Abstract
The genome is constantly attacked by genotoxic insults. DNA damage has long been established to cause cancer development through its mutagenic consequences. Conversely, DNA damage is induced during radiation- and chemotherapy to drive cells into apoptosis or senescence as outcomes of the DNA damage response (DDR). More recently, DNA damage has been recognized as a causal factor for the aging process. The causal role of DNA damage in aging and age-related diseases is illustrated by numerous congenital progeroid syndromes that are caused by mutations in genome maintenance pathways. The past two decades have brought rapid progress in the understanding of how DDR drives cancer development and causally contributes to the aging process. The DDR factor p53 turns out to not only take the centre stage during tumour development but also to play an important role in the aging process. Studies in metazoan models ranging from C. elegans to mammalian disease models have revealed cell autonomous and systemic DDR mechanisms that orchestrate adaptive responses that augment maintenance of the aging organism amid gradually accumulating DNA damage.
DNA damage drives the aging process
Aging is a nearly universal property of life forms ranging from single cell bacteria to humans. Fundamentally, aging is the default fate of life whose building blocks DNA, RNA, and proteins are constantly subjected to chemical alterations that impair their function. Albeit chemically more vulnerable than DNA, RNAs are usually rather rapidly turned over and the consequences of damaged RNA molecules temporarily restricted. Damaged proteins are either kept in shape by chaperones or are degraded and their amino acids recycled through the ubiquitin proteasome system or autophagy. Nonetheless, damaged proteins aggregate during aging and result in functional deterioration particularly in neurons where they give rise to dementia including the most prevalent Alzheimer’s disease. The consequences of DNA damage, however, are much more widespread as DNA contains the information for all RNA and proteins a cell produces.
It was estimated that tens of thousands of damaging events occur each day in every single one of our cells 1. The genotoxic attacks can originate from extrinsically inflicted radiation damage or chemicals as well as from endogenous sources such as metabolic byproducts or reactive oxygen species (ROS). Oxidative modifications and single strand breaks (SSBs) are the most frequently encountered lesions and are rapidly repaired by base excision repair (BER) and DNA ligases, respectively 2,3. Both systems rely on redundancy of several glycosylases that initiate BER and numerous ligases, as complete failure to repair those lesions is incompatible with life. Helix-distorting lesions are removed by nucleotide excision repair (NER), a highly complex repair pathway with intriguing connections to cancer and the aging process that we will discuss further below for their conceptually instructive character 4. Even highly cytotoxic double strand breaks (DSBs) and interstrand crosslinks (ICLs) occur on a daily basis. Those lesions are particularly dangerous because they impair the cell’s ability to replicate the DNA and segregate the chromosomes potentially giving rise to aneuploidy resulting in functional distortion of the genetic programs of the cell 5. The two major routes for DSB repair are non-homologous end joining (NHEJ) and homologous recombination (HR) but even in their complete absence there are still options to pull, through backup end joining or microhomology directed repair 6,7.
The distinction between the modes and results of NHEJ and HR make an interesting point about the constraints on genome maintenance in the context of other cellular functions. The rapid NHEJ process is employed mostly when cells are not replicating their DNA and when no undamaged repair template is available. Prior to the end joining the broken chromosome ends are resected potentially because a high-energy ionization event produces ROS that inflicts base damage adjacent to the break site as well as to aid the re-ligation. Given that only a fraction of the human genome encodes for genes the local loss of a DNA stretch might be tolerable in most instances 8,9. Speed at the expense of accuracy is particularly important when cells rapidly divide. HR, in contrast is highly accurate as it uses the homologous sequence as repair template. However, during replication HR might also boost genome instability when replication forks collapse amid the engagement of chromosomes in the HR process 10,11. HR is mostly employed during late S and within the G2 phases of the cell cycle 12. Some of the most prominent tumour suppressor genes are operating in the HR process. Mutations in the BRCA1 and BRCA2 genes predispose to breast and ovarian cancer and are both involved in HR. Interestingly, despite the backup DSB repair pathways, a combination of HR and NHEJ deficiency renders cells highly susceptible even to endogenous levels of DNA damage. The synthetic lethality of those two pathways led the way for tumour therapies that are personalized to HR defects, resulting e.g. from a BRCA1 mutation, and use inhibition of the NHEJ process 13. A clinical strategy for utilizing the sensitivity of BRCA1 deficient cells is the application of catalytic PARP1 inhibitors resulting in replication-associated DSBs that in the absence of functional HR cannot be resolved 14.
There are numerous instances where speed matters over accuracy. The speed argument goes particularly for cells that are engaged in replication. Even though the replicative DNA polymerases have a low error rate due to their proofreading activity that is accomplished through their exonuclease domain, occasional errors might slip through. Mismatch repair (MMR) scans the genome post replication for such errors by detecting structural aberrations of mispaired nucleotides 15. Some DNA lesions, for example helix-distorting lesions, cannot be circumvented by replicative DNA polymerases. In those cases translesion synthesis (TLS) polymerases take over 16. They handle the base pairing with less stringency but keep the replication forks going even though they tend to make mistakes.
Cancer and Aging: the flip side of DNA damage
There are two principally distinct outcomes of DNA damage. Erroneous repair can lead to mutations and chromosomal aberrations both of which are causal events in cancer development. In contrast, persistent DNA damage can block transcription and replication thus hampering cellular functionality and promoting cellular senescence and apoptosis. Consequently stem cell compartments become depleted, tissues degenerate, and homeostasis declines: ultimately, persistent DNA damage drives the aging process.
Erroneous repair can result from mistakes in placing the correct nucleotides, aberrant recombination, or lesion bypass and could alter the genetic information in numerous ways. When a mutation inactivates a tumour suppressor gene such as p53, the checkpoints that control the proliferation rate of a cell lose their function 17. When distinct sections of a chromosome are erroneously joined, the promoters might aberrantly drive the expression of an oncogene. A most prominent example for this is the MYC gene that is normally expressed in a highly controlled fashion to drive cell cycle entry. However, when the V(D)J recombination during B-cell maturation fails and the Eμ enhancer that normally boosts the expression of the immunoglobulin heavy chain gene is fused to the MYC gene the B cells are driven into uncontrolled proliferation 18. A recent study by the Nussenzweig lab revealed that even during interphase DSBs resulting from topoisomerase 2B activity at topological domain boundaries of chromatin results in chromosomal rearrangements observed in cancers 19.
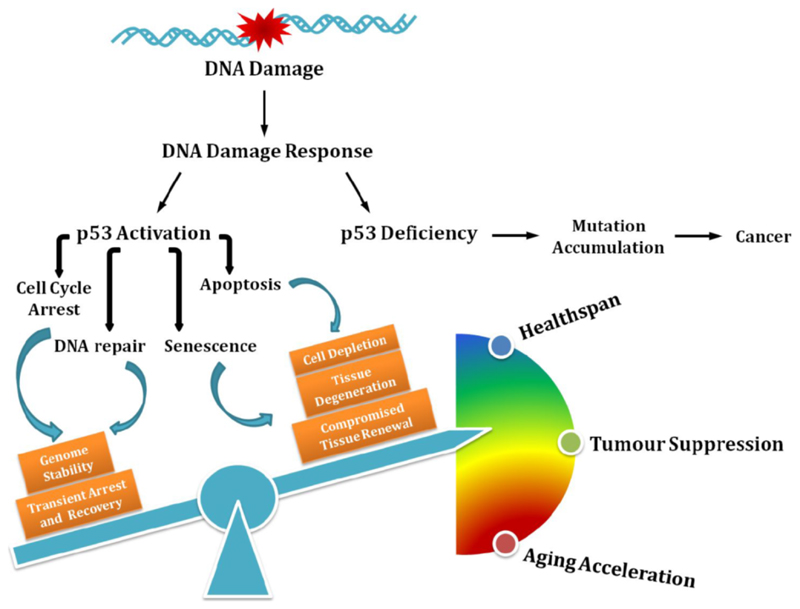
Through its mutagenic effect, DNA damage thus triggers tumour development. Importantly, the mutations need to affect genes that control the cell’s DDR as a functional DDR prevents the cells from uncontrolled proliferation. The DDR controls, through DNA damage checkpoints, under which circumstances cells may enter and proceed through the cell cycle 20. The tumour suppressor p53 plays an important role in controlling cellular proliferation in the context of DNA damage (Figure 1). Normally, p53 is a shortly lived protein whose activity is tightly regulated by various processes including transcriptional and translational control as well as posttranslational modifications (PTMs) 21. Moreover, p53 is rapidly ubiquitylated for example by MDM2 and consequently targeted for degradation by the UPS 22,23. However, in the presence of DNA damage, p53 is stabilized by DDR signalling. For instance, p53 is activated by several PTMs including phosphorylation on serine-15 by the phosphatidylinositol kinase-related kinases ataxia-telangiectasia mutated (ATM), and ATM and Rad3-related (ATR), or on serine-20 by the checkpoint kinase (CHK2) at the N-terminal transactivation domain, which interfere with the inhibitory interaction between MDM2 and p53, rendering p53 stabilization and accumulation. Specific DNA binding activity of p53 can be further enhanced or fine-tuned by phosphorylation and acetylation at the DNA-binding domain and C-terminal regulatory domain, modulating the distinct cell fate decisions 24–27. There are numerous regulative events that determine the quality of the p53 DDR response and certainly numerous more awaiting their discovery 28.
Figure. The influence of p53-mediated cell fate decision on cancer development and the aging process.
Defective p53 leads to accumulation of mutations that drive carcinogenesis; on the contrary, p53 regulates diverse outcomes of the DNA damage response, the fine-tuning of which balances healthspan, tumour suppression, and accelerated aging.
To exemplify in a simplistic way the outcomes of the p53 response, we can turn to its most ancestral form, which is the C. elegans p53-like CEP-1 protein 29,30.The homology to human p53 is mainly restricted to the DNA binding domain with most of the hot spot mutations that are associated with human cancers highly conserved. Also the residues forming the zinc finger are present in the nematode form. The most well characterized function of CEP-1 is the regulation of DNA damage-induced apoptosis in meiotic pachytene cells. The pachytene is the critical meiotic phase where recombination takes place. During meiotic recombination the SPO-11 endonuclease induces DSBs that are used for the exchange with the homologous chromosome. When the cells reach the late pachytene stage all DSBs should have been properly processed and the Holliday junctions resolved. When, however, DSBs are still present the meiotic recombination has failed and those cells couldn’t proceed to form genomically integer gametes. At this stage the CEP-1 mRNA gets translated as prior to this point in meiosis it has been repressed by the translational KH-motif RNA binding protein and Quaking homolog GLD-1 31. In addition to translational control, also the stability of CEP-1 is regulated through ubiquitylation 32. Once CEP-1 becomes available for activation through DDR triggered by the persistent DSBs it transcriptionally induces the two proapoptotic BH3-only domain genes egl-1 and ced-13 33–35. As in mammals, the BH3-only domain proteins inhibit Bcl2 that in worms is encoded by the ced-9 gene. CED-9 then alleviates its sequestration of the Apaf1-like CED-4, which in turn sets off the CED-3 caspase to seal the fate of the cells carrying unprocessed meiotic DSBs or those induced by ionizing radiation (IR) 36.
The apoptotic DDR regulation of p53 is highly conserved in humans where the BH3-only domain genes p53 up-regulated modulator of apoptosis (PUMA) and NOXA (named for ‘damage’) are induced in a similar fashion 37,38. In addition to triggering the apoptotic demise of genomically compromised cells, p53 also induces cell cycle arrest, most well characterized by the transcriptional activation of the cyclin-dependent kinase inhibitor 1A (CDKN1A) gene p21 that inhibits mitotic cyclin-dependent kinases (CDKs) 39. The cell cycle arrest is critical for allowing cells time to repair the damage, during which p53 enhances several DNA repair pathways to facilitate the clearance of DNA lesions 40. Not only is p53 involved in the NER by transcriptionally up-regulating the xeroderma pigmentosum complementation group C (XPC) and damage-specific DNA binding protein 2 (DDB2), both of which are important damage-recognizing factors required for initiating global-genome NER 41,42, but p53 has also been implicated in the transcriptional control of the MMR component human MutS homolog 2 (hMSH2) upon DNA damage 43,44. The expression of Fanconi anemia complementation group C (FANCC) gene, the lack of which causes an inherited DNA repair deficiency syndrome, is also closely related with the promoter abundance of p53 45. Beyond transcriptional regulation, p53 also modulates BER in a transcriptionally independent manner. The activities of the pivotal BER enzymes 8-oxoguanine glycosylase and AP endonuclease are augmented by direct interaction with p53 hence enhancing the efficiency of excising oxidative DNA lesions 46. Moreover, a sub-pool of ATM-phosphorylated p53 upon IR is found directly associated with the lesions, promoting DNA repair 47. In parallel to the repair pathways, p53 up-regulates the p53-controlled ribonucleotide reductase (p53R2) for supplementing sufficient nucleotides during DNA re-synthesis thereby further facilitating the DNA repair process 48.
Amid unrepairable damage, however, p53 may also induce cellular senescence thus permanently withdrawing cells from cycling yet keeping them metabolically active 49. The individual contributions of transient cell cycle arrest, cellular senescence and apoptosis to the tumour suppressor function of p53 might vary depending on the cell type and other tumour suppressor gene and oncogene mutations that may be present. However, we can again extract some conceptually instructive insights into the outcome of DDR. A failure to arrest the cell cycle either transiently or permanently amid DNA damage might fuel mutation rates and thus boost tumorigenesis. Even heavily genomically compromised cells might survive when p53 is dysfunctional.
Importantly, however, there is the other extreme of DDR. When p53 puts on the breaks too much, cells might not sufficiently proliferate to assure a physiological level of tissue regeneration (Figure 1). This can be detrimental particularly for tissues that require proliferative activity of their stem and progenitor cell compartments such as the hematopoietic system 50. In addition, extraneous apoptosis might result in loss of tissue integrity regardless of which cell type is affected. Cellular senescence not only impairs proliferation and thus tissue regeneration and homeostasis but beyond that impacts neighbouring cells and potentially even the organism through the senescence associated secretory phenotype (SASP) 51. SASP is a collection of heterogeneous cytokines that promote inflammation, tissue remodelling, and proliferation of recipient cells. In recent years, the contribution of senescent cells to aging has revealed the widespread physiological and pathological consequences of this cell fate. Highly pathological levels of cellular senescence have been observed in mice carrying mutations in the mitotic spindle checkpoint gene Bubr1 52. When, however, either the senescence pathway was abrogated by a mutation in the Ink4a encoded p16 gene or cells were eliminated when the p16 promoter was fused to a caspase that consequently eliminated senescent cells, the premature aging phenotype of the Bubr1 mutants was alleviated 53. Moreover, even the functional decline during normal aging could be slowed down by elimination of senescent cells 54. Currently, senolytic drugs are being tested such as Bcl-XL inhibitors that could selectively eliminate senescent cells and supposedly slow the aging process of the organism 55. However, it is important to notice that cellular senescence might also have positive functions as for example the SASP component PDGF-A was demonstrated to support wound repair in murine skin 56.
The outcome of enhanced p53 function has been first demonstrated in two different mutant mice that express hyperactive forms of p53 57,58. Tyner and colleagues generated the p53+/m mice, in which the m allele comprises a truncated form of p53 containing only exons 7-11 of the p53 coding sequence. With enhanced stability and transactivation activity of the m allele product, p53+/m mice were highly protected from tumours yet at the expense of accelerated aging 57. Further analysis of the haematopoietic stem cells (HSCs) revealed a reduced number of proliferating HSCs with age in p53+/m mice, confirming that increased p53 activity may lead to decreased production of progenitor and mature differentiated cells from the stem cells thereby contributing to the aging phenotype 59. In mice overexpressing another truncated isoform of p53, p44, signs of premature aging were observed as early as 4 months of age and, consistently, the mice displayed a low incidence of cancer 58. The fine balance between keeping the brakes on proliferation and controlling cellular survival has been kept in mice that carry an extra copy of the p53 gene, called the Super-p53 mice 60. These mice are protected from cancer development, likely because they are equipped with three p53 alleles that are therefore much less likely to lose heterozygocity. For extended lifespan, however, this was not sufficient and instead required an additional copy of the ARF tumour suppressor 61. The mice with increased basal level of p53 due to low expression levels of MDM2 (mdm2 puro/Δ7-12mice) were also cancer resistant yet with a normal lifespan 62, further echoing the notion that a balanced p53 levels is of great significance for tumour suppression without accelerated aging.
When it comes to the human, previous studies have revealed a Pro/Arg polymorphism at amino acid residue 72 of p53 that alters the potential of inducing apoptosis 63–65. A meta-analysis of the published literature has indicated that carriers of p53 Pro/Pro form, which is less potent in inducing apoptosis than p53 Arg/Arg form, had increased survival albeit higher mortality from cancer 65. Another independent study recruiting more than 9000 participants demonstrated an higher overall survival for p53 Pro/Pro carriers, and, most importantly, a increased survival after cancer or other life-threatening disease 66. However, contrasting results were obtained in a more recent study with even larger population 67, leaving it an open question whether hyper-active p53 plays an unfavourable role in human lifespan.
Persistent DNA damage: a driving force of aging
As DDR prevents tumourigenesis, its constitutive activation such as in hyperactive p53 mutants accelerates the aging process. The continuous activation of DDR, however, arises not only by genetic mutations that augment DDR, but can result from DNA lesions that are not repaired and thus persist. For instance critically shortened and thus unprotected telomeres are recognized as DNA-DSBs 68. Indeed, the inability of telomerase to maintain the telomere length cause premature aging through activating the DDR and p53 69,70. Mice that are lacking the catalytic subunit or the RNA component of telomerase have shorter lifespan with early onset of aging phenotypes, even though in mice these phenotypes require several generations of defective telomere extension to arrive at critical telomere shortening precipitating the progeroid pathologies 71,72. Premature replicative senescence also shortens lifespan in Ku80-/- mutant mice that lack functional NHEJ 73. Intriguingly, the accelerated aging phenotypes of both animal models can be alleviated by the loss of p53, underlining again the pivotal role of p53 in DDR-mediated premature aging 74,75.
Another prominent example for demonstrating the link between persistent DNA damage and premature aging comes from the autosomal genetic disorder Fanconi Anemia (FA) that is characterized by progressive bone marrow failure due to functional decline of hematopoietic stem and progenitor cells (HSPCs). In addition to their sensitivity to ICLs 76,77, FA cells are hyper-sensitive to oxidative stress with excessive levels of oxidative DNA damage 78,79. Ceccaldi et al. further demonstrated that the unresolved DNA damage leads to constitutive activation of p53 resulting in a p21-dependent G0/G1 cell-cycle arrest in HSPCs from FA patients. Remarkably, the depletion of p53 or p21 could alleviate the defects in HSPCs 80. Another example of the important role of p53 in controlling the DDR in HSPCs is provided by a mouse mutant that expresses the hypomorph p53515C allele in an Mdm2 deficient background. Here, p53 activation amid high ROS levels lead to exacerbated cell cycle arrest, senescence and apoptosis in HSPCs 81.
Not only in genetic diseases caused by mutations in genome stability factors, but also during normal aging, the ability of maintaining genome integrity declines especially in highly regenerative tissues such as the hematopoietic system. By comparing transcription profiles of purified HSCs from mice aged 2 to 21 months, Chambers and colleagues have reported that with age genes associated with stress response were up-regulated, while genes involved in maintaining genome integrity including DNA repair genes were down-regulated 82. Consistent with this observation, the efficiency of DNA repair in aged HSCs is limited, leading to gradual accumulation of DNA damage and thereby attenuating the self-renewal and tissue regenerative capacity of HSCs 83–86.
For understanding the role of persistent DNA damage in the aging process, congenital NER deficiencies have been particularly illuminating. NER recognizes helix-distorting lesions that are most prominently formed when short wavelength UV light links adjacent bases to form cyclobutane pyrimidine dimers (CPDs) and 6-4 pyrimidine-pyrimidone photopruducts (6-4PPs) 87. Indeed, CPDs are the primary instigator of UV-induced skin carcinogenesis 88. The lesions are recognized either by the scanning of global-genome (GG-) NER or when RNA polymerase II (RNAPII) stalls at a lesion and transcription-coupled (TC-) NER is activated. Both pathways then trigger a common NER pathway that verifies the damage, unwinds the double helix, incises on either side of the lesion, excises the stretch containing the damage, resynthesizes the gap and finally ligates the remaining gap.
Congenital defects affecting one or the other recognition pathway couldn’t possibly be more distinct 89. Xeroderma pigmentosum (XP) patients display pigmentation abnormalities, atrophic skin, and a several thousand fold elevated skin cancer susceptibility 90. Cockayne syndrome (CS) patients, on stark contrast suffer from postnatal growth retardation, neurological defects and display numerous manifestations of premature aging but remain cancer free 91,92. A number of NER genes are associated with XP but most clearly those involved in GG-NER show the most distinctive skin phenotypes and cancer susceptibility, while XP mutations associated with factors operating in the common NER pathway typically display in addition neurodegenerative symptoms 93. CS patients, in contrast, carry mutation in the CSA or CSB genes that operate in the initial steps of TC-NER. NER deficiencies are highly complex –on the molecular as well as on the pathological level– and not restricted to XP and CS alone, however, they are highly instructive as to the distinct outcomes of DDR 4. While GG-NER defects are mutagenic when unrepaired lesions pass through replication where they might result in replicative errors or genome instability when replication forks collapse, TC-NER defects result in stalling of RNAPII. At high levels of RNAPII stalling transcription is blocked and cells might no longer fulfil their function or might even apoptose. Therefore, while mutagenic DNA lesions fuel cancer development, persistent DNA lesions drive the aging process by hampering the DNA metabolism as replication and transcription are blocked.
Intriguingly, cells respond to transcription-blocking lesions already when they are detected at very low levels. Regardless of whether cells proliferate or are terminally differentiated, in response to those lesions they reduce the expression levels of genes that are involved in the somatic growth axis, particularly the Insulin-like growth factor-1 receptor (IGF-1R) and growth hormone receptor (GHR) 94. Both receptors are central elements of the somatic growth axis and their reduction has been established as bona fide longevity assurance mechanism that extends lifespan in mice that lack the pituitary or the GHR gene 95–97. Also progeroid (“premature aging-like”) mice that carry defects in NER show reduced somatic growth gene expression and dampened IGF-1 levels 98–100. It is likely that cells respond with attenuating the somatic growth axis to assure the survival of the organism when DNA damage accumulates 101. Indeed, stress resistance –a feature strongly correlated with longevity– was observed in cells experiencing transcription-blocking lesions as they became exquisitely resistance to oxidative stress 102, similarly to mice with reduced IGF-1R gene dosage 103.
The consequences of this type of DDR have been recently studied in the nematode C. elegans where the mechanisms of longevity have been most extensively investigated. Here, the FOXO transcription factor DAF-16, which is normally kept inactive by insulin-like signalling but –consistently with the IGF-1R dampening in mammals– is activated upon transcription-blocking DNA lesions, promotes the animals’ developmental growth and maintains the integrity and functionality of tissues in adult animals even when the DNA damage persists 104. DAF-16 comprises a bona fide longevity assurance factor and its involvement in DDR suggests a distinct mechanism of adaptations to genome instability: DNA damage tolerance by augmented maintenance of tissues, which is particularly relevant for cell types that are not proliferating such as the adult nematode’s cells outside of the germline that are entirely postmitotic. Therefore, there appear to be two types of longevity assurance mechanisms: DNA repair systems that prevent and delay the accumulation of DNA damage and lifespan regulators that determine the threshold to which DNA lesions are tolerated before they become detrimental. The latter are regulated by transcription factors such as DAF-16 that mediate gene expression programs that comprise stress resistance genes as well as developmental growth programs 104.
Intriguingly, the consequences DDR are not confined to the genomically compromised cell alone. Somatic growth signalling for instance is mediated through the secretion of IGF-1 and GH that have endocrine activities and SASP factors exert non-cell-autonomous effects 50. Also p53 has non-cell-autonomous consequences when for example it regulates the cytokine secretion of tumour cells that determine whether they are cleared by macrophages 105. In C. elegans innate immune factors that are induced in response to DNA damage in germ cells trigger a systemic stress resistance program that elevates somatic endurance thus extending reproductive lifespan to allow offspring generation once genome stability in the germ cells is reconstituted 106. It will be highly important to gain more insights into the non-cell-autonomous regulation of DDR and which role p53 might play in this process.
Alleviating DNA damage responses
Experiments discussed above particularly on additional p53 gene dosage as well as p53 ablation in a number of DNA repair deficiencies associated with exacerbated replicative senescence have indicated that a balance of DDR is important for cancer suppression and longevity and this balance could potentially be targeted for promoting health in old age (Figure 1). Also modulations of the consequences of DNA repair defects on cellular metabolism have been suggested to alleviate pathologies resulting from genome instability. Xrcc1 mutant micethat are defective in single-strand break repair show highly elevated protein poly ADP-ribosylation (PARylation) levels and develop neurological disorders characterized by ataxia. Interestingly, genetic ablation of Parp1 could restore ADP-ribose levels and reduce neuronal loss and alleviate the ataxia. One consequence of PARP1 activity is that excessive PARylation could dampen the NAD+ levels resulting in defective mitophagy, which could be alleviated by replenishment of NAD+ thus improving the health of progeroid NER and ATM mouse models 107,108. Elevated NAD+ levels have numerous consequences on cellular metabolism and could also promote longevity through Sirtuin-mediated activation of the mitochondrial unfolded protein response and FOXO signaling 109. In addition, NAD+ could also alleviate PARP1 inhibition mediated by the NAD+ binding protein DBC1 110. It is thus conceivable that intervention strategies could promote longevity and healthspan through affecting the activity of genome maintenance systems.
Concluding remarks
DNA damage is invariably occurring as a result of a plethora of endogenous and exogenous genotoxic insults. A complex arsenal of DNA repair mechanisms efficiently removes the lesions and ensures the maintenance of the soma during lifespans that were relevant for supporting offspring generation during the evolutionary history of a species. The capacity to keep genomes of somatic cells intact vanishes thereafter, leading to functional decline of cells and tissues. It is an intriguing and yet incompletely understood question how germ cells retain the levels of DNA repair accuracy that has allowed maintaining the gene pool of a species for many hundreds of millions of years. The aging organism responds to DNA damage by cell-autonomous and systemic DDR. The p53 gene is not only a most important tumour suppressor but also an orchestrator of an extensive network of DDR factors that are relevant for adaptation to DNA damage in the aging organism. Studies in metazoan model systems have begun to shed light on how signalling pathways and endocrine axes respond to the build-up of DNA damage with aging. Approaches such as combining proteome assessments with studies of PTMs and metabolic alterations have begun to shed new light on the complexity of DDR and the consequences of persistent DNA damage in the aging organism 111. It will be pivotal to further explore how longevity assurance mechanisms respond to DNA damage and how they influence the aging phenotype and the aetiology of aging-associated diseases.
Acknowledgments
H.O. received a fellowship from the Cologne graduate school on aging (CECAD-CGA). B.S acknowledges funding from the Deutsche Forschungsgemeinschaft (CECAD, SFB 829, SFB 670, and KFO 286), the European Research Council (ERC Starting grant 260383), Marie Curie (FP7 ITN CodeAge 316354, aDDRess 316390, MARRIAGE 316964), FLAG-ERA JTC 2015 (G-Immunomics, SCHU 2494/3-1), the German-Israeli Foundation (GIF 1104-68.11/2010), the Deutsche Krebshilfe (109453), the Bundesministerium für Bildung und Forschung (Sybacol FKZ0315893A-B), and the COST action (BM1408, GENiE).
Footnotes
H.O. and B.S. wrote the manuscript. The authors declare no competing interest.
References
- 1.De Bont R, van Larebeke N. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis. 2004;19(3):169–185. doi: 10.1093/mutage/geh025. [DOI] [PubMed] [Google Scholar]
- 2.Bauer NC, Corbett AH, Doetsch PW. The current state of eukaryotic DNA base damage and repair. Nucleic Acids Res. 2015;43(21):10083–10101. doi: 10.1093/nar/gkv1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008;9(8):619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 4.Edifizi D, Schumacher B. Genome Instability in Development and Aging: Insights from Nucleotide Excision Repair in Humans, Mice, and Worms. Biomolecules. 2015;5(3):1855–1869. doi: 10.3390/biom5031855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40(2):179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koole W, van Schendel R, Karambelas AE, et al. A Polymerase Theta-dependent repair pathway suppresses extensive genomic instability at endogenous G4 DNA sites. Nat Commun. 2014;5 doi: 10.1038/ncomms4216. 3216. [DOI] [PubMed] [Google Scholar]
- 7.Chang HHY, Pannunzio NR, Adachi N, Lieber MR. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat Rev Mol Cell Biol. 2017;18(8):495–506. doi: 10.1038/nrm.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Venter JC. The Sequence of the Human Genome. Science. 2001;291(5507):1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 9.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 10.Lambert S, Watson A, Sheedy DM, Martin B, Carr AM. Gross Chromosomal Rearrangements and Elevated Recombination at an Inducible Site-Specific Replication Fork Barrier. Cell. 2005;121(5):689–702. doi: 10.1016/j.cell.2005.03.022. [DOI] [PubMed] [Google Scholar]
- 11.Wolters S, Ermolaeva MA, Bickel JS, et al. Loss of Caenorhabditis elegans BRCA1 promotes genome stability during replication in smc-5 mutants. Genetics. 2014;196(4):985–999. doi: 10.1534/genetics.113.158295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hustedt N, Durocher D. The control of DNA repair by the cell cycle. Nat Cell Biol. 2016;19(1):1–9. doi: 10.1038/ncb3452. [DOI] [PubMed] [Google Scholar]
- 13.Torgovnick A, Schumacher B. DNA repair mechanisms in cancer development and therapy. Front Genet. 2015;6:157. doi: 10.3389/fgene.2015.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schoonen PM, Talens F, Stok C, et al. Progression through mitosis promotes PARP inhibitor-induced cytotoxicity in homologous recombination-deficient cancer cells. Nat Commun. 2017;8:1–13. doi: 10.1038/ncomms15981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukui K. DNA Mismatch Repair in Eukaryotes and Bacteria. J Nucleic Acids. 2010;2010(32):1–16. doi: 10.4061/2010/260512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sale JE, Lehmann AR, Woodgate R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat Rev Mol Cell Biol. 2012;13(3):141–152. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lane D, Levine A. p53 Research: The Past Thirty Years and the Next Thirty Years. Cold Spring Harbor Perspectives in Biology. 2010;2(12):a000893–a000893. doi: 10.1101/cshperspect.a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haluska FG, Finver S, Tsujimoto Y, Croce CM. The t(8; 14) chromosomal translocation occurring in B-cell malignancies results from mistakes in V-D-J joining. Nature. 1986;324(6093):158–161. doi: 10.1038/324158a0. [DOI] [PubMed] [Google Scholar]
- 19.Canela A, Maman Y, Jung S, et al. Genome Organization Drives Chromosome Fragility. Cell. 2017;170(3):507–511.e18. doi: 10.1016/j.cell.2017.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartek J, Lukas J. DNA damage checkpoints: from initiation to recovery or adaptation. Curr Opin Cell Biol. 2007;19(2):238–245. doi: 10.1016/j.ceb.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 21.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408(6810):307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 22.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69(7):1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 23.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Letters. 1997;420(1):25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 24.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer. 2003;3(3):155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 25.Ou Y-H, Chung P-H, Sun T-P, Shieh S-Y. p53 C-terminal phosphorylation by CHK1 and CHK2 participates in the regulation of DNA-damage-induced C-terminal acetylation. Mol Biol Cell. 2005;16(4):1684–1695. doi: 10.1091/mbc.E04-08-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mirzayans R, Andrais B, Scott A, Murray D. New insights into p53 signaling and cancer cell response to DNA damage: implications for cancer therapy. J Biomed Biotechnol. 2012;2012:170325. doi: 10.1155/2012/170325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reed SM, Quelle DE. p53 Acetylation: Regulation and Consequences. Cancers (Basel) 2014;7(1):30–69. doi: 10.3390/cancers7010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reinhardt HC, Schumacher B. The p53 network: cellular and systemic DNA damage responses in aging and cancer. Trends Genet. 2012;28(3):128–136. doi: 10.1016/j.tig.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Derry WB, Putzke AP, Rothman JH. Caenorhabditis elegans p53: role in apoptosis, meiosis, and stress resistance. Science. 2001;294(5542):591–595. doi: 10.1126/science.1065486. [DOI] [PubMed] [Google Scholar]
- 30.Schumacher B, Hofmann K, Boulton S, Gartner A. The C. elegans homolog of the p53 tumor suppressor is required for DNA damage-induced apoptosis. Curr Biol. 2001;11(21):1722–1727. doi: 10.1016/s0960-9822(01)00534-6. [DOI] [PubMed] [Google Scholar]
- 31.Schumacher B, Hanazawa M, Lee M-H, et al. Translational repression of C. elegans p53 by GLD-1 regulates DNA damage-induced apoptosis. Cell. 2005;120(3):357–368. doi: 10.1016/j.cell.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 32.Fernández-Majada V, Welz P-S, Ermolaeva MA, et al. The tumour suppressor CYLD regulates the p53 DNA damage response. Nat Commun. 2016;7 doi: 10.1038/ncomms12508. 12508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schumacher B, Schertel C, Wittenburg N, et al. C. elegans ced-13 can promote apoptosis and is induced in response to DNA damage. Cell Death Differ. 2005;12(2):153–161. doi: 10.1038/sj.cdd.4401539. [DOI] [PubMed] [Google Scholar]
- 34.Hofmann ER, Milstein S, Boulton SJ, et al. Caenorhabditis elegans HUS-1 is a DNA damage checkpoint protein required for genome stability and EGL-1-mediated apoptosis. Curr Biol. 2002;12(22):1908–1918. doi: 10.1016/s0960-9822(02)01262-9. [DOI] [PubMed] [Google Scholar]
- 35.Greiss S, Schumacher B, Grandien K, Rothblatt J, Gartner A. Transcriptional profiling in C. elegans suggests DNA damage dependent apoptosis as an ancient function of the p53 family. BMC Genomics. 2008;9:334. doi: 10.1186/1471-2164-9-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hofmann ER, Milstein S, Hengartner MO. DNA-damage-induced checkpoint pathways in the nematode Caenorhabditis elegans. Cold Spring Harb Symp Quant Biol. 2000;65:467–73. doi: 10.1101/sqb.2000.65.467. [DOI] [PubMed] [Google Scholar]
- 37.Shibue T, Takeda K, Oda E, et al. Integral role of Noxa in p53-mediated apoptotic response. Genes Dev. 2003;17(18):2233–2238. doi: 10.1101/gad.1103603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell. 2001;7(3):683–694. doi: 10.1016/s1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- 39.Deiry el WS, Tokino T, Velculescu VE, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75(4):817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 40.Williams AB, Schumacher B. p53 in the DNA-Damage-Repair Process. Cold Spring Harb Perspect Med. 2016;6(5) doi: 10.1101/cshperspect.a026070. a026070–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adimoolam S, Ford JM. p53 and DNA damage-inducible expression of the xeroderma pigmentosum group C gene. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(20):12985–12990. doi: 10.1073/pnas.202485699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hwang BJ, Ford JM, Hanawalt PC, Chu G. Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(2):424–428. doi: 10.1073/pnas.96.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scherer SJ, Maier SM, Seifert M, Hanselmann RG, Zang KD, Muller-Hermelink HK, Angel P, Welter C, Schartl M. p53 and c-Jun functionally synergize in the regulation of the DNA repair gene hMSH2 in response to UV. J Biol Chem. 2000;275(48):37469–37473. doi: 10.1074/jbc.M006990200. [DOI] [PubMed] [Google Scholar]
- 44.Scherer SJ, Welter C, Zang KD, Dooley S. Specific in vitro binding of p53 to the promoter region of the human mismatch repair gene hMSH2. Biochem Biophys Res Commun. 1996;221(3):722–728. doi: 10.1006/bbrc.1996.0663. [DOI] [PubMed] [Google Scholar]
- 45.Liebetrau W, Budde A, Savoia A, Grummt F, Hoehn H. p53 activates Fanconi anemia group C gene expression. Hum Mol Genet. 1997;6(2):277–283. doi: 10.1093/hmg/6.2.277. [DOI] [PubMed] [Google Scholar]
- 46.Achanta G, Huang P. Role of p53 in sensing oxidative DNA damage in response to reactive oxygen species-generating agents. Cancer Res. 2004;64(17):6233–6239. doi: 10.1158/0008-5472.CAN-04-0494. [DOI] [PubMed] [Google Scholar]
- 47.Rashid Al ST, Dellaire G, Cuddihy A, et al. Evidence for the direct binding of phosphorylated p53 to sites of DNA breaks in vivo. Cancer Research. 2005;65(23):10810–10821. doi: 10.1158/0008-5472.CAN-05-0729. [DOI] [PubMed] [Google Scholar]
- 48.Tanaka H, Arakawa H, Yamaguchi T, et al. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404(6773):42–49. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- 49.Johmura Y, Nakanishi M. Multiple facets of p53 in senescence induction and maintenance. Cancer Sci. 2016;107(11):1550–1555. doi: 10.1111/cas.13060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Behrens A, van Deursen JM, Rudolph KL, Schumacher B. Impact of genomic damage and ageing on stem cell function. Nat Cell Biol. 2014;16(3):201–207. doi: 10.1038/ncb2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rodier F, Coppe JP, Patil CK, et al. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat Cell Biol. 2009;11(8):973–979. doi: 10.1038/ncb1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baker DJ, Jeganathan KB, Cameron JD, et al. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet. 2004;36(7):744–749. doi: 10.1038/ng1382. [DOI] [PubMed] [Google Scholar]
- 53.Baker DJ, Wijshake T, Tchkonia T, et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479(7372):232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Childs BG, Durik M, Wijers ME, et al. Naturally occurring p16Ink4a-positive cells shorten healthy lifespan. Nature. 2016:1–20. doi: 10.1038/nature16932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chang J, Wang Y, Shao L, et al. Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nature Medicine. 2016;22(1):78–83. doi: 10.1038/nm.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Demaria M, Ohtani N, Youssef SA, et al. An Essential Role for Senescent Cells in Optimal Wound Healing through Secretion of PDGF-AA. Developmental Cell. 2014;31(6):722–733. doi: 10.1016/j.devcel.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tyner SD, Venkatachalam S, Choi J, et al. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415(6867):45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- 58.Maier B, Gluba W, Bernier B, et al. Modulation of mammalian life span by the short isoform of p53. Genes Dev. 2004;18(3):306–319. doi: 10.1101/gad.1162404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dumble M, Moore L, Chambers SM, et al. The impact of altered p53 dosage on hematopoietic stem cell dynamics during aging. Blood. 2007;109(4):1736–1742. doi: 10.1182/blood-2006-03-010413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Garcia-Cao I, Garcia-Cao M, Martin-Caballero J, et al. “Super p53” mice exhibit enhanced DNA damage response, are tumor resistant and age normally. EMBO J. 2002;21(22):6225–6235. doi: 10.1093/emboj/cdf595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Matheu A, Maraver A, Klatt P, et al. Delayed ageing through damage protection by the Arf/p53 pathway. Nature. 2007;448(7151):375–379. doi: 10.1038/nature05949. [DOI] [PubMed] [Google Scholar]
- 62.Mendrysa SM, O'Leary KA, McElwee MK, et al. Tumor suppression and normal aging in mice with constitutively high p53 activity. Genes Dev. 2006;20(1):16–21. doi: 10.1101/gad.1378506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matlashewski GJ, Tuck S, Pim D, et al. Primary structure polymorphism at amino acid residue 72 of human p53. Mol Cell Biol. 1987;7(2):961–963. doi: 10.1128/mcb.7.2.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dumont P, Leu JI-J, Pietra Della AC, George DL, Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet. 2003;33(3):357–365. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- 65.van Heemst D, Mooijaart SP, Beekman M, et al. Variation in the human TP53 gene affects old age survival and cancer mortality. Exp Gerontol. 2005;40(1–2):11–15. doi: 10.1016/j.exger.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 66.Ørsted DD, Bojesen SE, Tybjaerg-Hansen A, Nordestgaard BG. Tumor suppressor p53 Arg72Pro polymorphism and longevity, cancer survival, and risk of cancer in the general population. J Exp Med. 2007;204(6):1295–1301. doi: 10.1084/jem.20062476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kodal JB, Vedel-Krogh S, Kobylecki CJ, Nordestgaard BG, Bojesen SE. TP53 Arg72Pro, mortality after cancer, and all-cause mortality in 105,200 individuals. Sci Rep. 2017;7(1):336. doi: 10.1038/s41598-017-00427-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abdallah P, Luciano P, Runge KW, et al. A two-step model for senescence triggered by a single critically short telomere. Nat Cell Biol. 2009;11(8):988–993. doi: 10.1038/ncb1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.d'Adda di Fagagna F, Reaper PM, Clay-Farrace L, et al. A DNA damage checkpoint response in telomere-initiated senescence. Nature Publishing Group. 2003;426(6963):194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 70.Von Zglinicki T, Saretzki G, Ladhoff J, d'Adda di Fagagna F, Jackson SP. Human cell senescence as a DNA damage response. Mech Ageing Dev. 2005;126(1):111–117. doi: 10.1016/j.mad.2004.09.034. [DOI] [PubMed] [Google Scholar]
- 71.Blasco MA, Lee HW, Hande MP, et al. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91(1):25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 72.Rudolph KL, Chang S, Lee HW, et al. Longevity, stress response, and cancer in aging telomerase-deficient mice. Cell. 1999;96(5):701–712. doi: 10.1016/s0092-8674(00)80580-2. [DOI] [PubMed] [Google Scholar]
- 73.Vogel H, Lim DS, Karsenty G, Finegold M, Hasty P. Deletion of Ku86 causes early onset of senescence in mice. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(19):10770–10775. doi: 10.1073/pnas.96.19.10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chin L, Artandi SE, Shen Q, et al. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell. 1999;97(4):527–538. doi: 10.1016/s0092-8674(00)80762-x. [DOI] [PubMed] [Google Scholar]
- 75.Lim DS, Vogel H, Willerford DM, et al. Analysis of ku80-mutant mice and cells with deficient levels of p53. Mol Cell Biol. 2000;20(11):3772–3780. doi: 10.1128/mcb.20.11.3772-3780.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.de Winter JP, Joenje H. The genetic and molecular basis of Fanconi anemia. Mutat Res. 2009;668(1–2):11–19. doi: 10.1016/j.mrfmmm.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 77.Duxin JP, Walter JC. What is the DNA repair defect underlying Fanconi anemia? Curr Opin Cell Biol. 2015;37:49–60. doi: 10.1016/j.ceb.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Joenje H, Arwert F, Eriksson AW, de Koning H, Oostra AB. Oxygen-dependence of chromosomal aberrations in Fanconi's anaemia. Nature. 1981;290(5802):142–143. doi: 10.1038/290142a0. [DOI] [PubMed] [Google Scholar]
- 79.Pagano G, Talamanca AA, Castello G, et al. Oxidative stress in Fanconi anaemia: from cells and molecules towards prospects in clinical management. Biol Chem. 2012;393(1–2):11–21. doi: 10.1515/BC-2011-227. [DOI] [PubMed] [Google Scholar]
- 80.Ceccaldi R, Parmar K, Mouly E, et al. Bone marrow failure in Fanconi anemia is triggered by an exacerbated p53/p21 DNA damage response that impairs hematopoietic stem and progenitor cells. Cell Stem Cell. 2012;11(1):36–49. doi: 10.1016/j.stem.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Abbas HA, Maccio DR, Coskun S, et al. Mdm2 is required for survival of hematopoietic stem cells/progenitors via dampening of ROS-induced p53 activity. Cell Stem Cell. 2010;7(5):606–617. doi: 10.1016/j.stem.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chambers SM, Shaw CA, Gatza C, et al. Aging hematopoietic stem cells decline in function and exhibit epigenetic dysregulation. PLoS Biol. 2007;5(8):e201. doi: 10.1371/journal.pbio.0050201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rübe CE, Fricke A, Widmann TA, et al. Accumulation of DNA damage in hematopoietic stem and progenitor cells during human aging. PLoS ONE. 2011;6(3):e17487. doi: 10.1371/journal.pone.0017487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nijnik A, Woodbine L, Marchetti C, et al. DNA repair is limiting for haematopoietic stem cells during ageing. Nature. 2007;447(7145):686–690. doi: 10.1038/nature05875. [DOI] [PubMed] [Google Scholar]
- 85.Rossi DJ, Bryder D, Seita J, et al. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature. 2007;447(7145):725–729. doi: 10.1038/nature05862. [DOI] [PubMed] [Google Scholar]
- 86.Yahata T, Takanashi T, Muguruma Y, et al. Accumulation of oxidative DNA damage restricts the self-renewal capacity of human hematopoietic stem cells. Blood. 2011;118(11):2941–2950. doi: 10.1182/blood-2011-01-330050. [DOI] [PubMed] [Google Scholar]
- 87.Ribezzo F, Shiloh Y, Schumacher B. Systemic DNA damage responses in aging and diseases. Semin Cancer Biol. 2016;37–38:26–35. doi: 10.1016/j.semcancer.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jans J, Schul W, Sert YG, et al. Powerful skin cancer protection by a CPD-photolyase transgene. Curr Biol. 2005;15(2):105–115. doi: 10.1016/j.cub.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 89.Cleaver JE, Lam ET, Revet I. Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nat Rev Genet. 2009;10(11):756–768. doi: 10.1038/nrg2663. [DOI] [PubMed] [Google Scholar]
- 90.DiGiovanna JJ, Kraemer KH. Shining a light on xeroderma pigmentosum. J Invest Dermatol. 2012;132(3 Pt 2):785–796. doi: 10.1038/jid.2011.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cleaver JE, Bezrookove V, Revet I, Huang EJ. Conceptual developments in the causes of Cockayne syndrome. Mech Ageing Dev. 2013;134(5–6):284–290. doi: 10.1016/j.mad.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 92.Karikkineth AC, Scheibye-Knudsen M, Fivenson E, Croteau DL, Bohr VA. Cockayne syndrome: Clinical features, model systems and pathways. Ageing Res Rev. 2017;33:3–17. doi: 10.1016/j.arr.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kraemer KH, Patronas NJ, Schiffmann R, et al. Xeroderma pigmentosum, trichothiodystrophy and Cockayne syndrome: a complex genotype-phenotype relationship. Neuroscience. 2007;145(4):1388–1396. doi: 10.1016/j.neuroscience.2006.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Garinis GA, Uittenboogaard LM, Stachelscheid H, et al. Persistent transcription-blocking DNA lesions trigger somatic growth attenuation associated with longevity. Nat Cell Biol. 2009;11(5):604–615. doi: 10.1038/ncb1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bonkowski MS, Pamenter RW, Rocha JS, et al. Long-lived growth hormone receptor knockout mice show a delay in age-related changes of body composition and bone characteristics. J Gerontol A Biol Sci Med Sci. 2006;61(6):562–567. doi: 10.1093/gerona/61.6.562. [DOI] [PubMed] [Google Scholar]
- 96.Zhou Y, Xu BC, Maheshwari HG, et al. A mammalian model for Laron syndrome produced by targeted disruption of the mouse growth hormone receptor/binding protein gene (the Laron mouse) Proc Natl Acad Sci USA. 1997;94(24):13215–13220. doi: 10.1073/pnas.94.24.13215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liang H, Masoro EJ, Nelson JF, et al. Genetic mouse models of extended lifespan. Exp Gerontol. 2003;38(11–12):1353–1364. doi: 10.1016/j.exger.2003.10.019. [DOI] [PubMed] [Google Scholar]
- 98.Niedernhofer LJ, Garinis GA, Raams A, et al. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 2006;444(7122):1038–1043. doi: 10.1038/nature05456. [DOI] [PubMed] [Google Scholar]
- 99.van de Ven M, Andressoo JO, Holcomb VB, et al. Adaptive Stress Response in Segmental Progeria Resembles Long-Lived Dwarfism and Calorie Restriction in Mice. PLoS Genet. 2006;2(12):e192. doi: 10.1371/journal.pgen.0020192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.van der Pluijm I, Garinis GA, Brandt RM, et al. Impaired genome maintenance suppresses the growth hormone--insulin-like growth factor 1 axis in mice with Cockayne syndrome. PLoS Biol. 2006;5(1):e2. doi: 10.1371/journal.pbio.0050002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Schumacher B, Garinis GA, Hoeijmakers JHJ. Age to survive: DNA damage and aging. Trends Genet. 2008;24(2):77–85. doi: 10.1016/j.tig.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 102.Garinis GA, Uittenboogaard LM, Stachelscheid H, et al. Persistent transcription-blocking DNA lesions trigger somatic growth attenuation associated with longevity. Nat Cell Biol. 2009;11(5):604–615. doi: 10.1038/ncb1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Holzenberger M, Dupont J, Ducos B, et al. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. 2003;421(6919):182–187. doi: 10.1038/nature01298. [DOI] [PubMed] [Google Scholar]
- 104.Mueller MM, Castells-Roca L, Babu V, et al. DAF-16/FOXO and EGL-27/GATA promote developmental growth in response to persistent somatic DNA damage. Nat Cell Biol. 2014;16(12):1168–1179. doi: 10.1038/ncb3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xue W, Zender L, Miething C, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445(7128):656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ermolaeva MA, Segref A, Dakhovnik A, et al. DNA damage in germ cells induces an innate immune response that triggers systemic stress resistance. Nature. 2013;501(7467):416–420. doi: 10.1038/nature12452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fang EF, Scheibye-Knudsen M, Brace LE, et al. Defective Mitophagy in XPA via PARP-1 Hyperactivation and NAD. Cell. 2014;157(4):882–896. doi: 10.1016/j.cell.2014.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fang EF, Kassahun H, Croteau DL, et al. NAD+ Replenishment Improves Lifespan and Healthspan in Ataxia Telangiectasia Models via Mitophagy and DNA Repair. Cell Metabolism. 2016;24(4):566–581. doi: 10.1016/j.cmet.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mouchiroud L, Houtkooper RH, Moullan N, et al. The NAD(+)/Sirtuin Pathway Modulates Longevity through Activation of Mitochondrial UPR and FOXO Signaling. Cell. 2013;154(2):430–441. doi: 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li J, Bonkowski MS, Moniot S, et al. A conserved NAD(+) binding pocket that regulates protein-protein interactions during aging. Science. 2017;355(6331):1312–1317. doi: 10.1126/science.aad8242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Edifizi D, Nolte H, Babu V, et al. Multilayered Reprogramming in Response to Persistent DNA Damage in C. elegans. Cell Reports. 2017;20(9):2026–2043. doi: 10.1016/j.celrep.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]