Abstract
Objective
The Collaborative Cross (CC) is a mouse population model with diverse and reproducible genetic backgrounds used to identify novel disease models and genes that contribute to human disease. Since spontaneous tumour susceptibility in CC mice remains unexplored, we assessed tumour incidence and spectrum.
Design
We monitored 293 mice from 18 CC strains for tumour development. Genetic association analysis and RNA sequencing were used to identify susceptibility loci and candidate genes. We analysed genomes of patients with gastric cancer to evaluate the relevance of genes identified in the CC mouse model and measured the expression levels of ISG15 by immunohistochemical staining using a gastric adenocarcinoma tissue microarray. Association of gene expression with overall survival (OS) was assessed by Kaplan-Meier analysis.
Results
CC mice displayed a wide range in the incidence and types of spontaneous tumours. More than 40% of CC036 mice developed gastric tumours within 1 year. Genetic association analysis identified Nfκb1 as a candidate susceptibility gene, while RNA sequencing analysis of non-tumour gastric tissues from CC036 mice showed significantly higher expression of inflammatory response genes. In human gastric cancers, the majority of human orthologues of the 166 mouse genes were preferentially altered by amplification or deletion and were significantly associated with OS. Higher expression of the CC036 inflammatory response gene signature is associated with poor OS. Finally, ISG15 protein is elevated in gastric adenocarcinomas and correlated with shortened patient OS.
Conclusions
CC strains exhibit tremendous variation in tumour susceptibility, and we present CC036 as a spontaneous laboratory mouse model for studying human gastric tumourigenesis.
Keywords: tumor susceptibility, collaborative cross, gastric cancer, mouse model
Significance of this study.
What is already known on this subject?
Genetic susceptibility contributes to the development of common cancers.
Cancer susceptibility genes are identified using population-based model systems.
Rodents rarely develop spontaneous gastric cancer.
Engineered mouse models have contributed significantly to our current understanding of cancer biology but do not always accurately recapitulate de novo tumour development, nor do they represent human genetic diversity.
What are the new findings?
We characterised spontaneous tumour incidence across 18 strains of the Collaborative Cross population mouse model and found a wide range in the incidence and types of tumours.
We identified CC036 as a new mouse model for studying spontaneous human gastric tumourigenesis.
Genetic analysis identified the Nfκb1 locus significantly associated with gastric tumour susceptibility in CC036 mice and an additional 11 suggestive genetic loci.
Transcript profiling of mouse gastric tissues identified an inflammatory response signature that is significantly associated with overall survival (OS) in human gastric cancer patients.
The majority of human orthologues of mouse genes, which are differentially expressed in normal gastric tissue from CC036 mice, were preferentially altered by amplification or deletion in human gastric cancers and were significantly associated with OS of patients with gastric cancer.
We provide a novel model to study the role of Nfκb1 in inflammation-associated gastric tumourigenesis.
Significance of this study.
How might it impact on clinical practice in the foreseeable future?
The results suggest that inflammation and the Nfκb1 signalling pathway control gastric cancer susceptibility. Novel treatment options that target the molecular pathways of the gastritis-adenocarcinoma sequence should be explored.
Introduction
The development of human cancer has historically been attributed to the presence of rare mutations or germline polymorphisms in genes that have major effects on tumour growth or survival and their interaction with environmental factors.1 Identifying genes involved in cancer susceptibility has potential utility in risk management, leads to greater understanding of biological pathways involved in cancer development and elucidates how environmental factors exert their effects in combination with genetic variants.2
Genome-wide association studies (GWAS) have been successful for identifying genetic variants associated with susceptibility to many types of cancers.3 However, the genetic risk imposed by the variants identified so far accounts for only a minor proportion of familial clustering,4 implying that much of the genetic risk remains to be discovered. Moreover, GWAS have implicit limitations. Genetic heterogeneity in humans necessitates the collection of large numbers of samples from patients with cancer and control populations. Challenges in quantifying the variable levels of environmental carcinogens to which different individuals are exposed add a further dimension of uncertainty.
Mouse models are an important compliment to the study of cancer susceptibility. Studies using mice have revealed that tumour predisposition in different strains is controlled by multiple loci,4 5 which control fundamental processes such as tumour growth rate, ability to stimulate angiogenesis and the risk of malignant progression. These studies have led to the development of sophisticated genetically engineered mouse models of human cancer. Although these engineered models have contributed significantly to our current understanding of cancer biology, they do not always accurately recapitulate de novo tumour development, nor do they represent human genetic diversity. Thus, additional spontaneous models of tumour development are needed to identify novel cancer genes and mechanisms of tumour initiation and progression and to support preclinical therapeutic studies.
The Collaborative Cross (CC) mouse population model was established to model human genetic diversity by combining the genomes of eight diverse founder strains,6 capturing nearly 90% of the known variation present in laboratory mice.7 Collectively, many studies have shown that the CC panel consists of a population of mice with genetic and phenotypic diversity on par with human populations.8 9 However, spontaneous tumour susceptibility in CC mice remains unknown. In this study, we assessed the tumour incidence and spectrum in 18 CC strains and found diverse tumour susceptibilities. Most strikingly, within 1 year, CC036 mice spontaneously developed gastric tumours in the pyloric antrum with a sex-specific incidence of approximately 65% in women and 10% in men. Genetic and transcriptomic analyses of normal CC036 gastric tissues revealed many candidate genes that contribute towards gastric cancer (GC) susceptibility. These candidate genes significantly overlap with many known human GC genes, suggesting that CC036 is a new spontaneous mouse model for human gastric tumourigenesis.
Result
Tumour susceptibility across CC strains
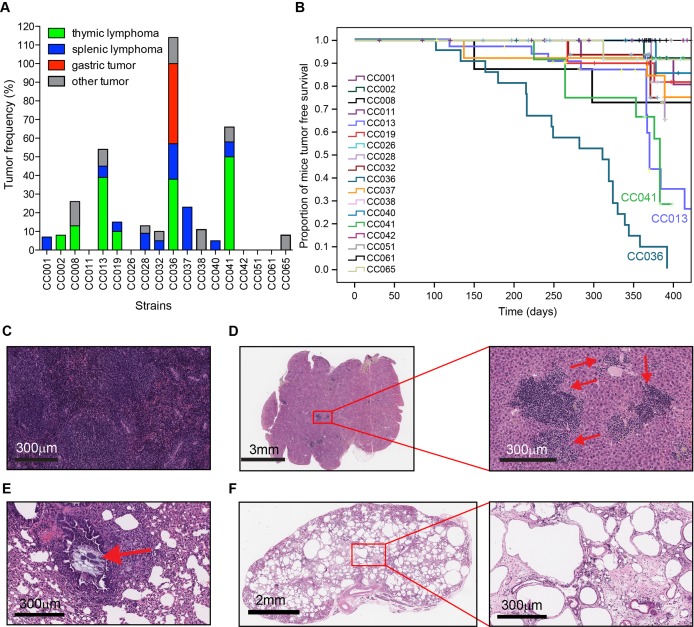
Survival was monitored up to 1 year in 18 CC strains (see online supplementary table 1), revealing a wide range in the incidence and types of spontaneous tumours. Tumours of the thymus and spleen were the most frequently observed abnormalities during necropsy across all strains (figure 1A). Fifty-seven per cent of CC013, 66% of CC041 and 100% of CC036 mice developed tumours within 1 year (figure 1B). Tumours of lymphoid origin, which commonly occurred in the three most tumour prone strains CC013 (42%), CC036 (67%) and CC037 (23%), frequently metastasised to other organs including lung and liver (figures 1C,D and 2C). Most striking was CC036, which was the most susceptible strain to spontaneous tumour development and is the focus of this study. All CC036 mice developed tumours within 1 year with lymphoid and stomach tumours being the most predominant (figures 1A and 2). Other strain-specific but non-tumour abnormalities were also observed (see online supplementary table 1). A large fraction of CC011 mice developed anal prolapse within 1 year (see online supplementary table 1), which is consistent with previous results.10 Pulmonary aspiration was frequently observed in CC040 mice (figure 1E, see online supplementary figure 1A,B), while polycystic kidneys were observed in CC013 and CC032 (figure 1F, see online supplementary figure 1C–E).
Figure 1.
Variation in disease susceptibility across 17 CC strains. (A) Tumour frequency and type across CC strains within 1 year. (B) Tumour-free survival across CC strains. (C) Representative H&E image of splenic lymphoma in CC037 mouse. (D) Representative H&E image of CC037 mouse liver (from C) showing numerous metastatic lymphoid lesions indicated by red arrows in enlargement. (E) Representative H&E image of CC040 mouse lung showing plant material obstructing a bronchus and surrounded by lymphocytic inflammation as indicated by the red arrow. (F) Representative H&E image of CC032 mouse kidney showing a polycystic phenotype. CC, Collaborative Cross.
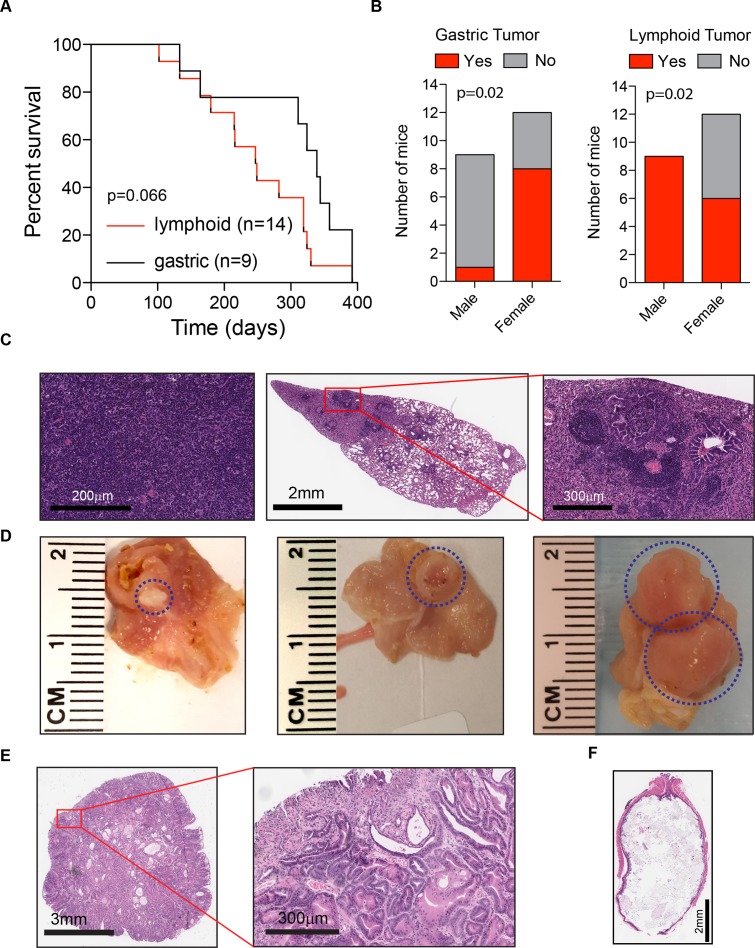
Figure 2.
Tumourigenicity in CC036 mice. (A) Latency of lymphoid and gastric tumours in CC036 mice. There is no significant difference in latency between two tumour types (p=0.066 obtained by log-rank test). (B) Incidence of lymphoid and gastric tumours in CC036 male and female mice. A significant sex difference was observed for gastric and lymphoid tumour development (p=0.02 obtained by Fisher’s exact test). (C) Representative H&E image of thymic lymphoma (left) and lung metastasis (middle and right). (D) Representative photographs of gastric tumours in CC036 mice. (E) Representative H&E image of gastric tumour showing hyperplastic glands with low-grade dysplasia. (F) Representative H&E image of oesophageal cross-section showing dilation and food matter obstructing the lumen.
CC036 mice are susceptible to spontaneous gastric tumours and thymic lymphoma
Detailed analysis of the tumour incidence in CC036 mice revealed that the time of onset of lymphoma development tended to be earlier than the onset of gastric tumours (p=0.066; figure 2A). The incidence of gastric tumours in female CC036 mice was approximately 67%, whereas in male CC036 mice, the incidence was only 11% (p=0.02; figure 2B). Conversely, the incidence of lymphoid tumours was 100% in male CC036 mice, while only 50% in female CC036 mice (p=0.02; figure 2B). The majority of lymphomas originated from the thymus (8 of 14 lymphomas), with 4 of 14 lymphomas originating from the spleen and 2 of 14 from the abdominal area (see online supplementary table 1). Five of 14 lymphomas showed metastatic lesions in distant organs including lung, liver and kidney (figure 2C, see online supplementary table 1). Histopathological analysis of gastric tumours revealed focal areas of low-grade dysplasia with increased architectural complexity, nuclear atypia and nuclear crowding in the pyloric antrum (figure 2D,E, see online supplementary figures 2A and 3A). In the tumour areas proliferation as measured by Ki-67 immunohistochemical staining was irregular with positive staining observed in regions with high nuclear/cytoplasmic ratios and negative staining in areas with low nuclear/cytoplasmic ratios, while Ki-67 staining was limited to the base of the crypts in adjacent normal tissue (see online supplementary figures 2B and 3B). To further characterise the lineages that are present in the tumour mass, we performed immunochistochemical and lectin staining for UEA1, HK-ATPase and TFF3 (see online supplementary figure 4). Tumours were positive for the surface mucous cell marker UEA1. Nests of UEA1 positive cells were also found invading beyond the muscularis mucosa into the submucosa (see online supplementary figure 4B). In approximately 25% of tumours, scattered cells stained lightly positive for the parietal cell marker HK-ATPase. TFF3 staining was largely negative, although few tumours showed scattered TFF3 positive cells. Mice with gastric tumours frequently showed oesophageal dilation due to tumour obstructed food from moving through the upper gastrointestinal tract (figure 2F). To exclude the possibility that gastric tumour development in CC0036 was associated with Helicobacter sp., we first characterised faecal gut microbiome composition from each of 18 strains at 12 weeks of age by 16S rRNA gene sequencing.9 The level of Helicobacter sp. was not different among strains from the same facility, although differences were observed between two facilities (see online supplementary figure 5A, additional details can be found in ref 9). We then characterised the stomach microbiome composition by 16S rRNA sequencing in six CC strains and showed a similar difference between strains based on animal facility as was seen in the faecal microbiome (see online supplementary figure 5B). The relative abundance level of Helicobacter sp. in CC036 is low (approximately 0.3%) similar to other strains from the same animal facility (see online supplementary figure 5C and table 2). These data lead us to conclude that genetic factors and/or the interaction between Helicobacter sp. and genetics may contribute to gastric tumour susceptibility in CC036 mice.
Genome-wide association analysis identified significant region on distal chromosome 3 associated with increased gastric tumour susceptibility
To identify genetic loci associated with CC036 gastric tumour susceptibility, we performed independent quantitative trait locus (QTL) analyses by interrogating 77 000 SNPs across the genome and identified 1 significant and 11 suggestive QTLs associated with gastric tumour susceptibility (figure 3A). The position of the peak significant SNP was at 135.8 Mb on Chromosome 3. To identify the CC founder contribution to this phenotype, we performed genome-wide association using a logistic regression model across haplotypes and showed that the CAST/EiJ founder haplotype on chromosome 3 contributed to the development of gastric tumours (figure 3B). We identified 13 candidate genes in this region including Nfκb1. The same haplotype analysis was performed across all suggestive loci (see online supplementary figure 6).
Figure 3.
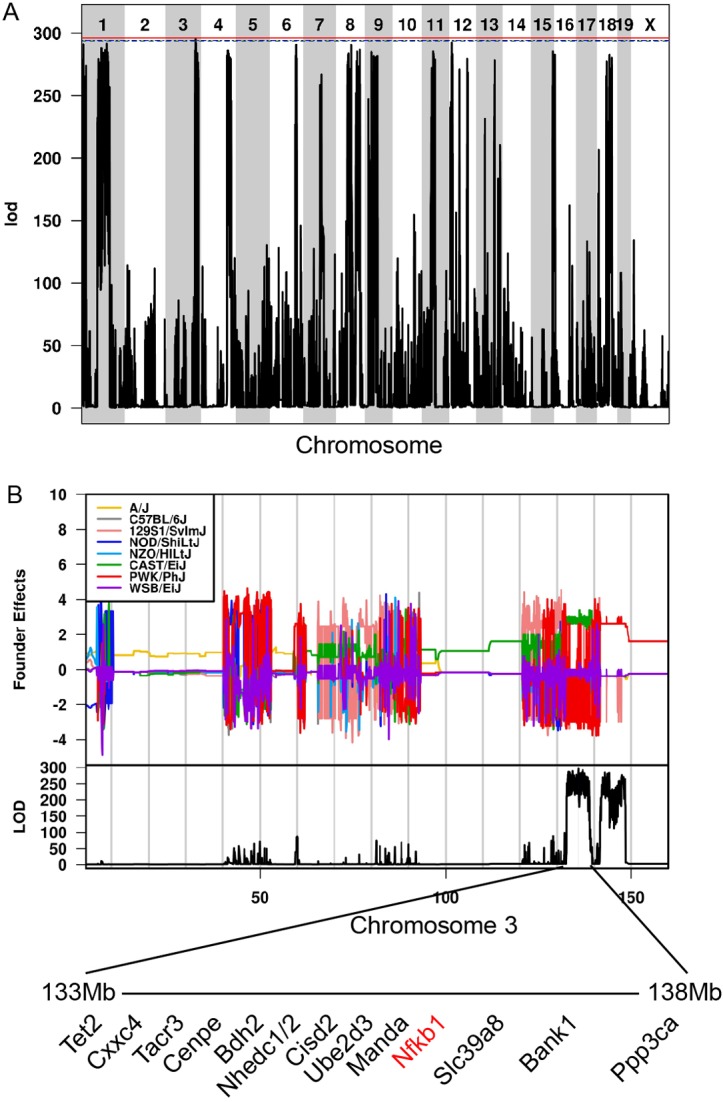
Identification of genetic loci associated with gastric cancer susceptibility in CC036. (A) Genome-wide association plot of CC036 gastric cancer susceptibility. Chromosomal position of mouse SNPs is shown on the x-axis. The logarithm of the odds (to the base 10) (lod) score of the association for each SNP is shown on the y-axis. (B) Founder haplotype contributions to gastric cancer susceptibility on chromosome 3. Candidate genes are listed below.
RNA-seq analysis reveals potential molecular mechanisms of gastric tumour susceptibility
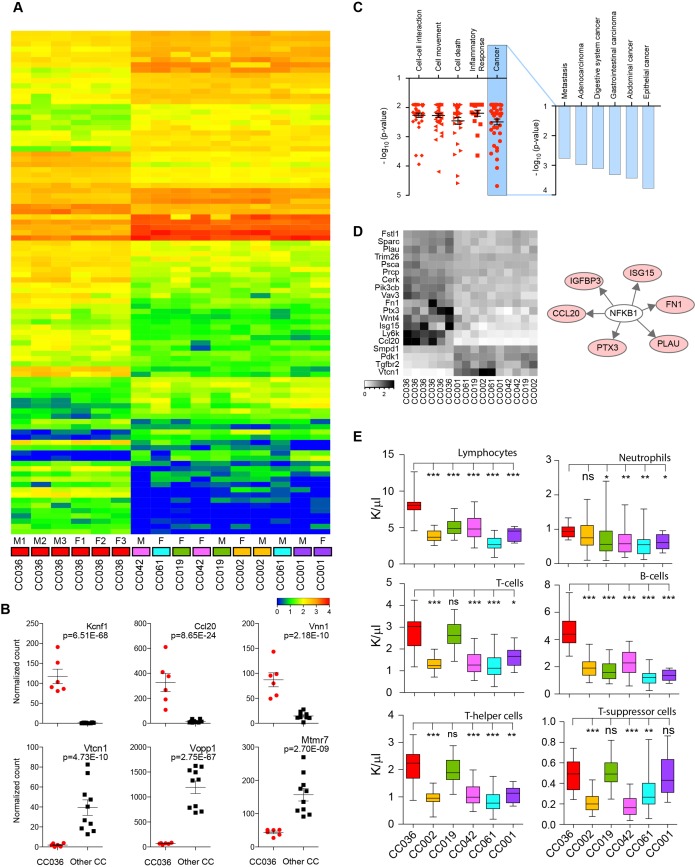
To gain insights into potential molecular mechanisms for gastric tumour susceptibility in CC036 mice, we performed RNA sequencing of gastric tissues from 12-week-old CC036 mice and five additional CC mouse strains not susceptible to gastric tumourigenesis (CC001, CC002, CC019, CC042 and CC061) as controls. At this age, the CC036 stomach mucosa appeared unremarkable with proliferation limited to the base of the crypts (see online supplementary figure 7A,B). For each strain, stomach tissues and corresponding RNA samples were obtained from three male and three female mice. For control strains, we pooled the male and female RNA samples respectively, whereas for the CC036 mice, RNA from each male and female mouse was independently sequenced and analysed. We identified 166 genes differentially expressed between CC036 and the control CC strains (fold-change 1.5 and adjusted p<0.05; figure 4A,B, see online supplementary table 3). Genes expressed at high levels in CC036 stomach compared with control CC strains include genes previously identified in gastrointestinal tract cancer such as Kcnf1, Ccl20 and Vnn1, whereas genes expressed at low levels in CC036 stomach compared with control CC strains include Vopp1, Vtcn1 and Mtmr7 (figure 4B).11–16 To further delineate the observed sexual dimorphism in gastric tumour incidence in CC036 mice, we directly compared gene expression levels in stomach tissues between female and male CC036 mice. Twenty-one genes were differentially expressed between stomach tissues of female and male CC036 mice (fold-change 1.5 and adjusted p<0.05; see online supplementary figure 8A). In addition to differences in expression of genes located on the sex chromosomes, we observed lower expression of Kruppel-like factor 4 (Klf4) and higher expression of the hepcidin encoding genes Hamp and Hamp2, which have been shown to play an important role in human GC development.17 18
Figure 4.
RNA sequencing and immune analysis in CC036 and five control CC strains. (A) Heatmap of differentially expressed genes in gastric tissue between CC036 and control CC strains (fold-change 1.5 and adjusted p<0.05). Red indicates higher expression and blue indicates lower expression. (B) Box plots of normalised sequence read counts for six genes significantly differentially expressed between CC036 and control CC strains. (C) Functional enrichment analysis of genes differentially expressed between CC036 and control CC strains using Ingenuity Pathway Analysis (IPA) (left panel). Detailed functional enrichment for cancer types and related phenotypes (right panel). (D) Heatmap of inflammatory-response gene signature in gastric tissue of CC036 and control CC strains. Black indicates higher expression, and white indicates lower expression. (E) Distribution of circulating lymphocytes, B cells (CD45R/B220), total T cells (CD3+), neutrophils, T-helper cells (CD3+/CD4+) and T-suppressor cells (CD3+/CD8+) in blood from CC036 and control CC strains. Error bars represent minimum and maximum values; the median is indicated with a horizontal bar inside each box. ***P<0.001, **p<0.01, *p<0.05; ns is not significant. P was obtained using non-parametric Mann-Whitney test. CC, Collaborative Cross.
Further bioinformatics analysis showed that 166 differentially expressed genes were significantly enriched in human cancer and inflammatory response related processes (figure 4C, left panel). Not surprisingly, we found significant enrichment for gastrointestinal tract cancer (figure 4C, right panel). The inflammatory response enrichment was comprised of 19 genes, 15 of which were expressed at higher levels in CC036 compared with control strains (figure 4D). Interestingly, 5 of these 19 genes are regulated by NFKB1 (figure 4D), consistent with our genome-wide association analysis that identified Nf κ b1 as a candidate regulator of gastric cancer susceptibility (figure 3B).
The immune system is thought to be a key regulator in cancer development. To investigate the immune composition in CC036 mice, we measured the relative abundance of specific immune cell populations using complete blood counts and flow cytometry to distinguish lymphocytes, B cells, T cells, CD4+ T-helper cells and CD8+ T-suppressor cells in CC036 and control CC strains. The relative abundance of specific immune cell populations was tightly constrained within each strain but varied across different strains (figure 4E). Interestingly, peripheral blood of CC036 mice had significantly higher levels of lymphocytes and neutrophils compared with the five control CC strains (p<0.005). Fluorescence-activated cell sorting (FACS) analysis showed that both the B cell, total T cell, T-helper and T-suppressor cell fractions were significantly higher in CC036 mice (figure 4E). Moreover, triplex immunohistochemical staining for CD3, CD11b and CD45R showed moderate levels of myeloid cell infiltrate (CD11b positive) in tumour areas, whereas only sparse CD11b positive staining was observed in normal CC036 gastric mucosa (see online supplementary figures 2C, 3C and 7C). Furthermore, blood of female CC036 mice showed significantly higher levels of white blood cells, lymphocytes and T cells compared with male CC036 mice (p<0.05; see online supplementary figure 8B and table 4). These results are consistent with the observed over-representation of genes involved in inflammatory responses in CC036 stomach tissues and which is conceivably associated with inflammation-associated tumourigenesis.
Evaluation of CC036 susceptibility gene orthologues in human GC
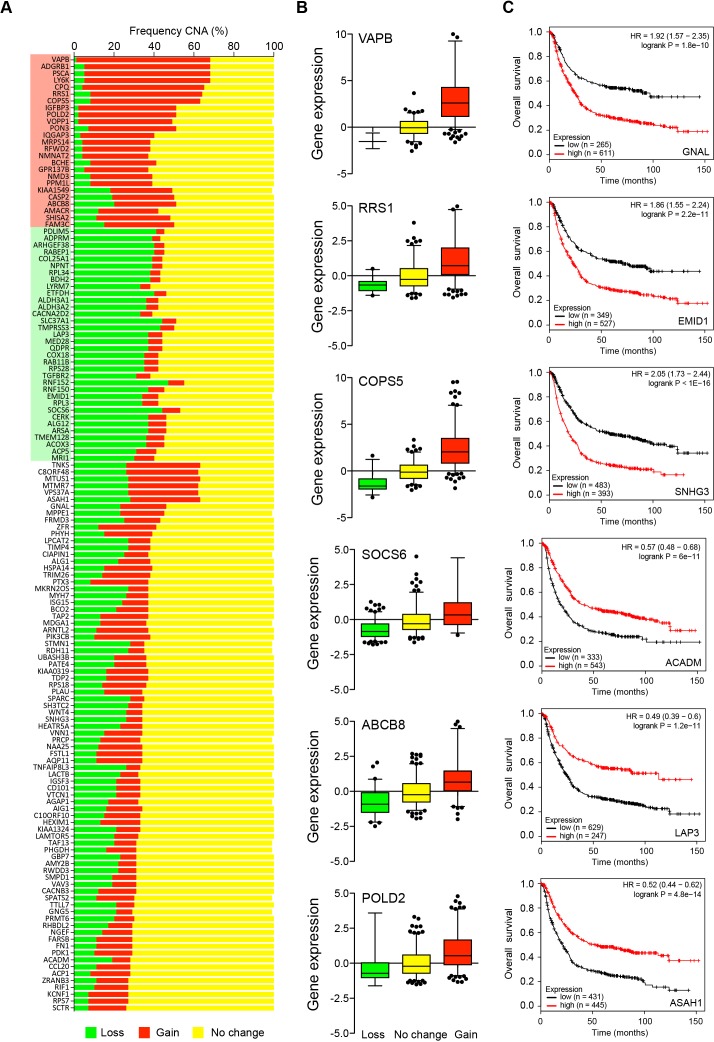
To assess the human relevance of the CC036 mouse model, we identified 140 human orthologues of the 166 mouse genes differentially expressed between CC036 and other CC strains (see online supplementary table 5). The Cancer Genome Atlas (TCGA) data were first used to evaluate the 140 human genes in human GC. We frequently observed DNA copy number alterations encompassing 59 of 140 genes in human GC where 25 genes were frequently increased in copy number and 34 genes frequently showed a decrease (figure 5A). Changes in DNA copy number are often observed in tumours, and DNA copy number aberrations are one of the mechanisms that can result in a change in gene expression in tumour progression.19 We observed significant associations between transcriptional expression levels and their copy number in 76 out of 140 genes (figure 5B, see online supplementary table 6).
Figure 5.
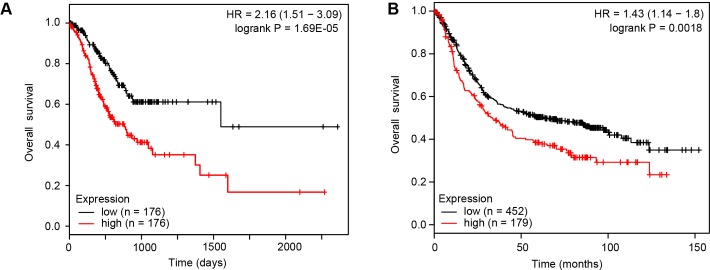
Evaluation of differentially expressed genes identified between CC036 and control strains in human gastric adenocarcinoma. (A) Frequency of copy number aberrations (CNAs) of human orthologues of differentially expressed mouse genes in human gastric cancers (TCGA). Red indicates increased DNA copy number (gain), green indicates decreased DNA copy number (loss) and yellow indicates no change. Red and green highlights of gene names indicate genes that are frequently gained and lost in human gastric cancer, respectively. (B) Representative box plots of the association between gene expression and CNA in human gastric cancers (TCGA). Red indicates increased DNA copy number (gain), green indicates decreased DNA copy number (loss) and yellow indicates no change. (C) Representative Kaplan-Meier plots for the association of gene expression with overall survival. Red indicates tumours with high transcript expression of the corresponding gene and black indicates low expression. Kaplan-Meier plots were obtained using a meta-analysis of 882 gastric cancer patients with a mean follow-up of 33 months available from Kaplan-Meier plotter. The p value represents the equality of survival curves based on a log-rank test. TCGA, The Cancer Genome Atlas.
To further assess the importance of 140 genes in human GC development, we evaluated their prognostic value for patients with GC in an integrated dataset.20 This analysis identified that 112 out of 140 genes were significantly associated with overall survival (OS) (adjusted p value <0.05; figure 5C, supplementary table 7). Fifty-two genes had an HR <1 (higher gene expression associated with good prognosis) and 60 genes had a HR >1 (higher gene expression associated with poor prognosis). We then specifically asked whether the 19-gene inflammatory response gene signature observed in CC036 stomach tissue (figure 4D) is associated with OS. Indeed, higher expression of the inflammatory response gene signature predicted poor OS in both TCGA (figure 6A) using SurvExpress21 and publicly available microarray data using Kaplan-Meier plotter20 (figure 6B).
Figure 6.
Inflammatory gene signature predicts overall survival in patients with stomach cancer. (A) Analysis of association of inflammatory gene signature in TCGA data using SurvExpress. (B) Meta-analysis of association of inflammatory gene signature in publicly available datasets using Kaplan-Meier plotter. The p value represents the equality of survival curves based on a log-rank test.
Impact of ISG15 on human GC patient survival
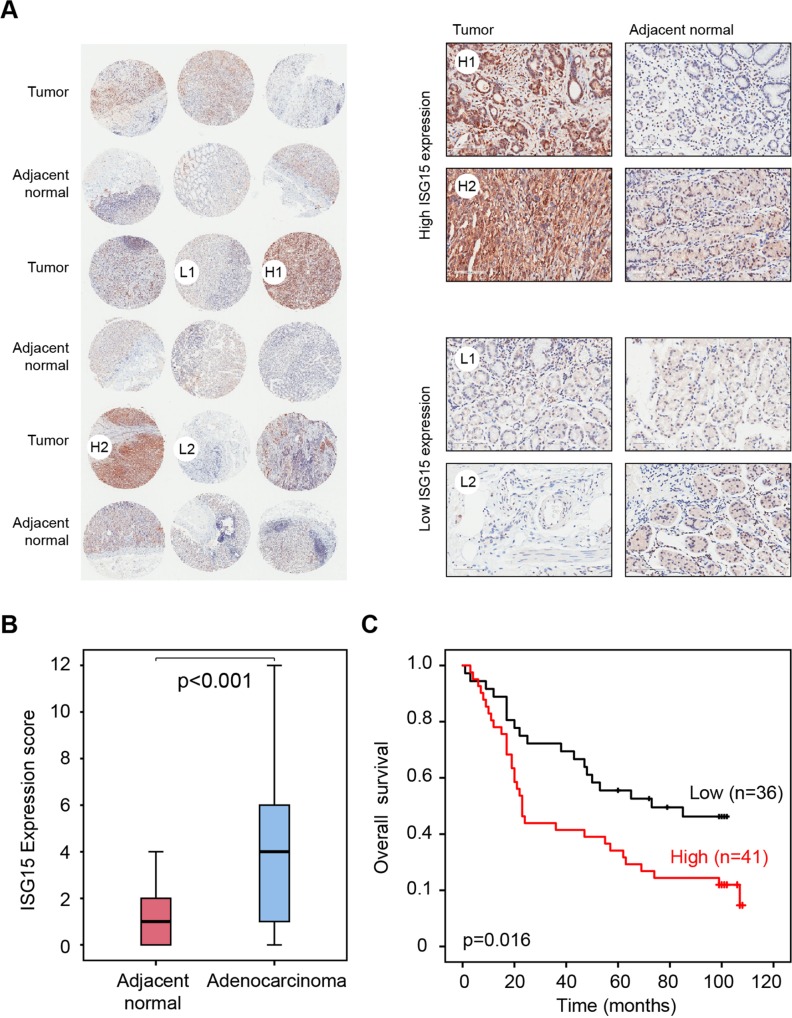
The interferon-stimulated gene 15 (ISG15), which we found to be a member of the 19-gene inflammatory response gene signature, is increasingly recognised for its role in cancers of the digestive system. We found that Isg15 was significantly upregulated in CC036 stomach tissues compared with control CC strains (log2 ratio=1.5; adjusted p=0.009; figure 4D and see online supplementary table 2). To further investigate the role of ISG15 in human GC, we examined the protein expression level of ISG15 using immunohistochemical (IHC) staining on a tissue microarray containing 84 gastric adenocarcinomas and 83 normal gastric tissues of which 76 were adjacent matched normal. A wide range of expression levels were observed across the GC (figure 7A). After quantifying ISG15 protein expression in the tumours and adjacent normal tissue sections we observed a significantly higher level of ISG15 in GC compared with adjacent normal stomach samples (p<0.001; figure 7B). Finally, we addressed whether ISG15 protein expression was associated with OS. The patient population was split based on the ISG15 IHC staining score (low: score ≤3; high: score >3). This analysis showed that patients with higher expression of ISG15 had a significantly worse survival compared with patients with lower expression (p value=0.016; figure 7C).
Figure 7.
ISG15 protein expression is increased in stomach adenocarcinoma and associated with poor prognosis. (A) Representative images of IHC staining of tissue microarray containing gastric cancer and adjacent normal tissue. Representative tumours with high expression are labelled H1 and H2 and tumours with low expression are labelled L1 and L2. (B) Box plot of ISG15 protein expression score in adenocarcinoma and adjacent normal tissue (p<0.001 obtained using Mann-Whitney test). (C) Association of ISG15 protein expression with overall survival. Red indicates tumours with a high ISG15 expression (score >3), and black indicates low expression (score ≤3). The p value represents the equality of survival curves based on a log-rank test.
Discussion
In this study, we assessed the spontaneous tumour incidence and spectrum in 18 CC strains. Our results show that tumour susceptibility is highly variable among different CC strains. Tumours of the thymus and spleen were the most frequently observed abnormalities. Our most exciting finding is strain CC036 that spontaneously develops gastric tumours. Histopathology and histochemistry of the tumours suggested that the majority were adenocarcinomas. GC is one of the most common malignancies in the world and ranks fifth in the incidence rate.22 GC is a polygenic disease, in which interactions of cancer genes with environmental risk factors or pathogens, such as Helicobacter pylori infection-associated inflammation, lead to early lesions of dysplasia within the gastric mucosa, and ultimately to the development of gastric adenocarcinoma. Animal models of GC have been developed to elucidate the molecular mechanisms underlying this disease.23 Although spontaneous gastric tumours have not been previously observed in mice, infection with Helicobacter sp. and carcinogen treatment have been used to establish mouse models that resemble human GC. For example, mice treated with N-methyl-N-nitrosourea (MNU) have been widely used to study gastric carcinogenesis.24 A combination of MNU treatment with Helicobacter felis infection resulted in rapid development of antral GC.25 Helicobacter is present in CC036 mice at levels similar or lower as compared with other CC strains raised in the same environment. Although we cannot exclude the possibility that Helicobacter sensitises the CC036 gastric tumour model, it appears that host genetics plays an important role. Additional studies using gnotobiotic techniques are necessary to dissect the role of Helicobacter in this model. Other mouse models of gastric cancer use transgenic mice such as the insulin–gastrin (INS-GAS) model.26 In these animals, human gastrin is regulated by the rat insulin 1 (Ins1) promoter leading to overexpression of gastrin in the pancreas resulting in gastric atrophy and intestinal metaplasia and eventually invasive gastric adenocarcinoma by 20 months of age. Infection with Helicobacter shortens the latency period.27 A number of other transgenetic or chemical models of gastric hyperplasia or tumourigenesis in mice have been developed (reviewed in ref 23). For example, mice mutant for Tff1, a gastric-specific tumour suppressor gene, rapidly developed gastric hyperplasia and a third of the mice went on to progress to dysplasia and carcinomas.28 Interestingly, our RNA sequencing analysis of gastric tissues from CC mice showed a trend for reduced expression of Tff1 in CC036 compared with the other CC strains (p value=0.016).
Juvenile polyposis syndrome is a rare autosomal dominant condition characterised by the presence of polyps along the gastrointestinal tract. Mutations in SMAD4 and BMPR1A can cause juvenile polyposis syndrome in 20% of cases.29 We found no polymorphisms in coding regions and no differences in gene expression of Smad4 and Bmpr1a in stomach tissues of 12-week-old CC036 mice (see online supplementary figure 9). Targeted DNA sequencing of Smad4 and Bmpr1a coding regions in CC036 stomach tumour tissues also did not reveal any mutations suggesting that tumour development in CC036 mice is not due to a mutation in these genes implicated in juvenile polyposis syndrome. This is the first CC mouse model for gastric tumourigenesis that can be used to dissect the set of genes and environmental components responsible for GC development to identify potential GC risk factors in humans.
In humans, GC incidence is approximately twice as high in men than women. However, rodents rarely develop spontaneous GC with the exception of cotton rats (Sigmodon hispidus), which develop spontaneous GC with an incidence of 25%–50% and the Z strain of the African rodent Mastomys natalensis.23 30 Interestingly, female predominance of GC was observed in cotton rats.30 These tumours develop secondary to hypergastrinaemia due to reduced intragastric pH. In addition, female C57BL/6 mice infected with Helicobacter felis showed significantly greater chronic inflammation with earlier onset compared with male mice.31 However, gastritis and development of gastrointestinal intraepithelial neoplasia in the INS-GAS model is male predominant.32 These differences are not well understood but likely depend on differences in immune response, sex hormones and possibly sex differences in the microbiome. For example, studies using the INS-GAS mouse model showed that oestradiol was protective in Helicobacter induced GC.33 34
We performed RNA sequencing of histologically normal gastric tissues from 12-week-old CC036 mice and five additional CC mouse strains not susceptible to gastric tumourigenesis to gain insights into potential molecular mechanisms for gastric tumour susceptibility. Genetic background can significantly influence gene expression levels in tissues, which has led to the hypothesis that tumour susceptibility gene signatures could be present in normal tissues prior to tumour induction.35 For example, analysis of human metastasis-predictive gene signatures in normal mouse tissues accurately categorised mice based on their metastatic potential supporting the presence of predictive signatures in normal tissues.36 Using this approach, we identified a 19-gene inflammatory response gene signature in normal CC036 stomach tissue that was associated with OS in human gastric cancer patients. ISG15, included in this 19-gene signature, encodes a member of the ubiquitin-like protein superfamily,37 and aberrant expression has been observed in gastric and other cancer types.38 Our previous work showed that combined oestrogen and progesterone treatment induced type 1 interferons, which modulate Isg15 expression in the mammary gland of oophorectomised mice.39 It is also known that ISG15 is induced by type I interferons as a primary response to diverse microbial and cellular stress stimuli. In addition, we observed upregulation of other inflammatory genes including the chemokine CCL20, which plays a role on mucosal surfaces in inflammation where it attracts immature dendritic cells and memory T cells.40 CCL20 is upregulated in Helicobacter pylori-associated gastritis, which can progress to GC.41 A direct comparison of gene transcript levels in stomach tissues of female and male CC036 mice showed lower expression of Klf4 and higher expression of the hepcidin genes Hamp and Hamp2 in female mice. Loss of Klf4 in mice has been shown to cause increased proliferation and altered differentiation of the adult gastric epithelium suggesting a critical role for Klf4 in maintaining gastric epithelial homeostasis.18 Hepcidin is critically important in regulation of systemic iron homeostasis and increased expression in gastric tumour tissues correlated with increasing tumour stages, which was associated with activation of JAK/STAT3 signalling.17 Our genetic analysis identified the Nfκb1 locus significantly associated with gastric tumour susceptibility in CC036 mice. Interestingly, human GWAS analysis also showed significant association between polymorphisms in NFκB1 and susceptibility of GC.42 NFκB1 transcriptionally regulates expression of genes involved in immune responses. Consistent with these observations, we found that four genes out of the 19-gene inflammatory signature are downstream targets of NFκB1, which includes ISG15 and CCL20. A recent study further supports our results showing that loss of Nfκb1 causes GC in mice with aberrant inflammation.43
In conclusion, we investigated tumour susceptibility across 18 CC strains and identified new spontaneous mouse models for the study of different malignancies including polycystic kidney disease, lymphoma and gastric tumourigenesis. The new gastric tumour model described has an average latency of approximately 1 year and may be mediated at least in part by an inflammatory signature that is evident at 2 months of age prior to the onset of disease. Our data show that susceptibility genes discovered through this model as well as the observed inflammatory response signature can be used as a biomarker to predict survival in human GC patients. Further investigations will need to focus on the progression of gastric tumours over time and whether disrupting the inflammatory signature early in life can prevent the development of GC.
Methods
Mice
CC mice were obtained from the Systems Genetics Core Facility at the University of North Carolina.44 Mice were acclimated at the Lawrence Berkeley National Laboratory for 8 weeks prior to the initiation of breeding. The study was carried out in strict accordance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The Animal Welfare and Research Committee at Lawrence Berkeley National Laboratory approved the animal use protocol. Tumour development was monitored for 1 year. Tumour tissue embedding and H&E staining were performed at the University of California San Francisco Mouse Pathology core facility, and tumour type was assessed by two pathologists.
QTL analysis
The frequency of stomach tumours in each CC line was used for genetic mapping. Forty per cent of mice from the CC036 strain developed stomach tumours. Although no other CC line developed stomach tumours, all other lines were coded at 1% to perform the analysis using gQTL, an online implementation of DOQTL for genetic analysis of CC data.45 Significance thresholds were determined using 1000 permutations and taking p=0.05.
Mouse blood analysis
Whole blood was collected into EDTA-coated tubes at 12 weeks of age in a cohort of 267 mice across 16 CC strains. Complete blood cell counts were acquired using a HemaVet950FS. Lymphocyte subpopulations were identified by FACS as described previously.9
Tissue microarray and immunohistochemical staining and scoring
A GC tissue microarray, with 84 tumour tissues and 83 normal stomach tissues of which 76 are adjacent matched normal, was purchased from Shanghai Outdo Biotech. Pathological grade and tumour-node-metastasis staging is based on the seventh staging classification of the American Joint Committee on Cancer and Union for International Cancer Control. Endogenous peroxidase activity was blocked with 3% hydrogen peroxide. Antigen retrieval was carried out in citrate buffer (10 mM, pH6) for 20 min at 92°C. Tissue sections were incubated with the primary ISG15 antibody (1:50, Cell Signaling Technology) for 16 hours at 4°C, and then incubated for 30 min at 37°C and subsequently with a secondary biotinylated antibody for 30 min at 37°C followed by incubation with streptavidin–peroxidase complex for 5 min at room temperature. An IHC staining score was assigned to each sample by a pathologist based on extent and intensity of staining (for extent: 0=no staining, 1=staining 1%–25%, 2=staining 26%–50%, 3=staining 51%–75% and 4=staining 76%–100%; for intensity: 0=no staining, 1=weak, 2=intermediate and 3=strong). The IHC staining score is calculated by multiplying the extent and intensity staining scores.
Statistical analysis
Differential gene expression analysis was conducted by comparing CC036 with the pooled results from the remaining five CC strains using DeSeq2. Gene-level counts were computed on independent biological replicates. Functional annotation of differentially expressed genes was performed using Ingenuity Pathway Analysis (Qiagen). Significance of the differences in blood counts across different CC strains of mice was assessed by non-parametric (Mann-Whitney) test. Genomic alterations and gene transcript levels for human GC in TCGA were obtained from cBioPortal.46 47 We used a rank-based non-parametric test (Kruskal-Wallis) to determine whether gene expression levels were significantly different between DNA copy number groups (p<0.05 was used as a threshold for significance). Association of differentially expressed mouse genes and OS in patients with GC was assessed using Kaplan-Meier plotter (http://kmplot.com) including Kaplan-Meier (KM) survival analysis, HR with 95% CIs and logrank p value for each gene.20 Association of the inflammatory response gene signature with OS was assessed using Kaplan-Meier plotter and SurvExpress.21
gutjnl-2018-316691supp001.xlsx (18.1KB, xlsx)
gutjnl-2018-316691supp002.jpg (2.8MB, jpg)
gutjnl-2018-316691supp003.jpg (2.2MB, jpg)
gutjnl-2018-316691supp004.jpg (4.5MB, jpg)
gutjnl-2018-316691supp005.jpg (3.6MB, jpg)
gutjnl-2018-316691supp006.jpg (747.8KB, jpg)
gutjnl-2018-316691supp007.xlsx (162.2KB, xlsx)
gutjnl-2018-316691supp008.jpg (2.3MB, jpg)
gutjnl-2018-316691supp009.jpg (5.5MB, jpg)
gutjnl-2018-316691supp010.xlsx (70.9KB, xlsx)
gutjnl-2018-316691supp011.jpg (439.3KB, jpg)
gutjnl-2018-316691supp012.xlsx (13KB, xlsx)
gutjnl-2018-316691supp013.xlsx (15.7KB, xlsx)
gutjnl-2018-316691supp014.xlsx (31.6KB, xlsx)
gutjnl-2018-316691supp015.xlsx (13.9KB, xlsx)
gutjnl-2018-316691supp016.jpg (885.8KB, jpg)
gutjnl-2018-316691supp017.pdf (99.9KB, pdf)
Acknowledgments
The authors would like to thank Richard Weiszmann and Ken Wan for quality control analysis of the RNA, preparing sequencing libraries and interfacing with the UC Berkeley’s QB3 Vincent J. Coates Genomics Sequencing Laboratory. We also thank Ben Booth for sequence data management, storage, visualisation and submission to the SRA and Chengfei Jiang for scanning pathological and IHC staining sections.
Footnotes
Contributors: PW, YW and Y-XZ: acquisition of data and analysis and interpretation of data. SAL: analysis and interpretation of data and statistical analysis. CMR and TW: immunohistochemical staining. KLW, XF, K-YJ and QS: pathological examination. CB: amplicon analysis and deposition. DWT: analysis and interpretation of data, critical revision of the manuscript for important intellectual content and statistical analysis. JKJ: obtained funding, microbiome analysis and critical revision of the manuscript for important intellectual content. SEC: RNA sequencing analysis and interpretation and critical revision of the manuscript for important intellectual content. XPZ: study concept and design, interpretation of data and critical revision of the manuscript for important intellectual content. J-HM and AMS: obtained funding, study concept and design, study supervision, acquisition of data, drafting of the manuscript, critical revision of the manuscript for important intellectual content and statistical analysis. All authors had access to the study data and reviewed and approved the final manuscript.
Funding: This work was primarily supported by Lawrence Berkeley National Laboratory Directed Research and Development programme funding (J-HM and AMS) and the Microbes to Biomes (M2B) Initiative (SEC, J-HM and AMS) under contract DE AC02-05CH11231. Additional support was provided by the Office of Naval Research under ONR contract N0001415IP00021 (JKJ, J-HM and AMS), by the National Natural Science Foundation of China (No 81802388) and the Natural Science Foundation from the Department of Science & Technology of Jiangsu Province (No. BK20180120) (PW) and by NIH training grant T32 OD011083 (KLW).
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: RNA sequencing data have been submitted to the Sequence Read Archive under accession number SUB3887143. 16S rRNA amplicon sequencing data is available on OSF (https://osf.io/9tycg/).
Patient consent for publication: Not required.
References
- 1. Balmain A. Cancer as a complex genetic trait: tumor susceptibility in humans and mouse models. Cell 2002;108:145–52. [DOI] [PubMed] [Google Scholar]
- 2. Simonds NI, Ghazarian AA, Pimentel CB, et al. Review of the Gene-Environment Interaction Literature in Cancer: What Do We Know? Genet Epidemiol 2016;40:356–65. 10.1002/gepi.21967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sud A, Kinnersley B, Houlston RS. Genome-wide association studies of cancer: current insights and future perspectives. Nat Rev Cancer 2017;17:692–704. 10.1038/nrc.2017.82 [DOI] [PubMed] [Google Scholar]
- 4. Witte JS, Visscher PM, Wray NR. The contribution of genetic variants to disease depends on the ruler. Nat Rev Genet 2014;15:765–76. 10.1038/nrg3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hunter KW. Mouse models of cancer: does the strain matter? Nat Rev Cancer 2012;12:144–9. 10.1038/nrc3206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Threadgill DW, Miller DR, Churchill GA, et al. The collaborative cross: a recombinant inbred mouse population for the systems genetic era. Ilar J 2011;52:24–31. 10.1093/ilar.52.1.24 [DOI] [PubMed] [Google Scholar]
- 7. Roberts A, Pardo-Manuel de Villena F, Wang W, et al. The polymorphism architecture of mouse genetic resources elucidated using genome-wide resequencing data: implications for QTL discovery and systems genetics. Mamm Genome 2007;18:473–81. 10.1007/s00335-007-9045-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mao JH, Langley SA, Huang Y, et al. Identification of genetic factors that modify motor performance and body weight using Collaborative Cross mice. Sci Rep 2015;5:16247 10.1038/srep16247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Snijders AM, Langley SA, Kim YM, et al. Influence of early life exposure, host genetics and diet on the mouse gut microbiome and metabolome. Nat Microbiol 2016;2:16221 10.1038/nmicrobiol.2016.221 [DOI] [PubMed] [Google Scholar]
- 10. Rogala AR, Morgan AP, Christensen AM, et al. The Collaborative Cross as a resource for modeling human disease: CC011/Unc, a new mouse model for spontaneous colitis. Mamm Genome 2014;25:95–108. 10.1007/s00335-013-9499-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Weidner P, Söhn M, Gutting T, et al. Myotubularin-related protein 7 inhibits insulin signaling in colorectal cancer. Oncotarget 2016;7:50490–506. 10.18632/oncotarget.10466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Raja UM, Gopal G, Shirley S, et al. Immunohistochemical expression and localization of cytokines/chemokines/growth factors in gastric cancer. Cytokine 2017;89:82–90. 10.1016/j.cyto.2016.08.032 [DOI] [PubMed] [Google Scholar]
- 13. Marshall KW, Mohr S, Khettabi FE, et al. A blood-based biomarker panel for stratifying current risk for colorectal cancer. Int J Cancer 2010;126:1177–86. 10.1002/ijc.24910 [DOI] [PubMed] [Google Scholar]
- 14. Gao C, Pang M, Zhou Z, et al. Epidermal growth factor receptor-coamplified and overexpressed protein (VOPP1) is a putative oncogene in gastric cancer. Clin Exp Med 2015;15:469–75. 10.1007/s10238-014-0320-7 [DOI] [PubMed] [Google Scholar]
- 15. Furukawa T, Kuboki Y, Tanji E, et al. Whole-exome sequencing uncovers frequent GNAS mutations in intraductal papillary mucinous neoplasms of the pancreas. Sci Rep 2011;1:161 10.1038/srep00161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Arigami T, Uenosono Y, Ishigami S, et al. Clinical significance of the B7-H4 coregulatory molecule as a novel prognostic marker in gastric cancer. World J Surg 2011;35:2051–7. 10.1007/s00268-011-1186-4 [DOI] [PubMed] [Google Scholar]
- 17. Zuo E, Lu Y, Yan M, et al. Increased expression of hepcidin and associated upregulation of JAK/STAT3 signaling in human gastric cancer. Oncol Lett 2018;15:2236–44. 10.3892/ol.2017.7574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Katz JP, Perreault N, Goldstein BG, et al. Loss of Klf4 in mice causes altered proliferation and differentiation and precancerous changes in the adult stomach. Gastroenterology 2005;128:935–45. 10.1053/j.gastro.2005.02.022 [DOI] [PubMed] [Google Scholar]
- 19. Snijders AM, Hermsen MA, Baughman J, et al. Acquired genomic aberrations associated with methotrexate resistance vary with background genomic instability. Genes Chromosomes Cancer 2008;47:71–83. 10.1002/gcc.20509 [DOI] [PubMed] [Google Scholar]
- 20. Szász AM, Lánczky A, Nagy Á, et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget 2016;7:49322–33. 10.18632/oncotarget.10337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aguirre-Gamboa R, Gomez-Rueda H, Martínez-Ledesma E, et al. SurvExpress: an online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS One 2013;8:e74250 10.1371/journal.pone.0074250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Torre LA, Siegel RL, Ward EM, et al. Global cancer incidence and mortality rates and trends–an update. Cancer Epidemiol Biomarkers Prev 2016;25:16–27. 10.1158/1055-9965.EPI-15-0578 [DOI] [PubMed] [Google Scholar]
- 23. Yu S, Yang M, Nam KT. Mouse models of gastric carcinogenesis. J Gastric Cancer 2014;14:67–86. 10.5230/jgc.2014.14.2.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yamamoto M, Furihata C, Ogiu T, et al. Independent variation in susceptibilities of six different mouse strains to induction of pepsinogen-altered pyloric glands and gastric tumor intestinalization by N-methyl-N-nitrosourea. Cancer Lett 2002;179:121–32. 10.1016/S0304-3835(02)00013-7 [DOI] [PubMed] [Google Scholar]
- 25. Tomita H, Takaishi S, Menheniott TR, et al. Inhibition of gastric carcinogenesis by the hormone gastrin is mediated by suppression of TFF1 epigenetic silencing. Gastroenterology 2011;140:879–91. 10.1053/j.gastro.2010.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang TC, Koh TJ, Varro A, et al. Processing and proliferative effects of human progastrin in transgenic mice. J Clin Invest 1996;98:1918–29. 10.1172/JCI118993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fox JG, Rogers AB, Ihrig M, et al. Helicobacter pylori-associated gastric cancer in INS-GAS mice is gender specific. Cancer Res 2003;63:942–50. [PubMed] [Google Scholar]
- 28. Lefebvre O, Chenard MP, Masson R, et al. Gastric mucosa abnormalities and tumorigenesis in mice lacking the pS2 trefoil protein. Science 1996;274:259–62. 10.1126/science.274.5285.259 [DOI] [PubMed] [Google Scholar]
- 29. Howe JR, Sayed MG, Ahmed AF, et al. The prevalence of MADH4 and BMPR1A mutations in juvenile polyposis and absence of BMPR2, BMPR1B, and ACVR1 mutations. J Med Genet 2004;41:484–91. 10.1136/jmg.2004.018598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kawase S, Ishikura H. Female-predominant occurrence of spontaneous gastric adenocarcinoma in cotton rats. Lab Anim Sci 1995;45:244–8. [PubMed] [Google Scholar]
- 31. Court M, Robinson PA, Dixon MF, et al. The effect of gender on Helicobacter felis-mediated gastritis, epithelial cell proliferation, and apoptosis in the mouse model. J Pathol 2003;201:303–11. 10.1002/path.1422 [DOI] [PubMed] [Google Scholar]
- 32. Lertpiriyapong K, Whary MT, Muthupalani S, et al. Gastric colonisation with a restricted commensal microbiota replicates the promotion of neoplastic lesions by diverse intestinal microbiota in the Helicobacter pylori INS-GAS mouse model of gastric carcinogenesis. Gut 2014;63:54–63. 10.1136/gutjnl-2013-305178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ohtani M, García A, Rogers AB, et al. Protective role of 17 beta -estradiol against the development of Helicobacter pylori-induced gastric cancer in INS-GAS mice. Carcinogenesis 2007;28:2597–604. 10.1093/carcin/bgm150 [DOI] [PubMed] [Google Scholar]
- 34. Ohtani M, Ge Z, García A, et al. 17 β-estradiol suppresses Helicobacter pylori-induced gastric pathology in male hypergastrinemic INS-GAS mice. Carcinogenesis 2011;32:1244–50. 10.1093/carcin/bgr072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Crawford NP, Hunter KW. New perspectives on hereditary influences in metastatic progression. Trends Genet 2006;22:555–61. 10.1016/j.tig.2006.07.009 [DOI] [PubMed] [Google Scholar]
- 36. Yang H, Crawford N, Lukes L, et al. Metastasis predictive signature profiles pre-exist in normal tissues. Clin Exp Metastasis 2005;22:593–603. 10.1007/s10585-005-6244-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ritchie KJ, Zhang DE. ISG15: the immunological kin of ubiquitin. Semin Cell Dev Biol 2004;15:237–46. 10.1016/j.semcdb.2003.12.005 [DOI] [PubMed] [Google Scholar]
- 38. Zuo C, Sheng X, Ma M, et al. ISG15 in the tumorigenesis and treatment of cancer: An emerging role in malignancies of the digestive system. Oncotarget 2016;7:74393–409. 10.18632/oncotarget.11911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Snijders AM, Langley S, Mao JH, et al. An interferon signature identified by RNA-sequencing of mammary tissues varies across the estrous cycle and is predictive of metastasis-free survival. Oncotarget 2014;5:4011–25. 10.18632/oncotarget.2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Frick VO, Rubie C, Keilholz U, et al. Chemokine/chemokine receptor pair CCL20/CCR6 in human colorectal malignancy: an overview. World J Gastroenterol 2016;22:833–41. 10.3748/wjg.v22.i2.833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wu YY, Tsai HF, Lin WC, et al. Upregulation of CCL20 and recruitment of CCR6+ gastric infiltrating lymphocytes in Helicobacter pylori gastritis. Infect Immun 2007;75:4357–63. 10.1128/IAI.01660-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li D, Wu C, Cai Y, et al. Association of NFKB1 and NFKBIA gene polymorphisms with susceptibility of gastric cancer. Tumour Biol 2017;39:101042831771710 10.1177/1010428317717107 [DOI] [PubMed] [Google Scholar]
- 43. O’Reilly LA, Putoczki TL, Mielke LA, et al. Loss of NF-κB1 Causes Gastric Cancer with Aberrant Inflammation and Expression of Immune Checkpoint Regulators in a STAT-1-Dependent Manner. Immunity 2018;48:570–83. 10.1016/j.immuni.2018.03.003 [DOI] [PubMed] [Google Scholar]
- 44. Welsh CE, Miller DR, Manly KF, et al. Status and access to the Collaborative Cross population. Mamm Genome 2012;23:706–12. 10.1007/s00335-012-9410-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Konganti K, Ehrlich A, Rusyn I, et al. gQTL: A Web Application for QTL Analysis Using the Collaborative Cross Mouse Genetic Reference Population. G3 2018;8:2559–62. 10.1534/g3.118.200230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–4. 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2018-316691supp001.xlsx (18.1KB, xlsx)
gutjnl-2018-316691supp002.jpg (2.8MB, jpg)
gutjnl-2018-316691supp003.jpg (2.2MB, jpg)
gutjnl-2018-316691supp004.jpg (4.5MB, jpg)
gutjnl-2018-316691supp005.jpg (3.6MB, jpg)
gutjnl-2018-316691supp006.jpg (747.8KB, jpg)
gutjnl-2018-316691supp007.xlsx (162.2KB, xlsx)
gutjnl-2018-316691supp008.jpg (2.3MB, jpg)
gutjnl-2018-316691supp009.jpg (5.5MB, jpg)
gutjnl-2018-316691supp010.xlsx (70.9KB, xlsx)
gutjnl-2018-316691supp011.jpg (439.3KB, jpg)
gutjnl-2018-316691supp012.xlsx (13KB, xlsx)
gutjnl-2018-316691supp013.xlsx (15.7KB, xlsx)
gutjnl-2018-316691supp014.xlsx (31.6KB, xlsx)
gutjnl-2018-316691supp015.xlsx (13.9KB, xlsx)
gutjnl-2018-316691supp016.jpg (885.8KB, jpg)
gutjnl-2018-316691supp017.pdf (99.9KB, pdf)