Abstract
Free-living amoebae are thought to represent an environmental niche in which amoeba-resistant bacteria may evolve towards pathogenicity. To get more insights into factors playing a role for adaptation to intracellular life, we characterized the transcriptomic activities of the emerging pathogen Mycobacterium abscessus in amoeba and murine macrophages (Mϕ) and compared them with the intra-amoebal transcriptome of the closely related, but less pathogenic Mycobacterium chelonae. Data on up-regulated genes in amoeba point to proteins that allow M. abscessus to resist environmental stress and induce defense mechanisms, as well as showing a switch from carbohydrate carbon sources to fatty acid metabolism. For eleven of the most upregulated genes in amoeba and/or Mϕ, we generated individual gene knock-out M. abscessus mutant strains, from which ten were found to be attenuated in amoeba and/or Mϕ in subsequence virulence analyses. Moreover, transfer of two of these genes into the genome of M. chelonae increased the intra-Mϕ survival of the recombinant strain. One knock-out mutant that had the gene encoding Eis N-acetyl transferase protein (MAB_4532c) deleted, was particularly strongly attenuated in Mϕ. Taken together, M. abscessus intra-amoeba and intra-Mϕ transcriptomes revealed the capacity of M. abscessus to adapt to an intracellular lifestyle, with amoeba largely contributing to the enhancement of M. abscessus intra-Mϕ survival.
Author summary
The emerging pathogen Mycobacterium abscessus differs from other rapidly growing mycobacteria by its ability to survive phagocytosis by human macrophages. These virulence traits render M. abscessus pathogenic in vulnerable hosts with structural lung disease. How patients become infected with M. abscessus remains unclear, as M. abscessus is not or only very rarely isolated from the environment. Co-culturing of M. abscessus and amoeba increases the virulence of the bacteria for subsequent mouse infection, but little is known about M. abscessus virulence in general. Until now, the prediction of M. abscessus virulence factors was essentially based on the comparative analysis of the genome of M. abscessus with that of Mycobacterium chelonae, a closely related mycobacterium that occasionally causes opportunistic human infections, but to a much lesser extent than M. abscessus. Here, by transcriptomic and functional approaches, we decipher a global view on M. abscessus adaptation to intracellular life.
Introduction
To date, most of the known mycobacterial species are environmental organisms found in soil [1], air [2] and water [3–5], and belong to the Rapid Growing Mycobacteria (RGM). In contrast, pathogenic mycobacteria mostly belong to the Slow Growing Mycobacteria (SGM), although some exceptions exist, as for example Mycobacterium abscessus, an emerging mycobacterial pathogen that is causing serious infections in patients with cystic fibrosis or other structural lung diseases. M. abscessus is member of the Mycobacterium chelonae complex, which includes M. abscessus, M. chelonae and Mycobacterium immunogenum. Together with Mycobacterium fortuitum, the M. chelonae complex members represent the main opportunistic pathogens among RGM [6–8].
Compared to other Non-Tuberculous Mycobacteria (NTM), recovery of M. abscessus from the environment is rare [9]. However, information from its genome sequence suggests the presence of the bacterium at the interface of soil, vegetation and water, an environment where free-living amoebae (FLA) are commonly found [10]. FLA have been isolated from habitats in common with mycobacteria [11,12] including cold-drinking water systems [13,14], hot water systems in hospitals and cooling towers [15]. FLA are ubiquitous organisms that feed on bacteria, and these bacteria have likely developed adaptations to the intracellular lifestyle to become Amoeba-Resistant Bacteria (ARB) [16,17]. Mycobacteria have been isolated from such habitats by amoebal enrichment techniques [18,19], suggesting that horizontal gene transfer and adaptation to an intracellular lifestyle might also take place in such an environment [20–22]. Finally, amoebae are often considered as an ancestral form of macrophages (Mϕ) sharing similar cellular structures and biological features [23–25].
M. abscessus has been shown to be resistant to amoeba phagocytosis and encystment, a property shared with all mycobacteria with the exception of the attenuated M. bovis BCG vaccine strain [16,26,27]. In addition, co-culture of M. abscessus with Acanthamoeba castellanii (Ac) increases its virulence when subsequently used for aerosol infection in the mouse model [27]. Similarly, co-culture of amoebae with M. avium was found to trigger M. avium virulence by enhancing both entry and intracellular multiplication of the bacterium [28]. The essential role of the ESX-4 M. abscessus type VII secretion system (T7SS) has also been demonstrated based on an intra-amoebal viability screen of M. abscessus, unraveling the active role of ESX-4 in intracellular survival of M. abscessus [29]. Taken together, it seems likely from reports in the literature that M. abscessus has contact to FLA as part of its natural lifecycle, which however is only poorly explored to date.
In order to gain deeper insights into this individual lifestyle of M. abscessus, we sought for ways to characterize the virulence traits and decided to study the transcriptional signatures of M. abscessus in Ac and Mϕ, in comparison to the ones of M. chelonae, with the aim of identifying potential adaptations of M. abscessus to intracellular life and to pathogenicity. Although amoebae and Mϕ share common features, it has been shown that an intra-amoebal life requires specific adaptations [29]. A full description and analysis of M. abscessus transcriptomes shall allow a complete picture of M. abscessus intracellular replication and survival mechanisms to be obtained, both in an amoeba environmental host and in Mϕ.
Results
Overall description of the M. abscessus intracellular transcriptomes
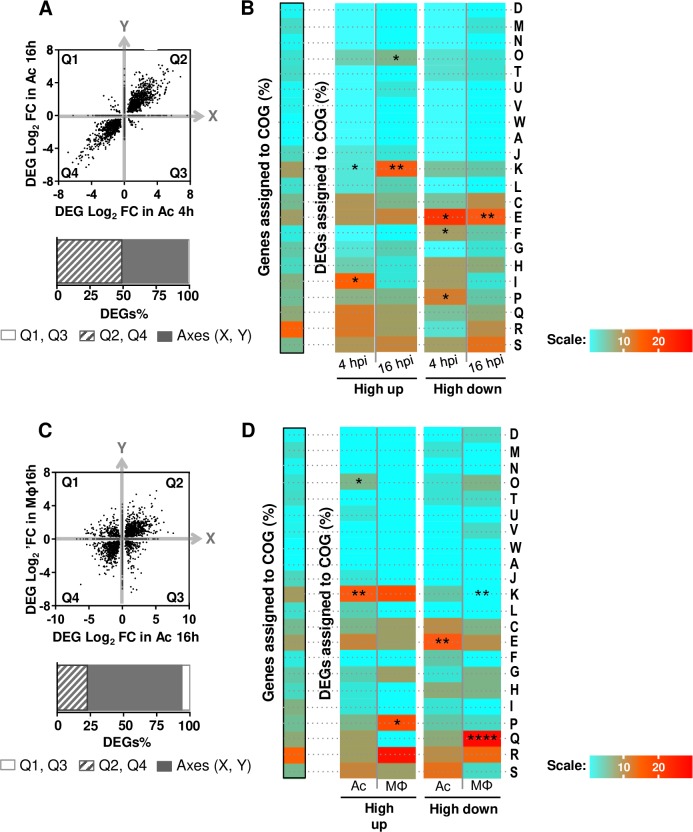
RNAseq data from M. abscessus planktonic or intracellular cultures (3 to 4 replicates per condition) were analyzed and compared to identify M. abscessus genes that were up- or down-regulated after Ac and Mϕ co-cultures. Differentially expressed genes (DEG) are thus defined as genes for which expression changes between intracellular and planktonic growth. Transcriptomes of M. abscessus in Ac 4 and 16 hours post-infection (hpi), in Mϕ 16 hpi and transcriptomes of M. chelonae in Ac 16 hpi were obtained and invariant genes were excluded from the analyses. Normalization and hierarchical clustering of normalized raw data confirmed the technical quality of transcriptomes (S1 Fig). DEGs were identified using the DESeq2 package [30] (S1 Table) and the fold change (FC) values were recorded. In particular, Log2 fold change (Log2FC) values from M. abscessus transcriptomes were then compared. In Ac, most DEGs up-regulated or down-regulated at 4 hpi were still up- or down-regulated at 16 hpi (Fig 1A). In order to detail the biological changes that correlated with M. abscessus intracellular regulation, we first grouped the DEGs into clusters of orthologues (Cluster of Orthologous Groups or COGs) [31]. Highly up-regulated genes (Log2FC > 4) in Ac were more frequently found in COG O (Post-translational modification, protein turnover, chaperones), COG K (Transcription) and COG I (Lipid transport and metabolism) compared to the genome reference, although this last category tended to be under-represented at 16 hpi, suggesting that the percentage of DEGs allocation for this category is lower than that from the genome allocation (Fig 1B). Comparatively, highly down-regulated genes (Log2FC < -4) were assigned to COG E and COG F (amino-acid and nucleotide transport and metabolism respectively) (Fig 1B).
Fig 1.
Description of M. abscessus transcriptomes in Ac (A, B) and Mϕ (C, D). Differentially expressed genes (DEGs) from comparisons of co-cultures of mycobacteria with A. castellanii (Ac) and macrophages (Mϕ) relative to planktonic growth were categorized according to their fold change (FC) expressed in Log2. DEGs Log2 fold change (FC) in Ac 4 hpi (X axis) are plotted against DEGs FC 16 hpi (Y axis) (A) DEGs Log2 FC in Ac 16 hpi (X axis) are also plotted against DEGs FC in macrophages (Mϕ) 16 hpi (Y axis) (C). Each dot on the graph corresponds to a DEG. DEGs from quadrant 2 (Q2) and 4 (Q4) are genes regulated in the same direction whereas DEGs from Q1 and Q3 are genes regulated in opposite direction. Dots on the plot axes (X, Y) are genes regulated in one condition only. Proportions of DEGs of Q1, Q2, Q3, Q4 and axis (X, Y) are quantified in the histogram. M. abscessus adaptations to Ac (B) and Mϕ (D) unraveled by COG categorization. Highly regulated genes (Log2FC > |4|) were assigned to COGs. The genome assignation (framed in black) serves as a reference for COG enrichment tests. Fisher’s exact tests were performed to compare the transcriptome sets and the genome set of gene assignments to COG. Assignments are expressed in percentages and depicted with a scale from light blue (lower percentages) to red (higher percentages). If the DEGs percentage for a COG category is significantly lower than that of the genome, the COG is under-represented. On the contrary, if the DEGs percentage for a COG category is significantly higher than that of the genome, the COG is over-represented. D: Cell cycle control, cell division, chromosome partitioning, M: Cell wall/membrane/envelope biogenesis, N: Cell motility, O: Post-translational modification, protein turnover, chaperones, T: Signal transduction mechanisms, U: Intracellular trafficking, secretion, and vesicular transport, V: Defense mechanisms, W: Extracellular structures, A: RNA processing and modification, J: Translation, ribosomal structure and biogenesis, K: Transcription, L: Replication, recombination and repair, C: Energy production and conversion, E: Amino acid transport and metabolism, F: Nucleotide transport and metabolism, G: Carbohydrate transport and metabolism, H: Coenzyme transport and metabolism, I: Lipid transport and metabolism, P: Inorganic ion transport and metabolism, Q: Secondary metabolites biosynthesis, transport and catabolism, R: General function prediction only, S: Function unknown. * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001.
At first glance, exposure of M. abscessus to the intra-amoebal and intra-macrophagic environments generated similar gene regulation patterns. Indeed, by comparing M. abscessus Ac-16hpi vs. Mϕ-16hpi transcriptomes, we show that only 20% of DEGs were regulated in the opposite direction (increased versus decreased and vice versa) and only 10% were specific to Ac or Mϕ (Fig 1C). The representation of DEGs according to their FC patterns highlighted that most DEGs showed low changes in both Ac and Mϕ (Log2FC < |2|) 16 hpi (S2A Fig). Nonetheless, in comparison to the situation in Mϕ, large variations of gene expression patterns were observed in Ac, the percentage of genes with a FC over 4 (Log2FC > |2|) being 4 times higher than in Mϕ (S2A Fig). In addition, up-regulated DEGs predominated in Ac in comparison to the in-Mϕ environment (S2B Fig). Thus, the intra-amoebal environment seems more prone to large variations and in particular to strong induction of genes compared to the in-Mϕ environment.
COG assignments highlighted the differences between M. abscessus high DEGs in Mϕ and Ac (Fig 1D). COG O, which was over-represented in the M. abscessus highly up-regulated genes in Ac, was more frequently associated with highly down-regulated genes in Mϕ (Fig 1B and 1D). By comparison, COG P (Inorganic ion transport and metabolism) was over-represented in the M. abscessus highly up-regulated genes in Mϕ only, potentially illustrating different adaptations to amoebal and Mϕ environments (Fig 1D).
Main biological pathway changes of M. abscessus in Ac and Mϕ
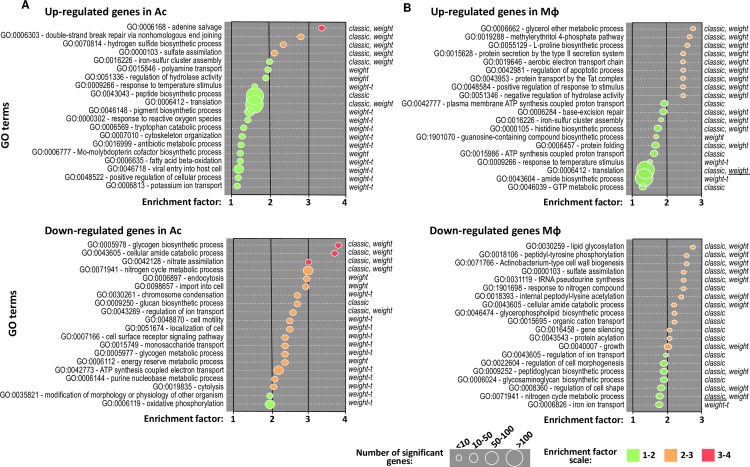
We performed a gene ontology (GO) enrichment analysis to further characterize the M. abscessus adaptions in Ac (Fig 2A) and Mϕ (Fig 2B). GO enrichment data were qualified by an enrichment factor (EF) (1 to 4) and a number of significantly enriched genes (from small (<10) to large (>100)) (Fig 2).
Fig 2. Gene ontology enrichment analyses applied on M. abscessus intracellular transcriptomes.
Differentially expressed genes in Ac (A) and in Mϕ (B) 16 hpi. GO enrichment analysis was performed with the topGO R package [72]. Enriched GOs are sorted according to their enrichment factor (EF), corresponding to the ratio of significant DEGs assigned to the GO over expected assigned DEGs to the GO as defined by topGO [72]. Enriched GOs are represented by circles which size is proportional to the amount of significant DEGs assigned. Positive statistical tests are given that face each GO. Method giving the smallest p-value is underlined.
In Ac, the most enriched up-regulated M. abscessus genes fell into polyamine transport (GO:0015846) to adenine salvage (GO:0006168), including small groups of genes involved in sulfur metabolism (sulfate assimilation pathway (SAP) (GO:0000103), hydrogen sulfide (H2S) biosynthetic pathway (GO:0070814) and detoxification (iron-sulfur cluster assembly (GO:0016226)) (Fig 2A, upper panel).
In Mϕ, M. abscessus up-regulated enriched genes fell into different GO in comparison to those up-regulated in Ac. L-proline biosynthetic process (GO:0055129); methylerythritol 4-phosphate MEP pathway (GO:0019288) and glycerol ether metabolic process (GO:0006662) were the most enriched (Fig 2B, upper panel). These GO are followed by the type II secretion system and notably the Tat (twin-arginine translocation) pathway.
In Ac M. abscessus infections, the most enriched down-regulated genes fell into the nitrate assimilation GO (GO:0042128), to glycogen biosynthetic process GO (GO:0005978) (Fig 2A, lower panel). In particular GO related to transport and metabolism of glucose were enriched (GO:0005977, GO:00015749, GO:0009250, GO:0005978) (Fig 2A, lower panel).
In Mϕ, M. abscessus down-regulated genes related to growth and parietal activities. From GO:0040007 corresponding to growth, up to GO:0030259 corresponding to lipid glycosylation, in addition to GO:0071941 (nitrogen cycle metabolism process), GO:0009259 (peptidoglycan synthesis) and GO:0022604 (regulation of cell morphogenesis), GO enrichment analysis indicated that M. abscessus slows down its energy-demanding metabolic processes and growth rate (Fig 2B, lower panel).
Taken together, these observations suggest that M. abscessus enters a slow-replicative state in Mϕ and dedicates its energy to detoxification and protein secretion into the host.
Regulation of the central carbon metabolism of M. abscessus in Ac and Mϕ
Following the GO enrichment analysis, we investigated the different M. abscessus up- and down-regulated genes from metabolic pathways in Ac or Mϕ. The major finding was that M. abscessus switches from a simple sugar-based carbon source to fatty acids inside Ac and Mϕ (Fig 3). The glycolysis/neoglucogenesis and pentose phosphate pathways were mostly down-regulated or unchanged during intracellular growth, whereas the β-oxidation of fatty acids was up-regulated in Ac and Mϕ. This switch was observed from the early time points after Ac infection. Fifteen genes predicted to encode enzymes necessary for the biochemical activation and β-oxidation of fatty acids were up-regulated in Ac and Mϕ, such as: fatty acid-coenzyme A (CoA) synthase (fadD3, 9, 10, 19); acyl-CoA dehydrogenase (fadE5, 14, 23–24, 27–29, 31); enoyl-CoA hydratase (echA19); hydroxy-butyryl-CoA dehydrogenase (fadB2) and acetyl-CoA transferase (fadA5, 6). Genes implicated in the synthesis of enzymes involved in the breakdown of cholesterol A and B rings were highly induced in Ac and Mϕ. β-oxidation of fatty acid and cholesterol breakdown result in the accumulation of propionyl-CoA that is detoxified by the methylmalonyl pathway. By-products of these 3 pathways and the GABA shunt feed the TCA cycle. The succinate generated by the TCA cycle enables the bacterium to deal with anaerobic respiration [32]. In addition, M. abscessus may detoxify glyoxylate by converting it into malate via the glyoxylate shunt (Fig 3).
Fig 3. Intracellular M. abscessus relies on fatty acid and cholesterol catabolism.
DEGs FC of genes implicated in Central Carbon Metabolism (CCM) is represented on a Heat Map ranging from blue (DOWN DEGs) to red (UP DEGs). On this Heat Map both M. abscessus intra-amoebal (Ac) and intra-macrophagic (Mϕ) DEGs are depicted.
Furthermore, down-regulation of the mycolate operon (MAB_2027-MAB_2039) (Table 1), encompassing the β-ketoacyl-ACP synthases (KasA and KasB) and β-ketoacyl synthases (MAB_2031 & MAB_2029), as well as the malonyl-CoA acyl carrier protein transacylase (MCAT) homolog (MAB_2034), revealed that intracellular M. abscessus undergoes starvation as previously described [33].
Table 1. Regulation of M. abscessus mycolate synthesis operon in Ac and Mϕ.
| Mmaa gene | Encoded protein | Mabsb gene | FC Ac 4 hpi | FC Ac 16 hpi |
FC Mϕ 16 hpi |
|---|---|---|---|---|---|
| MYCMA_RS13950 | Acyl carrier protein | MAB_2027 | -3.28 (3.15E-17) |
-2.91 (3.08E-07) |
NCc |
| MYCMA_RS13945 | 3-oxoacyl-ACP synthase (Kas B) | MAB_2028 | -2.68 (3.42E-10) |
-2.24 (8.40E-04) |
NC |
| MYCMA_RS13940 | Beta-ketoacyl synthase | MAB_2029 | -2.95 (8.03E-11) |
-4.48 (8.40E-04) |
NC |
| MYCMA_RS13935 | 3-oxoacyl-ACP synthase (Kas A) | MAB_2030 | -3.80 (8.03E-11) |
-3.15 (3.94E-03) |
-1.06 (5.38E-03) |
| MYCMA_RS13930 | Beta-ketoacyl synthase | MAB_2031 | -2.92 (1.02E-08) | NC | -1.85 (3.66E-03) |
| MYCMA_RS13925 | 3-oxoacyl-ACP reductase | MAB_2032 | -2.16 (1.92E-05) | NC | NC |
| MYCMA_RS13920 | Thioesterase | MAB_2033 | -2.09 (3.06E-04) |
NC | NC |
| MYCMA_RS13915 | Malonyl CoA-ACP transacylase | MAB_2034 | -2.09 (3.06E-04) |
NC | NC |
| MYCMA_RS13910 | Acyltransferase | MAB_2035 | NC | NC | NC |
| MYCMA_RS13905 | Membrane protein (MmpS) | MAB_2036 | -1.16 (1.36E-02) |
NC | -1.06 (6.55E-03) |
| MYCMA_RS13900 | Hypothetical protein (MmpL) | MAB_2037 | -0.68 (8.24E-03) |
-0.86 (8.24E-03) |
-0.72 (6.97E-07) |
| MYCMA_RS13895 | Transporter | MAB_2038 | NC | NC | NC |
| MYCMA_RS13890 | Lipase (LipH) | MAB_2039 | NC | 0.86 (1.61E-02) |
0.64 (7.73E-03) |
aMma gene: Mycobacterium massiliense gene (accession number NC_018150.2).
bMabs gene: Mycobacterium abscessus gene (accession number NC_010397.1).
cNC: no change in gene expression
P-values are indicted in brackets.
Regulation of putative virulence genes of M. abscessus in Ac and Mϕ
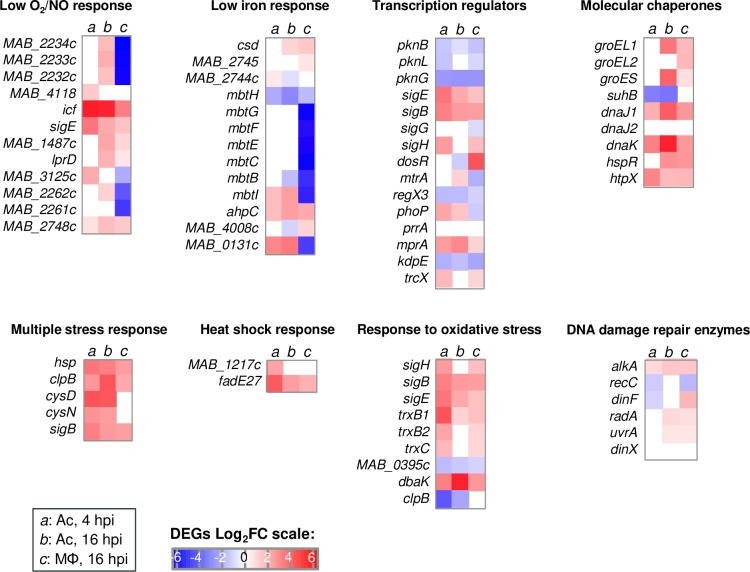
We assessed the regulation of M. abscessus genes that show orthologs in M. tuberculosis, which were known to be induced and to contribute to the cellular microbicidal defenses of the tubercle bacillus in Mϕ [34]. The results obtained were similar for infection of Ac and Mϕ, with a few exceptions in the response to low O2/NO and in the low iron response (Fig 4). Transcriptional regulators such as dosR, phoP and mtrA were regulated in the opposite direction in Ac and Mϕ, with phoP and mtrA only induced in Ac, and dosR exclusively induced in Mϕ. Other genes, known to contribute to the survival of the bacterium in response to oxidative stress [35], comprising ahpD, bcp, trxB1 and 2, and trxC genes [36], in addition to ahpC, were up-regulated in Ac and Mϕ.
Fig 4. Regulation of genes required for pathogenic mycobacteria survival in vitro.
Regulation of M. abscessus genes conserved in M. tuberculosis, known to be induced and to contribute to cellular microbicidal defenses of the tubercle bacillus in Mϕ [34] is represented on a Heat Map in a blue (repressed genes) to red (induced genes) color scale. On this Heat Map both M. abscessus intra-amoebal (Ac) and intra-macrophagic (Mϕ) DEGs are depicted and divided in categories: « broad transcription regulators » and genes implicated in the response to various intracellular stress (« Multiple stress response », « Heat Shock response », « Molecular chaperones », « DNA damage repair enzymes », « Response to low O2 / NO », « Low iron response », « Response to oxidative stress »).
Altogether, these analyses suggest that the induced sets of genes in Ac reflect the main adaptation signatures for resistance to intracellular stress that were also shown to be induced in Mϕ.
M. abscessus highly-induced genes in amoeba
Since we showed that the intra-amoebal environment is favorable to strong induction of a gene set that is thought to contribute to M. abscessus survival in phagocytic cells, the highest FC values of the M. abscessus transcriptome in Ac 4 and 16 hpi were chosen and then compared to the FC values obtained from the M. abscessus transcriptome in Mϕ and the M. chelonae transcriptome in Ac. This comparison highlighted 45 genes that were upregulated only in M. abscessus during infection of Ac (S2 Table), whereas we also found 38 genes that were most induced during infection of Mϕ (S3 Table). Based on these data sets, we selected the genes that were specifically induced in Ac, and constructed six different deletion mutants in the reference strain M. abscessus CIP 104536T (ΔOP1 to ΔOP6) (Table 2 and S2 Table). Moreover, based on the M. abscessus transcriptome data in Mϕ, we constructed additional five deletion mutants (ΔOP 7 to ΔOP11) targeting the most induced genes or genes implicated in the adaptation to intracellular stress (Table 2 and S3 Table).
Table 2. Deleted operons in M. abscessus CIP 104536T.
| Operon ID | Mabsa gene | Protein encoded | IPSb motif analysis on hypothetical proteins |
|---|---|---|---|
| OP1 (2231455) |
MAB_4664 | Hypothetical protein | No IPS |
| OP2 (395711) |
MAB_1242c | Hypothetical protein | No IPS |
| MAB_1243c | Hypothetical protein | IPS035568: ABC transporter. FecCD/TroCD-like | |
| MAB_1244c | Hypothetical protein | No IPS | |
| MAB_1245c | Hypothetical protein | No IPS | |
| MAB_1246c | Hypothetical protein | No IPS | |
| MAB_1247c | Hypothetical protein | IPS005531: Alkaline shock protein Asp23 | |
| OP3 (2230454) |
MAB_1517c | Probable O-methyltransferase Omt | |
| OP4 (396003) |
MAB_2649 | Putative membrane protein. MmpS family | |
| MAB_2650 | Putative membrane protein. MmpL family | ||
| OP5 (2231454) |
MAB_4663 | Hypothetical protein | No IPS |
| OP6 (2231514) |
MAB_4791c | Hypothetical protein | Twin-arginine translocation pathway. Signal sequence |
| OP7 (2229980) |
MAB_0086 | Taurine catabolism dioxygenase | |
| OP8 (395598) |
MAB_0734 | Hypothetical protein | Leukocidin/porin MspA superfamily (036435) |
| OP9 (396109) |
MAB_3132c | Membrane protein | No IPS |
| MAB_3133c | Hemin transporter | ||
| MAB_3134c | Transcriptional regulator | ||
| OP10 (2231416) |
MAB_4509c | Hypothetical protein | No IPS |
| OP11 (22311420) |
MAB_4532c | Hypothetical protein | N-acetyltransferase Eis (016181) |
aMabs gene: Mycobacterium abscessus gene (accession number NC_010397.1).
bIPS: InterProScan protein signature.
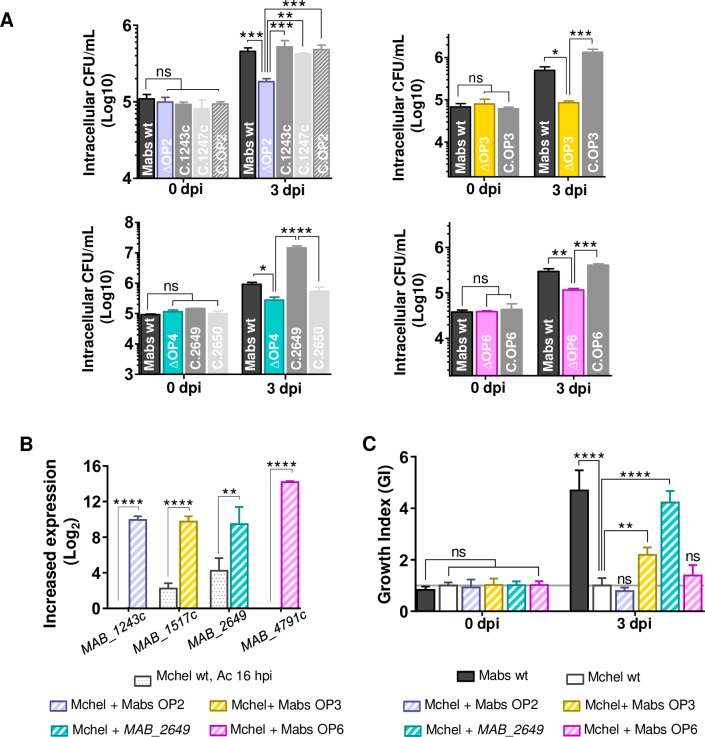
ΔOP1 to ΔOP6 strains were evaluated for their intracellular multiplication in Ac and in Mϕ (S3A Fig). All mutants were attenuated in Ac and Mϕ, except one (ΔOP5),(S3A Fig). All mutant strains had similar growth in vitro compared to the wild type strain (S3B Fig). Among the genes selected for mutagenesis, several (OP2 and 6) are absent from M. chelonae, or if present in the M. chelonae genome they were at least four times less induced (OP3 and OP4) in Ac. To demonstrate the contribution of those genes to M. abscessus virulence in macrophages, we first complemented the corresponding KO strains (ΔOP2, 3, 4 and 6). All strains recovered the wt phenotype except for the OP4 gene MAB_2650 potentially encoding an MmpL (Fig 5A). We further analyzed their contribution towards intracellular survival by overexpressing M. abscessus OP2, 3, 6 and MAB_2649 genes in M. chelonae (Fig 5B), whose growth is restricted in Mϕ in comparison to M. abscessus [27,37]. Only the overexpression of M. abscessus OP3 and OP4 (MAB_2649) increased M. chelonae survival in Mϕ (Fig 5C). By comparison, no increase in M. chelonae intracellular survival was observed when overexpressing OP2 and OP6 (Fig 5C).
Fig 5. Comparative transcriptomic analyses allow identifying genes required for M. abscessus survival in amoebae and Mϕ.
(A) Intracellular survival of 4 selected KO (ΔOP) and complemented strains in Mϕ. Mϕ were infected at 10 MOI and colony forming units (CFU) tests were performed 0 and 3 dpi. (B) Over-expression M. abscessus selected virulence genes in M. chelonae. FC values from M. chelonae transcriptome in Ac 16 hpi are compared to FC value mid-log phase cultures of M. chelonae strains overexpressing M. abscessus genes. (C) M. chelonae wt and M. chelonae overexpressing M. abscessus selected virulence genes replication in Mϕ relative to M. abscessus. Cells were infected at 10 MOI and colony forming units (CFU) tests were performed 0 and 3 dpi. The relative growth of each strain as compared to M. abscessus wt (Growth Index, GI) is given. All experiments were repeated twice or more, in duplicates (A) or triplicates (B). Statistical analyses were performed with GraphPad PRISM6. Histograms with error bars represent means ± SD. Differences between means were analyzed by ANOVA and the Tukey post-test allowing multiple comparisons to be performed. ns = non-significant. * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001.
M. abscessus highly-induced genes in Mϕ
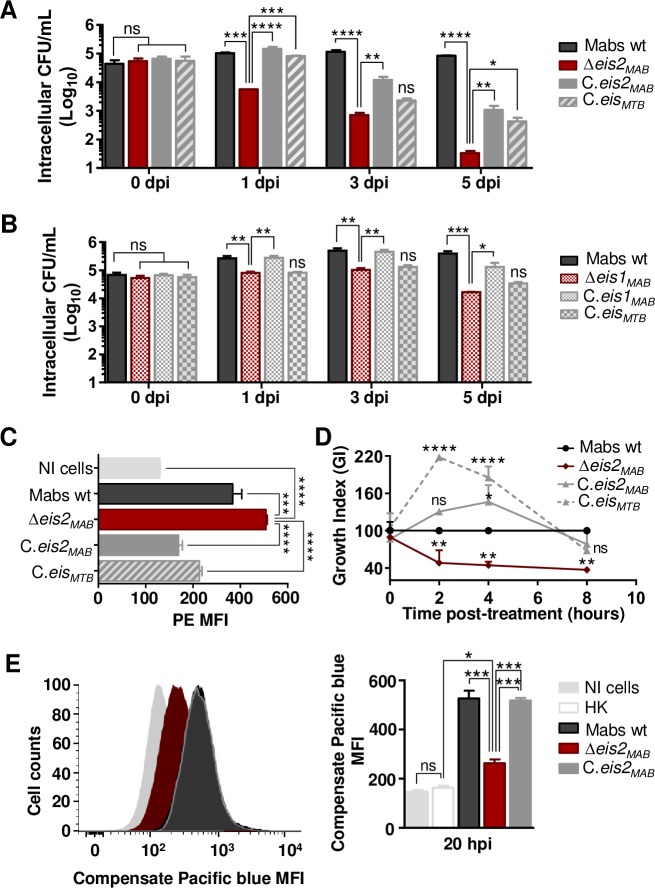
The observed defect in intracellular survival was noticed for all mutants from OP7 to OP11 (S3A Fig), the ΔOP11 mutant was particularly attenuated (GI<0.1). OP11 (MAB_4532c) KO strain tended to be eliminated by Mϕ while the growth of the KO strain was similar to the WT growth in vitro (S3B Fig). When complementing ΔOP11 strain with MAB_4532c, we restored the wt phenotype (Fig 6A). MAB_4532c encodes an Eis N-acetyl transferase protein, according to a motif analysis (InterProScan). Of interest, M. abscessus contains two eis genes, named eis1MAB (MAB_4124) and eis2MAB (MAB_4532c), whereas M. tuberculosis possesses a single eisMTB gene (Rv2416c), eis1MAB (MAB_4124) being the closest homolog by Bidirectional Best Hit (BBH) search. No conservation of synteny was observed between the respective genomic regions in M. abscessus and M. tuberculosis (S4A Fig). In contrast, the eis2MAB locus shows some similarity and conservation with the M. tuberculosis mmpL11 locus, with inverted and syntenic conservation of groups of genes (S4B Fig). The eisMTB locus is well-conserved in M. abscessus and corresponds to MAB_1619-MAB_1627 and MAB_1633-MAB_1637 regions (S4C Fig). Both eis1MAB and eis2MAB were found close to mmpL (brown arrows) and/or mmpS (orange arrow) genes (S4 Fig). In the M. abscessus eis2 locus, an mmpL gene (MAB_4529) was conserved in M. tuberculosis corresponding to mmpL11 (S4B Fig). To assess the function of M. abscessus eis genes, we first looked at the transcription and intracellular survival profiles and stated that unlike eis2MAB, eis1MAB was suppressed inside Mϕ (S5 Fig) and less impaired in its intracellular survival (Fig 6A and 6B). Complementation of Δeis1MAB and Δeis2MAB with eis1MAB or eis2MAB, respectively, allowed the recovery of the intracellular survival for both mutants (Fig 6A and 6B). We also performed transcomplementation of the Δeis1MAB and Δeis2MAB KO strains with the M. tuberculosis eis (eisMTB) variant. Of note, complementation of the mutants with the eisMTB gene allowed only partial restoration of the intracellular replicative phenotype for the Δeis2MAB mutant, but no restoration was observed for Δeis1MAB mutant (Fig 6A and 6B). Similar behaviors regarding apoptosis, necrosis, autophagy and phagosomal acidification were observed when comparing the wt M. abscessus strain with the Δeis2MAB mutant (S6 Fig). However, two major differences were observed. First, infection of Mϕ with the Δeis2MAB strain (at a MOI of 50) was associated with higher production of ROS by the cells and loss of eis2MAB also sensitized M. abscessus to ROS and notably to H2O2 (Fig 6C and 6D). Secondly, the Δeis2MAB mutant was unable to damage the phagosomal membrane and to provoke phagosome-cytosol contact as compared to the wt and complemented M. abscessus strains (Fig 6E).
Fig 6. M. abscessus eis2 gene is essential for survival in Mϕ and shares functions with M. tuberculosis eis conversely to M. abscessus eis1.
(A) Intracellular survival of M. abscessus eis2 KO strain (Δeis2MAB) and complementation in Mϕ. (B) Intracellular survival of M. abscessus eis1 KO strain (Δeis1MAB) and complementation in Mϕ. Mϕ were infected at 10 MOI and colony forming unit (CFU) tests were performed at several times post-infection (0, 1, 3 and 5 dpi). (C) Control of ROS production by M. abscessus Eis2. ROS production by Mϕ was assessed by flow cytometry with the Mitosox Red kit, 15 min post-infection at 50 MOI. (D) Sensitivity of M. abscessus eis2 KO strain to hydrogen peroxide (H2O2). Sensitivity to H2O2 was assessed by incubated bacterial cultures with H2O2 20 μM during 8 h. The amount of survival cells was determined by performing CFU tests at several hours post-infection (2, 4, 8 hpi). (E) Control of phagosomal rupture by M. abscessus Eis2. Phagosomal rupture was assessed by performing a FRET analysis as previously described [73]. Results are depicted as signal overlays per group with 1,000,000 events per condition acquired in not infected cells (NI cells), Heat killed M. abscessus (HK), wild-type M. abscessus (Mabs wt), KO strains (Δeis2MAB), KO strains complemented with eis2MAB (C.eis2MAB). All experiments were repeated twice or more in triplicates. Statistical analyses were performed with GraphPad PRISM6. Histograms with error bars represent means ± SD. Differences between means were analyzed by ANOVA and the Tukey post-test allowing multiple comparisons to be performed. ns = non-significant. * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001.
Discussion
The main objective of this work was to understand the genetic and molecular basis for the ability of smooth M. abscessus strains to withstand and survive in eukaryotic phagocytic cells. We focused our experiments on S variants as from previous work we know that this morphotype, in contrast to R variants, represents the most infectious and intracellular forms of M. abscessus strains, whereas the R form rather represents the extracellular versions of the bacteria [38]. Transcriptome analyses of M. abscessus 43S and M. chelonae strains infecting amoeba and Mϕ, respectively, revealed genes strongly induced during infection of phagocytes. Selected KO mutants constructed in M. abscessus CIP 104536T on the basis of the results from transcriptomic analyses of M. abscessus 43S allowed us to confirm that the genes, which were highlighted by our transcriptomic approaches were indeed required for mycobacterial survival in phagocytes, even in a different genetic M. abscessus strain background. These results are complementary to a previous Tn M. abscessus library viability screen in Ac, which had identified two other intracellular virulence factors, namely the type VII secretion system ESX-4 and the lipid transport protein MmpL8MAB [29,39]. The intracellular defects of strains that were deleted for genes highly induced in Ac (induced at least four times more compared to the intra-macrophagic transcriptome of M. abscessus and to the intra-Ac transcriptome of M. chelonae (OP1 to OP6)), suggest that the transcriptomic changes observed following a co-culture in amoebae reflected the response of M. abscessus in Mϕ. These results are in agreement with previous findings, suggesting that co-culturing of M. abscessus in amoebae enhances the virulence of M. abscessus in subsequent mouse infection experiments, likely through the induction of the phospholipase C-encoding plc virulence gene [27].
Most of the 6 loci (OP1 to OP6), studied, encode for hypothetical proteins, with the exception of the MAB_1517c gene that encodes a probable O-methyltransferase (OP3), and the MAB_2649 and MAB_2650 genes encoding MmpS and MmpL mycobacterial membrane proteins, respectively (OP4) (Table 2) [40].
At the M. abscessus OP2 locus, that mainly comprises genes of unknown function, we used a motif analysis to identify an ABC transporter, FecCD/TroCD-like in the MAB_1243c protein and an alkaline shock protein Asp23 in the MAB_1247c protein. A motif analysis performed on the M. abscessus OP6 locus shows that MAB_4791c encodes a protein implicated in the twin-arginine translocation pathway (see below).
Of note, it was also found that the over-induction of MAB_2649 and MAB_1517c in M. chelonae favors its replication in Mϕ, suggesting that the high induction of these two genes in amoeba may trigger M. abscessus virulence.
Finally, one of the most striking findings of our work, is the essential role of the eis2Mab gene in early resistance to the microbicidal action of Mϕ, via phagosomal membrane damage and cytosol contact, that might favor the intracellular survival of M. abscessus. Although M. abscessus possesses two Eis genes, there is no redundancy in their respective functions; the eis1Mab mutant presented a similar behavior to the wt strain in Mϕ, with only the loss of a log10 CFU at 5 dpi, compared to the quasi-total clearance of the eis2Mab mutant in Mϕ. Despite higher genomic identity between eis1MAB and eisMTB, the restoration of the phenotype when complemented with eisMTB was observed only for the eis2MAB mutant, demonstrating the similar role of eis2MAB to what is described for eisMTB in virulence. However the deletion of this gene in M. abscessus is more deleterious for the bacterium in Mϕ compared to the deletion of eisMTB [41,42]. EisMTB has been described as being important for M. tuberculosis survival inside Mϕ by controlling host cell apoptosis, autophagy, ROS production and innate immune defenses [43]. As also observed in M. tuberculosis [43], increasing the MOI (to 50) revealed further differences between eis2MAB KO and wt strains with regard to resistance to oxidized derivatives; however, this bacterial-load effect has not yet been observed for cell death mechanisms.
One of the peculiarities of the locus eis2MAB is that it shows similarities with a genomic region of M. tuberculosis, within which is also located the gene mmpL11. The potential counterpart in M. abscessus would be MAB_4529 (S5 Fig). Most of MmpL proteins are described as lipid transporters implicated in cell physiology and virulence [40,44]. M. abscessus has 27 MmpLs, twice as much as M. tuberculosis [40]. In M. tuberculosis, MmpL11 is implicated in heme iron acquisition [45] and transport of mycolic acid wax ester and long-chain triacylglycerols [46]. Three genes conserved in the M. abscessus eis2 locus encode for proteins belonging to lipid transport and metabolism pathways (COG I), which suggests, together with the conservation seen with the M. tuberculosis mmpL11 locus, that the M. abscessus eis2 locus might also participate in cell wall biogenesis [47].
Transcriptomic analysis revealed that nine M. abscessus genes, whose orthologues in M. tuberculosis contribute to virulence, were highly induced during infection of Mϕ (S2 Table). Among their gene products, WhiB7 and DevR-DevS are implicated in stress sensing [48]. WhiB7, a Fe-S cluster protein, was shown to be induced in response to perturbation in amino-acid metabolism, under reducing intracellular state, iron depletion and increased temperatures [49]. The 20-fold increase of M. abscessus whiB7 in Mϕ suggests that M. abscessus may undergo similar stresses in Mϕ as M. tuberculosis. The DevR response regulator of the histidine kinase DevS was also highly up-regulated. In M. tuberculosis, the devR-devS two-component system (also known as the DosR system) is activated in response to hypoxia [50]. Likewise, M. abscessus MAB_2562c, the orthologue of Rv0081, was induced 10-fold in Mϕ. A putative orthologue (MAB_1409c) of the dormancy response gene Rv1258c was also strongly induced in intra-macrophagic M. abscessus. The conserved alpha-ketoglutarate-dependent dioxygenase AlkB-encoding gene is thought to be involved in fatty acid metabolism, or in protection against DNA methylation. The aspC gene was induced 8-fold; AspC mediates nitrogen transfer from aspartate to glutamate, which in turn, together with glutamine, provides nitrogen to most of the biosynthesis pathways. This is thought to be essential in M. tuberculosis [51], while aspartate is required for mycobacterial virulence [52]. M. abscessus katA gene, which is conserved in M. avium and Listeria monocytogenes, is a catalase that degrades H2O2 into water and oxygen in a single reaction. Such a reaction, enabling resistance to oxidative metabolites, may be an important mechanism of bacillary survival within the host phagocyte [53]. M. abscessus eamA (MAB_0677c), which is thought to encode a drug/metabolite transporter, was induced in Mϕ. Two additional genes (MAB_3762 and MAB_3180) encoding proteins with an EamA domain were also highly induced. Finally, at the molecular function level, it appears that six of the most highly induced genes in M. abscessus in Mϕ encode acyl or N-acetyl transferase proteins playing a role in post-translational modifications.
The M. abscessus transcriptomes’ comparison in Ac or Mϕ allowed differences in metabolic adaptations to be highlighted. In Mϕ, M. abscessus enters a slow replicative stage, and activates the detoxification and protein secretion pathways. By comparison, in amoebae M. abscessus switches on protein synthesis, lipid transport and metabolism, transcription of genes involved in post-translational modifications (PTM), protein turnover and chaperones (COG O), reflecting a more active and replicative behavior as compared to a more persistent state in Mϕ. Actually, cell wall biogenesis including peptidoglycan and glycosaminoglycan biosynthetic processes were down-regulated in Mϕ. Similarly, mtrA, phoP and devR were differently regulated, with only devR up-regulated in Mϕ, confirming the switch towards a slow growth stage for M. abscessus in Mϕ.
Over-representation of the COG O (post-translational modification, protein turnover, molecular chaperone) category in M. abscessus infecting Ac indicates that M. abscessus may alter cellular processes during its interactions with host cells via PTM, as described in various pathogens [54–56]. Protein turnover does not only help in clearing of old proteins but also aids a fast adaptation to nutrient poor environments [57]. Molecular chaperones help pathogens override unfavorable conditions found in the host such as heat shock, oxidative and acid stresses [58]. They also contribute to the inhibition of lysosomal fusion and favor bacterial growth [58]. Molecular chaperones may therefore form a first line of defense and help consolidate pathogen virulence. Thus, over-representation of the COG O during infection of amoebae might reflect specific intracellular cues, mycobacteria face from the early time points post infection on.
In Ac, the most enriched GO is adenine salvage (GO:0006168) (Fig 2). This GO represents any process that generates adenine from derivatives without any de novo synthesis. Mycobacteria are able to limit the synthesis of this high energy demanding nucleotide [59]. Mycobacteria are also capable of scavenging free nitrogenous bases from the medium [59]. Under conditions of low energy availability or rapid multiplication, the salvage pathway may then be the main source of maintaining the nucleotide pool [59].
Sulfur metabolism (GO:0000103), hydrogen sulfide (H2S) biosynthetic pathway (GO:0070814) and detoxification via Fe-S cluster assembly proteins (GO:0016226) in addition to polyamine transport (GO:0015846), were also enriched by M. abscessus in Ac. In its reduced form, sulfur is used in the biosynthesis of the amino acid cysteine that is one of the prime targets for reactive nitrogen intermediates [60]. Those pathways might play a key role in M. abscessus survival in phagocytic cells, since genes involved in the metabolism of sulfur have consistently been identified as up-regulated in conditions that mimic the intra-macrophagic environment and during Mϕ infection for M. tuberculosis [36]. As for polyamines (cadaverine, putrescine and spermidine), they are known to have pleiotropic effects on cells via: their interaction with nucleic acids; a role in bacterial virulence by allowing mycobacterial escape from the phagolysosome; toxin activity or protection from oxidative and acid stress has also been demonstrated [61].
In Mϕ, glycerol ether metabolic process, MEP pathway and L-proline biosynthetic processes were the most enriched. Glycerol ether metabolic process corresponds to glycerophospholipids-seminolipids-plasmecholine metabolism and cellular amide biosynthetic processes. The MEP pathway is required for isoprenoid precursor biosynthesis [62]. A wide variety of monoterpenes and diterpenes belong to isoprenoid classes which function as toxins, growth inhibitors, or other secondary metabolites [63]. Finally, proline has been reported as an important factor in the adaptation of mycobacteria to slow growth rate and hypoxia [64]. It is believed that the proline-utilization pathway protects mycobacterial cells by detoxifying methylglyoxal, a by-product of endogenous glycerol metabolism [64] that can damage DNA and proteins within cells. Up-regulation of base-excision repair suggests that intracellular mycobacteria undergo DNA damage. Protein folding was also enriched, as well as the type II secretion system, which was enriched by more than two-fold. This secretion system promotes the specific transport of folded periplasmic proteins across a dedicated channel in the outer membrane, and it facilitates both Sec and Tat pathways to secrete proteins into the periplasm. Potential roles for SecA1 and SecA2 in M. tuberculosis dormancy has been reported while the Tat pathway was shown to contribute to virulence in Legionella pneumophila for instance, by aiding secretion of Phospholipase C [65], a virulence factor conserved in M. abscessus [21].
Both Ac and Mϕ were sensed as a stressful environment by M. abscessus, evidenced by the up-regulation of genes known to be involved in multiple stress responses. Induction of low O2 and low NO response genes confirm that hypoxic environments are encountered by M. abscessus both in Ac and Mϕ.
In conclusion, our findings confirm that the amoeba-induced genes play a role in potentiating the subsequent survival of M. abscessus in Mϕ. Both environments have commonalities, in terms of metabolic switches, especially to withstand the host response. It is likely that through such preparation during its intra-amoebic life that M. abscessus is able to withstand the noxious Mϕ environment, especially due to selected genes whose role has been highlighted during this work. The multiple leads opened during this work must now be followed to complete this viewpoint of synergistic potentiation of virulence conferred by the amoeba to M. abscessus, including the ultimate mechanisms of manipulation of the host's defense systems as seen with other intracellular pathogens.
Materials and methods
Bacterial strains, plasmids and growth conditions
A clinical isolate of M. abscessus subspecies massiliense smooth variant (43S) and M. chelonae type strain CCUG 47445 were used for the RNAseq experiments. Gene deletions were performed with CIP 104536T type smooth strain of M. abscessus subspecies abscessus. Both M. abscessus CIP 104536T strain and M. chelonae CCUG 47445 type strains were used to perform in vitro survival and complementation tests, while gene deletion experiments were performed with M. abscessus CIP 104536T. M. abscessus and M. chelonae strains were routinely grown aerobically at 37°C and 32°C respectively, in Middlebrook 7H9 medium (Sigma-Aldrich) supplemented with 0.2% glycerol, 1% glucose, and 250 mg/L kanamycin (Thermo Fisher Scientific) when necessary, with 25 mg/L zeocin (ThermoFisher Scientific) for the knockout strains, and with 25 mg/L zeocin plus 250 mg/L hygromycin (InvivoGen) for complemented strains. A. castellanii (ATCC 30010) was grown at room temperature without CO2 in peptone-yeast-extract-glucose (PYG) broth for the amplification of the strain. Mouse Mϕ J774.2 (Sigma) were grown and used as described [38,66].
Gene deletion and complementation
Deletion of genes was performed using the recombineering system as described previously [27,67]. Growth of the KO strains was checked by measuring the optical density of bacterial cultures in 7H9 medium supplemented with glycerol 0.2%. Complementation was performed after amplifying and cloning genes into the integrative plasmid pMVH361 as described [27].
RNA isolation and RNA sequencing
Approximately 107 cells were infected in 50 mL tubes, with low agitation, without CO2. Amoebae were infected at 100 MOI at 32°C in Page’s modified Neff’s amoeba saline (PAS) [68]. J774.2 Mϕ were infected at 50 MOI at 37°C in Dulbecco's Modified Eagle Medium (DMEM).
Cells were washed 3 times after 1 hour of infection and resuspended in medium supplemented with amikacin 250 μg/mL and incubated for 1 hour to eliminate extracellular bacteria. These relatively high MOI, employed for a limited time (1 h) were chosen to assure a sufficient infection rate of cells, as required for RNA sequencing. Three additional washes were performed and cells were resuspended in medium supplemented with amikacin 50 μg/mL for the rest of the infection. Amoebal cells were harvested 4 h and 16 hpi for intracellular M. abscessus RNA isolation and 16 hpi and for intracellular M. chelonae RNA isolation. Mϕ were harvested for intracellular M. abscessus RNA isolation 16 hpi. RNA isolation was performed as described [69]. Briefly, cells were lysed with a cold solution of guanidium thiocyanate (GTC), N-Lauryl-sarcosine, sodium citrate +/- Tween 80 plus β-mercaptoethanol. The lysates containing intracellular bacteria were collected, centrifuged and RNA was isolated from the bacterial pellets with TRIzol. The lysates were then transferred into 2 mL screw tubes containing zirconium beads and were conserved at -80°C for at least 1 day to allow inactivation of RNAses and cells dissolution. Bacteria cells were disrupted with a bead beater by performing to round at 6,500 rpm for 25 seconds, followed by one round at 6.500 rpm for 20 seconds. Two hundred μL of chloroform isoamyl alcohol were added and tubes were immediately mixed for 10 seconds. The mixture was centrifugated at 13,000 rpm for 15 minutes at 4°C. The RNA present in the upper phase was transferred to a fresh tube and precipitated by adding 0.8 volume of isopropanol. Tubes were inverted twice to allow precipitation and kept at -20°C for at least 2 hours. The precipitated RNA was then pelleted by centrifugation at 13,000 rpm for 30 min at 4°C. The pellet was washed with ethanol (70%) and centrifuged at 13,000 rpm for 10 min at 4°C. The washed pellet was air-dried, re-suspended in RNase-free water and stored at -80°C until cDNA library construction.
Control RNA was isolated from bacteria cells grown in amoeba (PAS buffer) or Mϕ co-culture medium (DMEM supplemented with 10% Fetal Bovine Serum respectively).
Biological replicates were prepared to allow statistical comparisons of infected and non-infected samples.
RNA treatments prior to library preparation and library preparation
RNA samples were treated with DNases (AMBION) to remove DNA contaminants, purified with the RNA MEGAclear kit (ThermoFisher), and depleted of ribosomal RNA with the riboZero kit (Illumina). RNA (total, depleted, purified) was checked on the Bioanalyser system (Agilent) for its quality and integrity. cDNA libraries were prepared with samples displaying a RNA integrity number above 7. RNA concentrations were measured using the nanodrop spectrophotometer (Thermo Scientific) and the Qubit fluorometer (Invitrogen). Libraries were prepared with the TruSeq Stranded RNA LT prep kit cDNA synthesis, set A (Illumina) which consists in: (1) RNA fragmentation, (2) 1st strand cDNA synthesis (Reverse transcriptase and random primers), (3) 2nd strand cDNA synthesis (removal of the RNA template and synthesis of a new strand with dUTP), (4) no end repair step, (5) adenylation of 3’ ends, (6) ligation of adapters and (7) enrichment of DNA fragments. Libraries were checked for concentration and quality on DNA chips with the Bioanalyzer Agilent. More precise and accurate quantification was performed with sensitive fluorescent-based quantitation assays ("Quant-It" assays kit and QuBit fluorometer, Invitrogen).
NGS sequencing and data analyses
Sequencing and statistical analyses were performed in the Transcriptome and Epigenome platform (PF2) of the Pasteur Institute, Paris, France. The cDNA libraries for strains M. abscessus subsp. massiliense 43S and M. chelonae CCUG 47445 were prepared and sequenced on an Illumina HiSeq 2500 system by performing an SRM run (SR: Single Read, PE: Paired-end Reads, M: multiplexed samples) of 51 cycles with 7 index bases read. The quality of the sequencing was assessed with the external FastQC program (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). After the trimming of adapter sequences and low-quality reads with cutadapt version 1.11, reads were aligned with RefSeq assemblies (M. abscessus subsp. massiliense strain GO06 assembly (GCF_000277775.2); M. chelonae CCUG 47445 assembly (GCF_001632805.1), using the Bowtie software version 0.12.7 (http://bowtie-bio.sourceforge.net/index.shtml) with defaults parameters. Genes were counted using featureCounts 1.4.6-p3 from Subreads package (parameters: -g gene -t ID -s 1). Differential analysis of gene expression was performed using the R software (version 3.3.1) and the Bioconductor packages DESeq2 (version 1.12.3) [30] using the default parameters and statistical tests for differential expression were performed applying the independent filtering algorithm. A generalized linear model was set in order to test for the differential expression between the biological conditions. For each pairwise comparison, raw p-values were adjusted for multiple testing according to the Benjamini and Hochberg (BH) procedure [70] and genes with an adjusted p-value lower than 0.05 were considered differentially expressed. Gene orthologs of M. massiliense and M. chelonae genes in the genome of the M. abscessus subsp. abscessus CIP 104536T reference strain were determined by Bi-directionnal Best Hit (BBH) searches using the Opscan software (http://wwwabi.snv.jussieu.fr/public/opscan/). Differentially expressed genes assignment to COGs was performed using the COG automatic Classification from the MicroScope database [71]. The percentage assignments were compared by performing Fisher’s exact tests. GO enrichment analyses were performed with the R software topGO package (Bioconductor) [72]. Protein signatures were addressed using InterProScan tool (https://www.ebi.ac.uk/interpro/search/sequence-search).
Quantitative real-time PCR (qRT-PCR)
qRT-PCR were performed with a CFX96 thermal cycler (Bio-Rad). Controls without reverse transcriptase were done on each RNA sample to rule out DNA contamination. The sigA gene was used as an internal control [27]. Each qRT-PCR was performed with three biological replicates.
In vitro survival assays
Survival of strains in amoebae and J774.2 Mϕ were performed as previously described [39]. Survival tests of KO strains were performed in duplicates three times. Confirmation of attenuated phenotypes and complementation tests were performed in triplicates three times.
Phagosome acidification and phagosomal escape assays
Phagosome acidification and phagosomal escape Fluorescence Energy Transfer (FRET) assays were conducted in J774.2 Mϕ as previously described [38,73].
Cell death, autophagy and ROS production assays
Mϕ death following infection with M. abscessus was assessed with the Dead Cell Apoptosis Kit with Annexin V FITC and PI for flow cytometry (ThermoFisher). Autophagy was assessed with the Premo Autophagy Tandem Sensor RFP-GFP-LC3B Kit (ThermoFisher). ROS production by J774.2 Mϕ was measured with the MitoSOX Red kit (ThermoFisher).
Infections were performed as previously described [39], at 50 MOI, except in the ROS production assay for which the cells were infected 15 min only.
Bacterial sensitivity to H2O2
Sensitivity to H2O2 was assessed by culturing the bacteria in 7H9 medium supplemented with glycerol 0,1% and H202 3% (Laboratoires Gilbert) (20 μM). CFU tests were performed at different times post-treatment (2 h, 4 h, 8 h) to determine the number of viable bacteria compared to the wt strain (Growth Index).
Supporting information
A. M. abscessus transcriptomes in A. castellanii 4 and 16 hpi. B. M. abscessus transcriptome in macrophages 16 hpi. C. M. chelonae transcriptome in A. castellanii 16 hpi. Hierarchical clustering of raw data (left panel) and transcriptome heatmaps (right panel) were depicted. Clustered were indicated by red and blue circles corresponding to raw data from intracellular bacteria and planktonic bacteria respectively. Hatched and filled circles correspond to 4 h and 16 h (co)-cultures respectively. Change in gene expression were depicted on the heatmap in a white to blue scale for repressed genes and a white to red scale for induced genes, the white color representing no change in gene expression.
(TIF)
A. Differentially expressed genes (DEGs) from comparisons of co-cultures with A. castellanii (Ac) and macrophages (Mϕ) relative to planktonic growth were categorized according to their fold change (FC) expressed in Log2. Low DEGs depict a FC < |2|, Med DEGs depict a FC between |2| and |4| and High DEGs depict a FC > than |4|. B. Ratio of UP DEGs over DOWN DEGs.
(TIF)
A. Intracellular survival of KO strains (ΔOP) in A. castellanii (Ac) and macrophages (Mϕ). Cells were infected at 10 MOI and colony forming units (CFU) tests were performed 0 and 3 dpi. The relative growth of each strain as compared to M. abscessus wt (Growth Index, GI) is given. B. KO strains growth in culture medium. The strains were cultured in 7H9 medium supplemented with glycerol 0.2% for seven days. Growth curves were obtained by measuring the cultures optical density each day. Experiments were repeated three times in triplicates. Statistical analyses were performed with GraphPad PRISM6. Histograms with error bars represent means ± SD. Differences between means were analyzed by ANOVA and the Tukey post-test allowing multiple comparisons to be performed. ns = non-significant, * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001.
(TIF)
A. Conservation of M. abscessus eis1 locus in M. tuberculosis. B. Conservation of M. abscessus eis2 locus in M. tuberculosis. C. Conservation of M. tuberculosis eis locus in M. abscessus. Bidirectional Best Hit (BBH) search was performed between M. abscessus and M. tuberculosis genomes with the Opscan software. BBHs were depicted by arrows filled with red, brown or orange. Brown arrows correspond to MmpL-encoding genes. Orange arrows correspond to MmpS-encoding genes. Greys bands link genes or groups of genes conserved in the two species.
(TIF)
Eis1MAB (left panel) and eis2MAB (right panel) expression in Mϕ was measured twice in triplicates by quantitative-real time PCR by normalization with sigA housekeeping gene.
(TIF)
A. Cell death. Mϕ death following infection with M. abscessus was assessed with the Dead Cell Apoptosis Kit with Annexin V FITC and PI for flow cytometry. B. Cell autophagy was measured Premo Autophagy Tandem Sensor RFP-GFP-LC3B Kit. At least 40 cells per condition were analyzed by confocal microscopy. To assess the number of autophagic particles per cell, cell nucleus was stained with Hoechst 33342 (blue spots). Representative images of autophagic particles were given. Stained cells with the premo-autophagy kit were either non-infected (NI cells) or infected with M. abscessus strains (Mabs wt and Δeis2MAB) or treated with chloroquine 30 μM for 48 h (+chloro) inhibiting autophagy or with HBSS solution for 2 hours (+HBSS) inducing autophagy. Autophagic particles are represented in red. Stained cells with the premo-autophagy kit were either non-infected (NI cells) or infected with M. abscessus strains (Mabs wt and Δeis2MAB) or treated with chloroquine 30 μM for 48 h (+chloro) inhibiting autophagy or with HBSS solution for 2 hours (+HBSS) inducing autophagy. Acidification of autophagosomes was determined by dividing GFP (sensitive to acidic pH) over RFP (no sensitive to acidic pH) fluorescence intensity. The number of autophagic particles per cell and acidification of autophagosomes were determined with the Fiji software. C. Phagosomal acidification was assessed as previously described [38]. Mϕ were infected at 10 (C) or 30 MOI (A and B). Histograms with error bars represent means ± SD. Differences between means were analyzed by ANOVA and the Tukey post-test allowing multiple comparisons to be performed. ns = non-significant. * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001.
(TIF)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We thank B. G. Marshall (Southampton University) and S. Gordon (University College, Dublin) for their careful reading of the manuscript and for giving valuable comments.
Data Availability
The raw data of the different transcriptomes are available on the GEO archive GSE124972 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE124972).
Funding Statement
V.D. was supported by French Cystic Fibrosis Patients Association Vaincre la Mucoviscidose (VLM) grant RF20150501377. Work in the laboratory of R.B. was supported by the Agence National de Recherche (ANR-10-LABX-62-IBEID, ANR-16-CE15-0003, and ANR-16- CE35-0009), VLM (grant RF20180502259), the Fondation pour la Recherche Médicale (DEQ20130326471), and the Institut Pasteur. The Transcriptome and Epigenome Platform is a member of the France Génomique consortium (ANR10‐NBS‐09‐08). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Lavania M, Katoch K, Katoch VM, Gupta AK, Chauhan DS, Sharma R, et al. Detection of viable Mycobacterium leprae in soil samples: Insights into possible sources of transmission of leprosy. Infect Genet Evol. 2008;8: 627–631. 10.1016/j.meegid.2008.05.007 [DOI] [PubMed] [Google Scholar]
- 2.Angenent LT, Kelley ST, St Amand A, Pace NR, Hernandez MT. Molecular identification of potential pathogens in water and air of a hospital therapy pool. Proc Natl Acad Sci U S A. National Academy of Sciences; 2005;102: 4860–5. 10.1073/pnas.0501235102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ben Salah I, Adékambi T, Drancourt M. Mycobacterium phocaicum in therapy pool water. Int J Hyg Environ Health. 2009;212: 439–444. 10.1016/j.ijheh.2008.10.002 [DOI] [PubMed] [Google Scholar]
- 4.Gomila M, Ramirez A, Gasco J, Lalucat J. Mycobacterium llatzerense sp. nov., a facultatively autotrophic, hydrogen-oxidizing bacterium isolated from haemodialysis water. Int J Syst Evol Microbiol. 2008;58: 2769–2773. 10.1099/ijs.0.65857-0 [DOI] [PubMed] [Google Scholar]
- 5.Guidotti TL, Ragain L, de Haas P, van Soolingen D. Communicating with healthcare providers. J Water Health. 2008;6: s53 10.2166/wh.2008.032 [DOI] [PubMed] [Google Scholar]
- 6.Griffith DE, Aksamit T, Brown-Elliott B a, Catanzaro A, Daley C, Gordin F, et al. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med. 2007;175: 367–416. 10.1164/rccm.200604-571ST [DOI] [PubMed] [Google Scholar]
- 7.Khan IUH, Selvaraju SB, Yadav JS. Method for rapid identification and differentiation of the species of the Mycobacterium chelonae complex based on 16S-23S rRNA gene internal transcribed spacer PCR-restriction analysis. J Clin Microbiol. 2005;43: 4466–72. 10.1128/JCM.43.9.4466-4472.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mignard S, Flandrois J-P. A seven-gene, multilocus, genus-wide approach to the phylogeny of mycobacteria using supertrees. Int J Syst Evol Microbiol. 2008;58: 1432–1441. 10.1099/ijs.0.65658-0 [DOI] [PubMed] [Google Scholar]
- 9.Thomson RM, Carter R, Tolson C, Coulter C, Huygens F, Hargreaves M. Factors associated with the isolation of Nontuberculous mycobacteria (NTM) from a large municipal water system in Brisbane, Australia. BMC Microbiol. 2013;13: 89 10.1186/1471-2180-13-89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ripoll F, Pasek S, Schenowitz C, Dossat C, Barbe V, Rottman M, et al. Non mycobacterial virulence genes in the genome of the emerging pathogen Mycobacterium abscessus. PLoS One. 2009;4: e5660 10.1371/journal.pone.0005660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas V, McDonnell G. Relationship between mycobacteria and amoebae: ecological and epidemiological concerns. Lett Appl Microbiol. 2007;45: 349–57. 10.1111/j.1472-765X.2007.02206.x [DOI] [PubMed] [Google Scholar]
- 12.Falkinham JO. Surrounded by mycobacteria: nontuberculous mycobacteria in the human environment. J Appl Microbiol. 2009;107: 356–67. 10.1111/j.1365-2672.2009.04161.x [DOI] [PubMed] [Google Scholar]
- 13.Eddyani M, De Jonckheere JF, Durnez L, Suykerbuyk P, Leirs H, Portaels F. Occurrence of free-living amoebae in communities of low and high endemicity for Buruli ulcer in southern Benin. Appl Environ Microbiol. 2008;74: 6547–53. 10.1128/AEM.01066-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomas V, Herrera-rimann K, Blanc DS, Greub G. Biodiversity of Amoebae and Amoeba-Resisting Bacteria in a Hospital Water Network. Appl Env Microbiol. 2006;72: 2428–2438. 10.1128/AEM.72.4.2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pagnier I, Raoult D, La Scola B. Isolation and identification of amoeba-resisting bacteria from water in human environment by using an Acanthamoeba polyphaga co-culture procedure. Environ Microbiol. 2008;10: 1135–1144. 10.1111/j.1462-2920.2007.01530.x [DOI] [PubMed] [Google Scholar]
- 16.Adékambi T, Ben Salah S, Khlif M, Raoult D, Drancourt M. Survival of environmental mycobacteria in Acanthamoeba polyphaga. Appl Environ Microbiol. 2006;72: 5974–5981. 10.1128/AEM.03075-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamrabet O, Mba Medie F, Drancourt M. Acanthamoeba polyphaga-enhanced growth of Mycobacterium smegmatis. PLoS One. 2012;7: e29833 10.1371/journal.pone.0029833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas V, Loret J-F, Jousset M, Greub G. Biodiversity of amoebae and amoebae-resisting bacteria in a drinking water treatment plant. Environ Microbiol. 2008;10: 2728–2745. 10.1111/j.1462-2920.2008.01693.x [DOI] [PubMed] [Google Scholar]
- 19.White CI, Birtles RJ, Wigley P, Jones PH. Mycobacterium avium subspecies paratuberculosis in free-living amoebae isolated from fields not used for grazing. Vet Rec. 2010;166: 401–402. 10.1136/vr.b4797 [DOI] [PubMed] [Google Scholar]
- 20.Gutierrez MC, Brisse S, Brosch R, Fabre M, Omaïs B, Marmiesse M, et al. Ancient Origin and Gene Mosaicism of the Progenitor of Mycobacterium tuberculosis. PLoS Pathog. Public Library of Science; 2005;1: e5 10.1371/journal.ppat.0010005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ripoll F, Pasek S, Schenowitz C, Dossat C, Barbe V, Rottman M, et al. Non mycobacterial virulence genes in the genome of the emerging pathogen Mycobacterium abscessus. PLoS One. 2009;4: e5660 10.1371/journal.pone.0005660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamrabet O, Merhej V, Pontarotti P, Raoult D, Drancourt M. The genealogic tree of mycobacteria reveals a long-standing sympatric life into free-living protozoa. PLoS One. 2012;7: e34754 10.1371/journal.pone.0034754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siddiqui R, Khan NA. Acanthamoeba is an evolutionary ancestor of macrophages: a myth or reality? Exp Parasitol. 2012;130: 95–7. 10.1016/j.exppara.2011.11.005 [DOI] [PubMed] [Google Scholar]
- 24.Barker J, Brown MRW. Trojan Horses of the microbial world: protozoa and the survival of bacterial pathogens in the environment. Microbiology. 1994;140: 1253–1259. 10.1099/00221287-140-6-1253 [DOI] [PubMed] [Google Scholar]
- 25.Greub G, Raoult D. Microorganisms resistant to free-living amoebae. Clin Microbiol Rev. 2004;17: 413–33. 10.1128/CMR.17.2.413-433.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salah IB, Ghigo E, Drancourt M. Free-living amoebae, a training field for macrophage resistance of mycobacteria. Clin Microbiol Infect. 2009;15: 894–905. 10.1111/j.1469-0691.2009.03011.x [DOI] [PubMed] [Google Scholar]
- 27.Bakala N’Goma JC, Le Moigne V, Soismier N, Laencina L, Le Chevalier F, Roux A-L, et al. Mycobacterium abscessus phospholipase C expression is induced during coculture within amoebae and enhances M. abscessus virulence in mice. Roy CR, editor. Infect Immun. American Society for Microbiology; 2015;83: 780–791. 10.1128/IAI.02032-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cirillo JD, Falkow S, Tompkins LS, Bermudez LE. Interaction of Mycobacterium avium with environmental amoebae enhances virulence. Infect Immun. 1997;65: 3759–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laencina L, Dubois V, Le Moigne V, Viljoen A, Majlessi L, Pritchard J, et al. Identification of genes required for Mycobacterium abscessus growth in vivo with a prominent role of the ESX-4 locus. Proc Natl Acad Sci. 2018;115: E1002–E1011. 10.1073/pnas.1713195115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15: 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tatusov RL, Galperin MY, Natale DA, Koonin E V. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28: 33–6. 10.1093/nar/28.1.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eoh H, Rhee KY. Multifunctional essentiality of succinate metabolism in adaptation to hypoxia in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2013;110: 6554–9. 10.1073/pnas.1219375110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jamet S, Quentin Y, Coudray C, Texier P, Laval F, Daffé M, et al. Evolution of Mycolic Acid Biosynthesis Genes and Their Regulation during Starvation in Mycobacterium tuberculosis. J Bacteriol. American Society for Microbiology (ASM); 2015;197: 3797–811. 10.1128/JB.00433-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mukhopadhyay S, Nair S, Ghosh S. Pathogenesis in tuberculosis: transcriptomic approaches to unraveling virulence mechanisms and finding new drug targets. FEMS Microbiol Rev. 2012;36: 463–485. 10.1111/j.1574-6976.2011.00302.x [DOI] [PubMed] [Google Scholar]
- 35.Sherman DR, Mdluli K, Hickey MJ, Barry CE, Stover CK. AhpC, oxidative stress and drug resistance in Mycobacterium tuberculosis. Biofactors. 1999;10: 211–7. [DOI] [PubMed] [Google Scholar]
- 36.Schnappinger D, Ehrt S, Voskuil MI, Liu Y, Mangan JA, Monahan IM, et al. Transcriptional Adaptation of Mycobacterium tuberculosis within Macrophages. J Exp Med. 2003;198: 693–704. 10.1084/jem.20030846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sousa S, Bandeira M, Carvalho PA, Duarte A, Jordao L. Nontuberculous mycobacteria pathogenesis and biofilm assembly. Int J Mycobacteriology. No longer published by Elsevier; 2015;4: 36–43. 10.1016/J.IJMYCO.2014.11.065 [DOI] [PubMed] [Google Scholar]
- 38.Roux A-L, Viljoen A, Bah A, Simeone R, Bernut A, Laencina L, et al. The distinct fate of smooth and rough Mycobacterium abscessus variants inside macrophages. Open Biol. The Royal Society; 2016;6: 160185 10.1098/rsob.160185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dubois V, Viljoen A, Laencina L, Le Moigne V, Bernut A, Dubar F, et al. MmpL8MAB controls Mycobacterium abscessus virulence and production of a previously unknown glycolipid family. Proc Natl Acad Sci U S A. 2018;115: E10147–E10156. 10.1073/pnas.1812984115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Viljoen A, Dubois V, Girard-Misguich F, Blaise M, Herrmann J-L, Kremer L. The diverse family of MmpL transporters in mycobacteria: from regulation to antimicrobial developments. Mol Microbiol. 2017;104 10.1111/mmi.13873 [DOI] [PubMed] [Google Scholar]
- 41.Wei J, Dahl JL, Moulder JW, Roberts EA, O’Gaora P, Young DB, et al. Identification of a Mycobacterium tuberculosis gene that enhances mycobacterial survival in macrophages. J Bacteriol. 2000;182: 377–84. 10.1128/jb.182.2.377-384.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu S, Barnes PF, Samten B, Pang X, Rodrigue S, Ghanny S, et al. Activation of the eis gene in a W-Beijing strain of Mycobacterium tuberculosis correlates with increased SigA levels and enhanced intracellular growth. Microbiology. Microbiology Society; 2009;155: 1272–81. 10.1099/mic.0.024638-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shin D-M, Jeon B-Y, Lee H-M, Jin HS, Yuk J-M, Song C-H, et al. Mycobacterium tuberculosis Eis Regulates Autophagy, Inflammation, and Cell Death through Redox-dependent Signaling. Deretic V, editor. PLoS Pathog. Public Library of Science; 2010;6: e1001230 10.1371/journal.ppat.1001230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chalut C. MmpL transporter-mediated export of cell-wall associated lipids and siderophores in mycobacteria. Tuberculosis (Edinb). 2016;100: 32–45. 10.1016/j.tube.2016.06.004 [DOI] [PubMed] [Google Scholar]
- 45.Owens CP, Chim N, Graves AB, Harmston CA, Iniguez A, Contreras H, et al. The mycobacterium tuberculosis secreted protein Rv0203 transfers heme to membrane proteins MmpL3 and MmpL11. J Biol Chem. 2013;288: 21714–21728. 10.1074/jbc.M113.453076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wright CC, Hsu FF, Arnett E, Dunaj JL, Davidson PM, Pacheco SA, et al. The Mycobacterium tuberculosis MmpL11 Cell Wall Lipid Transporter Is Important for Biofilm Formation, Intracellular Growth, and Nonreplicating Persistence. Ehrt S, editor. Infect Immun. 2017;85: e00131–17. 10.1128/IAI.00131-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamaryo-Botte Y, Rainczuk AK, Lea-Smith DJ, Brammananth R, van der Peet PL, Meikle P, et al. Acetylation of Trehalose Mycolates Is Required for Efficient MmpL-Mediated Membrane Transport in Corynebacterineae. ACS Chem Biol. 2015;10: 734–746. 10.1021/cb5007689 [DOI] [PubMed] [Google Scholar]
- 48.Kumar A, Farhana A, Guidry L, Saini V, Hondalus M, Steyn AJC. Redox homeostasis in mycobacteria: the key to tuberculosis control? Expert Rev Mol Med. Cambridge University Press; 2011;13: e39 10.1017/S1462399411002079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Geiman DE, Raghunand TR, Agarwal N, Bishai WR. Differential Gene Expression in Response to Exposure to Antimycobacterial Agents and Other Stress Conditions among Seven Mycobacterium tuberculosis whiB-Like Genes. Antimicrob Agents Chemother. 2006;50: 2836–2841. 10.1128/AAC.00295-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sherman DR, Voskuil M, Schnappinger D, Liao R, Harrell MI, Schoolnik GK. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha -crystallin. Proc Natl Acad Sci U S A. 2001;98: 7534–9. 10.1073/pnas.121172498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48: 77–84. 10.1046/j.1365-2958.2003.03425.x [DOI] [PubMed] [Google Scholar]
- 52.Gouzy A, Poquet Y, Neyrolles O. A central role for aspartate in Mycobacterium tuberculosis physiology and virulence. Front Cell Infect Microbiol. Frontiers Media SA; 2013;3: 68 10.3389/fcimb.2013.00068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Manca C, Paul S, Barry CE, Freedman VH, Kaplan G, Kaplan G. Mycobacterium tuberculosis catalase and peroxidase activities and resistance to oxidative killing in human monocytes in vitro. Infect Immun. American Society for Microbiology (ASM); 1999;67: 74–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ribet D, Cossart P. Pathogen-Mediated Posttranslational Modifications: A Re-emerging Field. Cell. Cell Press; 2010;143: 694–702. 10.1016/j.cell.2010.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Müller MP, Peters H, Blümer J, Blankenfeldt W, Goody RS, Itzen A. The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science. American Association for the Advancement of Science; 2010;329: 946–9. 10.1126/science.1192276 [DOI] [PubMed] [Google Scholar]
- 56.Parra J, Marcoux J, Poncin I, Canaan S, Herrmann JL, Nigou J, et al. Scrutiny of Mycobacterium tuberculosis 19 kDa antigen proteoforms provides new insights in the lipoglycoprotein biogenesis paradigm. Sci Rep. Nature Publishing Group; 2017;7: 43682 10.1038/srep43682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goldberg AL, St. John AC. Intracellular Protein Degradation in Mammalian and Bacterial Cells: Part 2. Annu Rev Biochem. Annual Reviews 4139 El Camino Way, P.O. Box 10139, Palo Alto, CA 94303–0139, USA; 1976;45: 747–804. 10.1146/annurev.bi.45.070176.003531 [DOI] [PubMed] [Google Scholar]
- 58.Neckers L, Tatu U. Molecular chaperones in pathogen virulence: emerging new targets for therapy. Cell Host Microbe. NIH Public Access; 2008;4: 519–27. 10.1016/j.chom.2008.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ducati RG, Breda A, Basso LA, Santos DS. Purine Salvage Pathway in Mycobacterium tuberculosis. Curr Med Chem. 2011;18: 1258–75. 10.2174/092986711795029627 [DOI] [PubMed] [Google Scholar]
- 60.Rhee KY, Erdjument-Bromage H, Tempst P, Nathan CF. S-nitroso proteome of Mycobacterium tuberculosis: Enzymes of intermediary metabolism and antioxidant defense. Proc Natl Acad Sci. 2005;102: 467–472. 10.1073/pnas.0406133102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shah P, Swiatlo E. MicroReview A multifaceted role for polyamines in bacterial pathogens. 2008; 10.1111/j.1365-2958.2008.06126.x [DOI] [PubMed] [Google Scholar]
- 62.Rohmer M, Knani M, Simonin P, Sutter B, Sahm H. Isoprenoid biosynthesis in bacteria: a novel pathway for the early steps leading to isopentenyl diphosphate. Biochem J. Portland Press Ltd; 1993;295: 517–24. 10.1042/bj2950517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gershenzon J, Dudareva N. The function of terpene natural products in the natural world. Nat Chem Biol. 2007;3: 408–414. 10.1038/nchembio.2007.5 [DOI] [PubMed] [Google Scholar]
- 64.Berney M, Weimar MR, Heikal A, Cook GM. Regulation of proline metabolism in mycobacteria and its role in carbon metabolism under hypoxia. Mol Microbiol. 2012;84: 664–81. 10.1111/j.1365-2958.2012.08053.x [DOI] [PubMed] [Google Scholar]
- 65.Rossier O, Cianciotto NP. The Legionella pneumophila tatB gene facilitates secretion of phospholipase C, growth under iron-limiting conditions, and intracellular infection. Infect Immun. American Society for Microbiology (ASM); 2005;73: 2020–32. 10.1128/IAI.73.4.2020-2032.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Le Moigne V, Belon C, Goulard C, Accard G, Bernut A, Pitard B, et al. MgtC as a Host-Induced Factor and Vaccine Candidate against Mycobacterium abscessus Infection. Ehrt S, editor. Infect Immun. 2016;84: 2895–2903. 10.1128/IAI.00359-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Medjahed H, Singh AK. Genetic manipulation of Mycobacterium abscessus. Curr Protoc Microbiol. 2010;Chapter 10: Unit 10D.2. 10.1002/9780471729259.mc10d02s18 [DOI] [PubMed] [Google Scholar]
- 68.Rowbotham TJ. Isolation of Legionella pneumophila from clinical specimens via amoebae, and the interaction of those and other isolates with amoebae. J Clin Pathol. 1983;36: 978–986. 10.1136/jcp.36.9.978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dubois V, Laencina L, Bories A, Le Moigne V, Pawlik A, Herrmann J-L, et al. Identification of Virulence Markers of Mycobacterium abscessus for Intracellular Replication in Phagocytes. J Vis Exp. 2018; 10.3791/57766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Benjamini, Hochberg. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Stat Soc Ser B. 1995;57: 289–300. [Google Scholar]
- 71.Vallenet D, Engelen S, Mornico D, Cruveiller S, Fleury L, Lajus A, et al. MicroScope: a platform for microbial genome annotation and comparative genomics. Database (Oxford). Oxford University Press; 2009;2009: bap021 10.1093/database/bap021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alexa A, Rahnenfuhrer J. topGO: Enrichment Analysis for Gene Ontology. R package version 2.36.0. 2019. [Google Scholar]
- 73.Simeone R, Sayes F, Song O, Gröschel MI, Brodin P, Brosch R, et al. Cytosolic access of Mycobacterium tuberculosis: critical impact of phagosomal acidification control and demonstration of occurrence in vivo. PLoS Pathog. 2015;11: e1004650 10.1371/journal.ppat.1004650 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. M. abscessus transcriptomes in A. castellanii 4 and 16 hpi. B. M. abscessus transcriptome in macrophages 16 hpi. C. M. chelonae transcriptome in A. castellanii 16 hpi. Hierarchical clustering of raw data (left panel) and transcriptome heatmaps (right panel) were depicted. Clustered were indicated by red and blue circles corresponding to raw data from intracellular bacteria and planktonic bacteria respectively. Hatched and filled circles correspond to 4 h and 16 h (co)-cultures respectively. Change in gene expression were depicted on the heatmap in a white to blue scale for repressed genes and a white to red scale for induced genes, the white color representing no change in gene expression.
(TIF)
A. Differentially expressed genes (DEGs) from comparisons of co-cultures with A. castellanii (Ac) and macrophages (Mϕ) relative to planktonic growth were categorized according to their fold change (FC) expressed in Log2. Low DEGs depict a FC < |2|, Med DEGs depict a FC between |2| and |4| and High DEGs depict a FC > than |4|. B. Ratio of UP DEGs over DOWN DEGs.
(TIF)
A. Intracellular survival of KO strains (ΔOP) in A. castellanii (Ac) and macrophages (Mϕ). Cells were infected at 10 MOI and colony forming units (CFU) tests were performed 0 and 3 dpi. The relative growth of each strain as compared to M. abscessus wt (Growth Index, GI) is given. B. KO strains growth in culture medium. The strains were cultured in 7H9 medium supplemented with glycerol 0.2% for seven days. Growth curves were obtained by measuring the cultures optical density each day. Experiments were repeated three times in triplicates. Statistical analyses were performed with GraphPad PRISM6. Histograms with error bars represent means ± SD. Differences between means were analyzed by ANOVA and the Tukey post-test allowing multiple comparisons to be performed. ns = non-significant, * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001.
(TIF)
A. Conservation of M. abscessus eis1 locus in M. tuberculosis. B. Conservation of M. abscessus eis2 locus in M. tuberculosis. C. Conservation of M. tuberculosis eis locus in M. abscessus. Bidirectional Best Hit (BBH) search was performed between M. abscessus and M. tuberculosis genomes with the Opscan software. BBHs were depicted by arrows filled with red, brown or orange. Brown arrows correspond to MmpL-encoding genes. Orange arrows correspond to MmpS-encoding genes. Greys bands link genes or groups of genes conserved in the two species.
(TIF)
Eis1MAB (left panel) and eis2MAB (right panel) expression in Mϕ was measured twice in triplicates by quantitative-real time PCR by normalization with sigA housekeeping gene.
(TIF)
A. Cell death. Mϕ death following infection with M. abscessus was assessed with the Dead Cell Apoptosis Kit with Annexin V FITC and PI for flow cytometry. B. Cell autophagy was measured Premo Autophagy Tandem Sensor RFP-GFP-LC3B Kit. At least 40 cells per condition were analyzed by confocal microscopy. To assess the number of autophagic particles per cell, cell nucleus was stained with Hoechst 33342 (blue spots). Representative images of autophagic particles were given. Stained cells with the premo-autophagy kit were either non-infected (NI cells) or infected with M. abscessus strains (Mabs wt and Δeis2MAB) or treated with chloroquine 30 μM for 48 h (+chloro) inhibiting autophagy or with HBSS solution for 2 hours (+HBSS) inducing autophagy. Autophagic particles are represented in red. Stained cells with the premo-autophagy kit were either non-infected (NI cells) or infected with M. abscessus strains (Mabs wt and Δeis2MAB) or treated with chloroquine 30 μM for 48 h (+chloro) inhibiting autophagy or with HBSS solution for 2 hours (+HBSS) inducing autophagy. Acidification of autophagosomes was determined by dividing GFP (sensitive to acidic pH) over RFP (no sensitive to acidic pH) fluorescence intensity. The number of autophagic particles per cell and acidification of autophagosomes were determined with the Fiji software. C. Phagosomal acidification was assessed as previously described [38]. Mϕ were infected at 10 (C) or 30 MOI (A and B). Histograms with error bars represent means ± SD. Differences between means were analyzed by ANOVA and the Tukey post-test allowing multiple comparisons to be performed. ns = non-significant. * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001.
(TIF)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
The raw data of the different transcriptomes are available on the GEO archive GSE124972 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE124972).