We read with interest the work by Jaeggi et al 1 and Paganinni et al 2 and commend their efforts. Despite differences in iron concentration, infants’ age and sequencing techniques, both studies demonstrate unfavourable iron effects on gut microbiota with decreased abundance of bifidobacteria and lactobacillus, and increased abundance of pathogenic bacteria in iron-deficient/anaemic Kenyan infants.
We have investigated changes in gut microbial composition due to iron fortification or supplementation in healthy, Swedish infants. Iron-sufficient infants at 6 months of age were randomly allocated to receive low-iron-fortified formula (1.2 mg Fe/day; n=24), high-iron-fortified formula (6.6 mg Fe/day; n=24) or no-added-iron formula with liquid ferrous sulfate supplementation (iron drops; 6.6 mg Fe/day; n=24) for 45 days. All participants gave their informed consent before inclusion through parents or legal guardians. Total iron intake was 1.2, 6.4 and 5.7 mg/day (all differences p<0.01) in the low-iron, high-iron and iron-drops group, respectively. Stool samples were collected before and after the intervention. We applied 16S rRNA gene amplicon sequencing of the V3–V4 region to profile the gut microbiome using Illumina MiSeq. We used QIIME3 to assess composition and diversity of gut microbiota and the DESeq2 package4 to investigate differences in relative abundance of gut bacteria among the groups. PICRUSt was used to predict metagenome functional content.5
Vaginally delivered infants (n=53) with paired stool samples were included in the analyses. There were no significant differences in anthropometrics or iron-related biomarkers among the randomisation groups; no adverse effects were reported (diarrhoea, increased rates of infections, other illnesses, etc), and growth was not affected (table 1).6
Table 1.
Baseline characteristics of the study participants and anthropometric and biochemical values at the 45-day follow-up.
| Low-iron formula | High-iron formula | Fe drops | |
| Participants (n) | 18 | 18 | 17 |
| Girls (n) | 7 | 9 | 11 |
| Birth weight (g)* | 3717±560 | 3548±425 | 3800±436 |
| Birth length (cm)* | 51.1±2.2 | 50.2±1.6 | 51.7±1.7 |
| Age at inclusion (months)* | 6.1±0.3 | 6.1±0.2 | 6.1±0.3 |
| Baseline | Follow-up | P values† | Baseline | Follow-up | P values† | Baseline | Follow-up | P values† | P values‡ | |
| Weight (kg)* | 8.3±1.0 | 9.1±1.1 | <0.001 | 8.0±1.2 | 8.8±1.1 | <0.001 | 8.4±0.9 | 9.2±0.9 | <0.001 | 0.49 |
| Length (cm)* | 68.4±2.4 | 71.3±2.7 | <0.001 | 67.4±2.8 | 69.9±2.6 | <0.001 | 68.2±2.3 | 71.7±3.9 | <0.001 | 0.26 |
| Hb (g/L)* | 111.6±6.0 | 110.2±9.0 | 0.71 | 112.2±7.0 | 112.9±5.9 | 0.62 | 118.0±11.5 | 112.2±5.8 | 0.06 | 0.51 |
| S-Fe (µmol/L)* | 9.5±4.2 | 9.5±4.3 | 0.66 | 9.7±3.8 | 8.7±3.6 | 0.42 | 8.8±4.5 | 9.6±3.6 | 0.64 | 0.78 |
| S-transferrin (g/L)* | 2.2±0.3 | 2.4±0.4 | 0.07 | 2.2±0.3 | 2.2±0.3 | 0.66 | 2.3±0.4 | 2.2±0.2 | 0.70 | 0.32 |
| S-ferritin (µg/L)§ | 89.4±44.7 | 61.2±32.5 | <0.001 | 72.3±40.7 | 70.5±47.0 | 0.81 | 109.3±85.8 | 92.2±62.9 | 0.14 | 0.17 |
| F-calprotectin (µg/g)¶ | 132 (71, 241) | 121 (55, 211) | NS** | 120 (59, 238) | 105 (62, 421) | NS** | 263 (104, 345) | 151 (109, 492) | NS** | NS††** |
Data are mean/geometric mean±SD or median (25th, 75th percentile).
*Mean ±SD.
†P values for within-group differences, paired-samples t-test.
‡P values for between-group differences, ANOVA.
§Geometric mean ±SD.
¶Median (25th, 75th percentile).
**P values for within-group differences, related-samples Wilcoxon signed-rank test.
††P values for between-group difference, independent-samples Kruskal-Wallis test.
F, faecal; Hb, haemoglobin; NS, not significant at p=0.05; S, serum.
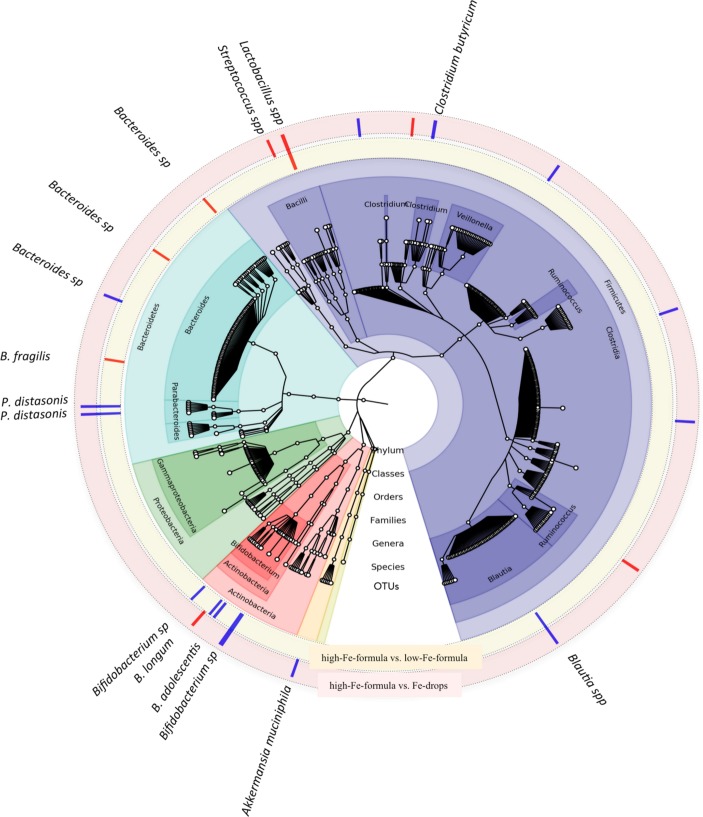
In this study, we confirm findings that consumption of high-iron formula is associated with decreased relative abundance of Bifidobacterium (p<0.001, 60% vs 78%) after only 45 days of intervention, but we did not detect enhanced growth of pathogenic bacteria. However, we were able to partly confirm previous findings regarding abundance of lactobacilli due to iron consumption. We found lower relative abundance of Lactobacillus sp (p<0.007, 8% vs 42%) in infants who received iron drops versus high-iron-formula group. Unexpectedly, we also found higher relative abundance of Lactobacillus sp (p<0.0002, 42% vs 32%) in high-iron compared with low-iron formula group; this result challenges the hypothesis that the mode of iron administration has a direct effect on lactobacilli colonisation in the gut. Furthermore, the iron-drops group had lower abundance of Streptococcus (p<0.0003, 0.2% vs 0.9%) but higher abundance of Clostridium (p<0.05, 25% vs 9%) and Bacteroides (p<0.02, 1.2% vs 0.9%) compared with the high-iron formula group (figure 1). In the present study, all groups received formula with added galacto-oligosaccharides (GOS) at 3.3 g/L. This prebiotic may mitigate adverse effects of iron fortification on gut microbiota,2 but in the iron-drops group, iron was administered apart from the formula meals. Thus, we cannot exclude a possible protective effect of GOS on the gut microbiota of infants in our study.
Figure 1.
Differences in gut bacterial composition depend on the concentration and administration mode of the consumed iron. In the cladogram, showing the results of the microbiome analysis over time, taxa are grouped on the basis of synapomorphy. The outermost small, white circles represent the 561 OTUs (operational taxonomic units). Differences in gut microbial composition between the high-Fe-formula group versus the low-Fe-formula group over time are presented in the yellow component around the cladogram, where blue bars represent lower relative abundance of bacteria in the high-Fe-formula group compared with the low-Fe-formula group and the red bars represent higher relative abundance in the high-Fe-formula group compared with the low-Fe-formula group, respectively. Differences in gut microbial composition between the high-Fe-formula group versus the Fe-drops group over time are presented in the red component around the cladogram, where the blue bars represent lower relative abundance of bacteria in the high-Fe-formula group and the red bars represent higher relative abundance in the high-Fe-formula group compared with the Fe-drops group, respectively. OTU, operational taxonomic unit.
As in the study by Paganinni et al,2 faecal calprotectin did not differ between the groups (table 1), but in our study, it correlated positively with Clostridium difficile in high-iron-formula (rSpearman=0.4, p<0.01) and iron-drops intervention groups (rSpearman=0.48, p<0.004). The bacterial function pathway related to Staphylococcus aureus infection (KEGG module 05150)5 was significantly lower in the iron-drops group compared with the low-iron-formula group (p=0.027). This is a novel finding which suggests that changes in bacterial composition due to administration of iron drops may reduce the protective response of the gut microbiota to bacterial infections. Nevertheless, no effects on the health of the participants were seen due to this.
To summarise, in healthy, non-anaemic Swedish infants, consumption of high-iron formula is associated with significantly lower abundance of bifidobacteria compared with low-iron formula, and administration of iron as drops, even in a dose comparable with the daily iron requirement and for a short time, leads to decreased relative abundance of lactobacilli and potentially increases susceptibility to bacterial infection.
Acknowledgments
We thank the families who participated in the study, research nurses Åsa Sundström and Camilla Steinvall Lindberg who helped during enrolment and data collection, and Stina Bäckman (FOI) for excellent assistance during the Illumina MiSeq run. We thank Richard Hurrell for constructive criticism of the manuscript.
Footnotes
Contributors: EAS-G, MD, OH, BL and TL designed the original study and EAS-G, MD, OH and TL conducted the study. TL was involved in planning of the study, analyses and interpretation of the data. KSS planned and performed laboratory work, analysed and interpreted data, and wrote the first draft of the manuscript. CL performed laboratory work and wrote the section on subjects and methods for the manuscript. AS assisted with bioinformatics, interpretation and visualisation of the data. CEW contributed to the discussion and provided intellectual input. All authors have read, provided critical comments and approved the final version of the letter.
Funding: This project was supported by the Umeå University Foundation for Medical Research and the regional agreement between Umeå University and Västerbotten County Council on cooperation in the fields of medicine, odontology and health (ALF). Semper AB, Sweden, generously provided the study formulae and the fruit-based and vegetable-based infant foods.
Disclaimer: The funding bodies had no role in designing or conducting the study, in the collection, management, analysis or interpretation of the data and had no input into the preparation, review or approval of the manuscript.
Competing interests: OH and BL are members of the Scientific Advisory Board of Semper and Hero. MD, OH, TL and BL have received research support from Semper and Hero unrelated to the present study. KSS, CL, AS, EAS-G and CEW declare no conflicts of interest related to the study.
Patient consent: Parental/guardian consent obtained.
Ethics approval: The Regional Ethical Review Board at Umeå University (Dnr 2010-213-31M).
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1. Jaeggi T, Kortman GA, Moretti D, et al. Iron fortification adversely affects the gut microbiome, increases pathogen abundance and induces intestinal inflammation in Kenyan infants. Gut 2015;64:731–42. 10.1136/gutjnl-2014-307720 [DOI] [PubMed] [Google Scholar]
- 2. Paganini D, Uyoga MA, Kortman GAM, et al. Prebiotic galacto-oligosaccharides mitigate the adverse effects of iron fortification on the gut microbiome: a randomised controlled study in Kenyan infants. Gut 2017;66:1956–67. 10.1136/gutjnl-2017-314418 [DOI] [PubMed] [Google Scholar]
- 3. Navas-Molina JA, Peralta-Sánchez JM, González A, et al. Advancing our understanding of the human microbiome using QIIME. Methods Enzymol 2013;531:371–444. 10.1016/B978-0-12-407863-5.00019-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Langille MG, Zaneveld J, Caporaso JG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 2013;31:814–21. 10.1038/nbt.2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Szymlek-Gay EA, Domellöf M, Hernell O, et al. Mode of oral iron administration and the amount of iron habitually consumed do not affect iron absorption, systemic iron utilisation or zinc absorption in iron-sufficient infants: a randomised trial. Br J Nutr 2016;116:1046–60. 10.1017/S0007114516003032 [DOI] [PubMed] [Google Scholar]