Abstract
Type I interferons (IFNs) (IFN-α, IFN-β) and type III IFNs (IFN-λ) share many properties, including induction by viral infection, activation of shared signaling pathways, and transcriptional programs. However, recent discoveries have revealed context-specific functional differences. Here, we provide a comprehensive review of type I and type III IFN activities, highlighting shared and distinct features from molecular mechanisms through physiological responses. Beyond discussing canonical antiviral functions, we consider the adaptive immune priming, anti-tumor, and autoimmune functions of IFNs. We discuss a model wherein type III IFNs serve as a front-line defense that controls infection at epithelial barriers while minimizing damaging inflammatory responses, reserving the more potent type I IFN response for when local responses are insufficient. In this context, we discuss current therapeutic applications targeting these cytokine pathways and highlight gaps in understanding of the biology of type I and type III IFNs in health and disease.
Introduction
Interferons (IFNs) are divided into three families (type I, type II, and type III) on the basis of sequence homology, which corresponds to the evolutionary relatedness, receptor usage, and functional activity of these cytokines. The type II IFN family includes a single member, IFN-γ, which has pro-inflammatory and immunomodulatory functions that are distinct from the type I and III IFNs; IFN-γ has been reviewed elsewhere (Alspach et al., 2018) and is not discussed herein. Type I IFNs originally were identified on the basis of their antiviral activity (Isaacs and Lindenmann, 1957; Isaacs et al., 1957), but subsequently were recognized to have anti-pro I iterative and immunomodulatory activities, as well as roles in modulating infection by non-viral pathogens. It was nearly 50 years later that type III IFNs were discovered (Kotenko et al., 2003; Sheppard et al., 2003), and initially it was unclear why the host would maintain seemingly redundant antiviral defense pathways. Although type I and type III IFNs are genetically distinct and use different receptors, they are induced by similar pathogen-sensing pathways and activate related antiviral, anti-proliferative, and immunomodulatory gene expression programs. The potential for type III IFNs to provide supplementary antiviral protection at epithelial surfaces was recognized early on, but it is now appreciated that type III IFNs might provide a front-line defense that confers less collateral damage than the more potent type I IFN response. This model of type III IFN action stems from an improved understanding of the cells and tissues in which type III IFNs exert their activity. For example, type III IFNs function broadly at anatomic barrier sites and have unique effects on hematopoietic cells, most strikingly neutrophils. We also now have a greater understanding of the regulatory mechanisms that distinguish type I and type III IFN signaling. Furthermore, the discovery of translation-independent effects of IFN signaling, as well as activation of non-canonical signaling pathways, suggests the potential for previously unappreciated mechanisms of IFN activity. Beyond spatial segregation of IFN signaling, we now have an improved understanding of the mechanisms by which IFN-stimulated genes (ISGs) exert their antiviral activities. Although excellent recent reviews are available about distinct aspects of the antiviral response induced by type I and type III IFN signaling (Crouse et al., 2015; García-Sastre, 2017; Hoffmann et al., 2015; Ingle et al., 2018; Kotenko and Durbin, 2017; Lazear et al., 2015b; Schoggins, 2018; Schreiber, 2017; Snell et al., 2017; Sorgeloos et al., 2013; Wack et al., 2015; Wells and Coyne, 2018), our goal is to provide a comprehensive overview of shared and unique activities of type I and type III IFNs. In addition to their canonical antiviral activities, we further consider other functions of IFNs, including adaptive immune priming, anti-tumor responses, and effects on autoimmunity. We focus on human and mouse IFNs, which are the best understood, but the diversity of IFNs found among vertebrates suggests that comparative immunology studies might reveal novel effector and regulatory mechanisms (Krause and Pestka, 2015). Since their discovery, IFNs have been harnessed for therapeutic applications with some clinical successes. Other promising applications might be revealed by gaining a more complete understanding of IFN structure, signaling activity, function, and regulation.
Type I and Type III IFNs
IFNs are part of the class II cytokine family, which also includes interleukin-10 (IL-10)-related cytokines (IL-10, IL-19, IL-20, IL-22, IL-24, and IL-26). Despite limited primary sequence homology, class II cytokines share a conserved structure comprised of six α-helices. Their receptors have two extracellular type III fibronectin domains that form the cytokine binding site (Renauld, 2003) (Figure 1). Tvoe I IFNs oriainally were discovered as secreted factors that rendered cells refractory to viral infections (Isaacs and Lindenmann, 1957; Isaacs et al., 1957). The molecular mechanisms underlying their antiviral activities became better defined when the genes for these cytokines were cloned in the 1980s and their cell surface receptors identified in the 1990s (reviewed in Vilcek, 2006). IFN gene families have evolved through gene duplication and divergence such that the complement of IFN genes varies among vertebrate species (Krause and Pestka, 2015). In humans and mice, the type I IFN family includes multiple IFN-α subtypes (13 in humans, 14 in mice) and single IFN-β, IFN-ε, IFN-κ, IFN-ω (humans), and IFN-ζ (mice) subtypes (Figure 2). All type I IFNs signal through a shared heterodimeric receptor, IFNAR, comprised of IFNAR1 and IFNAR2 subunits. Type I IFNs bind IFNAR2 with high affinity, then recruit the low-affinity IFNAR1, creating a signaling-competent ternary complex. Type I IFN genes lack introns (except for IFN-κ, which has one) and are clustered on human chromosome 9 and mouse chromosome 4. Although there is considerable redundancy among type I IFNs, differences in promoter sequences and biochemical properties contribute to distinct functional activities (Schreiber, 2017). Type III IFNs were discovered by two groups (Kotenko et al., 2003; Sheppard et al., 2003), and in humans include four subtypes, IFN-λ1 (IL-29), IFN-λ2 (IL-28A), IFN-λ3 (IL-28B), and IFN-λ4. IFN-λ4 was the last to be discovered, and is a pseudogene in many human populations (Prokunina-Olsson et al., 2013). In mice, the type III IFN family consists of IFN-λ2 and IFN-λ3; IFN-λ1 is a pseudogene and the genomic region encoding IFN-λ4 is absent (Kotenko and Durbin, 2017; Wack et al., 2015).
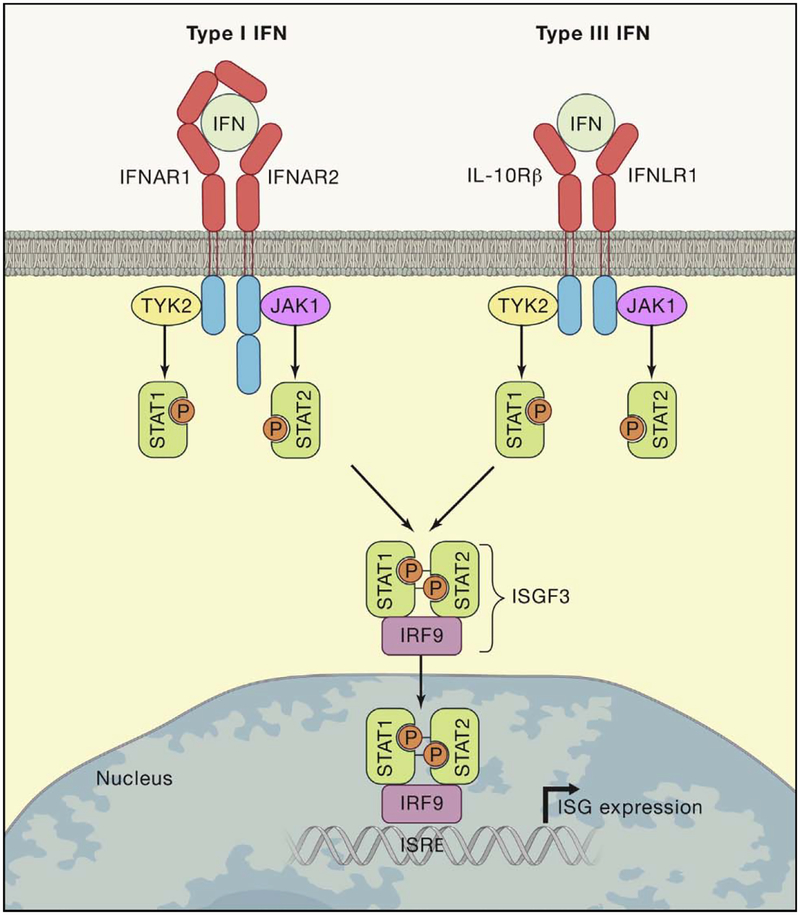
Figure 1. Canonical Type I and Type III IFN Signaling.
Type I and type III IFNs bind to distinct receptors but activate similar signaling pathways and transcriptional responses. The type I and type III IFN receptors are heterodimers comprised of IFNAR1 and IFNAR2 subunits or IFNLR1 and IL10Rβ subunits, respectively. IFNs first bind one receptor chain with high affinity (IFNAR2 or IFNLR1), then recruit a low-affinity receptor chain (IFNAR1 or IL10Rβ) to create a signaling-competent ternary complex. Receptor dimerization activates TYK2 and JAK1 kinases, which phosphorylate STAT1 and STAT2. Phosphorylated STAT1 and STAT2 heterodimers complex with IRF9 to produce the transcription factor ISGF3. ISGF3 binds to ISREs and promotes expression of hundreds of ISGs. Type I and type III IFNs also activate additional signaling pathways not depicted in this figure.
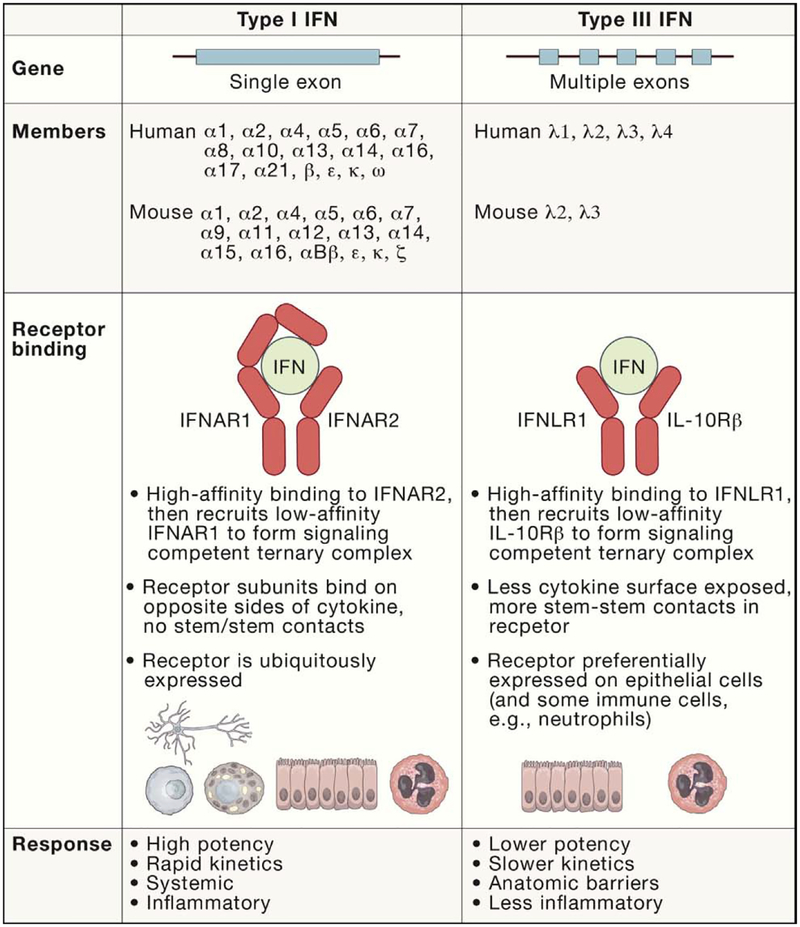
Figure 2. Comparison of Type I and Type III IFNs.
Although their signaling pathways and transcriptional responses have many similarities, some features distinguish type I and type III IFNs: (1) most type I IFN genes lack introns, whereas type III IFN genes have 5 or 6 exons; (2) the type I IFN family is larger, comprising 17 members in humans and 18 in mice, compared with 4 type III IFN members in humans and 2 in mice; (3) type I and type III IFNs bind distinct receptors. The type I IFN receptor (IFNAR) is ubiquitously expressed, whereas the type III IFN receptor (IFNLR) is expressed preferentially on epithelial cells, as well as neutrophils; (4) although the genes activated by type I and type III IFN signaling are similar, differences in cell type specificity and signaling kinetics result in distinct responses. The type I IFN response is more potent, rapid, and transient, whereas the type III IFN response is less potent, slower, and sustained. Many cell types respond to type I IFNs, resulting in a systemic response that is more inflammatory. In contrast, the type III IFN response is less inflammatory and concentrated at epithelial and barrier surfaces.
All type III IFNs signal through a shared heterodimeric receptor, IFNLR, comprised of IFNLR1 (also termed IL28Rα) and IL10Rβ. IFN-λ binds IFNLR1 with high affinity, and then recruits the low-affinity IL10Rβ (which is shared with other IL-10 family cytokines) to create a signaling-competent ternary complex. Type III IFN genes are clustered on human chromosome 19 and mouse chromosome 7 and share a 5-exon gene structure with other IL-10 family cytokines, although IFNL2 and IFNL3 have an additional 6th exon (Fox et al. 2009; Sabat, 2010). In humans, multiple polymorphisms in the IFNL locus are associated with clinical outcomes from hepatitis C virus (HCV) infection (Griffiths et al., 2015). Among these, a frameshift mutation in the promoter of IFNL4 results in the loss of IFN-λ4 production in many non-African populations and concomitant improved clearance of HCV (Prokunina-Olsson et al., 2013). The pseudogenization of IFN-λ4, along with selection for lower-potency variants, suggest IFN-λ4 signaling has been deleterious during human evolution (Bamford et al., 2018; Key et al., 2014).
IFN signaling forms the foundation of the vertebrate innate immune response to viral infections. ISG repertoire varies among species, but a core set of ~90 ISGs induced in diverse mammals suggests substantial conservation of the IFN-mediated antiviral response throughout mammalian evolution (Shaw et al., 2017). The significance of this system is evidenced by the variety of mechanisms by which viruses evade or antagonize IFN induction, signaling, or effector functions (García-Sastre, 2017; Hoffmann et al., 2015), as well as the susceptibility phenotypes observed in humans and mice with genetic disruptions in IFN production or signaling (Müller et al., 1994; Sancho-Shimizu et al., 2011). The role of IFN signaling during bacterial infections is more nuanced, given than IFN signaling protects against some bacterial infections and exacerbates disease caused by others (Boxx and Cheng, 2016).
IFN Signaling Pathways and the Antiviral Response
Despite their different receptors, the downstream signaling pathways and transcriptional responses activated by type I and III IFNs exhibit substantial overlap (Figure 3) (Kotenko and Durbin, 2017; Wack et al., 2015). Both type I and type III IFNs signal through the JAK-STAT pathway to activate the het-erotrimeric transcription factor complex ISGF3, comprised of phosphorylated STAT1 and STAT2, and interferon regulatory factor 9 (IRF9). Activated ISGF3 translocates to the nucleus and binds to IFN-stimulated response elements (ISREs) in the upstream promoter regions of ISGs, which encode proteins that act via a variety of mechanisms to restrict viral infection (reviewed in Schneider et al., 2014; Schoggins, 2014, 2018). Because ISGs generally are induced as part of a concerted transcriptional program, the effects of individual ISGs can be difficult to ascertain in isolation. However, recent work has identified molecular mechanisms behind the antiviral effects of prominent ISGs including viperin (Gizzi et al., 2018), interferon-induced protein with tetratricopeptide repeats 1 (IFIT1) (Daffis et al., 2010; Hyde et al., 2014; Pichlmair et al., 2011), Mx1 (Haller et al., 2015), PKR (Pfaller et al., 2011), OAS/RNASEL (Hornung et al., 2014), and IFI6 (Richardson et al., 2018). Because the signaling cascade is overlapping, many functional activities are shared between the two IFN families. However, distinct biological outcomes can result from differences in the magnitude and kinetics of signaling and in the types of cells that respond to type I versus type III IFNs.
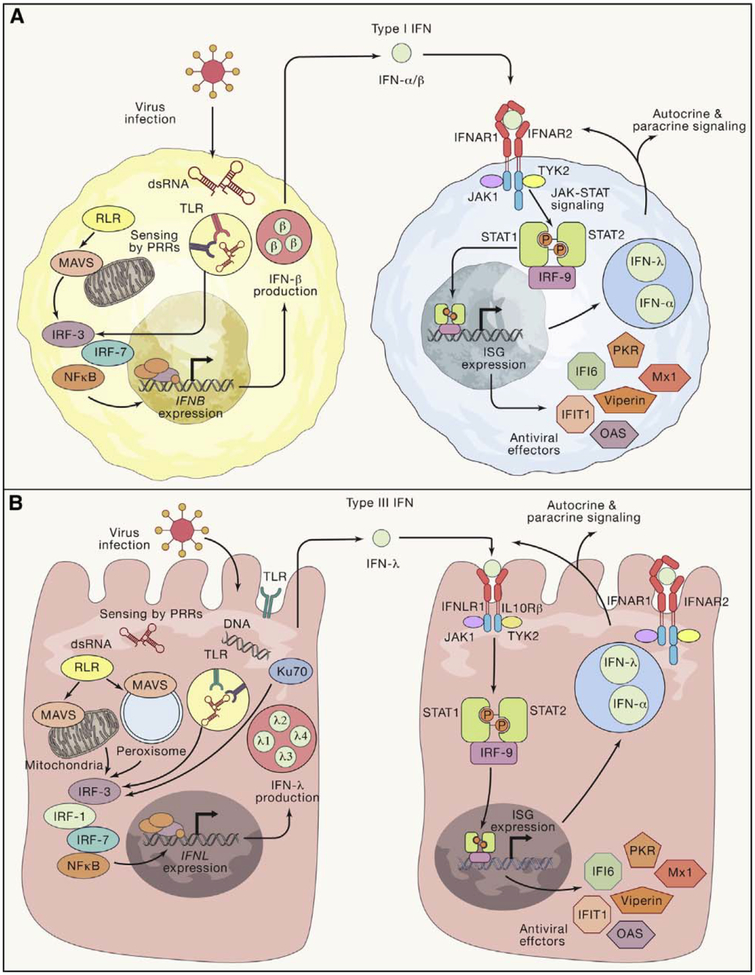
Figure 3. Induction and Antiviral Signaling of Type I and Type III IFNs.
(A and B) Type I IFNs (A) and type III IFNs(B) are both induced when viral infection is detected by PRRs including RIG-I like receptors (RLRs), Toll-like receptors (TLRs), and cGAS (not depicted). Differences in the subcellular localization of the PRR response can favor production of type III over type I IFNs (e.g., plasma membrane TLR4 signaling or peroxisomal MAVS signaling). PRR signaling activates IRF-family transcription factors which, together with NFκB, promote IFN expression. IFNs are secreted by infected cells and signal in a paracrine manner to uninfected cells to stimulate the production of ISGs, which act by a variety of mechanisms to induce an antiviral state. Among the ISGs induced by IFN signaling are other IFNs, resulting in a positive feedback loop of antiviral activity.
Recent structural studies of IFN-λ3 in complex with its heterodimeric receptor highlight differences in the ternary complexes of type I and III IFNs (Mendoza et al., 2017). The IFN-λ ternary complex has a distinct geometry compared with the type I IFN (IFN-ω) ternary complex (Thomas et al., 2011). In the IFN-ω complex, the two receptor chains bind on opposing faces of the cytokine, which contrasts with the binding interface of the IPN-λ3 ternary complex. Additionally, a large surface area of IFN-λ3 remains exposed in the ternary complex, whereas little surface area of IFN-ω is exposed in the ternary complex with IFNAR1 and IFNAR2 (Thomas et al., 2011). Lastly, the IFNLR1 and IL10Rβ chains make extensive stem-stem contacts that stabilize the complex; similar contacts between IFNAR1 and IFNAR2 in the type I IFN ternary complex were absent. In canonical type I IFN signaling, cytokines bind with high affinity to IFNAR2 and then form a ternary complex with IFNAR1 that mediates signaling. However, unlike IFN-α, IFN-β can form a high-affinity complex with IFNAR1, contributing to its potency and allowing for IFNAR2-independent IFN-β signaling and a distinct ISG profile (de Weerd et al., 2017; de Weerd et al., 2013). One determinant in the differential antiviral responses of type I and type III IFNs is the non-overlapping distribution of their respective receptors: IFNAR1 and IFNAR2 are expressed on virtually all nucleated cells, whereas IFNLR1 is expressed preferentially on epithelial cells. Accordingly, the antiviral effects of type III IFNs are especially evident at epithelial barriers, such as the gastrointestinal, respiratory, and reproductive tracts (Kotenko and Durbin, 2017; Lazear et al., 2015b; Wells and Coyne, 2018).
The signaling pathways induced by type I and III IFNs have subtle differences that contribute to their distinct functions. Although both signaling pathways use JAK1 and TYK2 kinases to trigger the formation of the ISGF3 complex, in certain cell types, type III IFNs also activate JAK2 signaling (Lee et al., 2012; Odendall et al., 2014). Furthermore, in addition to STAT1 and STAT2, type I IFNs can also signal through STAT3. In some cases, STAT3 negatively regulated type I IFN signaling by inhibiting STAT1-dependent gene activation (Ho and Ivashkiv, 2006; Wang et al., 2011), whereas in others, STAT3 contributed to type-I-IFN-mediated antiviral responses against influenza A virus (IAV) and vaccinia virus by inducing a subset of antiviral ISGs (Mahony et al., 2017). STAT3 also has been implicated in modulating type-III-IFN-mediated expression of microRNAs that mediate HCV replication or lung inflammatory responses (Aboulnasr et al., 2015; Cohen and Prince, 2013). In addition to the dominant JAK-STAT signaling response, both type I and type III IFNs can activate mitogen-activated protein kinase (MAPK) signaling (Platanias, 2005; Zhou et al., 2007). However, studies in intestinal organoid cultures found that the antiviral response induced by IFN-λ, but not IFN-β, was blocked by MAPK inhibitors, suggesting differential effects of STAT-independent signaling pathways (Pervolaraki et al., 2017). Other STAT-independent IFN activities include induction of neutrophil degranulation and tightening of cell-cell junctions in blood-brain barrier (BBB) endothelial cells, both of which occur in a translation-independent manner (Broggi et al., 2017; Lazear et al., 2015a).
Transcriptome profiling studies have established that the type I and type III signatures are overlapping. In general, type III IFNs are less potent than type I IFNs, and the ISGs induced by type III IFNs are a subset of those induced by type I IFNs (Crotta et al., 2013; Doyle et al., 2006; Marcello et al., 2006; Zhou et al., 2007). Increasing the amount of IFN-λ augments the number of ISGs to match that seen with type I IFNs, suggesting that the ISG repertoire reflects the magnitude of the signaling response. Indeed, antiviral ISGs such as MX1, viperin, and the IFITM, IFIT, and OAS family members, are induced by both type I IFN and type III IFNs, although the type I IFN response is more potent, especially at lower doses and earlier time points (Zhou et al., 2007). However, in more relevant primary cells and tissues, distinctions have emerged. For example, in human vaginal epithelial cells, a subset of ISGs (e.g., CXCL10, CXCL11, IFIT3, IFI30 and TDRD7) were uniquely or more highly induced by IFN-λ1 than by IFN-β (Caine et al., 2019). Because the study was carried out at one time point and with a single IFN dose, it is not known whether these differential ISG expression patterns would be sustained at earlier time points or with different IFN doses. Although the overall repertoire of genes induced by type I and III IFNs is shared, the kinetics of induction are distinct. ISG induction by type I IFNs peaks early and declines, whereas more sustained expression occurs with type III IFNs (Bolen et al., 2014; Jilg et al., 2014; Kohli et al., 2012; Marcello et al., 2006; Voigt and Yin, 2015). These effects might be linked to the rapid downregulation of type I IFN signaling by negative regulatory ISGs such as ISG15, USP18, and Tyro3, Axl, and Mer (TAM) receptors (François-Newton et al., 2011; Rothlin et al., 2007; Zhang et al., 2015). Mathematical modeling and experimental data suggest that the difference in kinetics between type I and type III IFNs is not due to receptor expression levels, but rather due to intrinsic qualities of the two signaling pathways (Pervolaraki et al., 2018).
Both type I and type III IFNs are induced after detection of pathogen-associated molecular patterns (PAMPs) and engagement and activation of cytosolic (e.g., RIG-I, MDA5, and cGAS) or endosomal (e.g., TLR3 and TLR4) pattern-recognition receptors (PRRs). For some PRRs, differences in the location of PAMP engagement can affect the type of IFN produced. TLR4 signaling in endosomes results in type I IFN production (Kagan et al., 2008), whereas TLR4 engagement at the plasma membrane induces type III IFNs. (Odendall et al., 2017). These distinct patterns of IFN induction might contribute to the protective activity of type III IFNs at epithelial barriers that continually encounter commensal microbiota and their PAMPs.
PRR signaling induces IFN expression by activating IRF transcription factors. In the classical model of type I IFN induction, PRR activation leads to IRF3 activation, followed by IFN-β induction (Honda et al., 2006; Paun and Pitha, 2007). IFN-β stimulates a first wave of ISG transcription, including the IFN-inducible transcription factor IRF7. Subsequent IRF7 activation then induces expression of multiple IFN-α subtypes, which mediate a second wave of ISG transcription. This model does not function in all cell types, however, and both IRF5 and IRF7 can participate in the first wave of type I IFN induction (Honda et al., 2005; Lazear et al., 2013). Analogous to type I IFNs, the type III IFNs also are induced by IRF3 and/or IRF7 (Osterlund et al., 2007). Additionally, at least one type III IFN, IFN-λ1, can be induced by IRF1 (Odendall et al., 2014), whereas IRF1 is not considered a primary driver of type I IFN expression (Reis et al., 1994). Other PRR signaling pathways, such as the cytosolic DNA sensor Ku70, also can uniquely induce type III IFNs compared with type I IFNs (Zhang et al., 2011). The in vivo biological relevance of these distinct IRF-mediated induction patterns still remains unclear, but might be linked to differences in temporal and spatial responses to viral infection (Pulverer et al., 2010).
Tissue-Specific Effects of IFN Signaling
Given the similar transcriptional responses induced by type I and type III IFNs, what is the purpose of maintaining seemingly redundant antiviral defense systems? Type III IFN signaling appears to serve as a first-line defense that is lower in magnitude, less inflammatory, and concentrated at anatomic barriers (reviewed in Kotenko and Durbin, 2017; Lazear et al., 2015b; Wack et al., 2015; Wells and Coyne, 2018). In this paradigm, only when the initial type III IFN-mediated defense is breached is it necessary to activate the more potent, but more inflammatory, systemic type I IFN response. This sequence allows immune signaling to be spatially segregated, and the effects of type III IFNs are most evident at epithelial barriers such as the gastrointestinal, respiratory, and female reproductive tracts, as well as tissue barriers such as the BBB and placenta. The constant exposure of external epithelial surfaces to commensal and pathogenic microbes necessitates a balance between protective and pathological immune responses. Type III IFN responsiveness at epithelial surfaces is related to the relatively high expression of IFNLR1 on epithelial cells. It also correlates with the density of peroxisomes in epithelial tissues, as these organelles favor type III IFN overtype I IFN production downstream of MAVS signaling (Odendall et al., 2014). Consistent with a key role for type III IFN signaling at barrier surfaces, a human primary immunodeficiency patient with a homozygous loss-of-function allele of IFNAR2 controlled naturally encountered pathogens sufficiently well to survive to more than one year of age. However, the patient succumbed to measles-mumps-rubella vaccination in which subcutaneous administration of live viruses bypassed epithelial barriers protected by type III IFNs (Duncan et al., 2015).
All epithelial surfaces encounter environmental microbes, but nowhere is the amount and complexity of exposure as great as in the gastrointestinal tract. Here, IFN responses must provide protection from microbial infection without initiating destructive inflammatory responses (reviewed in Ingle et al., 2018). The microbiome provides protection against viral infections and other intestinal insults, and its protective effects are mediated by IFN signaling (Baldridge et al., 2015; Kernbauer et al., 2014; Martin et al., 2018; Steed et al., 2017). Antiviral responses in the gut are dominated by type III rather than type I IFN signaling. This is a direct consequence of high IFNLR and low IFNAR expression on intestinal epithelial cells, which are targets for enteric viruses including norovirus, reovirus, rotavirus, and enteroviruses (Baldridge et al., 2017; Good et al., 2019; Johansson et al., 2007; Lin et al., 2016; Mahlakõiv et al., 2015; Nice et al., 2015; Pott et al., 2011). In contrast, lamina propria cells generally express greater levels of IFNAR than IFNLR, and type I IFN signaling restricts systemic spread of enteric viruses if they invade beyond the epithelium. In addition to their antiviral activity, type III IFNs serve important immunomodulatory functions in the gastrointestinal tract. For example, enteric-virus-induced type III IFNs activate an anti-inflammatory program in neutrophils, resulting in a STAT1- and translation-independent suppression of degranulation and reduction in reactive oxygen species (Broggi et al., 2017).
In contrast to the IFN-λ-specific responsiveness of intestinal epithelial cells, respiratory tract epithelial cells respond to both type I and type III IFNs (Hamming et al., 2013). Accordingly, both Ifnar1−/− and Ifnlr1−/−mice are more susceptible to infection with respiratory viruses, and treatment of mice or respiratory epithelial cells with type I or type III IFNs restricts viral replication (reviewed in Lazear et al., 2015b; Wells and Coyne, 2018). Nevertheless, recent studies have revealed location, cell type, and kinetic differences in how type I and type III IFNs control respiratory infections. After infection with IAV or other respiratory tract pathogens, epithelial cells rapidly produce type III IFNs, and type I IFNs were produced later and/or to a lesser extent (Crotta et al., 2013; Espinosa et al., 2017; Fox et al., 2015; Galani et al., 2017; Jewell et al., 2010; Okabayashi et al., 2011). Type III IFNs are important for controlling viral infection in the upper respiratory tract, whereas there is more functional redundancy between type I and III IFNs in the lower respiratory tract (Klinkhammer et al., 2018). Bone marrow chimera and conditional knockout experiments reveal that IFN signaling in epithelial cells is key to controlling IAV infection (Crotta et al., 2013; Galani et al., 2017). In contrast, control of Aspergillus fumigatus was mediated by IFN signaling in hematopoietic cells (Espinosa et al., 2017). These differing requirements for IFN signaling in the respiratory tract might reflect the target cells of different pathogens. For example, IAV infection is largely restricted to epithelial cells, whereas fungal spores are internalized by phagocytes. In contrast to its antiviral effects, IFN signaling can augment some bacterial infections (Boxx and Cheng, 2016), which can contribute to disease during viral-bacterial co-infection. For example, IAV-induced IFN promotes Streptococcus pneumonia colonization in the upper respiratory tract (Nakamura et al., 2011), and IFN induced by respiratory syncytial virus enhances Pseudomonas aeruginosa biofilm formation (HendricKs et al., 2016). IFN signaling shapes the inflammatory response to respiratory viruses, and has protective and pathological outcomes. For example, type I IFN signaling controls respiratory syncytial virus replication in the lungs, but also promotes inflammation-induced disease (Goritzka et al., 2014; Goritzka et al., 2015). Although neutrophils express ISGs in response to both type I and type III IFNs, inflammatory cytokines (e.g., Tnf, II1b, II6, and Ccl2) were induced predominantly by IFN-α and not IFN-λ. Accordingly, Ifnlr1−/− mice infected with IAV exhibited greater inflammatory burdens in the lungs than did wild-type (WT) or Ifnar1−/− mice (Galani et al., 2017). These observations support a model in which type III IFNs serve as the first-line response for controlling viral infection in respiratory tract epithelial cells while minimizing immune pathology.
The need to control infection while limiting inflammation is especially important in the central nervous system (CNS), which relies heavily on innate antiviral responses because leukocyte trafficking across the BBB is tightly regulated (reviewed in Klein and Hunter, 2017). Neurons, astrocytes, and microglia all produce and respond to type I IFNs (Delhaye et al., 2006; Drokhlyansky et al., 2017; Hwang and Bergmann, 2018), whereas type III IFNs do not appear to contribute substantially to the antiviral response within the CNS parenchyma (Lazear et al., 2015a; Sommereyns et al., 2008; Sorgeloos et al., 2013). In addition to inhibiting viral replication, IFNs independently control viral infection in the CNS by restricting BBB permeability and neuroinvasion. Both type I and type III IFNs induce cell junction tightening in brain microvasculature endothelial cells, and this response is independent of STAT1 signaling and new protein synthesis (Daniels et al., 2014; Lazear et al., 2015a). The BBB tightening effects of IFN signaling protect mice from viral neuroinvasion (Daniels et al., 2014; Douam et al., 2017; Lazear et al., 2015a). In addition to effects on endothelial cells, type I IFN signaling on astrocytes also modulates BBB permeability in a brain-region-specific manner (Daniels et al., 2017).
The barrier tightening effects of IFNs are not unique to the CNS, because they also tighten epithelial barriers in the respiratory and gastrointestinal tracts (LeMessurier et al., 2013; Odendall et al., 2017). Compromised epithelial barrier integrity contributes to disease progression in the autoimmune skin conditions psoriasis and atopic dermatitis. Of note, atopic dermatitis is associated with bacterial (e.g., Staphylococcus aureus) and viral (e.g., herpes simplex virus [HSV], human papilloma virus [HPV], and molluscum contagiosum poxvirus) infections of the skin, whereas psoriasis is not. Skin lesions from psoriasis patients exhibit high basal expression of ISGs and type III IFNs, but not type I IFNs, which might contribute to protection from cutaneous infections (Wolk et al., 2013). Conversely, type I IFNs contribute to inflammatory cascades that exacerbate psoriasis (reviewed in Grine et al., 2015), though the cytokine responses involved are complex, and it is unclear to what extent type III IFNs contribute to this process. Type I IFN responses in the skin might be affected by IFN-κ, which is produced specifically and constitutively by keratinocytes (LaFleur et al., 2001) but has a lower affinity for IFNAR2 than other type I IFNs (Harris et al., 2018). HPV epigenetically represses IFN-κ expression (Reiser et al., 2011; Rincon-Orozco et al., 2009), suggesting that IFN-κ might have a protective role in keratinocytes in the skin and female reproductive tract, although its antiviral properties have not been investigated extensively.
In primary human cervical and vaginal epithelial cells cultured ex vivo, both type I and type III IFNs induce ISG expression (Caine et al., 2019). Ifnar1−/− and Ifnlr1−/− mice both exhibit increased susceptibility to vaginal infection with HSV-2 or Zika virus (ZIKV), consistent with these cytokines eliciting protective antiviral responses in the female reproductive tract (Ank et al., 2008; Caine et al., 2019; Svensson et al., 2007; Yockey et al., 2016). Whereas IFN-α and IFN-β contribute to antiviral immunity in the female reproductive tract by mechanisms similar to those in other tissues, an additional type I IFN, IFN-ε, might have specific roles in the female reproductive tract. Unlike other IFNs, IFN-ε is regulated by hormonal status and is not induced by PRR signaling (Fung et al., 2013; Hermant et al., 2013). IFN-ε has a lower affinity for IFNAR2 and signals with lower potency than other type I IFNs, which might prevent excessive immune activation (Harris et al., 2018; Stifter et al., 2018). Nonetheless, IFN-ε induces ISG expression, restricts viral replication in cell culture, and protects mice from vaginal infection with HSV-2 or Chlamydia muridarum (Fung et al., 2013; Stifter et al., 2018; Tasker et al., 2016). The diminished potency of IFN-ε (as well as IFN-κ) compared with other type I IFNs highlights the potential for the induction of different functional outcomes even among IFNs signaling through the same receptor. In contrast to the protective effects of IFN-ε, IFN-β exacerbates Chlamydia disease in the genital tract by eliciting inflammatory immunopathology (Nagarajan et al., 2008).
The maternal-fetal interface is an anatomic barrier with complex immune regulation due to the need to protect the fetus from maternal pathogens while avoiding immune rejection of semi-allogeneic fetal tissue (reviewed in Ander et al., 2019; Yockey and Iwasaki, 2018). The interface between the maternal blood supply and fetal-derived placenta is formed by syncytiotrophoblasts, which are refractory to infection due in part to their constitutive secretion of cytokines including type III IFNs (Bayer et al., 2016; Corry et al., 2017). As maternally derived decidual cells respond to type I and type III IFNs, placenta-derived IFNs could induce an antiviral state on both sides of the maternal-fetal interface (Corry et al., 2017). Accordingly, recombinant IFN-λ2 administered to pregnant Ifnar1−/− mice induced ISGs in both placental and decidual cells (Bierne et al., 2012). In pregnant mice, Listeria monocytogenes infection induced Ifnl2 and Ifnl3 expression in placental and decidual tissue, but the effect of IFN signaling on fetal infection was not assessed (Bierne et al., 2012). Type III IFNs restrict transplacental transmission of ZIKV in mice but not viral burden in other tissues, suggesting a specific role at the maternal-fetal interface (Jagger et al., 2017). In contrast, type I IFNs have a dominant role in controlling ZIKV infection in most tissues in mice (Lazear et al., 2016). This results in part from species restrictions, including the inability of ZIKV to antagonize murine STAT2 and STING proteins (Ding et al., 2018; Gorman et al., 2018; Grant et al., 2016; Kumar et al., 2016). Accordingly, Ifnar1−/− mice exhibit enhanced ZIKV transplacental transmission and fetal pathology due to uncontrolled maternal viral replication (Jagger et al., 2017; Miner et al., 2016; Yockey et al., 2016). In addition to a protective antiviral response, type I IFN signaling might contribute to placental damage, fetal pathology, and fetal demise. Placental macrophages are susceptible to ZIKV and produce IFN-α (but not IFN-β or IFN-λ) in response to infection, providing a source of type I IFN within the infected placenta (Quicke et al., 2016). In human midgestation chorionic villus explants, IFN-β stimulation induced pathological morphological changes (syncytial knots and sprouts) (Yockey et al., 2018). Women with dysregulated type I IFN signaling (sustained IFN production or impaired receptor downregulation) exhibit poor pregnancy outcomes including pre-eclampsia as well as neurodevelopmental defects similar to those induced by congenital infection, altogether consistent with a role for dysregulated type I IFN responses in placental damage (Andrade et al., 2015; Crow and Manel, 2015; Meuwissen et al., 2016; Sanchis et al., 2005). In contrast, IFN-λ3 treatment did not elicit pathologic changes in human villus explants (Yockey et al., 2018), consistent with a protective antiviral role for type III IFN signaling in the placenta. The need to balance protective and pathological effects of type I IFN signaling during fetal development might continue into infancy. For example, the choroid plexus, which lines the brain ventricles, does not respond efficiently to type I IFN in neonates, leaving them vulnerable to HSV encephalitis. In contrast, this tissue efficiently mounts an IFN-dependent antiviral response in adults (Wilcox et al., 2016).
Immunomodulatory Effects of IFNs
Beyond their direct antiviral actions, type I IFNs regulate adaptive immune responses. Type I IFN signaling can promote or inhibit T cell priming depending on the temporal relationship between IFNAR signaling and T cell receptor (TCR) engagement (Crouse et al., 2015). IFNAR signaling after or coinciding with TCR stimulation promotes T cell proliferation, survival, and effector differentiation. However, IFN signaling before or without TCR stimulation elicits a negative regulatory effect, suppressing proliferation, and inducing apoptosis. These opposing effects of type I IFN signaling are thought to occur through recruitment of different STAT transcription factors (van Boxel-Dezaire et al., 2006); STAT1 is pro-inflammatory, anti-proliferative, and pro-apoptotic, whereas STAT3, STAT4, and STAT5 induce T cell survival and differentiation. Type I IFNs also can modulate T cell responses indirectly by (1) stimulating class I and II major histocompatibility complex (MHC) expression, (2) inducing expression of co-stimulatory molecules (e.g., CD80 and CD86) on antigen-presenting cells, (3) stimulating migration of antigen-presenting cells via increased C-C chemokine receptor type 5 (CCR5), CCR7, and lymphocyte function-associated antigen 1 (LFA-1) expression, (4) inducing negative regulators of natural killer (NK) cell activation and cytotoxicity (e.g., NKp46 and class I MHC), and (5) inducing dendritic cells (DCs) to produce chemokines (e.g., CXCL9 and CXCL10) and cytokines (e.g., IL-15) that enhance cross-presentation and promote T cell proliferation and survival (Crouse et al., 2015; Le Bon et al., 2003). Although type I IFNs activate T cell responses during acute infection, chronic IFN exposure can be detrimental to T cells that control pathogens. For example, during lymphocytic choriomeningitis virus (LCMV) infection, persistent type I IFN exposure caused DCs to produce the inhibitory cytokine IL-10 and cell surface protein PD-L1. Blockade of type I IFN signaling during the chronic phase resulted in enhanced IFN-γ responses, decreased expression of negative regulatory molecules, improved T cell immunity, and control of LCMV infection (Teijaro et al., 2013; Wilson et al., 2013). Remarkably, IFN-α and IFN-β have disparate effects: IFN-β blockade improves lymphoid structure, promotes lymphocyte migration, and mitigates T cell exhaustion, altogether promoting virus clearance. Conversely, blockade of IFN-α affected early viral dissemination but did not alter T cell exhaustion (Ng et al., 2015).
Type III IFNs also modulate T cell responses, but this effect is most likely indirect, perhaps via DCs, because T cells express minimal or no IFNLR1 (Zanoni et al., 2017). The addition of IFN-λ during peripheral blood mononuclear cell (PBMC) stimulation or a mixed lymphocyte reaction reduced the production of T helper 2 (Th2) cytokines (IL-4, IL-5, and IL-13) and increased production of IFN-γ (Dai et al., 2009; Jordan et al., 2007; Srinivas et al., 2008). Furthermore, increased IFN-λ production by PBMCs promoted Th1 skewing in response to a viral vaccine (Egli et al., 2014), and Ifnlr1−/− mice exhibited Th2 skewing and worse disease in an asthma model (Koltsida et al., 2011). Together, these findings indicate a Th1-skewing activity for type III IFNs. IFN-λ1 treatment also impaired differentiation of central memory into effector memory T cells. Administration of IFN-λ as an adjuvant for immunization of HIV gag DNA plasmid vaccine reduced the numbers and activity of regulatory T cells and increased the magnitude and quality of CD8+ T cell responses (Morrow et al., 2009). In mice, studies with acute LCMV infection suggested an inhibitory role of type III IFNs on T cells because Ifnlr1−/− mice had increased expansion of CD4+ and CD8+ T cells and enhanced memory T cell responses. However, IFN-λ signaling had a distinct effect during persistent LCMV infection including markedly diminished T cell responses observed in Ifnlr1−/− mice (Misumi and Whitmire, 2014). Disparate results also were observed in flavivirus models: there were no differences in the magnitude or quality of antigen-specific CD8+ T cell responses between WT and Ifnlr1−/− mice infected with West Nile virus, whereas loss of type III IFN signaling resulted in impaired T cell activation in the context of yellow fever virus infection (Douam et al., 2017; Lazear et al., 2015a). Thus, specific virus-host and immune cell interactions might determine the effect of type III IFN signaling on T cell proliferation and maturation.
The effect of type I IFN signaling on B cell and antibody responses varies depending on the context and the antigen. Type I IFN induces DCs to produce B cell stimulatory cytokines (e.g., B cell activating factor [BAFF] and a proliferation-inducing ligand [APRIL]), which enhance immunoglobulin (Ig) class switching (Le Bon et al., 2001; Litinskiy et al., 2002). Selective ablation of IFNAR1 in B cells impaired IFN-α-mediated stimulation of antibody responses and subclass switching, suggesting a direct effect of type I IFNs on B cells (Le Bon et al., 2006). However, in other contexts, type I IFNs can inhibit IL-7-dependent growth and survival of B cell precursors in vitro and ex vivo. Moreover, IFN-α treatment diminished bone marrow and splenic cellularity and markedly reduced numbers of B lineage cells (Lin et al., 1998). Consistent with a negative regulatory effect, during chronic LCMV infection, anti-IFNAR1 antibody treatment promoted survival and differentiation of LCMV-specific B cells and accelerated the generation of neutralizing antibodies (Fallet et al., 2016; Moseman et al., 2016; Sammicheli et al., 2016).
B cell and antibody responses most likely are regulated by type III IFN signaling, although the effects have not been consistent across models. IFN-λ1 augmented TLR-mediated activation of human B cells and IgG production (de Groen et al., 2015), and an IFN-λ3 adjuvant used with an HIV gag DNA plasmid vaccine enhanced HIV-specific IgG2a responses (Morrow et al., 2009). In contrast, recombinant IFN-λ3 inhibited IAV-stimulated Th2 cytokine release, B cell proliferation, and production of antiviral IgG (Egli et al., 2014). In West Nile virus and LCMV infection models, no differences in humoral responses were observed between WT and Ifnlr1−/− mice (Lazear et al., 2015a; Misumi and Whitmire, 2014).
Anti-tumor Effects of IFNs
Type I IFNs can act on tumors directly by blocking cell cycle progression and inducing apoptosis. They also have indirect anti-tumor activities, via priming immune cells to promote clearance and prevent metastasis (Booy et al., 2015; Borden, 2019; Di Trolio et al., 2015). In the context of chemotherapy and/or radiation therapy, type I IFN production by tumor and immune cells stimulates an adaptive (largely T cell) immune response against tumor-cell-associated antigens. This adjuvant effect of type I IFNs is needed because many cancers induce an exhausted and dysfunctional immune state inadequate for tumor clearance (Snell et al., 2017). Given their pro-apoptotic and immunomodulatory actions, type I IFNs were anticipated to be effective therapies against multiple malignancies (reviewed in Borden, 2019). Indeed, treatment with IFN-α (by itself or as part of combination therapy) has moderate success and clinical response for breast cancer, melanoma, and renal carcinoma (Budhwani et al., 2018). For melanoma in particular, IFN-α therapy has produced improvements in relapse-free and overall survival in large randomized trials (Kirkwood et al., 2001; Kirkwood et al., 1996). However, systemic type I IFN treatment has been limited as an anti-tumor modality by lack of efficacy for some tumors, propensity for adverse side effects, and poor tolerability of regimes for patients.
Type III IFNs also exert direct effects against cancer cells by inhibiting cell proliferation and promoting apoptosis (reviewed in Lasfar et al., 2016). However, because fewer cell types express IFNLR1 and respond to IFN-λ, it could serve as a more targeted anti-cancer therapy with diminished side effects compared to IFN-α. In mice, melanoma cells engineered to express IFN-λ2 exhibited greater rejection or slower growth (Lasfar et al., 2006). Type III IFNs inhibited growth of several different tumor cell lines and diminished local and metastatic tumor formation in mice including cancers from lung, liver, breast, and prostate (Abushahba et al., 2010; Lasfar et al., 2016; Li et al., 2008; Numasaki et al., 2007; Sato et al., 2006; Tezuka et al., 2012). Consistent with these findings, Ifnlr1−/− mice are more susceptible to sarcoma formation induced by chemical carcinogens as well as death in transplanted tumor models (Souza-Fonseca-Guimaraes et al., 2015). Type III IFNs can also act indirectly on tumors. IFN-λ suppressed tumor angiogenesis in a mouse model of melanoma by modulating the tumor microenvironment (Lasfar et al., 2006). IFN-λ also augments T cell and NK cell responses to multiple tumors including melanoma, lung adenocarcinoma, and breast cancer (Lasfar et al., 2016). A direct effect of IFN-λ signaling in NK cells was suggested after the failure of Ifnlr1−/− NK cells to suppress tumor growth in vivo (Souza-Fonseca-Guimaraes et al., 2015). Finally, IFN-λ signaling on mammary epithelial cells induced expression of the chemokine CXCL10, which recruits CD4+ T cells into the tumor microenvironment (Burkart et al., 2013). Because IFN-λ can alter tumorigenesis both directly and indirectly, it could have utility as an adjunctive anti-cancer therapy. Beyond these mechanisms, induction of IFNs in response to viral infections also can promote anti-tumor responses. Infection with an oncolytic vesicular stomatitis virus triggered IFN-λ expression in hematopoietic cells in vitro and sensitized melanoma to anti-tumor NK cell recognition and activation in vivo (Wongthida et al., 2010).
IFN Signaling and Autoimmunity
Unchecked responses to pathogens or PAMPs are associated with sustained innate immune signaling and autoimmune disease. Interferonopathies are caused by monogenic allele variants that result in chronic IFN signaling and severe inflammatory disease, such as Aicardi-Goutières syndrome or stimulator of interferon genes (STING)-associated vasculitis with onset in infancy syndrome, as well as other autoimmune diseases including systemic lupus erythematosus (SLE), rheumatoid arthritis (RA), and inflammatory vasculopathy. Mutations in several genes act by distinct mechanisms to cause sustained type I IFN signaling and resultant disease: (1) loss-of-function mutations leading to increased cytosolic DNA (TREX1 and SAMHD1) or RNA/DNA hybrid sensing (RNASEH2); (2) loss-of-function mutations leading to defects in RNA editing and aberrant recognition of self-RNA in the cytosol (ADAR1); (3) gain-of-function mutations in cytosolic RNA or DNA sensors leading to constitutive activation (MDA5 and STING); (4) loss-of-function mutations leading to altered unfolded protein responses and enhanced MAVS signaling (SKIV2L); and (5) loss-of-function mutations in negative regulators of IFNAR signaling (USP18) (Crow and Manel, 2015; Rodero and Crow, 2016; Yan, 2017).
The first evidence for a linkage between type I IFN signaling and autoimmunity came from patients receiving IFN therapy for chronic viral infections. A subset of these individuals developed signs of autoimmune diseases including SLE, RA, and polymyositis (Okanoue et al., 1996). Consistent with this observation, high amounts of serum IFN-α were present in SLE patients with disease activity, and genes in the IFN pathway (e.g., MDA5, TLR7, IRF5, and IRF7) are featured in clinical progression studies of SLE in humans (Thorlacius et al., 2018). Neutrophils and formation of extracellular traps (NETs) are observed in the kidneys of SLE patients, and type I IFN signaling can prime neutrophils to form NETs after stimulation with the complement factor C5a (Martinelli et al., 2004). The type I IFN response is also activated in patients with Sjögren’s syndrome, a chronic autoimmune disease that features focal lymphocytic infiltration of the exocrine glands, often causing dry eyes and dry mouth (Thorlacius et al., 2018).
The chronic skin disease psoriasis is characterized by epithelial cell hyperproliferation, impaired barrier function, and inflammation. Skin lesions from psoriasis patients exhibit high IFN-λ1 and ISG expression, which occurs in the absence of sustained expression of type I IFNs and appears to be driven by Th17 cells (Wolk et al., 2013). Analysis of IFN-λ-treated human keratinocytes revealed induction of the chemokines CXCL10 and CXCL11, which attract NK cells, CD8+ T cells, and CD4+ T cells (Witte et al., 2016). Elevated IFN-λ expression in psoriatic lesions was associated with upregulation of these chemokines compared with non-lesional skin from these patients or skin of healthy donors, both of which lack IFN-λ production (Witte et al., 2016).
The relationship between IFN-λ and autoimmune arthritis depends on the disease model. In the context of SLE, IFN-λ mRNA and protein amounts are higher in patients with active arthritis than in healthy controls, and serum IFN-λ amounts correlate with Th17 cytokine production and the extent of disease (Oke et al., 2017; Wu et al., 2011). RA patients had higher IFN-λ1 mRNA levels in PBMCs than healthy matched controls (Wu et al., 2013). However, in mice, treatment with IFN-λ reversed the development of collagen-induced arthritis by reducing numbers of pro-inflammatory Th17 and γδ T cells in the joints and inguinal lymph nodes (Blazek et al., 2015). This study showed that IFN-λ exerts an anti-inflammatory activity by restricting recruitment of IL-1β-expressing neutrophils; this result contrasts with the anti-inflammatory effects of IFN-λ in the intestine, where type III IFN signaling in neutrophils prevented granule release (Broggi et al., 2017).
Clinical Applications of Type I and Type III IFNs
The antiviral and immunomodulatory properties of type I IFNs have generated interest in their clinical use to control viral infections, enhance antigen presentation, and promote anti-tumor responses (Table 1). Although pre-clinical experiments suggest that type III IFNs might provide therapeutic benefits with diminished side effects compared with type I IFNs, no IFN-λ drug is approved for use in humans.
Table 1.
Clinical Applications of Type I and III IFNs
| Interferon | Form | Clinical Uses | Status | Reference |
|---|---|---|---|---|
| IFN-α-2a | recombinant | leukemia, melanoma, kaposi’s sarcoma | approved (Roferon-A) | (Cascinelli et al., 2001) |
| IFN-α-2a | pegylated | chronic HBV, chronic HCV, HPV (condylomata acuminata) | approved (Pegasys) | (McHutchison et al., 2009; Reichard et al., 1998) |
| IFN-α-2b | recombinant | leukemia, melanoma, multiple myeloma, carcinoid tumor, chronic HBV, chronic HCV, Bechet’s disease | approved (Intron-A) | (Janssen et al., 2005; Yao et al., 2017) |
| IFN-α-2b | pegylated | chronic HCV, melanoma | approved (Peglntron) | (Eggermont et al., 2008; Hansson et al., 2011; Lau et al., 2005; Manns et al., 2001; McHutchison et al., 2009) |
| IFN-β-1a | recombinant | multiple sclerosis | approved (Avonex, Rebif) | (Hauser et al., 2017; Kappos et al., 2015) |
| IFN-β-1a | pegylated | multiple sclerosis | approved (Plegridy) | (Khan et al., 2015; Newsome et al., 2016) |
| IFN-β-1b | recombinant | multiple sclerosis | approved (Betaseron, Actoferon) | (Bayas and Gold, 2003) |
| IFN-λ-1a | pegylated | chronic HDV | phase 2, completed 12/18 | |
| IFN-λ-1 | pegylated | chronic HCV | phase 1, completed 10/09 | |
| IFN-λ-1 | pegylated | HBV | phase 2, completed 12/13 | (Phillips et al., 2017) |
Since their initial discovery, there has been interest in harnessing the antiviral properties of IFNs for therapeutic use, and the greatest success to date was achieved with HCV. Prior to the advent of direct-acting antiviral therapy (which targets the HCV protease polymerase, and NS5A protein), IFN-α alone or in combination with ribavirin was the mainstay of anti-HCV therapy (Davis et al., 1989; Di Bisceglie et al., 1989; Heim, 2013). The mechanism of activity of IFN-α against HCV has remained somewhat uncertain, although it most likely is a combination of direct antiviral mechanisms and immunomodulatory effects on CD8+ T cell responses. Although iterative improvement in IFN-α treatment efficacy was achieved with pegylation and in combination with ribavirin, ultimately challenges including viral escape mechanisms, refractory IFN-α signaling in the liver, modest sustained virological response rates, and drug toxicity have relegated IFN-α treatment to a second-tier status in most developed countries (Sarasin-Filipowicz et al., 2008). The failure of IFN therapy against HCV is highest in patients with a pre-existing elevated ISG signature. The reduced responsiveness to IFN in some patients is thought to be due to altered IFN signaling pathways, epigenetic modification, and diminished transcription and/or translation responsiveness (Snell et al., 2017). Notwithstanding these limitations, pegylated IFN-α in combination with ribavirin remains a treatment option especially for patients with favorable IFNL genotypes (Huang et al., 2017). Although no IFN-λ drug is approved for use in humans, dose-ranging studies evaluated the safety, efficacy, and pharmacokinetics of pegylated IFN-λ therapy in chronic HCV infection (). A phase IIb study showed similar efficacy of pegylated IFN-λ1 treatment as for IFN-α treatment for chronic HCV infection (Muir et al., 2014; Muir et al., 2010). IFN-λ1 treatment was associated with improved or similar rates of virological response with fewer side effects than IFN-α in chronic HCV infection.
IFN-α treatment of chronic hepatitis B virus (HBV) infection was first reported in 1976, when four treated patients showed marked reductions or clearance in viral antigens (Greenberg et al., 1976). A 24-week treatment course of pegylated-IFN-α2A administered once weekly resulted in a higher response rate (24% versus 12%) than standard IFN-α, as defined by loss in HBV antigenemia, reductions in HBV DNA in blood, and improvement in serum liver enzymes (Cooksley et al., 2003). Based on its convenience, patient compliance, and likely efficacy, pegylated-IFN-α2A has replaced standard IFN-α therapy. Analogous to HCV treatment, the mechanism of action of IFN therapy against HBV has remained uncertain, but it most likely acts on multiple steps of the HBV life cycle as well as enhances NK and CD8+ T-cell-mediated clearance of infected cells. IFN-α in part inhibits HBV replication epigenetically by decreasing RNA transcription of pregenomic RNA and subgenomic RNA from covalently closed circular DNA (Belloni et al., 2012). As another potential mechanism, IFN-α induces APOBEC3G protein expression, which results in G to A hypermutation in HBV DNA and inhibition of replication (Suspène et al., 2005). Nonetheless, the standard 48-week pegylated-IFN-α2A therapy is difficult to tolerate (Terrault et al., 2016), and treatment is contraindicated in pregnant women and in those with advanced liver disease and decompensated cirrhosis. Following a dose ranging trial that confirmed its safety in humans (), a phase IIb trial showed that pegylated IFN-λ therapy also enhanced NK and CD8+ T cell effector responses that reduced HBV viral replication and antigenemia (Phillips et al., 2017). More recently, phase II trials with pegylated IFN-λ were initiated against chronic infection with hepatitis delta virus (HDV), which exacerbates HBV disease (). In preliminary results, patients in one of the pegylated IFN-λ treatment groups experienced a −2.4 log10 mean decline in HDV-RNA, and 4 out of 10 patients were HDV-RNA negative at the end of treatment.
Treatment of cancer-promoting infections such as HCV and HBV is an indirect means by which IFN therapies combat cancer, but the anti-tumor effects of IFNs have also been evaluated more directly. Before the development of more targeted cancer immunotherapies, type I IFNs alone or in combination with conventional chemotherapy were used to treat selected hematological malignancies (e.g., lymphoma, chronic myeloid leukemia, and hairy cell leukemia), and solid tumors (Kaposi’s sarcoma and renal cell carcinoma). Type I IFNs (IFN-α2A, IFN-α2B, and more recently, pegylated IFN-α2B) are still part of a therapeutic regimen against melanoma, principally as adjuvant therapy after surgical resection in high-risk patients. Type I IFNs are thought to exert their activity against melanoma by increasing the number of antigen-presenting cells infiltrating the tumor, decreasing the number of circulating regulatory T cells, altering the cytokine environment, changing the STAT1 and STAT3 signaling balance in tumor cells and host lymphocytes, and inducing self-recognition with auto-antibodies (Di Trolio et al., 2015). A meta-analysis of melanoma patients found that IFN-α improved disease-free outcome and survival, although the benefit was modest (Mocellin et al., 2010). In the future, IFN-α treatment might be combined with other immunotherapies to control or eliminate tumors and metastatic disease. This approach is premised on the ability of IFN-α to condition autologous DCs for adoptive cell immunotherapies and vaccines as well as the ability of type I IFNs to enhance NK and T cell activation, thereby augmenting drugs that promote immunogenic cell death or immune recognition of tumor neoantigens.
Although dysregulated IFN signaling can mediate some autoimmune conditions, type I IFNs have been used to treat multiple sclerosis (MS), a chronic, debilitating autoimmune disease characterized by inflammation in the CNS. In MS patients, genetic and/or environmental triggers induce activation and CNS migration of autoreactive T and B cells that respond to antigens similar to myelin and cause injury to myelin sheaths and oligodendrocytes, thereby altering neuronal functions. After an initial report of successful intrathecal therapy in ten patients (Jacobs et al., 1981), IFN-β became the first major drug class used for treatment of MS with its approval in 1993 (The IFNB Multiple Sclerosis Study Group, 1993). IFN-β therapy reduced the relapse rate, the development of brain lesions, and the progression of disability (Dhib-Jalbut and Marks, 2010). In one 21-year follow-up analysis of MS patients, IFN-β-1b therapy showed a 46.8% reduction in the long-term all-cause mortality compared with that in placebo (Goodin et al., 2012). Despite decades of clinical use, the mechanism of action of IFN-β in MS remains only partially understood (Jakimovski et al., 2018), and is thought to be related to the downregulation of the MHC class II expression on the antigen-presenting cells, induction of the regulatory T cells and the inhibitory cytokine IL-10, inhibition of T cell proliferation, and reduced adhesion molecule expression and inhibition of T cell migration across the BBB (Dhib-Jalbut and Marks, 2010).
Concluding Remarks
Type I and type III IFNs were first recognized for their antiviral activity, but it is now appreciated that they also have a multitude of immunomodulatory functions that influence tumor responses, autoimmune disease, and microbial infection. Although many of the activities induced by type I and type III IFNs overlap, spatial and kinetic differences in their responses allow type III IFNs to provide front-line protection at barrier surfaces, thereby minimizing the activation of the systemic type I IFN response and consequent immune pathology. In addition to epithelial cells, the range of cell types now known to respond to type III IFNs has expanded, and neutrophils are an example of a highly IFN-λ-responsive cell type that can exert activities through non-canonical pathways. IFN-λ-mediated neutrophil activation, as well as cell junction tightening by type I and type III IFNs, stands out for being STAT- and translation-independent. Along with activation of MAPK signaling by type I and type III IFNs and IFNAR2-independent signaling by IFN-β, this suggests that IFNs might have previously unappreciated activities that occur through distinct signaling pathways. Given that the dominant JAK-STAT signaling pathway activated by type I and type III IFNs is the same, it is unsurprising that they induce highly similar transcriptional responses. However, distinct transcriptional and functional responses might become more evident as these studies are extended to more physiologically relevant experimental systems, in which cell- or tissue-specific differences in signaling or effector molecules could influence the outcome of IFN signaling. It remains to be seen whether any specific ISGs are uniquely induced by type I versus type III IFNs, or whether differences in the type I and type III function will be fully attributable to differences in signaling potency and kinetics or tissue-specific responsiveness. Likewise, the specific functions of individual IFN subtypes that signal through the same receptor (e.g., 17 different human type I IFNs) remains unclear. The presence of large multigene IFN families is a conserved feature of vertebrates, even though particular IFN genes do not necessarily have orthologs in all species. In this review, we have discussed only human and mouse IFNs, but the number and diversity of IFN genes present in nature provides a wealth of opportunities to discover new IFN properties and biology.
ACKNOWLEDGMENTS
Research in the authors’ laboratories is supported in part by NIH grants R01AI139512 to H.M.L.; DP2AI117922 to J.W.S.; and R01AI073755, R01AI27828, and R01HD091218 to M.S.D.
Footnotes
DECLARATION OF INTERESTS
M.S.D. is a consultant for Inbios and Atreca and is on the Scientific Advisory Board of Moderna.
REFERENCES
- Aboulnasr F, Hazari S, Nayak S, Chandra PK, Panigrahi R, Ferraris P, Chava S, Kurt R, Song K, Dash A, et al. (2015). IFN-λ inhibits MiR-122 transcription through a Stat3-HNF4α inflammatory feedback loop in an IFN-α resistant HCV cell culture system. PLoS ONE 10, e0141655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abushahba W, Balan M, Castaneda I, Yuan Y, Reuhl K, Raveche E, de la Torre A, Lasfar A, and Kotenko SV (2010). Antitumor activity of type I and type III interferons in BNL hepatoma model. Cancer Immunol. Immunother 59, 1059–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alspach E, Lussier DM, and Schreiber RD (2018). Interferon γ and its important roles in promoting and inhibiting spontaneous and therapeutic cancer immunity. Cold Spring Harb. Perspect. Biol 11, a028480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ander SE, Diamond MS, and Coyne CB (2019). Immune responses at the maternal-fetal interface. Sci. Immunol 4, eaat6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade D, Kim M, Blanco LP, Karumanchi SA, Koo GC, Redecha P, Kirou K, Alvarez AM, Mulla MJ, Crow MK, et al. (2015). Interferon-α and angiogenic dysregulation in pregnant lupus patients who develop preeclampsia. Arthritis Rheumatol 67, 977–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ank N, Iversen MB, Bartholdy C, Staeheli P, Hartmann R, Jensen UB, Dagnaes-Hansen F, Thomsen AR, Chen Z, Haugen H, et al. (2008). An important role for type III interferon (IFN-lambda/IL-28) in TLR-induced antiviral activity. J. Immunol 180, 2474–2485. [DOI] [PubMed] [Google Scholar]
- Baldridge MT, Nice TJ, McCune BT, Yokoyama CC, Kambal A, Wheadon M, Diamond MS, Ivanova Y, Artyomov M, and Virgin HW (2015). Commensal microbes and interferon-λ determine persistence of enteric murine norovirus infection. Science 347, 266–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldridge MT, Lee S, Brown JJ, McAllister N, Urbanek K, Dermody TS, Nice TJ, and Virgin HW (2017). Expression of Ifnlrl on intestinal epithelial cells is critical to the antiviral effects of interferon lambda against norovirus and reovirus. J. Virol 91, e02079–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamford CGG, Aranday-Cortes E, Filipe LC., Sukumar S, Mair D, Filipe ADS, Mendoza JL, García KC, Fan S, Tishkoff SA, and McLauchlan J (2018). A polymorphic residue that attenuates the antiviral potential of interferon lambda 4 in hominid lineages. PLoS Pathog 14, e1007307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayas A, and Gold R (2003). Lessons from 10 years of interferon beta-1b (Betaferon/Betaseron) treatment. J. Neurol 250 (Suppl 4), IV3–IV8. [DOI] [PubMed] [Google Scholar]
- Bayer A, Lennemann NJ, Ouyang Y, Bramley JC, Morosky S, Marques ET Jr., Cherry S, Sadovsky Y, and Coyne CB (2016). Type III interferons produced by human placental trophoblasts confer protection against Zika virus infection. Cell Host Microbe 19, 705–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belloni L, Allweiss L, Guerrieri F, Pediconi N, Volz T, Pollicino T, Petersen J, Raimondo G, Dandri M, and Levrero M (2012). IFN-α inhibits HBV transcription and replication in cell culture and in humanized mice by targeting the epigenetic regulation of the nuclear cccDNA minichromosome. J. Clin. Invest 122, 529–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne H, Travier L, Mahlakõiv T, Tailleux L, Subtil A, Lebreton A, Paliwal A, Gicquel B, Staeheli P, Lecuit M, and Cossart P (2012). Activation of type III interferon genes by pathogenic bacteria in infected epithelial cells and mouse placenta. PLoS ONE 7, e39080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazek K, Eames HL, Weiss M, Byrne AJ, Perocheau D, Pease JE, Doyle S, McCann F, Williams RO, and Udalova IA (2015). IFN-λ resolves inflammation via suppression of neutrophil infiltration and IL-1β production. J. Exp. Med 212, 845–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolen CR, Ding S, Robek MD, and Kleinstein SH (2014). Dynamic expression profiling of type I and type III interferon-stimulated hepatocytes reveals a stable hierarchy of gene expression. Hepatology 59,1262–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booy S, Hofland L, and van Eijck C (2015). Potentials of interferon therapy in the treatment of pancreatic cancer. J. Interferon Cytokine Res 35,327–339. [DOI] [PubMed] [Google Scholar]
- Borden EC (2019). Interferons α and β in cancer: therapeutic opportunities from new insights. Nat. Rev. Drug Discov 18, 219–234. [DOI] [PubMed] [Google Scholar]
- Boxx GM, and Cheng G (2016). The roles of type I interferon in bacterial infection. Cell Host Microbe 19, 760–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broggi A, Tan Y, Granucci F, and Zanoni I (2017). IFN-λ suppresses intestinal inflammation by non-transiational regulation of neutrophil function. Nat. Immunol 18, 1084–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budhwani M, Mazzieri R, and Dolcetti R (2018). Plasticity of type I interferon-mediated responses in cancer therapy: from anti-tumor immunity to resistance. Front. Oncol 8, 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkart C, Arimoto K, Tang T, Cong X, Xiao N, Liu YC, Kotenko SV, Ellies LG, and Zhang DE (2013). Usp18 deficient mammary epithelial cells create an antitumour environment driven by hypersensitivity to IFN-λ and elevated secretion of Cxcl10. EMBO Mol. Med 5, 1035–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caine EA, Scheaffer SM, Arora N, Zaitsev K, Artyomov MN, Coyne CB, Moley KH, and Diamond MS (2019). Interferon lambda protects the female reproductive tract against Zika virus infection. Nat. Commun 10, 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascinelli N, Belli F, Mackie RM, Santinami M, Bufalino R, and Mora-bito A (2001). Effect of long-term adjuvant therapy with interferon alpha-2a in patients with regional node metastases from cutaneous melanoma: a randomised trial. Lancet 358, 866–869. [DOI] [PubMed] [Google Scholar]
- Cohen TS, and Prince AS (2013). Bacterial pathogens activate a common inflammatory pathway through IFNλ regulation of PDCD4. PLoS Pathog. 9, e1003682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooksley WG, Piratvisuth T, Lee SD, Mahachai V, Chao YC, Tanwan-dee T, Chutaputti A, Chang WY, Zahm FE, and Pluck N (2003). Peginterferon alpha-2a (40 kDa): an advance in the treatment of hepatitis B e antigen-positive chronic hepatitis B. J. Viral Hepat 10, 298–305. [DOI] [PubMed] [Google Scholar]
- Corry J, Arora N, Good CA, Sadovsky Y, and Coyne CB (2017). Organotypic models of type 111 interferon-mediated protection from Zika virus infections at the maternal-fetal interface. Proc. Natl. Acad. Sci. USA 114, 9433–9438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotta S, Davidson S, Mahlakoiv T, Desmet CJ, Buckwaiter MR, Albert ML, Staeheli P, and Wack A (2013). Type I and type III interferons drive redundant amplification loops to induce a transcriptional signature in influenza-infected airway epithelia. PLoS Pathog 9, e1003773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouse J, Kalinke U, and Oxenius A (2015). Regulation of antiviral T cell responses by type I interferons. Nat. Rev. Immunol 15, 231–242. [DOI] [PubMed] [Google Scholar]
- Crow YJ, and Manel N (2015). Aicardi-Goutières syndrome and the type I interferonopathies. Nat. Rev. Immunol 75, 429–440. [DOI] [PubMed] [Google Scholar]
- Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, Lin TY, Schneller S, Zust R, Dong H, et al. (2010). 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468, 452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Megjugorac NJ, Gallagher GE, Yu RY, and Gallagher G (2009). IFN-lambda1 (IL-29) inhibits GATA3 expression and suppresses Th2 responses in human naive and memory T cells. Blood 7 73, 5829–5838. [DOI] [PubMed] [Google Scholar]
- Daniels BP, Holman DW, Cruz-Orengo L, Jujjavarapu H, Durrant DM, and Klein RS (2014). Viral pathogen-associated molecular patterns regulate blood-brain barrier integrity via competing innate cytokine signals. MBio 5, e01476–e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels BP, Jujjavarapu H, Durrant DM, Williams JL, Green RR, White JP, Lazear HM, Gale M Jr., Diamond MS, and Klein RS (2017). Regional astrocyte IFN signaling restricts pathogenesis during neurotropic viral infection. J. Clin. Invest 127, 843–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis GL, Baiart LA, Schiff ER, Lindsay K, Bodenheimer HC Jr., Perrillo RP, Carey W, Jacobson IM, Payne J, and Dienstag JL; Hepatitis Interventional Therapy Group (1989). Treatment of chronic hepatitis C with recombinant interferon alfa. A multicenter randomized, controlled trial. N. Engl. J. Med 321, 1501–1506. [DOI] [PubMed] [Google Scholar]
- de Groen RA, Groothuismink ZM, Liu BS, and Boonstra A (2015). IFN-λ is able to augment TLR-mediated activation and subsequent function of primary human B cells. J. Leukoc. Biol 98, 623–630. [DOI] [PubMed] [Google Scholar]
- de Weerd NA, Vivian JP, Nguyen TK, Mangan NE, Gould JA, Braniff SJ, Zaker-Tabrizi L, Fung KY, Forster SC, Beddoe T, et al. (2013). Structural basis of a unique interferon-β signaling axis mediated via the receptor IFNAR1. Nat. Immunol 14, 901–907. [DOI] [PubMed] [Google Scholar]
- de Weerd NA, Matthews AY, Pattie PR, Bourke NM, Lim SS, Vivian JP, Rossjohn J, and Hertzog PJ (2017). A hot spot on interferon α/β receptor subunit 1 (IFNAR1) underpins its interaction with interferon-β and dictates signaling. J. Biol. Chem 292, 7554–7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaye S, Paul S, Blakqori G, Minet M, Weber F, Staeheli P, and Michiels T (2006). Neurons produce type I interferon during viral encephalitis. Proc. Natl. Acad. Sci. USA 103, 7835–7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhib-Jalbut S, and Marks S (2010). Interferon-beta mechanisms of action in multiple sclerosis. Neurology 74 (Suppl 1), S17–S24. [DOI] [PubMed] [Google Scholar]
- Di Bisceglie AM, Martin P, Kassianides C, Lisker-Melman M, Murray L, Waggoner J, Goodman Z, Banks SM, and Hoofnagle JH (1989). Recombinant interferon alfa therapy for chronic hepatitis C. A randomized, doubleblind, placebo-controlled trial. N. Engl. J. Med 321, 1506–1510. [DOI] [PubMed] [Google Scholar]
- Di Trolio R, Simeone E, Di Lorenzo G, Buonerba C, and Ascierto PA (2015). The use of interferon in melanoma patients: a systematic review. Cytokine Growth Factor Rev. 26, 203–212. [DOI] [PubMed] [Google Scholar]
- Ding Q, Gaska JM, Douam F, Wei L, Kim D, Balev M, Heller B, and Ploss A (2018). Species-specific disruption of STING-dependent antiviral cellular defenses by the Zika virus NS2B3 protease. Proc. Natl. Acad. Sci. USA 115, E6310–E6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douam F, Soto Albrecht YE, Hrebikova G, Sadimin E, Davidson C, Kotenko SV, and Ploss A (2017). Type III interferon-mediated signaling is critical for controlling live attenuated yellow fever virus infection in vivo. MBio 8, e00819–17, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle SE, Schreckhise H, Khuu-Duong K, Henderson K, Rosier R, Storey H, Yao L, Liu H, Barahmand-pour F, Sivakumar P, et al. (2006). Inter-leukin-29 uses a type 1 interferon-like program to promote antiviral responses in human hepatocytes. Hepatology 44, 896–906. [DOI] [PubMed] [Google Scholar]
- Drokhlyansky E, Göz Aytürk D, Soh TK, Chrenek R, O’Loughlin E, Madore C, Butovsky O, and Cepko CL (2017). The brain parenchyma has a type I interferon response that can limit virus spread. Proc. Natl. Acad. Sci. USA 114, E95–E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan CJ, Mohamad SM, Young DF, Skelton AJ, Leahy TR, Munday DC, Butler KM, Morfopoulou S, Brown JR, Hubank M, et al. (2015). Human 1FNAR2 deficiency: Lessons for antiviral immunity. Sci. Transi. Med 7, 307ra154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggermont AM, Suciu S, Santinami M, Testori A, Kruit WH, Marsden J, Punt CJ, Salès F, Gore M, Mackie R, et al. ; EORTC Melanoma Group (2008). Adjuvant therapy with pegylated interferon alfa-2b versus observation alone in resected stage III melanoma: final results of EORTC 18991, a randomised phase III trial. Lancet 372, 117–126. [DOI] [PubMed] [Google Scholar]
- Egli A, Santer DM, O’Shea D, Barakat K, Syedbasha M, Vollmer M, Baluch A, Bhat R, Groenendyk J, Joyce MA, et al. (2014). IL-28B is a key regulator of B- and T-cell vaccine responses against influenza. PLoS Pathog. 10, e1004556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa V, Dutta O, McElrath C, Du P, Chang YJ, Cicciarelli B, Pitler A, Whitehead I, Obar JJ, Durbin JE, et al. (2017). Type III interferon is a critical regulator of innate antifungal immunity. Sci. Immunol 2, eaan5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallet B, Narr K, Ertuna YI, Remy M, Sommerstein R, Cornille K, Kreutzfeldt M, Page N, Zimmer G, Geier F, et al. (2016). Interferon-driven deletion of antiviral B cells at the onset of chronic infection. Sci. Immunol 1, eaah6817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox BA, Sheppard PO, and O’Hara PJ (2009). The role of genomic data in the discovery, annotation and evolutionary interpretation of the interferon-lambda family. PLoS One 4, e4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JM, Crabtree JM, Sage LK, Tompkins SM, and Tripp RA (2015). Interferon lambda upregulates IDO1 expression in respiratory epithelial cells after influenza virus infection. J. Interferon Cytokine Res 35, 554–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- François-Newton V, Magno de Freitas Almeida G, Payelle-Brogard B, Monneron D, Pichard-García L, Piehler J, Pellegrini S, and Uzé G (2011). USP18-based negative feedback control is induced by type I and type III interferons and specifically inactivates interferon α response. PLoS ONE 6, e22200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung KY, Mangan NE, Cumming H, Horvat JC, Mayall JR, Stifter SA, De Weerd N, Roisman LC, Rossjohn J, Robertson SA, et al. (2013). Interferon-ε protects the female reproductive tract from viral and bacterial infection. Science 339,1088–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galani IE, Triantafyllia V, Eleminiadou EE, Koltsida O, Stavropoulos A, Manioudaki M, Thanos D, Doyle SE, Kotenko SV, Thanopoulou K, and Andreakos E (2017). Interferon-λ mediates non-redundant front-line antiviral protection against influenza virus infection without compromising host fitness. Immunity 46, 875–890.e6. [DOI] [PubMed] [Google Scholar]
- García-Sastre A (2017). Ten strategies of interferon evasion by viruses. Cell Host Microbe 22, 176–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gizzi AS, Grove TL, Arnold JJ, Jose J, Jangra RK, Garforth SJ, Du Q, Cahill SM, Dulyaninova NG, Love JD, et al. (2018). A naturally occurring antiviral ribonucleotide encoded by the human genome. Nature 558, 610–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good C, Wells AI, and Coyne CB (2019). Type III interferon signaling restricts enterovirus 71 infection of goblet cells. Sci. Adv 5, u4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodin DS, Ebers GG, Cutter G, Cook SD, O’Donnell T, Reder AT, Kremenchutzky M, Oger J, Rametta M, Beckmann K, and Knappertz V (2012). Cause of death in MS: long-term follow-up of a randomised cohort, 21 years after the start of the pivotal IFNβ−1b study. BMJ Open 2, e001972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goritzka M, Durant LR, Pereira C, Salek-Ardakani S, Openshaw PJ, and Johansson C (2014). Alpha/beta interferon receptor signaling amplifies early proinflammatory cytokine production in the lung during respiratory syncytial virus infection. J. Virol 88, 6128–6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goritzka M, Makris S, Kausar F, Durant LR, Pereira C, Kumagai Y, Culley FJ, Mack M, Akira S, and Johansson C (2015). Alveolar macrophage-derived type I interferons orchestrate innate immunity to RSV through recruitment of antiviral monocytes. J. Exp. Med 212, 699–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman MJ, Caine EA, Zaitsev K, Begley MC, Weger-Lucarelli J, Uccellini MB, Tripathi S, Morrison J, Yount BL, Dinnon KH 3rd, et al. (2018). An immunocompetent mouse model of Zika virus infection. Cell Host Microbe 23, 672–685.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant A, Ponia SS, Tripathi S, Balasubramaniam V, Miorin L, Souris-seau M, Schwarz MC, Sánchez-Seco MP, Evans MJ, Best SM, and García-Sastre A (2016). Zika virus targets human STAT2 to inhibit type I interferon signaling. Ceil Host Microbe 19, 882–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg HB, Pollard RB, Lutwick LI, Gregory PB, Robinson WS, and Merigan TC (1976). Effect of human leukocyte interferon on hepatitis B virus infection in patients with chronic active hepatitis. N. Engl. J. Med 295, 517–522. [DOI] [PubMed] [Google Scholar]
- Griffiths SJ, Dunnigan CM, Russell CD, and Haas JG (2015). The role of interferon-λ locus polymorphisms in hepatitis C and other infectious diseases. J. Innate Immun 7, 231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grine L, Dejager L, Libert C , and Vandenbroucke RE (2015). An inflammatory triangle in psoriasis: TNF, type I IFNs and IL-17. Cytokine Growth Factor Rev 26, 25–33. [DOI] [PubMed] [Google Scholar]
- Haller O, Staeheli P, Schwemmle M, and Kochs G (2015). Mx GTPases: dynamin-like antiviral machines of innate immunity. Trends Microbiol. 23, 154–163. [DOI] [PubMed] [Google Scholar]
- Hamming OJ, Terczyńska-Dyla E, Vieyres G, Dijkman R, Jørgensen SE, Akhtar H, Siupka P, Pietschmann T, Thiel V, and Hartmann R (2013). Interferon lambda 4 signals via the IFNλ receptor to regulate antiviral activity against HCV and coronaviruses. EMBO J. 32, 3055–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson J, Aamdal S, Bastholt L, Brandberg Y, Hernberg M, Nilsson B, Stierner U, and von der Maase H; Nordic Melanoma Cooperative Group (2011). Two different durations of adjuvant therapy with intermediate-dose interferon alfa-2b in patients with high-risk melanoma (Nordic IFN trial): a randomised phase 3 trial. Lancet Oncol. 12, 144–152. [DOI] [PubMed] [Google Scholar]
- Harris BD, Schreiter J, Chevrier M, Jordan JL, and Walter MR (2018). Human interferon-ε and interferon-κ exhibit low potency and low affinity for cell-surface IFNAR and the poxvirus antagonist B18R. J. Biol. Chem 293, 16057–16068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser SL, Bar-Or A, Comi G, Giovannoni G, Hartung HP, Hemmer B, Lublin F, Montalban X, Rammohan KW, Selmaj K, et al. ; OPERA I and OPERA II Clinical Investigators (2017). Ocrelizumab versus Interferon Beta-1a in Relapsing Multiple Sclerosis. N. Engl. J. Med 376, 221–234. [DOI] [PubMed] [Google Scholar]
- Heim MH (2013). 25 years of interferon-based treatment of chronic hepatitis C: an epoch coming to an end. Nat. Rev. Immunol 13, 535–542. [DOI] [PubMed] [Google Scholar]
- Hendricks MR, Lashua LP, Fischer DK, Flitter BA, Eichinger KM, Durbin JE, Sarkar SN, Coyne CB, Empey KM, and Bomberger JM (2016). Respiratory syncytial virus infection enhances Pseudomonas aeruginosa biofilm growth through dysregulation of nutritional immunity. Proc. Natl. Acad. Sci. USA 113, 1642–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermant P, Francius C, Clotman F, and Michiels T (2013). IFN-ε is constitutively expressed by cells of the reproductive tract and is inefficiently secreted by fibroblasts and cell lines. PLoS ONE 8, e71320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho HH, and Ivashkiv LB (2006). Role of STAT3 in type I interferon responses. Negative regulation of STAT1-dependent inflammatory gene activation. J. Biol. Chem 281, 14111–14118. [DOI] [PubMed] [Google Scholar]
- Hoffmann HH, Schneider WM, and Rice CM (2015). Interferons and viruses: an evolutionary arms race of molecular interactions. Trends Immunol. 36, 124–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, and Taniguchi T (2005). IRF-7 is the master regulator of type-l interferon-dependent immune responses. Nature 434, 772–777. [DOI] [PubMed] [Google Scholar]
- Honda K, Takaoka A, and Taniguchi T (2006). Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity 25, 349–360. [DOI] [PubMed] [Google Scholar]
- Hornung V, Hartmann R, Ablasser A, and Hopfner KP (2014). OAS proteins and cGAS: unifying concepts in sensing and responding to cytosolic nucleic acids. Nat. Rev. Immunol 14, 521–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Li MH, Hou M, and Xie Y (2017). Peginterferon alfa-2a for the treatment of chronic hepatitis C in the era of direct-acting antivirals. HBPD INT 16, 470–479. [DOI] [PubMed] [Google Scholar]
- Hwang M, and Bergmann CC (2018). Alpha/Beta Interferon (IFN-α/β) signaling in astrocytes mediates protection against viral encephalomyelitis and regulates IFN-γ-dependent responses. J. Virol 92, e01901–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde JL, Gardner CL, Kimura T, White JP, Liu G, Trobaugh DW, Huang C,Tonelli M, Paessler S, Takeda K, et al. (2014). A viral RNA structural element alters host recognition of nonself RNA. Science 343, 783–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingle H, Peterson ST, and Baldridge MT (2018). Distinct effects of type I and III interferons on enteric viruses. Viruses 10, E46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs A, and Lindenmann J (1957). Virus interference. I. The interferon. Proc. R. Soc. Lond. B Biol. Sci 147, 258–267. [PubMed] [Google Scholar]
- Isaacs A, Lindenmann J, and Valentine RC (1957). Virus interference. II. Some properties of interferon. Proc. R. Soc. Lond. B Biol. Sci 147, 268–273. [PubMed] [Google Scholar]
- Jacobs L, O’Malley J, Freeman A, and Ekes R (1981). Intrathecal interferon reduces exacerbations of multiple sclerosis. Science 214,1026–1028. [DOI] [PubMed] [Google Scholar]
- Jagger BW, Miner JJ, Cao B, Arora N, Smith AM, Kovacs A, Mysorekar IU, Coyne CB, and Diamond MS (2017). Gestational stage and IFN-λ signaling regulate ZIKV infection in utero. Cell Host Microbe 22, 366–376.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakimovski D, Kolb C, Ramanathan M, Zivadinov R, and Weinstock-Guttman B (2018). Interferon β for multiple sclerosis. Cold Spring Harb. Perspect. Med 8, a032003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen HL, van Zonneveld M, Senturk H, Zeuzem S, Akarca US, Cakaloglu Y, Simon C, So TM, Gerken G, de Man RA, et al. ; HBV 99–01 Study Group; Rotterdam Foundation for Liver Research (2005). Pegylated interferon alfa-2b alone or in combination with lamivudine for HBeAg-positive chronic hepatitis B: a randomised trial. Lancet 365,123–129. [DOI] [PubMed] [Google Scholar]
- Jewell NA, Cline T, Mertz SE, Smirnov SV, Flaño E, Schindler C, Grieves JL, Durbin RK, Kotenko SV, and Durbin JE (2010). Lambda interferon is the predominant interferon induced by influenza A virus infection in vivo. J. Virol 84, 11515–11522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jilg N, Lin W, Hong J, Schaefer EA, Wolski D, Meixong J, Goto K, Brisac C, Chusri P, Fusco DN, et al. (2014). Kinetic differences in the induction of interferon stimulated genes by interferon-α and interleukin 28B are altered by infection with hepatitis C virus. Hepatology 59, 1250–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson C, Wetzel JD, He J, Mikacenic C, Dermody TS, and Kelsall BL (2007). Type I interferons produced by hematopoietic cells protect mice against lethal infection by mammalian reovirus. J. Exp. Med 204,1349–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan WJ, Eskdale J, Srinivas S, Pekarek V, Keiner D, Rodia M, and Gallagher G (2007). Human interferon lambda-1 (IFN-lambda1/IL-29) modulates the Th1/Th2 response. Genes Immun. 8, 254–261. [DOI] [PubMed] [Google Scholar]
- Kagan JC, Su T, Horng T, Chow A, Akira S, and Medzhitov R (2008). TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-beta. Nat. Immunol 9, 361–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappos L, Wiendl H, Selmaj K, Arnold DL, Havrdova E, Boyko A, Kaufman M, Rose J, Greenberg S, Sweetser M, et al. (2015). Daclizumab HYP versus Interferon Beta-1a in Relapsing Multiple Sclerosis. N. Engl. J. Med 373, 1418–1428. [DOI] [PubMed] [Google Scholar]
- Kernbauer E, Ding Y, and Cad well K (2014). An enteric virus can replace the beneficial function of commensal bacteria. Nature 516, 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Key FM, Peter B, Dennis MY, Huerta-Sánchez E, Tang W, Prokunina-Olsson L, Nielsen R, and Andrés AM (2014). Selection on a variant associated with improved viral clearance drives local, adaptive pseudogenization of interferon lambda 4 (IFNL4). PLoS Genet. 10, e1004681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan UT, Tanasescu R, and Constantinescu CS (2015). PEGylated IFN-−1a in the treatment of multiple sclerosis. Expert Opin. Biol. Ther 15, 1077–1084. [DOI] [PubMed] [Google Scholar]
- Kirkwood JM, Strawderman MH, Emstoff MS, Smith TJ, Borden EC, and Blum RH (1996). Interferon alfa-2b adjuvant therapy of high-risk resected cutaneous melanoma: the Eastern Cooperative Oncology Group Trial EST 1684. J. Clin. Oncol 14, 7–17. [DOI] [PubMed] [Google Scholar]
- Kirkwood JM, Ibrahim JG, Sosman JA, Sondak VK, Agarwala SS, Ernstoff MS, and Rao U (2001). High-dose interferon alfa-2b significantly prolongs relapse-free and overall survival compared with the GM2-KLH/QS-21 vaccine in patients with resected stage IIB-III melanoma: results of intergroup trial E1694/S9512/C509801. J. Clin. Oncol 19, 2370–2380. [DOI] [PubMed] [Google Scholar]
- Klein RS, and Hunter CA (2017). Protective and pathological immunity during central nervous system infections. Immunity 46, 891–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinkhammer J, Schnepf D, Ye L, Schwaderlapp M,Gad HH, Hartmann R, Garcin D, Mahlakõiv T, and Staeheli P (2018). IFN-λ prevents influenza virus spread from the upper airways to the lungs and limits virus transmission. eLife 7, e33354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli A, Zhang X, Yang J, Russell RS, Donnelly RP, Sheikh F, Sherman A, Young H, Imamichi T, Lempicki RA, et al. (2012). Distinct and overlapping genomic profiles and antiviral effects of Interferon-λ and -α on HCV-infected and noninfected hepatoma cells. J. Viral Hepat 79, 843–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltsida O, Hausding M, Stavropoulos A, Koch S, Tzelepis G, Ubel C, Kotenko SV, Sideras P, Lehr HA, Tepe M, et al. (2011). IL-28A (IFN-λ2) modulates lung DC function to promote Th1 immune skewing and suppress allergic airway disease. EMBO Mol. Med 3, 348–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotenko SV, and Durbin JE (2017). Contribution of type III interferons to antiviral immunity: location, location, location. J. Biol. Chem 292, 7295–7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, Langer JA, Sheikh F, Dickensheets H, and Donnelly RP (2003). IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol 4, 69–77. [DOI] [PubMed] [Google Scholar]
- Krause CD, and Pestka S (2015). Cut, copy, move, delete: The study of human interferon genes reveal multiple mechanisms underlying their evolution in amniotes. Cytokine 76, 480–495. [DOI] [PubMed] [Google Scholar]
- Kumar A, Hou S, Airo AM, Limonta D, Mancinelli V, Branton W, Power C, and Hobman TC (2016). Zika virus inhibits type-I interferon production and downstream signaling. EMBO Rep. 17,1766–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFleur DW, Nardelli B, Tsareva T, Mather D, Feng P, Semenuk M, Taylor K, Buergin M, Chinchilla D, Roshke V, et al. (2001). Interferonkappa, a novel type I interferon expressed in human Keratinocytes. J. Biol. Chem 276, 39765–39771. [DOI] [PubMed] [Google Scholar]
- Lasfar A, Lewis-Antes A, Smirnov SV, Anantha S, Abushahba W, Tian B, Reuhl K, Dickensheets H, Sheikh F, Donnelly RP, et al. (2006). Characterization of the mouse IFN-lambda ligand-receptor system: IFN-lambdas exhibit antitumor activity against B16 melanoma. Cancer Res. 66, 4468–4477. [DOI] [PubMed] [Google Scholar]
- Lasfar A, Gogas H, Zloza A, Kaufman HL, and Kirkwood JM (2016). IFN-λ cancer immunotherapy: new kid on the block. Immunotherapy 8, 877–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau GK, Piratvisuth T, Luo KX, Marcellin P, Thongsawat S, Cooksley G, Gane E, Fried MW, Chow WC, Paik SW, et al. ; Peginterferon Alfa-23 HBeAg-Positive Chronic Hepatitis B Study Group (2005). Peginterferon Alfa-2a, lamivudine, and the combination for HBeAg-positive chronic hepatitis B. N. Engl. J. Med 352, 2682–2695. [DOI] [PubMed] [Google Scholar]
- Lazear HM, Lancaster A, Wilkins C, Suthar MS, Huang A, Vick SC, Clepper L, Thackray L, Brassil MM, Virgin HW, et al. (2013). IRF-3, IRF-5, and IRF-7 coordinately regulate the type I IFN response in myeloid dendritic cells downstream of MAVS signaling. PLoS Pathog. 9, e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear HM, Daniels BP, Pinto AK, Huang AC, Vick SC, Doyle SE, Gale M Jr., Klein RS, and Diamond MS (2015a). Interferon-λ restricts West Nile virus neuroinvasion by tightening the blood-brain barrier. Sci. Transi. Med 7, 284ra59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear HM, Nice TJ, and Diamond MS (2015b). Interferon-λ: immune functions at barrier surfaces and beyond. Immunity 43, 15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear HM, Govero J, Smith AM, Platt DJ, Fernandez E, Miner JJ, and Diamond MS (2016). A mouse model of Zika virus pathogenesis. Cell Host Microbe 19, 720–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bon A, Schiavoni G, D’Agostino G, Gresser I, Belardelli F, and Tough DF (2001). Type i interferons potently enhance humoral immunity and can promote isotype switching by stimulating dendritic cells in vivo. Immunity 14, 461–470. [DOI] [PubMed] [Google Scholar]
- Le Bon A, Etchart N, Rossmann C, Ashton M, Hou S, Gewert D, Borrow P, and Tough DF (2003). Cross-priming of CD8+ T cells stimulated by virus-induced type I interferon. Nat. Immunol 4,1009–1015. [DOI] [PubMed] [Google Scholar]
- Le Bon A, Thompson C, Kamphuis E, Durand V, Rossmann C, Kalinke U, and Tough DF (2006). Cutting edge: enhancement of antibody responses through direct stimulation of B and T cells by type I IFN. J. Immunol 176, 2074–2078. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Kim WJ, and Moon SK (2012). Role of the p38 MAPK signaling pathway in mediating interleukin-28A-induced migration of UMUC-3 cells. Int. J. Mol. Med 30, 945–952. [DOI] [PubMed] [Google Scholar]
- LeMessurier KS, Häcker H, Chi L, Tuomanen E, and Redecke V (2013). Type I interferon protects against pneumococcal invasive disease by inhibiting bacterial transmigration across the lung. PLoS Pathog. 9, e1003727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Lewis-Antes A, Huang J, Bafan M, and Kotenko SV (2008). Regulation of apoptosis by type III interferons. Cell Prolif. 41, 960–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Dong C, and Cooper MD (1998). Impairment of T and B cell development by treatment with a type I interferon. J. Exp. Med 187, 79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JD, Feng N, Sen A, Balan M, Tseng HC, McElrath C, Smirnov SV, Peng J, Yasukawa LL, Durbin RK, et al. (2016). Distinct roles of type I and type III interferons in intestinal immunity to homologous and heterologous rotavirus infections. PLoS Pathog. 12, e1005600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litinskiy MB, Nardelli B, Hilbert DM, He B, Schaffer A, Casali P, and Cerutti A (2002). DCs induce CD40-independent immunoglobulin class switching through BLyS and APRIL. Nat. Immunol 3, 822–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlakõiv T, Hernandez P, Gronke K, Diefenbach A, and Staeheli P (2015). Leukocyte-derived IFN-α/β and epithelial IFN-λ constitute a compartmentalized mucosal defense system that restricts enteric virus infections. PLoS Pathog. 11, e1004782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony R, Gargan S, Roberts KL, Bourke N, Keating SE, Bowie AG, O’Farrelly C, and Stevenson NJ (2017). A novel anti-viral role for STAT3 in IFN-α signalling responses. Cell. Mol. Life Sci 74,1755–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiftman M, Reindollar R, Goodman ZD, Koury K, Ling M, and Albrecht JK (2001). Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358, 958–965. [DOI] [PubMed] [Google Scholar]
- Marcello T, Grakoui A, Barba-Spaeth G, Machlin ES, Kotenko SV, MacDonald MR, and Rice CM (2006). Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation Kinetics. Gastroenterology 131, 1887–1898. [DOI] [PubMed] [Google Scholar]
- Martin PK, Marchiando A, Xu R, Rudensky E, Yeung F, Schuster SL, Kernbauer E, and Cadwell K (2018). Autophagy proteins suppress protective type I interferon signalling in response to the murine gut microbiota. Nat. Microbiol 3, 1131–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli S, Urosevic M, Daryadel A, Oberholzer PA, Baumann C, Fey MF, Dummer R, Simon HU, and Yousefi S (2004). Induction of genes mediating interferon-dependent extracellular trap formation during neutrophil differentiation. J. Biol. Chem 279, 44123–44132. [DOI] [PubMed] [Google Scholar]
- McHutchison JG, Lawitz EJ, Shiftman ML, Muir AJ, Galler GW, McCone J, Nyberg LM, Lee WM, Ghalib RH, Schiff ER, et al. ; IDEAL Study Team (2009). Peginterferon alfa-2b or alfa-2a with ribavirin for treatment of hepatitis C infection. N. Engl. J. Med 361, 580–593. [DOI] [PubMed] [Google Scholar]
- Mendoza JL, Schneider WM, Hoffmann HH, Vercauteren K, Jude KM, Xiong A, Moraga I, Horton TM, Glenn JS, de Jong YP, et al. (2017). Ttie IFN-λ-IFN-λR1-IL-10Rβ complex reveals structural features underlying type III IFN functional plasticity. Immunity 46, 379–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen ME, Schot R, Buta S, Oudesluijs G, Tinschert S, Speer SD, Li Z, van Unen L, Heijsman D, Goldmann T, et al. (2016). Human USP18 deficiency underlies type 1 interferonopathy leading to severe pseudo-TORCH syndrome. J. Exp. Med 213,1163–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miner JJ, Cao B, Govero J, Smith AM, Fernandez E, Cabrera OH, Garber C, Noll M, Klein RS, Noguchi KK, et al. (2016). Zika virus infection during pregnancy in mice causes placental damage and fetal demise. Cell 165, 1081–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misumi I, and Whitmire JK (2014). IFN-λ exerts opposing effects on T cell responses depending on the chronicity of the virus infection. J. Immunol 192, 3596–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocellin S, Pasquali S, Rossi CR, and Nitti D (2010). Interferon alpha adjuvant therapy in patients with high-risk melanoma: a systematic review and meta-analysis. J. Natl. Cancer Inst 102, 493–501. [DOI] [PubMed] [Google Scholar]
- Morrow MP, Pankhong P, Laddy DJ, Schoenly KA, Yan J, Cisper N, and Weiner DB (2009). Comparative ability of IL-12 and IL-28B to regulate Treg populations and enhance adaptive cellular immunity. Blood 113, 5868–5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseman EA, Wu T, de la Torre JC, Schwartzberg PL, and McGavem DB (2016). Type I interferon suppresses virus-specific B cell responses by modulating CD8+ T cell differentiation. Sci. Immunol 1, eaah3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir AJ, Shiffman ML, Zaman A, Yoffe B, de la Torre A, Flamm S, Gordon SC, Marotta P, Vierling JM, Lopez-Talavera JC, et al. (2010). Phase 1b study of pegylated interferon lambda 1 with or without ribavirin in patients with chronic genotype 1 hepatitis C virus infection. Hepatology 52, 822–832. [DOI] [PubMed] [Google Scholar]
- Muir AJ, Arora S, Everson G, Flisiak R, George J, Ghalib R, Gordon SC, Gray T, Greenbloom S, Hassanein T, et al. ; EMERGE study group (2014). A randomized phase 2b study of peginterferon lambda-1a for the treatment of chronic HCV infection. J. Hepatol 67, 1238–1246. [DOI] [PubMed] [Google Scholar]
- Müller U, Steinhoff U, Reis LF, Hemmi S, Pavlovic J, Zinkernagel RM, and Aguet M (1994). Functional role of type I and type II interferons in antiviral defense. Science 264, 1918–1921. [DOI] [PubMed] [Google Scholar]
- Nagarajan UM, Prantner D, Sikes JD, Andrews CW Jr., Goodwin AM, Nagarajan S, and Darville T (2008). Type I interferon signaling exacerbates Chlamydia muridarum genital infection in a murine model. Infect. Immun 76, 4642–4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Davis KM, and Weiser JN (2011). Synergistic stimulation of type I interferons during influenza virus coinfection promotes Streptococcus pneumoniae colonization in mice. J. Clin. Invest 121, 3657–3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsome SD, Kieseier BC, Arnold DL, Shang S, Liu S, Hung S, and Sabatella G (2016). Subgroup and sensitivity analyses of annualized relapse rate over 2 years in the ADVANCE trial of peginterferon beta-1a in patients with relapsing-remitting multiple sclerosis. J. Neurol 263, 1778–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng CT, Sullivan BM, Teijaro JR, Lee AM, Welch M, Rice S, Sheehan KC, Schreiber RD, and Oldstone MB (2015). Blockade of interferon Beta, but not interferon alpha, signaling controls persistent viral infection. Cell Host Microbe 17, 653–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nice TJ, Baldridge MT, McCune BT, Norman JM, Lazear HM, Artyomov M, Diamond MS, and Virgin HW (2015). Interferon-λ cures persistent murine norovirus infection in the absence of adaptive immunity. Science 347, 269–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numasaki M, Tagawa M, Iwata F, Suzuki T, Nakamura A, Okada M, Iwakura Y, Aiba S, and Yamaya M (2007). IL-28 elicits antitumor responses against murine fibrosarcoma. J. Immunol 178, 5086–5098. [DOI] [PubMed] [Google Scholar]
- Odendall C, Dixit E, Stavru F, Bierne H, Franz KM, Durbin AF, Boulant S, Gehrke L, Cossart P, and Kagan JC (2014). Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat. Immunol 75, 717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odendall C, Voak AA, and Kagan JC (2017). Type III IFNs are commonly induced by bacteria-sensing TLRs and reinforce epithelial barriers during infection. J. Immunol 199, 3270–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabayashi T, Kojima T, Masaki T, Yokota S, Imaizumi T, Tsutsumi H, Himi T, Fujii N, and Sawada N (2011). Type-III interferon, not type-I, is the predominant interferon induced by respiratory viruses in nasal epithelial cells. Virus Res. 760, 360–366. [DOI] [PubMed] [Google Scholar]
- Okanoue T, Sakamoto S, Itoh Y, Minami M, Yasui K, Sakamoto M, Nishioji K, Katagishi T, Nakagawa Y, Tada H, et al. (1996). Side effects of high-dose interferon therapy for chronic hepatitis C. J. Hepatol 25, 283–291. [DOI] [PubMed] [Google Scholar]
- Oke V, Brauner S, Larsson A, Gustafsson J, Zickert A, Gunnarsson I, and Svenungsson E (2017). IFN-λ1 withTh17 axis cytokines and IFN-α define different subsets in systemic lupus erythematosus (SLE). Arthritis Res. Ther 19, 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund PI, Pietilä TE, Veckman V, Kotenko SV, and Julkunen I (2007). IFN regulatory factor family members differentially regulate the expression of type III IFN (IFN-lambda) genes. J. Immunol 179, 3434–3442. [DOI] [PubMed] [Google Scholar]
- Paun A, and Pitha PM (2007). The IRF family, revisited. Biochimie 89, 744–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pervolaraki K, Stanifer ML, Münchau S, Renn LA, Albrecht D, Kurzhals S, Senis E, Grimm D, Schröder-Braunstein J, Rabin RL, and Boulant S (2017). Type I and type III interferons display different dependency on mitogen-activated protein kinases to mount an antiviral state in the human gut. Front. Immunol 8, 459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pervolaraki K, Rastgou Talemi S, Albrecht D, Bormann F, Bamford C, Mendoza JL, Garcia KC, McLauchlan J, Höfer T, Stanifer ML, and Boulant S (2018). Differential induction of interferon stimulated genes between type I and type III interferons is independent of interferon receptor abundance. PLoS Pathog. 14, e1007420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller CK, Li Z, George CX, and Samuel CE (2011). Protein Kinase PKR and RNA adenosine deaminase ADAR1: new roles for old players as modulators of the interferon response. Curr. Opin. Immunol 23, 573–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips S, Mistry S, Riva A, Cooksley H, Hadzhiolova-Lebeau T, Plavova S, Katzarov K, Simonova M, Zeuzem S, Woffendin C, et al. (2017). Peg-Interferon lambda treatment induces robust innate and adaptive immunity in chronic hepatitis B patients. Front. Immunol 8, 621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, Lassnig C, Eberle CA, Górna MW, Baumann CL, Burkard TR, Bürckstümmer T, Stefanovic A, Krieger S, Bennett KL, et al. (2011). IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat. Immunol 72, 624–630. [DOI] [PubMed] [Google Scholar]
- Platanias LC (2005). Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol 5, 375–386. [DOI] [PubMed] [Google Scholar]
- Pott J, Mahlaköiv T, Mordstein M, Duerr CU, Michiels T, Stockinger S, Staeheli P, and Hornef MW (2011). IFN-lambda determines the intestinal epithelial antiviral host defense. Proc. Natl. Acad. Sci. USA 708, 7944–7949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokunina-Olsson L, Muchmore B, Tang W, Pfeiffer RM, Park H, Dickensheets H, Hergott D, Porter-Gill P, Mumy A, Kohaar I, et al. (2013). A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat. Genet 45, 164–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulverer JE, Rand U, Lienenklaus S, Kugel D, Zietara N, Kochs G, Naumann R, Weiss S, Staeheli P, Hauser H, and Köster M (2010). Temporal and spatial resolution of type I and III interferon responses in vivo. J. Virol 84, 8626–8638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quicke KM, Bowen JR, Johnson EL, McDonald CE, Ma H, O’Neal JT, Rajakumar A, Wrammert J, Rimawi BH, Pulendran B, et al. (2016). Zika virus infects human placental macrophages. Ceil Host Microbe 20,83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichard O, Norkrans G, Frydén A, Braconier JH, Sönnerborg A, and Weiland O; The Swedish Study Group (1998). Randomised, double-blind, placebo-controlled trial of interferon alpha-2b with and without ribavirin for chronic hepatitis C. Lancet 357, 83–87. [DOI] [PubMed] [Google Scholar]
- Reis LF, Ruffner H, Stark G, Aguet M, and Weissmann C (1994). Mice devoid of interferon regulatory factor 1 (IRF-1) show normal expression of type I interferon genes. EM BO J. 73, 4798–4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser J, Hurst J, Voges M, Krauss P, Münch P, Iftner T, and Stubenrauch F (2011). High-risk human papillomaviruses repress constitutive kappa interferon transcription via E6 to prevent pathogen recognition receptor and antiviral-gene expression. J. Virol 85, 11372–11380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renauld JC (2003). Class II cytokine receptors and their ligands: key antiviral and inflammatory modulators. Nat. Rev. Immunol 3, 667–676. [DOI] [PubMed] [Google Scholar]
- Richardson RB, Ohlson MB, Eitson JL, Kumar A, McDougal MB, Boys IN, Mar KB, De La Cruz-Rivera PC, Douglas C, Konopka G, et al. (2018). A CRISPR screen identifies IFI6 as an ER-resident interferon effector that blocks flavivirus replication. Nat. Microbiol 3, 1214–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincon-Orozco B, Halec G, Rosenberger S, Muschik D, Nindl I, Bachmann A, Ritter TM, Dondog B, Ly R, Bosch FX, et al. (2009). Epigenetic silencing of interferon-kappa in human papillomavirus type 16-positive cells. Cancer Res. 69, 8718–8725. [DOI] [PubMed] [Google Scholar]
- Rodero MP, and Crow YJ (2016). Type I interferon-mediated monogenic autoinflammation: The type I interferonopathies, a conceptual overview. J. Exp. Med 213, 2527–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothlin CV, Ghosh S, Zuniga EI, Oldstone MB, and Lemke G (2007). TAM receptors are pleiotropic inhibitors of the innate immune response. Cell 131, 1124–1136. [DOI] [PubMed] [Google Scholar]
- Sabat R (2010). IL-10 family of cytokines. Cytokine Growth Factor Rev. 27, 315–324. [DOI] [PubMed] [Google Scholar]
- Sammicheli S, Kuka M, Di Lucia P, de Oya NJ, De Giovanni M, Fioravanti J, Cristofani C, Maganuco CG, Fallet B, Ganzer L, et al. (2016). Inflammatory monocytes hinder antiviral B cell responses. Sci. Immunol 1, eaah6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchis A, Cerveró L, Bataller A,Tortajada JL, Huguet J, Crow YJ., Ali M, Higuet LJ, and Martinez-Frías ML (2005). Genetic syndromes mimic congenital infections. J. Pediatr 146, 701–705. [DOI] [PubMed] [Google Scholar]
- Sancho-Shimizu V, Perez de Diego R, Jouanguy E, Zhang SY, and Casanova JL (2011). Inborn errors of anti-viral interferon immunity in humans. Curr. Opin. Virol 1, 487–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarasin-Filipowicz M, Oakeley EJ, Duong FH, Christen V, Terracciano L, Filipowicz W, and Heim MH (2008). Interferon signaling and treatment outcome in chronic hepatitis C. Proc. Natl. Acad. Sci. USA 105, 7034–7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A, Ohtsuki M, Hata M, Kobayashi E, and Murakami T (2006). Antitumor activity of IFN-lambda in murine tumor models. J. Immunol 176, 7686–7694. [DOI] [PubMed] [Google Scholar]
- Schneider WM, Chevillotte MD, and Rice CM (2014). Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol 32, 513–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW (2014). Interferon-stimulated genes: roles in viral pathogenesis. Curr. Opin. Virol 6, 40–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW (2018). Recent advances in antiviral interferon-stimulated gene biology. F1000Res. 7, 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber G (2017). The molecular basis for differential type I interferon signaling. J. Biol. Chem 292, 7285–7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw AE, Hughes J, Gu Q, Behdenna A, Singer JB, Dennis T, Orton RJ, Varela M, Gifford RJ, Wilson SJ, and Palmarini M (2017). Fundamental properties of the mammalian innate immune system revealed by multi-species comparison of type I interferon responses. PLoS Biol. 15, e2004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, Kuestner R, Garrigues U, Birks C, Roraback J, et al. (2003). IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat. Immunol 4,63–68. [DOI] [PubMed] [Google Scholar]
- Snell LM, McGaha TL, and Brooks DG (2017). Type I Interferon in Chronic Virus Infection and Cancer. Trends Immunol. 38, 542–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommereyns C, Paul S, Staeheli P, and Michiels T (2008). IFN-lambda (IFN-lambda) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 4, e1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorgeloos F, Kreit M, Hermant P, Lardinois C, and Michiels T (2013). Antiviral type I and type III interferon responses in the central nervous system. Viruses 5, 834–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza-Fonseca-Guimaraes F, Young A, Mittal D, Martinet L, Bruedigam C, Takeda K, Andoniou CE, Degli-Esposti MA, Hill GR, and Smyth MJ (2015). NK cells require IL-28R for optimal in vivo activity. Proc. Natl. Acad. Sci. USA 112, E2376–E2384, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Dai J, Eskdale J, Gallagher GE, Megjugorac NJ, and Gallagher G (2008). Interferon-lambdal (interleukin-29) preferentially down-regulates interleukin-13 over other T helper type 2 cytokine responses in vitro. Immunology 125, 492–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steed AL, Christophi GP, Kaiko GE, Sun L, Goodwin VM, Jain U, Esaulova E, Artyomov MN, Morales DJ, Holtzman MJ, et al. (2017). The microbial metabolite desaminotyrosine protects from influenza through type I interferon. Science 357, 498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stifter SA, Matthews AY, Mangan NE, Fung KY, Drew A, Tate MD, Soares da Costa TP, Hampsey D, Mayall J, Hansbro PM, et al. (2018). Defining the distinct, intrinsic properties of the novel type I interferon, IFNε. J. Biol. Chem 293, 3168–3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suspène R, Guétard D, Henry M, Sommer P, Wain-Hobson S, and Vartanian JP (2005). Extensive editing of both hepatitis B virus DNA strands by APOBEC3 cytidine deaminases in vitro and in vivo. Proc. Natl. Acad. Sci. USA 102, 8321–8326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson A, Bellner L, Magnusson M, and Eriksson K (2007). Role of IFN-alpha/beta signaling in the prevention of genital herpes virus type 2 infection. J. Reprod. Immunol 74, 114–123. [DOI] [PubMed] [Google Scholar]
- Tasker C, Subbian S, Gao P, Couret J, Levine C, Ghanny S, Soteropoulos P, Zhao X, Landau N, Lu W, and Chang TL (2016). IFN-ε protects primary macrophages against HIV infection. JCI Insight 1, e88255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teijaro JR, Ng C, Lee AM, Sullivan BM, Sheehan KC, Welch M, Schreiber RD, de la Torre JC, and Oldstone MB (2013). Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science 340, 207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrault NA, Bzowej NH, Chang KM, Hwang JP, Jonas MM, and Murad MH; American Association for the Study of Liver Diseases (2016). AASLD guidelines for treatment of chronic hepatitis B. Hepatology 63, 261–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezuka Y, Endo S, Matsui A, Sato A, Saito K, Semba K, Takahashi M, and Murakami T (2012). Potential anti-tumor effect of IFN-λ2 (IL-28A) against human lung cancer cells. Lung Cancer 78, 185–192. [DOI] [PubMed] [Google Scholar]
- The IFNB Multiple Sclerosis Study Group (1993). Interferon beta-1 b is effective in relapsing-remitting multiple sclerosis. I. Clinical results of a multicenter, randomized, double-blind, placebo-controlled trial. Neurology 43, 655–661. [DOI] [PubMed] [Google Scholar]
- Thomas C, Moraga I, Levin D, Krutzik PO, Podoplelova Y, Trejo A, Lee C, Yarden G, Vleck SE, Glenn JS, et al. (2011). Structural linkage between ligand discrimination and receptor activation by type I interferons. Cell 146, 621–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorlacius GE, Wahren-Herlenius M, and Rönnblom L (2018). An update on the role of type I interferons in systemic lupus erythematosus and Sjögren’s syndrome. Curr. Opin. Rheumatol 30, 471–481. [DOI] [PubMed] [Google Scholar]
- van Boxel-Dezaire AH, Rani MR, and Stark GR (2006). Complex modulation of cell type-specific signaling in response to type I interferons. Immunity 25, 361–372. [DOI] [PubMed] [Google Scholar]
- Vilcek J (2006). Fifty years of interferon research: aiming at a moving target. Immunity 25, 343–348. [DOI] [PubMed] [Google Scholar]
- Voigt EA, and Yin J (2015). Kinetic differences and synergistic antiviral effects between type I and type III interferon signaling indicate pathway independence. J. Interferon Cytokine Res 35, 734–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wack A, Terczyńska-Dyla E, and Hartmann R (2015). Guarding the frontiers: the biology of type III interferons. Nat. Immunol 16, 802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WB, Levy DE, and Lee CK (2011). STAT3 negatively regulates type I IFN-mediated antiviral response. J. Immunol 187, 2578–2585. [DOI] [PubMed] [Google Scholar]
- Wells AI, and Coyne CB (2018). Type III Interferons in Antiviral Defenses at Barrier Surfaces. Trends Immunol. 39, 848–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox DR, Folmsbee SS, Muller WJ, and Longnecker R (2016). The type I interferon response determines differences in choroid plexus susceptibility between newborns and adults in herpes simplex virus encephalitis. MBio 7, e00437–e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson EB, Yamada DH, Elsaesser H, Herskovitz J, Deng J, Cheng G, Aronow BJ, Karp CL, and Brooks DG (2013). Blockade of chronic type I interferon signaling to control persistent LCMV infection. Science 340, 202–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte E, Kokolakis G, Witte K, Warszawska K, Friedrich M, Christou D, Kirsch S, Sterry W, Volk HD, Sabat R, and Wolk K (2016). Interleukin-29 induces epithelial production of CXCR3A ligands and T-cell infiltration. J. Mol. Med. (Berl.) 94, 391–400. [DOI] [PubMed] [Google Scholar]
- Wolk K, Witte K, Witte E, Raftery M, Kokolakis G, Philipp S, Schönrich G, Warszawska K, Kirsch S, Prösch S, et al. (2013). IL-29 is produced by T(H)17 cells and mediates the cutaneous antiviral competence in psoriasis. Sci. Transi. Med 5, 204ra129. [DOI] [PubMed] [Google Scholar]
- Wongthida P, Diaz RM, Galivo F, Kottke T, Thompson J, Pulido J, Pavelko K, Pease L, Melcher A, and Vile R (2010). Type III IFN interleukin-28 mediates the antitumor efficacy of oncolytic virus VSV in immune-competent mouse models of cancer. Cancer Res. 70, 4539–4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Yang Q, Lourenco E, Sun H, and Zhang Y (2011). Interferon-lambdal induces peripheral blood mononuclear cell-derived chemokines secretion in patients with systemic lupus erythematosus: its correlation with disease activity. Arthritis Res. Ther 13, R88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Yang Q, Sun H, Li M, Zhang Y, and La Cava A (2013). Serum IFN-λ1 is abnormally elevated in rheumatoid arthritis patients. Autoimmunity 46, 40–43. [DOI] [PubMed] [Google Scholar]
- Yan N (2017). Immune diseases associated with TREX1 and STING dysfunction. J. Interferon Cytokine Res 37,198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao JC, Guthrie KA, Moran C, Strosberg JR, Kulke MH, Chan JA, LoConte N, McWilliams RR, Wolin EM, Mattar B, et al. (2017). Phase III prospective randomized comparison trial of depot octreotide plus interferon alfa-2b versus depot octreotide plus bevacizumab in patients with advanced carcinoid tumors: SWOG S0518. J. Clin. Oncol 35, 1695–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yockey LJ, and Iwasaki A (2018). Interferons and proinflammatory cytokines in pregnancy and fetal development. Immunity 49, 397–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yockey LJ, Varela L, Rakib T, Khoury-Hanold W, Fink SL, Stutz B, Szigeti-Buck K, Van den Pol A, Lindenbach BD, Horvath TL, and Iwasaki A (2016). Vaginal exposure to Zika virus during pregnancy leads to fetal brain infection. Cell 166, 1247–1256.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yockey LJ, Jurado KA, Arora N, Millet A, Rakib T, Milano KM, Hastings AK, Fikrig E, Kong Y, Horvath TL, et al. (2018). Type I interferons instigate fetal demise after Zika virus infection. Sci. Immunol 3, eaao1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanoni L, Granucci F, and Broggi A (2017). Interferon (IFN)-λ takes the helm: immunomodulatory roles of type III IFNs. Front. Immunol 8, 1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Brann TW, Zhou M, Yang J, Oguariri RM, Lidie KB, Imamichi H, Huang DW, Lempicki RA, Baseler MW, et al. (2011). Cutting edge: Ku70 is a novel cytosolic DNA sensor that induces type III rather than type I IFN. J. Immunol 186, 4541–4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Bogunovic D, Payelie-Brogard B, Francois-Newton V, Speer SD, Yuan C, Volpi S, Li Z, Sanal O, Mansouri D, et al. (2015). Human intracellular ISG15 prevents interferon-α/β over-amplification and auto-inflammation. Nature 517, 89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Hamming OJ, Ank N, Paludan SR, Nielsen AL, and Hartmann R (2007). Type III interferon (IFN) induces a type I IFN-like response in a restricted subset of cells through signaling pathways involving both the JaK-STAT pathway and the mitogen-activated protein kinases. J. Virol 81, 7749–7758. [DOI] [PMC free article] [PubMed] [Google Scholar]