Abstract
Hippocalcin (HPCA) is a neuron-specific calcium-binding protein predominantly expressed in the nervous system. In the present study, we demonstrate that HPCA regulates neuronal differentiation in SH-SY5Y cells. We observed that the expression level of HPCA was increased during neuronal differentiation. Depletion of HPCA inhibited both neurite outgrowth and synaptophysin (SYP) expression, whereas overexpression of HPCA enhanced neuronal differentiation. Interestingly, we also found that the expression of HPCA mRNA was modulated by miR-24-3p. Using a dual-luciferase assay, we showed that co-transfection of a plasmid containing the miR-24-3p binding site from the 3′-untranslated region (3′UTR) of the HPCA gene and an miR-24-3p mimic effectively reduced luminescence activity. This effect was abolished when miR-24-3p seed sequences in the 3′UTR of the HPCA gene were mutated. miR-24-3p expression was decreased during differentiation, suggesting that the decreased expression level of miR-24-3p might have upregulated mRNA expression of HPCA. As expected, upregulation of miR-24-3p by an miRNA mimic led to reduced HPCA expression, accompanied by diminished neuronal differentiation. In contrast, downregulation of miR-24-3p by an antisense inhibitor promoted neurite outgrowth as well as levels of SYP expression. Taken together, these results suggest that miR-24-3p is an important miRNA that regulates neuronal differentiation by controlling HPCA expression.
Electronic supplementary material
The online version of this article (10.1007/s00018-019-03290-3) contains supplementary material, which is available to authorized users.
Keywords: Hippocalcin, miR-24-3p, Neuronal differentiation, Synaptophysin, SH-SY5Y cells
Introduction
MicroRNAs (miRNAs) are small, highly conserved non-coding RNA molecules of approximately 22 nucleotides. They can modulate gene expression through complementary base pairing of the seed sequence (the 2–8 nucleotides from the 5′ end of the miRNA) located in the 3′-untranslated region (3′UTR) of the target mRNA, leading to the translational suppression and/or destabilization of the target mRNAs [1–3]. It is widely accepted that a single miRNA has the potential to inhibit the expression of hundreds of target mRNAs and, conversely, individual mRNAs are commonly targeted by multiple miRNAs [4]. Hence, miRNAs serve as key regulators in various biological processes, such as proliferation, differentiation, apoptosis, metabolism, and development [5–8]. In mammals, many miRNAs are spatially expressed in the brain, suggesting their essential roles in central nervous system development and function [9, 10]. The specific knockdown of Dicer, the key enzyme involved in processing miRNA precursors into mature miRNAs, leads to malformation of the midbrain and cerebellum and failure of neural crest and dopaminergic differentiation [11, 12]. Recent studies have also demonstrated the contribution of individual miRNAs to various aspects of synaptic development and plasticity. miR-132 is involved in activity-dependent structural and functional plasticity by targeting the Rho family GTPase-activating protein, p250GAP [13]. Brain-specific miR-124a is also critical for the differentiation and maturation of dentate gyrus neurons and retinal cone cells [14]. In addition, miR-335-5p plays a key role in spatial memory formation and hippocampal plasticity [15]. Therefore, miRNAs are key regulators of mammalian neurodevelopment and have been increasingly linked to neuronal differentiation and the translatability of mRNAs in neurons. Our previous study showed that HPCA expression was gradually increased during neuronal differentiation [16]. Therefore, we assumed that an miRNA would be associated with HPCA expression during neuronal differentiation. Using an miRNA Target Prediction algorithm, we found that only miR-24-3p had two potential binding sites to 3′UTR of HPCA among several miRNAs. miR-24-3p, which belongs to the miR-24 family, is a well-known cancer-associated miRNA that is differentially expressed in the initiation and progression in multiple kinds of tumors. Inhibition of miR-24-3p promotes osteogenic differentiation by targeting Smad5 [17]. miR-24-3p also facilitates cell proliferation and inhibits cell apoptosis in gastric cancer by targeting BCL2L11 [18]. However, there is no evidence of any association with neurons in miR-24-3p.
Hippocalcin (HPCA) is a high-affinity calcium-binding protein expressed most abundantly in pyramidal cells of the hippocampal CA1 region [19, 20]. HPCA is a member of the neuronal calcium sensor protein family. It has four EF-hand-structured Ca2+-binding domains and a myristoylation site in the N-terminus, which allow its translocation from the cytosol to the plasma membrane and/or perinuclear compartments in response to increased Ca2+ concentration [21–25]. HPCA acts as a molecular linker between calcium entry into neurons through N-methyl-d-aspartate receptors and the regulated endocytosis of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors [26]. During brain development, HPCA expression in hippocampal pyramidal cells abruptly increases, concurrent with synaptic formation [27]. Our previous study showed that HPCA increases NeuroD expression, resulting in neurite outgrowth during the differentiation of H19-7 cells [28]. Moreover, it was found to promote neuronal differentiation through activation of the PKCα/PLD1/SHP1 cascade, leading to the inhibition of astrocytic differentiation in cortical neural stem cells [16]. We also reported that HPCA stimulates astrocytic differentiation through the activation of STAT3 in hippocampal neural precursor cells [29]. However, the role of HPCA regulation by miRNAs in neural differentiation has not previously been studied.
Here, we showed that HPCA has an important role in the neuronal differentiation of SH-SY5Y cells. In addition, we identified miR-24-3p as a putative regulator of HPCA during neuronal differentiation. We observed that overexpression of miR-24-3p reduced HPCA mRNA levels and inhibited neuronal differentiation, whereas the inhibition of miR-24-3p had the opposite effects. Taken together, our findings suggest that the modulation of HPCA expression or miR-24-3p function may be used to regulate neuronal differentiation.
Materials and methods
Materials
For cell culture experiments, Dulbecco’s modified Eagle medium (DMEM) containing l-glutamine, high glucose concentration, and pyruvate and fetal bovine serum (FBS) were purchased from Gibco (Grand Island, NY, USA) and all-trans retinoic acid (RA) was purchased from Sigma-Aldrich (St Louis, MO, USA). A penicillin/streptomycin solution and trypsin/EDTA were purchased from WelGENE, Inc. (Daegu, Korea). Anti-HPCA (#ab24560) and anti-synaptophysin (SYP, #ab32127) antibodies were purchased from Abcam (Cambridge, UK), an anti-calnexin antibody (#ADI-SPA-860-F) was purchased from Enzo Life Sciences (Farmingdale, NY, USA), and an Alexa Fluor® 488-conjugated secondary goat anti-rabbit IgG (H + L) antibody (#A-11008) was purchased from Invitrogen (Carlsbad, CA, USA). All other chemicals were of analytical grade.
Cell culture and differentiation conditions
SH-SY5Y and HeLa cells were maintained in DMEM supplemented with 10% heat-inactivated FBS and 1% penicillin/streptomycin and incubated in a humidified atmosphere at 37 °C with 5% CO2. The medium was replaced every 2 days and cells were split before they reached confluence. SH-SY5Y cells were differentiated by treatment with 50 μM RA for 5 or 7 days. The culture medium was replaced on alternate days with fresh medium containing 50 μM RA.
RNA interference
For HPCA silencing experiments, an HPCA siRNA (ON-TARGET plus SMARTpool, #L-017429-02-0010) and a negative control siRNA (ON-TARGET plus Non-targeting pool, #D-001810-10-20) were purchased from Dharmacon (Lafayette, CO, USA). Transient siRNA transfections were performed in 12-well plates by introducing 100 nM HPCA siRNA or negative control siRNA into SH-SY5Y cells using Lipofectamine RNAiMAX transfection reagent (Invitrogen), according to the manufacturer’s protocol.
Retrovirus construction and transduction
HPCA cDNA was cloned into a retroviral vector containing IRES-EGFP. Virus particles were produced by transfecting the retrovirus packaging cell line, 293GPG, with the vector using Lipofectamine 3000 transfection reagent (Invitrogen) and harvesting supernatants containing viral particles after incubation for 48 h. For virus transduction, SH-SY5Y cells were incubated with a viral suspension (4 × 106 particles/mL) containing polybrene (1 μg/mL, δ-Aldrich) for 4 h, followed by transfer to fresh culture medium containing 50 μM RA.
Transfection of miRNA mimic and inhibitor
The miR-24-3p mimic and negative control miRNA were synthesized by Genolution (Seoul, Korea). A sequence-specific miR-24-3p inhibitor (cat. no. IH-300497-05-0010, miRIDIAN microRNA Human hsa-miR-24-3p—Hairpin Inhibitor) and its corresponding non-specific control (cat. no. IN-001005-01-20, miRIDIAN microRNA Hairpin Inhibitor Negative Control #1) were purchased from Dharmacon. The sequences of the miR-24-3p mimic and inhibitor are provided in Supplementary Table 1. HeLa or SH-SY5Y cells were seeded 1 day before transfection. Each mimic or inhibitor was transiently transfected into cells using Lipofectamine RNAiMAX transfection reagent, as described in the manufacturer’s instructions.
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cultured cells using RNAiso Plus (Takara Bio Inc., Ohtsu, Japan). cDNA was prepared for qPCR by reverse transcribing 300 ng of purified total RNA using GoScript™ Reverse Transcriptase and random primers (Promega Corporation, Madison, WI, USA). qPCR was performed using a SensiFAST™ SYBR No-ROX Kit (Bioline, London, UK) on a CFX Connect™ Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). The primers used for RT-qPCR are listed in Supplementary Table 1. Thermocycling conditions were 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Each sample was tested in duplicate and at least three samples obtained from independent experiments were analyzed. Relative quantification was carried out using the 2−ΔΔCt method. Gene expression was normalized to the internal control, GAPDH.
Western blotting assays
Cells were lysed in ice-cold hypotonic lysis buffer consisting of 10 mM Tris–HCl (pH 7.5), 10 mM NaCl, 10 mM EDTA, 0.5% Triton X-100, and Complete™ EDTA-free Protease Inhibitor Cocktail (Roche Diagnostics, Indianapolis, IN, USA). Protein samples (15–30 μg) were electrophoresed in NEXT GEL® 15% polyacrylamide gels (Amresco, Solon, OH, USA) and then transferred to PVDF membranes (Merck Millipore, Darmstadt, Germany). After blocking with 5% non-fat dried milk for 1 h, the membranes were incubated with rabbit anti-HPCA (1:500 dilution), rabbit anti-SYP (1:20,000 dilution), and rabbit anti-calnexin (1:2000 dilution) primary antibodies, followed by an HRP-conjugated secondary antibody (1:2000; Jackson ImmunoResearch, West Grove, PA, USA). Specific bands were detected by enhanced chemiluminescence (Thermo Fisher Scientific, Rockford, IL, USA) and were quantified using Image J software (v1.51 k, NIH, http://www.rsb.Info.nih.gov/ij/).
Immunofluorescence staining
Cells were initially fixed with 4% (w/v) paraformaldehyde in phosphate-buffered saline (PBS) for 20 min and then washed three times with 0.1% BSA in PBS at room temperature. After blocking with 10% normal goat serum in 0.1% BSA in PBS containing 0.3% Triton X-100 for 1 h at room temperature, cells were immunostained with a rabbit monoclonal anti-SYP primary antibody (1:200 dilution) at 4 °C overnight. Subsequently, cells were washed three times with PBS and then labeled with a 1:500 dilution of an Alexa Fluor® 488-conjugated goat anti-rabbit IgG (H + L) secondary antibody for 1 h, before mounting with Vectashield mounting medium (Vector Laboratories, Burlingame, CA, USA) containing 4,6-diamidino-2-phenylindole (DAPI). Immunoreactive cells were analyzed under an epifluorescence microscope (Nikon Instruments, Melville, NY, USA) at magnifications ranging from 20× to 40×.
Measurement of neurite outgrowth
Cells were cultured on coverslips in 24-well plates, fixed with 0.1% (w/v) picric acid/PBS containing 4% (w/v) paraformaldehyde, and incubated overnight at 4 °C with an anti-SYP antibody (1:200). After incubation with a 1:2000 dilution of Alexa Fluor® 488-conjugated goat anti-rabbit IgG secondary antibody, cells were mounted on slides with Vectashield. SYP-positive cells were photographed using an epifluorescence microscope. Neurite outgrowth was assessed by measuring the length of neurites in all positively identified neurite-bearing cells. The mean neurite length per cell was calculated using Image J software. The length of the primary neurite was defined as the distance from the soma to the tip of the branch. For this analysis, a cell was arbitrarily defined as neurite bearing when it bore at least one process equal to or longer than 25 μm (mean diameter of the cell body) [30]. For each graph, neurite length data were generated from randomly selected areas of at least five independent cultures from three independent experiments, and more than 100 cells were counted for each condition in each experiment.
Plasmid construction and dual-luciferase reporter assays
miRNA target sequences in the 3′UTR of the hHPCA gene were inserted using the XbaI restriction site in the pmirGLO vector (Promega Corporation, Cat. #E1330). The sequences targeted by miR-24-3p are summarized in Supplementary Table 1. The correct orientation and nucleotide sequence of the 3′UTR fragments in the plasmid constructs were further confirmed by sequencing analysis.
Dual-luciferase reporter plasmids were co-transfected with miRNA mimics or miRNA inhibitors into HeLa cells and after 48 h, cells were assayed for luciferase activity using the Dual-Glo® Luciferase Assay System (Promega Corporation, Cat. #E2920) and a luminometer (Promega Corporation), according to the manufacturer’s instructions. To correct for variations in transfection efficiency, firefly luciferase activity was normalized to Renilla luciferase activity. Firefly luciferase activity was calculated as the percent activity relative to luciferase activity in cells transfected with negative control miRNA mimics or inhibitors. For each transfection, luciferase activity was averaged from at least three replicates.
TaqMan RT-qPCR of miRNA
For the detection of endogenous miRNA expression, 10 ng of purified total RNA was reverse transcribed using a TaqMan® MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). The resulting cDNA was used amplified by PCR using a TaqMan® MicroRNA Assay (Applied Biosystems) according to the manufacturer’s instructions. cDNA amplification and detection of products were performed using a CFX Connect™ Real-Time PCR Detection System, with an initial denaturation at 95 °C for 10 min, followed by 40 cycles of amplification at 95 °C for 15 s and 60 °C for 1 min, before cooling. The threshold cycle (Ct) for miR-24-3p was automatically defined within the linear amplification phase and was normalized to the control, U6 snRNA, to calculate the ΔCt value. The relative difference in miR-24-3p expression levels (ΔΔCt) in the sorted cells was calculated and presented as fold induction (2−ΔΔCt).
Statistical analysis
All quantitative data are expressed as mean ± standard error of the mean (SEM). In each experiment, all measurements were performed at least in triplicate. Data were analyzed using a two-tailed, unpaired Student’s t test and values of P < 0.05 were considered statistically significant.
Results
HPCA was upregulated during the neuronal differentiation of SH-SY5Y cells
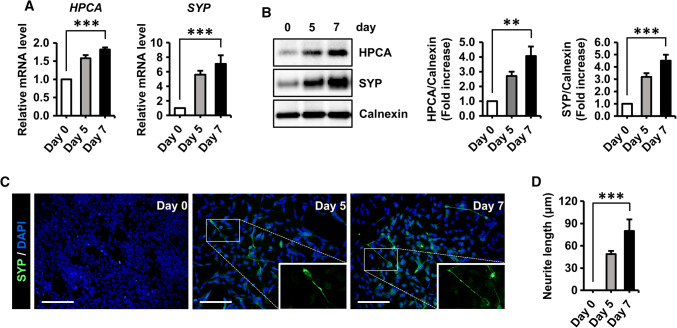
The human neuroblastoma cell line, SH-SY5Y, has been widely used as an in vitro model for neuroscience research because these cells are easily induced to differentiate by RA treatment [31–33]. To determine the involvement of HPCA in regulating human neuronal differentiation, SH-SY5Y cells were induced to differentiate in the presence of 50 μM RA for 5 or 7 days. As shown in Fig. 1a and b, the mRNA and protein levels of HPCA and SYP, a presynaptic neuronal marker, were gradually increased under differentiation conditions. Immunostaining showed a significant increase in the number of SYP-positive cells and neurite outgrowth (0 μm versus 79.9 ± 15.5 μm, P < 0.001, Fig. 1c, d) during differentiation. These data suggest that HPCA is involved in the neuronal differentiation of SH-SY5Y cells.
Fig. 1.
Differentiation induced expression of HPCA in SH-SY5Y cells. a, b SH-SY5Y cells were induced to differentiate by adding 50 μM all-trans retinoic acid (RA) for the indicated number of days. a The mRNA levels of the presynaptic marker, synaptophysin (SYP) and hippocalcin (HPCA), were determined by RT-qPCR. b Proteins were analyzed by western blotting with anti-HPCA, anti-SYP, and anti-calnexin antibodies. The graph shows mean densities as fold increases from five independent experiments (mean ± SEM). Band intensities were quantified using Quantity One® software. **P < 0.01, ***P < 0.001 compared with day 0 of differentiation. c Neuronal differentiation was induced by adding 50 μM RA for the indicated number of days. Cells were stained with DAPI (blue) and an anti-SYP antibody (green) to visualize neurite extensions. Scale bar, 100 μm. d Neurite lengths were measured in randomly selected areas in three independent experiments. ***P < 0.001 compared with day 0 of differentiation (mean ± SEM)
HPCA regulated the differentiation of SH-SY5Y cells
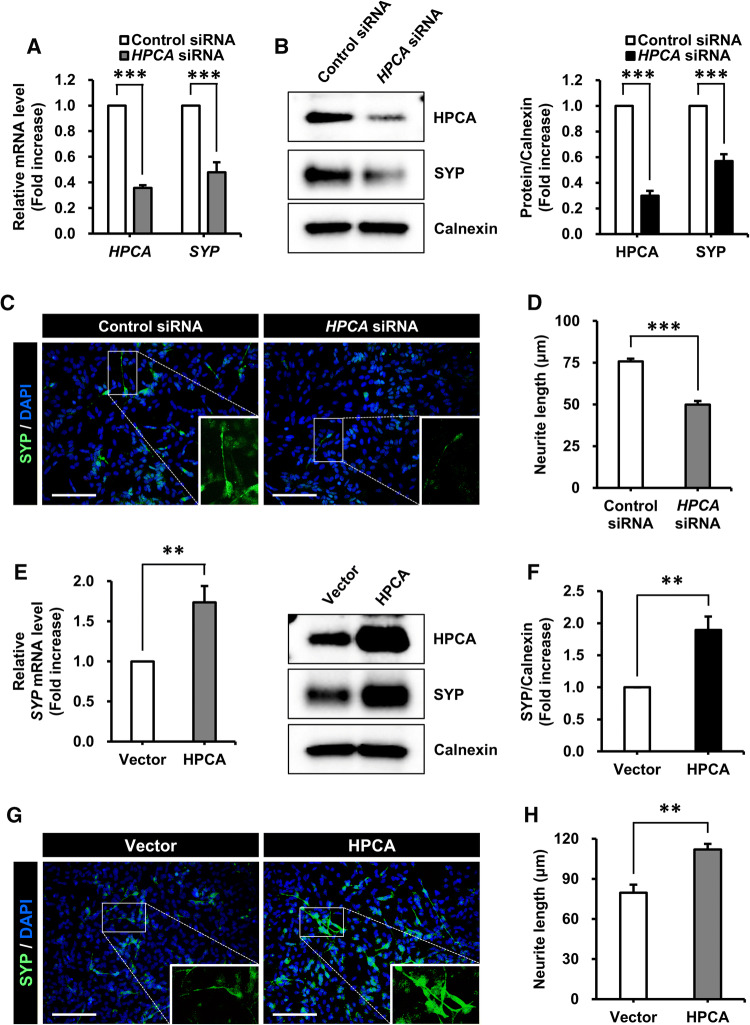
Previously, we reported that HPCA is critical for neuronal differentiation and neurite outgrowth in multiple cell types [16, 28]. To investigate whether HPCA was implicated in neuronal differentiation, either control siRNA or HPCA siRNA were transiently transfected into SH-SY5Y cells, which were then allowed to differentiate for 7 days. We showed that mRNA and protein levels of SYP were markedly attenuated by HPCA knockdown (Fig. 2a, b). To determine the role of HPCA in neuronal differentiation, we measured neurite length after 7 days of differentiation in the presence of HPCA siRNA. HPCA siRNA transfection significantly decreased neurite outgrowth (75.7 ± 1.6 μm versus 49.9 ± 2.2 μm, P < 0.001, Fig. 2c, d) when compared with control siRNA transfection, thus implicating HPCA in the regulation of SYP expression and neurite outgrowth during differentiation.
Fig. 2.
Effect of HPCA on the neuronal differentiation of SH-SY5Y cells. a, b SH-SY5Y cells were transiently transfected with control siRNA or HPCA siRNA, and then incubated for 7 days after the addition of RA. aHPCA and SYP mRNA levels were analyzed by RT-qPCR. b HPCA, SYP, and calnexin protein levels were determined by western blotting. The graph shows mean densities as fold increases from four independent experiments (mean ± SEM). Band intensities were quantified using Quantity One® software. ***P < 0.001 compared with the day 0 of differentiation. c Cells were transfected with control siRNA or HPCA siRNA and then induced to differentiate by adding RA. After 7 days, fixed cells were stained with an anti-SYP antibody (green) and DAPI (blue). Scale bar, 100 μm. d Neurite lengths were measured in randomly selected areas from five independent cultures. ***P < 0.001 compared with the control siRNA (mean ± SEM). e, f SH-SY5Y cells were transduced with pMSCV-IRES-EGFP or pMSCV-HPCA-Myc-IRES-EGFP and induced to differentiate for 7 days. eSYP mRNA levels were detected by RT-qPCR. f Cells were lysed and analyzed by western blotting with anti-HPCA, anti-SYP, and anti-calnexin antibodies. The graph shows mean densities as fold increases from five independent experiments (mean ± SEM). Band intensities were quantified with Quantity One® software. **P < 0.01 compared with day 0 of differentiation. g Immunofluorescence was used to visualize SYP (green) at day 7 of differentiation, with or without overexpression of HPCA. Nuclei were stained with DAPI (blue). Scale bar, 100 μm. h Neurite lengths were measured in randomly selected areas from five slides of each condition. **P < 0.01 compared with the vector control (mean ± SEM)
To confirm the role of HPCA in neuronal differentiation, cells were transduced with pMSCV-IRES-EGFP or pMSCV-HPCA-MYC-IRES-EGFP and induced to differentiate for 7 days. As shown in Fig. 2e, f, SYP mRNA and protein expression levels were significantly increased by HPCA overexpression. Moreover, HPCA markedly increased neurite outgrowth (79.7 ± 6.0 μm versus 112.0 ± 4.2 μm, P < 0.01, Fig. 2g, h) compared with the vector control. These findings suggest that HPCA is required for the neuronal differentiation of SH-SY5Y cells.
miRNA-24-3p targeted endogenous HPCA expression
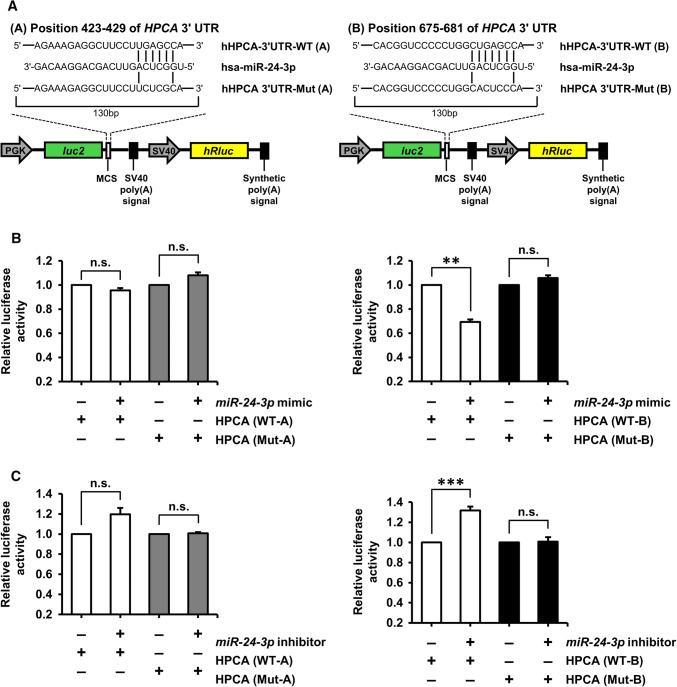
In the present study, we showed that HPCA expression was increased with the induction of differentiation. An important mode of post-transcriptional regulation is miRNA-mediated repression of mRNA transcripts. To identify miRNAs that target HPCA mRNA, we performed in silico screening using TargetScan (http://www.targetscan.org). The TargetScan database predicted two putative miR-24-3p binding sites in the 3′UTR of HPCA (positions 423–429 and 675–681 of HPCA 3′UTR, Fig. 3a), which suggested that miR-24-3p may serve as a suppressor of HPCA mRNA expression. To confirm that miR-24-3p bound to the HPCA mRNA 3′UTR, we constructed wild-type (WT-A and B) and mutant (Mut-A and B) plasmids encoding either the full or partial sequence of the 3′UTR of HPCA, including the predicted miR-24-3p target sites, and performed a luciferase reporter assay (Fig. 3a). Transfection with the miR-24-3p mimic specifically decreased the activity of a luciferase reporter gene fused to HPCA-3′UTR-WT-B (position 675–681: P < 0.01), but had no effect on HPCA-3′UTR-WT-A (position 423–429, Fig. 3b). The activity of the reporter gene fused to HPCA-3′UTR-Mut-B was unaffected by the presence of exogenous miR-24-3p (Fig. 3b).
Fig. 3.
Validation of HPCA as a direct target of miR-24-3p.a Map of representative bicistronic firefly/Renilla luciferase (FLuc/RLuc) plasmids containing HPCA mRNA 3′UTRs with putative miR-24-3p binding sites (WT) or mutated binding sites (Mut). b A control miRNA mimic or an miR-24-3p mimic was co-transfected with HPCA-3′UTR-WT or HPCA-3′UTR-Mut into HeLa cells. c A control miRNA inhibitor or an miR-24-3p inhibitor was co-transfected with HPCA-3′UTR-WT or HPCA-3′UTR-Mut into HeLa cells. Luciferase assays were performed to quantify the relative luciferase activity of FLuc normalized to the activity of RLuc. The relative activity of luciferase in control miRNA inhibitor-transfected cells was set to 1.0. **P < 0.01; ***P < 0.001; n.s. not significant
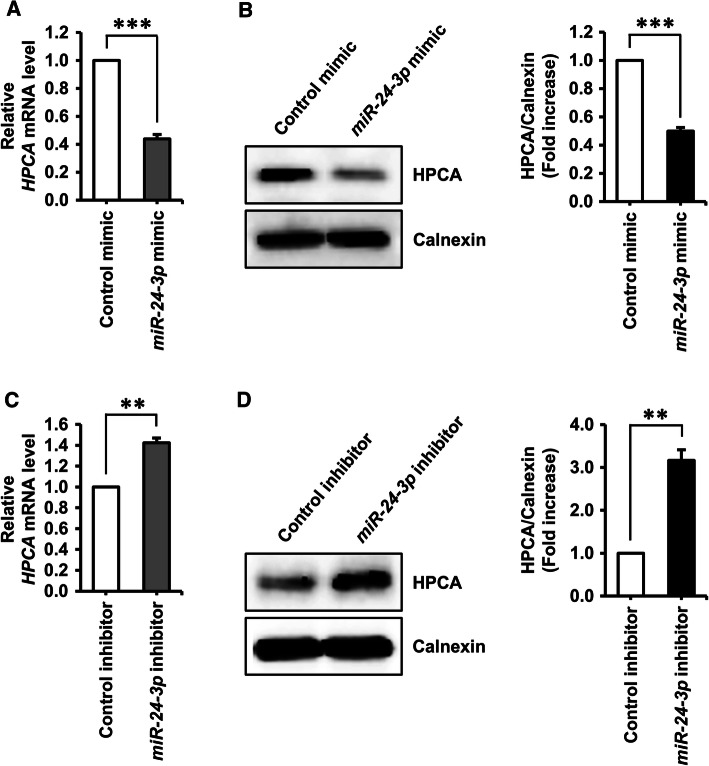
Next, we performed additional experiments using a specific inhibitor of miR-24-3p. Co-transfection of HPCA-3′UTR-WT-B with an miR-24-3p-specific inhibitor caused a significant increase (approximately 40%, P < 0.001) in luciferase activity, compared with the control inhibitor. Luciferase activity was not affected when HPCA-3′UTR-WT-A was co-transfected with an miR-24-3p-specific inhibitor (Fig. 3c). These results demonstrated that position 675–681 of the human HPCA 3′UTR is a functional binding site for miR-24-3p. Furthermore, we confirmed that miR-24-3p regulated endogenous HPCA expression. As shown in Fig. 4a, b, an miR-24-3p mimic decreased HPCA mRNA and protein levels, compared with a control mimic. By contrast, an miR-24-3p inhibitor significantly increased the levels of HPCA mRNA and protein (Fig. 4c, d). Taken together, these data indicated that miR-24-3p inhibited HPCA expression by binding to a single site present in the HPCA mRNA 3′UTR.
Fig. 4.
Identification of miR-24-3p as a potential upstream regulator of HPCA. a, b HeLa cells were transiently transfected with a control miRNA mimic or an miR-24-3p mimic for 2 days. aHPCA mRNA levels were detected by RT-qPCR. b Cells were lysed and analyzed by western blotting with anti-HPCA and anti-calnexin antibodies. The graph shows mean densities as fold increases from three independent experiments (mean ± SEM). Band intensities were quantified using Quantity One® software. ***P < 0.001 compared with the control miRNA mimic. c, d Cells were transfected with a control miRNA inhibitor or an miR-24-3p inhibitor for 3 days. cHPCA mRNA levels were analyzed by RT-qPCR. d Proteins were analyzed by western blotting with anti-HPCA and anti-calnexin antibodies. The graph shows mean densities as fold increases from three independent experiments (mean ± SEM). Band intensities were quantified with Quantity One® software. **P < 0.01 compared with the control miRNA inhibitor
miR-24-3p modulated neuronal differentiation by inhibiting HPCA in SH-SY5Y cells
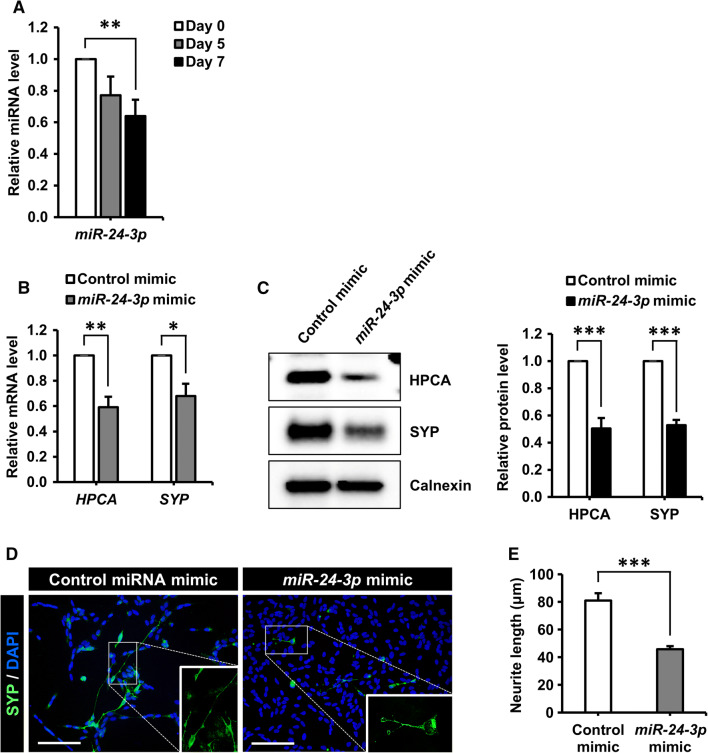
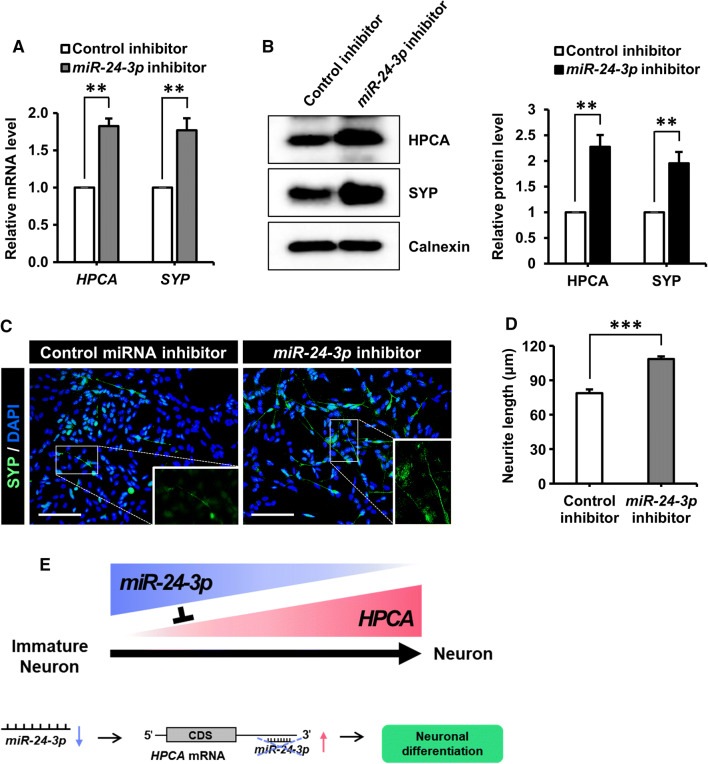
These findings prompted us to test whether the downregulation of miR-24-3p was responsible for the increased level of HPCA expression during neuronal differentiation. SH-SY5Y cells were differentiated and miR-24-3p levels were quantified by TaqMan qPCR assay (Fig. 5a). miR-24-3p levels were downregulated by approximately 40%, suggesting that decreased miR-24-3p expression may have been responsible for the increase in HPCA mRNA levels. To elucidate the role of miR-24-3p in the neurogenesis of SH-SY5Y cells, we transfected cells with an miR-24-3p mimic or a control mimic and then induced neuronal differentiation by RA treatment for 7 days. The miR-24-3p mimic suppressed SYP expression, as well as HPCA mRNA (Fig. 5b) and protein (Fig. 5c) levels. Moreover, the miR-24-3p mimic significantly decreased neurite outgrowth (81.0 ± 5.3 μm versus 45.7 ± 2.3 μm, P < 0.001, Fig. 5d, e) compared with the control mimic, indicating that miR-24-3p may regulate neuronal differentiation by targeting HPCA in SH-SY5Y cells. To further confirm the role of miR-24-3p in the neuronal differentiation of SH-SY5Y cells, we transfected cells with an miR-24-3p inhibitor or a control inhibitor. As expected, miR-24-3p inhibition promoted the levels of SYP and HPCA, when compared with the control inhibitor (Fig. 6a, b). Moreover, the miR-24-3p inhibitor significantly increased neurite outgrowth (78.7 ± 3.1 μm versus 108.4 ± 2.3 μm, P < 0.001, Fig. 6c, d) during the neuronal differentiation of SH-SY5Y cells. Taken together, these results demonstrated that miR-24-3p modulated neuronal differentiation by targeting HPCA expression in SH-SY5Y cells.
Fig. 5.
miR-24-3p regulates neuronal differentiation, presumably by reducing HPCA expression in SH-SY5Y cells. a Mature miR-24-3p levels during the differentiation of SH-SY5Y cells were quantified using TaqMan RT-qPCR. **P < 0.01 compared with day 0 of differentiation (mean ± SEM; n = 4). b, c Differentiation was induced in SH-SY5Y cells transfected with a control miRNA mimic or an miR-24-3p mimic. bHPCA and SYP mRNA levels were analyzed by RT-qPCR. c HPCA, SYP, and calnexin protein levels were determined by western blotting. The graph shows mean densities as fold increases from four independent experiments (mean ± SEM). Band intensities were quantified using Quantity One® software. *P < 0.05, **P < 0.01, ***P < 0.001 compared with the control miRNA mimic at day 7 of differentiation. d Cells were transfected with a control miRNA mimic or an miR-24-3p mimic and induced to differentiate for 7 days. Fixed cells were stained with an anti-SYP antibody (green) and DAPI (blue). Scale bar, 100 μm. e Neurite lengths were measured in randomly selected areas from three experiments (mean ± SEM). ***P < 0.001 compared with the control miRNA mimic at day 7 of differentiation
Fig. 6.
Inhibition of miR-24-3p promoted the neuronal differentiation of SH-SY5Y cells. a, b SH-SY5Y cells at day 2 of differentiation were transfected with a control miRNA inhibitor or an miR-24-3p inhibitor and were allowed to differentiate for 5 days. aHPCA and SYP mRNA levels during the differentiation period were detected by RT-qPCR. b Cells were lysed and analyzed by western blotting with anti-HPCA, anti-SYP, and anti-calnexin antibodies. The graph shows mean densities as fold increases from four independent experiments (mean ± SEM). Band intensities were quantified using Quantity One® software. **P < 0.01 compared with the control miRNA inhibitor at day 7 of differentiation. c Immunofluorescence was used to visualize SYP (green) at day 7 of differentiation with a control miRNA inhibitor or an miR-24-3p inhibitor. Nuclei were stained with DAPI (blue). Scale bar, 100 μm. d Neurite lengths were measured in random areas from five cultures (mean ± SEM). ***P < 0.001 compared with the control miRNA inhibitor. e The schematic model suggests that the inhibition of miR-24-3p promotes neuronal differentiation by increasing HPCA expression levels in SH-SY5Y cells
Discussion
HPCA has multiple functions in various cellular processes, including Ca2+ buffering, neuroprotection, and receptor-mediated endocytosis, by detecting and binding calcium to increase intracellular calcium influx [25, 26, 34]. We have previously shown that HPCA promotes NeuroD expression, resulting in neurite outgrowth during differentiation of H19-7 cells [28]. Furthermore, our previous study demonstrated that HPCA promotes neurogenesis by regulating the PKCα/PKD1/PLD1/SHP-1/STAT3 pathway in neural stem cells [16]. However, the molecular mechanisms involved in the regulation of HPCA expression during neuronal differentiation have not been studied yet. In the present study, we demonstrated a novel mechanism whereby HPCA expression is regulated by miR-24-3p in SH-SY5Y cells.
We observed that HPCA levels were markedly increased during the neuronal differentiation of SH-SY5Y cells. HPCA depletion significantly decreased neurite outgrowth and the expression of the neuronal marker, SYP, whereas overexpression of HPCA increased neurite outgrowth and SYP expression. These results strongly indicate that HPCA is required for the neuronal differentiation of SH-SY5Y cells. This is supported by a previous study showing that HPCA plays an important role in neuronal differentiation in various cell types, including H19-7 cells [28] and cortical neural stem cells [16].
miRNAs are key regulators of gene expression through the translational repression and/or degradation of target transcripts [35–37]. By focusing on miRNA-dependent gene regulation, we integrated bioinformatics-based predictions and found that the 3′UTR of HPCA contained two putative miR-24-3p binding sites. miRNAs partially bind to homologous sequences in the 3′UTR of their target genes [38, 39]. Using a dual-luciferase reporter system, we demonstrated that only the predicted miR-24-3p binding site at position 675-681 within the HPCA 3′UTR is a target for miR-24-3p. HPCA expression was downregulated by miR-24-3p at the mRNA and protein levels. However, it is not clear how miR-24-3p affects the gene expression levels of HPCA. Therefore, further studies will be focused on identifying mechanisms by which miR-24-3p regulates mRNA degradation and/or translation inhibition of HPCA.
Thus far, most research on miR-24-3p has focused on its function in cancer using in vitro model systems. For instance, up or downregulation of miR-24 is important for regulation of the growth and/or apoptosis of various cancer types, including breast cancer [40], lung adenocarcinoma [41], hepatocellular carcinoma [42], and colon cancer [43]. In the present study, we investigated its function in neuronal differentiation. miR-24-3p was found to modulate HPCA-mediated neurite outgrowth and neuronal marker expression in SH-SY5Y cells. However, since miR-24-3p mimics did not completely inhibit the expression of HPCA mRNA and protein levels, the possibility of other miRNAs involved in HPCA-mediated neuronal differentiation cannot be ruled out. Therefore, additional experiments are needed to identify HPCA target miRNAs and this will provide a more complete picture of HPCA regulation by miRNAs during neural development.
In conclusion, our results indicated that miR-24-3p directly targeted HPCA to regulate the neuronal differentiation of SH-SY5Y cells. To the best of our knowledge, this is the first demonstration of miR-24-3p inhibition promoting neuronal differentiation by increasing the expression levels of HPCA. Therefore, our findings suggested that miR-24-3p controlled neuronal differentiation by modulating HPCA expression and implicated the miR-24-3p-mediated regulation of HPCA expression as a potential therapeutic target for neuron development disorders in humans. The findings of the present study are summarized in Fig. 6e.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Science, ICT, and Future Planning, Republic of Korea (NRF-2018R1A1A1A05022185).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shin-Young Park, Phone: 82-2-2220-0610, FAX: 82-2-2220-2422, Email: ttokttoki@hanyang.ac.kr.
Joong-Soo Han, Phone: 82-2-2220-0623, FAX: 82-2-2220-2422, Email: jshan@hanyang.ac.kr.
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10(2):126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 3.Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336(6078):237–240. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 5.Luo W, Nie Q, Zhang X. MicroRNAs involved in skeletal muscle differentiation. J Genet Genomics. 2013;40(3):107–116. doi: 10.1016/j.jgg.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 6.Petri R, Malmevik J, Fasching L, Akerblom M, Jakobsson J. miRNAs in brain development. Exp Cell Res. 2014;321(1):84–89. doi: 10.1016/j.yexcr.2013.09.022. [DOI] [PubMed] [Google Scholar]
- 7.Stappert L, Roese-Koerner B, Brustle O. The role of microRNAs in human neural stem cells, neuronal differentiation and subtype specification. Cell Tissue Res. 2015;359(1):47–64. doi: 10.1007/s00441-014-1981-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ponnusamy M, Li PF, Wang K. Understanding cardiomyocyte proliferation: an insight into cell cycle activity. Cell Mol Life Sci. 2017;74(6):1019–1034. doi: 10.1007/s00018-016-2375-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miska EA, Alvarez-Saavedra E, Townsend M, Yoshii A, Sestan N, Rakic P, Constantine-Paton M, Horvitz HR. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5(9):R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lau P, Hudson LD. MicroRNAs in neural cell differentiation. Brain Res. 2010;1338:14–19. doi: 10.1016/j.brainres.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang T, Liu Y, Huang M, Zhao X, Cheng L. Wnt1-cre-mediated conditional loss of Dicer results in malformation of the midbrain and cerebellum and failure of neural crest and dopaminergic differentiation in mice. J Mol Cell Biol. 2010;2(3):152–163. doi: 10.1093/jmcb/mjq008. [DOI] [PubMed] [Google Scholar]
- 12.Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317(5842):1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wayman GA, Davare M, Ando H, Fortin D, Varlamova O, Cheng HY, Marks D, Obrietan K, Soderling TR, Goodman RH, Impey S. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc Natl Acad Sci USA. 2008;105(26):9093–9098. doi: 10.1073/pnas.0803072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanuki R, Onishi A, Koike C, Muramatsu R, Watanabe S, Muranishi Y, Irie S, Uneo S, Koyasu T, Matsui R, Cherasse Y, Urade Y, Watanabe D, Kondo M, Yamashita T, Furukawa T. miR-124a is required for hippocampal axogenesis and retinal cone survival through Lhx2 suppression. Nat Neurosci. 2011;14(9):1125–1134. doi: 10.1038/nn.2897. [DOI] [PubMed] [Google Scholar]
- 15.Capitano F, Camon J, Licursi V, Ferretti V, Maggi L, Scianni M, Del Vecchio G, Rinaldi A, Mannironi C, Limatola C, Presutti C, Mele A. MicroRNA-335-5p modulates spatial memory and hippocampal synaptic plasticity. Neurobiol Learn Mem. 2017;139:63–68. doi: 10.1016/j.nlm.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 16.Park SY, Yoon SN, Kang MJ, Lee Y, Jung SJ, Han JS. Hippocalcin Promotes Neuronal Differentiation and Inhibits Astrocytic Differentiation in Neural Stem Cells. Stem Cell Rep. 2017;8(1):95–111. doi: 10.1016/j.stemcr.2016.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Z, Sun Y, Cao S, Zhang J, Wei J. Downregulation of miR-24-3p promotes osteogenic differentiation of human periodontal ligament stem cells by targeting SMAD family member 5. J Cell Physiol. 2019;234(5):7411–7419. doi: 10.1002/jcp.27499. [DOI] [PubMed] [Google Scholar]
- 18.Zhang H, Duan J, Qu Y, Deng T, Liu R, Zhang L, Bai M, Li J, Ning T, Ge S, Wang X, Wang Z, Fan Q, Li H, Ying G, Huang D, Ba Y. Onco-miR-24 regulates cell growth and apoptosis by targeting BCL2L11 in gastric cancer. Protein Cell. 2016;7(2):141–151. doi: 10.1007/s13238-015-0234-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobayashi M, Takamatsu K, Saitoh S, Miura M, Noguchi T. Molecular cloning of hippocalcin, a novel calcium-binding protein of the recoverin family exclusively expressed in hippocampus. Biochem Biophys Res Commun. 1993;196(2):1017. doi: 10.1006/bbrc.1993.2351. [DOI] [PubMed] [Google Scholar]
- 20.Saitoh S, Takamatsu K, Kobayashi M, Noguchi T. Distribution of hippocalcin mRNA and immunoreactivity in rat brain. Neurosci Lett. 1993;157(1):107–110. doi: 10.1016/0304-3940(93)90654-4. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi M, Takamatsu K, Saitoh S, Noguchi T. Myristoylation of hippocalcin is linked to its calcium-dependent membrane association properties. J Biol Chem. 1993;268(25):18898–18904. [PubMed] [Google Scholar]
- 22.Braunewell KH, Gundelfinger ED. Intracellular neuronal calcium sensor proteins: a family of EF-hand calcium-binding proteins in search of a function. Cell Tissue Res. 1999;295(1):1–12. doi: 10.1007/s004410051207. [DOI] [PubMed] [Google Scholar]
- 23.O’Callaghan DW, Ivings L, Weiss JL, Ashby MC, Tepikin AV, Burgoyne RD. Differential use of myristoyl groups on neuronal calcium sensor proteins as a determinant of spatio-temporal aspects of Ca2+ signal transduction. J Biol Chem. 2002;277(16):14227–14237. doi: 10.1074/jbc.M111750200. [DOI] [PubMed] [Google Scholar]
- 24.O’Callaghan DW, Tepikin AV, Burgoyne RD. Dynamics and calcium sensitivity of the Ca2+/myristoyl switch protein hippocalcin in living cells. J Cell Biol. 2003;163(4):715–721. doi: 10.1083/jcb.200306042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aravind P, Chandra K, Reddy PP, Jeromin A, Chary KV, Sharma Y. Regulatory and structural EF-hand motifs of neuronal calcium sensor-1: Mg2+ modulates Ca2+ binding, Ca2+ -induced conformational changes, and equilibrium unfolding transitions. J Mol Biol. 2008;376(4):1100–1115. doi: 10.1016/j.jmb.2007.12.033. [DOI] [PubMed] [Google Scholar]
- 26.Palmer CL, Lim W, Hastie PG, Toward M, Korolchuk VI, Burbidge SA, Banting G, Collingridge GL, Isaac JT, Henley JM. Hippocalcin functions as a calcium sensor in hippocampal LTD. Neuron. 2005;47(4):487–494. doi: 10.1016/j.neuron.2005.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saitoh S, Takamatsu K, Kobayashi M, Noguchi T. Expression of hippocalcin in the developing rat brain. Brain Res Dev Brain Res. 1994;80(1–2):199–208. doi: 10.1016/0165-3806(94)90105-8. [DOI] [PubMed] [Google Scholar]
- 28.Oh DY, Cho JH, Park SY, Kim YS, Yoon YJ, Yoon SH, Chung KC, Lee KS, Han JS. A novel role of hippocalcin in bFGF-induced neurite outgrowth of H19-7 cells. J Neurosci Res. 2008;86(7):1557–1565. doi: 10.1002/jnr.21602. [DOI] [PubMed] [Google Scholar]
- 29.Kang MJ, Park SY, Han JS. Hippocalcin is required for astrocytic differentiation through activation of Stat3 in hippocampal neural precursor cells. Front Mol Neurosci. 2016;9:110. doi: 10.3389/fnmol.2016.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Radio NM, Mundy WR. Developmental neurotoxicity testing in vitro: models for assessing chemical effects on neurite outgrowth. Neurotoxicology. 2008;29(3):361–376. doi: 10.1016/j.neuro.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 31.Korecka JA, van Kesteren RE, Blaas E, Spitzer SO, Kamstra JH, Smit AB, Swaab DF, Verhaagen J, Bossers K. Phenotypic characterization of retinoic acid differentiated SH-SY5Y cells by transcriptional profiling. PLoS One. 2013;8(5):e63862. doi: 10.1371/journal.pone.0063862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheung YT, Lau WK, Yu MS, Lai CS, Yeung SC, So KF, Chang RC. Effects of all-trans-retinoic acid on human SH-SY5Y neuroblastoma as in vitro model in neurotoxicity research. Neurotoxicology. 2009;30(1):127–135. doi: 10.1016/j.neuro.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 33.Agholme L, Lindstrom T, Kagedal K, Marcusson J, Hallbeck M. An in vitro model for neuroscience: differentiation of SH-SY5Y cells into cells with morphological and biochemical characteristics of mature neurons. J Alzheimers Dis. 2010;20(4):1069–1082. doi: 10.3233/JAD-2010-091363. [DOI] [PubMed] [Google Scholar]
- 34.Mercer EA, Korhonen L, Skoglosa Y, Olsson PA, Kukkonen JP, Lindholm D. NAIP interacts with hippocalcin and protects neurons against calcium-induced cell death through caspase-3-dependent and -independent pathways. EMBO J. 2000;19(14):3597–3607. doi: 10.1093/emboj/19.14.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol. 2009;21(3):452–460. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 36.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12(2):99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 37.Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012;336(6078):233–237. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Finnegan EF, Pasquinelli AE. MicroRNA biogenesis: regulating the regulators. Crit Rev Biochem Mol Biol. 2013;48(1):51–68. doi: 10.3109/10409238.2012.738643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han X, Li Q, Liu C, Wang C, Li Y. Overexpression miR-24-3p repressed Bim expression to confer tamoxifen resistance in breast cancer. J Cell Biochem. 2019;120(8):12966–12976. doi: 10.1002/jcb.28568. [DOI] [PubMed] [Google Scholar]
- 41.Su C, Shi K, Cheng X, Han Y, Li Y, Yu D, Liu Z. Long noncoding RNA LINC00472 inhibits proliferation and promotes apoptosis of lung adenocarcinoma cells via regulating miR-24-3p/DEDD. Technol Cancer Res Treat. 2018;17:1533033818790490. doi: 10.1177/1533033818790490. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Fan JC, Zeng F, Le YG, Xin L. LncRNA CASC2 inhibited the viability and induced the apoptosis of hepatocellular carcinoma cells through regulating miR-24-3p. J Cell Biochem. 2018;119(8):6391–6397. doi: 10.1002/jcb.26479. [DOI] [PubMed] [Google Scholar]
- 43.Yin Y, Zhong J, Li SW, Li JZ, Zhou M, Chen Y, Sang Y, Liu L. TRIM11, a direct target of miR-24-3p, promotes cell proliferation and inhibits apoptosis in colon cancer. Oncotarget. 2016;7(52):86755–86765. doi: 10.18632/oncotarget.13550. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.