Cancer Fund and the NIHR GOSH BRC. J.A. and D.H. are funded by the GOSH CC and NIHR GOSH BRC. L.V.M. is funded by the Oak Foundation. The authors thank all participants and the CCLG Tissue Bank for access to samples and contributing CCLG Centres, including members of the ECMC Paediatric network. The CCLG Tissue Bank is funded by Cancer Research UK and CCLG.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ejca.2019.07.027.
Conflict of interest statement
There are no known conflicts of interest associated with this publication, and there has been no significant financial support for this work that could have influenced its outcome.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Fig. S1.
Validation metrics for version 2 of the paediatric panel. Comparison of quality control metrics in ten patient samples run in version 1 and version 2 of the paediatric panel: number of reads (A), percentage of unique on-target reads (B), percentage of mapped unique on-target reads (C) and percentage of duplicates (D). Correlation of allele frequencies obtained by NGS and ddPCR for cancer variants in horizon cell blends (76 SNPs and 28 indels) (E). Correlation of allele frequencies obtained by version 1 and version 2 for cancer variants in FFPE samples (60 SNPs and 2 indels) (F). NGS, next-generation sequencing; indels, insertions and deletions; VAF, variant allele frequency; FFPE, formalin-fixed paraffin-embedded; SNP, single nucleotide polymorphism; ddPCR, droplet digital polymerase chain reaction.
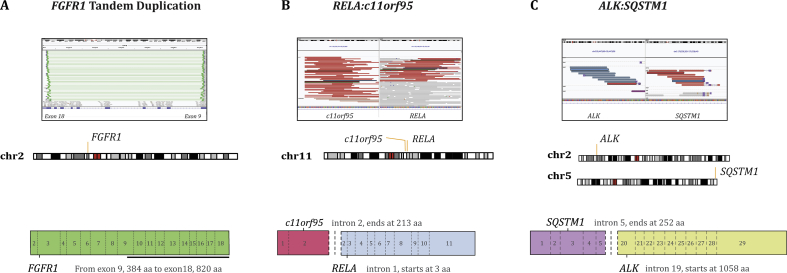
Fig. S2.
Illustration of structural variants detected by the panel. A snapshot of IGV showing spanning reads covering the structural variant region is shown on the top, and a cartoon illustration of the structural variant is displayed in the bottom. FGFR1 tandem duplication from exon 9 to exon 18 detected in a patient with glioma (A); fusion between exon 2 of c11orf95 and exon 2 of RELA detected in a patient with ependymoma (B); fusion between exon 5 of SQSTM1 and exon 20 of ALK detected in a patient with an inflammatory myofibroblastic tumour (C). IGV, integrative genomics viewer.
References
- 1.Fiore R.N., Goodman K.W. Precision medicine ethics: selected issues and developments in next-generation sequencing, clinical oncology, and ethics. Curr Opin Oncol. 2016;28(1):83–87. doi: 10.1097/CCO.0000000000000247. [DOI] [PubMed] [Google Scholar]
- 2.Tuff-Lacey A. A collaborative approach to enabling stratified cancer medicine in the UK. Drug Discov Today. 2015;20(12):1414–1418. doi: 10.1016/j.drudis.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 3.Middleton G. The National Lung Matrix Trial: translating the biology of stratification in advanced non-small-cell lung cancer. Ann Oncol. 2015;26(12):2464–2469. doi: 10.1093/annonc/mdv394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Do K., O'Sullivan Coyne G., Chen A.P. An overview of the NCI precision medicine trials-NCI MATCH and MPACT. Chin Clin Oncol. 2015;4(3):31. doi: 10.3978/j.issn.2304-3865.2015.08.01. [DOI] [PubMed] [Google Scholar]
- 5.Kim G. FDA approval summary: olaparib monotherapy in patients with deleterious germline BRCA-mutated advanced ovarian cancer treated with three or more lines of chemotherapy. Clin Cancer Res. 2015;21(19):4257–4261. doi: 10.1158/1078-0432.CCR-15-0887. [DOI] [PubMed] [Google Scholar]
- 6.Loong H.H. Crizotinib in the management of advanced-stage non-small-cell lung cancer. Future Oncol. 2015;11(5):735–745. doi: 10.2217/fon.14.314. [DOI] [PubMed] [Google Scholar]
- 7.Stagno F. Imatinib mesylate in chronic myeloid leukemia: frontline treatment and long-term outcomes. Expert Rev Anticancer Ther. 2016;16(3):273–278. doi: 10.1586/14737140.2016.1151356. [DOI] [PubMed] [Google Scholar]
- 8.Siegel R., Naishadham D., Jemal A. Cancer statistics. CA Cancer J Clin. 2012;62(1):10–29. doi: 10.3322/caac.20138. 2012. [DOI] [PubMed] [Google Scholar]
- 9.Mody R.J. Integrative clinical sequencing in the management of refractory or relapsed cancer in youth. J Am Med Assoc. 2015;314(9):913–925. doi: 10.1001/jama.2015.10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Worst B.C. Next-generation personalised medicine for high-risk paediatric cancer patients - the INFORM pilot study. Eur J Cancer. 2016;65:91–101. doi: 10.1016/j.ejca.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 11.Harttrampf A.C. Molecular screening for cancer treatment optimization (MOSCATO-01) in pediatric patients: a single-institutional prospective molecular stratification trial. Clin Cancer Res. 2017;23(20):6101–6112. doi: 10.1158/1078-0432.CCR-17-0381. [DOI] [PubMed] [Google Scholar]
- 12.Harris M.H. Multicenter feasibility study of tumor molecular profiling to inform therapeutic decisions in advanced pediatric solid tumors: the individualized cancer therapy (iCat) study. JAMA Oncol. 2016;2(5):608–615. doi: 10.1001/jamaoncol.2015.5689. [DOI] [PubMed] [Google Scholar]
- 13.Ortiz M.V. Integrating genomics into clinical pediatric oncology using the molecular tumor board at the memorial sloan kettering cancer center. Pediatr Blood Cancer. 2016;63(8):1368–1374. doi: 10.1002/pbc.26002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castel D. Histone H3F3A and HIST1H3B K27M mutations define two subgroups of diffuse intrinsic pontine gliomas with different prognosis and phenotypes. Acta Neuropathol. 2015;130(6):815–827. doi: 10.1007/s00401-015-1478-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwartzentruber J. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482(7384):226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 16.Peifer M. Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature. 2015;526(7575):700–704. doi: 10.1038/nature14980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Izquierdo E. Development of a targeted sequencing approach to identify prognostic, predictive and diagnostic markers in paediatric solid tumours. Oncotarget. 2017;8(67):112036–112050. doi: 10.18632/oncotarget.23000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Padovan-Merhar O.M. Enrichment of targetable mutations in the relapsed neuroblastoma genome. PLoS Genet. 2016;12(12) doi: 10.1371/journal.pgen.1006501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen B. Pediatric oncology provider views on performing a biopsy of solid tumors in children with relapsed or refractory disease for the purpose of genomic profiling. Ann Surg Oncol. 2016;23(Suppl 5):990–997. doi: 10.1245/s10434-016-5453-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson J.C. Detection of therapeutically targetable driver and resistance mutations in lung cancer patients by next-generation sequencing of cell-free circulating tumor DNA. Clin Cancer Res. 2016;22(23):5772–5782. doi: 10.1158/1078-0432.CCR-16-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rothe F. Plasma circulating tumor DNA as an alternative to metastatic biopsies for mutational analysis in breast cancer. Ann Oncol. 2014;25(10):1959–1965. doi: 10.1093/annonc/mdu288. [DOI] [PubMed] [Google Scholar]
- 22.Xu S. Circulating tumor DNA identified by targeted sequencing in advanced-stage non-small cell lung cancer patients. Cancer Lett. 2016;370(2):324–331. doi: 10.1016/j.canlet.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bettegowda C. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Mattos-Arruda L., Caldas C. Cell-free circulating tumour DNA as a liquid biopsy in breast cancer. Mol Oncol. 2016;10(3):464–474. doi: 10.1016/j.molonc.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alix-Panabieres C., Pantel K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov. 2016;6(5):479–491. doi: 10.1158/2159-8290.CD-15-1483. [DOI] [PubMed] [Google Scholar]
- 26.Kurihara S. Circulating free DNA as non-invasive diagnostic biomarker for childhood solid tumors. J Pediatr Surg. 2015;50(12):2094–2097. doi: 10.1016/j.jpedsurg.2015.08.033. [DOI] [PubMed] [Google Scholar]
- 27.Combaret V. Detection of tumor ALK status in neuroblastoma patients using peripheral blood. Cancer Med. 2015;4(4):540–550. doi: 10.1002/cam4.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panditharatna E. Clinically relevant and minimally invasive tumor surveillance of pediatric diffuse midline gliomas using patient-derived liquid biopsy. Clin Cancer Res. 2018;24(23):5850–5859. doi: 10.1158/1078-0432.CCR-18-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jimenez I. Circulating tumor DNA analysis enables molecular characterization of pediatric renal tumors at diagnosis. Int J Cancer. 2019;144(1):68–79. doi: 10.1002/ijc.31620. [DOI] [PubMed] [Google Scholar]
- 30.Chicard M. Whole-exome sequencing of cell-free DNA reveals temporo-spatial heterogeneity and identifies treatment-resistant clones in neuroblastoma. Clin Cancer Res. 2018;24(4):939–949. doi: 10.1158/1078-0432.CCR-17-1586. [DOI] [PubMed] [Google Scholar]
- 31.Allin D.M. Circulating tumour DNA is a potential biomarker for disease progression and response to targeted therapy in advanced thyroid cancer. Eur J Cancer. 2018;103:165–175. doi: 10.1016/j.ejca.2018.08.013. [DOI] [PubMed] [Google Scholar]
- 32.Chakravarty D. vol. 2017. JCO Precis Oncol; 2017. OncoKB: a precision oncology knowledge base. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forbes S.A. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45(D1):D777–D783. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grobner S.N. The landscape of genomic alterations across childhood cancers. Nature. 2018;555(7696):321–327. doi: 10.1038/nature25480. [DOI] [PubMed] [Google Scholar]
- 35.Gatz S.A. Chemotherapy responsiveness in a patient with multiply relapsed ameloblastic fibro-odontosarcoma of the maxilla. Pediatr Blood Cancer. 2015;62(11):2029–2032. doi: 10.1002/pbc.25627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee L., Gupta M., Sahasranaman S. Immune Checkpoint inhibitors: an introduction to the next-generation cancer immunotherapy. J Clin Pharmacol. 2016;56(2):157–169. doi: 10.1002/jcph.591. [DOI] [PubMed] [Google Scholar]
- 37.Kohsaka S. A recurrent neomorphic mutation in MYOD1 defines a clinically aggressive subset of embryonal rhabdomyosarcoma associated with PI3K-AKT pathway mutations. Nat Genet. 2014;46(6):595–600. doi: 10.1038/ng.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pajtler K.W. Molecular classification of ependymal tumors across all CNS compartments, histopathological grades, and age groups. Cancer Cell. 2015;27(5):728–743. doi: 10.1016/j.ccell.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whittaker S.R. A genome-scale RNA interference screen implicates NF1 loss in resistance to RAF inhibition. Cancer Discov. 2013;3(3):350–362. doi: 10.1158/2159-8290.CD-12-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Allen E.M. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 2014;4(1):94–109. doi: 10.1158/2159-8290.CD-13-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.European Proof-of Concept Therapeutic Stratification Trial of Molecular Anomalies in Relapsed or Refractory Tumours.https://clinicaltrials.gov/ct2/show/NCT02813135?term=esmart&rank=5.
- 42.Eleveld T.F. Relapsed neuroblastomas show frequent RAS-MAPK pathway mutations. Nat Genet. 2015;47(8):864–871. doi: 10.1038/ng.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.