Ths structure of the Fab fragment of the high-affinity anti-cocaine antibody h2E2 is reported both as the free Fab and in complex with the cocaine metabolite benzoylecgonine.
Keywords: high-affinity anti-cocaine antibody, antigen-binding fragment, crystal structure, benzoylecgonine complex, h2E2, Fab fragment
Abstract
A high-affinity anti-cocaine monoclonal antibody, designated h2E2, is entering phase 1 clinical trials for cocaine abuse therapy. To gain insight into the molecular details of its structure that are important for binding cocaine and cocaine metabolites, the Fab fragment was generated and crystallized with and without ligand. Structures of the unliganded Fab and the Fab fragment bound to benzoylecgonine were determined, and were compared with each other and with other crystallized anti-cocaine antibodies. The affinity of the h2E2 antibody for cocaine is 4 nM, while that of the cocaine metabolite benzoylecgonine is 20 nM. Both are higher than the reported affinity for cocaine of the two previously crystallized anti-cocaine antibodies. Consistent with cocaine fluorescent quenching binding studies for the h2E2 mAb, four aromatic residues in the CDR regions of the Fab (TyrL32, TyrL96, TrpL91 and TrpH33) were found to be involved in ligand binding. The aromatic side chains surround and trap the tropane moiety of the ligand in the complex structure, forming significant van der Waals interactions which may account for the higher affinity observed for the h2E2 antibody. A water molecule mediates hydrogen bonding between the antibody and the carbonyl group of the benzoyl ester. The affinity of binding to h2E2 of benzoylecgonine differs only by a factor of five compared with that of cocaine; therefore, it is suggested that h2E2 would bind cocaine in the same way as observed in the Fab–benzoylecgonine complex, with minor rearrangements of some hypervariable segments of the antibody.
1. Introduction
The ability of the mammalian adaptive immune system to generate high-affinity antibodies with specificities for virtually an unlimited number of antigens has long been exploited to develop a variety of therapeutic and diagnostic antibodies. Naturally occurring antigens generally consist of high-molecular-weight proteins, polypeptides, lipoproteins and lipopolysaccharides, while most low-molecular-weight biomolecules are not capable of generating an immune-system antibody response. However, low-molecular-weight molecules, such as commonly used drugs, can be manipulated into becoming immunogenic through linking them, as a hapten, to a larger immunogenic carrier protein in a manner that enables them to be recognized as active antigens by the immune system. Immunization protocols using hapten-carrier linked conjugates have enabled investigators to generate high-affinity antibodies that are specifically directed against non-immunogenic compounds such as heroin, cocaine, nicotine and phencyclidine, the usage and abuse of which pose considerable individual as well as public health problems. These antibodies have the potential to be effective therapeutics in reducing abuse and relapse behaviors by sequestering the target drug in the plasma, thereby dramatically reducing drug distribution to the central nervous system and blunting neurological effects (Carrera et al., 1995 ▸, 2000 ▸, 2001 ▸; Fox et al., 1996 ▸; Kvello et al., 2016 ▸; Cerny & Cerny, 2008 ▸; Kosten & Owens, 2005 ▸). Furthermore, the drug-binding Fab fragments or lower molecular weight scFv single-chain fragments derived from them, which have relatively fast elimination rates compared with intact antibodies, have the additional potential to be used as rescue treatment agents to assist detoxification in cases of acute overdoses of toxic drugs.
Human clinical trials using hapten-carrier vaccines that elicit endogenously produced polyclonal anti-nicotine and anti-cocaine antibodies have shown that a subset of vaccinated individuals can generate antidrug antibodies at levels sufficient to be therapeutically effective (Martell et al., 2005 ▸, 2009 ▸). Alternatively, the use of murine hybridoma or phage-display methodologies, along with recombinant DNA technologies, has enabled the production of biomanufactured murine–human chimeric, humanized and human monoclonal antibodies (mAbs) that can be administered in known doses to provide immediate, high-affinity, passive immunization responses. As potential immunotherapeutics for the treatment of cocaine abuse, the murine-derived chimeric mAb GNC92H2 (Carrera et al., 2005 ▸), the humanized mAb h2E2 (Norman et al., 2014 ▸) and the fully human anti-cocaine mAb GNCgzk (Treweek & Janda, 2012 ▸) have been generated and characterized with regard to their affinities and specificities for cocaine and its metabolites. Additionally, in rodent models the in vivo ability of anti-cocaine mAbs to (i) decrease brain cocaine concentrations by restricting its distribution from plasma, (ii) increase the drug levels needed to reinstate cocaine-dependent self-administration behavior in trained rats and (iii) substantially reduce cocaine toxicity/lethality have been well documented (Treweek & Janda, 2012 ▸; Norman et al., 2007 ▸, 2009 ▸; Norman & Ball, 2012 ▸; Wetzel et al., 2016 ▸). Based on such preliminary studies, the mAb h2E2 is entering phase 1 clinical trials for cocaine abuse therapy.
In order to gain a better understanding of the molecular nature of anti-cocaine antibody–drug binding interactions, the Fab fragments of two anti-cocaine antibodies, the chimeric mAb GNC92H2 (IgG, γ1, κ) and the murine anti-cocaine mAb M82G2 (IgG, γ1, κ), have been crystallized and their detailed three-dimensional structures have been elucidated (Larsen et al., 2001 ▸; Pozharski et al., 2005 ▸). While these two mAbs and h2E2 all have selectivity for cocaine, some of the specifics of their binding characteristics are distinct. The GNC92H2 and h2E2 mAbs are similar in that they both bind cocaine and the active derivative cocaethylene with comparable affinities, with low affinity for inactive cocaine metabolites such as ecgonine methyl ester and ecgonine. However, the h2E2 mAb is unique in that it has a higher affinity for cocaine, with a K d value of about 4 × 10−9 M (Paula et al., 2004 ▸), while GNC92H2 (Larsen et al., 2001 ▸) and M82G2 (Pozharski et al., 2005 ▸) have reported K d values of 4 × 10−7 and 1.4 × 10−7 M, respectively, K d values that are about 100-fold and 35-fold higher (lower affinity) than that of the h2E2 mAb utilized in the current study. Although both cocaine- and benzoylecgonine-bound Fab structures have been determined for the M82G2 mAb, its affinity for benzoylecgonine has not been reported. Antibody GNC92H2 has a 100-fold lower binding affiinity for benzoylecgonine than for cocaine (Larsen et al., 2001 ▸).
The binding-affinity values in and of themselves provide no basic information about the mechanism and details of ligand interactions with the binding site of the antibody. Therefore, detailed structural information is required to begin to understand the interactions needed for the high-affinity, selective binding of cocaine. In the present work, we have determined the crystal structure of the Fab fragment of h2E2 both as a complex with benzoylecgonine (BE) and as a methylated form of the Fab fragment in the absence of bound ligand. In addition to presenting, analyzing and comparing the h2E2 Fab crystal structures, we compare the binding of benzoylecgonine by h2E2 with that in two previously determined Fab–ligand complex structures of other anti-cocaine antibodies. This broadens the perspective on what, if any, commonalities exist with regard to how mAbs bind cocaine, as well as the limitations that may exist for generating mAbs with high-affinity and selective binding sites for low-molecular-weight antigens.
2. Materials and methods
2.1. Materials
Endoproteinase Lys-C (lysyl endoproteinase, Achromobacter protease I, Endo-Lys-C; catalog No. 129-02541) was obtained from Wako Pure Chemicals. HEPES buffer was from Sigma (catalog No. H-3375). NaCl was from Fisher Scientific. Borane–dimethyl complex (97%) was obtained from Sigma–Aldrich (catalog No. 180238) and 37% formaldehyde was from Sigma (catalog No. F8775). Concentrated formic acid, sequencing grade, was purchased from Fisher Scientific. Cocaine hydrochloride (catalog No. C-5776) was obtained from Sigma–Aldrich. GeneArt Seamless Cloning reagents (Invitrogen) were from Thermo Fisher Scientific. KOD polymerase (EMD Millipore, Billerica, Massachusetts, USA) was used for PCR amplification. Plasmid miniprep and PCR purification kits were from Qiagen (Germantown, Maryland, USA). Synthetic DNA and oligonucleotides were obtained from Integrated DNA Technologies (IDT; Coralville, Iowa, USA) and Blue Heron Biotech (Bothell, Washington, USA). Unless otherwise indicated, all other chemicals and reagents were from Sigma (St Louis, Missouri, USA) or Thermo Fisher Scientific (Waltham, Massachusetts, USA).
2.2. Expression from Escherichia coli and purification of recombinant h2E2 Fab
2.2.1. Vector construction
Amino-acid sequences for the h2E2 heavy-chain and light-chain variable domains as well as for the human lambda 2 constant domain were reverse-transcribed and optimized for expression in E. coli using the Blue Heron Biotech online codon-optimization tool (https://www.blueheronbio.com/codon-optimization). Synthetic DNA for the heavy- and light-chain variable domains was obtained from Blue Heron Biotech, and the lambda 2 constant domain was obtained as a gBlock from IDT. Coding regions for the h2E2 Fab were introduced into the plasmid pASK88-D1.3 (a kind gift from Arne Skerra, Technische Universität München, Germany) by stepwise replacement of the analogous regions of the D1.3 anti-lysozyme Fab using GeneArt Seamless Cloning. Firstly, the vector was PCR-amplified with primers that eliminated the light-chain constant domain, and the PCR product was then treated with DpnI to remove the plasmid template. The gBlock fragment coding for the human lambda 2 constant domain, which contained 16 bp vector overlaps, was assembled with the PCR-amplified vector using the GeneArt Seamless Cloning and Assembly Enzyme Mix (Invitrogen). The resulting vector was then amplified for assembly with the PCR-amplified VH coding region, and finally the process was repeated a third time to insert the VL domain. The pAKS88-D1.3 vector contained a human IgG1 CH1 domain that was identical to that of the h2E2 monoclonal antibody except for a 210Arg to Lys change near the C-terminal end of CH1. This domain was retained in the final h2E2 recombinant Fab expression vector. Cloned inserts were verified by Sanger sequence analysis (University of Chicago Comprehensive Cancer Center DNA Sequencing and Genotyping Facility, Chicago, Illinois, USA). The sequences of the PCR primers, sequencing primers and synthetic DNA are provided in Supplementary Tables S1–S4.
2.2.2. Expression and purification of recombinant h2E2 Fab (rFab)
E. coli strain JM83 (Yanisch-Perron et al., 1985 ▸) harboring pASK88-h2E2 and the helper plasmid pTum4 (Schlapschy et al., 2006 ▸; a kind gift from Arne Skerra, Technische Universität München, Germany) was grown in shake flasks at 37°C using M9 medium supplemented with glycerol, glucose, amino acids, trace metals and vitamins (Donnelly et al., 2006 ▸). Ampicillin (100 µg l−1) and chloramphenicol (30 µg l−1) were added to maintain the plasmids. At mid-log phase, the culture temperature was reduced to 25°C. Expression was then induced with anhydrotetracycline, which was added to a final concentration of 0.2 mg l−1. After overnight growth, the cells were harvested by centrifugation in 1 l centrifuge bottles. Each pellet was resuspended in 25 ml TES buffer (0.2 M Tris–HCl, 0.5 mM EDTA, 0.5 M sucrose pH 8.0) plus one cOmplete protease-inhibitor tablet (Roche) by shaking at 200 rev min−1 at 4°C. After the pellets had been completely resuspended, 50 µl of 100 mg ml−1 lysozyme and 25 ml ice-cold water were added and the suspension was incubated at 4°C for 1 h with agitation at 70 rev min−1. Spheroplasts were removed by centrifugation for 30 min at 30 000g and the supernatant constituted the periplasmic fraction. The rFab was purified from the periplasmic fraction by nickel-affinity chromatography on an ÄKTAexplorer 3D using an established protocol (Kim et al., 2004 ▸). The eluted protein was further purified by size-exclusion chromatography on a Superdex 200 (26/60) column in 20 mM HEPES, 100 mM NaCl pH 8.0. Fractions containing purified rFab were identified by SDS–PAGE under reducing conditions and were combined and concentrated to 18 mg ml−1 using a centrifugal filter unit. A culture volume of 8 l yielded approximately 3 mg recombinant h2E2 Fab (rFab).
2.3. Preparation and characterization of the Fab from the h2E2 mAb and reductive methylation of the Fab fragment
The Fab fragment of the h2E2 anti-cocaine mAb was generated by Endo-Lys-C digestion followed by purification, as described previously (Kirley & Norman, 2015 ▸). The Fab fragment was reductively methylated on lysine residues as a way to aid crystallization, basically as described previously (Walter et al., 2006 ▸; Tan et al., 2014 ▸). For more details, see the supplementary information.
To ensure that methylation did not negatively affect the binding of cocaine to the Fab fragment, the methylated Fab fragment was examined for cocaine binding using the fluorescence assay reported previously (Kirley & Norman, 2015 ▸). Cocaine binding to the Fab and mFab was measured by monitoring the change in intrinsic protein tyrosine and tryptophan fluorescence upon titration with ligand at 20°C. Briefly, measurements of intrinsic protein fluorescence and quenching of that fluorescence by cocaine were made using a Hitachi F-2000 fluorescence spectrophotometer. 2 ml Fab solutions in Tris-buffered saline (TBS) were analyzed and titrated with ligands, measuring the emission at 330 nm with excitation at both 280 and 295 nm after each addition of ligand.
The K d for cocaine of the methyl-Fab was measured to be 1.2 ± 0.3 nM (using tryptophan-selective excitation at 295 nm). This value is actually slightly less (a slightly higher affinity) than we reported for the unmodified Fab fragment using the same methodology (3.8 ± 1.6 nM; Kirley & Norman, 2015 ▸).
2.4. Crystallization and diffraction data collection
2.4.1. Fab–benzoylecgonine complex
The purified and concentrated rFab (18 mg ml−1) was mixed with cocaine hydrochloride (to a final cocaine concentration of 10 mM) and the mixture was incubated on ice for 2h. The Fab/cocaine mixture was then screened for crystallization conditions with a Mosquito liquid dispenser (TTP Labtech, Cambridge, Massachusetts, USA) using the sitting-drop vapor-diffusion technique in 96-well CrystalQuick plates (Greiner Bio-One, Monroe, North Carolina, USA). Several commercially available crystallization screens were used. For each condition, 0.4 µl Fab/cocaine solution and 0.4 µl crystallization formulation were mixed and the mixture was equilibrated against 135 µl crystallization formulation in the well at 16°C. Diffraction-quality crystals appeared after two months under a condition consisting of 2% Tacsimate pH 7, 0.1 M HEPES pH 7.5, 20% PEG 3350 (condition G10 of PEG/Ion HT from Hampton Research, Aliso Viejo, California, USA). Before data collection, the crystals were treated with a cryoprotectant composed of the crystallization reagent plus 25%(v/v) glycerol. X-ray diffraction data were collected at 100 K on the 19ID beamline of the Structural Biology Center at the Advanced Photon Source (APS), Argonne National Laboratory using SBCcollect (Rosenbaum et al., 2006 ▸). The best data set was processed to 2.63 Å with the HKL-3000 suite (Minor et al., 2006 ▸; Table 1 ▸).
Table 1. Crystallographic and data-collection parameters.
Values in parentheses are for the highest resolution shell.
| rFab–BE complex | Free mFab | |
|---|---|---|
| Unit-cell parameters (Å, °) | a = 70.05, b = 85.84, c = 164.4, α = 82.9, β = 81.8, γ = 71.3 | a = 110.8, b = 128.4, c = 91.20, β = 125.3 |
| Space group | P1 | C2 |
| No. of molecules in asymmetric unit | 8 | 2 |
| V M (Å3 Da−1) | 2.5 | 2.9 |
| Solvent content (%) | 51 | 57 |
| Wavelength (Å) | 0.97915 | 0.97919 |
| Resolution (Å) | 38.6–2.63 (2.68–2.63) | 27.8–2.15 (2.19–2.15) |
| No. of unique reflections | 104134 | 54989 |
| R merge | 0.119 (0.805) | 0.054 (0.869) |
| R meas | 0.140 (0.970) | 0.062 (1.009) |
| R p.i.m. | 0.075 (0.537) | 0.030 (0.506) |
| CC1/2 | 0.983 (0.572) | 0.998 (0.639) |
| Multiplicity | 3.5 (3.1) | 4.1 (3.7) |
| Completeness (%) | 98.3 (98.5) | 97.9 (96.2) |
| Mean I/σ(I) | 13.9 (1.6) | 32.7 (2.3) |
| Wilson B factor (Å2) | 45.7 | 44.7 |
2.4.2. Unliganded (free) Fab
Purified and concentrated methyl-Fab (mFab; 10 mg ml−1) was screened at 16°C using the commercial crystallization screens MCSG-1, MCSG-2, MCSG-3 and MCSG-4 (Microlytic Inc.). The crystallization drops were set up using a Mosquito liquid dispenser as described above. An initial hit was observed in condition A7 from MCSG-2. Further optimization was carried out manually using the hanging-drop vapor-diffusion method by setting up crystallization drops at room temperature consisting of 1 µl protein solution and 1 µl well solution using Linbro plates equilibrated against a volume of 0.5 ml. Crystals usually appeared within a day and grew to full size in a week. The crystal used for data collection was obtained from Wizard II condition No. 36 diluted with water (8% PEG 3000, 80 mM phosphate–citrate pH 4.2, 160 mM NaCl). The crystals were unstable (breaking apart and dissolving rapidly) when transferred to cryoprotectant solution consisting of the reservoir solution supplemented with 25% glycerol or ethylene glycol. Therefore, a crystal from the mother liquor was directly transferred to liquid nitrogen. In situ annealing of the crystal for 10 s alleviated the initially observed problem of ice rings. Data collection was carried out on the 19BM beamline at APS. The crystallographic parameters are included in Table 1 ▸ and the refinement statistics are given in Table 2 ▸.
Table 2. Refinement statistics.
| rFab–BE complex | Free mFab | |
|---|---|---|
| Program used | Phenix | Phenix |
| Resolution range (Å) | 38.60–2.63 | 27.8–2.15 |
| No. of reflections | 104051 | 54930 |
| R factor | 0.185 | 0.214 |
| R free † | 0.244 | 0.254 |
| No. of non-H atoms | ||
| Protein | 24869 | 6169 |
| Heteroatoms | 220 | 206 |
| Waters | 173 | 111 |
| Mean B factor (Å2) | ||
| Protein | 61.7 | 74.1 |
| Heteroatoms | 51.5 | 71.0 |
| Waters | 47.4 | 55.5 |
| R.m.s.d.s | ||
| Bonds (Å) | 0.004 | 0.003 |
| Bond angles (°) | 0.72 | 0.60 |
| Ramachandran plot (%) | ||
| Favored regions | 95.3 | 95.6 |
| Outliers | 0.4 | 0 |
| PDB code | 6nfn | 6nex |
The test set consisted of 5% of the reflections.
2.5. Structure determination and refinement
2.5.1. Fab–benzoylecgonine complex
The rFab–benzoylecgonine complex crystals belonged to space group P1. The data could not be scaled in a higher symmetry space group. The structure of the complex was solved by the molecular-replacement method using MOLREP (Vagin & Teplyakov, 2010 ▸) within the HKL-3000 suite (Minor et al., 2006 ▸). The VLVH pair of the Fab from the hH35 antibody Fab–human hepsin complex structure (Koschubs et al., 2012 ▸; PDB entry 3t2n) was first used as a search template. The search resulted in the location of eight VLVH pairs in one asymmetric unit of the crystal with a correlation coefficient of 35.3%. After several cycles of rigid-body and position/displacement refinements, the above model containing eight VLVH pairs were fixed while the CLCH1 pair from the same Fab (PDB entry 3t2n) was used as a search template, resulting in the locations of all eight CLCH1 pairs in the asymmetric unit with a correlation coefficient of 53.6%. After several cycles of refinement, automatic model rebuilding with Buccaneer (Cowtan, 2008 ▸) within the HKL-3000 package was performed, which led to the building of a model with correlation coefficients of 92% for the main chain and 79% for the side chains of all eight Fabs. The completion of the structural model, including the addition of ligand molecules, was performed manually using Coot (Emsley et al., 2010 ▸). The final refinement cycles were performed with Phenix (Adams et al., 2010 ▸). The structure solution by molecular replacement placed two clusters of four Fabs each. Within each cluster of four Fabs there is an approximate fourfold symmetry between them.
2.5.2. Unliganded Fab
The structure was determined by molecular replacement using Phaser (McCoy et al., 2007 ▸). The Fab portion of the h2E2 rFab–benzoylecgonine complex structure (this work) was used as the search structure. The final structure was refined by Phenix to a resolution of 2.15 Å with an R factor and R free of 0.214 and 0.254, respectively. The crystallographic parameters are included in Table 1 ▸, whereas the refinement statistics are shown in Table 2 ▸. In the case of the unliganded mFab crystals the two Fabs per asymmetric unit are related by pseudo-twofold symmetry but were refined independently.
A schematic view of the interactions of benzoylecgonine and h2E2 Fab was generated by LigPlot+ (Laskowski & Swindells, 2011 ▸). All other structural figures were generated with PyMOL (http://pymol.org). Elbow-angle calculations were performed using the web server at http://proteinmodel.org/AS2TS/RBOW/index.html (Stanfield et al., 2006 ▸).
3. Results
3.1. Expression of recombinant h2E2 Fab in E. coli
Recombinant h2E2 Fab (termed rFab) was produced with the E. coli expression vector pASK88 (Schiweck & Skerra, 1995 ▸). In this vector, the two polypeptides of the Fab are expressed as an operon under the tightly regulated control of the tetA promoter, and secretion of the heavy- and light-chain components is directed to the periplasm for proper folding and assembly via the OmpA and PhoA signal peptides (Skerra, 1994a ▸,b ▸). A His6 affinity tag is appended to the C-terminal end of the heavy chain to simplify purification. The h2E2 expression cassette was constructed using E. coli codon- and expression-optimized synthetic DNA. Fragments coding for the 2E2 VH, VL and human lambda 2 constant domains were assembled in pASK88 using seamless cloning techniques. pASK88 carries a human IgG1 heavy-chain constant domain 1 (CH1γ1), which is identical to that in the h2E2 antibody except for the presence of Lys at position 210 (h2E2 has Arg at this position). For the purpose of this study, this residue was not changed as it was not expected to affect the structure or function of the binding site of the antibody. h2E2 rFab was expressed in shake flasks with an E. coli strain carrying the helper plasmid pTUM4 to facilitate the proper folding and assembly of the polypeptides in the periplasm (Schlapschy et al., 2006 ▸). After purification, the yield ranged from 0.3 to 0.4 mg rFab per litre of shake-flask culture.
3.2. Crystallization and structure determination of rFab in the presence of cocaine
Crystallization trials were carried out with the rFab alone and in the presence of cocaine. After two months, diffraction-quality crystals were produced in one condition (2% Tacsimate pH 7, 0.1 M HEPES pH 7.5, 20% PEG 3350) in a drop containing rFab and cocaine. The rFab complex crystals belonged to space group P1, with unit-cell parameters a = 70.05, b = 85.84, c = 164.4 Å, α = 82.9, β = 81.8, γ = 71.3°. The structure was determined by the molecular-replacement method as outlined in Section 2 and was refined to 2.63 Å resolution, with a final R factor of 0.185 and R free of 0.244. Table 1 ▸ lists the data-collection parameters and Table 2 ▸ shows the refinement parameters. The crystals contained eight copies of the rFab complex per asymmetric unit.
Although the Fab complex crystals were obtained from a drop containing Fab and cocaine, the structure revealed that the hapten observed bound to the Fab in the crystals was BE and not cocaine. In all eight copies of the complex that were refined independently in the crystal, the electron-density map shows a carboxyl group at the C2 position of the tropane ring system but not a methyl ester, consistent with BE and not cocaine. In five of the copies of the Fab complex molecule in the crystal, the electron density supported the modeling of a water molecule near the carboxyl group of benzoylecgonine.
Non-enzymatic hydrolysis of the methyl ester group of cocaine is efficient at ambient temperature and at pH values above 5.5 (Das Gupta, 1982 ▸; Warner & Norman, 2000 ▸; Murray & Al-Shora, 1978 ▸). Although the above-reported conditions are not the same as those used in our crystallization trials, the complex crystals were observed after two months at 16°C employing neutral pH conditions (pH 7.5). The hydrolysis of the methyl ester group of cocaine resulted in the formation of BE, which subsequently complexed with the Fab and produced crystals. No crystals were observed early in the experiment when the cocaine would have been intact.
3.3. Overall structure of the h2E2 Fab–benzoylecgonine complex
The amino-acid sequence of the variable domains of h2E2 Fab is shown in Fig. 1 ▸. The structure of the h2E2 Fab shows the typical immunoglobulin light (L) and heavy (H) chains organized with the two variable domains, VH and VL, related by a pseudo-twofold axis and the two constant domains, CH1 and CL, related by another pseudo-twofold axis. The BE binding site is provided by the three complementarity-determining regions (CDRs) of VH and VL domains, termed H1, H2, H3 and L1, L2, L3 (see Fig. 1 ▸ for the sequence; Kabat numbering is shown). There are eight copies of the Fab–BE complex in the asymmetric unit of the crystal, which were independently refined without any noncrystallographic symmetry imposed on them. The electron density is of good quality except for one segment connecting the β-strands in the constant domain of the heavy chain, residues 127–135, that was not modeled. The electron density at the C-terminal end of the H chain was also of poor quality in some of the Fab molecules in the asymmetric unit. The H-chain electron density was visible to Pro213, and the rest of the chain, including the His6 tag at the C-terminus, was presumed to be disordered since there was no discernable electron density in the maps corresponding to this sequence. The light-chain electron density was visible to Pro211, with the last four C-terminal residues absent from the electron-density maps.
Figure 1.
The amino-acid sequence of the variable domains of h2E2 Fab. The Kabat numbering for the variable-domain residues is shown. The CDRs are indicated in bold. Residues with contacts within 4.0 Å of the ligand are boxed.
3.4. Fab–benzoylecgonine interactions
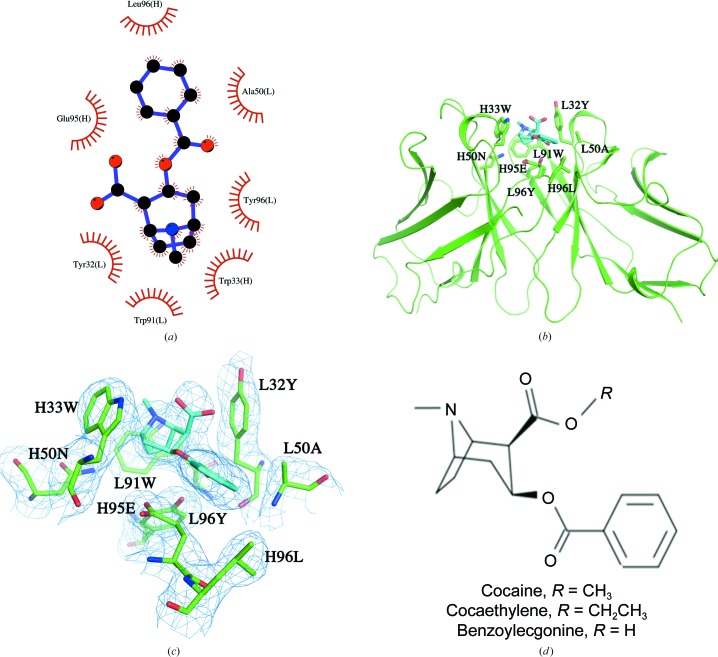
The benzoylecgonine moiety was observed to bind in a shallow groove formed by the CDR loops of the Fab, which contain an impressive array of aromatic side chains, as shown in Fig. 2 ▸. Benzoylecgonine was bound to the CDR loops of the Fab entirely through van der Waals interactions, without any direct hydrogen bonds or charge–charge interactions. The interactions between benzoylecgonine and the Fv are the same in all eight Fabs in the complex crystals, suggesting that the binding site is rigid and was not affected by the crystal packing. The residues contacting BE from the Fab include light-chain CDR loops L1 (Tyr32), L2 (Ala50) and L3 (Trp91 and Tyr96) and heavy-chain CDR loops H1 (Trp33), H2 (Asn50) and H3 (Glu95 and Leu96), resulting in a total of 65 contacts within a 4.0 Å cutoff distance. Although GlyL49 is not considered to be part of CDR L1, it does contact the ligand. The BE tropane ring system is surrounded by aromatic side chains of TyrL32, TrpL91, TyrL96 and TrpH33. One cation–π interaction can be described for TrpH33, for which the center of the aromatic ring is at a distance of 4.0 Å from the positively charged N atom of the tropane ring. Interestingly, the benzoyl ring of BE makes no contacts with the aromatic side chains of the Fab; instead, the contacts were provided by GlyL49, AlaL50, GluH95 and LeuH96.
Figure 2.
(a) A schematic view of the interactions of benzoylecgonine and h2E2 Fab generated by LigPlot+ (Laskowski & Swindells, 2011 ▸). (b) A view of the complex structure showing the binding-site interactions between benzoylecgonine (C atoms in cyan) and the variable-region residues of the h2E2 Fab (C atoms in green). The ligand and the binding residues from the Fab are shown as stick drawings, whereas the VL and VH domains are shown as a Cα carton in green. (c) A view of the electron density observed in a 2mF o − DF c OMIT map is shown for benzoylecgonine and the surrounding residues contoured at 1σ. (d) Chemical representation of cocaine and its analogs benzoylecgonine and cocaethylene.
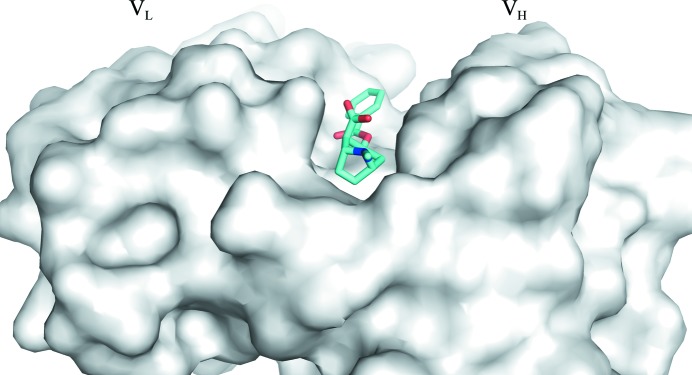
The carboxylic acid group of BE is fully exposed to the solvent with no contacts with the Fab within 4 Å (see Figs. 2 ▸ b and 3 ▸). A water molecule mediates hydrogen-bond interactions between the carbonyl group of the BE benzoyl ester and the main-chain O atom of TyrL32 and between the main-chain N atom of AlaL50 and the side-chain hydroxyl group of TyrL96.
Figure 3.
A surface (gray) rendering of h2E2 Fab showing the binding site of benzoylecgonine (stick drawing) located in a surface groove.
3.5. Structure of the unliganded (or free) Fab
Crystallization trials carried out with the purified, unliganded Fab fragment produced by proteolysis of the h2E2 antibody resulted in poor-quality crystals under many conditions containing PEG as a precipitant. Extensive attempts to optimize these conditions did not prove successful. Subsequently, the purified Fab fragment was subjected to reductive methylation of lysines as described in Section 2. The resulting Fab sample is termed mFab. This facilitated the crystallization of the Fab fragment. Importantly, lysine methylation of the Fab did not diminish its affinity for cocaine. The unliganded mFab crystals belonged to space group C2, with unit-cell parameters a = 110.8, b = 128.4, c = 91.2 Å, β = 125.3°. The structure was determined by molecular replacement using the Fab portion of the structure of the benzoylecgonine complex.
3.6. Structural comparison of the free Fab and the complex
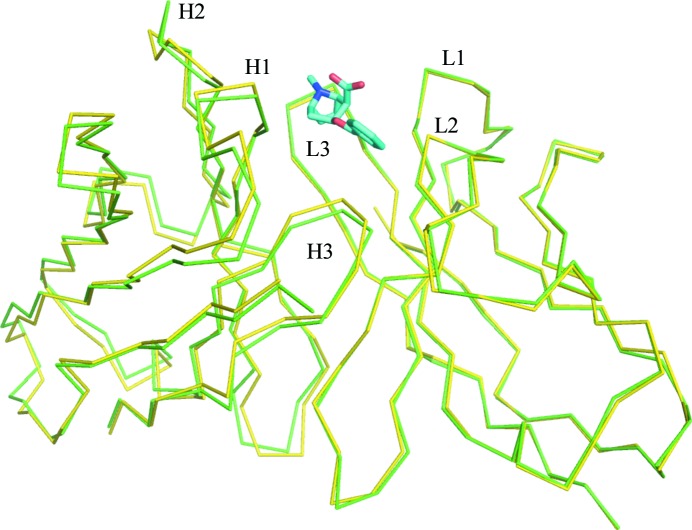
A structural overlap of the variable domains of unliganded mFab and the rFab–BE complex is shown in Fig. 4 ▸. Since the free and complex Fabs crystallized in different unit cells and space groups, comparison between them is complicated by the effects of crystal packing on local conformations and different elbow angles. Caution should be taken in the interpretation of the changes that are observed. Comparison of the Fv (both VL and VH) of one free Fab (chains L and H) with one of the complexed Fabs (chains A and B) with the closest elbow angle resulted in an r.m.s.d. of 0.6 Å, whereas the same comparison with the other Fab molecule found in the free Fab crystal shows an r.m.s.d. of 0.7 Å. When the individual variable domains are compared (i.e. VL versus VL and VH versus VH), the r.m.s.d.s are 0.3 and 0.7 for the VL domains and 0.4 and 0.6 for the VH domains, respectively, for the two Fabs found in the free Fab crystals. The CDR loops did not show any larger deviations compared with other parts of the variable domains. Fig. 4 ▸ highlights the deviations observed in the respective VH domains when the two VL domains are superimposed, reflecting a minor variation in the VL/VH association between the free Fab and the complexed Fab. The elbow angles observed for the Fab molecules in the crystals of both the complex and the unliganded structures are presented in Supplementary Table S5.
Figure 4.
A view of the Cα trace of the structural overlap of unliganded and liganded h2E2 Fab. The light-chain V domains are superposed to illustrate the deviations in the heavy-chain variable domain. Complex/liganded Fab is in green and free/unliganded Fab is in yellow. The ligand is shown as a stick drawing (C atoms in cyan, O atoms in red, N atoms in blue). The VH domain is on the left and the VL domain is on the right. The CDR loops are labeled.
4. Discussion
As mentioned in Section 1, two structures of antibody Fab–cocaine complexes have previously been published for the anti-cocaine antibodies M82G2 and GNC92H2. The antibody GNC92H2 was generated using benzoylecgonine conjugated to keyhole limpet hemocyanin (KLH) via a six-carbon adipic acid linker at the C2 ester site as an antigen. The mAb 2E2, like GNC92H2, was raised to benzoylecgonine (BE) coupled to KLH but linked via an amide bond rather than an ester, using 1,4-diaminobutane as a linker (see Supplementary Fig. S1). The difference between 2E2 and h2E2 is that in the humanized version the constant domain of the mouse lambda light chain is replaced by the human light-chain constant domain. The M82G2 antibody was also raised against a BE hapten but linked at the p-benzoyl position via a short thiocyanate coupler to bovine testes β-galactosidase. In addition, the structure of the cocaine-degrading catalytic antibody 7A1 Fab has been reported in complex with cocaine (Zhu et al., 2006 ▸). The structure of the 7A1 Fab–cocaine complex (PDB entry 2ajv) shows that cocaine is bound on a surface groove similar to the h2E2 complex, but the dissociation constant for cocaine binding for this antibody was not reported.
4.1. Crystallization
It is important to note the differences in the two h2E2 Fab samples (rFab versus mFab) used in the present study, which probably affected their crystallization behavior. The two protein samples differ at the C-terminal end of the heavy chain. The C-terminal amino-acid sequence of the heavy chain of the Fab obtained by proteolytic cleavage of the IgG is 210RVEPKSCDK218, whereas the sequence of the heavy chain in the E. coli-produced rFab is 210KVEPKSCHHHHHH222. The former sample modified further by reductive methylation of lysines (mFab) was used in the production of apo Fab crystals, whereas the latter sample was used for the Fab–BE complex.
Although reductive methylation of the h2E2 Fab produced by cleavage of the mAb allowed the identification of conditions for crystallization of the unliganded protein, no crystals were produced in the presence of cocaine under the same crystallization conditions. This implies that the Fab structure bound to cocaine (or benzoylecgonine) is not compatible with the packing of molecules in this crystal form. Indeed, in the apo crystals the ligand-binding site is involved in crystal contacts in one of the two Fab molecules in the asymmetric unit. The side chains of the ligand-binding residues Tyr34 and Trp93 of the light chain form hydrogen bonds to the main-chain atoms of a crystallographically related molecule (see Supplementary Table S6 for details). In addition, the aromatic side chain of Trp93 forms cation–π interaction with a lysine residue across the crystallographic interface. Not surprisingly, attempts to soak cocaine into apo Fab crystals were not successful. Transfer of the apo crystals to mother liquor containing cocaine resulted in the immediate breakup and dissolution of the crystals. Future experiments will involve the identification of a new crystal form of the apo Fab in which the binding site is not occluded and a search for conditions for h2E2 Fab–cocaine complex crystallization at low pH (≤5) that inhibit the hydrolysis of cocaine.
4.2. Hapten recognition by h2E2
As predicted by the previous computational docking modeling study of cocaine binding by the 2E2 Fab (Lape et al., 2010 ▸), benzoylecgonine binds not in a pocket but in a groove situated between the VH and VL domains. The recognition of the benzoylecgonine ligand by the h2E2 Fab is primarily driven by aromatic side chains trapping the tropane ring system. This is consistent with the large quenching of h2E2 mAb tyrosine and tryptophan fluorescence, which was used to measure the binding of cocaine and BE to intact h2E2 as well as to the h2E2 Fab fragment (Kirley & Norman, 2015 ▸). In the conformation of benzoylecgonine observed in the h2E2 Fab complex structure, the methyl group of the ester which would be present in cocaine or the ethylene group which would be present in cocaethylene would be fully exposed to solvent and would not contact the Fab (see Figs. 2 ▸ b and 3 ▸). The BE-binding residues or the crystal contacts observed in the present Fab complex structure do not preclude the possibility of cocaine binding to the Fab in exactly the same orientation. No steric conflicts were observed when benzoylecgonine in the binding site is replaced by cocaine in silico (see the surface rendition of the binding site in Fig. 3 ▸, which shows the carboxyl group exposed to solvent) in any of the eight independent Fab molecules. However, the origin of the observed selectivity of h2E2 for cocaine and cocaethylene relative to benzoylecgonine (K d of 1 nM for cocaethylene and 4 nM for cocaine compared with 20 nM for benzoylecgonine) is not apparent from this structure (Kirley & Norman, 2015 ▸). Cocaine/cocaethylene binding could be accompanied by minor changes in the conformation of the h2E2 CDR loops. It is unlikely that cocaine and cocaethylene have a completely different binding orientation compared with benzoylecgonine, especially considering the relatively small differences in the binding energies deduced from the abovementioned K d values (a 0.8 kcal mol−1 gain in free energy in the case of cocaine compared with BE and a 1.8 kcal mol−1 gain in free energy in the case of cocaethylene compared with BE).
4.3. Comparison of the h2E2 Fab complex with the Fab complexes of the M82G2 and GNC92H2 anti-cocaine antibodies
The antibodies M82G2 (Pozharski et al., 2005 ▸) and GNC92H2 (Larsen et al., 2001 ▸) bind cocaine with a much lower affinity (micromolar dissociation constants) compared with h2E2 (nanomolar dissociation constant). All three antibodies consist of IgG1 heavy chains, but the previously reported antibodies both have a kappa light chain, while h2E2 contains the less common lambda light chain and therefore has a VL domain sequence that is quite distinct from the kappa light chains of the other two anti-cocaine mAbs. In the case of the M82G2 Fab, structures of the free Fab (PDB entry 1rfd), cocaine-bound Fab (PDB entry 1q72) and benzoylecgonine-bound Fab (PDB entry 1qyg) have been deposited in the Protein Data Bank. Pozharski et al. (2005 ▸) extensively discussed the Fab–cocaine binding interactions of the M82G2 (PDB entry 1q72) and GNC92H2 (PDB entry 1i7z; Larsen et al., 2001 ▸) antibodies in view of their similar binding affinities and noted diversity of ligand recognition.
The binding site is a surface groove in both the h2E2 and M82G2 Fabs, as opposed to a deep pocket in the case of the GNC92H2 Fab. Different cocaine binding modes are observed in these antibodies owing to the differences in the CDR loop structures and in the placement of the aromatic residues in their respective heavy and light chains, which play an important role in recognition. A simple comparison of the number of van der Waals contacts between the ligand and the Fab shows that the number is larger for the M82G2 and GNC92H2 Fab complexes (Table 3 ▸) compared with the h2E2 Fab–benzoylecgonine complex. No hydrogen bonds (distance between non-H atoms of ≤3.2 Å between the ligand and the Fab) were observed between cocaine and the Fab in the two previously published complexes, but two hydrogen bonds were present between benzoylecgonine and Fab in the case of the M82G2 Fab complex (PDB entry 1qyg). Water-mediated hydrogen bonds were observed between the Fab and ligands in all structures (Table 3 ▸). Detailed calculations are warranted in order to understand the basis of the difference in affinities observed between the previously reported Fab complexes and that of h2E2.
Table 3. Comparison of the number of van der Waals contacts and hydrogen bonds (the distance cutoff between non-H atoms is ≤3.2 Å) between the ligand (cocaine or benzoylecgonine) and the Fab in various complex structures.
The numbers of water molecules mediating hydrogen bonds between the ligand and the Fab are included in a separate column.
| Fab complex | No. of contacts (≤4.0 Å) | No. of hydrogen bonds | No. of water molecules mediating hydrogen bonds |
|---|---|---|---|
| h2E2 (benzoylecgonine; this study) | 66 | 0 | 1 |
| PDB entry 1qyg (benzoylecgonine; Pozharski et al., 2005 ▸) | 103 | 2 | 2 |
| PDB entry 1q72 (cocaine; Pozharski et al., 2005 ▸) | 93 | 0 | 1 |
| PDB entry 1i7z (cocaine; Larsen et al., 2001 ▸) | 68 | 0 | 1 |
5. Conclusion
The benzoylecgonine moiety is bound to a surface groove and its recognition by the h2E2 mAb is driven by the aromatic side chains of the Fab surrounding and trapping the tropane ring system of the ligand, forming significant van der Waals interactions that probably contribute to the nanomolar affinity observed. The smaller metabolites of cocaine such as ecgonine methyl ester and ecgonine are probably discriminated against by the antibody owing to the smaller hydrophobic surface presented by these metabolites to the antibody. It is likely that cocaine will bind in the same position as benzoylecgonine but accompanied by minor rearrangements of the hypervariable segments that could result in interactions with the methyl ester group of cocaine. Future refinements of the current study will require crystallization under conditions that inhibit the non-enzymatic hydrolysis of ester groups for the generation of Fab–cocaine and Fab–cocaethylene complex crystals. In addition, computational studies of binding free energies using free-energy perturbation methods may lead to a clearer understanding of the ligand-dependent binding properties observed for this antibody.
6. Related literature
The following references are cited in the supporting information for this article: Fishwild et al. (1996 ▸).
Supplementary Material
PDB reference: Fab fragment of anti-cocaine antibody h2E2, 6nex
PDB reference: bound to benzoylecgonine, 6nfn
Detailed information on methods. DOI: 10.1107/S2053230X19013608/rl5180sup1.pdf
Acknowledgments
The Structural Biology Center beamlines are supported by the US Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357. The authors declare the following conflict of interest: Drs Norman and Ball are named as inventors on issued patents on the matter and the use of the h2E2 anti-cocaine monoclonal antibody.
Funding Statement
This work was funded by National Institutes of Health grant U01DA039550 to Andrew B. Norman.
References
- Adams, P. D., Afonine, P. V., Bunkóczi, G., Chen, V. B., Davis, I. W., Echols, N., Headd, J. J., Hung, L.-W., Kapral, G. J., Grosse-Kunstleve, R. W., McCoy, A. J., Moriarty, N. W., Oeffner, R., Read, R. J., Richardson, D. C., Richardson, J. S., Terwilliger, T. C. & Zwart, P. H. (2010). Acta Cryst. D66, 213–221. [DOI] [PMC free article] [PubMed]
- Carrera, M. R., Ashley, J. A., Parsons, L. H., Wirsching, P., Koob, G. F. & Janda, K. D. (1995). Nature (London), 378, 727–730. [DOI] [PubMed]
- Carrera, M. R., Ashley, J. A., Wirsching, P., Koob, G. F. & Janda, K. D. (2001). Proc. Natl Acad. Sci. USA, 98, 1988–1992. [DOI] [PMC free article] [PubMed]
- Carrera, M. R., Ashley, J. A., Zhou, B., Wirsching, P., Koob, G. F. & Janda, K. D. (2000). Proc. Natl Acad. Sci. USA, 97, 6202–6206. [DOI] [PMC free article] [PubMed]
- Carrera, M. R., Trigo, J. M., Wirsching, P., Roberts, A. & Janda, K. D. (2005). Pharmacol. Biochem. Behav. 81, 709–714. [DOI] [PubMed]
- Cerny, E. H. & Cerny, T. (2008). Expert Opin. Investig. Drugs, 17, 691–696. [DOI] [PubMed]
- Cowtan, K. (2008). Acta Cryst. D64, 83–89. [DOI] [PMC free article] [PubMed]
- Das Gupta, V. (1982). Int. J. Pharm. 10, 249–257.
- Donnelly, M. I., Zhou, M., Millard, C. S., Clancy, S., Stols, L., Eschenfeldt, W. H., Collart, F. R. & Joachimiak, A. (2006). Protein Expr. Purif. 47, 446–454. [DOI] [PMC free article] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Fishwild, D. M., O’Donnell, S. L., Bengoechea, T., Hudson, D. V., Harding, F., Bernhard, S. L., Jones, D., Kay, R. M., Higgins, K. M., Schramm, S. R. & Lonberg, N. (1996). Nature Biotechnol. 14, 845–851. [DOI] [PubMed]
- Fox, B. S., Kantak, K. M., Edwards, M. A., Black, K. M., Bollinger, B. K., Botka, A. J., French, T. L., Thompson, T. L., Schad, V. C., Greenstein, J. L., Gefter, M. L., Exley, M. A., Swain, P. A. & Briner, T. J. (1996). Nature Med. 2, 1129–1132. [DOI] [PubMed]
- Kim, Y., Dementieva, I., Zhou, M., Wu, R., Lezondra, L., Quartey, P., Joachimiak, G., Korolev, O., Li, H. & Joachimiak, A. (2004). J. Struct. Funct. Genomics, 5, 111–118. [DOI] [PMC free article] [PubMed]
- Kirley, T. L. & Norman, A. B. (2015). Hum. Vaccin. Immunother. 11, 458–467. [DOI] [PMC free article] [PubMed]
- Koschubs, T., Dengl, S., Dürr, H., Kaluza, K., Georges, G., Hartl, C., Jennewein, S., Lanzendörfer, M., Auer, J., Stern, A., Huang, K.-S., Packman, K., Gubler, U., Kostrewa, D., Ries, S., Hansen, S., Kohnert, U., Cramer, P. & Mundigl, O. (2012). Biochem. J. 442, 483–494. [DOI] [PubMed]
- Kosten, T. R. & Owens, S. M. (2005). Pharmacol. Ther. 108, 76–85. [DOI] [PubMed]
- Kvello, A. M., Andersen, J. M., Oiestad, E. L., Morland, J. & Bogen, I. L. (2016). J. Pharmacol. Exp. Ther. 358, 181–189. [DOI] [PubMed]
- Lape, M., Paula, S. & Ball, W. J. (2010). Eur. J. Med. Chem. 45, 2291–2298. [DOI] [PMC free article] [PubMed]
- Larsen, N. A., Zhou, B., Heine, A., Wirsching, P., Janda, K. D. & Wilson, I. A. (2001). J. Mol. Biol. 311, 9–15. [DOI] [PubMed]
- Laskowski, R. A. & Swindells, M. B. (2011). J. Chem. Inf. Model. 51, 2778–2786. [DOI] [PubMed]
- Martell, B. A., Mitchell, E., Poling, J., Gonsai, K. & Kosten, T. R. (2005). Biol. Psychiatry, 58, 158–164. [DOI] [PubMed]
- Martell, B. A., Orson, F. M., Poling, J., Mitchell, E., Rossen, R. D., Gardner, T. & Kosten, T. R. (2009). Arch. Gen. Psychiatry, 66, 1116–1123. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Minor, W., Cymborowski, M., Otwinowski, Z. & Chruszcz, M. (2006). Acta Cryst. D62, 859–866. [DOI] [PubMed]
- Murray, J. B. & Al-Shora, H. I. (1978). J. Clin. Pharm. Ther. 3, l–6.
- Norman, A. B. & Ball, W. J. (2012). Immunotherapy, 4, 335–343. [DOI] [PMC free article] [PubMed]
- Norman, A. B., Gooden, F. C. T., Tabet, M. R. & Ball, W. J. (2014). Drug Metab. Dispos. 42, 1125–1131. [DOI] [PMC free article] [PubMed]
- Norman, A. B., Norman, M. K., Buesing, W. R., Tabet, M. R., Tsibulsky, V. L. & Ball, W. J. (2009). J. Pharmacol. Exp. Ther. 328, 873–881. [DOI] [PMC free article] [PubMed]
- Norman, A. B., Tabet, M. R., Norman, M. K., Buesing, W. R., Pesce, A. J. & Ball, W. J. (2007). J. Pharmacol. Exp. Ther. 320, 145–153. [DOI] [PubMed]
- Paula, S., Tabet, M. R., Farr, C. D., Norman, A. B. & Ball, W. J. (2004). J. Med. Chem. 47, 133–142. [DOI] [PubMed]
- Pozharski, E., Moulin, A., Hewagama, A., Shanafelt, A. B., Petsko, G. A. & Ringe, D. (2005). J. Mol. Biol. 349, 570–582. [DOI] [PubMed]
- Rosenbaum, G., Alkire, R. W., Evans, G., Rotella, F. J., Lazarski, K., Zhang, R.-G., Ginell, S. L., Duke, N., Naday, I., Lazarz, J., Molitsky, M. J., Keefe, L., Gonczy, J., Rock, L., Sanishvili, R., Walsh, M. A., Westbrook, E. & Joachimiak, A. (2006). J. Synchrotron Rad. 13, 30–45. [DOI] [PMC free article] [PubMed]
- Schiweck, W. & Skerra, A. (1995). Proteins, 23, 561–565. [DOI] [PubMed]
- Schlapschy, M., Grimm, S. & Skerra, A. (2006). Protein Eng. Des. Sel. 19, 385–390. [DOI] [PubMed]
- Skerra, A. (1994a). Gene, 141, 79–84. [DOI] [PubMed]
- Skerra, A. (1994b). Gene, 151, 131–135. [DOI] [PubMed]
- Stanfield, R. L., Zemla, A., Wilson, I. A. & Rupp, B. (2006). J. Mol. Biol. 357, 1566–1574. [DOI] [PubMed]
- Tan, K., Kim, Y., Hatzos-Skintges, C., Chang, C., Cuff, M., Chhor, G., Osipiuk, J., Michalska, K., Nocek, B., An, H., Babnigg, G., Bigelow, L., Joachimiak, G., Li, H., Mack, J., Makowska-Grzyska, M., Maltseva, N., Mulligan, R., Tesar, C., Zhou, M. & Joachimiak, A. (2014). Methods Mol. Biol. 1140, 189–200. [DOI] [PMC free article] [PubMed]
- Treweek, J. B. & Janda, K. D. (2012). Mol. Pharm. 9, 969–978. [DOI] [PMC free article] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Walter, T. S., Meier, C., Assenberg, R., Au, K. F., Ren, J., Verma, A., Nettleship, J. E., Owens, R. J., Stuart, D. I. & Grimes, J. M. (2006). Structure, 14, 1617–1622. [DOI] [PMC free article] [PubMed]
- Warner, A. & Norman, A. B. (2000). Ther. Drug Monit. 22, 266–270. [DOI] [PubMed]
- Wetzel, H. N., Tsibulsky, V. L. & Norman, A. B. (2016). Drug Alcohol Depend. 168, 287–292. [DOI] [PMC free article] [PubMed]
- Yanisch-Perron, C., Vieira, J. & Messing, J. (1985). Gene, 33, 103–119. [DOI] [PubMed]
- Zhu, X., Dickerson, T. J., Rogers, C. J., Kaufmann, G. F., Mee, J. M., McKenzie, K. M., Janda, K. D. & Wilson, I. A. (2006). Structure, 14, 205–216. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: Fab fragment of anti-cocaine antibody h2E2, 6nex
PDB reference: bound to benzoylecgonine, 6nfn
Detailed information on methods. DOI: 10.1107/S2053230X19013608/rl5180sup1.pdf