Summary
In pursuit of food, hungry animals mobilize significant energy resources and overcome exhaustion and fear. How need and motivation control the decision to continue or change behavior is not understood. Using a single fly treadmill, we show that hungry flies persistently track a food odor and increase their effort over repeated trials in the absence of reward suggesting that need dominates negative experience. We further show that odor tracking is regulated by two mushroom body output neurons (MBONs) connecting the MB to the lateral horn. These MBONs, together with dopaminergic neurons and Dop1R2 signaling, control behavioral persistence. Conversely, an octopaminergic neuron, VPM4, which directly innervates one of the MBONs, acts as a brake on odor tracking by connecting feeding and olfaction. Together, our data suggest a function for the MB in internal state-dependent expression of behavior that can be suppressed by external inputs conveying a competing behavioral drive.
Keywords: goal-directed behavior, olfactory system, dopamine, mushroom body, Drosophila melanogaster, octopamine, persistence, foraging, DopR2, learning
Graphical Abstract
Highlights
-
•
Hunger motivates persistent food odor tracking even without reward
-
•
Two synaptically connected MBONs, -γ1pedc>αβ and -α2sc, regulate odor tracking
-
•
Octopamine neurons connect feeding and counteract MBON and odor tracking
-
•
Dopaminergic neurons and Dop1R2 signaling promote persistent tracking
What drives behavioral persistence versus quitting? Sayin et al. propose that circuit modules in the fly’s learning center and dopamine drive gradually increasing food odor tracking, which can be efficiently suppressed by extrinsic, but directly innervating, feeding-related neuromodulatory neurons.
Introduction
Flexibility is an important factor in an ever in-flux environment, where scarcity and competition are the norm. Without persistence to achieve its goals, however, an animal’s strive to secure food, protect its offspring, or maintain its social status is in jeopardy. Therefore, sensory cues related to food or danger often elicit strong impulses. However, these impulses must be strictly controlled to allow for coherent goal-directed behavior and to permit behavioral transitions when sensible. Inhibition of antagonistic behavioral drives at the cognitive and physiological level has been proposed as a major task of a nervous system (Bari and Robbins, 2013). Which sensory cues and ultimately which behaviors are prioritized and win depends on the animal’s metabolic state, internal motivation, and current behavioral context. How this is implemented at the level of individual neurons, circuit motifs, and mechanisms remains an important open question.
Like most animals, energy-deprived flies prioritize food seeking and feeding behavior. To find food, flies can follow olfactory or visual cues over long distances (see for instance Álvarez-Salvado et al., 2018, Root et al., 2011). External gustatory cues provide information about the type and quality of the eventually encountered food. However, only internal nutrient levels will provide reliable feedback about the quality and quantity of a food source and ultimately suppress food-seeking behaviors (Corrales-Carvajal et al., 2016, Dethier and Goldrich-Rachman, 1976, Mann et al., 2013, Thoma et al., 2016). Therefore, food odor, the taste of food, and post-ingestive internal feedback signals induce sequential and partly antagonistic behaviors (Mann et al., 2013, Thoma et al., 2016). Interestingly, chemosensory and internal feedback systems typically mediated by distinct neuromodulators appear to converge in the mushroom body (MB) (Cohn et al., 2015, Kim et al., 2017, Krashes et al., 2009, Lewis et al., 2015, Tsao et al., 2018). How neurons and neural circuits signal and combine external and internal cues to maintain or suppress competing behavioral drives is not well understood.
In mammals, norepinephrine (NE) released by a brain stem nucleus, the locus coeruleus, has been implicated in controlling the balance between persistence and action selection (Berridge and Waterhouse, 2003, Schwarz and Luo, 2015). The potential functional counterpart of NE in insects could be octopamine (OA). Flies lacking OA indeed show reduced arousal, for instance upon starvation (LeDue et al., 2016, Li et al., 2016, Zhang et al., 2013). Additionally, OA neurons (OANs) gate appetitive memory formation of odors (Burke et al., 2012, Perry and Barron, 2013) and also modulate taste neurons and feeding behavior (LeDue et al., 2016, Wang et al., 2016, Youn et al., 2018). OANs are organized in distinct clusters and project axons to diverse higher brain regions in a cell type-specific manner (Busch et al., 2009, Busch and Tanimoto, 2010). The precise roles and important types of OA and NE neurons in state-dependent action selection remain to be elucidated.
Similar to NE and OA, dopamine (DA) is being studied in many aspects of behavioral adaptation and flexibility. Different classes of DA neurons (DANs) innervating primarily the MB signal negative or positive context (Aso and Rubin, 2016, Burke et al., 2012, Cohn et al., 2015, Lewis et al., 2015, Liu et al., 2012, Plaçais et al., 2012, Riemensperger et al., 2005), novelty (Hattori et al., 2017), forgetting (Berry et al., 2012, Berry et al., 2015), or even wrong predictions (Felsenberg et al., 2017, Felsenberg et al., 2018).
Here, we took advantage of the small number and discrete organization of neuromodulatory neurons in the fly brain to analyze the mechanistic relationship between motivation-dependent persistence in one behavior and the decision to disengage and change to another behavior. Using a single fly spherical treadmill assay, we find that hungry flies increase their effort to track a food odor with every unrewarded trial. We show that MB output through two identified MBONs (MBON-γ1pedc>α/β and MBON-α2sc) is required for persistent odor tracking. MBON-α2sc provides a MB connection to the lateral horn (LH), where it can modify innate food odor attraction as recently shown (Dolan et al., 2018). Furthermore, we pinpoint a specific type of OAN, VPM4 (ventral paired medial), which connects feeding centers directly to MBON-γ1pedc>α/β and disrupts food odor tracking. Finally, our experimental data suggest that persistent tracking depends on DANs, including PPL1-γ1pedc, and signaling through dopamine receptor Dop1R2 in αβ-type KCs. Based on our results, we propose that MB output and a direct external input, depending on internal state and motivation, gradually promote or interrupt ongoing behavior.
Results
Flies Persistently Track Attractive Food Odors in the Absence of Reward
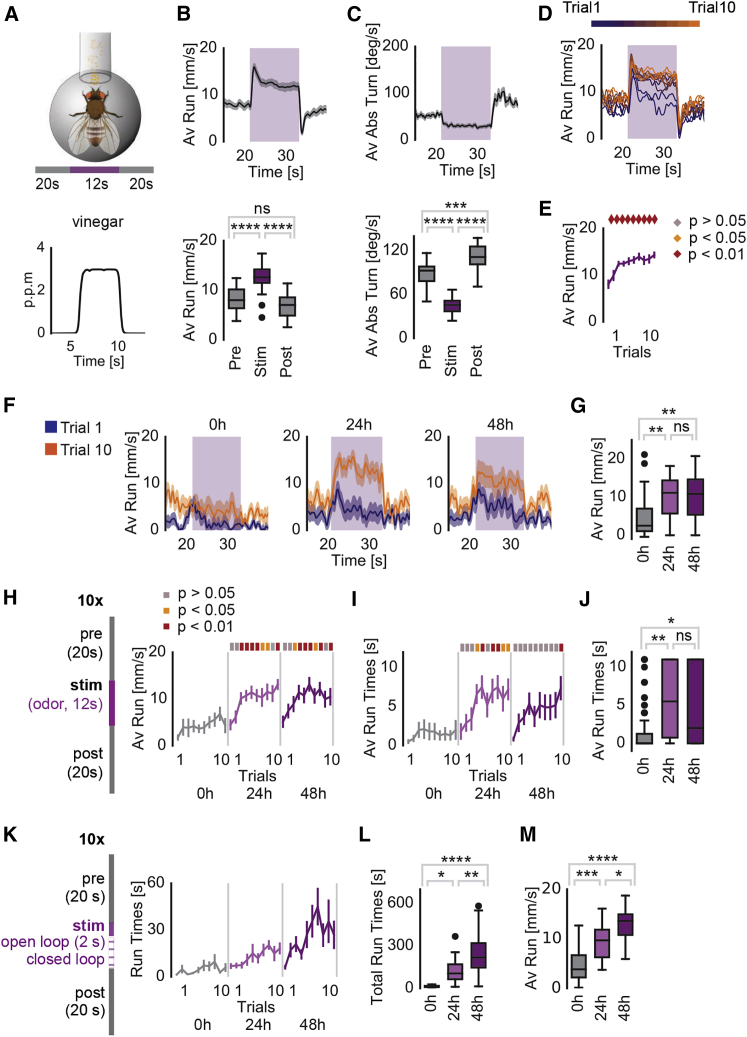
To study the mechanisms underpinning continuing versus stopping of a behavior, we devised a spherical treadmill assay with a tethered fly exposed to a low-speed frontal air stream or odor stimulus allowing us to repeatedly stimulate the animal with odorant at a precise concentration and duration (Figure 1A). Upon an initial period of 3 min of habituation, we recorded running speed, turns, and stops during a 20 s pre-stimulus, a 12 s stimulus, and a 20 s post-stimulus period (Figure 1A). To test persistence in odor tracking, the protocol was repeated 10 times with variable inter-stimulus intervals. We used 3 ppm of vinegar odor as a highly attractive cue to analyze the odor tracking behavior of a hungry fly (24 h starvation). Flies ran on average at a speed of 7.3 mm/s during the pre-stimulus periods (Figure 1B). During odor stimulation, flies sped up significantly and reached speeds of 12.4 mm/s on average (Figure 1B). Upon cessation of the odor stimulus, flies showed a strong offset behavior with stopping before regaining an average speed of 6.8 mm/s (Figure 1B). In addition to changing speed, the flies suppressed turning and were headed straighter suggesting that they were indeed tracking the odorant (Figure 1C). The loss of the olfactory cue at the end of the stimulation period led to a significant increase in turning behavior (Figures 1C and S1A), suggesting that the flies were searching for the stimulus as previously observed for fly larvae, adult flies (Álvarez-Salvado et al., 2018, Gomez-Marin et al., 2011), and other animals including humans (Porter et al., 2007). Interestingly, this behavior evolved over 10 trials (Figures 1D and 1E). Although flies showed an initial acceleration at stimulus onset already during the first 3 trials, they did not persistently track the odors at high speed for more than a fraction of the stimulus time (Figure 1D). With increasing number of trials, however, the flies ran over longer distances and more frequently for the entire stimulus time of 12 s (Figure S1B). In addition, they ran faster with each trial and suppressed turning more efficiently (Figures 1D and S1C). Some of the increase in speed continued also between odor stimulations from trial 1 to 10 (Figures 1D and S1C). These data show that flies reliably track food odors by suppressing turning behavior and increasing speed, and that these behaviors intensify over trial number.
Figure 1.
Persistent Odor Tracking Is Motivated by Hunger
(A) Top: Spherical-treadmill assay for olfactory stimuli.
(B) Top: Average running speed with SEM (mm/s) of 18 wild-type Canton S flies under repeated vinegar exposure for 10 trials. Shaded areas represent the odor exposure duration. Bottom: Average running speeds of flies during vinegar exposure were significantly higher compared to the speeds observed during pre- and post-stimulation periods.
(C) Top: Average absolute turning speed with SEM (deg/s) of 18 flies under repeated vinegar exposure for 10 trials. Bottom: The absolute turning speed under vinegar was significantly lower than the turning speed recorded in pre- and post-stimulation periods.
(D) Average running speed with SEM of 18 wild-type flies over time for each of the individual 10 trials.
(E) Comparison of average running over trials.
(F) Average running speed with SEM of fed (0 h) and hungry (24 and 48 h) flies during trial 1 and trial 10 of repeated vinegar exposure.
(G and H) Average running speed with SEM during odor stimulation during trial 1 to 10 for fed, 24 h and 48 h starved flies. The boxplot (G) displays the Tukey’s post hoc analysis for the main group effect.
(I and J) Average running bout times with SEM during open-loop odor exposure for fed and food-deprived flies over 10 trials. The boxplot (J) displays the Tukey’s post hoc analysis for the main group effect.
(K) Left: Schematics for the closed-loop assay. Right: Average running bout times with SEM during closed-loop odor exposure for differentially food-deprived flies over 10 trials.
(L) Total running times of fed and starved flies during odor stimulation.
(M) Average summed running bout times during 10 trials for all groups in closed-loop experiments under vinegar exposure.
For all analyses, statistical notations are as follows: ns, p > 0.05; ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001. In all panels, error bars denote SEM.
Any change in the fly’s environment, including a change in airflow or wind, could induce forward running. To test this, we analyzed the behavior of mutants of the essential olfactory co-receptor Orco, which is required to detect vinegar (Semmelhack and Wang, 2009). Orco mutant flies showed a significantly reduced reaction to vinegar stimulation as compared to heterozygous controls upon stimulus onset (Figure S1D). These results show that the animal’s reaction depends on the detection of a sensory stimulus such as an appetitive odor.
We next asked whether the valence of the stimulus influenced odor tracking behavior in our assay. Frontal stimulation with CO2, which elicits aversion in laboratory assays with walking flies (Bräcker et al., 2013, Suh et al., 2004), led to the opposite behavior compared to the behavior elicited by vinegar in hungry animals (Figure S1E). Flies slowed down and significantly increased their turning to left and right consistent with odor aversion (Figure S1E). A similar avoidance behavior was observed in a tethered flying fly assay with frontal odor stimulation (Badel et al., 2016). Of note, however, CO2 is not always aversive; freely flying flies are attracted to this gas in a state-dependent manner (van Breugel et al., 2018).
Taken together, these data show that targeted food odor tracking behavior increases over time even in the absence of a food reward. Loss of the odor stimulus increases turning, indicating search behavior.
Tracking Intensity Depends on Hunger State
Numerous studies have addressed how reward increases an animal’s attraction to a sensory cue. Less is known that explains why an animal maintains and even improves a not yet successful behavior. In healthy animals, the interest in food is regulated by their need to acquire calories and nutrients. This was also evident in our assay: fed flies did not show any persistence in food odor tracking (Figures 1F–1J). By contrast, 24 and 48 h starved flies showed persistent tracking behavior (Figures 1F–1J). To better investigate the influence of hunger on the fly’s persistence, we changed the assay from an open to a closed loop configuration, and allowed the fly to control the offset of the odorant by stopping to run (Figures 1K–1M). Fed flies tracked the odor for 13.3 s, while 24 h starved flies followed the odor for 122.4 s, on average (Figure 1L). Interestingly, 48 h starved flies showed a higher persistence than 24 h starved animals and tracked the odor for up to 248.2 s over ten trials, on average (Figure 1L). It is unlikely that the increase in running times is due simply to improved motor skills over trials, where the best skills, and hence fastest running would be expected to always occur at the last trials; however, among all recorded trials the longest trials of each fly were distributed across all trial numbers (1–10) including trials as early as number 2 (Figure S1F). Moreover, the same experiment with only air during stimulus phase did not induce long and fast tracking (Figure S1G–S1J). Finally, not only tracking time but also tracking speed depended on starvation time, with 48 h starved animals running on average faster than 24 h starved flies during odor stimulation (Figure 1M). These results demonstrate that starvation specifically and gradually changes the animal’s behavioral expression and persistence in tracking a food odor.
Specific MB Output Neurons Control Food Odor Tracking
Recent work implicated the MB in metabolic state-dependent modulation of innate olfactory behaviors (Bräcker et al., 2013, Grunwald Kadow, 2019, Lewis et al., 2015, Tsao et al., 2018).
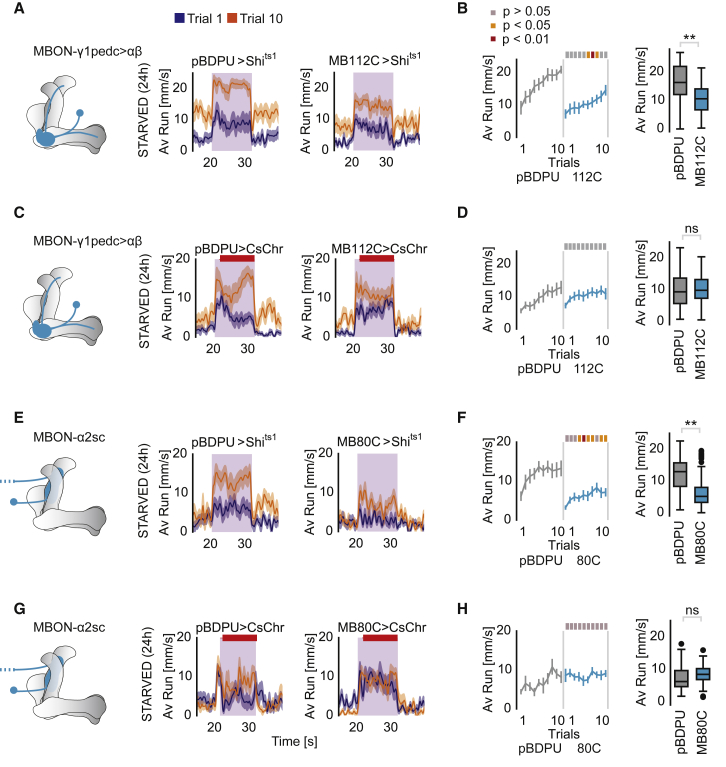
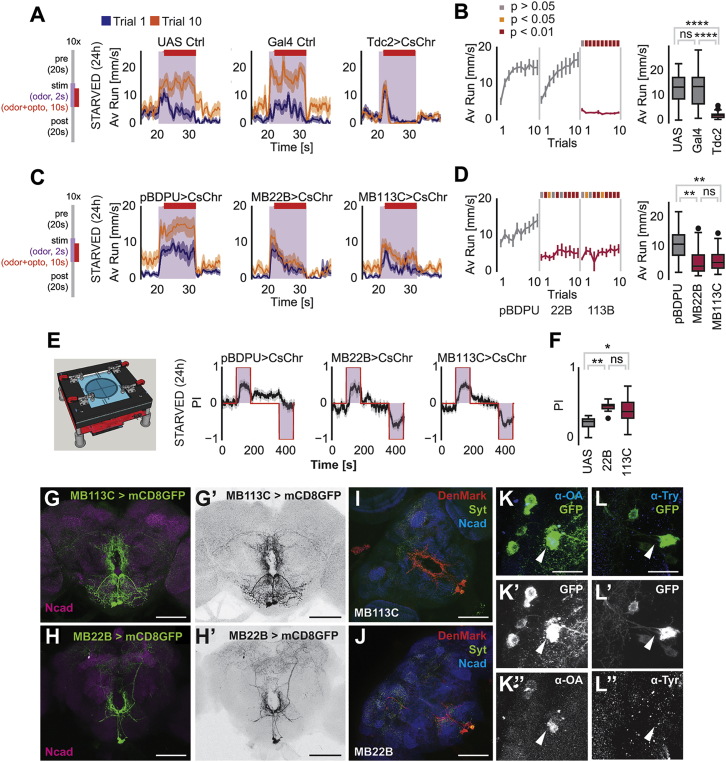
We, therefore, next addressed whether the MB network contributes to hunger-induced persistent food odor tracking. In a screen of Split-Gal4 lines that target MBONs for a reduction in attraction to vinegar with the T-maze assay, we identified MBON line MB112C, which expresses in MVP2/MBON-γ1pedc>α/β neurons (Aso et al., 2014b) (Figure S2A). This result was in line with recent work using a different food finding assay (Perisse et al., 2016, Tsao et al., 2018). On the ball, inactivation of MBON-γ1pedc>α/β’s synaptic output using overexpression of the effector Shibirets1 (shits1) significantly reduced olfactory tracking speed in the starved animal (Figures 2A and 2B). In order to test whether activation of MBON-γ1pedc>α/β was sufficient to induce odor tracking, we expressed the red-shifted channel rhodopsin CsChrimson under the control of MB112C (MB112C>CsChrimson), and stimulated the flies with 12 s odor stimulus, but overlapped it, 2 s after odor onset, with a 10 s red-light stimulus (Figures 2C and 2D). Activation of MBON-γ1pedc>α/β induced tracking in the presence of an odor stimulus in fed animals, but did not further increase tracking in the starved fly, indicating that this neuron is already active in the hungry animal (Figures 2C, 2D, and S2B–S2D). Light alone also induced some forward running, albeit to a lesser extent than odor alone (Figures S2E–S2G).
Figure 2.
Two Synaptically Connected MBONs Are Required for Odor Tracking
(A) Running speeds during stimulus phase in trial 1 and trial 10 upon blocking synaptic output of MVP2/MBON-γ1pedc>αβ (MB112C>UAS-Shibirets1) at non-permissive temperature compared to control with empty-Gal4 (pBDPU-Gal4>UAS-Shibirets1).
(B) Average running speeds during stimulus phase over trials.
(C) Running speeds during acute MBON-γ1pedc>αβ activation with CsChrimson (MB112-Gal4>UAS-CsChrimson) in starved flies compared to controls (pBDPU-Gal4>UAS-CsChrimson).
(D) Running speeds during stimulus phase over trials. (E) Running speeds during stimulus phase in trial 1 and trial 10 upon blocking synaptic output of MBON-α2sc (MB80C) compared to controls (pBDPU-Gal4>UAS-Shibirets1).
(F) Running speeds during stimulus phase over trials.
(G) Running speeds in starved flies when the MB80C neuron was activated via optogenetics (MB80C>UAS-CsChrimson) (controls: pBDPU-Gal4>UAS-CsChrimson).
(H) Running speeds during stimulus phase over trials.
For all analyses, statistical notations are as follows: ns, p > 0.05; ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001. In all panels, error bars denote SEM.
MBON-γ1pedc>α/β projects from the MB γ1/peduncular region again to KCs and other MBONs providing output from α and β lobes (Takemura et al., 2017). Among the primary neurons innervated by MBON-γ1pedc>α/β is MBON-α2sc (labeled by line MB080C) (Takemura et al., 2017), which is also required for food odor attraction (Figure S2A). Indeed, inactivation of MBON-α2sc output (MB080C>shits1) resulted in a strong reduction in average food odor tracking in the ball assay (Figures 2E and 2F). Moreover, the trial-by-trial increase was reduced (Figures 2EF). While the phenotype is stronger for MBON-α2sc inactivation than for MBON-γ1pedc>α/β, these data are consistent with the hypothesis that MBON-γ1pedc>α/β is among the neurons providing input to MBON-α2sc during odor tracking. Furthermore, optogenetic activation of MBON-α2sc induced faster and more persistent tracking at odor trial 1, but not at trial 10, compared to controls (Figures 2G and 2H; 2-way RM ANOVA ptrial x group interaction = 0.0390), supporting a role for MBON-α2sc in regulating tracking intensity.
Together, the data implicate two MBONs in odor tracking over multiple trials.
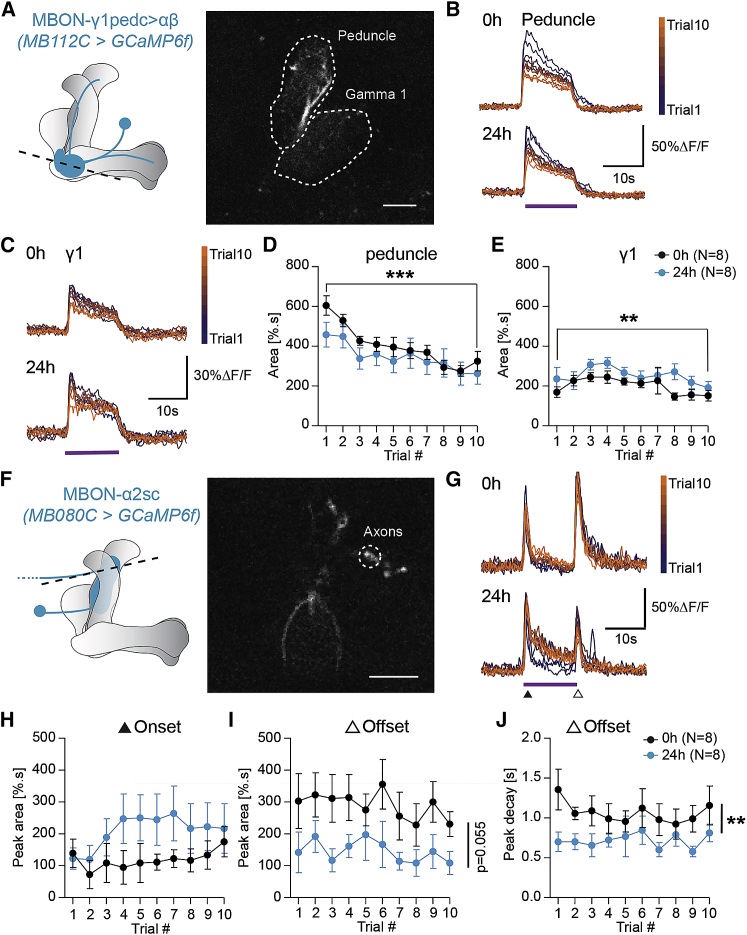
MBON-γ1pedc>α/β Shows Trial-Dependent Odor Responses
Given the requirement of MBON-γ1pedc>α/β and -α2sc, we predicted that their response to repeated odor experience should gradually change over trials. Thus, we imaged odor responses using GCaMP6f calcium imaging in vivo and stimulated the fly as done during the behavioral experiment (Figures 3A and 3E). We quantified two regions of interest for MBON-γ1pedc>α/β, the peduncle—the region immediately downstream of the odor input site (calyx)—and the γ1 lobe (Figures 3A–3E). Odor responses in the peduncle region gradually decreased from trial 1 to 10 (Figures 3B and 3D). Responses in the γ1 dendrites also showed some significant changes over trials, which followed a different pattern. The calcium signal first increased until trial 3 or 4 before going back to or below its baseline level (Figures 3C and 3E). Interestingly, MBON-γ1pedc>α/β receives differential KC input in the peduncle versus γ1-lobe region. While γ-KCs provide γ1-input, the peduncle region receives input exclusively from α/β-KCs (Takemura et al., 2017). These data indicate that MBON-γ1pedc>α/β either is intrinsically sensitive to the number of prior odor experiences or receives trial-dependent olfactory input, for instance by α/β-KCs and/or through dopaminergic modulation.
Figure 3.
MBONs Show Trial- and Feeding State-Dependent Odor Responses
(A) Left: Scheme of MBON-γ1pedc>αβ (MB112C) innervating the MB lobes. Right: MB112C>GCaMP6f expression in an in vivo two-photon preparation. ROIs are marked by dashed lines (scale bar 10 μm).
(B) Average traces of odor responses (purple line) of MB112C peduncle dendrites from trial 1 to 10.
(C) Average traces of odor responses of MB112C γ1 dendrites from trial 1 to 10.
(D) Quantification of the area under the fluorescence response during the stimulus period in the peduncle region from trial 1 to 10.
(E) Quantification of the area under the fluorescence response during the stimulus period in the γ1 region from trial 1 to 10.
(F) Left: Scheme of MBON-α2sc (MB80C) innervating the MB lobes. Right: MB80C>GCaMP6f expression in an in vivo two-photon preparation. The ROI is marked by dashed lines (scale bar 10 μm).
(G) Average traces of odor responses of MB80C axons from trial 1 to 10.
(H, I) Quantification of the area under the fluorescence responses during the stimulus period and after the stimulus period.
(J) Quantification of the offset peak decay time constant over trials.
For all analyses, statistical notations are as follows: ns, p > 0.05; ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001. In all panels, error bars denote SEM.
Next, we recorded odor responses of MBON-α2sc using the same imaging setup and observed an interesting response to the 12 s odor stimulus in each trial; an initial transient calcium peak (i.e., on-response) was followed by a strong decrease and, upon odor stimulus end, a strong re-bound response (i.e., off-response; Figure 3G). This type of response is consistent with a relief of inhibitory input at the end of the stimulus. However, we did not observe a significant effect of trial number on MBON-α2sc. Still, while the responses remained relatively stable over trials (Figures 3H–3J), we observed a significantly longer off-response decay time between fed and hungry animals (Figure 3J).
Taken together, while these data point to a complex relationship between neuronal activity and behavior, possibly due to the inability of the fly to express behavior during imaging, they also provide evidence that these MBONs are indeed modulated by the number of odor experiences and also by the animal’s metabolic state.
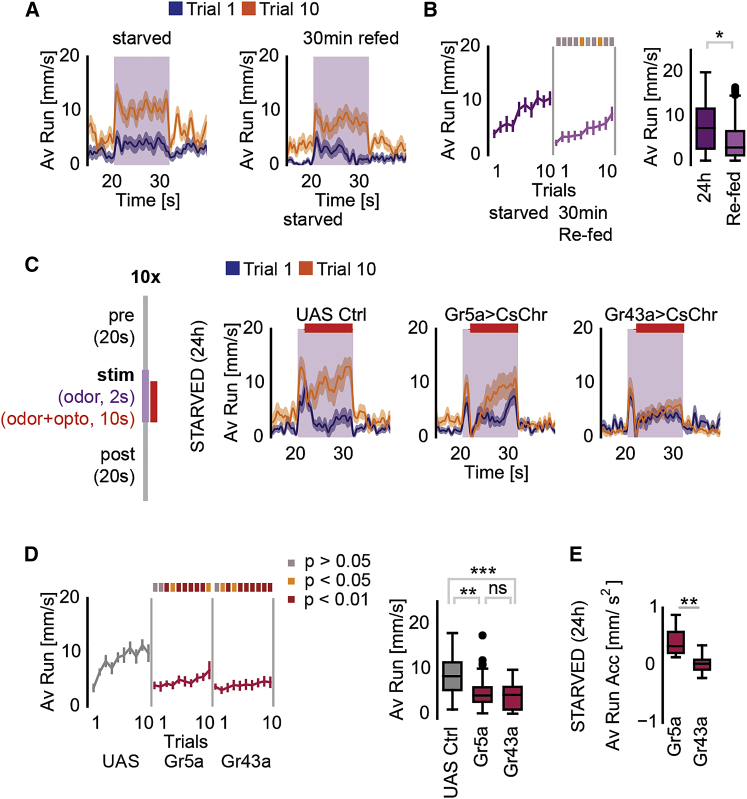
Sugar Neuron Activation Suppresses Odor Tracking
Our behavioral data implicated hunger or feeding state as a main motivator for persistent odor tracking. To test this assumption more directly, we re-fed 24 h-starved flies for only 30 min with standard fly food just prior to the assay. This re-feeding significantly suppressed food odor tracking, in particular during the first few trials of the assay (Figures 4A and 4B). Therefore, feeding-induced cues such as post-ingestion signals appear to suppress food odor tracking in our assay as also shown for other paradigms (Corrales-Carvajal et al., 2016, Mann et al., 2013, Thoma et al., 2016). To test the ability of the taste of food or internal sugar to suppress food odor tracking with high temporal precision and during the continued presence of odor, we expressed CsChrimson and optogenetically activated two sets of sugar taste neurons (Figures 4C and 4D). First, activation of exclusively peripheral (labellar and tarsal) gustatory receptor (Gr) 5a-expressing sweet taste neurons was employed to mimic the taste of a potential calorie source (Thoma et al., 2016). Next, Gr43a neurons were activated to mimic ingested sugar, because Gr43a is not only expressed in peripheral taste neurons but has also been shown to function as internal sugar sensor in the brain (Miyamoto et al., 2012) and potentially in the gut (Park and Kwon, 2011). Gr5a or Gr43a neuron activation led to an immediate stop of odor tracking in spite of continued odor stimulation, suggesting that animals that reach a food source prioritize short-range or internal feeding-related over long-range signals (Figures 4C and 4D). However, although light-stimulation of Gr5a neurons continued, flies reverted their behavior and quickly resumed odor tracking and reached high speed again at the end of each of the trials (Figures 4C–4E). Similarly, Inagaki et al. have previously reported that prolonged optogenetic activation of sugar taste neurons (i.e., Gr5a) only transiently induces proboscis extension, which ceases much before the end of the light stimulation (Inagaki et al., 2014). By contrast, Gr43a neuron activation lastingly reduced the fly’s tracking behavior consistent with mimicking of high internal sugar (Figures 4C–4E). From these results, we conclude that a meal, presumably through post-ingestive signals, but not just the taste of food, lastingly suppresses food odor tracking.
Figure 4.
Taste and Food Suppress Odor Tracking
(A) Average running speeds of 24 h and 24 h, 30 min re-fed animals at trial 1 and trial 10.
(B) Running speeds over trials in 24 h starved and 30 min re-fed flies compared to 24 h starved animals. The boxplot displays the main group effect.
(C) Left: Schematics of the concurrent odor and optogenetic-activation protocol. Right: Gr5a>UAS-CsChrimson and Gr43a>UAS-CsChrimson flies compared to control (pBDPU-Gal4>UAS-CsChrimson).
(D) Running speed over trials during simultaneous odor and light activation. The boxplot displays the main group effect.
(E) Average running activity displayed as boxplots for flies expressing CsChrimson in Gr5a or Gr43a neurons (Gr5a-Gal4;UAS-CsChrimson and Gr43a-Gal4;UAS-CsChrimson).
For all analyses, statistical notations are as follows: ns, p > 0.05; ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001. In all panels, error bars denote SEM.
Octopamine Inhibits Tracking in Hungry Flies
Based on our results, we sought to identify the neural circuits that coordinate and prioritize sensory information to enable feeding stimuli to override odor-stimulated behavior.
OANs were previously implicated in appetitive olfactory learning (Burke et al., 2012, Schröter et al., 2007, Schwaerzel et al., 2003) and modulation of feeding-related behaviors (LeDue et al., 2016, Youn et al., 2018, Zhang et al., 2013). Therefore, we tested whether acute activation of all OANs resulted in tracking suppression by using optogenetic activation via the Tdc2-Gal4 driver paired with the odor stimulus (Figures 5A and 5B). Activation (Tdc2>CsChrimson) fully suppressed odor tracking and led to immediate and persistent slowing down or stopping (Figures 5A and 5B). To gain more evidence that Tdc2+ neuron activation suppressed food finding behavior and represented something attractive to the animal, we used a custom-built 4-arm olfactory choice assay (Figure S3A). Optogenetic activation of Tdc2+ neurons alone or in the presence of vinegar odor in the same quadrant resulted in a significantly higher dwell time of the flies in the illuminated quadrants (Figures S3A–S3D). Based on these data, OANs appear to be good candidates for enabling the animal to prioritize the short-range exploration and exploitation of a food source over odor-induced long-range food search.
Figure 5.
A Subset of Octopaminergic Neurons Inhibit Odor Tracking
(A) Acute optogenetic activation of octopaminergic neurons. CsChrimson was expressed in octopaminergic neurons by Tdc2>UAS-CsChrimson (Controls: UAS Ctrl: +>UAS-CsChrimson, Gal4 Ctrl: Tdc2-Gal4>+). Running speeds during trial 1 and trial 10.
(B) Evolution of average running speeds for Tdc2>UAS-CsChrimson flies during odor exposure over trials. The boxplot displays the main group effect.
(C) Acute optogenetic activation of VPM neurons. MB22B harbors VPM3 and VPM4 neurons, whereas MB113C labels only VPM4 (Control: pBDPU-Gal4>UAS-CsChrimson). Running speeds during trial 1 and trial 10.
(D) Average running speeds for MB22B>UAS-CsChrimson and MB113C>UAS-CsChrimson flies during odor exposure. The boxplot displays the main group effect.
(E) Left: Scheme of optogenetic and olfactory behavioral test arena. Right: Average preference index during optogenetic activation of octopaminergic neurons under vinegar exposure.
(F) Activation of VPMs compared to genetic controls.
(G–J) Expression patterns and polarity of MB22B (H, H′, and J) and MB113C (G, G′, and J) split-Gal4 lines. SEZ (subesophageal zone); PEZ (periesophageal zone)
(K–L″) VPM4 neurons (MB113C>mCD8GFP) express octopamine.
For all analyses, statistical notations are as follows: ns, p > 0.05; ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001. In all panels, error bars denote SEM.
A Specific Subtype of OA Neurons Antagonizes Food Odor Tracking
The ventral cluster of Tdc2+ neurons represents one of the largest and contains several types of OANs such as the VUM (ventral unpaired medial) and VPM (ventral paired medial) type neurons, which again contain different anatomically distinct types of neurons (Busch and Tanimoto, 2010).
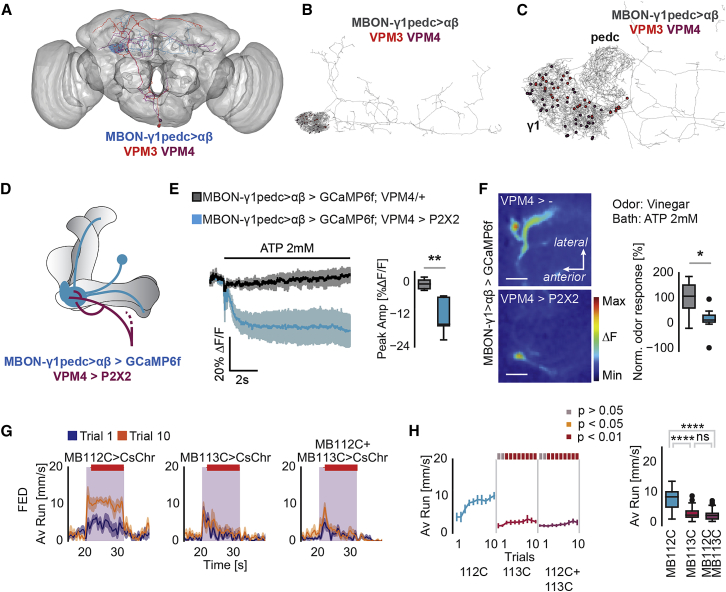
To pinpoint the exact neuron(s) capable of suppressing odor tracking, we screened several candidate OAN lines by optogenetic activation. Activation of OANs labeled by two Split-Gal4 lines, MB113C and MB22B, labeling two neuron types of the VPM cluster, VPM3 and VPM4 (Figures 5G–5J), strongly suppressed food odor tracking (Figures 5C and 5D) and led to a sustained decrease in speed during light plus odor stimulation (Figure 5D). Genetic controls and flies carrying the same transgenes, but which were stimulated only with odor and not light, behaved like control flies (Figures S3E and S3F). Inhibition of the synaptic output of these VPM neurons in fed or starved flies did not result in a change in food odor tracking (Figures S3G–S3J), showing that these neurons are not directly involved in the execution of tracking behavior or olfactory processing. In addition, activation of VPM3/VPM4 in the 4-arm olfactory choice assay significantly increased the time flies spent on the light and odor quadrant (Figures 5E and 5F). This result supports our interpretation that OA-VPM3/VPM4 neurons suppress long-distance food search behavior.
Line MB113C and MB22B overlap only in one neuron type, VPM4, (Aso et al., 2014a), indicating that this neuron was central to the observed behavior. Antibody staining against octopamine confirmed the categorization as OAN (Figures 5K and 5L). Interestingly, in line with our findings, a recent study showed that VPM4 neurons promote feeding and modulate peripheral sugar taste neurons in hungry flies (Youn et al., 2018). Based on this previous work and our results, it appears that OA-VPM4 neurons suppress odor tracking behavior and promote feeding related behaviors.
VPM4 Directly Inhibits MBON-γ1pedc>α/β
VPM3/VPM4 neurons extend dendrites within the sub- and periesophageal zones and send projections to the MB and other higher brain regions (Figures 5I and 5J). Interestingly, at the level of the MB lobes, the innervation pattern of MBON-γ1pedc>α/β overlaps partially with VPM4 in the γ1 MB lobe, indicating that VPM4 and MBON-γ1pedc>α/β might be directly connected (Figure S4A). Moreover, recent analysis of the MB connectome in the Drosophila larva revealed an octopaminergic neuron, named OAN-γ1, forming direct synapses onto MBON-γ1/γ2 neurons, which are proposed to mediate feed-forward inhibition similar to MBON-γ1pedc>α/β (Eichler et al., 2017).
We investigated the possibility of similar connectivity using a recently published EM volume of an entire female adult Drosophila brain (Zheng et al., 2018). Reconstruction of MBON-γ1pedc>α/β and subsequent sampling of its synaptic inputs to the γ1 compartment quickly led to the identification of two putatively aminergic neurons (based on the presence of dense core vesicles). Further reconstructions identified these neurons morphologically to be VPM3 and VPM4 (Figure 6A). Both neurons form numerous synaptic contacts onto MBON-γ1pedc>α/β dendrites exclusively in the γ1 compartment with VPM4 making about 50% more synapses than VPM3 (Figures 6B, 6C, and S4B–S4D). In addition, MBON-γ1pedc>α/β also made a smaller number of reciprocal synaptic contacts back to VPM3 but not to VPM4 (Figure S4B). We found no evidence that VPMs contact MBON-γ1pedc>α/β in other regions of the MB lobes, although we cannot completely rule this out.
Figure 6.
VPM4 Modulates MVP2-Dependent Tracking
(A–C) EM reconstruction reveals synaptic connections between MBON-γ1pedc>αβ and VPM3 and VPM4.
(A) Skeletons of EM reconstruction of MBON-γ1pedc>αβ (blue), VPM3 (red), and VPM4 (purple) on the neuropil of a whole fly brain.
(B) Red (VPM3) and purple (VPM4) indicate the synapses between VPMs and MBON-γ1pedc>αβ, respectively.
(C) Higher magnification of B.
(D) Scheme showing VPM4 and MBON-γ1pedc>αβ neurons at the level of the mushroom body and the genetic combination of transgenes expressed in the fly used for the experiment.
(E) Left: Average traces of GCaMP fluorescence in MBON-γ1pedc>αβ dendrites upon ATP application on brain in an in vivo preparation. Right: Boxplots displaying peak amplitude of % ρF/F GCaMP fluorescence in MBON-γ1pedc>αβ/MVP2 upon ATP application.
(F) Left: Representative pseudocolored images displaying GCaMP fluorescence in MBON-γ1pedc>αβ during odor stimulation (12 s vinegar) upon ATP application in an in vivo preparation. Right: Boxplots displaying normalized (to genetic control) responses (area under the curve) to 12 s vinegar and ATP stimulation in MBON-γ1pedc>αβ/MVP2.
(G) Epistasis experiment for VPM4 and MBON-γ1pedc>αβ suggesting that VPM4 suppresses MBON-γ1pedc>αβ induced odor tracking. Running speed at trial 1 and 10.
(H) Running speeds over trials during odor stimulation period.
For all analyses, statistical notations are as follows: ns, p > 0.05; ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001. In all panels, error bars denote SEM.
Next, we expressed the ATP-sensitive mammalian channel P2X2 (Yao et al., 2012) in VPM4 to test the effect of activating this OAN on MBON-γ1pedc>α/β (Figure 6D). We monitored GCaMP6f fluorescence in MBON-γ1pedc>α/β upon adding ATP to an in vivo brain preparation (Figures 6D and 6E). Addition of ATP led to a significant decrease in baseline GCaMP fluorescence in MBON-γ1pedc>α/β neurons in the presence of P2X2 in VPM4 but not in controls (Figure 6E). Similarly, activation of VPM4 through ATP also reduced the odor-evoked GCaMP6f signal in MBON-γ1pedc>α/β neurons (Figure 6F). These results provide strong evidence for an inhibitory synaptic connection between VPM4 and MBON-γ1pedc>α/β.
Consistent with a direct synaptic connection, we found that activation of VPM4 and inactivation of MBON-γ1pedc>α/β showed similar phenotypes (i.e., reduction of odor tracking). Therefore, we argued that activation of VPM4 could potentially override the activation of MBON-γ1pedc>α/β and inhibit odor tracking. To test this, we combined MB112C- and MB113C-Gal4 with UAS-CsChrimson and applied the same protocol as above. Activation of only MBON-γ1pedc>α/β led to significant odor tracking in response to vinegar in the fed fly as expected (Figures 6G, 6H, S4E, and S4F). Concurrent activation of both neurons (VPM4 and MBON-γ1pedc>α/β) showed that VPM4 activation with the Split-Gal4 line MB113C was sufficient to suppress the effect of activation of MBON-γ1pedc>α/β (Figures 6G, 6H, S4E, and S4F). This could occur through the direct synaptic interaction of these two neurons, although other mechanisms cannot be excluded.
We conclude that OA-VPM4 arbitrates between odor-mediated food search and the rewarding experience of feeding by promoting taste-induced consumption behavior (Youn et al., 2018) and simultaneously suppressing food odor tracking.
Dopaminergic Neurons Are Necessary for Persistent Odor Tracking
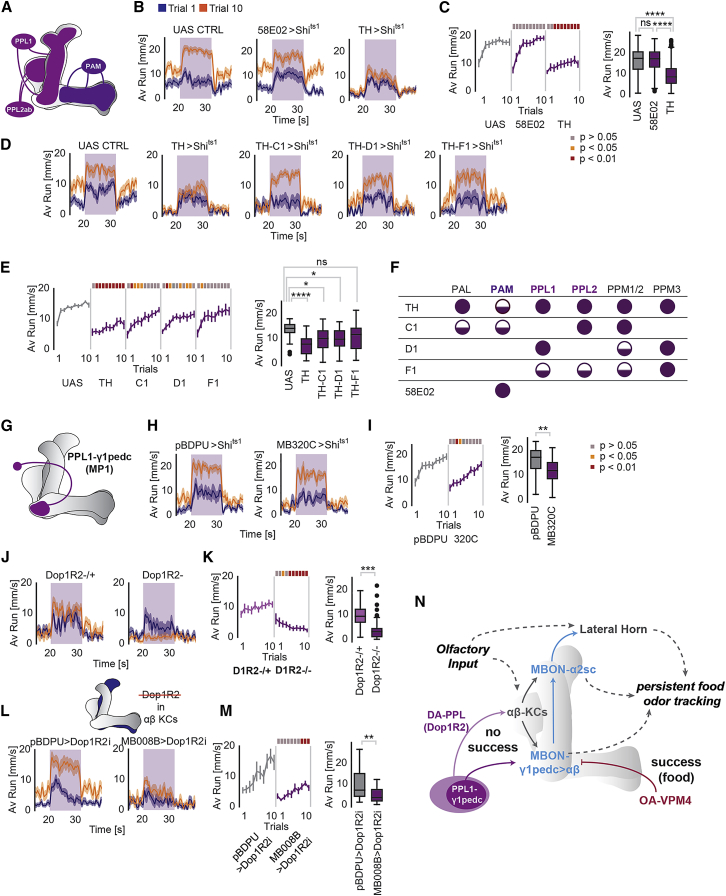
In lieu of a reward or activation of sugar or VPM neurons, flies continue to track food odor. Given the important role of specific MBONs in this behavior, we tested whether dopamine could be involved. Two major subsets of DANs previously implicated in odor-guided behavior exist in the fly brain: the protocerebral anterior medial (PAM) and the protocerebral posterior lateral (PPL1) cluster (Aso et al., 2014a). In addition, a number of smaller DAN subsets exist in the fly (Mao and Davis, 2009) (Figure 7A). While the PAM cluster mediates positive experiences during classical conditioning, PPL1 neurons transmit painful stimuli such as electric shock or heat to the MB (Burke et al., 2012, Galili et al., 2011, Liu et al., 2012). The transgenic line TH-Gal4 labels all cells in the PPL1 and PPL2ab clusters, as well as only one DAN from the PAM group. Thermogenetic activation of these ‘TH+’ neurons can substitute an aversive stimulus and induce negative memories (Aso et al., 2012).
Figure 7.
Persistence in Odor Tracking Depends on Dopamine Signaling
(A) Scheme of dopaminergic neurons innervating MB lobes.
(B) Running speed of hungry flies at trial 1 and 10 (24 h starved) with inactivated output of either PAM DANs (58E02-Gal4>UAS-Shits1) or TH+/PPL DANs (TH-Gal4>UAS-Shits1).
(C) Average running speed over 10 trials for hungry flies (24 h starved) with inactivated output of PAM DANs (58E02-Gal4>UAS-Shits1) or TH+ DANs (TH-Gal4>UAS-Shits1) compared to control. The boxplot displays the main group effect.
(D) Running speed during trial 1 and 10 of hungry flies (24 h starved) with inactivated output of different subsets of DANs within the TH-Gal4 positive DAN cluster.
(E) Average running speed during odor stimulation over 10 trials of hungry flies (24 h starved) with inactivated output of different DAN/TH+ subsets. The boxplot displays the main group effect.
(F) Table of TH-Gal4 transgenes in different clusters of DANs.
(G) Scheme displaying PPL1-γ1pedc (MP1) DAN innervating the MB.
(H) Running speed during trial 1 and 10 of hungry flies (24 h starved) with inactivated output of PPL1-γ1pedc (MP1) compared to controls.
(I) Average running speed during odor stimulation over 10 trials of hungry flies (24 h starved) with inactivated output of PPL1-γ1pedc (MP1) compared to controls. The boxplot displays the main group effect.
(J) Running speed during trial 1 and 10 of hungry flies (24 h starved) lacking the Dop1R2 receptor gene and heterozygous controls.
(K) Average running speed during odor stimulation over 10 trials of hungry flies (24 h starved) without Dop1R2 compared to heterozygous controls. The boxplot displays the main group effect.
(L) Running speed during trials 1 and 10 for hungry flies (24 h starved) with Dop1R2 receptor knockdown in αβ Kenyon cells (MB008B>Dop1R2i) compared to controls (pBDPU>Dop1R2i).
(M) Average running speeds over 10 trials for hungry flies (24 h starved) with Dop1R2 receptor knockdown in αβ Kenyon cells (MB008B>Dop1R2i) compared to controls (pBDPU>Dop1R2i). The boxplot displays the main group effect.
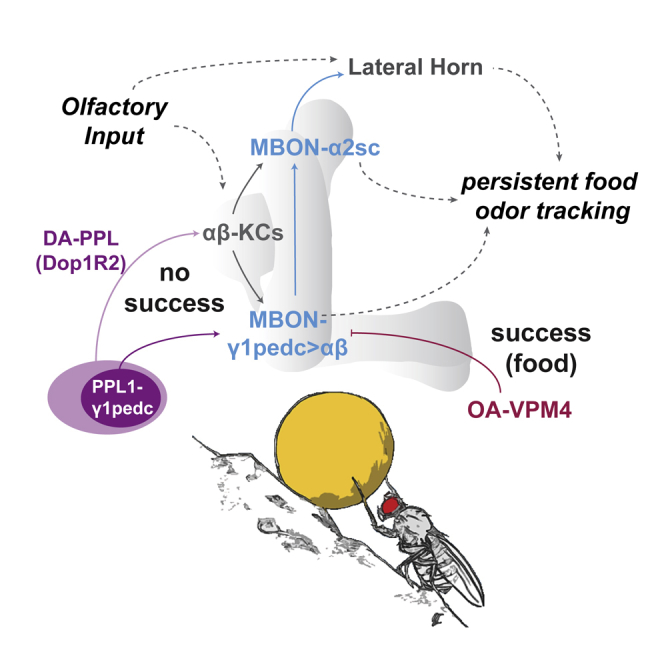
(N) Model of neurons implicated in persistent odor tracking (see main text for details).
For all analyses, statistical notations are as follows: ns, p > 0.05; ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001. In all panels, error bars denote SEM.
Inactivation of TH+ neuron output by overexpression of shibire under the control of TH-Gal4 (TH>shits1) changed the fly’s behavior significantly (Figure 7B). While TH-shits1 flies accelerated at the beginning of each trial in response to odorant, their behavior did not evolve from trial 1 to trial 10 (Figures 7B and 7C). As a permanent lack of dopamine affects motor activity and startle behavior (Riemensperger et al., 2011), we tested whether TH-shits1 flies were simply not able to run as fast as controls. To this end, we measured the flies’ spontaneous running speed in the absence of odor at the elevated temperature of 35°C, as this stimulates fast running in control flies (Figures S5A and S5B). Here, TH-shits1 flies behaved indistinguishably from control flies and ran at speeds of 20 and even 30 mm/s (Figure S5B). These data suggest that motor deficits are not responsible for the reduction in odor tracking persistence upon inactivation of TH+ DANs.
In contrast to TH+ neuron inactivation, inactivation of all PAM neurons with the line 58E02-Gal4 driving UAS-shits1 had no effect on the fly’s tracking behavior and persistence as compared to control flies (Figures 7B and 7C). These data posit that a subset of DANs, in particular those involved in aversive memory formation, are required to drive increased odor tracking with every non-rewarded trial.
TH-Gal4 labels a number of DANs in the fly brain, including some outside the MB (Mao and Davis, 2009). We sought to further narrow down the type of DAN that mediates persistence in odor tracking. We screened a number of transgenic lines expressing either in single types or smaller subsets of DANs (Aso and Rubin, 2016, Liu et al., 2012) (Figures 7E–7H and S7C–S7E). Inhibition of most single PPL1 DANs did not result in a significant difference in tracking behavior compared to controls (Figures S5D and S5E). However, inhibition of some TH+ neuron subsets using Gal4-transgenic lines generated by enhancer-bashing of the original TH-Gal4 enhancer significantly decreased persistent odor tracking (Figures 7D–7F). Furthermore, driving shits1 expression in PPL1-γ1pedc (or MP1, MB320C-Gal4), previously shown to modulate MBON-γ1pedc>α/β in a hunger state-dependent manner (Figures 7G–7I) (Pavlowsky et al., 2018, Perisse et al., 2016, Tsao et al., 2018), also significantly dampened the flies’ average tracking behavior compared to control (Figures 7H and 7I). Altogether, although we cannot exclude a contribution of DANs outside of the MB network, these results suggest that subsets of PPL1 and PPL2 neurons are involved in maintaining and increasing olfactory tracking but also that no single subset alone mediates the full effect.
Dop1R2 in Kenyon Cells Regulates Persistent Tracking
Having implicated dopamine in persistent odor tracking, we next sought to identify the mechanism. The best studied dopamine receptors in the fly are D1-like receptors Dop1R1 (or Dumb, dDA1, DopR1) and Dop1R2 (or Damb, DopR2). Prior work suggested that Dop1R1 mediates memory acquisition, and Dop1R2 facilitates forgetting; while Dop1R1 induces stronger cAMP-dependent signaling, Dop1R2 signaling initiates higher calcium levels in the post-synaptic neuron through a different G-protein than induced by Dop1R1 (Berry et al., 2012, Himmelreich et al., 2017). Interestingly, Dop1R2 has also been implicated in the formation of long-lasting aversive memory induced through a protocol of repeated pairings of odor with electric shocks (Plaçais et al., 2012, Plaçais et al., 2017). We analyzed mutant flies of both receptors on the fly ball to test their persistence in odor tracking. Dop1R1 mutants performed non-significantly differently from heterozygous controls and reached the same average speed during stimulus periods (Figures S5F and S5G). By contrast, Dop1R2 mutant animals displayed a strongly reduced speed and tracked the odorant less persistently (Figures 7J and 7K). Their average speed during stimulus periods was significantly reduced, although their speed during the first stimulus period (dark blue curve, Figure 7J) was not different from heterozygous controls (Figures 7J and 7K). Instead, the significant reduction in stimulus speed resulted from a gradual decrease of running speed and length in response to the stimulus (Figure 7K). As both types of MBONs involved in persistent odor tracking receive input by αβ-type KCs, we next asked whether Dop1R2 signaling was required in these neurons by knocking down Dop1R2 expression by RNAi specifically in these neurons (MB008B>Dop1R2i). This was indeed the case. Flies without Dop1R2 in αβ-type KCs showed a strong decrease in persistent odor tracking over trials compared to controls (Figures 7L and 7M). These data implicate Dop1R2 as an important mediator of tracking persistence in the absence of reward, a function perhaps in line with a role of this receptor in maintaining memory of past, possibly negative, experience (see for instance [Dolan et al., 2018, Plaçais et al., 2013, Séjourné et al., 2011]).
Discussion
What drives gradually increasing persistence in behavior? For the fly, we propose a model by which a circuit module of KCs, MBONs, and DANs drive gradually increasing odor tracking, which can be efficiently suppressed by extrinsic MBON-innervating feeding-related OANs (Figure 7N). Behavioral persistence has been previously analyzed in flies in a different context. For instance, courtship of fly males and copulation with a female are maintained by dopaminergic neurons in the ventral nerve cord, where they counteract GABAergic neurons (Crickmore and Vosshall, 2013). In that scenario, DANs in the ventral nerve cord maintain an ongoing behavior and prevent that the male disengages prematurely before successful insemination.
Our experimental data also implicate DANs, primarily from within the PPL1 (e.g., PPL1-γ1pedc) and PPL2ab clusters, and Dop1R2 signaling. In particular, inactivation of synaptic output of DANs positive for TH-Gal4 as well as loss of Dop1R2 in αβ-type KCs reduced the increase in odor tracking from trial to trial, while not affecting the speed at first odor stimulation. These data suggest that TH+ DANs promote goal-directed movement, i.e., odor tracking, through a Dop1R2-dependent mechanism in KCs.
We find that MBON-γ1pedc>αβ, which receives dopaminergic input by PPL1-γ1pedc (Aso et al., 2014a, Aso et al., 2014b, Yamagata et al., 2015), is required for odor tracking (see also Tsao et al., 2018). Moreover, we also observed a trial-to-trial decrease in odor response of this MBON, matching the dopamine-induced synaptic depression previously observed in MBONs upon learning (Cohn et al., 2015, Hige et al., 2015, Owald et al., 2015). Notably, PPL1-γ1pedc activates Dop1R2 in MBON-γ1pedc>αβ, a signal recently found to be critical for appetitive long-term memory (Pavlowsky et al., 2018). Nevertheless, it appears that, in addition to PPL1-γ1pedc, other DANs regulate behavioral persistence by modulating in particular αβ-KCs. It is intriguing to speculate about a common function of Dop1R2 in the formation of long-lasting aversive memory induced by repeatedly pairing odor with an aversive experience (Plaçais et al., 2012, Plaçais et al., 2017) and the behavior studied here—increased and persistent expression of a behavior induced by the experience of repeated failure to reach a goal.
Our experimental data further implicated MBON-α2sc, which is connected to MBON-γ1pedc>αβ (Takemura et al., 2017). Our calcium imaging data are consistent with an inhibitory interaction between the two MBONs. However, some of our behavioral data and prior imaging data (Perisse et al., 2016) do not support an inhibitory connection. Furthermore, MBON-γ1pedc>αβ projects to other brain regions and downstream targets, and similarly MBON-α2sc receives additional inputs—all of which could be equally or more important for persistent behavior than a direct connection between these two MBONs. Finally, some DANs respond to movement (Aimon et al., 2019), including PPL1-γ2α’1/MV1 (Berry et al., 2015). Although we did not find an essential role of this particular neuron in odor tracking persistence, movement might contribute to the activity of MBONs responding the odorant.
Remarkably, MBON-α2sc connects the MB to neurons within the LH (Dolan et al., 2018). Thus, we speculate that the LH might assign an odor to its corresponding behavioral category, such as “food-related” for vinegar (Dolan et al., 2018), while the MB acts as a top-down control to gauge the expression of an innate behavior (i.e., tracking an appetitive odor) according to state and experience.
Our behavioral data led us to propose a circuit model (Figure 7N). Using computational modeling, we have tested whether the MB network including DANs and MBONs could, in theory, produce the observed behavior (Figures S5H–S5O; STAR Methods). Indeed, we find that a simplified recurrent circuit of KCs, DANs, and MBONs (Eichler et al., 2017, Takemura et al., 2017) can account for the observed behavioral persistence and also the measured MBON-γ1pedc>αβ odor responses. While this model cannot replace experimental evidence, it forms a useful theoretical framework for future studies on the role of the MB in behavioral persistence.
Based on our present data and computational predictions, we propose a model by which the recurrent circuit architecture of the MB, in addition to storing information for future behavior, is ideally suited to maintain and gradually change ongoing behavior, for instance by modulating output of the LH, according to the animal’s internal state and needs.
The use of an olfactory treadmill has allowed us to dissect the different aspects of a food search. In particular, how does food and feeding suppress food search if the sensory cue, the odor, is still present? OA-VPM4 connects feeding centers (i.e., SEZ) directly with odor tracking-promoting MBON-γ1pedc>αβ and inhibits its activity suggesting an inhibitory connection between VPM4 and the MBON. Nevertheless, we cannot exclude that OA-VPM4 signals through multiple mechanisms including OA and possibly other neurotransmitters. In addition, a recent study showed that activation of VPM4 promotes proboscis extension to sugar (Youn et al., 2018). Although a direct role in taste detection through pharynx or labellum appears unlikely (data not shown and Youn et al., 2018), it is possible that feeding behavior itself (e.g., lymphatic sugar, food texture, activity of feeding muscles) are detected and/or promoted by these neurons and then brought to the MB. We propose that VPM4 is a direct mediator between olfactory-guided food search and the rewarding experience of feeding and related behavior.
Conclusions
Our data provide a neural circuit mechanism empowering flies to express and prioritize behavior in a need- and state-dependent manner.
It is exciting to speculate that fundamentally similar circuit motifs might exist in NE and DA neuron-containing circuits in the mammalian brain, governing the organization of behavior in a flexible and context-dependent manner by integrating internal and external context. For instance, noradrenergic neurons of the brainstem nucleus of the solitary tract (NST) receive taste information, and input from the gastrointestinal tracts, lungs, and heart (Carleton et al., 2010). Neurons in the NST project to multiple brain regions including the amygdala, hypothalamus, and insular cortex (Carleton et al., 2010), all of which receive internal state as well as other sensory information.
Our data in the fly provide an experimental and theoretical framework for a better understanding of the fundamental circuit mechanisms underpinning neuromodulation of context-dependent behavioral persistence and withdrawal.
STAR-Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Mouse Alexa488 | Molecular Probes | AB_221544 |
| Anti-Mouse Alexa633 | Molecular Probes | AB_141431 |
| Anti-Rabbit Alexa568 | Molecular Probes | AB_141416 |
| Anti-Rabbit Alexa633 | Molecular Probes | AB_2535731 |
| Anti-Rat Alexa568 | Molecular Probes | AB_141874 |
| Mouse monoclonal anti-ChAT | Yasuyama et al., 1995 | N/A |
| Mouse monoclonal anti-OA | Jena Bioscience | AB_2315000 |
| Rabbit polyclonal anti-dsRed | Clontech | AB_10013483 |
| Rabbit polyclonal anti-Tyr | Millipore | AB_11215460 |
| Rat monoclonal anti-GFP [3H9] | Chromotek | AB_10773374 |
| Rat monoclonal anti-Ncadherin | DSHB | AB_528121 |
| Experimental Models: Organisms/Strains | ||
| D.mel/Canton-S | Bloomington DSC | FlyBase: FBst0064349 |
| D.mel/Dop1R1attP | Gift from Vanessa Ruta | N/A |
| D.mel/Dop1R2attP | Keleman et al., 2012 | FlyBase: FBal0283280 |
| D.mel/GMR58E02-Gal4 | Bloomington DSC | FlyBase: FBst0041347 |
| D.mel/GMR64C08-Gal4 | Bloomington DSC | FlyBase: FBst0039299 |
| D.mel/GMR95A10-LexA | Bloomington DSC | FlyBase: FBst0061633 |
| D.mel/Gr43a-Gal4 | Miyamoto et al., 2012 | FlyBase: FBti0168340 |
| D.mel/Gr5a-Gal4 | Bloomington DSC | FlyBase: FBst0057592 |
| D.mel/LexAop2-mCD8-GFP | Bloomington DSC | FlyBase: FBst0056182 |
| D.mel/LexAop-P2X2 | Bloomington DSC | FlyBase: FBst0076030 |
| D.mel/MB10B | Janelia RC | FlyBase: FBst0068293 |
| D.mel/MB112C | Janelia RC | FlyBase: FBst0068263 |
| D.mel/MB113C | Janelia RC | FlyBase: FBst0068264 |
| D.mel/MB11B | Janelia RC | FlyBase: FBst0068294 |
| D.mel/MB131B | Janelia RC | FlyBase: FBst0068265 |
| D.mel/MB152B | Janelia RC | FlyBase: FBst0068266 |
| D.mel/MB185B | Janelia RC | FlyBase: FBst0068267 |
| D.mel/MB18B | Janelia RC | FlyBase: FBst0068296 |
| D.mel/MB210B | Janelia RC | FlyBase: FBst0068272 |
| D.mel/MB22B | Janelia RC | FlyBase: FBst0068298 |
| D.mel/MB242A | Janelia RC | FlyBase: FBst0068307 |
| D.mel/MB27B | Janelia RC | FlyBase: FBst0068301 |
| D.mel/MB28B | Janelia RC | N/A |
| D.mel/MB298B | Janelia RC | FlyBase: FBst0068309 |
| D.mel/MB2B | Janelia RC | FlyBase: FBst0068305 |
| D.mel/MB310C | Janelia RC | FlyBase: FBst0068313 |
| D.mel/MB320C | Janelia RC | FlyBase: FBst0068253 |
| D.mel/MB355B | Janelia RC | N/A |
| D.mel/MB364B | Janelia RC | FlyBase: FBst0068318 |
| D.mel/MB370B | Janelia RC | FlyBase: FBst0068319 |
| D.mel/MB371B | Janelia RC | FlyBase: FBst0068383 |
| D.mel/MB399B | Janelia RC | FlyBase: FBst0068369 |
| D.mel/MB417B | Janelia RC | FlyBase: FBst0068321 |
| D.mel/MB418B | Janelia RC | FlyBase: FBst0068322 |
| D.mel/MB419B | Janelia RC | FlyBase: FBst0068323 |
| D.mel/MB433B | Janelia RC | FlyBase: FBst0068324 |
| D.mel/MB434B | Janelia RC | FlyBase: FBst0068325 |
| D.mel/MB438B | Janelia RC | FlyBase: FBst0068326 |
| D.mel/MB461B | Janelia RC | FlyBase: FBst0068327 |
| D.mel/MB463B | Janelia RC | FlyBase: FBst0068370 |
| D.mel/MB477B | Janelia RC | FlyBase: FBst0068328 |
| D.mel/MB504B | Janelia RC | FlyBase: FBst0068329 |
| D.mel/MB50B | Janelia RC | Flybase: FBst0068365 |
| D.mel/MB51B | Janelia RC | FlyBase: FBst0068275 |
| D.mel/MB52B | Janelia RC | N/A |
| D.mel/MB542B | Janelia RC | FlyBase: FBst0068372 |
| D.mel/MB543B | Janelia RC | FlyBase: FBst0068335 |
| D.mel/MB549C | Janelia RC | FlyBase: FBst0068373 |
| D.mel/MB552B | Janelia RC | N/A |
| D.mel/MB57B | Janelia RC | FlyBase: FBst0068277 |
| D.mel/MB58B | Janelia RC | FlyBase: FBst0068278 |
| D.mel/MB5B | Janelia RC | FlyBase: FBst0068306 |
| D.mel/MB60B | Janelia RC | FlyBase: FBst0068279 |
| D.mel/MB74C | Janelia RC | FlyBase: FBst0068282 |
| D.mel/MB80C | Janelia RC | FlyBase: FBst0068285 |
| D.mel/MB82C | Janelia RC | FlyBase: FBst0068286 |
| D.mel/MB83C | Janelia RC | FlyBase: FBst0068287 |
| D.mel/MB8B | Janelia RC | FlyBase: FBst0068291 |
| D.mel/MB93C | Janelia RC | FlyBase: FBst0068289 |
| D.mel/MB9B | Janelia RC | FlyBase: FBst0068292 |
| D.mel/pBDP-Gal4U | Bloomington DSC | FlyBase: FBst0068384 |
| D.mel/Tdc2-Gal4 | Bloomington DSC | FlyBase: FBst0009313 |
| D.mel/TH-C1-Gal4 | Liu et al., 2012 | FlyBase: FBtp0083567 |
| D.mel/TH-D1-Gal4 | Liu et al., 2012 | FlyBase: FBtp0083568 |
| D.mel/TH-F1-Gal4 | Liu et al., 2012 | FlyBase: FBtp0083570 |
| D.mel/TH-Gal4 | Bloomington DSC | FlyBase: FBst0008848 |
| D.mel/UAS-CsChrimson | Bloomington DSC | FlyBase: FBst0055134 |
| D.mel/UAS-DenMark | Bloomington DSC | FlyBase: FBst0033062 |
| D.mel/UAS-GCaMP6f | Bloomington DSC | FlyBase: FBst0042747 |
| D.mel/UAS-mCD8-GFP | Bloomington DSC | FlyBase: FBst0030001 |
| D.mel/UAS-Shibirets1 | Bloomington DSC | FlyBase: FBst0044222 |
| D.mel/UAS-syt-GFP | Bloomington DSC | FlyBase: FBst0006926 |
| D.mel/w1118 | Bloomington DSC | FlyBase: FBst0003605 |
| D.mel/UAS-Dop1R2i | Bloomington DSC | FlyBase: FBti0157409 |
| Software and Algorithms | ||
| Matplotlib 1.4.2 | Matplotlib | https://matplotlib.org |
| Numpy 1.8 | Numpy | https://numpy.org |
| Prism 6 and 7 | GraphPad | https://www.graphpad.com/scientific-software/prism |
| Python 2.7 | Python | https://www.python.org |
| Pyvttbl 0.5.2.2 | Github | https://github.com/rogerlew/pyvttbl |
| Scipy.stats 0.14 | Scipy | https://scipy.org |
| pCLAMP 10.3 | Molecular Devices | https://www.moleculardevices.com/ |
| Igor Pro 6.37 | Wave Metrics | https://www.wavemetrics.com/ |
| NeuroMatic 3.0 | Rothman and Silver, 2018 | http://neuromatic.thinkrandom.com/index.html |
| LAS AF E6000 and LAS X | Leica Microsystems | https://www.leica-microsystems.com/ |
| FV10-ASW | Olympus | https://www.olympus-lifescience.com/en/ |
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ilona C. Grunwald Kadow (ilona.grunwald@tum.de).
Experimental Model and Subject Details
Flies were raised at 25°C, 60% humidity, with a 12/12 hours light/dark cycle on a standard cornmeal media. For optogenetic experiments, adult flies were collected at eclosion, kept under blue light only conditions (470nm, 0,05 μW/mm2) on an all-trans-retinal supplemented food (1:250). Fly lines were obtained from the Bloomington stock centers or directly from Janelia Research Campus.
Method Details
Spherical Treadmill behavioral assay
The spherical treadmill was built according to (Seelig et al., 2010) with several modifications to accommodate olfactory instead of visual stimulation protocols. Fly tethering was performed under cold anesthesia and flies were immediately transferred onto the treadmill. After 3 min of acclimatization, experiments were initialized and controlled via a custom-written Python program. Flies that failed to acclimatize and reach a minimum speed of 1.5 mm/s before the first stimulus were discarded. An experiment consisted of 10 consecutive trials (with the exception of 6 trials for CO2 experiments to prevent anesthesia), which were separated by semi-randomized intervals of 60 ± 2-20 s. Each trial was recorded for a minimum of 52 s. The recording was divided into pre-stimulation (20 s), stimulation (dependent on experimental procedure) and post-stimulation (30 s) periods. For open-loop experiments, the stimulation period was 12 s. The closed-loop experiments utilized a short open-loop phase (2 s), followed by a closed-loop phase. In the closed-loop phase, the fly controlled the odor stimulation length, i.e., the odor channel was kept open as long as the online speed criteria were met (> 0 mm/s for 100 ms). The online speed data acquisition rate in the cardinal directions was ∼4kHz for all experiments. The recorded speed data were down-sampled to 10 Hz by summation. Butterworth filtering was employed in all open-loop data after down-sampling. All data analyses were performed with Python 2.7, numpy 1.8, scipy.stats (0.14), pyvttbl (0.5.2.2) and GraphPad Prism 7. Running and absolute turning average speeds were calculated as averages of 100 ms data points collected at cardinal directions in the respective phases of a trial. To minimize the impact of tethering artifacts, 100 ms data points for absolute turning speeds of each single trial were filtered with average absolute turning values, which were computed from the whole length of a respective trial. Average run time was defined as the initial uninterrupted running (speed > 0 mm/s) bout time length upon odor contact. Average run activity was measured as the fraction of odor stimulation time, where flies showed running speed higher than 0 mm/s. A stop was defined as not moving (0 mm/s) for at least 100 ms. Average run plots display millimeters run per second averaged across the entire stimulus period, and therefore also reflect early stopping (before the end of stimulus speed) and slower speeds. This form of display was most frequently used in figure panels to illustrate the change in speed and track length over trials in a combined manner. Data visualization was done with matplotlib (1.4.2). Optogenetic activation on the ball was achieved by using a single high-power mounted LED at 617 nm, calibrated at 30 W/mm2 (M617, Thorlabs). Light stimulation in the absence of frontal air or odor stimulation induced some attraction and forward running toward the light source. This attraction was independent of the genotype of the animal and was seen also in wild-type Canton S flies. Similarly, simultaneous odor and light stimulation had no effect on Gal4 control flies. For appetitive olfactory stimulus delivery on the treadmill, a custom-made PTFE (Teflon) 4 mm tube was used and stationed at 3 mm distance from the tethered fly. The air speed was set to 100 mL/min via a Natec Sensors mass-flow controller. A balsamic vinegar solution (Alnatura Aceto Balsamico, Germany) was prepared daily at 20% v/v dilution in 100 mL Schott bottles. The vinegar concentration was measured with a miniPID (Aurora Scientific, miniPID 200B) and Arduino Uno at 100 Hz. PID recordings were filtered with Butterworth. The miniPID was calibrated with ethyl-butyrate according to (Semmelhack and Wang, 2009). For CO2 delivery, a CO2 stream was injected via a PTFE syringe inserted into the 100 mL/min pressured air main stream. The auxiliary line carried pure CO2 at 50 mL/min.
Optogenetic and olfactory 4-arm maze
The 4-arm arena is based on the optogenetics-only arena described in (Aso et al., 2014b). The air/odor delivery was achieved via 4 passive solvent channels (Schott bottles containing Millipore water or vinegar solution 20% v/v) and a rotary pump (Thomas G12/01-4 EB). The rotary pump (∼200 mL/min) was connected to an outlet at the arena center. The negative pressure generated by the pump facilitates drawing headspace in the passive solvent channels. For rapid switching between odor channels and target quadrants, a set of solenoid valves (Festo MFH-3-MF) were used. For optogenetics, a custom assembled LED array (Amber SMD PLCC2) was utilized to stimulate each quadrant of the arena at 617nm. The arena was illuminated via IR-LEDs and experiments were recorded with a CMOS camera (FLIR Flea3 MP Mono). The behavioral analysis expressed as preference index ((number of flies in stimulus quadrants - number of flies in non-stimulus quadrants) / total number of flies). Hardware control and data acquisition was achieved via Arduino Mega and in-house MATLAB scripts. Examples of tracking data can be seen in Videos S1 and S2.
Arena Preference Data of Empty>CsChrimson Control Flies to LED Only, Related to Figure 5 Video displays control flies in optogenetic arena stimulated with red light LED.
Arena Preference Data of Tdc2>CsChrimson Control Flies to LED Only, Related to Figure 5 Video displays experimental flies expressing CsChrimson in OANs in optogenetic arena stimulated with red light LED.
T-maze
The two-choice population assay or T-maze was performed as previously described in (Lewis et al., 2015). Briefly, flies were tested in groups of ∼60 in a non-aspirated T-maze and were allowed 1 min to respond to stimuli. Experimentation was carried out in climate-controlled boxes at 32°C and 60% rH. A preference index (PI) was calculated by subtracting the number of flies on the air side by the number of flies on the stimulus side and normalizing by the total number of flies. Statistical analysis was performed using the Kruskal-Wallis’ test and the Dunn’s multiple comparisons post hoc test using GraphPad Prism 6.
Immunohistochemistry
Adult (4-7 days old) fly brains were dissected, fixed and stained as described previously (Lewis et al., 2015). All microscopy was performed at a Leica SP8 confocal microscope. Images were processed using ImageJ and Adobe Photoshop.
Electron microscopy and connectomics analysis
Reconstructions are based on an ssTEM (serial section transmission electron microscope) dataset comprising an entire adult fly brain (Zheng et al., 2018). Neuron skeletons were manually reconstructed using CATMAID (https://catmaid.readthedocs.org) (Saalfeld et al., 2009, Schneider-Mizell et al., 2016). MBON-γ1pedc>αβ was initially identified by sampling downstream of KCs in the γ1/peduncle compartment. VPM3 and VPM4 were found by semi-random sampling of synaptic inputs of MBON-γ1pedc>αβ in the γ1 compartment. For identification, their microtubule-containing backbones were reconstructed and compared with published light-level data (Aso et al., 2014a, Busch et al., 2009). Subsequently, their axonal branches in the γ1 compartment were reconstructed to completion and synaptic sites were annotated. Synaptic connections described here represent fast, chemical synapses matching previously described typical criteria: thick black active zones, pre- (e.g., T-bar, vesicles) and postsynaptic membrane specializations (Prokop and Meinertzhagen, 2006). Visualization and analysis were performed using open-source R (https://github.com/jefferis/nat and https://github.com/jefferis/elmr; ((Manton et al., 2014)) and Python (https://github.com/schlegelp/pymaid) libraries. Please see also Videos S3 and S4 for details on imaged synapses.
. EM Data, Synapse of VPM3 Neuron, Related to Figure 6 Video displays synapse of VPM3 neuron.
EM Data, Synapse of VPM4 Neuron, Related to Figure 6 Video displays synapse of VPM4 neuron.
Ex vivo and in vivo Calcium Imaging
Calcium imaging experiments were performed on 4-8 days old MB112C-Gal4;UAS-GCaMP6f flies, and MB80C-Gal4;UAS-GCaMP6f (for in vivo, odor stimulation), and on 4-7 days old R95A10-lexA,lexAop-P2X2;MB112C-Gal4;UAS-GCaMP6f flies and their controls +,lexAop-P2X2;MB112C-Gal4;UAS-GCaMP6f (for in vivo, ATP and/or odor stimulation).
Preparations of flies for in vivo experiments were prepared as previously described (Bräcker et al., 2013). Preparations were imaged using a Leica DM6000FS fluorescent microscope equipped with a 40x water immersion objective and a Leica DFC 9000 GT fluorescent camera or a Olympus FV1000 two-photon system with BX61WI microscope and a 40x objective and a mode-locked T:Sapphire Mai Tai DeepSee laser at 910 nm to excite the GCaMP6 fluorescence. All images were acquired with either the Leica LAS X or the Olympus FV10-ASW image acquisition software. In order to minimize the brain movement of in vivo preparations under the two-photon microscope, a drop of 1% low melting temperature agarose (NuSieveGTG, Lonza) in imaging buffer maintained at 37°C was added to the exposed brain. A custom-made odor delivery system with mass flow controllers was used for vinegar stimulation. The odor was delivered in a continuous airstream (1000 mL/min) through a 8-mm Teflon tube placed ∼1 cm away from the fly. In vivo two-photon time series data were acquired at a rate of ∼4 frames/s with 175 × 175 pixel resolution.
For experiments including stimulation of P2X2 with ATP, ATP was either added to the buffer on top of the brain of a living fly to a final concentration of 2 mM or, when paired with odor stimulations, using a custom-built perfusion system (∼2 mL/min, 2mM). Images were acquired at a rate of 20 frames/s (static conditions) or 16 frames/s, with 8x8 binning.
Changes in fluorescence intensity were measured in manually drawn regions of interest (ROI) using the LAS AF E6000 Lite, LAS X or the Olympus FV10-ASW software. Relative changes in fluorescence intensity were defined as ΔF/F = 100∗ (Fi – F0)/F0 for the i frames after stimulation. Fluorescence background, F0, is the average fluorescence of 5 frames (∼1 s; in vivo, two-photon), 20 frames (∼1 s; in vivo, ATP static) or 17 frames (∼1 s; in vivo, ATP perfused).
Computational model
We assumed that the KC and DAN populations have neuronal activity governed by the following equations:
where denotes KC activity and denotes DAN activity. During odor presentation, the input is set to one, otherwise it is zero. is the connectivity strength of KC-KC and KC-DAN synapses (Figure S5J, gray), and corresponds to DAN-DAN and DAN-KC (Figure S5J, black). The time constant of integration for each population, , was fixed according KC responses observed in response to prolonged stimulus presentation (Galili et al., 2014). It is important to mention that this system can be analytically solved in the case of constant input. In matrix form with , we can rewrite the equation as:
where is the identity matrix. This linear equation can be solved in terms of the eigenvalues and corresponding eigenvectors of , and the main property of this system is the re-scaling of the effective time constant of neuronal activity projected onto the eigenmodes, . Notice that for the effective time constant goes to infinity, resulting in a perfect integration of the input without decay. Therefore, for values of connectivity with , KCs and DANs will maintain persistent activity even after stimulus offset. We explored this property of the recurrent network to generate the persistent activity consistent with behavioral observations (see Figure 1).
To directly compare model output of persistent neural activity to experimental observations of persistent running, we first applied a transfer function to convert the normalized sum of KC and DAN activity to MBON activity in the same range as the data (Figure 3D). Specifically, we considered an exponential transfer function:
where is a multiplicative constant and the exponent. Then, to obtain running speeds, we applied another exponential transfer function to the MBON activity.
Model fitting and parameters
We used the data from Figures 1E and 3D to fit our model’s parameters by minimizing the mean squared error between the model output (predicted MBON activity and running speed) and the data (average fluorescence change of the peduncle region and measured average running speed, respectively). We fitted the two parameters of both exponential transfer functions. For perturbations up to 20% to the sum we still observed consistent persistent activity in the network (data not shown). For the results reported in Figure S5 we fixed the perturbation to 10%. The fitted parameters of the exponential transfer function in Figure S5O are and of Figure S5M are . The remaining parameters were fixed: = 12 s, , and .
Quantification and Statistical Analysis
Behavioral analysis
Statistical analyses were performed in Graphpad 7, and Python 2.7 with Pyvttbl (0.5.2.2) and Scipy.stats (0.14) packages. All results of the statistical analysis can be found in Table S1. Outliers were removed from all calculations with ROUT (Graphpad, Q = 5%) analysis. Average speeds and run times, as described in the ‘Spherical Treadmill behavioral assay’ section above, were calculated for the indicated time windows (pre, stim, post). Baseline running speeds were analyzed before initial odor stimulation (see Table S1). Average run speeds show fly performance from odor onset until odor termination per trial or over all trials as indicated.
In Figures 1B and 1C, wild-type CS data were analyzed under different time windows (before, during and after odor exposure) for run and turn speeds with one-way ANOVA and Tukey’s multiple comparisons as a post hoc test. To further inspect how run speeds over ten trials changed for the same dataset, the run speeds during the odor stimulation phase in the later trials were compared to the first trial in Dunnett’s multiple comparisons test, after one-way ANOVA test (Figure 1E). During the closed-loop analyses, average run times did not conform normality. Therefore, total run times were calculated for individual flies and used in subsequent statistical analysis. Total run times, and run speeds were compared with one-way ANOVA and Tukey’s multiple post hoc comparisons (Figures 1M and 1N).
In all subsequent spherical treadmill experiments (except S2G), two-way repeated-measures ANOVA was used. The genotypes were considered independent factors. Only when the same set of flies were analyzed under different conditions (Figure S1E: Air versus CO2), both two factors (trials and conditions) were considered as the repeating factors in two-way repeated-measures ANOVA.
Main genotype or condition effects, independent of trial-to-trial differences, were depicted in boxplots. p values in the boxplots were inferred directly from two-way repeated-measures ANOVA for two groups (Figures 2B, 2D, 2F, 2H, 4E, 7I, 7K, 7M, S1D, S1E, S2D, and S2F) and from Tukey’s multiple comparisons for more than two groups (Figures 1F, 1J, 4B, 5B, 5D, 6H, 7C, S5D, S5F, S5H, and S6E; for Figures 7E, S7C, and S7E only, Dunnett’s test). Trial-to-trial post hoc comparisons were shown in line graphs, where each genotype was contrasted to relevant control group for each ten trials individually (i.e., trial 1 test group versus trial 1 control group etc.). When a single experimental group was present, Sidak’s multiple comparisons were utilized (Figures 2B, 2D, 2F, 2H, 4E, 7I, 7K, 7M, S1D, S1E, S2D, and S2F). In other conditions where more than one experimental group was measured against a control group, Dunnett’s multiple comparisons test were used (Figures 1G, 1H, 4B, 5B, 5D, 6H, 7C, 7E, S5D, S5F, S5H, S6E, S7C, and S7E).
Exceptions to the two-way analyses were as follows (Figures 1M, 1N, and S2G). During the closed-loop analyses (Figures 1M and 1N), average run times did not conform normality. Therefore, total run times were calculated for individual flies and used in subsequent statistical analysis. Total run times, and run speeds were compared with one-way ANOVA and Tukey’s multiple post hoc comparisons (Figures 1M and 1N). In Figure S2G, only one repeating factor was tested. Consequently, one-way repeated-measures ANOVA was employed and succeeded by a Dunnett’s multiple comparisons post hoc test for all trials.
Baseline speed comparisons were handled via Welch’s t test for two groups and one-way ANOVA and Tukey’s multiple post hoc comparisons for three groups (Table S1). All post hoc tests were chosen according to Graphpad’s recommendations. For all analyses, statistical notations are as follows: ‘ ns ’ p > 0.05, ’ ∗ ‘ p < 0.05, ‘ ∗∗ ’ p < 0.01, ‘ ∗∗∗ ‘ p < 0.001, ‘ ∗∗∗∗ ‘ p < 0.0001. In all panels, error bars denote SEM.
Ex vivo and in vivo Calcium Imaging
ΔF/F traces were analyzed using the NeuroMatic 3.0 package for IgorPro 6.37 (WaveMetrics). Figure 3: The area under the curve (AUC) was computed for the entire stimulus period (MBON-γ1pedc>αβ; Figures 3A–3D), for the onset peak odor response (between 0 and 5 s after stimulus onset; MBON-α2sc; Figures 3F–3H) and for the offset peak odor response (between 0 and 20 s after stimulus offset, the longest peak duration observed in the dataset; MBON-α2sc; Figure 3I). The offset peak decay was defined as the time constant of a single exponential fit of the peak decay (Figure 3J). AUCs and decay values were plotted over trials and compared using two-way repeated-measure ANOVA (Prism 7, GraphPad). For Figure 6D: The negative peak amplitude was computed as the average fluorescence for 5 s, 20 s after ATP application. Data were compared using an unpaired t test with Welch’s correction for unequal variances (Prism 7, GraphPad). For Figure 6F: Data are expressed as the difference of AUC before and after ATP bath application, normalized by the average difference. Data were compared using an unpaired t test (Prism 7, GraphPad). Pseudo-colored images were generated using a custom-written MATLAB program and ImageJ.
Data and Code Availability
Source data and analysis code supporting the current study have not been deposited in a public repository, but are available from the corresponding author on request.
Acknowledgments
We would like to acknowledge Lasse Bräcker, Jörg Henninger, Christian Schmid, and Stefan Precht for their important technical contributions. We are grateful to Vivek Jayaraman and Armin Bahl for help with the treadmill assay. We thank Anja B. Friedrich and Heidi Miller-Mommerskamp for help with anatomy and general fly husbandry. Thanks to Vanessa Ruta, Yoshinori Aso, and Gerald Rubin for sharing fly lines prior to publication. We thank Richard Axel, Matthieu Louis, Hiromu Tanimoto, Scott Waddell, Silke Sachse, Nicolas Gompel, and members of the Grunwald Kadow lab for very valuable and helpful comments on the manuscript. We further thank S. Waddell, R. Roberts, N. Massood-Panah, and I. J. Alifor for their contributions to the EM tracing.
This work was funded by the Max Planck Society (to J.G. and I.C.G.K.) and the Technical University of Munich (to J.G. and I.G.K.), an ERC starting grant (637472, to I.C.G.K.), the Marie Curie Training Network (FliAct to I.C.G.K.), a Capes-Humboldt postdoctoral fellowship (to M.W.) and the German Research Foundation (CRC870 (A04), INST 95/1382-1, and INST 95/1419-1, to I.C.G.K.). All EM reconstruction work was supported by the Howard Hughes Medical Institute (to D.D.B.), the Wellcome Trust (203261/Z/16/Z), an ERC Consolidator grant (649111) and core support from the MRC (MC- U105188491) to G.S.X.E.J.
Author Contributions
S.S. designed and built the spherical treadmill assay with help and input by I.C.G.K. S.S. carried out all behavioral analysis on the treadmill assay with the exception of Figures 7L, 7M, 4A, and 4B, which were done by J.-F.D.B. J.-F.D.B. and K.P.S. designed and carried out all imaging experiments. M.E.W. and J.G. developed theoretical models which were critical to develop the experimental logic of this project. L.P.L. designed and built the 4-arm maze and carried out the T-maze screen for MBON lines involved in vinegar attraction. L.-M.F. and B.G. carried out and analyzed the 4-arm maze experiments. P.S., A.E.-S., C.B.F., N.S., S.A.C.-S., J.S.L., D.D.B., M.C., and G.S.X.E.J. carried out and analyzed all experiments relating to the EM reconstruction. S.S. and I.C.G.K. conceptualized the study with input by J.-F.D.B. I.C.G.K. oversaw and directed the study and wrote the manuscript with contributions of all authors.
Declaration of Interests
The authors declare no competing interests.
Published: August 27, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.neuron.2019.07.028.
Supplemental Information
Figures S1–S5
Summary Table of All Statistical Analysis and Results, Related to Figures 1–7 and S1–S5 Table contains statistical results and statistic methods for all results presented.
Article plus Supplemental Information
References
- Aimon S., Katsuki T., Jia T., Grosenick L., Broxton M., Deisseroth K., Sejnowski T.J., Greenspan R.J. Fast near-whole-brain imaging in adult Drosophila during responses to stimuli and behavior. PLoS Biol. 2019;17:e2006732. doi: 10.1371/journal.pbio.2006732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez-Salvado E., Licata A.M., Connor E.G., McHugh M.K., King B.M., Stavropoulos N., Victor J.D., Crimaldi J.P., Nagel K.I. Elementary sensory-motor transformations underlying olfactory navigation in walking fruit-flies. eLife. 2018;7:e37815. doi: 10.7554/eLife.37815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso Y., Rubin G.M. Dopaminergic neurons write and update memories with cell-type-specific rules. eLife. 2016;5:e16135. doi: 10.7554/eLife.16135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso Y., Herb A., Ogueta M., Siwanowicz I., Templier T., Friedrich A.B., Ito K., Scholz H., Tanimoto H. Three dopamine pathways induce aversive odor memories with different stability. PLoS Genet. 2012;8:e1002768. doi: 10.1371/journal.pgen.1002768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso Y., Hattori D., Yu Y., Johnston R.M., Iyer N.A., Ngo T.T., Dionne H., Abbott L.F., Axel R., Tanimoto H., Rubin G.M. The neuronal architecture of the mushroom body provides a logic for associative learning. eLife. 2014;3:e04577. doi: 10.7554/eLife.04577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso Y., Sitaraman D., Ichinose T., Kaun K.R., Vogt K., Belliart-Guérin G., Plaçais P.Y., Robie A.A., Yamagata N., Schnaitmann C. Mushroom body output neurons encode valence and guide memory-based action selection in Drosophila. eLife. 2014;3:e04580. doi: 10.7554/eLife.04580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badel L., Ohta K., Tsuchimoto Y., Kazama H. Decoding of context-dependent olfactory behavior in Drosophila. Neuron. 2016;91:155–167. doi: 10.1016/j.neuron.2016.05.022. [DOI] [PubMed] [Google Scholar]
- Bari A., Robbins T.W. Inhibition and impulsivity: behavioral and neural basis of response control. Prog. Neurobiol. 2013;108:44–79. doi: 10.1016/j.pneurobio.2013.06.005. [DOI] [PubMed] [Google Scholar]
- Berridge C.W., Waterhouse B.D. The locus coeruleus-noradrenergic system: modulation of behavioral state and state-dependent cognitive processes. Brain Res. Brain Res. Rev. 2003;42:33–84. doi: 10.1016/s0165-0173(03)00143-7. [DOI] [PubMed] [Google Scholar]
- Berry J.A., Cervantes-Sandoval I., Nicholas E.P., Davis R.L. Dopamine is required for learning and forgetting in Drosophila. Neuron. 2012;74:530–542. doi: 10.1016/j.neuron.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry J.A., Cervantes-Sandoval I., Chakraborty M., Davis R.L. Sleep facilitates memory by blocking dopamine neuron-mediated forgetting. Cell. 2015;161:1656–1667. doi: 10.1016/j.cell.2015.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräcker L.B., Siju K.P., Varela N., Aso Y., Zhang M., Hein I., Vasconcelos M.L., Grunwald Kadow I.C. Essential role of the mushroom body in context-dependent CO2 avoidance in Drosophila. Curr. Biol. 2013;23:1228–1234. doi: 10.1016/j.cub.2013.05.029. [DOI] [PubMed] [Google Scholar]
- Burke C.J., Huetteroth W., Owald D., Perisse E., Krashes M.J., Das G., Gohl D., Silies M., Certel S., Waddell S. Layered reward signalling through octopamine and dopamine in Drosophila. Nature. 2012;492:433–437. doi: 10.1038/nature11614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch S., Tanimoto H. Cellular configuration of single octopamine neurons in Drosophila. J. Comp. Neurol. 2010;518:2355–2364. doi: 10.1002/cne.22337. [DOI] [PubMed] [Google Scholar]
- Busch S., Selcho M., Ito K., Tanimoto H. A map of octopaminergic neurons in the Drosophila brain. J. Comp. Neurol. 2009;513:643–667. doi: 10.1002/cne.21966. [DOI] [PubMed] [Google Scholar]
- Carleton A., Accolla R., Simon S.A. Coding in the mammalian gustatory system. Trends Neurosci. 2010;33:326–334. doi: 10.1016/j.tins.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn R., Morantte I., Ruta V. Coordinated and compartmentalized neuromodulation shapes sensory processing in Drosophila. Cell. 2015;163:1742–1755. doi: 10.1016/j.cell.2015.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrales-Carvajal V.M., Faisal A.A., Ribeiro C. Internal states drive nutrient homeostasis by modulating exploration-exploitation trade-off. eLife. 2016;5:e19920. doi: 10.7554/eLife.19920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crickmore M.A., Vosshall L.B. Opposing dopaminergic and GABAergic neurons control the duration and persistence of copulation in Drosophila. Cell. 2013;155:881–893. doi: 10.1016/j.cell.2013.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethier V.G., Goldrich-Rachman N. Anesthetic stimulation of insect water receptors. Proc. Natl. Acad. Sci. USA. 1976;73:3315–3319. doi: 10.1073/pnas.73.9.3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan M.J., Belliart-Guerin G., Bates A.S., Frechter S., Lampin-Saint-Amaux A., Aso Y., Roberts R.J.V., Schlegel P., Wong A., Hammad A. Communication from learned to innate olfactory processing centers is required for memory retrieval in Drosophila. Neuron. 2018;100:651–668. doi: 10.1016/j.neuron.2018.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler K., Li F., Litwin-Kumar A., Park Y., Andrade I., Schneider-Mizell C.M., Saumweber T., Huser A., Eschbach C., Gerber B. The complete connectome of a learning and memory centre in an insect brain. Nature. 2017;548:175–182. doi: 10.1038/nature23455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenberg J., Barnstedt O., Cognigni P., Lin S., Waddell S. Re-evaluation of learned information in Drosophila. Nature. 2017;544:240–244. doi: 10.1038/nature21716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenberg J., Jacob P.F., Walker T., Barnstedt O., Edmondson-Stait A.J., Pleijzier M.W., Otto N., Schlegel P., Sharifi N., Perisse E. Integration of parallel opposing memories underlies memory extinction. Cell. 2018;175:709–722. doi: 10.1016/j.cell.2018.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili D.S., Lüdke A., Galizia C.G., Szyszka P., Tanimoto H. Olfactory trace conditioning in Drosophila. J. Neurosci. 2011;31:7240–7248. doi: 10.1523/JNEUROSCI.6667-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili D.S., Dylla K.V., Lüdke A., Friedrich A.B., Yamagata N., Wong J.Y., Ho C.H., Szyszka P., Tanimoto H. Converging circuits mediate temperature and shock aversive olfactory conditioning in Drosophila. Curr. Biol. 2014;24:1712–1722. doi: 10.1016/j.cub.2014.06.062. [DOI] [PubMed] [Google Scholar]
- Gomez-Marin A., Stephens G.J., Louis M. Active sampling and decision making in Drosophila chemotaxis. Nat. Commun. 2011;2:441. doi: 10.1038/ncomms1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunwald Kadow I.C. State-dependent plasticity of innate behavior in fruit flies. Curr. Opin. Neurobiol. 2019;54:60–65. doi: 10.1016/j.conb.2018.08.014. Published online September 13, 2018. [DOI] [PubMed] [Google Scholar]
- Hattori D., Aso Y., Swartz K.J., Rubin G.M., Abbott L.F., Axel R. Representations of Novelty and Familiarity in a Mushroom Body Compartment. Cell. 2017;169:956–969.e17. doi: 10.1016/j.cell.2017.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hige T., Aso Y., Modi M.N., Rubin G.M., Turner G.C. Heterosynaptic Plasticity Underlies Aversive Olfactory Learning in Drosophila. Neuron. 2015;88:985–998. doi: 10.1016/j.neuron.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelreich S., Masuho I., Berry J.A., MacMullen C., Skamangas N.K., Martemyanov K.A., Davis R.L. Dopamine Receptor DAMB Signals via Gq to Mediate Forgetting in Drosophila. Cell Rep. 2017;21:2074–2081. doi: 10.1016/j.celrep.2017.10.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki H.K., Panse K.M., Anderson D.J. Independent, reciprocal neuromodulatory control of sweet and bitter taste sensitivity during starvation in Drosophila. Neuron. 2014;84:806–820. doi: 10.1016/j.neuron.2014.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keleman K., Vrontou E., Kruttner S., Yu J.Y., Kurtovic-Kozaric A., Dickson B.J. Dopamine neurons modulate pheromone responses in Drosophila courtship learning. Nature. 2012;489:145–149. doi: 10.1038/nature11345. [DOI] [PubMed] [Google Scholar]
- Kim H., Kirkhart C., Scott K. Long-range projection neurons in the taste circuit of Drosophila. eLife. 2017;6:e23386. doi: 10.7554/eLife.23386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krashes M.J., DasGupta S., Vreede A., White B., Armstrong J.D., Waddell S. A neural circuit mechanism integrating motivational state with memory expression in Drosophila. Cell. 2009;139:416–427. doi: 10.1016/j.cell.2009.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDue E.E., Mann K., Koch E., Chu B., Dakin R., Gordon M.D. Starvation-Induced Depotentiation of Bitter Taste in Drosophila. Curr. Biol. 2016;26:2854–2861. doi: 10.1016/j.cub.2016.08.028. [DOI] [PubMed] [Google Scholar]
- Lewis L.P., Siju K.P., Aso Y., Friedrich A.B., Bulteel A.J., Rubin G.M., Grunwald Kadow I.C. A Higher Brain Circuit for Immediate Integration of Conflicting Sensory Information in Drosophila. Curr. Biol. 2015;25:2203–2214. doi: 10.1016/j.cub.2015.07.015. [DOI] [PubMed] [Google Scholar]
- Li K., Nakajima M., Ibanez-Tallon I., Heintz N. A Cortical Circuit for Sexually Dimorphic Oxytocin-Dependent Anxiety Behaviors. Cell. 2016;167:60–72. doi: 10.1016/j.cell.2016.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Plaçais P.Y., Yamagata N., Pfeiffer B.D., Aso Y., Friedrich A.B., Siwanowicz I., Rubin G.M., Preat T., Tanimoto H. A subset of dopamine neurons signals reward for odour memory in Drosophila. Nature. 2012;488:512–516. doi: 10.1038/nature11304. [DOI] [PubMed] [Google Scholar]
- Mann K., Gordon M.D., Scott K. A pair of interneurons influences the choice between feeding and locomotion in Drosophila. Neuron. 2013;79:754–765. doi: 10.1016/j.neuron.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manton J.D., Ostrovsky A.D., Goetz L., Costa M., Rohlfing T., Jefferis G.S.X.E. Combining genome-scale Drosophila 3D neuroanatomical data by bridging template brains. bioRxiv. 2014 [Google Scholar]
- Mao Z., Davis R.L. Eight different types of dopaminergic neurons innervate the Drosophila mushroom body neuropil: anatomical and physiological heterogeneity. Front. Neural Circuits. 2009;3:5. doi: 10.3389/neuro.04.005.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto T., Slone J., Song X., Amrein H. A fructose receptor functions as a nutrient sensor in the Drosophila brain. Cell. 2012;151:1113–1125. doi: 10.1016/j.cell.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owald D., Felsenberg J., Talbot C.B., Das G., Perisse E., Huetteroth W., Waddell S. Activity of defined mushroom body output neurons underlies learned olfactory behavior in Drosophila. Neuron. 2015;86:417–427. doi: 10.1016/j.neuron.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.H., Kwon J.Y. Heterogeneous expression of Drosophila gustatory receptors in enteroendocrine cells. PLoS ONE. 2011;6:e29022. doi: 10.1371/journal.pone.0029022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlowsky A., Schor J., Placais P.Y., Preat T. A GABAergic feedback shapes dopaminergic input on the Drosophila mushroom body to promote appetitive long-term memory. Curr. Biol. 2018;28:1783–1793. doi: 10.1016/j.cub.2018.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perisse E., Owald D., Barnstedt O., Talbot C.B., Huetteroth W., Waddell S. Aversive Learning and Appetitive Motivation Toggle Feed-Forward Inhibition in the Drosophila Mushroom Body. Neuron. 2016;90:1086–1099. doi: 10.1016/j.neuron.2016.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry C.J., Barron A.B. Neural mechanisms of reward in insects. Annu. Rev. Entomol. 2013;58:543–562. doi: 10.1146/annurev-ento-120811-153631. [DOI] [PubMed] [Google Scholar]
- Plaçais P.Y., Trannoy S., Isabel G., Aso Y., Siwanowicz I., Belliart-Guérin G., Vernier P., Birman S., Tanimoto H., Preat T. Slow oscillations in two pairs of dopaminergic neurons gate long-term memory formation in Drosophila. Nat. Neurosci. 2012;15:592–599. doi: 10.1038/nn.3055. [DOI] [PubMed] [Google Scholar]
- Plaçais P.Y., Trannoy S., Friedrich A.B., Tanimoto H., Preat T. Two pairs of mushroom body efferent neurons are required for appetitive long-term memory retrieval in Drosophila. Cell Rep. 2013;5:769–780. doi: 10.1016/j.celrep.2013.09.032. [DOI] [PubMed] [Google Scholar]
- Plaçais P.Y., de Tredern É., Scheunemann L., Trannoy S., Goguel V., Han K.A., Isabel G., Preat T. Upregulated energy metabolism in the Drosophila mushroom body is the trigger for long-term memory. Nat. Commun. 2017;8:15510. doi: 10.1038/ncomms15510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter J., Craven B., Khan R.M., Chang S.J., Kang I., Judkewitz B., Volpe J., Settles G., Sobel N. Mechanisms of scent-tracking in humans. Nat. Neurosci. 2007;10:27–29. doi: 10.1038/nn1819. [DOI] [PubMed] [Google Scholar]
- Prokop A., Meinertzhagen I.A. Development and structure of synaptic contacts in Drosophila. Semin. Cell Dev. Biol. 2006;17:20–30. doi: 10.1016/j.semcdb.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Riemensperger T., Völler T., Stock P., Buchner E., Fiala A. Punishment prediction by dopaminergic neurons in Drosophila. Curr. Biol. 2005;15:1953–1960. doi: 10.1016/j.cub.2005.09.042. [DOI] [PubMed] [Google Scholar]
- Riemensperger T., Isabel G., Coulom H., Neuser K., Seugnet L., Kume K., Iché-Torres M., Cassar M., Strauss R., Preat T. Behavioral consequences of dopamine deficiency in the Drosophila central nervous system. Proc. Natl. Acad. Sci. USA. 2011;108:834–839. doi: 10.1073/pnas.1010930108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Root C.M., Ko K.I., Jafari A., Wang J.W. Presynaptic facilitation by neuropeptide signaling mediates odor-driven food search. Cell. 2011;145:133–144. doi: 10.1016/j.cell.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman J.S., Silver R.A. NeuroMatic: an integrated open-source software toolkit for acquisition, analysis and simulation of electrophysiological data. Front. Neuroinform. 2018;12:14. doi: 10.3389/fninf.2018.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saalfeld S., Cardona A., Hartenstein V., Tomancak P. CATMAID: collaborative annotation toolkit for massive amounts of image data. Bioinformatics. 2009;25:1984–1986. doi: 10.1093/bioinformatics/btp266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider-Mizell C.M., Gerhard S., Longair M., Kazimiers T., Li F., Zwart M.F., Champion A., Midgley F.M., Fetter R.D., Saalfeld S. Quantitative neuroanatomy for connectomics in Drosophila. eLife. 2016;5:e12059. doi: 10.7554/eLife.12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröter U., Malun D., Menzel R. Innervation pattern of suboesophageal ventral unpaired median neurones in the honeybee brain. Cell Tissue Res. 2007;327:647–667. doi: 10.1007/s00441-006-0197-1. [DOI] [PubMed] [Google Scholar]
- Schwaerzel M., Monastirioti M., Scholz H., Friggi-Grelin F., Birman S., Heisenberg M. Dopamine and octopamine differentiate between aversive and appetitive olfactory memories in Drosophila. J. Neurosci. 2003;23:10495–10502. doi: 10.1523/JNEUROSCI.23-33-10495.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz L.A., Luo L. Organization of the locus coeruleus-norepinephrine system. Curr. Biol. 2015;25:R1051–R1056. doi: 10.1016/j.cub.2015.09.039. [DOI] [PubMed] [Google Scholar]
- Seelig J.D., Chiappe M.E., Lott G.K., Dutta A., Osborne J.E., Reiser M.B., Jayaraman V. Two-photon calcium imaging from head-fixed Drosophila during optomotor walking behavior. Nat. Methods. 2010;7:535–540. doi: 10.1038/nmeth.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séjourné J., Plaçais P.Y., Aso Y., Siwanowicz I., Trannoy S., Thoma V., Tedjakumala S.R., Rubin G.M., Tchénio P., Ito K. Mushroom body efferent neurons responsible for aversive olfactory memory retrieval in Drosophila. Nat. Neurosci. 2011;14:903–910. doi: 10.1038/nn.2846. [DOI] [PubMed] [Google Scholar]
- Semmelhack J.L., Wang J.W. Select Drosophila glomeruli mediate innate olfactory attraction and aversion. Nature. 2009;459:218–223. doi: 10.1038/nature07983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh G.S., Wong A.M., Hergarden A.C., Wang J.W., Simon A.F., Benzer S., Axel R., Anderson D.J. A single population of olfactory sensory neurons mediates an innate avoidance behaviour in Drosophila. Nature. 2004;431:854–859. doi: 10.1038/nature02980. [DOI] [PubMed] [Google Scholar]
- Takemura S.Y., Aso Y., Hige T., Wong A., Lu Z., Xu C.S., Rivlin P.K., Hess H., Zhao T., Parag T. A connectome of a learning and memory center in the adult Drosophila brain. eLife. 2017;6:e26975. doi: 10.7554/eLife.26975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma V., Knapek S., Arai S., Hartl M., Kohsaka H., Sirigrivatanawong P., Abe A., Hashimoto K., Tanimoto H. Functional dissociation in sweet taste receptor neurons between and within taste organs of Drosophila. Nat. Commun. 2016;7:10678. doi: 10.1038/ncomms10678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao C.H., Chen C.C., Lin C.H., Yang H.Y., Lin S. Drosophila mushroom bodies integrate hunger and satiety signals to control innate food-seeking behavior. eLife. 2018;7:e35264. doi: 10.7554/eLife.35264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Breugel F., Huda A., Dickinson M.H. Distinct activity-gated pathways mediate attraction and aversion to CO2 in Drosophila. Nature. 2018;564:420–424. doi: 10.1038/s41586-018-0732-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.P., Lin Y.Q., Zhang L., Wilson Y.A., Oyston L.J., Cotterell J., Qi Y., Khuong T.M., Bakhshi N., Planchenault Y. Sucralose promotes food intake through NPY and a neuronal fasting response. Cell Metab. 2016;24:75–90. doi: 10.1016/j.cmet.2016.06.010. [DOI] [PubMed] [Google Scholar]
- Yamagata N., Ichinose T., Aso Y., Plaçais P.Y., Friedrich A.B., Sima R.J., Preat T., Rubin G.M., Tanimoto H. Distinct dopamine neurons mediate reward signals for short- and long-term memories. Proc. Natl. Acad. Sci. USA. 2015;112:578–583. doi: 10.1073/pnas.1421930112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z., Macara A.M., Lelito K.R., Minosyan T.Y., Shafer O.T. Analysis of functional neuronal connectivity in the Drosophila brain. J. Neurophysiol. 2012;108:684–696. doi: 10.1152/jn.00110.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuyama K., Kitamoto T., Salvaterra P.M. Immunocytochemical study of choline acetyltransferase in Drosophila melanogaster: an analysis of cis-regulatory regions controlling expression in the brain of cDNA-transformed flies. J. Comp. Neurol. 1995;361:25–37. doi: 10.1002/cne.903610103. [DOI] [PubMed] [Google Scholar]
- Youn H., Kirkhart C., Chia J., Scott K. A subset of octopaminergic neurons that promotes feeding initiation in Drosophila melanogaster. PLoS ONE. 2018;13:e0198362. doi: 10.1371/journal.pone.0198362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Branch A., Shen P. Octopamine-mediated circuit mechanism underlying controlled appetite for palatable food in Drosophila. Proc. Natl. Acad. Sci. USA. 2013;110:15431–15436. doi: 10.1073/pnas.1308816110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z., Lauritzen J.S., Perlman E., Robinson C.G., Nichols M., Milkie D., Torrens O., Price J., Fisher C.B., Sharifi N. A complete electron microscopy volume of the brain of adult Drosophila melanogaster. Cell. 2018;174:730–743. doi: 10.1016/j.cell.2018.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Arena Preference Data of Empty>CsChrimson Control Flies to LED Only, Related to Figure 5 Video displays control flies in optogenetic arena stimulated with red light LED.
Arena Preference Data of Tdc2>CsChrimson Control Flies to LED Only, Related to Figure 5 Video displays experimental flies expressing CsChrimson in OANs in optogenetic arena stimulated with red light LED.
. EM Data, Synapse of VPM3 Neuron, Related to Figure 6 Video displays synapse of VPM3 neuron.
EM Data, Synapse of VPM4 Neuron, Related to Figure 6 Video displays synapse of VPM4 neuron.
Figures S1–S5
Summary Table of All Statistical Analysis and Results, Related to Figures 1–7 and S1–S5 Table contains statistical results and statistic methods for all results presented.
Article plus Supplemental Information
Data Availability Statement
Source data and analysis code supporting the current study have not been deposited in a public repository, but are available from the corresponding author on request.