Abstract
A potential mechanism for the global distribution of waterborne pathogens is through carriage by the migratory waterbirds. However, this mode of transmission has yet been confirmed epidemiologically. Here, we conducted whole genome sequencing of Vibrio spp. collected from waterbirds, sediments, and mollusks in the estuary of the Liaohe River in China to investigate this transmission mode. We found that a V. parahaemolyticus strain isolated from a waterbird was clonally related to the other V. parahaemolyticus strains obtained from the sediments and mollusks, and three V. mimicus strains isolated from bird feces were genomically related to those found in the mollusks and upstream groundwater, suggesting that the bird-carried Vibrio strains were acquired through the direct predation of the local mollusks. Surprisingly, two bird-carried V. parahaemolyticus strains belonging to the same clone were identified in Panjin and Shanghai, which are over 1,150 km apart, and another two were found at two locations 50 km apart, further supporting that waterbirds are capable of carrying and disseminating these pathogens over long distances. Our results provide the first evidence of direct transmission from mollusks to waterbirds and confirm that waterbirds act as disseminating vehicles of waterborne pathogens. Effective surveillance of migratory waterbirds along their routes will be valuable for predicting future epidemics of infectious diseases.
Subject terms: Phylogenetics, Water microbiology
Introduction
Vibrio parahaemolyticus is a Gram-negative comma-shaped bacterium and a leading cause of seafood-related gastroenteritis1. Infection by V. parahaemolyticus is often associated with the consumption of raw or undercooked seafood2. In the USA alone, V. parahaemolyticus is estimated to cause approximately 35,000 human illnesses each year3. V. parahaemolyticus serogroup O3:K6 has been recognized to cause pandemic-level disease and has been intensively investigated4. The serogroup emerged in India in 1996 and was rapidly spread to other Asian countries and other continents4. In South America, O3:K6 was first reported in Peru in 19965 and then rapidly spread to Chile in 1997 and the USA in 1998, to Brazil in 2001 and to Mexico in 20046.
V. parahaemolyticus and other Vibrio species have been found widely in various environments, including estuaries and rivers. Extensive studies have identified the natural reservoirs of V. parahaemolyticus and other Vibrio species7,8. Invertebrates such as chironomids, copepods, and oysters are suggested to be the natural reservoirs for the Vibrio species. Evidence of the transmission of Vibrio species from fish to great cormorants has also recently been published9.
As early as the 1970s, pathogenic Vibrio species were isolated from the faces of various waterbird species, including Ardea herodias (great blue heron) and Larus delawarensis (ridge-billed gull)10. However, the public health significance of waterbird-carried Vibrio species has not attracted the deserved attention of the scientific community, and the avenue through which the waterbirds acquire the pathogens remains poorly understood. Laviad-Shitrit et al.11 found that both Vibrio species and Aeromonas species were abundant in many types of birds in Israel, suggesting that migratory waterbirds are potential disseminators of waterborne pathogens. However, the roles that southward autumn and northward spring bird migrations might play in the transmission of waterborne pathogens and the evolutionary origins of these pathogens have rarely been examined. There have been no molecular epidemiological studies of the origins of waterbird-carried Vibrio species. Whether migratory birds mediate the spread of these pathogens remains largely unknown.
Yingkou and Panjin are located along the estuary of the Liaohe River in North China (Fig. 1). In 2017, we isolated several multidrug-resistant (MDR) Vibrio species from fish farms in the upstream reaches of the Liaohe River (Fig. 1). To evaluate the impact of the aquacultural activities on the downstream river, we sampled sediments, mollusks, and waterbird feces in Panjin and Yingkou. Unexpectedly, abundant V. parahaemolyticus and other pathogens were identified in the waterbird feces and mollusks. In this study, we investigated the evolutionary origins of the Vibrio species carried by waterbirds using whole-genome sequencing.
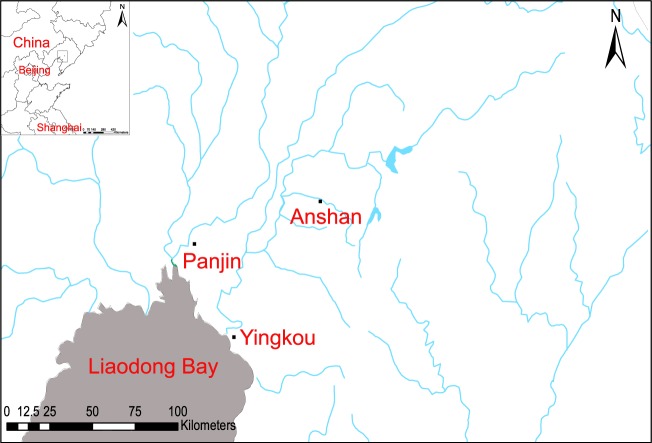
Figure 1.
Sampling sites in this study. The sampling positions in Anshan, Panjin, and Yingkou are indicated in the square. The sampling sites were mapped by the ArcGIS Desktop 10.2 software (http://desktop.arcgis.com/).
Results
Epidemiological investigation and identification of waterborne bacterial pathogens on fish farms
In August 2017, massive outbreaks of ornamental fish disease occurred on several fish farms in Anshan. An epidemiological investigation showed that all the sampled fish farms bred their own brood fish, without importing of parent fish from other sources, which excluded the possible input of pathogens from external sources. A total of 60 fish feed samples from three fish farms were analyzed microbiologically but no Vibrio sp. was isolated. Groundwater was the only source of the water used for aquaculture. The fish farmers pumped the groundwater into their aquaculture systems without any treatment or disinfection because groundwater is usually considered to be of high quality suitable for aquaculture. Three Vibrio mimicus strains were isolated from fish and groundwater in a fish farm (Table 1). A follow-up investigation showed that several tons of wastewater from fish ponds were discharged into the river after the clean-up of the outbreak-related ponds, which might have resulted in the transmission of the pathogens from Anshan into the estuary downstream. Antibiotics, sulfamethoxazole, florfenicol, fluoroquinolone, sulfonamide, and tetracycline were used widely for the treatment of fish diseases in these farms. Concerns were raised on antibiotic resistance development as the outbreaks occurred despite the use of various antibiotics to treat the infections.
Table 1.
General features of strains isolated in this study.
| Sampling site | Sources | Species | Strain Name | Isolation date | Sequence type |
|---|---|---|---|---|---|
| Anshan | Fish | V. mimicus | VM14 | Oct-17 | / |
| Underground water | V. mimicus | VM34 | Mar-18 | / | |
| Underground water | V. mimicus | VM61 | Jun-18 | / | |
| Yingkou | Mollusk | V. parahaemolyticus | YK38 | Oct-17 | ST1823 |
| Sediment | V. parahaemolyticus | YK39 | Oct-17 | ST1823 | |
| Sediment | V. parahaemolyticus | YK40 | Oct-17 | ST1823 | |
| Sediment | V. parahaemolyticus | YK13 | Oct-17 | ST2006 | |
| Sediment | V. parahaemolyticus | YK17 | Oct-17 | ST2007 | |
| Mollusk | V. parahaemolyticus | YK41 | Oct-17 | ST1823 | |
| Mollusk | V. parahaemolyticus | YK32 | Oct-17 | ST236 | |
| Mollusk | V. parahaemolyticus | YK34 | Oct-17 | ST1498 | |
| Mollusk | V. mimicus | VM20 | Mar-18 | / | |
| Mollusk | V. parahaemolyticus | YK51 | Jun-18 | ST415 | |
| Mollusk | V. parahaemolyticus | YK45 | May-18 | ST1823 | |
| Bird feces (Common Greenshank) | V. parahaemolyticus | YK33 | Oct-17 | ST2008 | |
| Bird feces (Common Greenshank) | V. scophthalmi | YK47 | Oct-17 | / | |
| Bird feces (Eurasian Curlew) | V. parahaemolyticus | YK49 | Oct-17 | ST1823 | |
| Bird feces (Black-tailed Godwit) | V. mimicus | VM27 | Apr-18 | / | |
| Bird feces (Black-tailed Godwit) | V. mimicus | VM37 | Apr-18 | / | |
| Bird feces (Black-tailed Godwit) | V. mimicus | VM41 | Apr-18 | / | |
| Panjin | Estuary | V. parahaemolyticus | PJ42 | Oct-17 | ST1823 |
| Estuary | V. parahaemolyticus | PJ43 | Oct-17 | ST1823 | |
| Sediment | V. parahaemolyticus | PJ44 | Oct-17 | ST1823 | |
| Bird feces (Common Greenshank) | V. parahaemolyticus | PJ37 | Oct-17 | ST2008 | |
| Bird feces (Common Greenshank) | V. parahaemolyticus | PJ18 | Oct-17 | ST415 | |
| Bird feces (Common Greenshank) | V. parahaemolyticus | PJ35 | Oct-17 | ST3 | |
| Shanghai | Bird feces (Common Greenshank) | V. parahaemolyticus | SH50 | Oct-18 | ST415 |
Isolation of vibrio species in the estuary of liaohe river
To investigate the impact of wastewater discharge on the river downstream, especially on a migratory waterbird sanctuary, we conducted a field investigation of the downstream river delta region. A total of 180 bird fecal samples were collected in the cities of Shanghai, Yingkou and Panjin (Table S1). Mollusk and sediment samples were also collected from the intertidal zones at Yingkou and Panjin. The monitoring of the environmental variables in Anshan, Yingkou and Panjin showed that seawater in Yingkou had highest concentration of dissolved organic carbon (22.3 mg L−1), total phosphorus (67 μg L−1), and nitrite (0.45 mg L−1) (Table S2).
In total, 27 strains of Vibrio species were isolated in this study, 70.4% of which were V. parahaemolyticus. In Yingkou, 10 V. parahaemolyticus strains and one V. mimicus strain were isolated from the sediment and mollusk samples, whereas two strains of V. parahaemolyticus were recovered from bird feces. In addition, three V. mimicus strains and one V. scophthalmi strain were also isolated from bird feces.
In Panjin, three V. parahaemolyticus strains were isolated from bird feces, and another three were isolated from sediments or estuary seawater. In Shanghai, one V. parahaemolyticus strain was isolated out of 25 bird feces sampled.
Phylogenetic and evolutionary analysis of V. parahaemolyticus from bird feces
To determine the origins of V. parahaemolyticus in bird feces, we sequenced the 19 V. parahaemolyticus strains (Table S3). In silico multilocus sequence typing (MLST) of the V. parahaemolyticus strains identified eight sequence types (STs) among the 19 strains. Nine strains isolated from mollusks, estuary and sediments from Panjin and Yingkou were all assigned to ST1823, whereas the strains PJ18, YK51, PJ35, YK32, YK34, YK13, and YK17were identified as ST415, ST415, ST3, ST236, ST1498, ST2006 and ST2007, respectively (Table 1). YK33 and PJ37 both belonged to ST2008. In addition, the SH50 strain obtained from Shanghai was also identified as ST415.
All except three new STs were found in the PubMLST database with various frequencies. ST3 is a recognized pandemic ST with the highest frequency in the database (249/2887, 8.63%). ST1823, ST415, ST236 and ST1498 were presented in the database with much lower frequencies of 0.48%, 1.5%, 0.07% and 0.1%, respectively (Table S4).
We further selected the publicly available genomes for phylogenetic and evolutionary analyses based on two criteria. First, the genomes of the strains phylogenetically close to the sequenced strains on MLST typing were selected. Second, the strains that occurred geographically close to the sampling sites or at sites overlapping bird migration routes were selected for comparison.
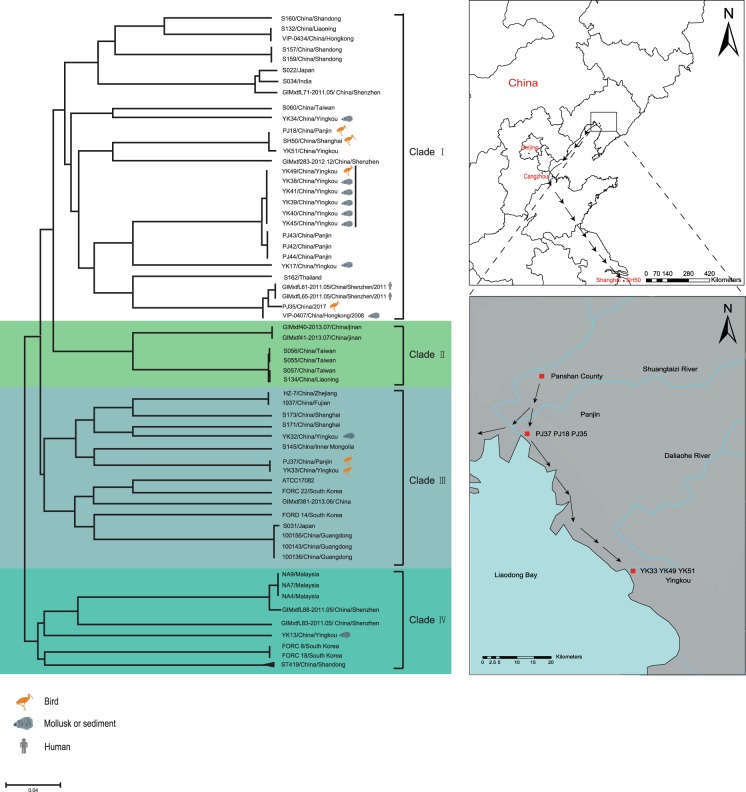
Finally, we incorporated 47 publicly available genomes for the phylogenetic analysis to examine the origins of our strains (Table S5). The recombinant SNPs were removed by the recombination detection program (RDP) as described in the supplemental file (Table S6, Appendix 1). Only non-recombinant core genome SNPs were used for phylogenetic inference. The ST1823 genome tree is shown in Fig. 2. The nine ST1823 strains from Yingkou and Panjin formed two separate clusters differing by 77 SNPs (Fig. S1), indicating that they belonged to two different lineages but might share a most recent common ancestor circulating in the river delta region. Notably, strain YK49, which was isolated from a bird (Eurasian curlew), differed by two SNPs from the other ST1823 strains isolated from mollusks or sediments in Yingkou (Fig. S1). Strikingly, two bird-carried ST415 strains, PJ18 and SH50 (differing by ten SNPs), were found in Panjin and Shanghai (over 1100 km apart), respectively. These two strains differ by 1,129 SNPs from another ST415 strain, YK51, which was obtained from the mollusks in Yingkou. In addition, two bird-carried strains, YK33 and PJ37, differing by two SNPs, were also isolated from the same species of waterbirds (common greenshank) in two different locations 50 km apart, which intersect the migration route of the common greenshank (Fig. 2). Other mollusk-derived strains were dispersed throughout the genome tree (Fig. 2).
Figure 2.
Geophylogeny of V. parahaemolyticus genomes and the migration route of the common greenshank from Panjin to Yingkou in autumn. left: maximum likelihood phylogenies of 66 V. parahaemolyticus genomes. Bootstrapping was performed with 1,000 replicates. Right: The isolation sites of bird-carried V. parahaemolyticus. The sampling sites were mapped by the ArcGIS Desktop 10.2 software (http://desktop.arcgis.com/). Precise migration route of the common greenshank from Panjin to Yingkou in autumn 2017 is indicated in arrows.
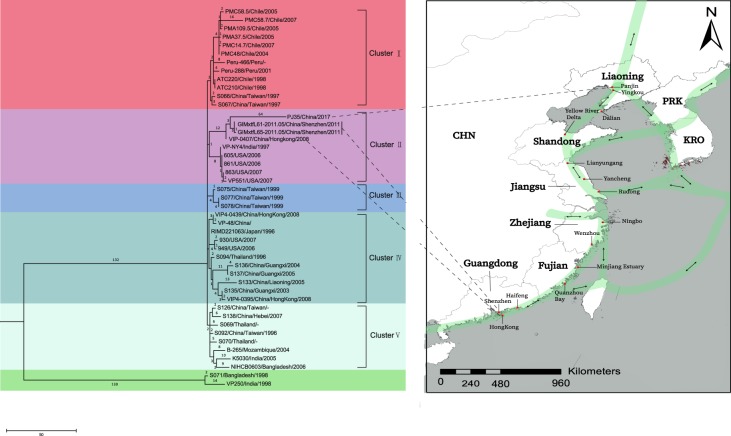
For the sole ST3 strain, PJ35, isolated from a waterbird in Panjin, we included 44 other ST3 genomes in the analysis (Fig. 3). The phylogenetic tree showed that PJ35 was genomically most closely related to the two Shenzhen strains and one Hong Kong strain with 64 strain-specific SNPs.
Figure 3.
The divergence of 44 ST3 V. parahaemolyticus genomes. Left: maximum parsimony tree of 44 ST3 V. parahaemolyticus genomes. Homoplasy index (HI) was 0.0 for the maximum parsimony tree. The numbers above the branches indicate the numbers of SNPs. Right: migration route of the common greenshank, with stopovers at Shenzhen, Haifeng, Quanzhou Bay, Minjiang Estuary, Rudong, Yancheng, Lianyungang, Yellow River Delta, Yingkou, and Panjin. The sampling sites were mapped by the ArcGIS Desktop 10.2 software (http://desktop.arcgis.com/).
Origins of V. mimicus from waterbird feces
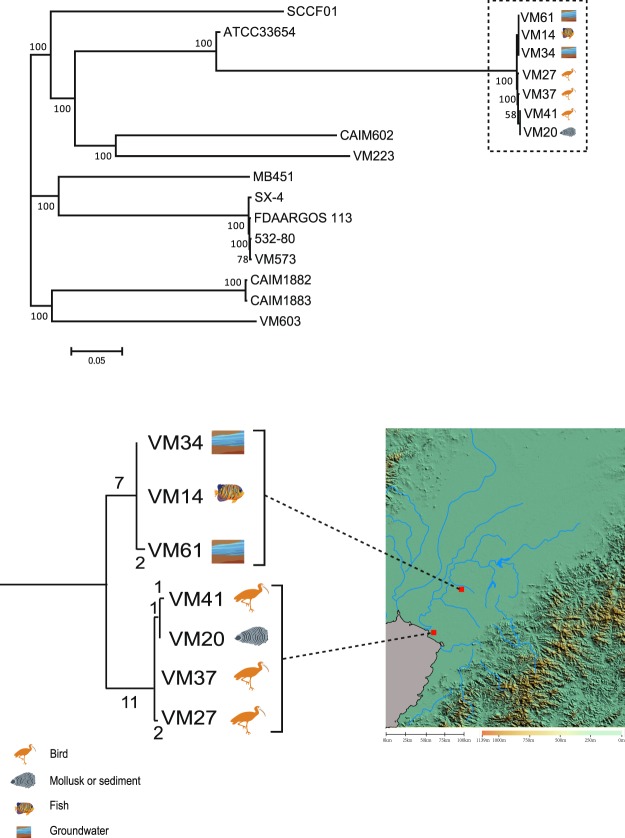
We next determined the phylogenetic relationships of the seven V. mimicus strains from this study and 12 V. mimicus sequences from the publicly available genomes (Table S7). The three bird-carried V. Mimicus strains clustered with those isolated from sediments or groundwater from the same area (Fig. 4).
Figure 4.
Phylogenetic relationships of V. mimicus strains. Top: Maximum-likelihood phylogenies of 24 V. mimicus strains. The bootstrap was performed with 1000 replicates. The unit of the scale bar indicates the evolutionary distance in substitutions per nucleotide. Down: Geophylogeny of seven sequenced V. mimicus strains. The numbers above the branches indicate the numbers of SNPs. Locations of the strains obtained from Anshan and Yingkou are indicated on the right side. The satellite imagery of the investigated region was created by Global Mapper v20 (http://www.bluemarblegeo.com/products/global-mapper.php).
The four Yingkou and the three Anshan strains were closely related, differing by only up to 18 SNPs, indicating that they might share a most recent common ancestor (Fig. 4). The strains VM14, VM34, and VM61 (from Anshan) belonged to the same cluster which was repeatedly isolated from underground water, with seven branch-specific SNPs. The strains VM20, VM27, VM37, and VM41 (from Yingkou) formed another cluster with 11 branch-specific SNPs.
Antibiotic resistance genes (ARGs) in the sequenced strains
Genomic analysis confirmed that most of the V. parahaemolyticus strains carried blaCARB-47 (except for YK13 with a blaCARB-48 gene). A few of the sequenced V. parahaemolyticus strains carried additional ARGs, such as dfrA31 and qnrC. Seven sequenced V. mimicus strains from birds and underground water all carried the resistance genes aph(3″)-Ib, aph(6)-Id, qnrVC4, floR, sul2, tet(59), and dfrA6 on their chromosomes (Table S8).
Modeling the dynamics of the V. parahaemolyticus-carrying birds
We used the intertidal zone of the estuary of the Liaohe River in Yingkou as the site for modelling the dynamics of the V. parahaemolyticus-carrying birds.
To set the modeling parameters, the numbers of common greenshank that stopped over in the investigated region were counted every week. To estimate the sizes of the prey population(mollusks), the dynamics of mollusk density in the sediment were determined with the method described by Finke et al.12.
The prevalence of V. parahaemolyticus in mollusks (∏) was estimated by an equation with three parameters (see Eq. (1) in Methods): the total pool size (total number of mollusks in all investigated sampling sites), the number of positive pools, and the number of pools tested. In our case, the number of pools tested was 138. As all of the samples came from six different sampling times (each time consisted of 25 sampling points) and the average number of mollusks at each sampling point was 660 individuals, the size of the pools was 6 × 25 × 660 = 99,000. Because six V. parahaemolyticus strains were isolated from the mollusks, the number of positive pools was six. When we input these parameters into Eq. 1, the prevalence of V. parahaemolyticus-infected mollusks was calculated to be 4.5 × 10−7 (95% confidence interval: 1.64 × 10−7 to 9.77 × 10−7).
We also observed the dynamics of the mollusks and the waterbirds (common greenshank) populations from August 2017 to November 2017. As shown in Fig. S2A, there was a slight decline in the population density of the mollusks during this period, after which it was maintained at approximately 660 individuals/m2. To simplify the model, we assumed that the total biomass of the mollusks was nearly constant. At the end of August, there were only a few hundred common greenshanks in Yingkou; the population peaked at approximately 1,300 birds, which then gradually left Yingkou after October 2017. The number of common greenshanks that arrived and stayed in Yingkou showed a nearly normal distribution (Fig. S2B). Therefore, these data allowed us to predict the dynamics of the V. parahaemolyticus-carrying common greenshank using Eq. (2) (see Methods). Overall, the V. parahaemolyticus-carrying common greenshank population remained at a relatively low level, with a maximum of 95 individuals predicted to carry V. parahaemolyticus in the studied region.
Discussion
Genomic analysis of the bird-carried Vibrio species confirmed that migratory birds are disseminators of Vibrio species
Using high-resolution genome sequencing, we determined the precise origin of Vibrio species in birds. Our results demonstrated that several Vibrio strains isolated from bird feces were clonally related to the strains found in the mollusks in the local area. Therefore, the role of migratory birds in the transmission of Vibrio species must be reevaluated. The dissemination of Vibrio species by migratory birds is rarely considered13, and very little is known about the role that migratory birds play in these processes. Although it has recently been recognized that the dissemination of antibiotic resistance can be mediated by migratory birds14, the potential of migratory birds to disseminate Vibrio species had been neglected by the scientific community for more than three decades, until 2008 in a study by Halpern et al. (2008) which suggested that the dispersal of Vibrio species by waterbird may be attributable to their direct predation of chironomids and copepods15. Pretzer et al. (2017) also reported that many V. cholerae isolates from European countries were genetically related to the strains present in a European lake (Neusiedler See)16. Therefore, they speculated that the long-distance transfer of V. cholerae strains was probably via migratory birds. However, this mode of transmission lacked solid epidemiological evidence. In this study, a V. parahaemolyticus ST3 strain was isolated from a fecal sample of a common greenshank that was genomically closely related to two human clinical strains from Shenzhen (South China) and one strain isolated from an oyster in Hong Kong. The finding provides an exceptional clue for the pandemic expansion of ST3 V. parahaemolyticus populations across the globe. Since it was first identified in Japan (where it was spread from Indonesia), the mode through which this pandemic strain spread to China and arrived on the American continent has remained a matter for speculation. Many studies have suggested that the transmission of Vibrio populations can be mediated by the movement of ocean currents or human activities5. However, the movement of ocean currents is in the opposite direction to the course of the epidemic expansion in the American continent (Fig. S3). The explosive spread of the ST3 clone over hundreds of kilometers of coastline along the Pacific coast of South America since 1996 also cannot be explained simply by human activity16. The identification of this ST3 strain and other STs in bird feces suggests that the transmission of V. parahaemolyticus by migratory birds is another important factor, filling the gaps in the hypothesis described above.
In addition, the isolation of the V. parahaemolyticus ST1823 strains from both bird feces (Eurasian curlew) and mollusks also provides solid evidence that bird predation of local mollusks is an important source of Vibrio species. In addition, two closely related V. parahaemolyticus strains, YK33 and PJ37, were also isolated from a migratory bird (common greenshank) in two locations 50 km distant apart.
Another striking finding was that two bird-carried ST415 strains, PJ18 and SH50, were found in Panjin and Shanghai over 1100 km apart, respectively, further supporting the proposition that waterbirds mediates the long-distance transmission of V. parahaemolyticus. In addition, these two strains were genomically related to another ST415 strain which was obtained from the mollusks. The pubmlst data showed that the ST415 strain was not commonly identified in North China, the origin of ST415 V. parahaemolyticus in Yingkou remains unknown. One plausible possibility is that bird feces containing ST415 strains contaminated the seafood in Yingkou, resulting in its detection in the mollusks. Therefore, this ST could become a pandemic clone and poses a potential threat to human health. Because many V. parahaemolyticus STs (such as ST36) have recently been shown to be transmitted across countries, it is likely that some of these STs will be carried by migratory birds to become pandemic clones. Modeling the dynamics of V. parahaemolyticus-carrying birds also indicated that the presence of V. parahaemolyticus in birds, albeit infrequently, is likely to contaminate V. parahaemolyticus-free seafood and thus poses a potential threat to public health.
In this study, we also identified an extraordinary example of how V. mimicus strains can be moved between distant areas. Vibrio mimicus isolates were repeatedly isolated from underground water in the upstream reaches of the Liaohe River, which are probably the sources of contamination of the river delta region in Yingkou. More importantly, we found the same V. mimicus clone in the underground water in Anshan and in bird feces in Yingkou, which directly supported the hypothesis that the predation of mollusks and zooplankton by migratory birds played an essential role in the dissemination of V. mimicus.
Limitations of this study
One of the limitations of this study was that we only collected samples from three major waterbird species. Apart from the common greenshank, black-tailed godwit, and Eurasian curlew, other waterbird species were also abundant at the sampling sites, including the marsh sandpiper, common sandpiper, common redshank, common snipe, and Eurasian oystercatcher, all of which are long-distance migratory birds that depend on mollusks as their main food source. Therefore, the transmission of Vibrio species from mollusks to birds is probably a general model for most waterbird and the total carriage of pathogens by waterbirds is likely to be much higher. Another limitation of this study was that we did not collect enough samples nationally to reconstruct the spatial and temporal transmission patterns of bird-carried V. parahaemolyticus. To what extent this mode of transmission contributes to the global dissemination of V. parahaemolyticus and other Vibrio species remains to be assessed. In future studies, we will investigate the prevalence of Vibrio species at other sites along the migration routes.
Very few Vibrio species was isolated from the fish and water sampled in this study. This might be associated with the methodology used for the enrichment of Vibrio species. As our main purpose was to obtain Vibrio species from bird feces, water samples were not filtered, which might not have recovered vibrios living in zooplankton. In addition, the numbers of fish samples were too small to recover enough Vibrio species. However, previous studies suggested that fish might be the nature reservoirs and vectors of Vibrio spp.9, and waterbirds can be contaminated with vibrios by feeding on fish17. Further studies need to focus on the epidemiological links between Vibrio species in waterbirds and Vibrio species in contaminated fish.
Public health significance of waterbird migration
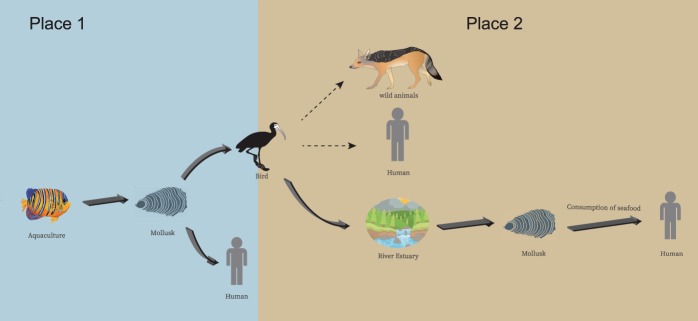
Mollusks, crustaceans, crabs, and oysters are the common food resources of waterbirds, and the majority of waterbird species are long-distance migratory birds. Millions of birds in Liaoning travel between their winter homes (extending from South China to Australia) and their summer breeding grounds (Siberia) each year, and the stopover sites in these migrations include most coastal regions of China, the Korean Peninsula, and Japan. For instance, the common greenshank is widely distributed from the UK to Siberia as its breeding ground. They stop over at Panjin and Yingkou from late August to early November and return to their winter grounds in Australia and Southeast Asia. Thus, the migratory common greenshank might acquire Vibrio species through predation along its migration route and carry them to its next destination. The identification of another ST415 strain in Yingkou also highlighted that mollusks were the source of the Vibrio species isolated from the feces of the waterbirds. Bird feces containing these pathogens would contaminate the environment and seafood at the next destination and thus pose a potential threat to human health. This large-scale migration occurs twice a year, constantly promoting the dissemination of pathogens. During this process, these birds may also be caught by wild animals or humans, leading to further pathogen spread (Fig. 5).
Figure 5.
Possible spread of Vibrio species from mollusks to migratory waterbird.
The findings of this study provide solid support for the hypothesis that Vibrio species are potentially spread across countries and continents by rivers and the migration of birds. From a public health perspective, the migration of waterbirds across national and intercontinental borders provides a mechanism for the global dispersal of bacterial pathogens.
The results herein also identified antibiotic-resistance genes (ARGs) in waterborne pathogens collected from the fish farms. The development of MDR may be associated with the overuse of these antibiotics in aquaculture. The ARGs were likely transferred by waterborne pathogens along the river. Interestingly, we found that the MDR V. mimicus strains originating from Anshan had established their population in the estuary of the Liaohe River and been transmitted to the waterbird-inhabited stopover sites, which confirmed a dissemination pattern of ARGs from the upstream river to the estuary and subsequently transferred to birds. The findings underscore that the aquacultural activity in the upstream river may greatly promote the spread of ARGs and trigger a chain reaction of antibiotic resistance transfer in the ecological systems in the estuary region. Moreover, because migratory birds may carry MDR bacteria and transport them to regions far from their original habitats, multidrug resistance is also transmitted, albeit infrequently, to places that traditionally have no public awareness of disease prevention17.
The roles of wild birds in the spread of avian flu and tick-borne viruses are well recognized18,19. Therefore, the migration of birds is probably a general route of pathogen transmission. This requires global attention. To prevent the rapid spread of pathogens, public health authorities must strengthen their active surveillance of birds and monitor their exact migration routes. Based on the information presented here, the public should reduce their direct and indirect contact with birds in the migration season and minimize the potential exposure of birds to these pathogens.
Conclusions
Our study suggest that the migration of birds is a significant mechanism in the long-distance dissemination of V. parahaemolyticus and V. mimicus. Waterbirds may be the missing link in understanding the long-distance transmission of bacterial pathogens across continents. These findings confirm that bacterial pathogens can be transmitted from mollusks to migratory birds in estuaries. They also extend our understanding of the mechanisms underlying the long-distance spread of bacterial pathogens, which is very important in the prevention of pandemics. Effective surveillance of waterbirds should be undertaken to monitor the dissemination of waterborne pathogens along their migration routes, which will also provide valuable information for predicting future epidemics of infectious diseases.
Materials and Methods
Sampling
From August 30 to October 27, 2017, and from March 13 to April 24, 2018, 66 water samples were collected from fish farms in Anshan and the nearby groundwater (Fig. 1). At the same time, in-field observations were conducted in the intertidal zones at Panjin, Yingkou and Shanghai to capture the moment of excretion for individual birds and record the bird species. A total of 180 samples of fresh feces from seven types of birds, including bean goose (Anser fabalis), white-front goose (Anser erythropus), Saunders’s Gull (:Larus saundersi), red-billed gull (Chroicocephalus novaehollandiae scopulinus), common greenshank (Tringa nebularia), Eurasian curlew (Numenius arquata) and black-tailed godwit (Limosa limosa) were collected (Table S1); and 170 sediment samples and 77 mollusk samples were collected from Panjin and Yingkou. All the samples were sent to the laboratory within 12 h of collection. In addition, a range of environmental variables (water temperature, pH, total phosphorus, dissolved organic carbon, and nitrite) were determined according to Kirschner et al.20.
Isolation and identification of the Vibrio species in water, sediment, and bird fecal samples
The sediments, bird feces and water samples were aseptically disaggregated and precultured in alkaline peptone water at 37 °C for 12 h. Afterwards, one loop of culture was spread directly in triplicate onto thiosulfate citrate bile salts (TCBS) agar plates and incubated at 37 °C for 24 h. The yellow and green colonies on the TCBS, which were suspected to be Vibrio species, were subcultured in Luria broth (LB) to produce pure cultures. For bacterial identification, a partial sequence of 16 S rRNA was amplified with PCR using the primers 27 F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492 R (5′-GGTTACCTTGTTACGACTT-3′)21. The amplicons with a size of approximately 1400 bp were sequenced with Sanger sequencing technology at Shanghai Jingtong Biotech Co. Ltd. The similarity analysis of the 16S rRNA gene sequence was performed using the BLASTn program on NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to confirm the bacterial species.
Genome sequencing and de novo assembly
Genomic DNA was extracted from overnight bacterial cultures grown ontrypticase soy agar, and fragmented and tagged for multiplexing with the Nextera XT DNA Sample Preparation Kit (Illumina). The tagged DNAs were sequenced with the Illumina HiSeq. 2500 platform at Beijing Novogene Bioinformatics Technology Co., Ltd. The FASTQ reads were quality trimmed with Trimmomatic (v0.36)22, and bases with a PHRED score of <30 were removed from the trailing end. To obtain the draft genomes, the contigs were assembled de novo with SPAdes version 3.023.
In silico MLST
In silico MLST of V. parahaemolyticus was performed with the MLST 1.8 server at the Center for Genomic Epidemiology (https://cge.cbs.dtu.dk//services/MLST/)24.
Identification of SNPs and phylogenetic inferences
The genomes of ATCC 33654, and RIMD221063 were used as the reference genomes to call the SNPs for V. mimicus, and V. parahaemolyticus, respectively. A total of 47 publicly available V. parahaemolyticus genomes were obtained from GenBank. Using these and those of the 27 sequenced strains, stringent SNP calling was performed with a custom pipeline, as described previously, to guarantee that only genuine SNPs were included in the analysis25. The V. parahaemolyticus core genome SNPs were extracted based on the previously defined V. parahaemolyticus core genome26. Recombination events were confirmed with RDP327. Each recombinant site was confirmed with at least three different analytical methods. Fifteen V. mimicus genomes available from GenBank were also used to investigate the evolutionary relationships of the V. mimicus strains sequenced in this study (Table S5). The phylogenetic trees were constructed with the maximum likelihood method using RAxML 7.2.828. Generalized Time Reversible (GTR) with Gamma rates (G) and Invariant sites (I) (GTR + G + I) model was used. For closely related genomic strains, maximum parsimony algorithms were used to precisely identify the SNPs that differed among them, in PAUP 4.029.
Gene content analysis of V. mimicus and V. parahaemolyticus
RAST sever was used to annotate the sequences of each genome determined with next-generation sequencing30. ARGs were identified with ResFinder31.
Modeling the dynamics of V. parahaemolyticus-carrying birds
We used the intertidal zone of Yingkou in the estuary of the Liaohe River as the location for modeling the dynamics of V. parahaemolyticus-carrying birds.
To set the modeling parameters, the numbers of common greenshank stopover points in the investigated region were counted every week. To estimate the size of the prey population (mollusks), the dynamics of the mollusk density in the sediment were determined with the method described by Finke GR et al.13.
If we let k be the pool size, x the number of positive pools, and m the number of pools tested, and the prevalence of V. parahaemolyticus-infected mollusks is denoted ∏, the MLE of V. parahaemolyticus prevalence in mollusks (∏) can be described as suggested by Cowling et al.32 as follows:
| 1 |
The details of the modeling inference can be found in the Supplemental File (Appendix 2).
If we let H and B(t) be the sizes of the prey population (mollusks) and the predator population (common greenshank), respectively, the number of V. parahaemolyticus-infected birds can be described as follows:
| 2 |
where P denotes the prevalence of V. parahaemolyticus in the mollusks, ands indicates the daily predation rate. If we assume that P and s are constant values, then s × H is the number of mollusks that can be caught by one bird daily, and P × s × H is the number of V. parahaemolyticus-infected mollusks that can be caught by one bird per day.
Supplementary information
Acknowledgements
We thank Dr. Kang Chen for assistance with modeling. We would like to thank the Mr. Yixiang Zhang and Mr. Xudong Shen for the bird migration information and sampling assistance. This research was funded by the National Natural Science Foundation of China (81903372) and National Key R&D Program of China (2017YFD0701700).
Author contributions
S.F. and R.J.L. conceived and designed the experiments. Sampling and microbiological analysis were performed by J.H., Q.Y., S.Y. and Y.W. Data analysis and the draft of the manuscript were performed by S.F., R.L. and Y. L.
Data availability
The raw sequencing data were submitted to GenBank (NCBI) under the BioProject No. PRJNA496566.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Songzhe Fu, Jingwei Hao and Qian Yang.
Contributor Information
Songzhe Fu, Email: fusongzhe@hotmail.com.
Ruijun Li, Email: liruijun@dlou.edu.cn.
Supplementary information
is available for this paper at 10.1038/s41598-019-52791-5.
References
- 1.Bonninjusserand M, et al. Vibrio species involved in seafood-borne outbreaks (Vibrio cholerae, V. parahaemolyticus and V. vulnificus): review of microbiological versus recent molecular detection methods in seafood products. Crit. Rev. Food. Sci. Nutr. 2017;28:1–14. doi: 10.1080/10408398.2017.1384715. [DOI] [PubMed] [Google Scholar]
- 2.Letchumanan V, Yin WF, Lee LH, Chan KG. Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from retail shrimps in Malaysia. Front. Microbiol. 2015;6:33. doi: 10.3389/fmicb.2015.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scallan E, et al. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 2011;17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nair GB, et al. Global dissemination of Vibrio parahaemolyticus serotype O3:K6 and its serovariants. Clin. Microbiol. Rev. 2007;20:39. doi: 10.1128/CMR.00025-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gil AI, et al. O3:K6 serotype of Vibrio parahaemolyticus identical to the global pandemic clone associated with diarrhea in Peru. Int. J. Infect. Dis. 2007;11:324–8. doi: 10.1016/j.ijid.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Velazquez-Roman J, Leónsicairos N, Canizalez-Roman A. Pandemic Vibrio parahaemolyticus O3:K6 on the American continent. Front. Cell. Infect.Microbial. 2013;3:110. doi: 10.3389/fcimb.2013.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lutz C, Erken M, Noorian P, Sun S, Mcdougald D. Environmental reservoirs and mechanisms of persistence of Vibrio cholerae. Front. Microbiol. 2013;4:375. doi: 10.3389/fmicb.2013.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vezzulli L, et al. Climate influence on Vibrio and associated human diseases during the past half-century in the coastal North. Atlantic. P. Natl. A. Sci. Iedia.A. 2016;113:E5062. doi: 10.1073/pnas.1609157113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laviad-Shitrit S, et al. Great cormorants (Phalacrocorax carbo) as potential vectors for the dispersal of Vibrio cholerae. Sci. Rep. 2017;7:7973. doi: 10.1038/s41598-017-08434-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee JV, Bashford DJ, Donovan TJ, Furniss AL, West PA. The incidence of Vibrio cholerae in water, animals and birds in Kent, England. J. Appl.Microbiol. 1982;52:281–291. doi: 10.1111/j.1365-2672.1982.tb04852.x. [DOI] [PubMed] [Google Scholar]
- 11.Laviad-shitrit S, Izhaki I, Arakawa E, Halpern M. Wild waterfowl as potential vectors of Vibrio cholerae and Aeromonas species. Trop. Med. Int. Health. 2018;23:758–764. doi: 10.1111/tmi.13069. [DOI] [PubMed] [Google Scholar]
- 12.Finke GR, Bozinovic F, Navarrete SA. A mechanistic model to study the thermal ecology of a southeastern Pacific dominant intertidal mussel and implications for climate change. Physiol. Biochemzool. 2009;82:303. doi: 10.1086/599321. [DOI] [PubMed] [Google Scholar]
- 13.Cardoso, et al. A comprehensive survey of Aeromonas sp. and Vibrio sp. in seabirds from southeastern Brazil: outcomes for public health. J. Appl. Microbiol. 2018;124:1283–1293. doi: 10.1111/jam.13705. [DOI] [PubMed] [Google Scholar]
- 14.Allen HK, et al. Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev.Microbiol. 2010;8:251–259. doi: 10.1038/nrmicro2312. [DOI] [PubMed] [Google Scholar]
- 15.Halpern M, Senderovich Y, Izhaki I. Water birds—The Missing Link in Epidemic and Pandemic Cholera Dissemination? Plos. Pathog. 2008;4:e1000173. doi: 10.1371/journal.ppat.1000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pretzer C, et al. High genetic diversity of Vibrio cholerae in the European lake Neusiedler See is associated with intensive recombination in the reed habitat and the long-distance transfer of strains. Environ.Microbiol. 2017;19:328–344. doi: 10.1111/1462-2920.13612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Senderovich Y, Izhaki I, Halpern M. Fish as reservoirs and vectors of Vibrio cholerae. PLoS One. 2010;5:e8607. doi: 10.1371/journal.pone.0008607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yun Y, et al. Phylogenetic Analysis of Severe Fever with Thrombocytopenia Syndrome Virus in South Korea and Migratory Bird Routes Between China, South Korea, and Japan. Am. J. Trop. Med. Hyg. 2015;93:468. doi: 10.4269/ajtmh.15-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu Y, Peng G, Ben W, Si Y. Southward autumn migration of water birds facilitates cross-continental transmission of the highly pathogenic avian influenza H5N1. virus. Sci. Rep. 2016;6:30262. doi: 10.1038/srep30262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirschner AK, et al. Vibrio cholerae non-O1/non-O139 strains in a large alkaline lake in Austria: dependence on temperature and dissolved organic carbon quality. Appl. Environ.Microbiol. 2008;74:2004–2015. doi: 10.1128/AEM.01739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bosshard PP, Santini Y, GrãTer D, Stettler R, Bachofen R. Bacterial diversity and community composition in the chemocline of the meromictic alpine Lake Cadagno as revealed by 16S rDNA analysis. FEMS.Microbiol.Ecol. 2000;31:173–182. doi: 10.1111/j.1574-6941.2000.tb00682.x. [DOI] [PubMed] [Google Scholar]
- 22.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bankevich A, et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Bio. l. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larsen MV, et al. Multilocus Sequence Typing of Total-Genome-Sequenced Bacteria. J. Clin. Microbiol. 2012;50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan CHS, Octavia S, Sintchenko V, Lan R. SnpFilt: A pipeline for reference-free assembly-based identification of SNPs in bacterial genomes. Comput. Biol. Chem. 2016;65:178. doi: 10.1016/j.compbiolchem.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalezescalona N, Jolley KA, Reed E, Martinezurtaza J. Defining a Core Genome Multilocus Sequence Typing Scheme for the Global Epidemiology ofVibrio parahaemolyticus. J. Clin. Microbiol. 2017;55:1682–1697. doi: 10.1128/JCM.00227-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin D, Rybicki E. RDP: detection of recombination amongst aligned sequences. Bioinformatics. 2000;16:562–563. doi: 10.1093/bioinformatics/16.6.562. [DOI] [PubMed] [Google Scholar]
- 28.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 29.Swofford DL. Paup - a Computer-Program for Phylogenetic Inference Using Maximum Parsimony. J. Gen. Physiol. 1993;102:A9–A9. [Google Scholar]
- 30.Aziz RK, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zankari E, et al. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cowling DW, Gardner IA, Johnson WO. Comparison of methods for estimation of individual level prevalence based on pooled samples. Prev. Vet. Med. 1999;39:211–25. doi: 10.1016/S0167-5877(98)00131-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing data were submitted to GenBank (NCBI) under the BioProject No. PRJNA496566.