To the Editor:
Respiratory tract inflammation in chronic obstructive pulmonary disease (COPD) is known to be driven by neutrophils, lymphocytes, and T-helper cell type 1 cells, whereas inflammation in asthma is primarily mediated by T-helper cell type 2 cells and eosinophils (1). In patients with asthma, an increased eosinophil count is associated with an increased exacerbation risk and improved disease control with inhaled corticosteroid use. Similarly, evidence suggests that 20–40% of patients with stable COPD have elevated sputum and blood eosinophils that are associated with exacerbations and response to steroid treatment (2). Eosinophilic inflammation in COPD may also be a target for therapy with drugs used to treat eosinophilic asthma, as suggested by a recent trial of mepolizumab (anti–IL-5 monoclonal antibody) that showed a reduction in the exacerbation rate in patients with COPD and high blood eosinophil counts (3). The transcriptomic profiles of eosinophils in patients with eosinophilic asthma showed differences compared with eosinophils from healthy subjects (4). Whether there are COPD-specific changes in eosinophil gene expression is not known. Eosinophils become partially activated before they migrate to sites of inflammation, so eosinophil counts represent the overall levels of eosinophils and not necessarily the activation state of eosinophils. We hypothesized that eosinophils from subjects with eosinophilic COPD would be activated, similar to those obtained from patients with eosinophilic asthma but different from those obtained from control subjects. To test this hypothesis, we performed whole-blood gene expression profiling of subjects with COPD and control smokers in the COPDGene study, and compared the results with previously reported results from gene expression profiling studies of eosinophilic asthma and COPD (4–6), as well as known markers of eosinophil activation (7).
COPDGene is a multicenter observational study that enrolled 10,192 smokers with and without COPD. COPD was defined by a postbronchodilator ratio of forced expiratory volume in 1 second (FEV1) to forced vital capacity (FVC) < 0.7 with FEV1 < 80% predicted; smokers without COPD had FEV1/FVC ≥ 0.7 and FEV1 ≥ 80%. Eosinophilic COPD was defined based on an eosinophil count of ≥300 cells/μl (8). An unselected subset of 535 COPDGene subjects underwent whole-blood RNA sequencing via previously described methods (9). Differential gene expression analyses were performed with the limma voom R package (10) (data supplement).
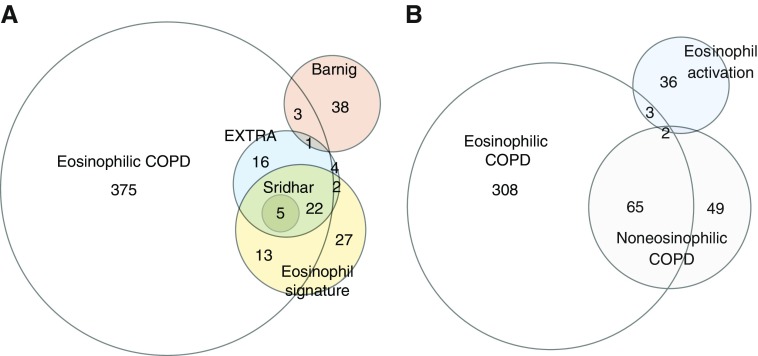
The study population included 231 subjects with COPD and 224 control subjects with normal spirometry, after the exclusion of seven subjects who were taking oral steroids. Patients with COPD were older and more likely to be non-Hispanic white individuals, with a greater lifetime smoking history and inhaled corticosteroid use (Table E1 in the data supplement). Subjects with COPD had higher neutrophil counts and lower lymphocyte counts than the control subjects, but there was no difference in eosinophil counts or in the proportion of subjects with eosinophil counts of ≥300 cells/μl (Table E1). We performed a differential expression analysis comparing eosinophilic versus noneosinophilic COPD (n = 49 vs. 182) and found 505 differentially expressed genes at a false discovery rate of <0.05 (File E1). We compared the differentially expressed genes with 1) known eosinophil signature genes used in Sridhar and colleagues’ study (6), 2) genes that correlated with blood eosinophil count in pediatric and adult patients with asthma in the EXTRA (Omalizumab in Severe Allergic Asthma Inadequately Controlled With Standard Therapy) study (5), 3) differentially expressed genes from peripheral blood of patients with COPD treated with benralizumab (Sridhar and colleagues’ study) (6), and 4) differentially expressed eosinophil genes from patients with eosinophilic asthma (Barnig and colleagues’ study) (4) (File E2). As expected, the top differentially expressed genes were known to be expressed in eosinophils, including SMPD3 (sphingomyelin phosphodiesterase 3), SLC29A1 (solute carrier family 29 member 1), CLC (Charcot-Leyden crystal galactin), and IL5RA (interleukin 5 receptor subunit α) (Figures 1 and 1A). Furthermore, upregulated genes in eosinophilic COPD overlapped with 44 out of 50 eosinophilic asthma genes from the EXTRA study that predicted response to anti–IL-13 (lebrikizumab) (P < 5 × 10−70) and five out of five genes from Sridhar and colleagues’ study (6) (P < 3 × 10−9), whereas four genes overlapped with eosinophilic asthma genes from Barnig and colleagues’ study (4) (P < 0.01). A pathway analysis demonstrated enrichment in immune-related functions (Table 1).
Figure 1.
Euler diagram comparing gene expression signatures from eosinophilic chronic obstructive pulmonary disease (COPD) and previously published datasets. Overlapping genes were all upregulated in eosinophilic COPD. (A) Number of overlapping genes from subjects with eosinophilic COPD and eosinophilic asthma (EXTRA) (4), benralizumab-responsive COPD genes (6), and eosinophilic signature genes. (B) Eosinophil-associated genes from subjects with eosinophilic COPD show a significant overlap with known markers of eosinophil activation.
Table 1.
Eosinophilic Chronic Obstructive Pulmonary Disease–associated Genes and Pathways in the COPDGene Study
| Analysis | Comparison | Number of Differentially Expressed Genes | Top Differentially Expressed Genes | Pathway Analysis |
|---|---|---|---|---|
| Differential expression | Eosinophilic vs. noneosinophilic COPD | 505 | SMPD3 | GO:0050865 regulation of cell activation |
| SLC29A1 | GO:0001655 urogenital system development | |||
| PIK3R6 | GO:0022407 regulation of cell–cell adhesion | |||
| PRSS33 | GO:0044262 cellular carbohydrate metabolic process | |||
| CLC | ||||
| Eosinophilic COPD vs. eosinophilic controls | 0 | — | — | |
| Eosinophilic COPD exacerbators vs. nonexacerbators | 3 | NUDT5 | — | |
| ANKRD40 | ||||
| BCCIP | ||||
| Eosinophil-associated genes | Eosinophilic COPD (n = 49) | 378 | ACSM3 | GO:0030099 myeloid cell differentiation |
| IL5RA | GO:0046434 organophosphate catabolic process | |||
| ADORA3 | GO:1903706 regulation of hemopoiesis | |||
| TEC | ||||
| CYSLTR2 | ||||
| Noneosinophilic COPD (n = 182) | 114 | SIGELC8 | NA | |
| IL5RA | ||||
| ADGRE4P | ||||
| OLIG2 | ||||
| SLC29A1 | ||||
| Eosinophilic control subjects (n = 33) | 0 | — |
Definition of abbreviations: ACSM3 = acyl-CoA synthetase medium chain family member 3; ADGRE4P = adhesion G protein-coupled receptor E4, Pseudogene; ADORA3 = adenosine A3 receptor; ANKRD40 = ankyrin repeat domain 40; BCCIP = BRCA2 and CDKN1A interacting protein; CLC = Charcot-Leyden crystal galectin; COPD = chronic obstructive pulmonary disease; COPDGene = genetic epidemiology of COPD; CYSLTR2 = cysteinyl leukotriene receptor 2; GO = gene ontology; IL5RA = IL-5 receptor subunit α; OLIG2 = oligodendrocyte transcription factor 2; NA = not applicable; NUDT5 = nudix hydrolase 5; PIK3R6 = phosphoinositide-3-kinase regulatory subunit 6; PRSS33 = serine protease 33; SIGELC8 = sialic acid-recognizing Ig-like lectin 8; SLC29A1 = solute carrier family 29 member 1 (Augustine Blood Group); SMPD3 = sphingomyelin phosphodiesterase 3; TEC = Tec Protein tyrosine kinase.
We compared gene expression in subjects with eosinophilic COPD (n = 14) versus those without a reported COPD exacerbation in the prior year (n = 35), and found that three genes were differentially expressed (Table 1). NUDT5 (nudix hydrolase 5), which was downregulated in subjects with eosinophilic COPD and exacerbations, is a transforming growth factor-β–responsive eosinophilic gene that is downregulated upon transforming growth factor-β stimulation (11). The differential expression of eosinophil-expressed genes in subjects with eosinophilic COPD suggests that there could be functional differences in the eosinophils that may explain differences in clinical phenotypes (i.e., exacerbations).
Contrary to our hypothesis, we did not find any statistically differentially expressed genes between subjects with eosinophilic COPD and eosinophilic control subjects (Table 1). The sensitivity to detect gene expression differences in whole blood may be reduced in a comparison of two groups with elevated eosinophils. Therefore, to further characterize the eosinophil transcriptome in COPD, we identified genes associated with the eosinophil count by performing multivariable linear regression of gene expression (File E3) (10). There were 378 eosinophil-associated genes in eosinophilic COPD and 114 eosinophil-associated genes in noneosinophilic COPD. In contrast, in eosinophilic control subjects, eosinophil-associated genes were not identified after false discovery rate correction, which may be a result of reduced power due to the small number of subjects. Eosinophil-associated genes in eosinophilic COPD were enriched for known markers of eosinophil activation (IL5RA, SEMA7A [semaphorin 7A], RNASE3 [ribonuclease A family member 3], CD9, and IL2RA [IL-2 receptor subunit α]) (P < 3 × 10−4), which have been associated with active eosinophilic inflammation and asthma (7). Eosinophil-associated genes from eosinophilic COPD did not show a statistically significant overlap with markers of eosinophil activation (P = 0.50) (Figure 1B).
These results show that eosinophilic COPD and eosinophilic asthma share a similar blood transcriptomic profile with overlapping eosinophil-specific gene expression. The transcriptomic signature of eosinophilic COPD is not only driven by the increased level of eosinophils but also may demonstrate functional differences in the eosinophils themselves, as suggested by the differential expression of eosinophil-associated genes depending on the phenotype and disease status. A limitation of our study is that the RNA sequencing was performed on whole blood, so it may not fully reflect lung eosinophil gene expression. Future studies on isolated eosinophils from sputum or lung tissue will be necessary to confirm the results in lung-specific eosinophilic populations.
In conclusion, we found that subjects with eosinophilic COPD had a blood gene expression profile that was enriched for eosinophilic gene expression and activated eosinophil marker genes similar to those observed in subjects with eosinophilic asthma, which may support “precision medicine” trials to repurpose biologic therapies for asthma in a well-defined eosinophilic COPD subgroup.
Supplementary Material
Footnotes
Supported by National Institutes of Health grants R01HL125583, R01HL130512, R01HL124233, R01HL126596, U01HL089897, U01HL089856, T32HL007427, and P01HL132825. The COPDGene project is also supported by the COPD Foundation through contributions made to an industry advisory board comprised of AstraZeneca, Boehringer Ingelheim, GlaxoSmithKline, Novartis, Pfizer, Siemens, and Sunovion.
This letter has a data supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Author disclosures are available with the text of this letter at www.atsjournals.org.
Contributor Information
Collaborators: for the COPDGene Investigators
References
- 1.Barnes PJ. Immunology of asthma and chronic obstructive pulmonary disease. Nat Rev Immunol. 2008;8:183–192. doi: 10.1038/nri2254. [DOI] [PubMed] [Google Scholar]
- 2.Bafadhel M, Pavord ID, Russell REK. Eosinophils in COPD: just another biomarker? Lancet Respir Med. 2017;5:747–759. doi: 10.1016/S2213-2600(17)30217-5. [DOI] [PubMed] [Google Scholar]
- 3.Pavord ID, Chanez P, Criner GJ, Kerstjens HAM, Korn S, Lugogo N, et al. Mepolizumab for eosinophilic chronic obstructive pulmonary disease. N Engl J Med. 2017;377:1613–1629. doi: 10.1056/NEJMoa1708208. [DOI] [PubMed] [Google Scholar]
- 4.Barnig C, Alsaleh G, Jung N, Dembélé D, Paul N, Poirot A, et al. Circulating human eosinophils share a similar transcriptional profile in asthma and other hypereosinophilic disorders. PLoS One. 2015;10:e0141740. doi: 10.1371/journal.pone.0141740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choy DF, Jia G, Abbas AR, Morshead KB, Lewin-Koh N, Dua R, et al. Peripheral blood gene expression predicts clinical benefit from anti-IL-13 in asthma. J Allergy Clin Immunol. 2016;138:1230–1233, e8. doi: 10.1016/j.jaci.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 6.Sridhar S, Liu H, Pham TH, Damera G, Newbold P. Modulation of blood inflammatory markers by benralizumab in patients with eosinophilic airway diseases. Respir Res. 2019;20:14. doi: 10.1186/s12931-018-0968-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johansson MW. Activation states of blood eosinophils in asthma. Clin Exp Allergy. 2014;44:482–498. doi: 10.1111/cea.12292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yun JH, Lamb A, Chase R, Singh D, Parker MM, Saferali A, et al. COPDGene and ECLIPSE Investigators. Blood eosinophil count thresholds and exacerbations in patients with chronic obstructive pulmonary disease. J Allergy Clin Immunol. 2018;141:2037–2047, e10. doi: 10.1016/j.jaci.2018.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parker MM, Chase RP, Lamb A, Reyes A, Saferali A, Yun JH, et al. RNA sequencing identifies novel non-coding RNA and exon-specific effects associated with cigarette smoking. BMC Med Genomics. 2017;10:58. doi: 10.1186/s12920-017-0295-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen ZJ, Hu J, Esnault S, Dozmorov I, Malter JS. RNA Seq profiling reveals a novel expression pattern of TGF-β target genes in human blood eosinophils. Immunol Lett. 2015;167:1–10. doi: 10.1016/j.imlet.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.