The apo-to-holo transition of an enzyme allows insights into the structural hinges that accommodate different ligands within its active site. Here, such a hinge is elucidated in Toxoplasma gondii prolyl-tRNA synthetase and shown in its different possible conformations in two apo forms and two inhibitor-bound forms.
Keywords: toxoplasmosis, prolyl-tRNA synthetase, enzyme–inhibitor complexes, comparative crystallography, drug design, r.m.s.d. per residue profile, apo–holo transistions
Abstract
Prolyl-tRNA synthetase (PRS) is a member of the aminoacyl-tRNA synthetase family that drives protein translation in cells. The apicomplexan PRSs are validated targets of febrifugine (FF) and its halogenated derivative halofuginone (HF). PRSs are of great interest for drug development against Plasmodium falciparum and Toxoplasma gondii. In this study, structures of apo and FF-bound T. gondii (TgPRS) are revealed and the dynamic nature of the conformational changes that occur upon FF binding is unraveled. In addition, this study highlights significant conformational plasticity within two different crystal structures of apo PRSs but not within drug-bound PRSs. The apo PRSs exist in multi-conformational states and manifest pseudo-dimeric structures. In contrast, when FF is bound the PRS dimer adopts a highly symmetrical architecture. It is shown that TgPRS does not display extant fold switching, in contrast to P. falciparum PRS, despite having over 65% sequence identity. Finally, structure-comparison analyses suggest the utility of r.m.s.d. per residue (r.m.s.d./res) as a robust tool to detect structural alterations even when the r.m.s.d. is low. Apo TgPRS reveals FF/HF-induced rigidity and this work has implications for drug-design studies that rely on the apo structures of target proteins.
1. Introduction
The Chinese herb Dichroa febrifuga has been used to treat malaria for many centuries (Koepfli et al., 1947 ▸). The active compound in the herb was discovered to be a quinazolinone-type alkaloid later named febrifugine (FF; Coatney et al., 1950 ▸). As FF is highly toxic, a less toxic derivative of FF called halofuginone (HF) was developed (Hewitt et al., 1952 ▸). The inhibitory target of FF and HF was later determined to be prolyl-tRNA synthetase (PRS), a member of the aminoacyl-tRNA synthetase (aaRS) family (Kikuchi et al., 2006 ▸; Keller et al., 2012 ▸; Son et al., 2013 ▸); Zhou et al., 2013 ▸; Jain et al., 2014 ▸, 2015 ▸; Jain, Yogavel et al., 2017 ▸). Parasite aaRSs, i.e. aaRSs that are encoded in the genome of parasites for protein translation, are being studied as valuable targets in multiple pathogenic organisms that cause malaria, leishmaniasis, toxoplasmosis, cryptosporidiosis and coccidiosis (Bhatt et al., 2009 ▸; Jackson et al., 2011 ▸; Khan et al., 2011 ▸, 2013 ▸, 2014 ▸; Hoepfner et al., 2012 ▸; Gowri et al., 2012 ▸; Koh et al., 2012 ▸, 2013 ▸; Hoen et al., 2013 ▸; Pasaje et al., 2016 ▸; Jain et al., 2014 ▸, 2015 ▸; Pham et al., 2014 ▸; Hussain et al., 2015 ▸; Herman et al., 2015 ▸; Kato et al., 2016 ▸; Sonoiki et al., 2016 ▸; Sharma et al., 2016 ▸; Palencia et al., 2016 ▸; Jain, Yogavel et al., 2017 ▸; Jain, Sharma et al., 2017 ▸; Das et al., 2018 ▸; Manickam et al., 2018 ▸; Nachiappan et al., 2018 ▸). Toxoplasma gondii is an obligate intracellular parasite that is the cause of toxoplasmosis. We previously solved the crystal structure of HF-bound T. gondii PRS (TgPRS) at ∼2.2 Å resolution and showed that HF inhibited T. gondii with an EC50 of 0.94 nM, while FF did so with an EC50 of ∼4 nM (Jain et al., 2014 ▸, 2015 ▸; Jain, Yogavel et al., 2017 ▸). HF acts by mimicking the PRS substrates l-proline and the adenine moiety of the CCA arm of tRNA; it thus occupies two substrate-binding pockets (Kikuchi et al., 2006 ▸; Keller et al., 2012 ▸; Son et al., 2013 ▸; Zhou et al., 2013 ▸; Jain et al., 2014 ▸, 2015 ▸; Jain, Yogavel et al., 2017 ▸).
TgPRS is a homodimeric enzyme whose final product is a tRNA molecule charged with its cognate amino acid proline (Jain et al., 2015 ▸; Jain, Yogavel et al., 2017 ▸; Jain, Sharma et al., 2017 ▸). Here, we have solved two crystal structures of TgPRS in the apo state and in the FF-bound state. We provide an analysis of the conformational heterogeneity observed within two distinct apo TgPRS structures. We also reveal substantial conformational changes that occur upon FF binding in TgPRS. Conformationally heterogeneous landscapes in aaRS proteins have been studied previously in the context of the KMSKS loop of tyrosine-tRNA synthetases (Datt & Sharma, 2014 ▸). Relatedly, between 0.5% and 4% of proteins are known to exhibit extant fold-switching (Porter & Looger, 2018 ▸). Extant fold-switching occurs as a response to cellular stimuli that include, but are not limited to, the binding of cofactors, ligands or drugs, their concentrations, pH shifts or functional necessity (Porter & Looger, 2018 ▸). In particular, class D fold-switching is defined as when the oligomeric states of the apo and the holo forms are identical but the corresponding constituent secondary-structure elements differ. The apicomplexan PRS from Plasmodium falciparum has been shown to belong to the class D fold-switchers (Jain et al., 2014 ▸, 2015 ▸; Jain, Yogavel et al., 2017 ▸; Porter & Looger, 2018 ▸). Our current work provides new insights into the extant fold-switching phenomenon, as we observe no evidence for this in TgPRS (Porter & Looger, 2018 ▸). This is surprising as T. gondii and P. falciparum are highly related apicomplexan parasites and the two PRSs share ∼64.6% sequence identity.
We observed high symmetry within monomeric PRS protomers when bound to FF, but not in monomeric apo PRSs. Our data provide evidence for ligand-induced fit as well as for drug-induced rigidity in TgPRS when bound to FF. Structural comparisons of the apo and FF-bound structures suggest that induced fit and conformational selection, which are two widely accepted mechanisms for protein–ligand interactions, now regarded as within the extended conformational selection model, are manifest in TgPRS. Finally, structure-comparison analyses based on the r.m.s.d. per residue (r.m.s.d./res) show that this tool is more robust for revealing structural alterations than the use of the r.m.s.d. alone.
2. Methods
2.1. Protein purification, crystallization and structure determination
Purification and crystallization of TgPRS was performed in accordance with previously published methods (Jain et al., 2014 ▸, 2015 ▸; Jain, Yogavel et al., 2017 ▸). Diffraction-quality crystals of apo TgPRS were obtained in Morpheus (Molecular Dimensions) condition D3, while crystals of the TgPRS–FF–AMPPNP complex grew in Morpheus II (Molecular Dimensions) condition E9. Diffraction data were collected on the PROXIMA 1 and 2A beamlines at Synchrotron SOLEIL, France. A total of 3600 frames were collected in 0.1° oscillation steps with 0.1 s exposure per frame. The data were processed and scaled using DIALS (Winter et al., 2018 ▸) and the relevant statistics are summarized in Table 1 ▸. The structures were solved by molecular replacement, with apo TgPRS (PDB entry 5xif; Jain, Yogavel et al., 2017 ▸) as a search model for the apo structure. The structure of TgPRS–HF–AMPPNP (PDB entry 5xiq; Jain, Yogavel et al., 2017 ▸) was used as a search model for TgPRS–FF–AMPPNP. All structures were solved with Phenix (Adams et al., 2010 ▸). X-ray refinement restraint parameters were generated for the ligand febrifugine using the grade web server (http://grade.globalphasing.org). These were subsequently refined using phenix.refine. NCS was not used during refinement, but TLS was. The atomic structures were subjected to refinement cycles using simulated annealing (Cartesian) for three cycles. This was used to remove model bias. After each step, the models were manually adjusted so there was coherence in 2F o − F c and F o − F c electron-density maps. Coot (Emsley et al., 2010 ▸) was used for all rebuilding. The presence of the ligands was confirmed using OMIT maps (Fig. 1 ▸). Model quality was assessed using the MolProbity server (Chen et al., 2010 ▸). Atomic cooordinates and structure factors have been deposited in the PDB under accession codes 6aa0 and 6a88.
Table 1. Summary of data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell.
| Apo TgPRS (PDB entry 6aa0) | TgPRS–FF–AMPPNP (PDB entry 6a88) | |
|---|---|---|
| Data collection | ||
| Beamline | PROXIMA 2A, SOLEIL | PROXIMA 1, SOLEIL |
| Wavelength (Å) | 0.980066 | 0.97857 |
| Detector type | EIGER X 9M | PILATUS 6M |
| Crystal-to-detector distance (mm) | 394.0 | 393.4 |
| Oscillation (°) | 0.1 | 0.1 |
| Exposure (s) | 0.1 | 0.1 |
| No. of images | 3600 | 3600 |
| Data-processing software | xia2/DIALS | xia2/DIALS |
| a, b, c (Å) | 113.0, 113.0, 119.8 | 76.9, 90.8, 93.0 |
| α, β, γ (°) | 90, 90, 120 | 89.9, 80.2, 75.5 |
| Space group | P31 | P1 |
| Resolution (Å) | 97.91–3.20 (3.28–3.20) | 42.83–2.60 (2.67–2.60) |
| R merge | 0.163 (0.847) | 0.108 (0.602) |
| R meas | 0.172 (0.89) | 0.128 (0.73) |
| No. of unique reflections | 47193 (3515) | 73015 (5337) |
| Completeness (%) | 100 (100) | 98.8 (98.1) |
| Multiplicity | 10.8 (10.7) | 3.4 (3.2) |
| 〈I/σ(I)〉 | 10.3 (2.7) | 7.6 (2.5) |
| CC1/2 | 0.996 (0.844) | 0.987 (0.675) |
| Refinement | ||
| Resolution | 69.9–3.0 (3.09–3.03) | 39.9–2.38 (2.40–2.38) |
| No. of reflections | ||
| Work set | 52197 (2611) | 90423 (2739) |
| Test set | 2782 (140) | 4608 (128) |
| R work/R free | 0.229/0.268 | 0.184/0.227 |
| Model quality | ||
| R.m.s.d., bond lengths (Å) | 0.005 | 0.007 |
| R.m.s.d., bond angles (°) | 0.649 | 0.719 |
| Ramachandran statistics | ||
| Favoured (%) | 92.77 | 96.96 |
| Allowed (%) | 5.94 | 2.78 |
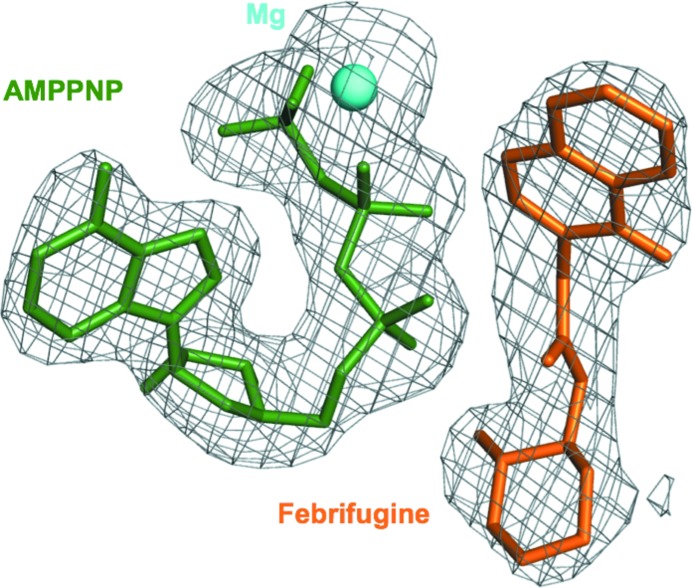
Figure 1.
Electron-density fit of FF and AMPPNP. A composite OMIT difference Fourier map (F o – F c) generated at 2.6 Å and contoured at 1.7σ showing bound FF (coral), AMPPNP (dark green) and the single Mg2+ ion (cyan).
2.2. Structural analyses
In this study, we first analyzed two distinct apo TgPRS structures: Apo1 (PDB entry 5xif) and Apo2 (PDB entry 6aa0). In addition, we studied the TgPRS–HF–AMPPNP co-crystal structure (Holo1; PDB entry 5xiq) and the TgPRS–FF–AMPPNP co-crystal structure (Holo2; PDB entry 6a88). All relevant space-group information is shown in Table 2 ▸. We attempted to decrease the bias and be as cautious as possible when analyzing the differences between the two apo structures. Apo1 crystallized in space group P1211, whereas the structure reported here, Apo2, crystallized in space group P31. The unit-cell dimensions have increased in Apo2 and this is most likely to be due to looser crystal packing in this case. Therefore, the analysis was based on the protomers in the biological unit rather than on the entire unit cell. Interestingly, however, the two holo structures both crystallized in space group P1 with equivalent unit-cell parameters. This in itself is further proof of the concept of the apo protein being highly flexible and developing drug-induced rigidity upon ligand interaction. A sequence alignment mapping structural differences (electron-density quality and conformational variability) was computed for Apo1 versus Apo2 and for Holo1 versus Holo2 (Supplementary Fig. S1). An alignment of Apo2 versus Holo2 was also computed in the same manner (Supplementary Fig. S2).
Table 2. Unit-cell parameters and space group along with R work/R free values and resolution information for all PDB entries compared in this study.
| TgPRS, Apo1 (PDB entry 5xif) | TgPRS, Apo2 (PDB entry 6aa0) | TgPRS–HF–AMPPNP, Holo1 (PDB entry 5xiq) | TgPRS–FF–AMPPNP, Holo2 (PDB entry 6a88) | |
|---|---|---|---|---|
| a, b, c (Å) | 76.7, 86.4, 97.2 | 113.0, 113.0, 119.8 | 76.9, 90.9, 92.9 | 76.9, 90.8, 93.0 |
| α, β, γ (°) | 90, 106.5, 90 | 90, 90, 120 | 89.9, 80.9, 75.8 | 89.9, 80.2, 75.5 |
| Space group | P1211 | P31 | P1 | P1 |
| Resolution† (Å) | 53.41–2.48 (2.57–2.48) | 97.91–3.20 (3.28–3.20) | 50.00–2.19 (2.24–2.19) | 42.83–2.60 (2.67–2.60) |
| R work/R free | 0.193/0.249 | 0.229/0.268 | 0.165/0.207 | 0.184/0.227 |
Values in paretheses are for the highest resolution shell.
Both dimers were compared for r.m.s.d. differences and the aggregate scores displayed a variability of 0.4–0.8 Å between them. Considering ∼1 Å to be a cutoff for biologically significant differences between two structures, it was deemed that what is true for the AB dimer also holds for the CD dimer. Therefore, only the AB chains are discussed as representative models of the structures.
We computed the r.m.s.d. for the protein backbone using the GESAMT tool from the CCP4 suite (Winn et al., 2011 ▸; Krissinel, 2012 ▸). This was performed for the eight chains from the four dimers, namely Apo1A, Apo1B, Apo2A, Apo2B, Holo1A, Holo1B, Holo2A and Holo2B. Furthermore, an r.m.s.d./res study was performed and a scatter plot was constructed. R.m.s.d./res values were computed using the structure-alignment tool in UCSF Chimera and are indicative of the conformational alterations that a particular residue undergoes.
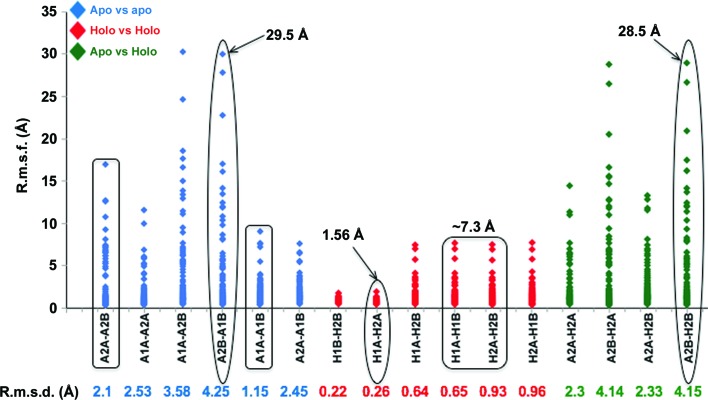
The backbone r.m.s.d. results within each cluster are arranged in a table: apo–apo in blue, holo–holo in red and apo–holo in green (Fig. 2 ▸ and Supplementary Fig. S3). The scatter plot for r.m.s.d./res is presented in the same order as for the r.m.s.d.s (Fig. 2 ▸ and Supplementary Fig. S3). Each dot in the graph depicts the r.m.s.d./res between the corresponding residues in the compared PDB pairs. Thus, the peaks in the histogram indicate higher relative displacement for any given residue pair.
Figure 2.
R.m.s.d. versus r.m.s.d./res analysis of Apo1/2 versus Holo1/2. R.m.s.d./res variance graphs of the combinations considered in r.m.s.d. analyses are shown. The highest data points representing drastic displacement of the corresponding residues in significant pairs are displayed. The rectangles cover the data sets pointing towards dimeric symmetry within the apo and holo enzymes, while ovals highlight further examined data sets. The corresponding r.m.s.d. values computed from GESAMT are noted below the graph. R.m.s.d./res is labelled r.m.s.f. (fluctuation per residue) to avoid confusion.
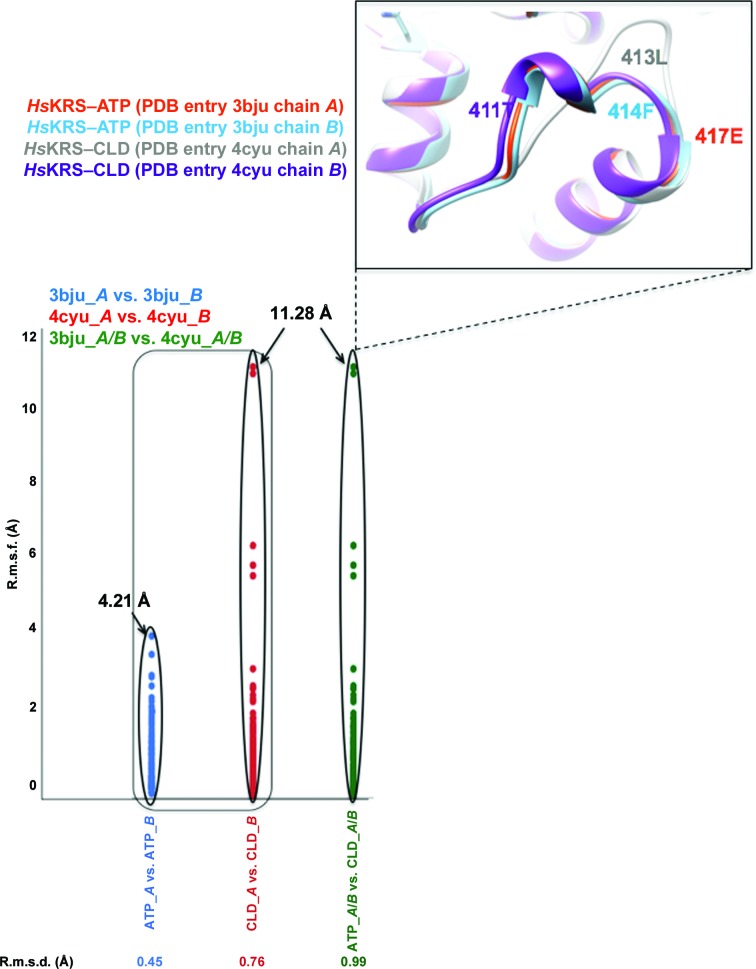
As a control for our r.m.s.d.–r.m.s.d./res study, we performed the same analyses on a pair of Homo sapiens lysyl-tRNA synthetase (HsKRS) structures bound to ATP (PDB entry 3bju) and to cladosporin (PDB entry 4ycu). Our observations here serve as a proof of concept that r.m.s.d./res is a superior tool to r.m.s.d. when comparing structures that may display small/large conformational changes. R.m.s.d. is the arithmetic mean of the cumulative Cα displacement values within the corresponding residues. R.m.s.d./res, on the other hand, is a better way of visualizing the differences between any two structures as the whole breadth of corresponding residue deviations are evident from such an analysis (Figs. 2 ▸ and 3 ▸).
Figure 3.
R.m.s.d. versus r.m.s.d./res analysis of ATP-bound and cladosporin (CLD)-bound forms of HsKRS (PDB entry 3bju versus PDB entry 4ycu). The rectangle covers data sets that point towards dimeric asymmetry within the ATP-bound and CLD-bound enzymes, while the ovals highlight the drastic differences observed between them. The corresponding r.m.s.d. values computed from GESAMT are noted below the graph. R.m.s.d./res is labelled r.m.s.f. (fluctuation per residue) to avoid confusion.
2.3. Fold-switching and mechanism of drug binding
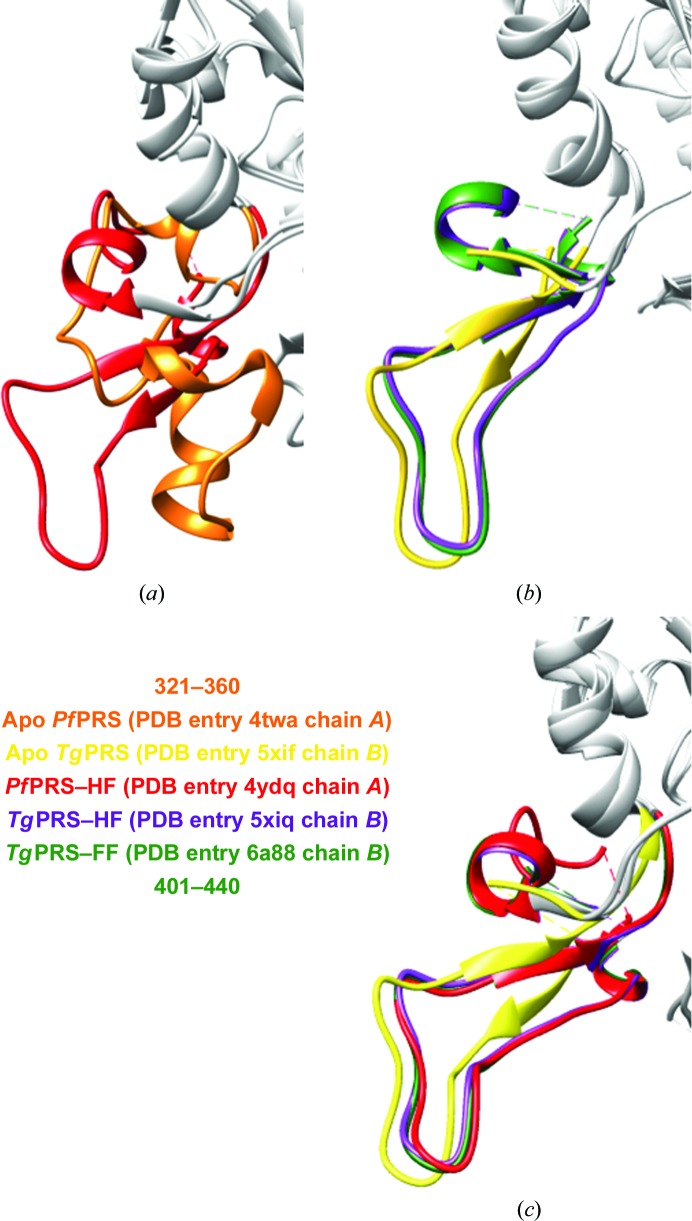
The apo and HF-bound complexes of P. falciparum PRS (PfPRS) have been attested to display fold-switching, which has been observed for related PRSs (Fig. 4 ▸ a; Porter & Looger, 2018 ▸). We therefore investigated this for apo and drug-bound structures of TgPRS (Fig. 4 ▸ b). Furthermore, we also analyzed the dimerization loop in the Holo1 and Holo2 dimers in the asymmetric unit. All figures were produced using UCSF Chimera (Pettersen et al., 2004 ▸)
Figure 4.
Evidence for fold-switching in PfPRS but not in TgPRS. (a) An overlay of monomers of apo PfPRS (PDB entry 4twa; orange) and PfPRS–HF (PDB entry 4ydq; red). The region that displays fold-switching (residues 321–360) is coloured and the rest of the backbone is shown in grey. (b) An overlay of monomers of apo TgPRS (PDB entry 5xif; lime yellow), TgPRS–HF (PDB entry 5xiq; purple) and TgPRS–FF (PDB entry 6a88; forest green). The corresponding residues to this region of PfPRS in TgPRS (residues 401–440) are coloured and the rest of the backbone ribbon is shown in grey. (c) The corresponding residues of apo TgPRS (PDB entry 5xif; lime yellow), PfPRS–HF (PDB entry 4ydq; red), TgPRS–HF (PDB entry 5xiq; purple) and TgPRS–FF (PDB entry 6a88; forest green) are almost entirely superimposable.
3. Results and discussion
In this study, the co-crystal structure of TgPRS with FF and adenylyl-imidodiphosphate (AMPPNP) (Holo2) was resolved to 2.6 Å resolution (Table 1 ▸; Fig. 5 ▸). A new apo TgPRS structure (Apo2) was also resolved to 3.2 Å resolution (Table 1 ▸). Direct conformational comparisons were performed with the help of the previously published apo TgPRS (Apo1; PDB entry 5xif) and TgPRS–HF (Holo1; PDB entry 5xiq) structures at 2.4 and 2.1 Å resolution, respectively (Jain, Yogavel et al., 2017 ▸). These four structures therefore provided us with a platform to assess any conformational changes upon drug (FF/HF) binding in TgPRS.
Figure 5.
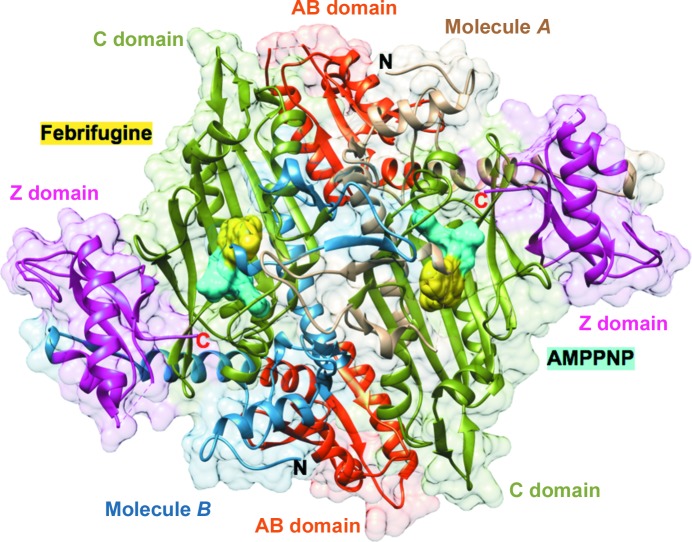
TgPRS–FF–AMPPNP in the asymmetric unit of the crystal. A dimer with bound FF (yellow) and AMPPNP (labeled ANP; cyan). The catalytic (C) domains (residues 434–602), anticodon-binding (AB) domains (residues 621–717) and C-terminal zinc-binding-like (Z) domains (residues 744–832) are labeled, as are the protein termini.
3.1. Apo1 versus Apo2
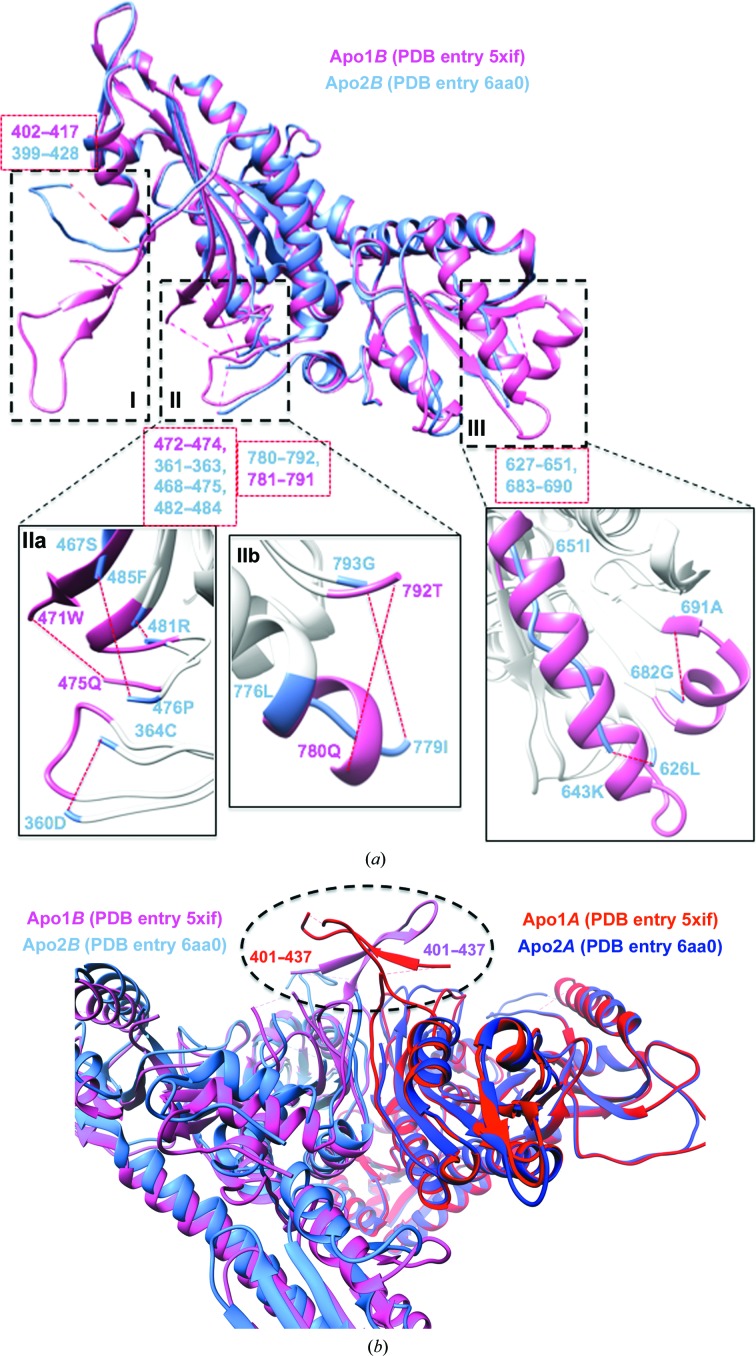
Firstly, we undertook a comparison of the Apo1 and Apo2 structures (Fig. 6 ▸), in which we observed two key points: the disordered sections in both generally overlap, suggesting higher degrees of freedom in some structural elements within the apoenzyme states and that conformational variability is a function that is more strongly expressed at the boundaries of disordered sections. The r.m.s.d.s of Apo1A versus Apo1B and of Apo2A versus Apo2B are of the order of ∼1.15 and ∼2.1 Å, respectively, indicating that the Apo2 dimer is possibly more asymmetric than the Apo1 dimer between the A and B monomers (Fig. 2 ▸, Supplementary Fig. S3). Thus, the apo TgPRSs appear to adopt pseudo-dimeric scaffolds. Our r.m.s.d./res analysis emphasizes this point as dynamics between the two different apo TgPRS structures and within each apo pseudo-dimer are evident (Fig. 2 ▸ and Supplementary Fig. S3). There are numerous structural differences between Apo1B and Apo2B: highly disordered regions and regions that adopt altered conformations (Fig. 6 ▸ a). The regions revealed to be highly plastic from the alignment include residues 399–437, 469–475, 627–651, 683–690 and 781–791 (Supplementary Fig. S1). Pro429 is strikingly mobile, with an r.m.s.d./res of ∼29 Å when Apo1B and Apo2B or Apo2B and Holo2B are compared. The sections that adopt altered conformations are 429–438 with an average r.m.s.d./res of 29.5 Å, 643–651 with an average r.m.s.d./res of 13.7 Å and 776–779 with an average r.m.s.d./res of 2.4 Å (Figs. 2 ▸ and 6 ▸ a and Supplementary Fig. S3). We observed conformational alterations from α-helix to coil-like at residues 643–651 and 776–779.
Figure 6.
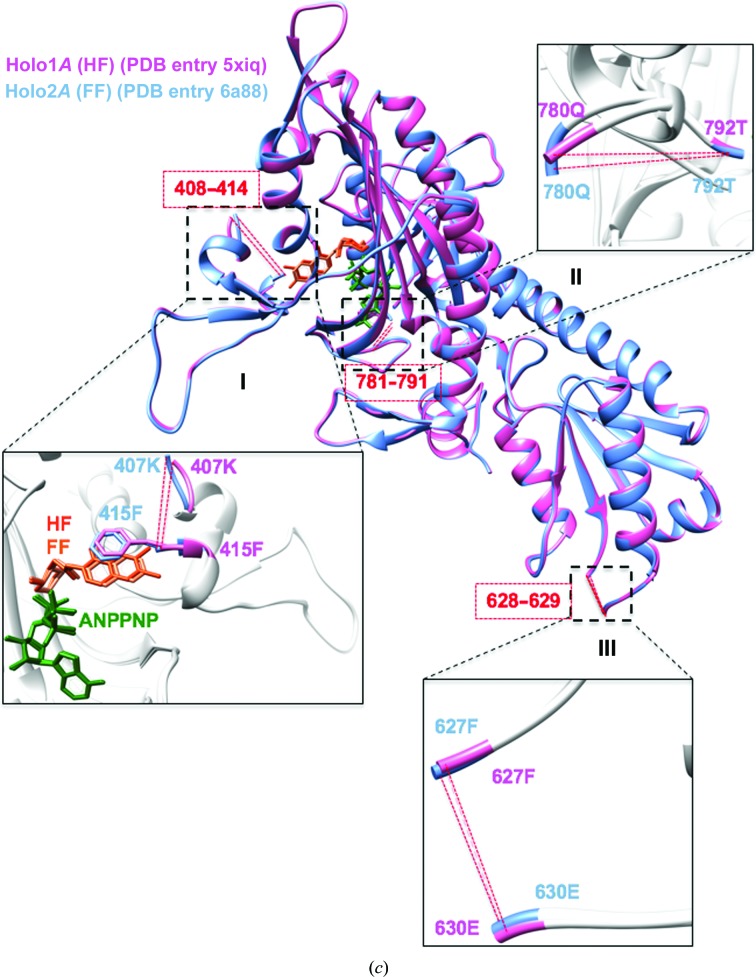
Structural overlays of apo and holo TgPRS. (a) Superposition of the Apo1B (PDB entry 5xif; hot pink) and Apo2B (PDB entry 6aa0; cornflower blue) TgPRS structures with structural differences highlighted. Close-up views of the key differences are shown in I, II and III. The missing residues in Apo1B are in regions I (402–417), IIa (472–474) and IIb (781–791). In contrasting, Apo2B has missing residues in regions I (399–428), IIa (361–363, 468–475 and 482–484), IIb (780–792) and III (627–651 and 683–690). (b) Structural comparison of the dimerization hinge among apo TgPRS structures. The dimerization regions (401–437) are ordered in Apo1 but are missing in Apo2. Apo1A, red; Apo1B, hot pink; Apo2A, purple; Apo2B, cornflower blue. (c) (c) Structural overlay of the HF-complexed Holo1A (PDB entry 5xiq; hot pink) and FF-complexed Holo2A (PDB entry 6aa8; cornflower blue) TgPRS structures with structural similarities highlighted. The alignment is divided into three sections: I, II and III. Missing elements (408–414, 628–629 and 781–791) are shown in red.
The solvent content of Apo1 is 53.84% and that of Apo2 is 62.4%. Indeed, it is likely that the conformational differences arise owing to these solvent-content differences, although no discernible influence of crystal packing could be attributed to the conformational differences. Thus, the above analyses revealed significant structural malleability even within apo PRS, which was suggestive of varied global conformations in ligand-free states of TgPRS.
Symmetry within the two protomers of the PRS homodimer has been considered to be a component of a half-site reactivity mechanism (Larson et al., 2012 ▸). Half-site reactivity in the context of homodimers can impact k cat (Larson et al., 2012 ▸; Fang et al., 2015 ▸). This suggests the possibility of structural elements that enable two asymmetric protomers to further dimerize and gain order. Our examination of TgPRS involving inspection of the structures identified loop 401–437 as the dimerization hinge, and there is a corresponding loop in PfPRS (Jain et al., 2014 ▸, 2015 ▸). This hinge is intrinsically disordered between the Apo1 and Apo2 TgPRS, indicating a higher degree of freedom for this hinge on the basis of electron density. With the exception of Pro429 in Apo1A, the integrity of this dimerization interface is intact in both protomers of Apo1 (Fig. 6 ▸ b).
3.2. Holo1 versus Holo2
On the other hand, comparisons of Holo1 and Holo2 confirm that they exhibit high symmetry within the constituent protomers in the dimers. The r.m.s.d.s of Holo1A versus Holo1B and Holo2A versus Holo2B are ∼0.65 and ∼0.93 Å, respectively (Fig. 2 ▸ and Supplementary Fig. S3). The highest extent of structural alteration (r.m.s.d./res) is only ∼7.3 Å compared with 29.5 Å between the two apo structures (Fig. 2 ▸ and Supplementary Fig. S3). Indeed, Holo1A and Holo2A display insignificant structural differences. Considering that both FF and HF belong to the same chemical class, it is unsurprising that there is a high similarity in structural integrity between FF- and HF-bound PRS structures (Figs. 6 ▸ c and 7 ▸). Interestingly, this fidelity is maintained even through different organisms: the active sites of TgPRS–HF, TgPRS–FF and PfPRS–HF are perfectly superimposable (Figs. 4 ▸ c and 6 ▸ c and Supplementary Fig. S1). Binding HF uses two Mg2+ ions, whereas binding FF requires only one. Besides this, there is no difference in the interaction between TgPRS or PfPRS and AMPPNP (Figs. 4 ▸ c and 6 ▸ c and Supplementary Fig. S1; Jain, Yogavel et al., 2017 ▸).
Figure 7.
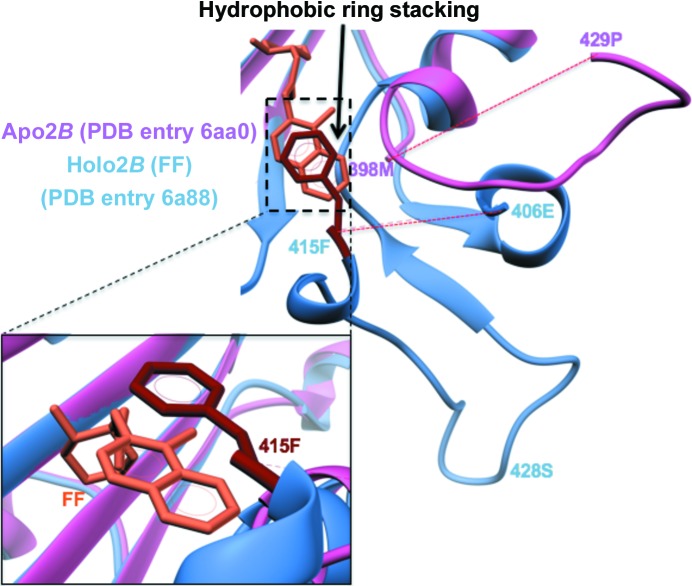
FF-induced active-site loop ordering in TgPRS. The mode of FF binding is similar to that of HF (Jain, Yogavel et al., 2017 ▸). Drug-induced loop stabilization of residues 399–428 and π-stacking of Phe415 is clearly visible. In Apo2B, this loop is disordered.
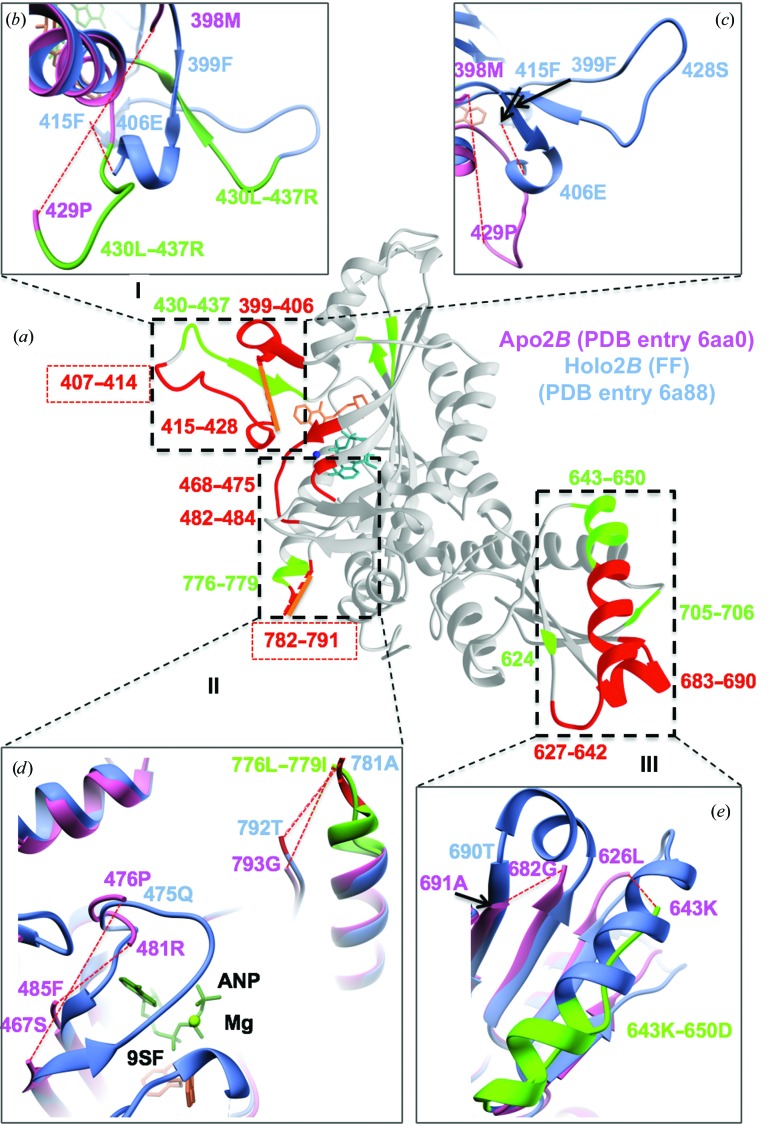
3.3. Apo2 versus Holo2
Based on the r.m.s.d. and r.m.s.d./res plots (Fig. 2 ▸ and Supplementary Fig. S3), we next decided to compare the Apo2B and Holo2B TgPRS (Figs. 7 ▸ and 8 ▸). The alignment analysis sheds light on the structural transitions that occur between the apo and holo states of TgPRS (Supplementary Fig. S2). Sequence alignment reveals high flexibility of the 399–438, 627–650 and 776–792 regions between the apo and holo states of TgPRS. It also reveals that the binding sites for FF and AMPPNP are disordered in the apo state of TgPRS (Figs. 6 ▸ a, 7 ▸ and Supplementary Fig. S2). For ease of understanding, only the holo structure is displayed and the residues that are missing or in a different conformation in Apo2B are marked correspondingly (Fig. 8 ▸). The conformational changes occur between residues 429 and 438 with an average r.m.s.d./res of 28.5 Å, between residues 642 and 650 with an average r.m.s.d./res of 9.9 Å and between residues 776 and 779 with an average r.m.s.d./res of 3.0 Å (Fig. 2 ▸ and Supplementary Fig. S3). The deviation in the position of loop 429–438 is evident in Fig. 8 ▸(b). Additionally, the conformational alteration of the α-helix cap residues 776–779 is displayed in Fig. 8 ▸(d). The shift in conformation from α-helix to coil-like for residues 643–650 is indicative of plasticity of the structural elements (Fig. 8 ▸ e).
Figure 8.
Structural differences between the Apo2B and Holo2B TgPRS structures. (a) FF-complexed Holo2B (dark grey) is divided into three sections, I, II and III, based on its structural differences from Apo2B. Only the missing residues (red) and those that adopt altered conformations (lime green) in Apo2B have been overlaid on Holo2B, while the common missing links are represented by a solid orange line. (b) Close-up view of the conformationally heterogeneous 430–437 loop in section I. In Apo2B, this loop is in a disordered conformation when compared with FF-complexed Holo2B. (c) Missing residues in Apo2B (399–428) and FF-complexed Holo2B (407–414) within section I are marked. Arrows represent residues in the background (Phe415 and Phe399). (d) Section II depicts the regions 468–475, 482–484 and 780–792 that are missing in Apo2B. Residues 782–791 are missing in FF-complexed Holo2B. The drastic conformational change in the motif 776–779 is also depicted. (e) Close-up view of section III showing the missing regions (627–642 and 683–690) in Apo2B along with the conformational differences within the region 643–650.
3.4. Heterogeneity within protomers
Additionally, we analyzed the extent of structural heterogeneity within the two protomers of apo TgPRS. To cover the entire spectrum, we chose the Apo1B versus Apo2B pair for deeper examination (Fig. 6 ▸ a). This pair showed an r.m.s.d. of 4.25 Å but r.m.s.d./res values as high as ∼29.5 Å (Fig. 2 ▸ and Supplementary Fig. S3). Holo versus holo pairs showed high rigidity and concurrence, with none exceeding an r.m.s.d./res of ∼7.3 Å, with the lowest alterations observed in Holo1B versus Holo2B, with r.m.s.d./res values as low as ∼1.0 Å (Figs. 6 ▸ c and 7 ▸ and Supplementary Fig. S3). To remove bias from our own study, we thus chose to further examine Holo1A and Holo2A, which have an r.m.s.d. of 0.26 Å and a maximum r.m.s.d./res of 1.56 Å. Apo2B versus Holo2B shows a backbone r.m.s.d. of 4.15 Å but an r.m.s.d./res exceeding ∼28 Å (Figs. 2 ▸ and 8 ▸ and Supplementary Fig. S3).
3.5. Evidence for a lack of extant fold-switching in TgPRS and for an extended-conformational selection model
In the light of a recent publication, we also confirmed fold-switching in apo and HF-bound structures of PfPRS (Porter & Looger, 2018 ▸; Fig. 4 ▸ a). Intriguingly, when the TgPRS apo and drug-bound structures were compared, no evidence of fold-switching was observed (Figs. 4 ▸ b and 8 ▸ c). We noticed no influence of lattice contacts on the ability to fold-switch in the apo/holo protein transitions for TgPRS.
Our work also reiterates the conformational selection model for TgPRS–drug interactions, as one apo TgPRS structure adopts a conformation akin to the FF-bound TgPRS (the same is true for HF-bound TgPRS; Figs. 2 ▸ and 4 ▸ c). In addition, there is significant induced fit upon drug binding as evident visually and by r.m.s.f. analysis (Figs. 2 ▸, 7 ▸ and 8 ▸).
3.6. Advantages of r.m.s.d./res over r.m.s.d.
We propose that r.m.s.d./res analysis is more revealing about global structural changes than r.m.s.d. comparisons (Figs. 2 ▸ and 3 ▸). While the maximal r.m.s.d./res values for the apo TgPRS structures are 8.6 and 16.6 Å for Apo1 and Apo2, respectively, those for the holo TgPRSs are ∼7.3 Å (Fig. 2 ▸ and Supplementary Fig. S3). This divergence itself is proof of significant structural transitions consequent to the envelopment of FF by TgPRS. The plastic nature of the apo conformations can be contrasted with the rigid framework of the drug-bound enzyme using r.m.s.d./res data (Fig. 2 ▸ and Supplementary Fig. S3). Additionally, we recently extended the r.m.s.d.–r.m.s.d./res analyses to another set of aaRS structures bound to ATP at 2.3 Å resolution and cladosporin at 2.1 Å resolution (Chhibber-Goel & Sharma, 2019 ▸). R.m.s.d.s for ATP-bound Homo sapiens lysyl-tRNA synthetase (HsKRS) structures remain below ∼1 Å and the maximum r.m.s.d./res increases to >11 Å, indicating a large conformational change in limited regions of KRSs (Fig. 3 ▸ and Supplementary Fig. S4). The inset in Fig. 3 ▸ shows the region that differs conformationally: from Thr411 to Glu417 in HsKRS. Thus, local and global changes in protein structures in various states of ligand engagement can be reliably assessed via a combination of r.m.s.d. and r.m.s.d./res analyses. The analysis of TgPRS apo/holo structures presented here highlights key issues of structural variance in enzyme states that are vital for consideration in structure-based drug development.
4. Conclusions
The TgPRS enzyme is flexible in ligand-free states, as evident from the two apo structures. Further, upon engagement with FF/HF the enzyme adopts a highly ordered structure in the two protomers that constitute the biological dimer. In contrast to PfPRS, the close evolutionary homologue TgPRS does not display extant fold-switching. This work therefore highlights the conformational heterogeneity inherent within apicomplexan PRSs.
Supplementary Material
PDB reference: T. gondii prolyl-tRNA synthetase, apo form, 6aa0
PDB reference: complex with febrifugine and ATP analog, 6a88
Supplementary Figures. DOI: 10.1107/S2053230X19014808/ow5017sup1.pdf
Acknowledgments
The authors thank the beamline staff at PROXIMA 1 and PROXIMA 2A for assistance during data collection at SOLEIL. We thank Dr Rai for inspiration.
Funding Statement
This work was funded by Department of Biotechnology, Ministry of Science and Technology grants PR6303 and PR13636. Indo-French Centre for the Promotion of Advanced Research (IFCPAR/CEFIPRA) grant . Department of Science and Technology, India grant SB/S2/JCB-41/2013 to Amit Sharma.
References
- Adams, P. D., Afonine, P. V., Bunkóczi, G., Chen, V. B., Davis, I. W., Echols, N., Headd, J. J., Hung, L.-W., Kapral, G. J., Grosse-Kunstleve, R. W., McCoy, A. J., Moriarty, N. W., Oeffner, R., Read, R. J., Richardson, D. C., Richardson, J. S., Terwilliger, T. C. & Zwart, P. H. (2010). Acta Cryst. D66, 213–221. [DOI] [PMC free article] [PubMed]
- Bhatt, T. K., Kapil, C., Khan, S., Jairajpuri, M. A., Sharma, V., Santoni, D., Silvestrini, F., Pizzi, E. & Sharma, A. (2009). BMC Genomics, 10, 644. [DOI] [PMC free article] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Chhibber-Goel, J. & Sharma, A. (2019). Proteins, 87, 730–737. [DOI] [PubMed]
- Coatney, G. R., Cooper, W. C., Culwell, W. B., White, W. C. & Imboden, C. A. (1950). J. Natl Malar. Soc. 9, 183–186. [PubMed]
- Das, P., Babbar, P., Malhotra, N., Sharma, M., Jachak, G. R., Gonnade, R. G., Shanmugam, D., Harlos, K., Yogavel, M., Sharma, A. & Reddy, D. S. (2018). J. Med. Chem. 61, 5664–5678. [DOI] [PubMed]
- Datt, M. & Sharma, A. (2014). J. Struct. Funct. Genomics, 15, 45–61. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Fang, P., Han, H., Wang, J., Chen, K., Chen, X. & Guo, M. (2015). Chem. Biol. 22, 734–744. [DOI] [PMC free article] [PubMed]
- Gowri, V. S., Ghosh, I., Sharma, A. & Madhubala, R. (2012). BMC Genomics, 13, 621. [DOI] [PMC free article] [PubMed]
- Herman, J. D., Pepper, L. R., Cortese, J. F., Estiu, G., Galinsky, K., Zuzarte-Luis, V., Derbyshire, E. R., Ribacke, U., Lukens, A. K., Santos, S. A., Patel, V., Clish, C. B., Sullivan, W. J., Zhou, H., Bopp, S. E., Schimmel, P., Lindquist, S., Clardy, J., Mota, M. M., Keller, T. L., Whitman, M., Wiest, O., Wirth, D. F. & Mazitschek, R. (2015). Sci. Transl. Med. 7, 288ra77. [DOI] [PMC free article] [PubMed]
- Hewitt, R. I., Wallace, W. S., Gill, E. R. & Williams, J. H. (1952). Am. J. Trop. Med. Hyg. 1, 768–772. [PubMed]
- Hoen, R., Novoa, E. M., López, A., Camacho, N., Cubells, L., Vieira, P., Santos, M., Marin-Garcia, P., Bautista, J. M., Cortés, A., Ribas de Pouplana, L. & Royo, M. (2013). Chembiochem, 14, 499–509. [DOI] [PubMed]
- Hoepfner, D., McNamara, C. W., Lim, C. S., Studer, C., Riedl, R., Aust, T., McCormack, S. L., Plouffe, D. M., Meister, S., Schuierer, S., Plikat, U., Hartmann, N., Staedtler, F., Cotesta, S., Schmitt, E. K., Petersen, F., Supek, F., Glynne, R. J., Tallarico, J. A., Porter, J. A., Fishman, M. C., Bodenreider, C., Diagana, T. T., Movva, N. R. & Winzeler, E. A. (2012). Cell Host Microbe, 11, 654–663. [DOI] [PMC free article] [PubMed]
- Hussain, T., Yogavel, M. & Sharma, A. (2015). Antimicrob. Agents Chemother. 59, 1856–1867. [DOI] [PMC free article] [PubMed]
- Jackson, K. E., Habib, S., Frugier, M., Hoen, R., Khan, S., Pham, J. S., de Pouplana, L. R., Royo, M., Santos, M. A. S., Sharma, A. & Ralph, S. A. (2011). Trends Parasitol. 27, 467–476. [DOI] [PubMed]
- Jain, V., Kikuchi, H., Oshima, Y., Sharma, A. & Yogavel, M. (2014). J. Struct. Funct. Genomics, 15, 181–190. [DOI] [PubMed]
- Jain, V., Sharma, A., Singh, G., Yogavel, M. & Sharma, A. (2017). ACS Infect. Dis. 3, 281–292. [DOI] [PubMed]
- Jain, V., Yogavel, M., Kikuchi, H., Oshima, Y., Hariguchi, N., Matsumoto, M., Goel, P., Touquet, B., Jumani, R. S., Tacchini-Cottier, F., Harlos, K., Huston, C. D., Hakimi, M. A. & Sharma, A. (2017). Structure, 25, 1495–1505. [DOI] [PubMed]
- Jain, V., Yogavel, M., Oshima, Y., Kikuchi, H., Touquet, B., Hakimi, M. A. & Sharma, A. (2015). Structure, 23, 819–829. [DOI] [PubMed]
- Kato, N., Comer, E., Sakata-Kato, T., Sharma, A., Sharma, M., Maetani, M., Bastien, J., Brancucci, N. M., Bittker, J. A., Corey, V., Clarke, D., Derbyshire, E. R., Dornan, G. L., Duffy, S., Eckley, S., Itoe, M. A., Koolen, K. M. J., Lewis, T. A., Lui, P. S., Lukens, A. K., Lund, E., March, S., Meibalan, E., Meier, B. C., McPhail, J. A., Mitasev, B., Moss, E. L., Sayes, M., Van Gessel, Y., Wawer, M. J., Yoshinaga, T., Zeeman, A. M., Avery, V. M., Bhatia, S. N., Burke, J. E., Catteruccia, F., Clardy, J. C., Clemons, P. A., Dechering, K. J., Duvall, J. R., Foley, M. A., Gusovsky, F., Kocken, C. H. M., Marti, M., Morningstar, M. L., Munoz, B., Neafsey, D. E., Sharma, A., Winzeler, E. A., Wirth, D. F., Scherer, C. A. & Schreiber, S. L. (2016). Nature (London), 538, 344–349.
- Keller, T. L., Zocco, D., Sundrud, M. S., Hendrick, M., Edenius, M., Yum, J., Kim, Y. J., Lee, H. K., Cortese, J. F., Wirth, D. F., Dignam, J. D., Rao, A., Yeo, C. Y., Mazitschek, R. & Whitman, M. (2012). Nature Chem. Biol. 8, 311–317. [DOI] [PMC free article] [PubMed]
- Khan, S., Garg, A., Camacho, N., Van Rooyen, J., Kumar Pole, A., Belrhali, H., Ribas de Pouplana, L., Sharma, V. & Sharma, A. (2013). Acta Cryst. D69, 785–795. [DOI] [PubMed]
- Khan, S., Sharma, A., Belrhali, H., Yogavel, M. & Sharma, A. (2014). J. Struct. Funct. Genomics, 15, 63–71. [DOI] [PubMed]
- Khan, S., Sharma, A., Jamwal, A., Sharma, V., Pole, A. K., Thakur, K. K. & Sharma, A. (2011). Sci. Rep. 1, 188. [DOI] [PMC free article] [PubMed]
- Kikuchi, H., Yamamoto, K., Horoiwa, S., Hirai, S., Kasahara, R., Hariguchi, N., Matsumoto, M. & Oshima, Y. (2006). J. Med. Chem. 49, 4698–4706. [DOI] [PubMed]
- Koepfli, J. B., Mead, J. F. & Brockman, J. A. (1947). J. Am. Chem. Soc. 69, 1837. [DOI] [PubMed]
- Koh, C. Y., Kim, J. E., Napoli, A. J., Verlinde, C. L. M. J., Fan, E., Buckner, F. S., Van Voorhis, W. C. & Hol, W. G. J. (2013). Mol. Biochem. Parasitol. 189, 26–32. [DOI] [PMC free article] [PubMed]
- Koh, C. Y., Kim, J. E., Shibata, S., Ranade, R. M., Yu, M., Liu, J., Gillespie, J. R., Buckner, F. S., Verlinde, C. L. M. J., Fan, E. & Hol, W. G. J. (2012). Structure, 20, 1681–1691. [DOI] [PMC free article] [PubMed]
- Krissinel, E. (2012). J. Mol. Biochem. 1, 76–85. [PMC free article] [PubMed]
- Larson, E. T., Kim, J. E., Napuli, A. J., Verlinde, C. L. M. J., Fan, E., Zucker, F. H., Van Voorhis, W. C., Buckner, F. S., Hol, W. G. J. & Merritt, E. A. (2012). Acta Cryst. D68, 1194–1200. [DOI] [PMC free article] [PubMed]
- Manickam, Y., Chaturvedi, R., Babbar, P., Malhotra, N., Jain, V. & Sharma, A. (2018). Drug Discov. Today, 23, 1233–1240. [DOI] [PubMed]
- Nachiappan, M., Jain, V., Sharma, A., Yogavel, M. & Jeyakanthan, J. (2018). Int. J. Biol. Macromol. 120, 1379–1386. [DOI] [PubMed]
- Palencia, A., Liu, R.-J., Lukarska, M., Gut, J., Bougdour, A., Touquet, B., Wang, E.-D., Li, X., Alley, M. R. K., Freund, Y. R., Rosenthal, P. J., Hakimi, M.-A. & Cusack, S. (2016). Antimicrob. Agents Chemother. 60, 5817–5827. [DOI] [PMC free article] [PubMed]
- Pasaje, C. F. A., Cheung, V., Kennedy, K., Lim, E. E., Baell, J. B., Griffin, M. D. W. & Ralph, S. A. (2016). Sci. Rep. 6, 27531. [DOI] [PMC free article] [PubMed]
- Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C. & Ferrin, T. E. (2004). J. Comput. Chem. 25, 1605–1612. [DOI] [PubMed]
- Pham, J. S., Dawson, K. L., Jackson, K. E., Lim, E. E., Pasaje, C. F. A., Turner, K. E. C. & Ralph, S. A. (2014). Int. J. Parasitol. Drugs Drug Resist. 4, 1–13. [DOI] [PMC free article] [PubMed]
- Porter, L. L. & Looger, L. L. (2018). Proc. Natl. Acad. Sci. USA, 115, 5968–5973. [DOI] [PMC free article] [PubMed]
- Sharma, A., Sharma, M., Yogavel, M. & Sharma, A. (2016). PLoS Negl. Trop. Dis. 10, e0005084. [DOI] [PMC free article] [PubMed]
- Son, J., Lee, E. H., Park, M., Kim, J. H., Kim, J., Kim, S., Jeon, Y. H. & Hwang, K. Y. (2013). Acta Cryst. D69, 2136–2145. [DOI] [PubMed]
- Sonoiki, E., Palencia, A., Guo, D., Ahyong, V., Dong, C., Li, X., Hernandez, V. S., Zhang, Y.-K., Choi, W., Gut, J., Legac, J., Cooper, R., Alley, M. R. K., Freund, Y. R., DeRisi, J., Cusack, S. & Rosenthal, P. J. (2016). Antimicrob. Agents Chemother. 60, 4886–4895. [DOI] [PMC free article] [PubMed]
- Winn, M. D., Ballard, C. C., Cowtan, K. D., Dodson, E. J., Emsley, P., Evans, P. R., Keegan, R. M., Krissinel, E. B., Leslie, A. G. W., McCoy, A., McNicholas, S. J., Murshudov, G. N., Pannu, N. S., Potterton, E. A., Powell, H. R., Read, R. J., Vagin, A. & Wilson, K. S. (2011). Acta Cryst. D67, 235–242. [DOI] [PMC free article] [PubMed]
- Winter, G., Waterman, D. G., Parkhurst, J. M., Brewster, A. S., Gildea, R. J., Gerstel, M., Fuentes-Montero, L., Vollmar, M., Michels-Clark, T., Young, I. D., Sauter, N. K. & Evans, G. (2018). Acta Cryst. D74, 85–97. [DOI] [PMC free article] [PubMed]
- Zhou, H., Sun, L., Yang, X. L. & Schimmel, P. (2013). Nature (London), 494, 121–124. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: T. gondii prolyl-tRNA synthetase, apo form, 6aa0
PDB reference: complex with febrifugine and ATP analog, 6a88
Supplementary Figures. DOI: 10.1107/S2053230X19014808/ow5017sup1.pdf