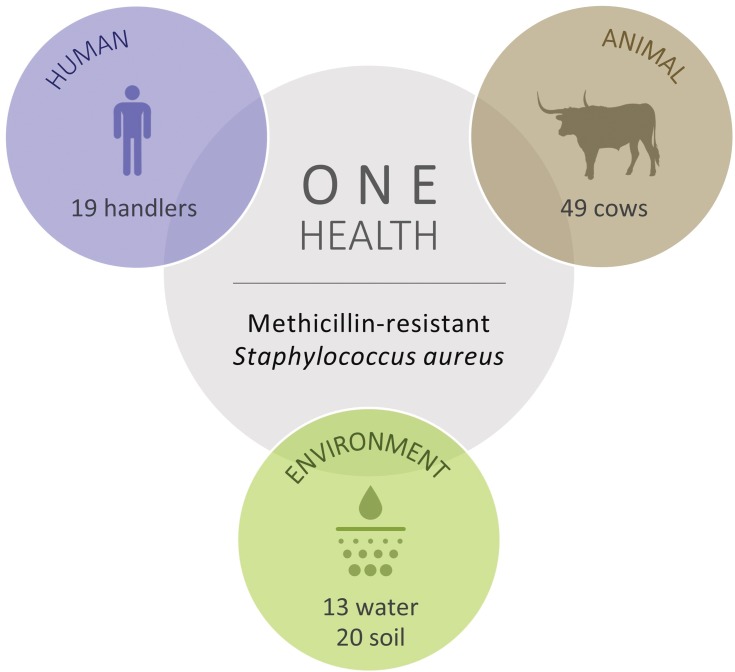
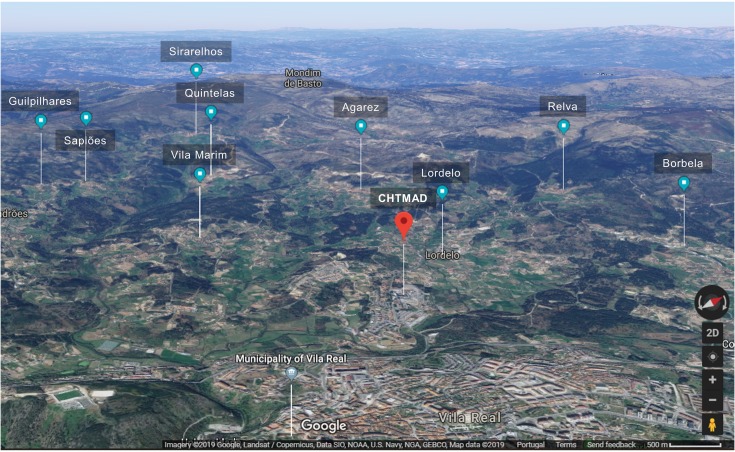
Antimicrobial resistance (AMR) is acknowledged today as one of the most concerning threats to global human health since the future and confines of modern medicine continue to be defined and shaped by the success of antimicrobial therapy.1 As resistant bacteria may spread without recognizing human-animal or geographic borders, many major international organizations have stated that the rising threat of AMR must be addressed though a concerted and multisectoral One Health approach involving humans, animals and the environment. Staphylococcus aureus is a hardy, adaptable, opportunistic pathogenic agent that is able thrive in diverse environments.2–4 It is often present in the natural flora of the human and animal nose and skin, being also recovered from food, food production systems and the environment, causing a range of illnesses from minor skin infections and food poisoning to life-threatening diseases.2,3,5 Methicillin‐resistant Staphylococcus aureus (MRSA) represents a global pandemic threat being resistant to most beta-lactam antibiotics and to other major antimicrobial drug classes.2,3 MRSA was first established in the healthcare setting, emerging afterwards in the community, animals and in food of animal origin, disclosing novel MRSA reservoirs.4,5 Presently, S. aureus represents a leading cause of livestock infection, resulting in great economic losses in the food industry.4–6 Livestock-associated MRSA (LA-MRSA) has also been reported among human patients who have been in contact with infected or colonized animals, being the causing agent of sepsis, endocarditis, osteomyelitis, pneumonia and other major invasive infections.3,5,7 MRSA in animals was first reported in a case of bovine mastitis in the early 1970s and since then MRSA has been progressively identified in farm animals and in humans contacting with these animals, revealing MRSA as a major veterinary and zoonotic pathogen.4,7 In order to follow a One Health approach to determine the prevalence and transmission of MRSA in cattle, mouth and nose swabs of 49 healthy cows (11 Friesians and 28 cross-breeds) were recovered together with 19 human, 13 water and 20 soil samples from the animals’ handlers and environments (Figure 1). Sampling was performed from February to April 2019 in 9 different locations within a radius of approximately four kilometers from the CHTMAD hospital center in Vila Real, Portugal (Vila Marim, Sirarelhos, Lordelo, Sapiões, Guilpilhares, Quintelas, Relva, Borbela and Agarez; Figure 2). Mouth and nose swabs were obtained from all consenting handlers in regular contact with the sampled animals; water and soil samples were collected from different points of each corresponding drinking trough and cowshed, respectively. Swabs were collected into Stuart transport media; water was recovered into 500 mL PET flasks with 60 mg/L sodium thiosulphate and soil was gathered into appropriate zipper seal sample bags. Samples were processed in the same day or stored at 4°C for a maximum of 24 h. Swabs and soil samples were incubated into Brain Heart Infusion broth with 6.5% (w/v) NaCl for 48 h at 37°C and after seeded on Oxacillin Resistance Screening Agar Base (ORSAB) supplemented with 2 mg/L oxacillin. Water samples were filtered through 47 mm 0.2 μm filters that were further placed on ORSAB plates with 2 mg/L oxacillin. All plates were incubated for 24 to 48 h at 37°C and after screened for presumptive MRSA colonies. Livestock and livestock production systems are known to be potential reservoirs for MRSA clones with the capacity to cross the species barrier, endure host-adaptive evolution, and become established in human populations worldwide as successful epidemic lineages.3,6 Hence, it was expected that MRSA strains could be recovered among the animal, human and environmental samples screened, that would allow the study of the occurrence and transmission of different MRSA lineages and their resistance determinants. However, no presumptive MRSA colonies were recovered from the total of 101 samples screened in selective ORSAB media. Since cattle and their respective handlers and environments are ordinarily colonized with MRSA,4,7 the observed absence of this major human and animal pathogen represents a serendipitous result, with a very positive impact in food safety and public health. Although the proximity to the main hospital center of the region, samples were collected from extensive production systems in higher mountain rural areas. These systems mainly use natural resources and do not regularly use antimicrobials in subtherapeutic doses, which would result in lower levels of antibiotic pressure selecting for MRSA. Through a One Health approach, this study revealed that the screened Friesian and crossbreed cattle, their handlers and their surrounding environments do not represent reservoirs for MRSA, revealing that the studied cattle from Vila Real, Portugal are, in this aspect, safe from the zoonotic point of view. Nevertheless, studying the prevalence and transmission of methicillin-susceptible S. aureus harboring resistance determinants for other important antimicrobial drugs would be interesting as well as the extension of the study to other bacterial species that also represent major global AMR threats.
Figure 1.
Total number of cattle, human, water and soil samples screened for methicillin‐resistant Staphylococcus aureus (MRSA).
Figure 2.
Satellite 3D image of the 9 different sampling locations in relation to the CHTMAD hospital centre in Vila Real, Portugal.
Acknowledgments
This work was funded by the R&D Project CAREBIO2 - Comparative assessment of antimicrobial resistance in environmental biofilms through proteomics - towards innovative theranostic biomarkers, with reference NORTE-01-0145-FEDER-030101 and PTDC/SAU-INF/30101/2017, financed by the European Regional Development Fund (ERDF) through the Northern Regional Operational Program (NORTE 2020) and the Foundation for Science and Technology (FCT). This work was supported by the Associate Laboratory for Green Chemistry - LAQV which is financed by national funds from FCT/MCTES (UID/QUI/50006/2019). Vanessa Silva is supported by national funds through FCT/MCTES and by the European Social Fund through POCH/FSE under the PhD grant SFRH/BD/137947/2018.
Ethical Statements
This work was approved Institutionally by University of Trás-os-Montes and Alto Douro Board and written informed consent was obtained from all subjects.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Ahmad M, Khan AU. Global economic impact of antibiotic resistance: a review. J Glob Antimicrob Resist. 2019. doi: 10.1016/j.jgar.2019.05.024 [DOI] [PubMed] [Google Scholar]
- 2.Gajdács M. The continuing threat of methicillin-resistant Staphylococcus aureus. Antibiotics. 2019;8(2):52. doi: 10.3390/antibiotics8020052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dweba CC, Zishiri O, El Zowalaty M. Methicillin-resistant Staphylococcus aureus: livestock-associated, antimicrobial, and heavy metal resistance. Infect Drug Resist. 2018;11:2497–2509. doi: 10.2147/IDR.S175967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aires-de-Sousa M. Methicillin-resistant Staphylococcus aureus among animals: current overview. Clin Microbiol Infect. 2017;23(6):373–380. doi: 10.1016/j.cmi.2016.11.002 [DOI] [PubMed] [Google Scholar]
- 5.Boswihi SS, Udo EE. Methicillin-resistant Staphylococcus aureus: an update on the epidemiology, treatment options and infection control. Curr Med Res Pract. 2018;8(1):18–24. doi: 10.1016/j.cmrp.2018.01.001 [DOI] [Google Scholar]
- 6.Spoor LE, McAdam PR, Weinert LA, et al. Livestock origin for a human pandemic clone of community-associated methicillin-resistant Staphylococcus aureus. Baquero F, ed. MBio. 2013;4(4):e00356–13. doi: 10.1128/mBio.00356-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goerge T, Lorenz MB, van Alen S, Hübner N-O, Becker K, Köck R. MRSA colonization and infection among persons with occupational livestock exposure in Europe: prevalence, preventive options and evidence. Vet Microbiol. 2017;200:6–12. doi: 10.1016/j.vetmic.2015.10.027 [DOI] [PubMed] [Google Scholar]