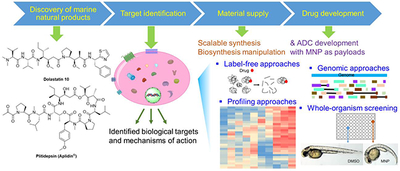
Abstract
Marine natural products represent novel and diverse chemotypes that serve as templates for the discovery and development of therapeutic agents with distinct mechanisms of action. These genetically encoded compounds produced by an evolutionary optimized biosynthetic machinery are usually quite complex and can be difficult to recreate in the laboratory. The isolation from the source organism results in limited amount of material; however, the development of advanced NMR technologies and dereplication strategies has enabled the structure elucidation on small scale. In order to rigorously explore the therapeutic potential of marine natural products and advance them further, the biological characterization has to keep pace with the chemical characterization. The limited marine natural product supply has been a serious challenge for thorough investigation of the biological targets. Several marine drugs have reached the markets or are in clinical trials, where those challenges have been overcome, including through the development of scalable syntheses. However, the identification of mechanisms of action of marine natural products early in the discovery process is potentially game changing, since effectively linking marine natural products to potential therapeutic applications in turn triggers motivation to tackle challenging syntheses and solve the supply problem. An increasing number of sensitive technologies and methods have been developed in recent years, some of which have been successfully applied to marine natural products, increasing the value of these compounds with respect to their biomedical utility. In this review, we discuss advances in overcoming the bottlenecks in marine natural product research, emphasizing on the development and advances of diverse target identification technologies applicable for marine natural product research.
Keywords: marine natural products, first-in-class pharmacological agents, target identification approaches, material supply strategies
Graphical abstract
1. Introduction
Historically, natural products served as important drug leads against a wide array of human diseases. From 1981 to 2014, among all of the 1211 approved small-molecule therapeutic agents, natural product related drugs could account for almost 65% of the marketed drugs, which were either used unaltered or developed based on the naturally occurring structures.[1] In recent years, the interest in marine sources has intensified, in part due to the awareness of the tremendous biodiversity in the marine environment viewed as a guarantee for new chemistry and biomedical potential, and in part due to the drastic advances in both research techniques and strategies to address the key issues hampering marine natural product-based drug discovery and development[2,3] Initial genome sequencing of marine natural product-producing microorganisms suggest that most marine natural products (MNPs) remain to be discovered.[4] MNPs have been well recognized as powerful bioweapons to fill the pipeline of drug leads for the pharmaceutical industry. As MNPs possess unique structural architectures covering broad chemical space, they continuously serve as promising templates to inspire drug development against a variety of diseases. Besides their clinical utility, MNPs are also applied as probes in advancing basic biomedical research for the study of biological pathways and exploring unconventional biological space for drug discovery. This review is not intended to be comprehensive, but highlights accomplishments in the development of advanced technologies to explore the biological targets of MNPs and the representative achievements using MNPs to target diverse therapeutically relevant biological space.
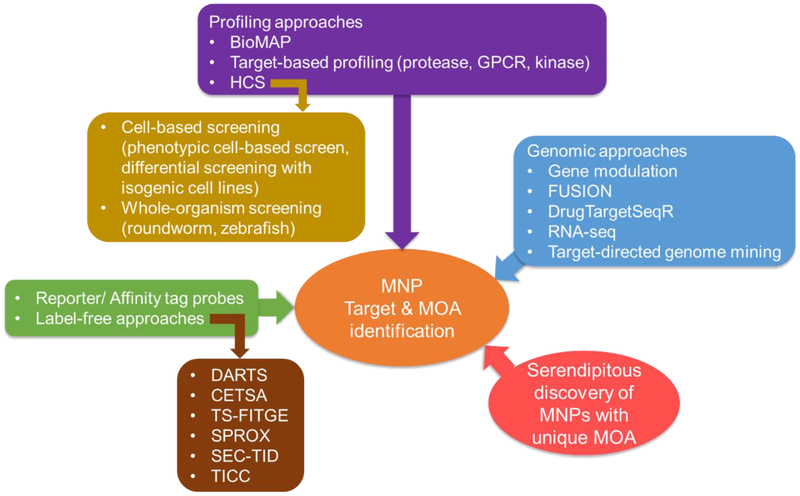
To continue to fuel the drug pipeline with MNPs (Chapter 2) and successfully compete with complementary drug discovery approaches, MNP drug discovery research requires addressing three major challenges: (1) how to identify novel molecules; (2) how to solve the supply issues and (3) how to correlate bioactivity with underlying target/MOA. Advances have been made in all these areas (Figure 1). The increased investigation and annotation of marine biological (genetic) diversity has been yielding a wealth of genome sequence information indicating biosynthetic potential that mostly remains to be translated into chemicals, as many compounds are only predicted based on biosynthetic gene cluster (BGC) sequence data. However, a steady number of chemically diverse structures has been characterized, in large part due to improvements of analytical capabilities and development of dereplication techniques, including nanoscale NMR[5] and molecular networking,[6] respectively, which will not be discussed here. More relevant to this review, advances in exploring the therapeutic potential of existing MNPs are intimately tied to solving the supply problem to enable a rigorous biological evaluation and catalyze development efforts (Chapter 3). All MNPs possess biological functions and may have biomedical potential. However, there has to be an incentive to develop scalable methods. The incentive is the bioactivity linked to a direct therapeutic target (binding partner) or at least tractable mechanism of action tied to a disease process and that can provide biomarkers for preclinical and clinical studies. Novel target identification methods and techniques to capture MOA with minimal amount of material are potential game changers as they can be implemented early in the discovery process, even before a synthesis route has been developed (Chapter 4). The diversity of MNPs allows for the discovery of potential first-in-class therapeutic agents with common drug targets (proteins) and unusual biological targets like lipid membrane/ sterols and provides fundamental options for the development of effective antibody-drug conjugates (ADCs) (Chapter 5). The following chapters should illustrate representative achievements to provide multifaceted insight into the current stages of research to advance MNPs and connect them with their therapeutically relevant biological targets.
Figure 1.
Three bottlenecks and corresponding research highlights in marine natural product drug discovery.
2. Drugs and Drug Candidates from the Ocean
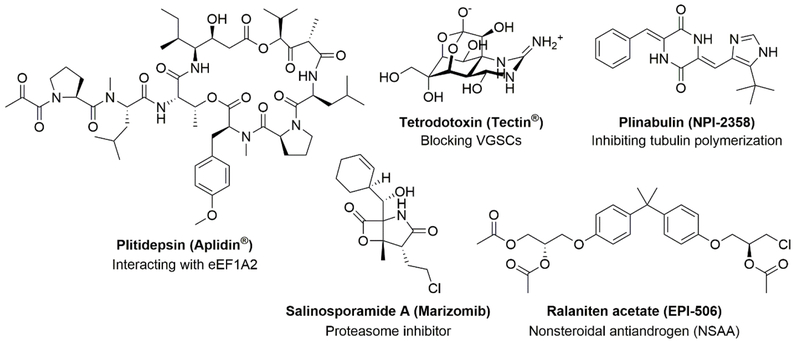
Through decades of intense efforts in both academia and the pharmaceutical industry, ten marine-derived drugs (Figure 2) have successfully reached the market, among which five are for the treatment of cancer, including cytarabine (Cytosar-U®), trabectedin (ET-743, Yondelis®), eribulin mesylate (Halaven®) and the antibody-drug conjugates (ADCs) brentuximab vedotin (Adcetris®) and polatuzumab vedotin (Polivy®), three are for the treatment of hypertriglyceridemia (fatty acid type drugs; Lovaza®, Vascepa® and Epanova®), two are for anti-virus treatment, including vidarabine (Vira-A®) and iota-carrageenan (Carragelose®) and one is for the amelioration of severe chronic pain, namely ziconotide (Prialt®).
Figure 2.
Timeline for drugs from the ocean.
Until now, over twenty MNPs as drug candidates have been investigated in different phases of clinical trials and a plethora of drug leads are under extensive preclinical development for various applications. [7,8] Newman and Cragg’s recent review on drugs and drug candidates from marine sources should be consulted by interested readers. [8] Due to their potency, some MNPs also serve as payloads for ADCs, as demonstrated for the marketed brentuximab vedotin and polatuzumab vedotin (Figures 2 and 13). MNPs increasingly inspire the development of novel pharmacotherapeutics for various applications. Here we briefly highlight several structurally and functionally diverse molecules reaching different stages of clinical trials to further display the rich arsenal of bioweapons offered by the marine environment.
The cyclic depsipeptide plitidepsin (Aplidin®, first-in-class, Figure 3) was originally isolated from the marine tunicate Aplidium albicans and is known to interact with eukaryotic elongation factor 1A2 (eEF1A2) in tumor cells. It has reached Phase III clinical trial for the treatment of relapsed/refractory multiple myeloma in combination with dexamethasone.[9,10]
Figure 3.
Drug candidates from the ocean reaching clinical trials.
Tetrodotoxin (Tectin®, Figure 3), probably the most well-known marine toxin, is a guanidine derivative with a highly oxygenated carbon skeleton from puffer fish (specifically the liver, ovaries and skin) of the Tetraodontidae family, which was believed to accumulate via the marine food chain and/or derive from certain endo-symbiotic bacteria, such as Vibrio spp. and Pseudomonas spp.[11] Mechanism studies indicated tetrodotoxin exerted its effect by competitively blocking voltage-gated sodium channels (VGSCs) in a highly selective manner. Tetrodotoxin binding to VGSCs in nerve cell membranes prevents depolarization and propagation of action potentials and leads to the loss of sensation.[12] More recently, it has been extensively used as a chemical tool to functionally characterize VGSCs and has been applied as an analgesic agent against pathologic pain.[11,13] Two Phase III safety and efficacy studies for management of moderate to severe inadequately controlled cancer-related pain were completed in 2012 and results are awaiting to be reported.[8]
Plinabulin (NPI-2358, Figure 3) is a synthetic analog of halimide, a naturally-occurring aromatic alkaloid derived from the marine fungus Aspergillus sp. CNC139, which was collected from the waters off the Philippine Islands (halimide is also known as phenylahistin, isolated from terrestrial strains, Aspergillus ustus NSC-F037 and NSC-F038).[14,15] Targeting near the colchicine binding site on tubulin monomer and thus inhibiting tubulin polymerization, this molecule could lead to destabilization of the tumor vascular network, reduce tumor blood flow, and thereby cause tumor regression. [16,17] Plinabulin is now under investigation in Phase III trials to assess its application in combination with docetaxel in patients with advanced non-small cell lung cancer due to its function as a vascular-disrupting agents and its apoptotic effect on tumor cells.
Salinosporamide A (Marizomib, Figure 3) was isolated from marine bacteria Salinispora tropica and Salinispora arenicola and identified as a potent proteasome inhibitor.[18] Ever since the success of bortezomib, the first approved therapeutic proteasome inhibitor as anticancer agent, the proteasome has received growing interest as a target for the development of anticancer agents.[19] More importantly, in 2012, another proteasome inhibitor carfilzomib (Kyprolis®), an analog of epoxomicin (initially isolated from an Actinomycetes strain),[20] was approved as an anticancer drug, suggesting the potential of natural proteasome inhibitors as therapeutic agents. Salinosporamide A entered phase I clinical trials for the treatment of multiple myeloma only three years after its discovery.[19,21] To examine the safety, pharmacokinetics and pharmacodynamics of salinosporamide A, a phase II clinical trial in patients with relapsed/ refractory multiple myeloma has been completed.[22]
Ralaniten acetate (EPI-506, Figure 3) is an experimental nonsteroidal antiandrogen (NSAA), which is a prodrug of MNP EPI-002 and a derivative of MNP EPI-067. It targets the androgen receptor (AR) N terminus (NTD) and can block the AR signaling and has entered phase II clinical trial for treatment of metastatic castration-resistant prostate cancer (mCRPC).[23]
3. Research Highlights in Overcoming the Supply Problem to Advance Marine Natural Products for Therapeutic Applications
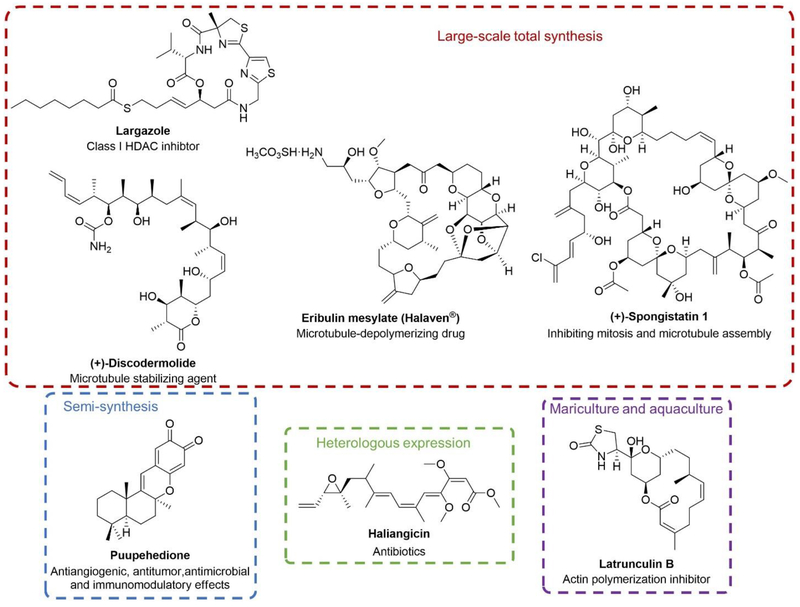
The successful translation of MNPs into commercial drugs further demonstrates the potential of MNPs as promising leads for the development of therapeutic agents. Even though there are bottlenecks obstructing the development process, significant advances have been made to overcome the related issues in MNP research. The isolation of therapeutically promising MNPs from natural sources is often not reproducible and even serendipitous, so the material supply of MNPs is always a critical problem, which would affect the structure elucidation and prevent the comprehensive biological investigation of the MNPs. With the development of various research strategies in synthesis and biosynthesis, the limited supply issue has been greatly improved. For example, total synthesis, especially large-scale synthesis of complex MNPs, has provided plentiful material supply and valid chemistry evidence for thorough biological investigation to promote the MNP research. For example, the large-scale total synthesis of largazole (Figure 4), a class I histone deacetylase (HDAC) inhibitor isolated from marine cyanobacteria originally believed to be Symploca sp.[24–26] and later reclassified as Caldora penicillata,[27,28] has been established and decagrams of the target compound was obtained as final product in 21% overall yield over eight steps.[29] Eribulin mesylate (Halaven®, Figure 3) is a synthetic analog of MNP halichondrin B, which was originally isolated from sponge Halichondria okadai in 1986. It is a microtubule-depolymerizing drug approved by FDA for the treatment of metastatic breast cancer.[30] The supply problem for eribulin mesylate was solved by developing scalable synthetic methods, even though over 60 steps are required.[31,32] (+)-Discodermolide (Figure 4) was first isolated from marine sponge Discodermia dissoluta in 1990[33] and it was reported to stabilize microtubules.[34,35] (+)-Discodermolide was proceeded into phase I clinical trial in 2002, the results further confirmed the activity of (+)-discodermolide as a microtubule stabilizer. Since there is minimal success in supplying enough material from other approaches, the material of (+)-discodermolide for thorough clinical studies is from large-scale total synthesis. Thus, intense efforts have been made by different research groups and companies and a number of synthetic methods have been developed.[36–42] Smith and his co-workers have established and optimized the total synthesis of (+)-discodermolide and meanwhile completed the gram-scale synthetic compound production to enable convenient supply for clinical investigations. [43–46] In addition, Smith et al. designed and developed the total synthesis of spongistatin 1 (Figure 4),[47,48] a MNP originally isolated from a sponge of the genus Spongia. [49] Spongistatin 1 is one of the most potent tumor cell growth inhibitors,[48] and it has been reported to inhibit mitosis and microtubule assembly.[50] Spongistatin 1 was reported to represent a promising therapeutic agent for the treatment of leukemic tumor cells which exhibit chemoresistance due to overexpression of X-linked inhibitor of apoptosis protein (XIAP).[51]
Figure 4.
Examples of MNPs where the supply problem was solved by development of scalable total syntheses (largazole, eribulin mesylate (Halaven®), (+)-discodermolide and (+)-spongistatin 1), semisynthesis (puupehedione), heterologous expression (haliangicin), and mariculture (latrunculin B).
With the utilization of compounds readily isolated from natural sources, semisynthesis also serves as a pivotal solution to the limited supply of MNPs. For instance, an enantiospecific and concise semisynthetic route of MNP puupehedione (Figure 4) that was originally isolated from Verongid sponge,[52] was described.[53] The semisynthesis was achieved from the natural product sclareolide within seven steps. Puupehediones exhibit a variety of biological activities including antiangiogenic, antitumor, antimicrobial and immunomodulatory effects. [54] Sufficient supply of puupehedione through semisynthesis allows for complete and in-depth study to extend the biomedical applications of puupehediones. Moreover, heterologous expression of biosynthetic gene clusters represents an emerging approach to enable MNP material supply. Successful heterologous production of various compounds have been reported, including polyketides, nonribosomal peptides and hybrid nonribosomal peptide–polyketides and isoprenoids,[55] although scalability is not necessarily straightforward. An interesting example regarding the heterologous expression of MNPs is haliangicin (Figure 4), the first marine myxobacterial antibiotic from the halophilic myxobacterium Halicmgium ochraceum SMP-2.[56] The biosynthetic gene cluster (BGC) of haliangicin was identified first and then expressed heterologously in Myxococcus xanthus to afford the haliangicin in a highly efficient manner, resulting in a 10-fold increase in the production and 3-fold increase in the growth rate. [57]
Besides, mariculture is another solution to the MNP supply problem. Bergman et al. reported the extended culture at sea of three sponge species Negombata magnifica, which produces latrunculin B (Figure 4), a cell-permeable actin polymerization inhibitor and confirmed this technology to be an appropriate approach for the accumulation of MNPs.[58] Apart from the approaches mentioned above, fermentation is also applied to provide MNP supply for scientific research and clinical investigations. Although the proteasome inhibitor salinosporamide A (Figure 3) has been synthesized, its ongoing clinical investigations mainly rely on large-scale saline fermentation for continuous supply.
4. Advances in Characterization of Target and Mechanism of Action
Although MNPs are clearly excellent starting points for the development of pharmacotherapeutics, one major challenge is to annotate the bioactivity and unveil the underlying mechanism to translate them into suitable biomedical applications. Among all the established target identification methods, the classical strategy to address this problem is to design a probe with reporter/affinity tag, where the natural product serves as a warhead. However, these strategies require extensive efforts to design feasible probes, conduct structure-activity relationship (SAR) studies, optimize natural product structures and validate resulted hits, among which are likely false positive targets.[59,60] To overcome these limitations, numerous label-free approaches were developed to probe the biological targets of various compounds without requirement of chemical modification and could be performed in high-throughput manner.[61,62] These strategies not only save efforts in chemical synthesis and structural modification, but also avoid the risk of altering the activity and molecular behavior of natural products as well as obviate the immediate need for SAR studies prior to target identification campaigns. Moreover, all the strategies mentioned below enable the identification of multiple targets for a certain compound, which would certainly expedite the determination of mechanisms of action (on-target) and the elucidation of potential side effects (off-target) or relevant pleiotropic pharmacology. More importantly, it is very difficult to identify the biological target and mechanism of action (MOA) of a compound with very small amount of material, which is very common in the case of MNPs. This bottleneck can be overcome by the following and other emerging approaches.
4.1. Label-free Approaches
4.1.1. Drug Affinity Responsive Target Stability (DARTS)
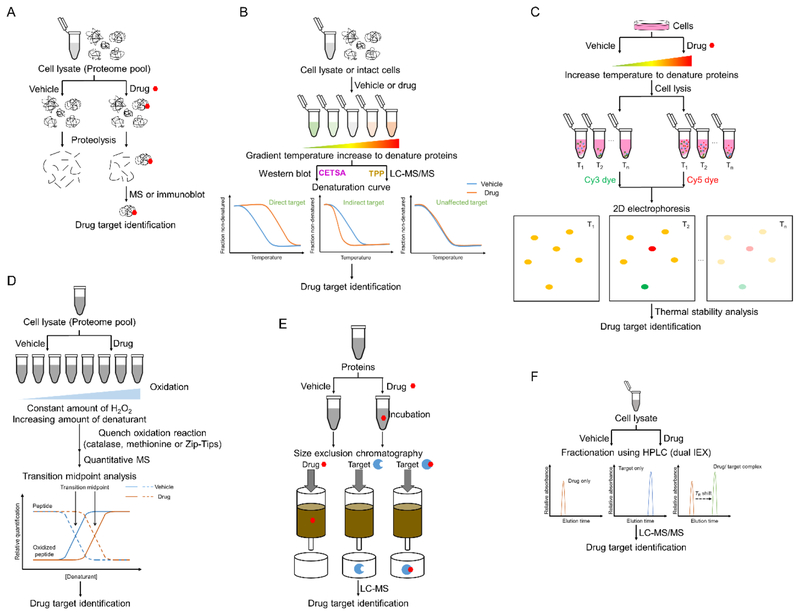
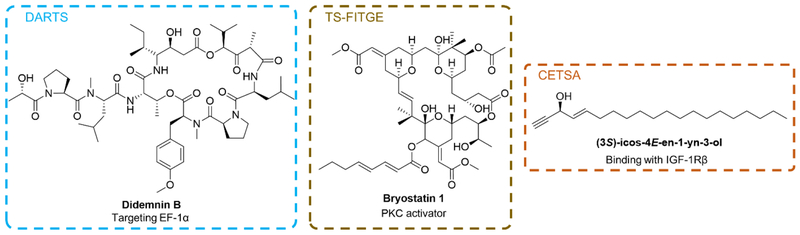
Drug affinity responsive target stability (DARTS) was developed to identify the molecular targets of small-molecule drugs and study protein-ligand interactions (Figure 6A). It is based on the principle that a protein becomes more stabilized and less susceptible to proteolysis when it is ligand/drug-bound than when it is ligand/drug-free. This reduction in the protease susceptibility is specific to the target protein(s) regardless of affinity. In addition, DARTS can utilize any small molecule in its native form without further chemical modifications or SAR studies.[63] Generally, protein or whole-cell lysates are obtained before incubation with small-molecule drugs or vehicle control, followed by proteolysis and then immunoblot to identify drug targets. [64] The feasibility of DARTS was demonstrated using human Jurkat cells after treatment of didemnin B (Figure 5). Didemnin B is an analog of plitidepsin (Aplidin®, Figure 3) and it was reported to inhibit protein synthesis by targeting the guanine nucleotide-binding elongation factor EF-1α.[65] DARTS revealed a much stronger protected band corresponding to EF-1α in the didemnin B treated Jurkat cell proteolyzed extracts compared to that in the vehicle treated group,[66] suggesting the practical applicability of DARTS for complex MNP target identification.
Figure 6.
Illustrative schemes of label-free technologies. A. Drug affinity responsive target stability (DARTS); B. Cellular thermal shift assay (CETSA); C. Thermal stability shift-based fluorescence difference in two-dimensional gel electrophoresis (TS-FITGE); D. Stability of proteins from rates of oxidation (SPROX); E. Size-exclusion chromatography for target identification (SEC-TID); F. Target identification by chromatography co-elution (TICC).
Figure 5.
Structures of MNPs where label-free approaches were used for target identification or validation including didemnin B (DARTS), bryostatin 1 (TS-FITGE) and (3S)-icos-4E-en-1-yn-3-ol (CETSA).
4.1.2. Cellular Thermal Shift Assay (CETSA)
Drug-target interaction can also be assessed by a cellular thermal shift assay (CETSA), which takes advantage of the biophysical principle of ligand-induced thermal stabilization of target proteins, producing a thermal shift to a higher temperature in its melting curve (Figure 6B).[67] In this method, cells are cultured with or without drug treatment and the resulting aliquots are heated to different temperatures to induce protein denaturation, remaining soluble proteins are then extracted and quantified. Quantification of the soluble target proteins at each temperature is achieved using Western blot to generate the protein thermal melting curve, which enables determination of thermal stability and identification of ligand-induced thermal shifts.[67] For example, an acetylenic alcohol, (3S)-icos-4E-en-1-yn-3-ol (Figure 5), was isolated from the marine sponge Cribrochalina vasculum and it was reported to induce tumor-specific apoptosis in non-small cell lung cancer cells. [68] CETSA of this acetylenic alcohol revealed that this alcohol can specifically bind with the insulin growth factor-1 receptor β (IGF-1Rβ), demonstrating the therapeutic potential of this compound for the treatment of cancer, especially non-small cell lung carcinoma (NSCLC). [68,69] To study the effect of drugs on the thermal profile of proteins on a proteome-wide scale, thermal proteome profiling (TPP) was developed by combing CETSA with multiplexed quantitative mass spectrometry (Figure 6B).[70] TPP exhibits promising applicability in identifying both therapeutically relevant targets and off-target protein binding to discover off-target toxicity or pleiotropic pharmacology. [71] Moreover, comparison of the thermal profiles obtained after drug treatment of intact cells with cell extracts enables to distinguish direct ligand/drug-target binding from downstream effectors, which could serve as a means to establish mechanistic biomarkers. However, since CETSA and TPP are both dependent on the extraction of proteins in the detergent-free buffer, they are not suitable for proteins not soluble under these conditions, such as membrane proteins.[61,70] To overcome this critical limitation, Savitski and Drewes et al. developed an optimized TPP approach to extend its application for ligand-membrane protein interactions with the involvement of a mild detergent during cell extraction.[72] Denaturation temperature shift was observed for ATP-binding transmembrane proteins during their following assessment.
4.1.3. Thermal Stability Shift-based Fluorescence Difference in Two-dimensional Gel Electrophoresis (TS-FITGE)
Another label-free method to identify targets from a proteome-wide scale is TS-FITGE (thermal stability shift-based fluorescence difference in two-dimensional gel electrophoresis) (Figure 6C). Similar to CETSA, TS-FITGE detects the thermal stability shift of the target protein upon ligand engagement; but unlike CETSA, TS-FITGE takes advantage of the intra-gel quantification of each protein spot resulting from 2D gel electrophoresis to reveal the thermal stability change, with following inter-gel image analysis for hit confirmation. [73] Using this method, the protein kinase C α (PKCα) was confirmed to be the target of bryostatin 1 (Figure 5), a MNP from the marine bryozoan Bugiila neritina,[74] suggesting the applicability of TS-FITGE to identify membrane proteins as active biological targets-.[74] Bryostatin 1 is demonstrated to be a potent activator of protein kinase C (PKC),[75] and it entered phase II clinical trials for assessment of its effect on various diseases, including cancer, HIV and Alzheimer’s disease (AD). [8] Moreover, using TS-FITGE, nucleophosmin (NPM) was identified as a new protein target of hordenine, which is a natural product that can induce in vitro translation.[73]
4.1.4. Other Approaches
In addition to the label-free approaches applied for the target identification of natural products from marine species, several more methods have been developed to complement the biological target identification process applied to different compounds so far. Even though the generality and applicability of these methods in the target identification of MNPs have not been verified yet, they are amenable approaches to be applied to MNPs, especially because they only require a small amount of material.
SPROX (stability of proteins from rates of oxidation) takes advantage of mass spectrometry- to measure protein thermodynamic stability change for target identification (Figure 6D).[62] It uses hydrogen peroxide in the presence of a denaturant in different concentrations to oxidize proteins[76] and measures thermodynamic changes of protein-folding reactions to detect ligand-target interactions. [77] Using SPROX, two known proteins, cyclophilin A and UDP-glucose-4-epimerase, were identified as biological targets of cyclosporine A, which is an immunosuppressive medication in organ transplantation.[77]
Another approach to identify targets for small-molecule drugs is a platform named SEC-TID (size-exclusion chromatography for target identification) (Figure 6E). SEC-TID is developed on the basis of size-exclusion chromatography in 384-well format to resolve drug-target interactions. [78] In this method, small-molecule drugs are mixed with recombinant or purified protein targets first, drug-target interactions are then resolved using size-exclusion column chromatography. Subsequently, small-molecule drugs bound to their targets are quantified using LC-MS.[62] This approach was utilized to elucidate the target of the Wnt inhibitor XAV939 as tankyrase 1. [79] Moreover, novel molecular interaction between the tumor-vascular disrupting agent vadimezan/ASA404 and farnesyl pyrophosphate synthase, as well as the interaction between the diuretic mefruside and carbonic anhydrase XIII, were identified using this strategy.
The emerging awareness of polypharmacology led to the establishment of an applicable approach for drug target identification, namely TICC (target identification by chromatography coelution) (Figure 6F). TICC monitors the interactions of small-molecule compounds with protein targets using non-denaturing HPLC coupled to LC-MS. TICC was developed based on the rationale that upon binding to a protein, a shift would occur to the chromatographic retention time of a compound. The identification of this characteristic shift leads to the deconvolution of the biological targets.[80] A cytoplasmic delta (24)-sterol C-methyltransferase, Erg6p, was identified as the biologically relevant secondary target of antifungal compound 4513-0042. Moreover, a possible off-target of the dopamine receptor agonist A77636 was identified as well. [80] TICC is suitable for the detection of protein targets with low abundance and low affinity, but it can only detect noncovalent hydrophobic protein-ligand interactions.[81]
The label-free approaches for target identification avoid chemical modification of the test compounds, which is especially beneficial for MNPs that are usually characterized by complex structures and challenging synthesis of functional tagged compounds for affinity chromatography.
4.2. Genomic Approaches
Besides the traditional target-/cell-based high-throughput screening strategies, which only focus on selected targets/cellular properties, modern screening approaches incorporate genome-wide analysis to correlate MNPs with biological targets or at least mechanisms in an unbiased fashion.[82]
4.2.1. Gene Dosage Screens
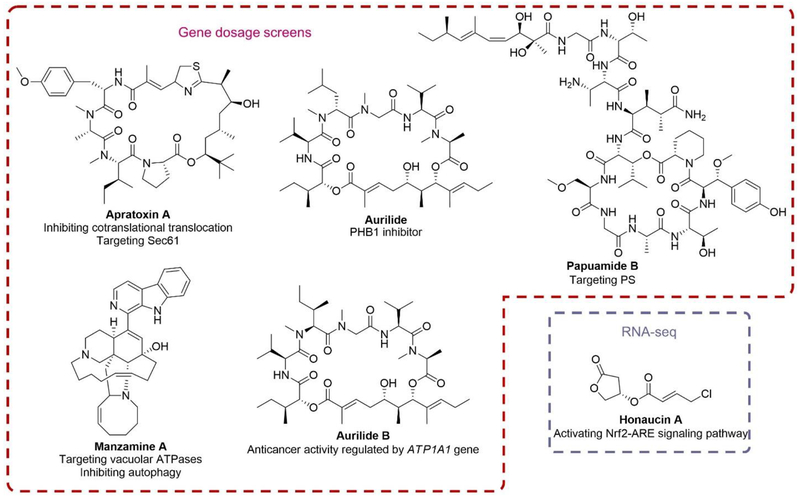
Systematically modulating gene product levels via cDNA overexpression and/or si/sh/miRNA-mediated mRNA knockdown has been increasingly utilized to elucidate the targets and mechanisms of natural product action. Gene dosage screens on a genomic scale in mammalian cells have been first undertaken with apratoxin A (Figure 7), a MNP produced by cyanobacterium Moorea bouillonii.[83] Genomic cDNA overexpression indicated that apratoxin A interrupts fibroblast growth factor receptor (FGFR)-mediated and signal transducer and activator of transcription (STAT) 3 signaling pathway.[84] Later apratoxin A was demonstrated to be an inhibitor of cotranslational translocation of proteins destined for the secretory pathway, such as receptor tyrosine kinases and growth factors, with its direct target identified as the central subunit of the protein translocation channel Sec61.[85–87] In addition, the protein target of aurilide (Figure 7), a polyketide-peptide hybrid isolated from the sea hare Dolabella auricularia,[88] was demonstrated to be prohibitin 1 (PHB1) using an affinity matrix prepared by immobilizing an aurilide-based biotin probe on avidin beads. Treatment of aurilide was reported to induce a mitochondria morphological change from tubular to fragmented forms, similar to the effects of siRNA-mediated PHB1 knockdown, which further validates the inactivation of PHB1 by aurilide.[89] On the other hand, in a recent report, gene ATP1A1 was identified to regulate the cytotoxicity of aurilide B (Figure 7), an analog of aurilide from marine cyanobacterium Lyngbya majuscula Gomont (Oscillatoriaceae), via a pooled and barcoded short-hairpin RNA (shRNA) library screen.[90]
Figure 7.
Structures of MNPs that were subjected to various genomics based mechanism studies, including gene dosage screens (apratoxin A, aurilide, aurilide B, manzamine A and papuamide B), RNA-seq (honaucin A).
An advanced genetic approach utilizing reducing or increasing gene dosage for target identification is chemogenomic profiling.[91] One practical strategy is named haploinsufficiency profiling (HIP), which identifies targets and pathways directly affected by the drug based on the observation that lowering a single gene dosage from two copies to one copy would cause increased sensitivity to drug treatment in diploid yeast.[92,93] The homozygous deletion profiling (HOP) is a complementary approach to identify functionally related genes that buffer the targeted pathway. [94] In this strategy, both copies of a certain gene would be deleted. Manzamine A (Figure 7) is an alkaloid isolated from sponges Haliclona sp.,[95] Pellina sp.,[96] and Xestospongia sp.[97] and has been demonstrated to exhibit various bioactivities, including antimalarial,[98] antibacterial[96] and anticancer[99] activities. It is also demonstrated to decrease the formation of single cells, abolish cell migration and sensitize pancreatic cancer cells to apoptosis induced by TRAIL (TNF-related apoptosis inducing ligand).[100] Chemogenomic profiling conducted in S. cerevisiae indicated that manzamine A (Figure 7) prevents autophagy in pancreatic cancer cells by targeting vacuolar ATPase.[101] Moreover, Andersen and Boone et al. reported a compendium composed of a number of chemical-genetic interaction profiles, resulting from exposure of yeast haploid deletion mutants to a small library of natural product extracts and pure compounds to identify hypersensitivity.[102] Extracts or pure compounds with similar MOAs were clustered together using a method utilizing hierarchical clustering and factorgram to reveal association of a compound or gene with more than one group. As a result, the biological target of papuamide B (Figure 7), a cytotoxic cyclic depsipeptide with anti-HIV activity originally isolated from sponge Theonella mirabilis,[103] was identified as phosphatidylserine (PS).[102]
Besides the successful application of gene dosage manipulation methods in the target identification process of MNPs, other genomic/transcriptomic screening technologies have been developed to decipher complex ligand/drug-target interactions.
4.2.2. Functional Signature Ontology (FUSION)
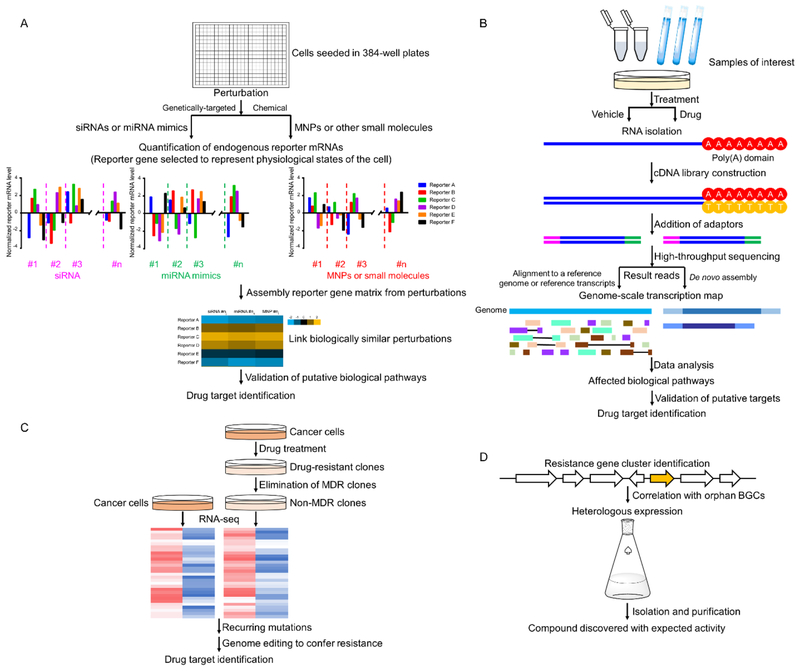
To properly develop therapeutic agents targeting disease-related mechanisms, an approach named FUSION (functional signature ontology) was recently developed to identify biological targets and MOA for MNPs (Figure 8A). A certain set of genes will display characteristic expression signatures from screening of siRNAs, miRNA mimics and MNPs. These signatures were assembled together into a similarity matrix to generate FUSION maps, which link bioactive compounds to specific biological processes.[104] During the assessment, several endogenous reporters of a specific biological process or disease will be selected first to quantify the perturbation after treatment. With this strategy, the putative MOA and potential therapeutic applications for several MNPs were successfully unraveled. In addition, this strategy is further highlighted by the successful application in target identification for didemnin B (Figure 5). The FUSION strategy indicated that didemnin B exerted its effects through activation of mTORC1.[105] Subsequent investigations regarding the MOA of didemnin B turned out to be consistent with published results, which demonstrated that didemnin B induced apoptosis by targeting both PPT1 (palmitoyl-protein thioesterase 1) and eEF1A1 (eukaryotic elongation factor 1A1).[106, 107]
Figure 8.
Flowcharts for genomic target identification approaches either in disease relevant cell types (A–C) or the producing organism (D). A: Functional signature ontology (FUSION); B: DrugTargetSeqR; C: RNA-sequencing (RNA-seq); D: Target-directed mining of bacterial genomes encoding MNP production.
4.2.3. RNA Sequencing (RNA-seq)
RNA-seq is a high-throughput strategy of transcriptome profiling using deep-sequencing approaches (Figure 8B).[108] It measures the expression of numerous genes at the same time and provides insights into multiple biological processes. RNA-seq can not only detect known transcripts, but also investigate the entire transcriptome in a high-throughput and quantitative manner.[108,109] In addition, RNA-seq is demonstrated to exhibit superior resolution to expose the fine transcriptome structure with a single nucleotide resolution.[109] The advancement of RNA-seq has been adopted in MNP research for potential target identification and especially mechanism studies. For example, the polysaccharide from the red alga Gracilariopsis lemaneiformis (PGL) was reported to significantly decrease proliferation and change morphology of lung cancer cells. The intrinsic mechanism and the genes modulated by PGL were revealed through transcriptome analysis using RNA-seq. The sequencing results indicate that PGL affects the apoptosis- and cell cycle-related gene expression, consistent with reported apoptosis and cell cycle arrest caused by PGL treatment.[110] Honaucin A (Figure 7), a MNP isolated from cyanobacterium Leptolyngbya crossbyana,[111] was demonstrated to activate Nrf2-ARE (nuclear erythroid 2-related factor 2-antioxidant response element) signaling pathway to exert antiinflammatory activity after high-throughput transcriptomic analysis by RNA-seq.[112]
4.2.4. DrugTargetSeqR
DrugTargetSeqR was developed to identify biological targets of drugs and other bioactive small-molecule compounds. It takes advantage of the repeated drug resistance related mutations in cancer identified by high-throughput sequencing and additionally incorporates CRISPR/Cas9-based genome editing and computational mutation discovery to reveal the targets of compounds of interest (Figure 8C). It was also developed to overcome some limitations inherited in gene manipulation methods, since the gene knockdown and phenotype correlation often fail due to differences between cumulative biological effects of protein knockdown and fast-acting chemical modulators.[113] To perform DrugTargetSeqR, resistant cancer cells are generated by treating the cells with test compounds, which will be further isolated to get rid of the MDR clones. The selected resistant cell clones and the parental cancer cells are then analyzed by RNA-seq. Mutations that specifically arise in response to drug treatment are identified. CRISPR/Cas9-based genome editing is adopted to determine the genes responsible for the recurring resistance of a specific drug, followed by biological investigation to validate the target. This approach was applied to ispinesib and YM155, two anticancer agents that entered phase II clinical trials,[114,115] to unveil mechanisms likely to cause drug resistance. Through integration with reporter gene assays or phenotypic screens, this approach displays potential to be applicable for more diverse compounds including non-cytotoxic agents.[113]
4.2.5. Genome Mining to Explore Biological Targets
Along with the advance achieved in exploring the biological targets of MNPs using different genomic approaches in disease-relevant cell types, there are parallel advances facilitating genome mining of MNP producing organisms/bacteria as a new method to identify natural products with therapeutically relevant biological targets, particularly antibiotics. Genome mining toward specific targets is a new approach (Figure 8D) developed to forecast the biological function of natural products without a priori knowledge of these compounds utilizing presumptive resistance genes encoding target-modified proteins with orphan biosynthetic gene clusters. Using this strategy, Moore et al. discovered antibiotic producing gene clusters that produce fatty acid synthase (FAS) inhibitors belonging to the thiotetronic acid family from the marine actinomycete Salinispora. The gene cluster function was validated by direct cloning as well as heterologous expression. This methodology could be utilized to identify new gene clusters responsible for antibiotics production and could provide a helpful genome mining tool to discover new antibiotics.[116] Moreover, ARTS (Antibiotic Resistant Target Seeker) was developed to allow for efficient genome mining using Internet resources. ARTS incorporates target-directed genome mining and prediction of gene clusters for antibiotic production to serve as an information platform to identify potential or reported gene cluster targets.[117] In a recent report, Müller et al. described the discovery of topoisomerase inhibitors from myxobacteria following a self-resistance guided strategy derived from genome mining. Genes encoding biosynthesis of a topoisomerase inhibitor were identified first and subsequent activation of this gene cluster led to the discovery of a new group of compounds named pyxidicyclines.[ 118] Müller et al. further claimed the potential of target-driven genome mining in identifying new functional gene clusters and discovering new natural products in myxobacteria.[118] The successful application of target-directed genome mining in both marine bacterium and myxobacteria indicates the potential of this approach in predicting and identifying natural products with specific MOAs and demonstrates its practical value in different organisms, which would further enlarge the scope of novel chemical entity discovery from terrestrial and marine sources.
4.3. Profiling Methods
4.3.1. Antibiotic mode of action profile (BioMAP)
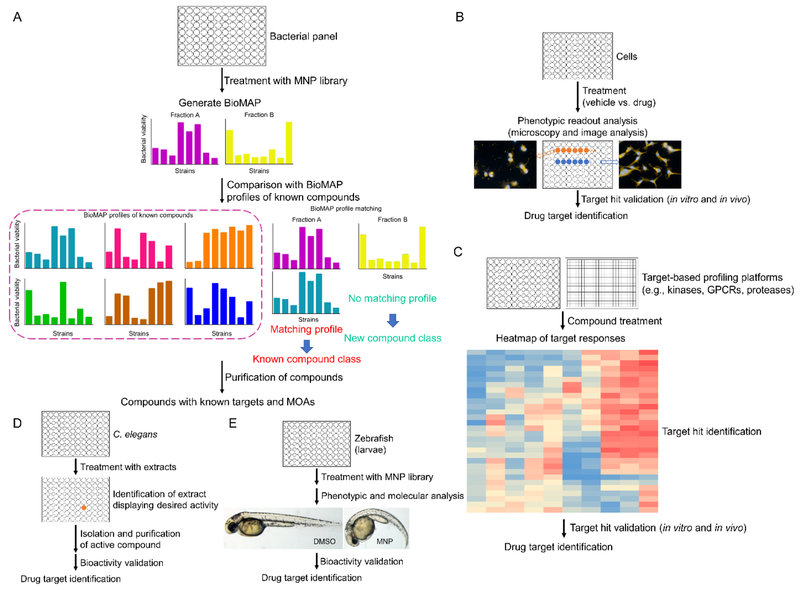
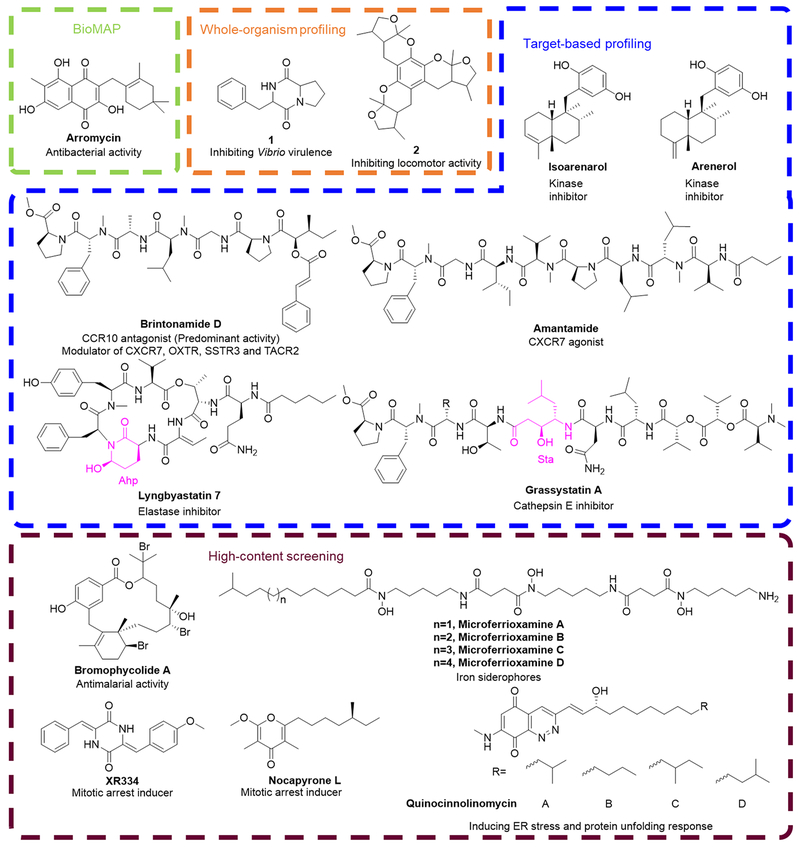
The NCI-60 human tumor cell line screen has served as a reliable platform for the discovery of new compounds with potent anticancer activity and MOA identification through biological response pattern comparison. [119] Similarly, BioMAP (Figure 10A) was established based on the hypothesis that antibiotics from the same structure class would generate similar biological activity profiles across a panel of bacterial pathogens, which can be used to classify unknown compounds by structural class. BioMAP provides a new profiling strategy for the discovery of new antibiotics with unique biological fingerprints. For example, arromycin (Figure 9) was isolated from a marine-derived actinobacterial library after BioMAP characterization and it was demonstrated to exhibit an antibacterial profile that did not match those of the training set included in this profiling. [120] Arromycin processes an unprecedented carbon skeleton for naphthoquinone antibiotics, it is the first reported naphthoquinone scaffold with a 2,4,4-trimethylcyclohexenyl C ring. Even though the exact MO A of arromycin’s antibacterial activity remains unclear, its discovery validates the feasibility of BioMAP screen for the identification of structurally novel compounds as starting points for further medicinal chemistry investigation. [120]
Figure 10.
Schematic figure for profiling methods. A: BioMAP; B: High-content screening (HCS); C: Target-based profiling; D: C. elegans-based screening system; E: Zebrafish-based screening for impact on development and behavior.
Figure 9.
Structures of selected MNPs subjected to profiling methods, BioMAP (arromycin), target-based profiling (isoarenarol, arenerol, brintonamide D, amantamide, grassystatin A and lyngbyastatin 1), high-content screening (bromophycolide A, microferrioxamine A–D, XR334, nocapyroneL and quinocinnolinomycins A–D) and whole-organism screening (diketopiperazine 1 and compound 2). Pharmacophores for protease inhibitors are highlighted.
4.3.2. Target-based profiling
The increasing prevalence of phenotypic screening has greatly promoted method development to assist in the target identification processes. Meanwhile, direct target-based profiling methods (Figure 10B) still play a pivotal role in discovering new compounds targeting potential therapeutic targets and determining the biological role of MNPs. Representative target-based profiling platforms were developed on the basis of prevalent therapeutic targets, including kinases, G protein-coupled receptors (GPCRs) and proteases. Kinases play a crucial role in cell regulation including signal transmission, cell differentiation, proliferation, metabolism and apoptosis. [121] High-throughput kinase profiling allows a parallel approach by interrogating compounds against numerous targets in a single screen.[122] As an effort to discover new kinase inhibitors, a series of marine invertebrate extracts were screened against a panel of protein kinases. As a result, isoarenarol (Figure 9) and arenarol (Figure 9) from sponge Dysidea arenaria Bergquist were identified to be potent inhibitors of several therapeutically important kinases.[123] GPCRs represent the most common signal transduction system and the most intensively studied therapeutic targets since they are involved in diverse fundamental physiological functions. The dysfunction of GPCRs is implicated in diseases such as cancer, diabetes, hypertension and heart failure. [124] From profiling against a panel of 241 GPCR targets (agonist, antagonist and orphan), brintonamide D (Figure 9) was identified as a GPCR modulator, which can function as a C-X-C chemokine receptor type 7 (CXCR7) agonist as well as an antagonist of C-C chemokine receptor type 10 (CCR10), oxytocin receptor (OXTR), somatostatin receptor type 3 (SSTR3) and tachykinin receptor 2 (TACR2). Among all the GPCR targets, brintonamide D exhibits more than 10-fold potency toward CCR10 as an antagonist compared to other hits. [124] Recently, the discovery of amantamide (Figure 9) was reported and it was characterized as a GPCR agonist that can selectively activate the activity of CXCR7.[125] Proteases represent potential drug targets for various diseases including cancer, cardiovascular disorders as well as diseases resulting from parasites and viruses. [126] Target-based screening against a panel of proteases has led to the identification of several protease inhibitors from marine cyanobacteria. For instance, grassystatin A (Figure 9) has been reported to selectively inhibit aspartic protease cathepsin E and, to a lesser extent, cathepsin D after screening against a diverse set of 59 proteases.[127] Lyngbyastatin 7 (Figure 9) was found to exhibit elastase inhibitory activity, protect bronchial epithelial cells against elastase-induced antiproliferation and abrogate the elastase-triggered induction of pro-inflammatory cytokine expression. [128,129]
4.3.3. High-Content Screening (HCS)
High-content screening (HCS) is an image-based screening strategy that incorporates high-throughput techniques to collect quantitative data from complex biological systems (Figure 10C). HCS technology has been integrated into all the different aspects of drug discovery, including primary compound screening, SAR screening and evaluation of ADME (absorption, distribution, metabolism and excretion) properties. It has been considered as a mainstream technology in the pharmaceutical industry. [130] And it is certainly adopted by academia to discover new compounds with interesting biological activities, such as in the MNP field. Roch et al. reported a newly developed live cell-imaging platform for the discovery of new MNPs and investigation of their antimalaria effects on Plasmodium falciparum, which is regarded as the most aggressive malaria parasite. [131] They have taken advantage of this platform to discover new parasite inhibitors from a MNP library. The screening results identified bromophycolide A (Figure 9), isolated from red alga Callophycus serratus,[132] to exhibit potent antimalarial activities in the parasite while little cytotoxicity on the blood cells of the host. [131]
With the advancement of HCS, the combination of high-throughput screening technologies with imaging technique accompanied with automated microscopy has led to the establishment of CP (cytological profiling). CP utilizes imaging with automated microscopy in a high-throughput manner to quantify cells with a variety of descriptors. [133] This approach was applied to prioritize MNP extracts or fractions with interesting biological activity before further purification and following structure elucidation, leading to the discovery microferrioxamines A–D (Figure 9) as iron siderophores.[134] Moreover, using CP as a strategy to the discovery of MNPs with a specific MOA from a microbially derived extract library revealed two constituents from a marine sediment sample, as mitotic arrest inducers. These two constituents were then identified as XR334 and nocapyrone L (Figure 9), which function through disruption of tubulin dynamics and calcium channel function, respectively.[135]
To further explore the potential of HCS and enhance the probability to discovery novel compounds, an upgraded screening platform termed Compound Activity Mapping was developed by incorporating image-based CP information with metabolomics analysis.[136] This new technology can predict the constituents and MOA of bioactive components for very complex natural product extract or fraction library. For proof of principle, this strategy was applied to a marine bacterial sediment extract library for the discovery of quinocinnolinomycins A–D (Figure 9), which were identified to cause ER (endoplasmic reticulum) stress and induce the unfolding of proteins.
4.3.4. Whole-Organism Screening
With the aim of obtaining insight into the in vivo biological activity and to provide more predictable information about the potential therapeutic uses of a natural product, whole-organism models have been employed in medium/high-throughput screening at an early stage of marine biodiscovery.[137] Two representative examples utilizing roundworm (Caenorhabditis elegans) and zebrafish (Danio rerio), the most conventionally used simple model organisms (beyond yeast), are described here to indicate the pivotal role of an in vivo model in biomedical research and pharmaceutical development.
Caenorhabditis elegans has become a widely used animal model increasingly incorporated into various stages of drug discovery to reveal the biomedical relevance of natural products/synthetic molecules.[138,139] Using a C. elegans-based model (Figure 10D) for Vibrio alginolyticus infection, 256 extractions derived from sponge (Haliclona spp.) associated bacteria were screened to identify bioactive compounds that inhibit the virulence of Vibrio species and rescue the affected worms.[140] The initial screening identified the extract of Alcaligenes faecalis as complex nature product mixture capable of rescuing V. alginolyticus without significant toxicity. The following purification efforts led to the identification of a diketopiperazine (1, Figure 9) as the active constituent. The subsequent in vitro assays indicated that this compound solely inhibited the virulence of V. alginolyticus by suppressing quorum sensing without affecting its proliferation. In vivo assays further showed the newly discovered molecule could reduce the intestinal colonization of Vibrio species, improve pharyngeal pumping (food intake) of C. elegans and disrupt the formation of biofilm.
To date, numerous compounds have been discovered with readily available bioactivity using zebrafish as model.[141] This system proved particularly valuable for identifying novel neuroactive molecules against central nervous system diseases. As traditional in vitro assays fail to model brain complexity, and in vivo mice/rats assays were less compatible for high-throughput screening (high expense and ethical issues), zebrafish behavior-based screening (Figure 10E) is considered as an approach with predictive power.[142,143] Through profiling a library of xyloketals and isoprenyl phenyl ethers derived from marine mangrove fungus, a compound (2, Figure 9) was identified as a potent inhibitor of locomotor activity in larval zebrafish. Several derivatives were synthesized and evaluated at whole organism level using a pentylenetetrazole (PTZ)-induced epilepsy model in zebrafish. One parent compound together with three modified derivatives possessed the ability to attenuate PTZ-induced locomotor hyperactivity, revealing their potential for development as anti-epilepsy drug candidates. In addition, largazole (Figure 3) was validated as an antiangiogenic agent in a hypoxia-inducible factor (HIF)-dependent manner in human cells in vivo in zebrafish using a genetic model with activated HIF.[144]
4.4. Multidimensional Screening Platforms
In addition to the strategy incorporating genetic/chemical perturbation, isogenic cell lines and genetic zebrafish model systems have been employed together to pursue druggable targets/active constituents from a library of siRNAs (targeting 7784 genes)/commercial compounds (4720 small molecules) and MNPs simultaneously affecting multiple cancer pathways. [144] As oncogenic KRAS and hypoxia-inducible factor (HIF) pathways shared significant overlap in terms of gene expression signatures, molecules targeting both pathways were hypothesized to benefit cancer treatment in improving efficacy and lowering the risk of resistance. Two MNPs, dolastatin 10 (microtubule-depolymerizing agent) and largazole (class I HDAC inhibitor), were rediscovered as hits in modulating both KRAS and HIF pathways by displaying differential cytotoxicity between parental HCT116 and HCT1 16HIF-1α−/−/HIF-2α−/− and HCT116WT KRAS cells. For proof-of-concept studies, largazole was chosen for further in vitro and in vivo evaluation. At the transcriptional, protein expression, cellular as well as whole-organismal level, largazole was proven to exert antiproliferative and anti angiogenic effects in a HIF-dependent manner. Largazole treatment phenocopied the colgate (hdac1) mutant, validating HDAC as the target in zebrafish and specifically abrogated HIF-induced angiogenesis in vhl mutants. This multidimensional screening platform not only enabled the identification of molecules/targets with desired selectivity, but also offered molecular insights with an in vivo dimension in a rapid and targeted fashion.
All the approaches discussed in Chapter 4 that are applicable for the target identification of MNPs are summarized in Figure 11.
Figure 11.
Summarized approaches and advances in identifying targets and MOAs of MNPs.
5. Marine Natural Products as First-in-Class Pharmacological Agents
All the approaches (Figure 11) described in Chapter 4 are amenable to protein target identification and most drugs including MNP-derived drugs target proteins. In many cases, revealing the biological target of a molecule is first attempted by recognition of structural features resembling known pharmacophores. For example, largazole[25,26] (Figure 3) was recognized as a prodrug and the active species, largazole thiol, is proposed to interact with Zn2+-dependent HDAC isoforms since the thiol motif is similar to that of other HDAC inhibitors, such as FK228.[25,26] Compounds containing amino-hydroxy-piperidone (Ahp) moiety frequently display serine protease inhibitory activity such as lyngbyastatin 7 (Figure 9), while molecules with embedded statine unit [(3S,4S)-4-amino-3-hydroxy-6-methylheptanoic acid, Sta] like grassystatins A (Figure 9) are characteristic aspartic protease inhibitors. In addition to the protein targeted MNPs as first-in-class pharmacological agents, the incredible diversity of MNP has enabled the production and discovery of compounds targeting non-protein biological entities.
5.1. Marine Natural Products Targeting DNA Functionality
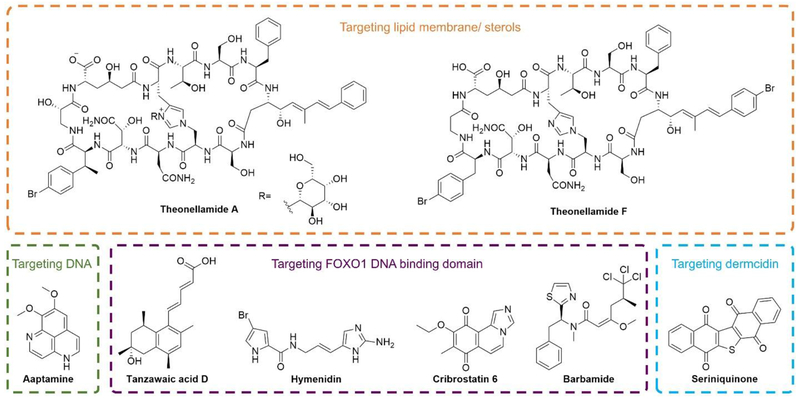
Some of the drugs listed in Chapter 2 target DNA at the level of replication and transcription and other drugs through intercalation. For example, cytarabine (Cytosar-U®, Figure 2) and vidarabine (Vira-A®, Figure 2) can both interfere with DNA synthesis,[145] while trabectedin (ET-743, Yondelis®, Figure 2) has been reported to interfere with activated transcription[146] and induce double-strand DNA breaks. [147] Moreover, aaptamine (Figure 12), a natural product commonly isolated from marine sponge Aaptos, exhibits potent DNA binding activity, which could be utilized for fragment-based drug design. [148] In addition, the transcription factor forkhead box O1 (FOXO1) was reported to negatively regulate the epidermal growth factor receptor (EGFR) signaling pathway. Stabilization of the FOXO1-DNA complex by small-molecule compounds could overcome drug resistance in anti-EGFR-based lung cancer therapy. High-throughput screening against a marine natural product library identified four hits as FOXO1-DNA stabilizers to selectively bind to the FOXO1 DNA-binding domain (DBD). The four hits are tanzawaic acid D (fungus Penicillium steckii, Figure 12), hymenidin (sponge Stylissa carteri, Figure 12), cribrostatin 6 (sponge Cribrochalina vasculum, Figure 12) and barbamide (Figure 12).
Figure 12.
Structures of MNPs with wide-ranging biological targets including DNA (aaptamine), FOXO1 DNA-binding domain (tanzawaic acid D, hymenidin, cribrostatin 6 and barbamide), lipid/sterol (theonellamides A and F) and new protein target dermcidin (seriniquinone).
5.2. Marine Natural Products Targeting Unusual Biological Space
The diversity of MNPs allows for the discovery of compounds with novel structural characteristics, as well as the identification of compounds with unique MOAs, thus enabling the discovery of MNPs targeting unusual biological space. For example, theonellamide A (Figure 12) was originally isolated from the marine sponge Theonella sp.[149] and this bicyclic dodecapeptide was reported to exert antifungal activity while in the meantime display moderate cytotoxicity toward mammalian cells. In-depth study regarding the interaction of theonellamide A with the membrane revealed that theonellamide A binds to the membrane surface through interaction with sterols and the accumulation of theonellamide A could cause morphological change of the local membrane. With this unique characteristic, theonellamide-based probes would enable visualization of sterol-containing domains in living cells, which could further unveil the dynamic changes in membrane morphology.[150] Yoshida et al.[151] reported the chemogenomic profiling of theonellamide F (Figure 12), a MNP originally isolated from sponge Theonella sp..[152] The results indicated that theonellamide F would induce overproduction of 1,3-β-D-glucan in a Rho1-dependent manner. Thus, theonellamides were further characterized as sterol-binding compounds to cause membrane damage and activate Rho1-dependent 1,3-β-D-glucan synthesis.
In addition to the lipid membrane/sterol targeting compounds, MNPs provide an opportunity to discover promising therapeutic agents with unique biological targets. Recently, a new natural product, seriniquinone (Figure 12), was isolated from the marine bacterium Serinicoccus and it was demonstrated to exhibit selective activity over a set of melanoma cell lines within the NCI-60 screen panel. Seriniquinone is the first compound reported to target dermcidin overexpressed in melanoma cells. Emerging data suggest that dermcidin plays a growing role in stabilizing cancer, targeting dermcidin by seriniquinone provides a new cell-specific approach to initiate autophagy and apoptotic cell death.[153]
5.3. Antibody Conjugation as Enabling Technology for Marine Natural Products
MNPs have been productive resources for cytotoxic compounds as drug leads. However, their exquisite potency may cause undesired off-target cytotoxicity. Antibody-targeted chemotherapy is a therapeutic strategy that utilizes a cytotoxic agent linked to a monoclonal antibody (mAb) that can specifically recognize a tumor-associated antigen, the complex used in this strategy is ADCs, which has become a representative class of therapeutic agents for the treatment of cancer. The mAb delivers the highly cytotoxic agent to tumor cells over normal healthy cells to achieve specificity and targeted cancer cell killing.[154,155] Currently, five ADCs have been approved by FDA in clinical application, including Mylotarg® (gemtuzumab ozogamicin) for the treatment of acute myeloid leukemia (AML),[156] Adcetris® (brentuximab vedotin) for the treatment of relapsed or refractory Hodgkin lymphoma (HL) and systemic anaplastic large cell lymphoma (ALCL),[157] Besponsa® (inotuzumab ozogamicin) for the treatment of B-cell acute lymphoblastic leukemia (ALL),[158] Kadcyla® (ado-trastuzumab emtansine) for the treatment of metastatic breast cancer[159] and Polivy® (polatuzumab vedotin-piiq) in combination with bendamustine and a rituximab product for the treatment of relapsed or refractory diffuse large B-cell lymphoma (DLBCL).[160] Considering the successes of Adcetris® (brentuximab vedotin) and Polivy® (polatuzumab vedotin-piiq), the ADC conjugates of monomethyl auristatin E (MMAE), a derivative of dolastatin 10 (Figure 13), a natural product isolated from marine cyanobacterium Caldora penicillata,[27] more efforts were made to seek for potent cytotoxins for the treatment of cancer in ADC format. For example, microtubule-targeting agent eribulin has been developed as ADC payload with humanized anti-human folate receptor alpha (FRA) antibody farletuzumab. This ADC has been reported to be a promising treatment for FRA-positive cancer.[161] Recently, a highly toxic proteasome inhibitor, carmaphycin B (Figure 13), a natural product isolated from the marine cyanobacterium Symploca sp.,[162] was engineered to function as an ADC warhead. This investigation resulted a potent cytotoxic analog of carmaphycin B with a suitable linker for conjugation as ADC and reveal additional underlying SAR principles of this compound class for further optimization (Figure 13).[155]
Figure 13.
Structures of dolastatin 10/MMAE, eribulin, carmaphycin B and their derived ADCs.
6. Conclusion
The advances in MNP research over the past years assisted with the sample prioritization to focus to identify novel structures and the spectroscopic characterization of novel MNPs, but also enabled effective ways to solve the MNP supply. However, the biological characterization of novel (and oftentimes structurally complex) MNPs is usually a challenging problem, especially since usually only small quantities are isolated and not all compounds would be subjected to total synthesis or scale-up efforts, unless a promising bioactivity or novel target has been identified. Various technologies have been developed to surpass this problem.
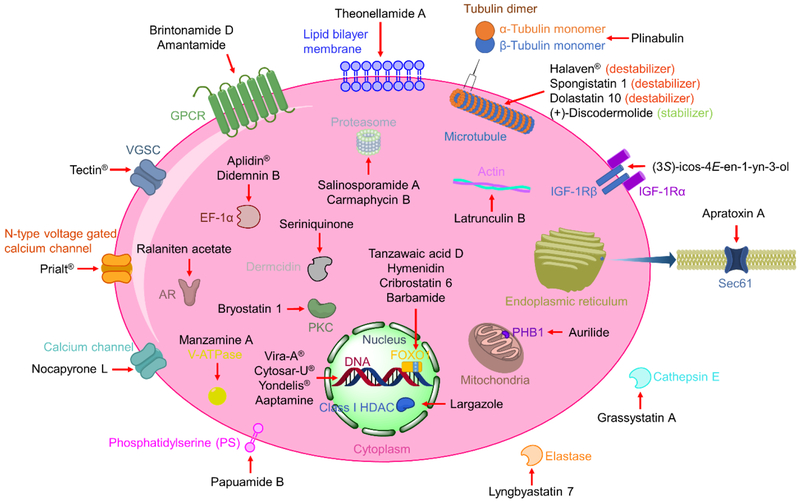
The utilization of label-free approaches in identifying the biological targets of MNPs has saved the efforts to modify the MNP, which is an advantage considering the commonly small amount of isolated compounds and usually complex structures. Genomic approaches provide options to investigate the biological role of MNPs from a genome-wide aspect, especially target-directed genome mining of producing organisms is expected to become increasingly productive when seeking to identify antibiotic targets. The more general profiling methods further facilitate the target identification process through direct target identification and phenotypic screening. In addition to the intensively studied therapeutic target proteins such as enzymes and GPCRs, MNPs have been reported to target unusual biological space, including lipid membrane components and proteins with unique biological functionality (Figure 14). The development of MNP-derived ADCs further demonstrates the potential of MNPs in different forms. Taken together, all the approaches discussed could be applied to identify the biological role of MNPs and we expect wider applications of these approaches to discover MNPs with unique MOAs. We anticipate that, through the increased development of assay systems to assess the biological roles of MNPs, we will witness the discovery of more first-in-class or most-potent-in-class drugs from marine sources.
Figure 14.
Representative MNPs and their cellular (and extracellular) targets.
According to the record in the MarinLit database, over 30,000 compounds have been reported from marine sources. Even though a decrease in the discovery rate of novel natural products has been observed while the rate and absolute number of the discovery of natural products have increased in the past years, the development of innovative discovery methods will continue to produce compounds with novel chemical characteristics.[163] More importantly, the advancement of target identification technologies guarantees in-depth investigation of MNPs that have been reported but the biological role of which has not been fully understood yet. The improved biological annotations of these MNPs would further facilitate the identification of potential drug candidates/ leads. Therefore, we expect that marine sources will continuously provide a rich supply for promising first-in-class therapeutic agents with new structural and biological characteristics.
Acknowledgements
We acknowledge the National Institutes of Health for financial support of our MNP research (grant R01CA172310).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interest
The authors declare the following competing financial interest(s): H.L. is a co-founder of Oceanyx Pharmaceuticals, Inc., which has licensed patents and patent applications related to marine natural products mentioned in this review and is negotiating additional licenses (apratoxins, largazole).
References
- [1].Newman DJ, Cragg GM, Natural products as sources of new drugs from 1981 to 2014, J Nat Prod 79(3) (2016) 629–661. [DOI] [PubMed] [Google Scholar]
- [2].Gerwick WH, Moore BS, Lessons from the past and charting the future of marine natural products drug discovery and chemical biology, Chem Biol 19(1) (2012) 85–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Montaser R, Luesch H, Marine natural products: a new wave of drugs?, Future Med Chem 3(12)(2011)1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Schorn MA, Alanjary MM, Aguinaldo K, Korobeynikov A, Podell S, Patin N, Lincecum T, Jensen PR, Ziemert N, Moore BS, Sequencing rare marine actinomycete genomes reveals high density of unique natural product biosynthetic gene clusters, Microbiology 162(12) (2016) 2075–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Molinski TF, NMR of natural products at the ‘nanomole-scale’, Nat Prod Rep 27(3) (2010) 321–329. [DOI] [PubMed] [Google Scholar]
- [6].Yang JY, Sanchez LM, Rath CM, Liu X, Boudreau PD, Bruns N, Glukhov E, Wodtke A, de Felicio R, Fenner A, Wong WR, Linington RG, Zhang L, Debonsi HM, Gerwick WH, Dorrestein PC, Molecular networking as a dereplication strategy, J Nat Prod 76(9) (2013) 1686–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Martins A, Vieira H, Gaspar H, Santos S, Marketed marine natural products in the pharmaceutical and cosmeceutical industries: tips for success, Mar Drugs 12(2) (2014) 1066–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Newman DJ, Cragg GM, Drugs and drug candidates from marine sources: an assessment of the current “state of play”, Planta Med 82(9-10) (2016) 775–789. [DOI] [PubMed] [Google Scholar]
- [9].Alonso-Álvarez S, Pardal E, Sánchez-Nieto D, Navarro M, Caballero MD, Mateos MV, Martín A, Plitidepsin: design, development, and potential place in therapy, Drug Des Devel Ther 11(2017)253–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Losada A, Muñoz-Alonso MJ, García C, Sánchez-Murcia PA, Martínez-Leal JF, Domínguez JM, Lillo MP, Gago F, Galmarini CM, Translation elongation factor eEF1A2 is a novel anticancer target for the marine natural product plitidepsin, Sci Rep 6 (2016) 35100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lago J, Rodríguez LP, Blanco L, Vieites JM, Cabado AG, Tetrodotoxin, an Extremely Potent Marine Neurotoxin: Distribution, Toxicity, Origin and Therapeutical Uses, Mar Drugs 13(10) (2015)6384–6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bane V, Lehane M, Dikshit M, O’Riordan A, Furey A, Tetrodotoxin: chemistry, toxicity, source, distribution and detection, Toxins 6(2) (2014) 693–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Nieto FR, Cobos EJ, Tejada M, Sánchez-Fernández C, González-Cano R, Cendán CM, Tetrodotoxin (TTX) as a therapeutic agent for pain, Mar Drugs 10(2) (2012) 281–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kanoh K, Kohno S, Asari T, Harada T, Katada J, Muramatsu M, Kawashima H, Sekiya H, Uno I, (−)-phenylahistin: a new mammalian cell cycle inhibitor produced by aspergillus ustus, Bioorg Med Chem Lett 7(22) (1997) 2847–2852. [Google Scholar]
- [15].Gomes N, Lefranc F, Kijjoa A, Kiss R, Can Some marine-derived fungal metabolites become actual anticancer agents?, Mar Drugs 13(6) (2015) 3950–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Nicholson B, Lloyd G, Miller B, Palladino M, Kiso Y, Hayashi Y, Neuteboom S, NPI-2358 is a tubulin-depolymerizing agent: in-vitro evidence for activity as a tumor vascular-disrupting agent, Anti-Cancer Drugs 17(1) (2006) 25–31. [DOI] [PubMed] [Google Scholar]
- [17].Singh A, Bandi M, Raje N, Richardson P, Palladino M, Chauhan D, Anderson K, A novel vascular disrupting agent plinabulin triggers JNK-mediated apoptosis and inhibits angiogenesis in multiple myeloma cells, Blood 117(21) (2011) 5692–5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Feling RH, Buchanan GO, Mincer TJ, Kauffman CA, Jensen PR, Fenical W, Salinosporamide A: a highly cytotoxic proteasome inhibitor from a novel microbial source, a marine bacterium of the new genus Salinospora, Angew Chem Int Ed Engl 42(3) (2003) 355–357. [DOI] [PubMed] [Google Scholar]
- [19].Gulder T, Moore B, Salinosporamide Natural Products: Potent 20 S Proteasome Inhibitors as Promising Cancer Chemotherapeutics, Angew Chem Int Ed 49(49) (2010) 9346–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Meng L, Mohan R, Kwok BH, Elofsson M, Sin N, Crews CM, Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity, Proc Natl Acad Sci U S A 96(18) (1999) 10403–10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chauhan D, Catley L, Li G, Podar K, Hideshima T, Velankar M, Mitsiades C, Mitsiades N, Yasui H, Letai A, Ovaa H, Berkers C, Nicholson B, Chao T, Neuteboom S, Richardson P, Palladino M, Anderson K, A novel orally active proteasome inhibitor induces apoptosis in multiple myeloma cells with mechanisms distinct from Bortezomib, Cancer Cell 8(5) (2005) 407–419. [DOI] [PubMed] [Google Scholar]
- [22].Richardson P, Spencer A, Cannel P, Harrison S, Catley L, Underhill C, Zimmerman T, Hofmeister C, Jakubowiak A, Laubach J, Palladino M, Longenecker A, Lay A, Wear S, Lloyd G, Hannah A, Reich S, Spear M, Anderson K, Phase 1 clinical evaluation of twice-weekly marizomib (npi-0052), a novel proteasome inhibitor, in patients with relapsed/refractory multiple myeloma (MM), Blood 118(21) (2011) 140–141. [Google Scholar]
- [23].Andersen R, Sponging off nature for new drug leads, Biochem Pharmacol 139 (2017) 3–14. [DOI] [PubMed] [Google Scholar]
- [24].Liu Y, Salvador LA, Byeon S, Ying Y, Kwan JC, Law BK, Hong J, Luesch H, Anticolon cancer activity of largazole, a marine-derived tunable histone deacetylase inhibitor, J Pharmacol Exp Ther 335(2) (2010) 351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ying Y, Taori K, Kim H, Hong J, Luesch H, Total synthesis and molecular target of largazole, a histone deacetylase inhibitor, J Am Chem Soc 130(26) (2008) 8455–8459. [DOI] [PubMed] [Google Scholar]
- [26].Hong J, Luesch H, Largazole: from discovery to broad-spectrum therapy, Nat Prod Rep 29(4) (2012) 449–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Engene N, Tronholm A, Salvador-Reyes LA, Luesch H, Paul VJ, Caldora penicillata gen. nov., comb. nov. (cyanobacteria), a pantropical marine species with biomedical relevance, J Phycol 51(4) (2015) 670–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Salvador-Reyes LA, Engene N, Paul VJ, Luesch H, Targeted natural products discovery from marine cyanobacteria using combined phylogenetic and mass spectrometric evaluation, J Nat Prod 78(3) (2015) 486–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Chen Q, Chaturvedi P, Luesch H, Process development and scale-up total synthesis of largazole, a potent class I histone deacetylase inhibitor, Org Process Res Dev 22(2) (2018) 190–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Dybdal-Hargreaves NF, Risinger AL, Mooberry SL, Eribulin mesylate: mechanism of action of a unique microtubule-targeting agent, Clin Cancer Res 21(11) (2015) 2445–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Austad B, Benayoud F, Calkins T, Campagna S, Chase C, Choi H, Christ W, Costanzo R, Cutter J, Endo A, Fang F, Hu Y, Lewis B, Lewis M, McKenna S, Noland T, Orr J, Pesant M, Schnaderbeck M, Wilkie G, Abe T, Asai N, Asai Y, Kayano A, Kimoto Y, Komatsu Y, Kubota M, Kuroda H, Mizuno M, Nakamura T, Omae T, Ozeki N, Suzuki T, Takigawa T, Watanabe T, Yoshizawa K, Process Development of Halaven®: Synthesis of the C14-C35 fragment via iterative nozaki-hiyama-kishi reaction-williamson ether cyclization, Synlett 24(3) (2013) 333–337. [Google Scholar]
- [32].Yu M, Zheng W, Seletsky B, From micrograms to grams: scale-up synthesis of eribulin mesylate, Nat Prod Rep 30(9) (2013) 1158–1164. [DOI] [PubMed] [Google Scholar]
- [33].Gunasekera S, Gunasekera M, Longley R, Schulte G, Discodermolide - a new bioactive polyhydroxylated lactone from the marine sponge discodermia-dissoluta, J Org Chem 55(16) (1990) 4912–4915. [Google Scholar]
- [34].Hung DT, Chen J, Schreiber SL, (+)-Discodermolide binds to microtubules in stoichiometric ratio to tubulin dimers, blocks taxol binding and results in mitotic arrest, Chem Biol 3(4) (1996) 287–293. [DOI] [PubMed] [Google Scholar]
- [35].terHaar E, Kowalski R, Hamel E, Lin C, Longley R, Gunasekera S, Rosenkranz H, Day B, Discodermolide, a cytotoxic marine agent that stabilizes microtubules more potently than taxol, Biochemistry 35(1) (1996) 243–250. [DOI] [PubMed] [Google Scholar]
- [36].Nerenberg J, Hung D, Somers P, Schreiber S, Total synthesis of the immunosuppressive agent (−)-discodermolide, J Am Chem Soc 115(26) (1993) 12621–12622. [Google Scholar]
- [37].Smith A, Qiu Y, Jones D, Kobayashi K, Total synthesis of (−)-discodermolide, J Am Chem Soc 117(48) (1995) 12011–12012. [Google Scholar]
- [38].Harried S, Yang G, Strawn M, Myles D, Total synthesis of (−)-discodermolide: An application of a chelation-controlled alkylation reaction, J Org Chem 62(18) (1997) 6098–6099. [Google Scholar]
- [39].Paterson I, Florence G, Gerlach K, Scott J, Total synthesis of the antimicrotubule agent (+)-discodermolide using boron-mediated aldol reactions of chiral ketones, Angew Chem Int Ed 39(2) (2000) 377–380. [DOI] [PubMed] [Google Scholar]
- [40].Mickel S, Niederer D, Daeffler R, Osmani A, Kuesters E, Schmid E, Schaer K, Gamboni R, Chen W, Loeser E, Kinder F, Konigsberger K, Prasad K, Ramsey T, Repic J, Wang R, Florence G, Lyothier I, Paterson I, Large-scale synthesis of the anti-cancer marine natural product (+)-discodermolide. Part 5: Linkage of fragments C1-6 and C7-24 and finale, Org Process Res Dev 8(1) (2004) 122–130. [Google Scholar]
- [41].Arefolov A, Panek J, Crotylsilane reagents in the synthesis of complex polyketide natural products: total synthesis of (+)-discodermolide, J Am Chem Soc 127(15) (2005) 5596–5603. [DOI] [PubMed] [Google Scholar]
- [42].de Lemos E, Poree F, Bourin A, Barbion J, Agouridas E, Lannon M, Commercon A, Betzer J, Pancrazi A, Ardisson J, Total Synthesis of Discodermolide: Optimization of the Effective Synthetic Route, Chem Eur J 14(35) (2008) 11092–11112. [DOI] [PubMed] [Google Scholar]
- [43].Smith A, Kaufman M, Beauchamp T, LaMarche M, Arimoto H, Gram-scale synthesis of (+)-discodermolide, Org Lett 1(11) (1999) 1823–1826. [DOI] [PubMed] [Google Scholar]
- [44].Smith A, Beauchamp T, LaMarche M, Kaufman M, Qiu Y, Arimoto H, Jones D, Kobayashi K, Evolution of a gram-scale synthesis of (+)-discodermolide, J Am Chem Soc 122(36) (2000) 8654–8664. [Google Scholar]
- [45].Smith A, Freeze B, Brouard I, Hirose T, A practical improvement, enhancing the large-scale synthesis of (+)-discodermolide: A third-generation approach, Org Lett 5(23) (2003) 4405–4408. [DOI] [PubMed] [Google Scholar]
- [46].Smith A, Freeze B, Xian M, Hirose T, Total synthesis of (+)-discodermolide: A highly convergent fourth-generation approach, Org Lett 7(9) (2005) 1825–1828. [DOI] [PubMed] [Google Scholar]
- [47].Smith A, Zhu W, Shirakami S, Sfouggatakis C, Doughty V, Bennett C, Sakamoto Y, Total synthesis of (+)-spongistatin 1. An effective second-generation construction of an advanced EF Wittig salt, fragment union, and final elaboration, Org Lett 5(5) (2003) 761–764. [DOI] [PubMed] [Google Scholar]
- [48].Smith A, Tomioka T, Risatti C, Sperry J, Sfouggatakis C, Gram-scale synthesis of (+)-spongistatin 1: Development of an improved, scalable synthesis of the F-ring subunit, fragment union, and final elaboration, Org Lett 10(19) (2008) 4359–4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Pettit G, Cichacz Z, Gao F, Herald C, Boyd M, Schmidt J, Hooper J, Isolation and structure of spongistatin-1, J Org Chem 58(6) (1993) 1302–1304. [Google Scholar]
- [50].Bai R, Cichacz Z, Herald C, Pettit G, Hamel E, Spongistatin-1, a highly cytotoxic, sponge-derived, marine natural product that inhibits mitosis, microtubule assembly, and the binding of vinblastine to tubulin, Mol Pharmacol 44(4) (1993) 757–766. [PubMed] [Google Scholar]
- [51].Schyschka L, Rudy A, Jeremias I, Barth N, Pettit G, Vollmar A, Spongistatin 1: a new chemosensitizing marine compound that degrades XIAP, Leukemia 22(9) (2008) 1737–1745. [DOI] [PubMed] [Google Scholar]
- [52].Hamann M, Scheuer P, Kellyborges M, Biogenetically diverse, bioactive constituents of a sponge, order Verongida - bromotyramines and sesquiterpene-shikimate derived metabolites, J Org Chem 58(24) (1993) 6565–6569. [Google Scholar]
- [53].Wang HS, Li HJ, Nan X, Luo YY, Wu YC, Enantiospecific Semisynthesis of Puupehedione-Type Marine Natural Products, J Org Chem 82(23) (2017) 12914–12919. [DOI] [PubMed] [Google Scholar]
- [54].Martinez-Poveda B, Quesada A, Medina M, Pleiotropic Role of Puupehenones in Biomedical Research, Marine Drugs 15(10) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Zhang H, Boghigian BA, Armando J, Pfeifer BA, Methods and options for the heterologous production of complex natural products, Nat Prod Rep 28(1) (2011) 125–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Fudou R, Iizuka T, Sato S, Ando T, Shimba N, Yamanaka S, Haliangicin, a novel antifungal metabolite produced by a marine myxobacterium 2. Isolation and structural elucidation, J Antibiot 54(2) (2001) 153–156. [DOI] [PubMed] [Google Scholar]
- [57].Sun Y, Feng Z, Tomura T, Suzuki A, Miyano S, Tsuge T, Mori H, Suh J, Iizuka T, Fudou R, Ojika M, Heterologous production of the marine myxobacterial antibiotic haliangicin and its unnatural analogues generated by engineering of the biochemical pathway, Sci Rep 6 (2016) 22091–22101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Bergman O, Mayzel B, Anderson MA, Shpigel M, Hill RT, Ilan M, Examination of marine-based cultivation of three demosponges for acquiring bioactive marine natural products, Mar Drugs 9(11) (2011) 2201–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Schenone M, Dancik V, Wagner B, Clemons P, Target identification and mechanism of action in chemical biology and drug discovery, Nat Chem Biol 9(4) (2013) 232–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Kawatani M, Osada H, Affinity-based target identification for bioactive small molecules, Medchemcomm 5(3) (2014) 277–287. [Google Scholar]
- [61].Chang J, Kim Y, Kwon HJ, Advances in identification and validation of protein targets of natural products without chemical modification, Nat Prod Rep 33(5) (2016) 719–730. [DOI] [PubMed] [Google Scholar]
- [62].Li J, Xu H, West G, Jones L, Label-free technologies for target identification and validation, MedChemComm 7(5) (2016) 769–777. [Google Scholar]
- [63].Lomenick B, Jung G, Wohlschlegel JA, Huang J, Target identification using drug affinity responsive target stability (DARTS), Curr Protoc Chem Biol 3(4) (2011) 163–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Lomenick B, Olsen RW, Huang J, Identification of direct protein targets of small molecules, ACS Chem Biol 6(1) (2011) 34–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Crews C; Collins J; Lane W; Snapper M; Schreiber S, GTP-dependent finding of the antiproliferative agent didemnin to elongation-factor 1-alpha, J Biol Chem 269(22) (1994) 15411–15414. [PubMed] [Google Scholar]
- [66].Lomenick B, Hao R, Jonai N, Chin RM, Aghajan M, Warburton S, Wang J, Wu RP, Gomez F, Loo JA, Wohlschlegel JA, Vondriska TM, Pelletier J, Herschman HR, Clardy J, Clarke CF, Huang J, Target identification using drug affinity responsive target stability (DARTS), Proc Natl Acad Sci U S A 106(51) (2009) 21984–21989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Molina D, Jafari R, Ignatushchenko M, Seki T, Larsson E, Dan C, Sreekumar L, Cao Y, Nordlund P, Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay, Science 341(6141) (2013) 84–87. [DOI] [PubMed] [Google Scholar]
- [68].Zovko A, Viktorsson K, Haag P, Kovalerchick D, Farnegardh K, Alimonti A, Ilan M, Carmeli S, Lewensohn R, Marine sponge cribrochalina vasculum compounds activate intrinsic apoptotic signaling and inhibit growth factor signaling cascades in non-small cell lung carcinoma, Mol Cancer Ther 13(12) (2014) 2941–2954. [DOI] [PubMed] [Google Scholar]
- [69].Zovko A, Novak M, Hååg P, Kovalerchick D, Holmlund T, Färnegårdh K, Ilan M, Carmeli S, Lewensohn R, Viktorsson K, Compounds from the marine sponge Cribrochalina vasculum offer a way to target IGF-1R mediated signaling in tumor cells, Oncotarget 7(31) (2016) 50258–50276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Savitski MM, Reinhard FB, Franken H, Werner T, Savitski MF, Eberhard D, Martinez Molina D, Jafari R, Dovega RB, Klaeger S, Kuster B, Nordlund P, Bantscheff M, Drewes G, Tracking cancer drugs in living cells by thermal profiling of the proteome, Science 346(6205)(2014) 1255784. [DOI] [PubMed] [Google Scholar]
- [71].Martinez Molina D, Nordlund P, The cellular thermal shift assay: a novel biophysical assay for in situ drug target engagement and mechanistic biomarker studies, Annu Rev Pharmacol Toxicol 56 (2016) 141–161. [DOI] [PubMed] [Google Scholar]
- [72].Reinhard FB, Eberhard D, Werner T, Franken H, Childs D, Doce C, Savitski MF, Huber W, Bantscheff M, Savitski MM, Drewes G, Thermal proteome profiling monitors ligand interactions with cellular membrane proteins, Nat Methods 12(12) (2015) 1129–1131. [DOI] [PubMed] [Google Scholar]
- [73].Park H, Ha J, Koo JY, Park J, Park SB, Label-free target identification using in-gel fluorescence difference, Chem Sci 8(2) (2017) 1127–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Pettit G, Herald C, Doubek D, Herald D, Arnold E, Clardy J, Isolation and structure of bryostatin-1, J Am Chem Soc 104(24) (1982) 6846–6848. [Google Scholar]
- [75].Nelson TJ, Sun MK, Lim C, Sen A, Khan T, Chirila FV, Alkon DL, Bryostatin effects on cognitive function and PKCε in Alzheimer’s disease Phase Ha and expanded access trials, J Alzheimers Dis 58(2) (2017) 521–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].West G, Tang L, Fitzgerald M, Thermodynamic analysis of protein stability and ligand binding using a chemical modification- and mass-spectrometry based strategy, Anal Chem 80(11) (2008) 4175–4185. [DOI] [PubMed] [Google Scholar]
- [77].West GM, Tucker CL, Xu T, Park SK, Han X, Yates JR, Fitzgerald MC, Quantitative proteomics approach for identifying protein-drug interactions in complex mixtures using protein stability measurements, Proc Natl Acad Sci U S A 107(20) (2010) 9078–9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Salcius M, Bauer A, Hao Q, Li S, Tutter A, Raphael J, Jahnke W, Rondeau J, Bourgier E, Tallarico J, Michaud G, SEC-TID: A label-free method for small-molecule target identification, J Biomol Screen 19(6) (2014) 917–927. [DOI] [PubMed] [Google Scholar]
- [79].Tian X, Hou W, Fang Y, Fan J, Tong H, Bai S, Chen Q, Xu H, Li Y, XAV939, a tankyrase 1 inhibitior, promotes cell apoptosis in neuroblastoma cell lines by inhibiting Wnt/beta-catenin signaling pathway, Journal of Experimental & Clinical Cancer Research 32 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Chan J, Vuckovic D, Sleno L, Olsen J, Pogoutse O, Havugimana P, Hewel J, Bajaj N, Wang Y, Musteata M, Nislow C, Emili A, Target Identification by Chromatographic Co-elution: Monitoring of Drug-Protein Interactions without Immobilization or Chemical Derivatization, Mol Cell Proteomics 11(7) (2012) M111-016642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Ziegler S, Pries V, Hedberg C, Waldmann H, Target Identification for Small Bioactive Molecules: Finding the Needle in the Haystack, Angew Chem Int Ed 52(10) (2013) 2744–2792. [DOI] [PubMed] [Google Scholar]
- [82].Luesch H, Towards high-throughput characterization of small molecule mechanisms of action, Mol Biosyst 2(12) (2006) 609–620. [DOI] [PubMed] [Google Scholar]
- [83].Luesch H, Yoshida WY, Moore RE, Paul VJ, Corbett TH, Total structure determination of apratoxin A, a potent novel cytotoxin from the marine cyanobacterium Lyngbya majuscula, J Am Chem Soc 123(23) (2001) 5418–5423. [DOI] [PubMed] [Google Scholar]
- [84].Luesch H, Chanda SK, Raya RM, DeJesus PD, Orth AP, Walker JR, Izpisúa Belmonte JC, Schultz PG, A functional genomics approach to the mode of action of apratoxin A, Nat Chem Biol 2(3) (2006) 158–167. [DOI] [PubMed] [Google Scholar]
- [85].Liu Y, Law BK, Luesch H, Apratoxin a reversibly inhibits the secretory pathway by preventing cotranslational translocation, Mol Pharmacol 76(1) (2009) 91–104. [DOI] [PubMed] [Google Scholar]
- [86].Paatero AO, Kellosalo J, Dunyak BM, Almaliti J, Gestwicki JE, Gerwick WH, Taunton J, Paavilainen VO, Apratoxin kills cells by direct blockade of the sec61 protein translocation channel, Cell Chem Biol 23(5) (2016) 561–566. [DOI] [PubMed] [Google Scholar]
- [87].Huang KC, Chen Z, Jiang Y, Akare S, Kolber-Simonds D, Condon K, Agoulnik S, Tendyke K, Shen Y, Wu KM, Mathieu S, Choi HW, Zhu X, Shimizu H, Kotake Y, Gerwick WH, Uenaka T, Woodall-Jappe M, Nomoto K, Apratoxin A shows novel pancreas-targeting activity through the binding of sec 61, Mol Cancer Ther 15(6) (2016) 1208–1216. [DOI] [PubMed] [Google Scholar]
- [88].Suenaga K, Mutou T, Shibata T, Itoh T, Fujita T, Takada N, Hayamizu K, Takagi M, Irifune T, Kigoshi H, Yamada K, Aurilide, a cytotoxic depsipeptide from the sea hare Dolabella auricularia: isolation, structure determination, synthesis, and biological activity, Tetrahedron 60(38) (2004) 8509–8527. [Google Scholar]
- [89].Sato S, Murata A, Orihara T, Shirakawa T, Suenaga K, Kigoshi H, Uesugi M, Marine natural product aurilide activates the OPA1-mediated apoptosis by binding to prohibitin, Chem Biol 18(1) (2011) 131–139. [DOI] [PubMed] [Google Scholar]
- [90].Takase S, Kurokawa R, Arai D, Kanemoto Kanto K, Okino T, Nakao Y, Kushiro T, Yoshida M, Matsumoto K, A quantitative shRNA screen identifies ATP1A1 as a gene that regulates cytotoxicity by aurilide B, Sci Rep 7(1) (2017) 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Giaever G, Chu A, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, Arkin A, Astromoff A, El Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian K, Flaherty P, Foury F, Garfinkel D, Gerstein M, Gotte D, Guldener U, Hegemann J, Hempel S, Herman Z, Jaramillo D, Kelly D, Kelly S, Kotter P, LaBonte D, Lamb D, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi S, Revuelta J, Roberts C, Rose M, Ross-Macdonald P, Scherens B, Schimmack G, Shafer B, Shoemaker D, Sookhai-Mahadeo S, Storms R, Strathern J, Valle G, Voet M, Volckaert G, Wang C, Ward T, Wilhelmy J, Winzeler E, Yang Y, Yen G, Youngman E, Yu K, Bussey H, Boeke J, Snyder M, Philippsen P, Davis R, Johnston M, Functional profiling of the Saccharomyces cerevisiae genome, Nature 418(6896) (2002) 387–391. [DOI] [PubMed] [Google Scholar]
- [92].Giaever G, Shoemaker DD, Jones TW, Liang H, Winzeler EA, Astromoff A, Davis RW, Genomic profiling of drug sensitivities via induced haploinsufficiency, Nat Genet 21(3) (1999) 278–283. [DOI] [PubMed] [Google Scholar]
- [93].Giaever G, Flaherty P, Kumm J, Proctor M, Nislow C, Jaramillo DF, Chu AM, Jordan MI, Arkin AP, Davis RW, Chemogenomic profiling: identifying the functional interactions of small molecules in yeast, Proc Natl Acad Sci U S A 101(3) (2004) 793–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, Proctor M, St Onge RP, Tyers M, Koller D, Altman RB, Davis RW, Nislow C, Giaever G, The chemical genomic portrait of yeast: uncovering a phenotype for all genes, Science 320(5874) (2008) 362–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Sakai R, Higa T, Manzamine-A, a novel antitumor alkaloid from a sponge, J Am Chem Soc 108(20) (1986) 6404–6405. [Google Scholar]
- [96].Nakamura H, Deng S, Kobayashi J, Ohizumi Y, Tomotake Y, Matsuzaki T, Hirata Y, Keramamine-A and -B, novel antimicrobial alkaloids from the Okinawan marine sponge Pellina sp, Tetrahedron Lett 28(6) (1987) 621–624. [Google Scholar]
- [97].Edrada RA, Proksch P, Wray V, Witte L, Müller WE, Van Soest RW, Four new bioactive manzamine-type alkaloids from the Philippine marine sponge Xestospongia ashmorica, J Nat Prod 59(11) (1996) 1056–1060. [DOI] [PubMed] [Google Scholar]
- [98].Ang K, Holmes M, Higa T, Hamann M, Kara U, In vivo antimalarial activity of the beta-carboline alkaloid manzamine A, Antimicrob Agents Chemother 44(6) (2000) 1645–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Lin LC, Kuo TT, Chang HY, Liu WS, Hsia SM, Huang TC, Manzamine A exerts anticancer activity against human colorectal cancer cells, Mar Drugs 16(8) (2018) 252–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Guzmán EA, Johnson JD, Linley PA, Gunasekera SE, Wright AE, A novel activity from an old compound: Manzamine A reduces the metastatic potential of AsPC-1 pancreatic cancer cells and sensitizes them to TRAIL-induced apoptosis, Invest New Drugs 29(5) (2011) 777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Kallifatidis G, Hoepfner D, Jaeg T, Guzmán EA, Wright AE, The marine natural product manzamine A targets vacuolar ATPases and inhibits autophagy in pancreatic cancer cells, Mar Drugs 11(9) (2013) 3500–3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Parsons A, Lopez A, Givoni I, Williams D, Gray C, Porter J, Chua G, Sopko R, Brost R, Ho C, Wang J, Ketela T, Brenner C, Brill J, Fernandez G, Lorenz T, Payne G, Ishihara S, Ohya Y, Andrews B, Hughes T, Frey B, Graham T, Andersen R, Boone C, Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast, Cell 126(3) (2006) 611–625. [DOI] [PubMed] [Google Scholar]
- [103].Ford P, Gustafson K, McKee T, Shigematsu N, Maurizi L, Pannell L, Williams D, de Silva E, Lassota P, Allen T, Van Soest R, Andersen R, Boyd M, Papuamides A-D HIV-inhibitory and cytotoxic depsipeptides from the sponges Theonella mirabilis and Theonella swinhoei collected in Papua New Guinea, J Am Chem Soc 121(25) (1999) 5899–5909. [Google Scholar]
- [104].Potts MB, Kim HS, Fisher KW, Hu Y, Carrasco YP, Bulut GB, Ou YH, Herrera-Herrera ML, Cubillos F, Mendiratta S, Xiao G, Hofree M, Ideker T, Xie Y, Huang LJ, Lewis RE, MacMillan JB, White MA, Using functional signature ontology (FUSION) to identify mechanisms of action for natural products, Sci Signal 6(297) (2013) ra90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Potts MB, McMillan EA, Rosales TI, Kim HS, Ou YH, Toombs JE, Brekken RA, Minden MD, MacMillan JB, White MA, Mode of action and pharmacogenomic biomarkers for exceptional responders to didemnin B, Nat Chem Biol 11(6) (2015) 401–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Meng L, Sin N, Crews CM, The antiproliferative agent didemnin B uncompetitively inhibits palmitoyl protein thioesterase, Biochemistry 37(29) (1998) 10488–10492. [DOI] [PubMed] [Google Scholar]
- [107].Hetherington AM, Sawyez CG, Sutherland BG, Robson DL, Arya R, Kelly K, Jacobs RL, Borradaile NM, Treatment with didemnin B, an elongation factor 1A inhibitor, improves hepatic lipotoxicity in obese mice, Physiol Rep 4(17) (2016) e12963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Wang Z, Gerstein M, Snyder M, RNA-Seq: a revolutionary tool for transcriptomics, Nat Rev Genet 10(1) (2009) 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Khatoon Z, Figler B, Zhang H, Cheng F, Introduction to RNA-Seq and its applications to drug discovery and development, Drug Dev Res 75(5) (2014) 324–330. [DOI] [PubMed] [Google Scholar]
- [110].Kang Y, Li H, Wu J, Xu X, Sun X, Zhao X, Xu N, Transcriptome profiling reveals the antitumor mechanism of polysaccharide from marine algae gracilariopsis lemaneiformis, PLoS One 11(6) (2016) e0158279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Choi H, Mascuch SJ, Villa FA, Byrum T, Teasdale ME, Smith JE, Preskitt LB, Rowley DC, Gerwick L, Gerwick WH, Honaucins A-C, potent inhibitors of inflammation and bacterial quorum sensing: synthetic derivatives and structure-activity relationships, Chem Biol 19(5) (2012) 589–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Mascuch SJ, Boudreau PD, Carland TM, Pierce NT, Olson J, Hensler ME, Choi H, Campanale J, Hamdoun A, Nizet V, Gerwick WH, Gaasterland T, Gerwick L, Marine natural product honaucin A attenuates inflammation by activating the Nrf2-ARE pathway, J Nat Prod 81(3) (2018) 506–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Kasap C, Elemento O, Kapoor TM, DrugTargetSeqR: a genomics- and CRISPR-Cas9-based method to analyze drug targets, Nat Chem Biol 10(8) (2014) 626–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Lee CW, Bélanger K, Rao SC, Petrella TM, Tozer RG, Wood L, Savage KJ, Eisenhauer EA, Synold TW, Wainman N, Seymour L, A phase II study of ispinesib (SB-715992) in patients with metastatic or recurrent malignant melanoma: a National Cancer Institute of Canada Clinical Trials Group trial, Invest New Drugs 26(3) (2008) 249–255. [DOI] [PubMed] [Google Scholar]
- [115].Clemens MR, Gladkov OA, Gartner E, Vladimirov V, Crown J, Steinberg J, Jie F, Keating A, Phase II, multicenter, open-label, randomized study of YM155 plus docetaxel as first-line treatment in patients with HER2-negative metastatic breast cancer, Breast Cancer Res Treat 149(1) (2015) 171–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Tang X, Li J, Millán-Aguiñaga N, Zhang JJ, O’Neill EC, Ugalde JA, Jensen PR, Mantovani SM, Moore BS, Identification of thiotetronic acid antibiotic biosynthetic pathways by target-directed genome mining, ACS Chem Biol 10(12) (2015) 2841–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Alanjary M, Kronmiller B, Adamek M, Blin K, Weber T, Huson D, Philmus B, Ziemert N, The Antibiotic Resistant Target Seeker (ARTS), an exploration engine for antibiotic cluster prioritization and novel drug target discovery, Nucleic Acids Res 45(W1) (2017) W42–W48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Panter F, Krug D, Baumann S, Muller R, Self-resistance guided genome mining uncovers new topoisomerase inhibitors from myxobacteria, Chem Sci 9(21) (2018) 4898–4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Bai R, Paull K, Herald C, Malspeis L, Pettit G, Hamel E, Halichondrin-B And Homohalichondrin-B, Marine natural-products binding in the vinca domain of tubulin - discovery of tubulin-based mechanism of action by analysis of differential cytotoxicity data, J Biol Chem 266(24) (1991) 15882–15889. [PubMed] [Google Scholar]
- [120].Wong W, Oliver A, Linington R, Development of Antibiotic Activity Profile Screening for the Classification and Discovery of Natural Product Antibiotics, Chem Biol 19(11) (2012) 1483–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Skropeta D, Pastro N, Zivanovic A, Kinase inhibitors from marine sponges, Mar Drugs 9(10) (2011) 2131–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Goldstein DM, Gray NS, Zarrinkar PP, High-throughput kinase profiling as a platform for drug discovery, Nat Rev Drug Discov 7(5) (2008) 391–397. [DOI] [PubMed] [Google Scholar]
- [123].Yoo H, Leung D, Sanghara J, Daley D, van Soest R, Andersen R, Isoarenarol, a new protein kinase inhibitor from the marine sponge Dysidea arenaria, Pharm Biol 41(4) (2003) 223–225. [Google Scholar]
- [124].Al-Awadhi FH, Gao B, Rezaei MA, Kwan JC, Li C, Ye T, Paul VJ, Luesch H, Discovery, Synthesis, pharmacological profiling, and biological characterization of brintonamides A-E, novel dual protease and GPCR modulators from a marine cyanobacterium, J Med Chem 61(14) (2018) 6364–6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Liang X, Luo D, Yan JL, Rezaei MA, Salvador-Reyes LA, Gunasekera SP, Li C, Ye T, Paul VJ, Luesch H, Discovery of amantamide, a selective CXCR7 agonist from marine cyanobacteria. Org Lett 21(6) (2019) 1622–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Drag M, Salvesen GS, Emerging principles in protease-based drug discovery, Nat Rev Drug Discov 9(9) (2010) 690–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Kwan JC, Eksioglu EA, Liu C, Paul VJ, Luesch H, Grassystatins A-C from marine cyanobacteria, potent cathepsin E inhibitors that reduce antigen presentation, J Med Chem 52(18) (2009) 5732–5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Taori K, Matthew S, Rocca JR, Paul VJ, Luesch H, Lyngbyastatins 5-7, potent elastase inhibitors from Floridian marine cyanobacteria, lyngbya spp, J Nat Prod 70(10) (2007) 1593–1600. [DOI] [PubMed] [Google Scholar]
- [129].Luo D, Chen QY, Luesch H, Total synthesis of the potent marine-derived elastase inhibitor lyngbyastatin 7 and in vitro biological evaluation in model systems for pulmonary diseases, J Org Chem 81(2) (2016) 532–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Zanella F, Lorens JB, Link W, High content screening: seeing is believing, Trends Biotechnol 28(5) (2010) 237–245. [DOI] [PubMed] [Google Scholar]
- [131].Cervantes S, Stout PE, Prudhomme J, Engel S, Bruton M, Cervantes M, Carter D, Tae-Chang Y, Hay ME, Aalbersberg W, Kubanek J, Le Roch KG, High content live cell imaging for the discovery of new antimalarial marine natural products, BMC Infect Dis 12 (2012) 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Kubanek J, Prusak AC, Snell TW, Giese RA, Hardcastle KI, Fairchild CR, Aalbersberg W, Raventos-Suarez C, Hay ME, Antineoplastic diterpene-benzoate macrolides from the Fijian red alga callophycus serratus, Org Lett 7(23) (2005) 5261–5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Woehrmann MH, Bray WM, Durbin JK, Nisam SC, Michael AK, Glassey E, Stuart JM, Lokey RS, Large-scale cytological profiling for functional analysis of bioactive compounds, Mol Biosyst 9(11) (2013) 2604–2617. [DOI] [PubMed] [Google Scholar]
- [134].Schulze C, Bray W, Woerhmann M, Stuart J, Lokey R, Linington R, “Function-first” lead discovery: mode of action profiling of natural product libraries using image-based screening, Chem Biol 20(2) (2013) 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Ochoa J, Bray W, Lokey R, Linington R, Phenotype-guided natural products discovery using cytological profiling, J Nat Prod 78(9) (2015) 2242–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Kurita K, Glassey E, Linington R, Integration of high-content screening and untargeted metabolomics for comprehensive functional annotation of natural product libraries, Proc Natl Acad Sci U S A 112(39) (2015) 11999–12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Strange K, Drug discovery in fish, flies, and worms, Ilar Journal 57(2) (2016) 133–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Kaletta T, Hengartner M, Finding function in novel targets: c-elegans as a model organism, Nat Rev Drug Discov 5(5) (2006) 387–398. [DOI] [PubMed] [Google Scholar]
- [139].O’Reilly L, Luke C, Perlmutter D, Silverman G, Pak S, C. elegans in high-throughput drug discovery, Adv Drug Deliver Rev 69 (2014) 247–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Durai S, Vigneshwari L, Balamurugan K, Caenorhabditis elegans-based in vivo screening of bioactives from marine sponge-associated bacteria against Vibrio alginolyticus, J Appl Microbiol 115(6) (2013) 1329–1342. [DOI] [PubMed] [Google Scholar]
- [141].Crawford A, Esguerra C, de Witte P, Fishing for drugs from nature: Zebrafish as a technology platform for natural product discovery, Planta Medica 74(6) (2008) 624–632. [DOI] [PubMed] [Google Scholar]
- [142].Bruni G, Rennekamp A, Velenich A, McCarroll M, Gendelev L, Fertsch E, Taylor J, Lakhani P, Lensen D, Evron T, Lorello P, Huang X, Kolczewski S, Carey G, Caldarone B, Prinssen E, Roth B, Keiser M, Peterson R, Kokel D, Zebrafish behavioral profiling identifies multitarget antipsychotic-like compounds, Nat Chem Biol 12(7) (2016) 559–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Long S, Liang F, Wu Q, Lu X, Yao X, Li S, Li J, Su H, Pang J, Pei Z, Identification of marine neuroactive molecules in behaviour-based screens in the larval zebrafish, Mar Drugs 12(6) (2014) 3307–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Bousquet MS, Ma JJ, Ratnayake R, Havre PA, Yao J, Dang NH, Paul VJ, Carney TJ, Dang LH, Luesch H, Multidimensional screening platform for simultaneously targeting oncogenic KRAS and hypoxia-inducible factors pathways in colorectal cancer, ACS Chem Biol 11(5) (2016) 1322–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Cozzarelli NR, The mechanism of action of inhibitors of DNA synthesis, Annu Rev Biochem 46 (1977) 641–668. [DOI] [PubMed] [Google Scholar]
- [146].Minuzzo M, Marchini S, Broggini M, Faircloth G, D’Incalci M, Mantovani R, Interference of transcriptional activation by the antineoplastic drug ecteinascidin-743, Proc Natl Acad Sci U S A 97(12) (2000) 6780–6784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Soares DG, Escargueil AE, Poindessous V, Sarasin A, de Gramont A, Bonatto D, Henriques JA, Larsen AK, Replication and homologous recombination repair regulate DNA double-strand break formation by the antitumor alkylator ecteinascidin 743, Proc Natl Acad Sci U S A 104(32) (2007) 13062–13067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Bowling JJ, Pennaka HK, Ivey K, Wahyuono S, Kelly M, Schinazi RF, Valeriote FA, Graves DE, Hamann MT, Antiviral and anticancer optimization studies of the DNA-binding marine natural product aaptamine, Chem Biol Drug Des 71(3) (2008) 205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Matsunaga S, Fusetani N, Theonellamides A-E, cytotoxic bicyclic peptides, from a marine sponge theonella sp, J J Org Chem 60(5) (1995) 1177–1181. [Google Scholar]
- [150].Espiritu R, Cornelio K, Kinoshita M, Matsumori N, Murata M, Nishimura S, Kakeya H, Yoshida M, Matsunaga S, Marine sponge cyclic peptide theonellamide A disrupts lipid bilayer integrity without forming distinct membrane pores, Biochim Biophys Acta Biomembranes 1858(6) (2016) 1373–1379. [DOI] [PubMed] [Google Scholar]
- [151].Nishimura S, Arita Y, Honda M, Iwamoto K, Matsuyama A, Shirai A, Kawasaki H, Kakeya H, Kobayashi T, Matsunaga S, Yoshida M, Marine antifungal theonellamides target 3beta-hydroxysterol to activate Rho1 signaling, Nat Chem Biol 6(7) (2010) 519–526. [DOI] [PubMed] [Google Scholar]
- [152].Matsunaga S, Fusetani N, Hashimoto K, Walchli M, Theonellamide-F - a novel antifungal bicyclic peptide from a marine sponge theonella sp, J Am Chem Soc 111(7) (1989) 2582–2588. [Google Scholar]
- [153].Trzoss L, Fukuda T, Costa-Lotufo L, Jimenez P, La Clair J, Fenical W, Seriniquinone, a selective anticancer agent, induces cell death by autophagocytosis, targeting the cancer-protective protein dermcidin, Proc Natl Acad Sci U S A 111(41) (2014) 14687–14692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].DiJoseph JF, Armellino DC, Boghaert ER, Khandke K, Dougher MM, Sridharan L, Kunz A, Hamann PR, Gorovits B, Udata C, Moran JK, Popplewell AG, Stephens S, Frost P, Damle NK, Antibody-targeted chemotherapy with CMC-544: a CD22-targeted immunoconjugate of calicheamicin for the treatment of B-lymphoid malignancies, Blood 103(5) (2004)1807–1814. [DOI] [PubMed] [Google Scholar]
- [155].Almaliti J, Miller B, Pietraszkiewicz H, Glukhov E, Naman CB, Kline T, Hanson J, Li X, Zhou S, Valeriote FA, Gerwick WH, Exploration of the carmaphycins as payloads in antibody drug conjugate anticancer agents, Eur J Med Chem 161 (2019) 416–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Stark A, FDA approves Mylotarg for treatment of acute myeloid leukemia, FDA New Release, (2017, September 01), retrived from https://www.fda.gov/news-events/press-announcements/fda-approves-mylotarg-treatment-acute-myeloid-leukemia.
- [157].Deng C, Pan B, O’Connor OA, Brentuximab vedotin, Clin Cancer Res 19(1) (2012) 22–27. [DOI] [PubMed] [Google Scholar]
- [158].Simon S, FDA approves Besponsa (Inotuzumab ozogamicin) for acute lymphoblastic leukemia in adults. American Cancer Society, (2017, August 18), retrived from https://www.cancer.org/latest-news/fda-approves-besponsa-inotuzumab-ozogamicin-for-all.html. [Google Scholar]
- [159].Kadcyla improves survival in women diagnosed with advanced-stage her2-positive disease. Breastcancer.org, (2013, October 01), retrived from https://www.breastcancer.org/research-news/20131001. [Google Scholar]
- [160].Kayata E, FDA approves Polatuzumab vedotin-piiq combination for DLBCL patients, Cancer Network, (2019, June 11), retrived from https://www.cancemetwork.com/article/fda-approves-polatuzumab-vedotin-piiq-combination-dlbcl-patients. [Google Scholar]
- [161].Cheng X, Li J, Tanaka K, Majumder U, Milinichik AZ, Verdi AC, Maddage CJ, Rybinski KA, Fernando S, Fernando D, Kuc M, Furuuchi K, Fang F, Cienaka T, Grasso L, Albone EF, MORAb-202, an antibody-drug conjugate utilizing humanized anti-human FRα farletuzumab and the microtubule-targeting agent eribulin, has potent antitumor activity, Mol Cancer Ther 17(12) (2018) 2665–2675. [DOI] [PubMed] [Google Scholar]
- [162].Pereira AR, Kale AJ, Fenley AT, Byrum T, Debonsi HM, Gilson MK, Valeriote FA, Moore BS, Gerwick WH, The carmaphycins: new proteasome inhibitors exhibiting an α,β-epoxyketone warhead from a marine cyanobacterium, ChemBioChem 13(6) (2012) 810–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Pye CR, Bertin MJ, Lokey RS, Gerwick WH, Linington RG, Retrospective analysis of natural products provides insights for future discovery trends, Proc Natl Acad Sci U S A 114(22) (2017) 5601–5606. [DOI] [PMC free article] [PubMed] [Google Scholar]