Abstract
The development of next-generation therapies for neuropsychiatric illness will likely rely on a precise and accurate understanding of human brain dynamics. Toward this end, researchers have focused on collecting large quantities of neuroimaging data. For simplicity, we will refer to large cross-sectional neuroimaging studies as broad studies and to intensive longitudinal studies as deep studies. Recent progress in identifying illness subtypes and predicting treatment response in neuropsychiatry has been supported by these study designs, along with methods bridging machine learning and network science. Such methods combine analytic power, interpretability, and direct connection to underlying theory in cognitive neuroscience. Ultimately, we propose a general framework for the treatment of neuropsychiatric illness relying on the findings from broad and deep studies combined with basic cognitive and physiologic measurements.
Introduction
Neuropsychiatric illness has widespread and devastating effects on populations around the world, affecting approximately 20% of individuals in the U.S. alone [1]. Converging evidence from genetic, behavioral, and neuroimaging [2,3] studies has demonstrated overlapping pathological features in these disorders, suggesting that both common and unique pathophysiological mechanisms underlie clinical symptoms such as anxiety, depression, and psychosis. Accordingly, the classic notion of discrete psychiatric syndromes defined by clinical symptoms [4] is being challenged by more biologically and empirically driven models that link brain and behavior [5]. High rates of comorbidity between disorders hamper the identification of generalizable pathophysiological principles, similar to those that allow us to understand dysfunction of less complex internal organs. The dearth of such principles may partially explain the fact that a large cohort of patients do not respond to psychotherapy, psychopharmacologics [6], and brain stimulation protocols [7•]. Indeed, a marked consequence of the brain’s vast complexity is the existence of many distinct and overlapping pathways for cognitive function and dysfunction, constituting a major challenge in developing accurate diagnoses and predicting individual responses to treatment.
How, if ever, can we elucidate and intervene on these overlapping pathophysiological mechanisms that underlie neuropsychiatric illness? Recent efforts toward this aim have focused on the acquisition of human neuroimaging data sets with samples of unprecedented size [8•,9–11]. These so-called broad studies provide an excellent picture of between-individual or population-level variance, allowing the prediction of treatment response from high-dimensional neuroimaging and affective phenotypes based on methods from network neuroscience and machine learning [12••]. The widespread application of such methods has been facilitated by advances in computer processing power and repurposing of graphics processing units (GPUs) for machine learning. In complementary efforts, researchers have also collected data with repeated measures on a small cohort or single individual. These so-called deep studies have generated insights into the substantial within-individual variation in neuroimaging phenotypes that occurs on the scale of days, weeks, and months [13•,14,15]. Daily changes in neuroimaging phenotypes have also been linked to variability in behavioral and affective profiles [16], suggesting that temporal derivatives of neuroimaging phenotypes may contain unique, neuropsychiatrically relevant information. As such, both broad and deep studies have uniquely contributed to our understanding of healthy neurophysiology and neuropsychopathology.
While initial progress has been made through these unique forms of big data, neuropsychiatry still remains far from the goal of using generalizable principles to develop and deliver treatment. In this review, we begin by describing the results of broad and deep studies in more detail, along with methods well-suited for each study type. Next, we describe a framework to maximize the clinical translatability of broad and deep neuroimaging studies. Specifically, we posit that broad studies can inform models that identify who would benefit from intervention and how to intervene, while deep studies can inform models that suggest when to intervene. Network science and machine learning serve as the foundations for these models and will undoubtedly play a critical role in the coming generation of neuropsychiatric care.
Informing diagnosis and treatment through large population-level studies
Within the past decade, the neuroimaging community has seen the emergence of broad neuroimaging studies with historically large sample sizes (Figure 1). The Human Connectome Project [9], the UK Biobank [8•], the Philadelphia Neurodevelopmental Cohort [11], and Enhancing Neuroimaging Genetics through Meta-Analysis (ENIGMA) [10] have each generated valuable insights into the relations among brain structure, brain function, and behavior. Importantly, analyses of these data have increasingly relied on methods from network neuroscience [17•], an emerging field that provides elegant approaches for the quantitative description of complex multivariable phenotypes in brain anatomy and physiology. In large-scale brain networks, one can succinctly capture the collective role of several regions simultaneously through node-level metrics, such as the participation coefficient [18,19], controllability [20], hubness, nodal efficiency [21], and weighted degree [18] (Box 1). These metrics can be readily computed using freely available code [22,23]. Notably, these statistics are influenced by the topology of the entire network, change dynamically over time in functional networks [24], and are altered in neuropsychiatric disease [25–27]. To combine brain network models with clinical, behavioral, genetic, and cognitive data requires the use of multivariate statistical approaches that acknowledge the complexity of each of these data types by jointly accounting for their covariance structure [8•,28]. Sparse canonical correlation analysis (sCCA) [29] and partial least squares (PLS) [30] are two examples of such multivariate statistical methods that are well-suited to identify covariance patterns between brain networks and high dimensional behavioral, clinical, and genetic data (Figure 1).
Figure 1.
Broad and deep neuroimaging studies. The neuroimaging community has seen the rise of studies with increasingly large sample sizes (broad studies) and increasingly intensive sampling (deep studies). Broad studies (left) typically involve cross-sectional sampling of a specific population. Multivariate statistics and deep neural-network classifiers are two examples of methods that are well-suited to identify high-dimensional patterns between brain network, behavioral, clinical, and genetic phenotypes. Ultimately, such models might lead to the automated classification of neuropsychiatric illness based on neurobehavioral phenotypes, along with predictions of responses to various treatment options. Deep studies (right) typically involve intensive, repeated sampling of a small number of individuals longitudinally over days, months, or years. Multilayer network models can capture how the interaction between different components of brain activity, transcriptomes, metabolomes, or behavior changes over time. Here (middle right), each node represents a component and the time-varying edges represent their time-dependent interactions. Such models could prove particularly powerful for delineating how variability in individual networks over time affects treatment response.
Box 1. Modularity
Complex networks often contain non-trivial clustering in the form of modularity, in which groups of nodes exist that are more densely connected with each other than with nodes in other modules [18,31].
Network density
A fully dense network is one in which a connection exists between every possible pair of nodes. The density of a network is the number of existing connections divided by the number of possible connections.
Participation coefficient
Participation coefficient quantifies the extent to which a node sits on the boundary of multiple modules [18,19,31,32], poised to coordinate activity between functional systems of the brain [33].
Weighted degree
The weighted degree, or strength, of a node is the sum of its connection weights. Weighted degree can be further broken down into within-module and between-module degree, referring to the strength of a node’s connections to nodes in the same module or in other modules.
Hubs
Hubs are brain regions with unique roles in structural and functional networks due to their many, and often diverse, connections with other brain regions. Hubs are often disrupted in neuropsychiatric illness [34,26]. Hubs can be defined in several ways [18,19,32,35,36], often relying on a balance between participation coefficient and within-module degree.
Nodal efficiency
Nodal efficiency is a measure related to the average number of nodes that must be traversed to go from a given node to all other nodes [21,37]. This quantifies how an individual node contributes to the small world properties of brain networks [38].
Controllability
Unlike the above metrics, which quantify the static topological role of nodes in a network, network control theory [17•,23] uses a dynamical systems perspective to quantify the ability of each node to support transitions between states of activity. Two common metrics are average and modal controllability, which capture the ability of regional input to drive nearby or distant state transitions [23,39]. These principles have been explored in neuropsychiatric illness [25,40], over development [41], and across species [42].
Machine learning classifiers have demonstrated clear promise for neuropsychiatric diagnosis [43••], even with unimodal neuroimaging data. In a multisite study (n = 941), the ENIGMA schizophrenia working group utilized consensus-based classifiers to distinguish individuals with schizophrenia from healthy controls with 76% accuracy using structural MRI alone [44]. However, the clinical utility of such classifiers may not be realized until they are able to distinguish a particular disorder from a heterogeneous clinical population rather than healthy controls. Results from studies based on multivariate statistics suggest that the inclusion of clinical and behavioral data may help classifiers resolve this heterogeneity. In a large multisite study (n = 1188), CCA was used to define biotypes of major depressive disorder (MDD) based on resting state functional connectivity and clinical symptoms, allowing for diagnosis of depression with 85–90% accuracy in a replication set and prediction of positive response to transcranial magnetic stimulation (TMS) [12••]. The model was also able to distinguish MDD from schizophrenia more easily than from generalized anxiety disorder, reflecting the varying degrees of overlap in neurobehavioral phenotypes between different forms of mental illness. Importantly, individual patient data from independent samples can be fed into these models to generate priors for clinicians. In the near future, these models are likely to become increasingly powerful as open data sharing practices facilitate the growth of training data sets [45]. Such efforts will be critical for generating low dimensional representations of clinical symptoms and network measures of brain structure and function that are useful in the diagnosis and sub-diagnosis of disease, and in the selection of interventions and treatments (Figure 1).
Harnessing individual differences and within-subject dynamics
In contrast to broad studies, which leverage large sample sizes to make inferences about individuals in a defined population, deep studies are particularly suited for investigating the interdependencies between a diverse range of phenotypes that might vary meaningfully over time in single individuals (Figure 1). Perhaps the most impressive deep study is the MyConnectome project [13•], which is the first to describe the existence and nature of a complex interactome between resting state functional connectivity, transcriptomics, metabolomics, food intake, and behavior over the course of 532 days. It is interesting to consider the potential for such an interactome to inform the development of targeted neuromodulatory interventions that depend on the state of the brain at the time of stimulation [46]. Indeed, daily variation in brain network connectivity could confound the effects of stimulation, leading to mixed responses to such treatments for depression [7•]. To better understand these temporal variations, one can consider using multilayer network models, which can identify changes in network structure over time by taking into account the interactions between network components and the interactions within each network component with time [47] (Figure 1). One can also use linear autoregressive models, Hidden Markov Models, or Long Short-Term Memory recurrent neural networks [48] to predict how a complex, interacting system evolves over time. While the level of depth reported in the MyConnectome project is currently impractical for patient care, it illustrates the complex origins of day-to-day individual variation and — alongside other intensive sampling studies — offers useful benchmarks to inform future data collection [14,49,50••,51].
Typical approaches for ‘parcellation,’ – obtaining representative signals within anatomically [52] or functionally [33] similar regions or ‘parcels’ – tend to rely on registering brain images to a common template space. However, performing targeted manipulations of distributed cognitive systems that exhibit dysfunction in neuropsychiatric illness demands exceptional precision in mapping brain network architecture and function. Thus, the growing focus on subject-specific parcellation to define these parcels independently for each participant or patient is a critical complement to the intensive sampling of deep studies (Figure 2) [14,15,53,54]. Constructing subject-specific parcellations builds on historical work in tumor resection, where neurosurgeons and anesthesiologists perform patient-specific functional mapping of language and motor circuits with fMRI, pharmacology, and electrical stimulation [55,56]. When seeking to map all circuits across the entire brain, one would focus on mapping individual differences in functional topography that might hold diagnostic and prognostic value, with methods that do not depend on warping subject-level volumes to an average brain [50••,57]. Recently, such individualized parcellation techniques have been combined with resting state fMRI to identify novel subnetworks within the default mode network (DMN) [58], a system that has been broadly implicated in virtually every neuropsychiatric illness [59–62]. These observations motivate further studies of individual differences in the distribution of cortical real estate between particular functional networks [63] and their finer subdivisions in the context of neuro-psychiatric illness (Figure 2). In these efforts, deep neuroimaging studies will be particularly important, by providing sufficient data to use subject-specific parcellations. This approach will account for — rather than average over — individual network topographies (Figure 2). Resolving individual differences in spatial topography will facilitate an accurate study of the neural basis of temporal fluctuations in individual symptoms. The richly sampled temporal dimension of deep studies adds a layer of complexity untouched by most broad studies and the individual-oriented methodology improves the accuracy of patient-specific predictions. Ultimately, meta-analysis of deep studies might inform a generalizable approach, if not pathophysiological principles, for making individual predictions of the optimal treatment as a function of time and a more easily measurable subset of variables.
Figure 2.
Subject-specific parcellation uncovers individualized topography of functional networks. (a) Sample depiction of the default mode network in a group-level parcellation, which warps subject volumes to standard space, potentially averaging over important differences in functional network topography. Yellow overlay indicates activity, while colored lines indicate parcel boundaries. (b) Sample depiction of subject-specific activation map (yellow overlay) with group-level parcellation borders (colored lines) overlayed, illustrating the possible variation in subject-specific functional topography.
Using network models to link intensive behavioral assessment with neuroimaging findings
A key counterpart to accurately interpreting changes in functional brain dynamics over time in a patient cohort is the ability to concurrently measure changes in behavior, emotions, and mood — core symptoms of neuropsychiatric illness, which are typically assessed retrospectively in the clinical setting. Experience-sampling (ES) encompasses the measurement of these factors, in addition to physiology, in real time through the use of personal data recording tools [64]. Subject-specific symptom networks can be constructed by computing cross-correlations between measures of different emotions over time, quantifying the cofluctuation of psychiatric symptoms [65•,66,67] (Figure 3). Additionally, directed networks can be constructed using pairwise regression between time-lagged measures, capturing the temporal precedence of symptom fluctuations [68]. Higher order features of these symptom graphs [65•], such as network density (Box 1), are greater in individuals with MDD than in healthy controls [69], suggesting that the temporally dynamic interplay between symptoms may be altered by disease processes. ES also lends itself well to the study of substance use disorders, in which daily emotional variability can trigger relapse [70].
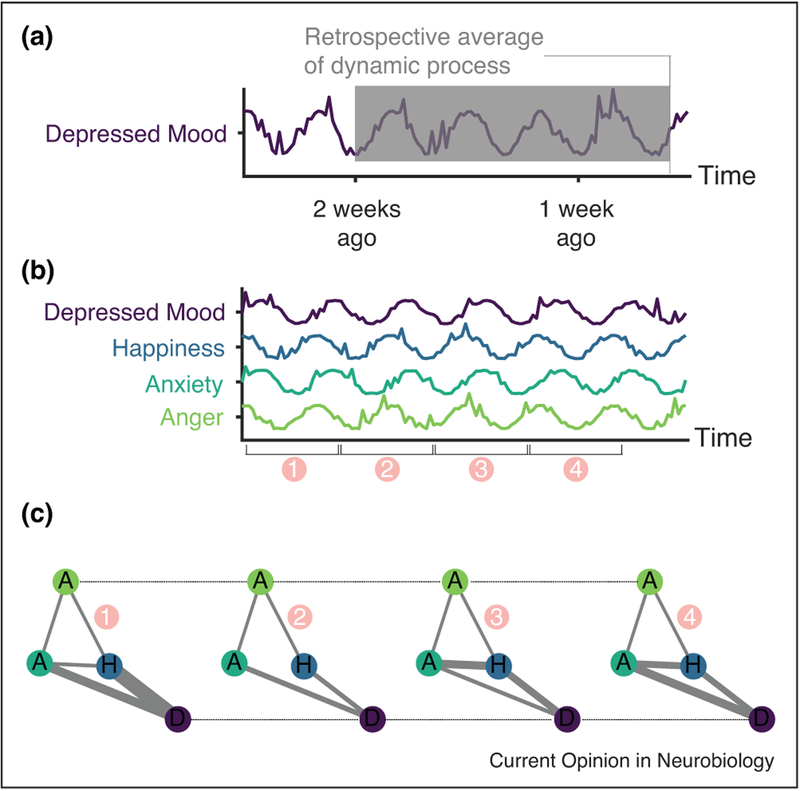
Figure 3.
Measuring mood dynamics with experience sampling. (a) Illustration of symptom measurement by retrospective report, demonstrating how scales of mood symptoms that ask for retrospective reporting average over rich mood dynamics and are subject to recall bias [72–74]. (b) Example time series of mood measurements from the Profile of Mood States [76]. Spikes in the time series indicate rapid changes in mood induced by brief events. The numbering on the x-axis indicates windowing for network construction. (c) Illustration of multilayer emotional network construction from subsequent windows of time series shown in panel (b). Nodes represent mood features, with the letter label corresponding to features in (b), solid gray edges represent the correlation between mood features within a particular time window, and dashed black edges link mood features together across time. Constructing such a network facilitates the application of numerous methods for analyzing the temporal dynamics of multivariate relationships [47].
Major limitations of ES are the burden of repeated assessment on participants and the potential for the process of ES itself to influence symptoms (i.e. reactivity [71]). Nevertheless, commonly used forms [72–74] for evaluating mental health utilize retrospective reporting, assuming stationarity in these dynamic phenotypes [75] (Figure 3). Schizophrenia, for example, is characterized by a lack of insight and poor working memory, and therefore real-time assessment may be more likely to accurately capture cognitive and emotional state than single-shot clinical evaluations or self-report measures. Thus, ES is a highly promising approach for identifying neural correlates of symptom dynamics in complex, overlapping neuropsychiatric pathologies.
The use of ES has begun to enter into neuroimaging studies, though not with the same force as the machine learning techniques described above. In schizophrenia patients, corticostriatal task activation and reduced motor activity were found to predict negative symptoms [77]. Similarly, physiological signs of autonomic dysfunction acquired through wearable technology were associated with positive symptom severity [78]. In a study of patients with anorexia nervosa, reward circuit activity was related to longitudinal body-mass index measurements and body-related rumination [79]. Notably, this particular study used group-level parcellation techniques, indicative of a common disconnect between the use of cutting edge methods in social science and those in neuroscience.
Despite these intriguing findings, no parallels have yet been drawn between basic ES measures, network models of psychiatric symptoms, and structural or functional brain networks. Functional brain network dynamics have been extensively characterized [24,80], and there are likely rich relationships with behavioral and symptom dynamics, as suggested by the MyConnectome project. One could gain traction on these relationships using multilayer network construction with subsequent community detection [47] to draw parallels between dynamic functional networks and symptom networks. Brain regions with high inter-scan variability in functional connectivity and high within-scan community change, that is, flexibility [16], may confer similar variability onto behavior phenotypes.
The use of advanced machine learning techniques for time series analysis, such as recurrent neural networks and Hidden Markov Models, as well as unsupervised multivariate statistical methods, are promising underexplored avenues for finding covariance between complex neural and behavioral phenotypes in neuropsychatric illness. Furthermore, ES could explain temporal variance in cortical excitability [81,82], an important factor in TMS response [83••], and allow for its targeted control. While targeted neuromodulatory treatment paradigms are currently being refined, with the aid of findings from broad studies, ES provides us with useful methods that will help to identify the optimal time in a disease course to deliver these treatments.
Conclusion
Across many academic disciplines, the use of machine learning techniques, often informed by network theory, has skyrocketed in the last decade, concordant with the collection of data with larger (broad) and more intensive (deep) samples. Both broad and deep studies provide the neuroscience community with unique opportunities to advance the diagnosis and treatment of neuropsychiatric illness, with the aid of network science and machine learning. Broad studies allow for network analysis followed by dimensionality reduction and classification for identifying meaningful symptom-neuropathology correspondence and predicting treatment responses. Deep studies demonstrate the importance of individual variability and provide a framework for understanding and manipulating complex, individual phenomes. Experience sampling provides the tools for acquiring intensive repeated physiologic and behavioral measures, the network models of which may have critical unexplored neural correlates. Ultimately, a model using priors derived from broad studies, patient-specific neuroimaging data, and symptom networks might predict the optimal timing and type of treatment for individual patients in real time based on a subset of measurements captured through a personal device. The merger of these techniques has the potential to usher in a next-generation approach to psychiatric care and contribute to our fundamental understanding of the complex relationship between mind, body, and behavior.
Acknowledgements
D.S.B., D.L.S., and E.J.C. acknowledge support from the John D. and Catherine T. MacArthur Foundation, the Alfred P. Sloan Foundation, the ISI Foundation, the Paul Allen Foundation, the Army Research Laboratory (W911NF-10-2-0022), the Army Research Office (Bassett-W911NF-14-1-0679, Grafton-W911NF-16-1-0474, DCIST-W911NF-17-2-0181), the Office of Naval Research, the National Institute of Mental Health (2-R01-DC-009209-11, R01-MH112847, R01-MH107235, R21-M MH-106799), the National Institute of Child Health and Human Development (1R01HD086888-01), National Institute of Neurological Disorders and Stroke (R01 NS099348), and the National Science Foundation (BCS-1441502, BCS-1430087, NSF PHY-1554488 and BCS-1631550). The content is solely the responsibility of the authors and does not necessarily represent the official views of any of the funding agencies. The authors would like to thank Urs Braun for helpful comments on this manuscript.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Substance Abuse and Mental Health Services Administration: Key substance use and mental health indicators in the United States: Results from the 2016 National Survey on Drug Use and Health. Tech. Rep. 1, Center for Behavioral Health Statistics and Quality, Substance Abuse and Mental Health Services Administration, Rockville, MD: 2017:10:. Retrieved from https://www.samhsa.gov/data/. [Google Scholar]
- 2.Goodkind M, Eickhoff SB, Oathes DJ, Jiang Y, Chang A, Jones-Hagata LB, Ortega BN, Zaiko YV, Roach EL, Korgaonkar MS et al. : Identification of a common neurobiological substrate for mental illness. JAMA Psychiatry 2015, 72:305–315 10.1001/jamapsychiatry.2014.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaczkurkin AN, Moore TM, Calkins ME, Ciric R, Detre JA,Elliott MA, Foa EB, Garcia de la Garza A, Roalf DR, Rosen A et al. : Common and dissociable regional cerebral blood flow differences associate with dimensions of psychopathology across categorical diagnoses. Mol Psychiatry 2017. 10.1038/mp.2017.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.APA AAP: Diagnostic and Statistical Manual of Mental Disorders (DSM-5®). 5th ed. Arlington, VA: American Psychiatric Association; 2013. [Google Scholar]
- 5.Insel TR: The NIMH research domain criteria (RDoC) project: Precision medicine for psychiatry. Am J Psychiatry 2014, 171:395–397 10.1176/appi.ajp.2014.14020138. [DOI] [PubMed] [Google Scholar]
- 6.Gaynes BN, Warden D, Trivedi MH, Wisniewski SR, Fava M, Rush AJ: What did STAR*D teach us? Results from a large-scale, practical, clinical trial for patients with depression. Psychiatric Serv 2009, 60:1439–1445 10.1176/ps.2009.60.11.1439. [DOI] [PubMed] [Google Scholar]
- 7.•.Rachid F: Maintenance repetitive transcranial magnetic stimulation (rTMS) for relapse prevention in with depression: a review. Psychiatry Res 2018, 262:363–372 10.1016/j.psychres.2017.09.009. [DOI] [PubMed] [Google Scholar]; This review provides a thorough overview of studies examining the safety and efficacy of maintenance repetitive TMS in acute responders to repetitive TMS. The variable success in response to maintenance treatment reflects the importance of understanding temporal fluctuations in symptoms.
- 8.•.Miller KL, Alfaro-Almagro F, Bangerter NK, Thomas DL, Yacoub E, Xu J, Bartsch AJ, Jbabdi S, Sotiropoulos SN, Andersson JL et al. : Multimodal population brain imaging in the UK Biobank prospective epidemiological study. Nat Neurosci 2016, 19:1523–1536 10.1038/nn.4393. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper describes the collection and initial analysis of data from the UK Biobank. Mass univariate testing as well as multivariate statistics are employed to reveal population associations between brain structure, function and clinical data.
- 9.Van Essen DC, Smith SM, Barch DM, Behrens TE, Yacoub E, Ugurbil K: The WU-Minn human connectome project: an overview. NeuroImage 2013, 80:62–79 10.1016/j.neuroimage.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson PM, Stein JL, Medland SE, Hibar DP, Vasquez AA, Renteria ME, Toro R, Jahanshad N, Schumann G, Franke B et al. : The ENIGMA Consortium: large-scale collaborative analyses of neuroimaging and genetic data. Brain Imaging Behav 2014, 8:153–182 10.1007/s11682-013-9269-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Satterthwaite TD, Elliott MA, Ruparel K, Loughead J, Prabhakaran K, Calkins ME, Hopson R, Jackson C, Keefe J, Riley M et al. : Neuroimaging of the philadelphia neurodevelopmental cohort. NeuroImage 2014, 86:544–553 10.1016/j.neuroimage.2013.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.••.Drysdale AT, Grosenick L, Downar J, Dunlop K, Mansouri F, Meng Y, Fetcho RN, Zebley B, Oathes DJ, Etkin A et al. : Resting-state connectivity biomarkers define neurophysiological subtypes of depression. Nat Med 2017, 23:28–38 10.1038/nm.4246. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study applies unsupervised learning and hierarchical clustering to multivariate symptom and brain network measurement to identify bio-types of depression which predict response to repetitive TMS. The authors demonstrate out-of-sample replicability of this method for diagnosing depression and distinguishing mood disorders from psychotic disorders.
- 13.•.Poldrack RA, Laumann TO, Koyejo O, Gregory B, Hover A, Chen MY, Gorgolewski KJ, Luci J, Joo SJ, Boyd RL et al. : Long-term neural and physiological phenotyping of a single human. Nat Commun 2015, 6:8885 10.1038/ncomms9885. [DOI] [PMC free article] [PubMed] [Google Scholar]; This publication describes initial results from the MyConnectome project, which is perhaps the most intensive individual studies to date. The study demonstrates daily variability in functional connectome structure and a complex network of interaction between gene expression, metabolomics, behavior and brain networks.
- 14.Laumann TO, Gordon EM, Adeyemo B, Snyder AZ, Joo SJ, Chen MY, Gilmore AW, McDermott KB, Nelson SM, Dosenbach NU et al. : Functional system and areal organization of a highly sampled individual human brain. Neuron 2015, 87:658–671 10.1016/j.neuron.2015.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finn ES, Shen X, Scheinost D, Rosenberg MD, Huang J, Chun MM, Papademetris X, Constable RT: Functional connectome fingerprinting: identifying individuals using patterns of brain connectivity. Nat Neurosci 2015, 18:1664–1671 10.1038/nn.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Betzel RF, Satterthwaite TD, Gold JI, Bassett DS: Positive affect, surprise, and fatigue are correlates of network flexibility. Sci Rep 2017, 7:520 10.1038/s41598-017-00425-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.•.Bassett DS, Sporns O: Netw Neurosci 2017. 10.1038/nn.4502. [DOI] [PMC free article] [PubMed]; Network neuroscience is an interdisciplinary field at the intersection of graph theory, network science, neuroscience, psychology and physics. This review highlights key network methods and emphasizes the importance of considering that the brain has components that appear functionally independent but ultimately acts as an integrated system.
- 18.Sporns O, Betzel RF: Modular brain networks. Annu Rev Psychol 2016, 67:613–640 10.1146/annurev-psych-122414-033634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bertolero MA, Yeo BTT, Bassett DS, D’Esposito M: A mechanistic model of connector hubs, modularity and cognition. Nat Hum Behav 2018, 2:765–777 10.1038/s41562-018-0420-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu S, Pasqualetti F, Cieslak M, Telesford QK, Alfred BY, Kahn AE, Medaglia JD, Vettel JM, Miller MB, Grafton ST et al. : Controllability of structural brain networks. Nat Commun 2015:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Latora V, Marchiori M: Efficient behavior of small-world networks. Phys Rev Lett 2001, 87:198701 10.1103/PhysRevLett.87.198701. [DOI] [PubMed] [Google Scholar]
- 22.Rubinov M, Sporns O: Complex network measures of brain connectivity: uses and interpretations. NeuroImage 2010, 52:1059–1069 10.1016/J.NEUROIMAGE.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 23.Gu S, Pasqualetti F, Cieslak M, Telesford QK, Yu AB, Kahn AE, Medaglia JD, Vettel JM, Miller MB, Grafton ST et al. : Controllability of structural brain networks. Nat Commun 2015, 6 10.1038/ncomms9414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shine JM, Bissett PG, Bell PT, Koyejo O, Balsters JH, Gorgolewski KJ, Moodie CA, Poldrack RA: The dynamics of functional brain networks: integrated network states during cognitive task performance. Neuron 2016, 92:544–554 10.1016/j.neuron.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeganathan J, Perry A, Bassett DS, Roberts G, Mitchell PB, Breakspear M:: Fronto-limbic dysconnectivity leads to impaired brain network controllability in young people with bipolar disorder and those at high genetic risk. NeuroImage: Clin 2018, 19:71–81 10.1016/j.nicl.2018.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Den Heuvel MP, Sporns O, Collin G, Scheewe T, Mandl RC, Cahn W, Goni J, Pol HE, Kahn RS: Abnormal rich club organization and functional brain dynamics in schizophrenia. JAMA Psychiatry 2013, 70:783–792 10.1001/jamapsychiatry.2013.1328. [DOI] [PubMed] [Google Scholar]
- 27.Li M, Chen Z, Li T: Small-world brain networks in schizophrenia. Shanghai Arch Psychiatry 2012, 24:322–327 10.3969/j.issn.1002-0829.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith SM, Nichols TE, Vidaurre D, Winkler AM, Behrens TE, Glasser MF, Ugurbil K, Barch DM, Van Essen DC, Miller KL: A positive-negative mode of population covariation links brain connectivity, demographics and behavior. Nat Neurosci 2015, 18:1565–1567 10.1038/nn.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Avants BB, Cook PA, Ungar L, Gee JC, Grossman M: Dementia induces correlated reductions in white matter integrity and cortical thickness: a multivariate neuroimaging study with sparse canonical correlation analysis. NeuroImage 2010, 50:1004–1016 10.1016/j.neuroimage.2010.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krishnan A, Williams LJ, McIntosh AR, Abdi H: Partial Least Squares (PLS) methods for neuroimaging: a tutorial and review. NeuroImage 2011, 56:455–475 10.1016/j.neuroimage.2010.07.034 Multivariate Decoding and Brain Reading. [DOI] [PubMed] [Google Scholar]
- 31.Sporns O: Contributions and challenges for network models in cognitive neuroscience. Nat Neurosci 2014, 17:652–660 10.1038/nn.3690. [DOI] [PubMed] [Google Scholar]
- 32.Bertolero MA, Yeo BTT, D’Esposito M: The diverse club. Nat Commun 2017, 8:1277 10.1038/s41467-017-01189-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas Yeo BT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, Roffman JL, Smoller JW, Zollei L, Polimeni JR et al. : The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol 2011, 106:1125–1165 10.1152/jn.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buckner RL, Sepulcre J, Talukdar T, Krienen FM, Liu H, Hedden T, Andrews-Hanna JR, Sperling RA, Johnson KA: Neurobiology of disease cortical hubs revealed by intrinsic functional connectivity: mapping, assessment of stability, and relation to Alzheimer’s disease. 2009. 10.1523/JNEUROSCI.5062-08.2009. [DOI] [PMC free article] [PubMed]
- 35.van den Heuvel MP, Sporns O: Rich-club organization of the human connectome. J Neurosci 2011, 31:15775–15786 10.1523/JNEUROSCI.3539-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gordon EM, Lynch CJ, Gratton C, Petersen SE, Dosenbach NUF, Nelson Correspondence SM: Three distinct sets of connector hubs integrate human brain function. Cell Rep 2018, 24:1687–1695e4 10.1016/j.celrep.2018.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hilger K, Ekman M, Fiebach CJ, Basten U: Efficient hubs in the intelligent brain: nodal efficiency of hub regions in the salience network is associated with general intelligence. Intelligence 2017, 60:10–25 10.1016/J.INTELL.2016.11.001. [DOI] [Google Scholar]
- 38.Bassett DS, Bullmore E: Small-world brain networks. Neuroscientist 2006, 12:512–523 10.1177/1073858406293182. [DOI] [PubMed] [Google Scholar]
- 39.Pasqualetti F, Zampieri S, Bullo F: Controllability metrics, limitations and algorithms for complex networks. IEEE Trans Control Netw Syst 2014, 1:40–52 10.1109/TCNS.2014.2310254. [DOI] [Google Scholar]
- 40.Khambhati AN, Davis KA, Lucas TH, Litt B, Bassett DS: Virtual cortical resection reveals push-pull network control preceding seizure evolution. Neuron 2016, 91:1170–1182 10.1016/j.neuron.2016.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang E, Giusti C, Baum GL, Gu S, Pollock E, Kahn AE, Roalf DR, Moore TM, Ruparel K, Gur RC et al. : Developmental increases in white matter network controllability support a growing diversity of brain dynamics. Nat Commun 2017, 8 10.1038/s41467-017-01254-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim J, Soffer JM, Kahn AE, Vettel JM, Pasqualetti F, Bassett DS: Topological principles of control in dynamical network systems. arXiv 2017, 1702:354. [Google Scholar]
- 43.••.Arbabshirani MR, Plis S, Sui J, Calhoun VD: Single subject prediction of brain disorders in neuroimaging: promises and pitfalls. NeuroImage 2017, 145:137–165 10.1016/j.neuroimage.2016.02.079. [DOI] [PMC free article] [PubMed] [Google Scholar]; This review article provides exhaustive coverage (240 papers) of the efforts to date in diagnosing neuropsychiatric illness at the patient level by applying supervised learning methods to neuroimaging data. The authors discuss the use of several machine learning methods and emphasize the distinction between statistically significant group-level effects and diagnostic accuracy.
- 44.Rozycki M, Satterthwaite TD, Koutsouleris N, Erus G, Doshi J, Wolf DH, Fan Y, Gur RE, Gur RC, Meisenzahl EM et al. : Multisite machine learning analysis provides a robust structural imaging signature of schizophrenia detectable across diverse patient populations and within individuals. Schizophrenia Bull 2017. 10.1093/schbul/sbx137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poldrack RA, Gorgolewski KJ: Making big data open: data sharing in neuroimaging. Nat Neurosci 2014, 17:1510–1517 10.1038/nn.3818. [DOI] [PubMed] [Google Scholar]
- 46.Silvanto J, Pascual-Leone A: State-dependency of transcranial magnetic stimulation. Brain Topogr 2008, 21:1–10 10.1007/s10548-008-0067-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muldoon SF, Bassett DS: Network and multilayer network approaches to understanding human brain dynamics. Philos Sci 2016, 83:710–720 10.1086/687857. [DOI] [Google Scholar]
- 48.Längkvist M, Karlsson L, Loutfi A: A review of unsupervised feature learning and deep learning for time-series modeling. Pattern Recogn Lett 2014, 42:11–24 10.1016/J.PATREC.2014.01.008. [DOI] [Google Scholar]
- 49.Anderson JS, Ferguson MA, Lopez-Larson M, Yurgelun-Todd D: Reproducibility of single-subject functional connectivity measurements. Am J Neuroradiol 2011, 32:548–555 10.3174/ajnr.A2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.••.Gordon EM, Laumann TO, Gilmore AW, Newbold DJ, Greene DJ, Berg JJ, Ortega M, Hoyt-Drazen C, Gratton C, Sun H et al. : Precision functional mapping of individual human brains. Neuron 2017, 95 10.1016/j.neuron.2017.07.011 791–807.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study examines individual variability in resting state functional neuroimaging using data from 10 highly sampled individuals. The findings demonstrate close correspondence between resting state networks, task activation and structural features and also suggest a need for longer resting state scan times.
- 51.Poldrack RA: Precision neuroscience: dense sampling of individual brains. Neuron 2017, 95:727–729 10.1016/j.neuron.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 52.Cammoun L, Gigandet X, Meskaldji D, Thiran JP, Sporns O,Do KQ, Maeder P, Meuli R, Hagmann P: Mapping the human connectome at multiple scales with diffusion spectrum MRI. J Neurosci Methods 2012, 203:386–397 10.1016/j.jneumeth.2011.09.031. [DOI] [PubMed] [Google Scholar]
- 53.Chong M, Bhushan C, Joshi AA, Choi S, Haldar JP, Shattuck DW, Spreng RN, Leahy RM: Individual parcellation of resting fMRI with a group functional connectivity prior. NeuroImage 2017, 156:87–100 10.1016/j.neuroimage.2017.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Honnorat N, Satterthwaite TD, Gur RE, Gur RC, Davatzikos C: sGraSP: a graph-based method for the derivation of subject-specific functional parcellations of the brain. J Neurosci Methods 2017, 277:1–20 10.1016/j.jneumeth.2016.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weng HH, Noll KR, Johnson JM, Prabhu SS, Tsai YH, Chang SW, Huang YC, Lee JD, Yang JT, Yang CT et al. : Accuracy of presurgical functional MR imaging for language mapping of brain tumors: a systematic review and meta-analysis. Radiology 2017, 286:162971 10.1148/radiol.2017162971. [DOI] [PubMed] [Google Scholar]
- 56.Dierker D, Roland JL, Kamran M, Rutlin J, Hacker CD, Marcus DS, Milchenko M, Miller-Thomas MM, Benzinger TL, Snyder AZ et al. : Resting-state functional magnetic resonance imaging in presurgical functional mapping: sensorimotor localization. Neuroimaging Clin N Am 2017, 27:621–633 10.1016/j.nic.2017.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kong R, Li J, Orban C, Sabuncu MR, Liu H, Schaefer A, Sun N, Zuo XN, Holmes AJ, Eickhoff SB et al. : Spatial topography of individual-specific cortical networks predicts human cognition, personality, and emotion. Cereb Cortex 2018. 10.1093/cercor/bhy123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Braga RM, Buckner RL: Parallel interdigitated distributed networks within the individual estimated by intrinsic functional connectivity. Neuron 2017, 95:457–471e5 10.1016/j.neuron.2017.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anticevic A, Repovs G, Barch DM: Working memory encoding and maintenance deficits in schizophrenia: Neural evidence for activation and deactivation abnormalities. Schizophrenia Bull 2013, 39:168–178 10.1093/schbul/sbr107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Posner J, Song I, Lee S, Rodriguez CI, Moore H, Marsh R, Blair Simpson H: Increased functional connectivity between the default mode and salience networks in unmedicated adults with obsessive–compulsive disorder. Hum Brain Mapp 2017, 38:678–687 10.1002/hbm.23408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Coutinho JF, Fernandesl SV, Soares JM, Maia L, Gonçalves ÓF, Sampaio A: Default mode network dissociation in depressive and anxiety states. Brain Imaging Behav 2016, 10:147–157 10.1007/s11682-015-9375-7. [DOI] [PubMed] [Google Scholar]
- 62.Padmanabhan A, Lynch CJ, Schaer M, Menon V: The default mode network in autism. Biol Psychiatry: Cogn Neurosci Neuroimaging 2017, 2:476–486 10.1016/j.bpsc.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Satterthwaite TD, Davatzikos C: Towards an individualized delineation of functional neuroanatomy. Neuron 2015, 87:471–473 10.1016/j.neuron.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hektner JM, Schmidt JA, Csikszentmihalyi M: Experience sampling method: measuring the quality of everyday life. Sage 2007. [Google Scholar]
- 65.•.Borsboom D:: A network theory of mental disorders. World Psychiatry 2017, 16:5–13 10.1002/wps.20375. [DOI] [PMC free article] [PubMed] [Google Scholar]; This review describes an approach by which network models can describe how psychiatric symptoms influence one another within an individual. It also introduces the possibility of understanding illness states in terms of response to external factors given a set of symptom network dynamics
- 66.Borsboom D, Cramer AO, Schmittmann VD, Epskamp S, Waldorp LJ: The small world of psychopathology. PLoS ONE 2011, 6:e27407 10.1371/journal.pone.0027407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Costantini G, Richetin J, Preti E, Casini E, Epskamp S, Perugini M: Stability and variability of personality networks. A tutorial on recent developments in network psychometrics. Pers Individ Diff 2017. 10.1016/j.paid.2017.06.011. [DOI] [Google Scholar]
- 68.Bringmann LF, Pe ML, Vissers N, Ceulemans E, Borsboom D, Vanpaemel W, Tuerlinckx F, Kuppens P: Assessing temporal emotion dynamics using networks. Assessment 2016, 23:425–435 10.1177/1073191116645909. [DOI] [PubMed] [Google Scholar]
- 69.Pe ML, Kircanski K, Thompson RJ, Bringmann LF, Tuerlinckx F, Mestdagh M, Mata J, Jaeggi SM, Buschkuehl M, Jonides J et al. : Emotion-network density in major depressive disorder. Clin Psychol Sci 2015, 3:292–300 10.1177/2167702614540645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jones A, Tiplady B, Houben K, Nederkoorn C, Field M: Do daily fluctuations in inhibitory control predict alcohol consumption? An ecological momentary assessment study. Psychopharmacology 2018, 235:1–10 10.1007/s00213-018-4860-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rowan PJ, Cofta-Woerpel L, Mazas CA, Vidrine JI, Reitzel LR, Cinciripini PM, Wetter DW: Evaluating reactivity to ecological momentary assessment during smoking cessation. Exp Clin Psychopharmacol 2007, 15:382–389 10.1037/1064-1297.15.4.382. [DOI] [PubMed] [Google Scholar]
- 72.Beck AT, Steer RA, Brown GK: Beck depression inventory-II. San Antonio 1966, 78:490–498 10.1002/9780470479216.corpsy0113. [DOI] [Google Scholar]
- 73.Kroenke K, Spitzer RL: The PHQ-9: a new depression diagnostic and severity measure. Psychiatric Ann 2002, 32:509–515 10.3928/0048-5713-20020901-06. [DOI] [Google Scholar]
- 74.Kay S, Fiszbein A: Bulletin LOS undefined 1987: the positive and negative syndrome scale (PANSS) for schizophrenia. PsycnetApaOrg 1987, 13:261. [DOI] [PubMed] [Google Scholar]
- 75.Schwarz N: Retrospective and concurrent self-reports: the rationale for real-time data capture. Sci Real-time Data Capture: Self-reports Health Res 2007, 11:11–26. [Google Scholar]
- 76.Terry PC, Lane AM, Fogarty GJ: Construct validity of the profile of mood states — adolescents for use with adults. Psychol Sport Exerc 2003, 4:125–139 10.1016/S1469-0292(01)00035-8. [DOI] [Google Scholar]
- 77.Kluge A, Kirschner M, Hager OM, Bischof M, Habermeyer B, Seifritz E, Walther S, Kaiser S: Combining actigraphy, ecological momentary assessment and neuroimaging to study apathy in patients with schizophrenia. Schizophrenia Res 2017, 195:176–182 10.1016/j.schres.2017.09.034. [DOI] [PubMed] [Google Scholar]
- 78.Cella M, Okruszek Ł, Lawrence M, Zarlenga V, He Z, Wykes T: Using wearable technology to detect the autonomic signature of illness severity in schizophrenia. Schizophrenia Res 2017, 195:537–542 10.1016/j.schres.2017.09.028. [DOI] [PubMed] [Google Scholar]
- 79.Seidel M, King JA, Ritschel F, Boehm I, Geisler D, Bernardoni F, Holzapfel L, Diestel S, Diers K, Strobel A et al. : The real-life costs of emotion regulation in anorexia nervosa: a combined ecological momentary assessment and fMRI study. Transl Psychiatry 2018, 8:28 10.1038/s41398-017-0004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Preti MG, Bolton TA, Van De Ville D: The dynamic functional connectome: state-of-the-art and perspectives. NeuroImage 2017, 160:41–54 10.1016/j.neuroimage.2016.12.061. [DOI] [PubMed] [Google Scholar]
- 81.Rogasch NC, Daskalakis ZJ, Fitzgerald PB: Cortical inhibition, excitation, and connectivity in schizophrenia: a review of insights from transcranial magnetic stimulation. Schizophrenia Bull 2014, 40:685–696 10.1093/schbul/sbt078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bestmann S, Krakauer JW: The uses and interpretations of the motor-evoked potential for understanding behaviour. Exp Brain Res 2015, 233:679–689 10.1007/s00221-014-4183-7. [DOI] [PubMed] [Google Scholar]
- 83.••.Sun Y, Blumberger DM, Mulsant BH, Rajji TK, Fitzgerald PB, Barr MS, Downar J, Wong W, Farzan F, Daskalakis ZJ: Magnetic seizure therapy reduces suicidal ideation and produces neuroplasticity in treatment-resistant depression. Transl Psychiatry 2018, 8:253 10.1038/s41398-018-0302-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper demonstrates that individual variability in frontal cortical excitability is a strong predictor of response to magnetic seizure therapy. This technique might hold enormous promise if intra-individual variability in cortical excitability can be exploited to improve treatment response.