Abstract
Purpose:
Recent efforts have demonstrated that radiomic features extracted from the peritumoral region, the area surrounding the tumor parenchyma, have clinical utility in various cancer types. However, as like any radiomic features, peritumoral features may also be unstable and/or non-reproducible. Hence, the purpose of this study was to assess the stability and reproducibility of computed tomography (CT) radiomic features extracted from the peritumoral regions of lung lesions where stability was defined as the consistency of a feature by different segmentations, and reproducibility was defined as the consistency of a feature to image acquisition.
Methods:
Stability was measured utilizing the “Moist run” dataset and reproducibility was measured utilizing the Reference Image Database to Evaluate Therapy Response test-retest dataset. Peritumoral radiomic features were extracted from incremental distances of 3–12 mm outside the tumor parenchyma segmentation. A total of 264 statistical, histogram and texture radiomic features were assessed from the selected peritumoral region-of-interests. All features (except wavelet texture features) were extracted using standardized algorithms defined by the Image Biomarker Standardization Initiative. Stability and reproducibility of features were assessed using concordance correlation coefficient. The clinical utility of stable and reproducible peritumoral features were tested in three previously published lung cancer datasets using overall survival as the endpoint.
Results:
Features found to be stable and reproducible, regardless of the peritumoral distances, included statistical, histogram and a subset of texture features suggesting that these features are less affected by changes size or shape differences of the peritumoral region due to different segmentations and image acquisitions. The stability and reproducibility of 3D Laws and wavelet texture features were inconsistent across all peritumoral distances. The analyses also revealed that a subset of features were consistently stable irrespective of the initial parameters (e.g., seed point) for a given segmentation algorithm. No significant differences were found for stability for features that were extracted from region-of-interests (ROIs) bounded by a lung parenchyma mask versus ROIs that were not bounded by a lung parenchyma mask (i.e., peritumoral regions that were allowed to extend outside of lung parenchyma). After testing the clinical utility of peritumoral features, stable and reproducible features were shown to be more likely to create repeatable models than unstable and non-reproducible features.
Conclusions:
This study identified a subset of stable and reproducible CT radiomic features extracted from the peritumoral region of lung lesions. The stable and reproducible features identified in this study could be applied to a feature selection pipeline for CT radiomic analyses. According to our findings, top performing features in models for overall survival are most likely to be stable and reproducible hence, it may be best practice to utilize them to achieve repeatable studies and reduce the creation of overfit models.
Keywords: Radiomics, lung cancer, CT, stability, reproducibility, quantitative imaging
Introduction
Radiomics is the process of converting standard-of-care medical images into quantitative image-based data that can subsequently analyzed using conventional biostatistics, machine learning methods, and artificial intelligence1. Conventional radiomic features based on shape, size, intensity, and texture are typically extracted from the intratumoral region-of-interest (ROI) to quantify the cancer phenotype2. These radiomic features, of which many are beyond visual acuity, have shown to be significantly associated with cancer detection, diagnosis, prognosis, prediction of response to treatment, and monitoring of disease status3–7. However, there has been a renewed interest in quantitative characterization of the peritumoral region, the area immediately surrounding the tumor parenchyma, since this region is involved in immune infiltration, blood and lymphatic vascular networks, and stromal inflammation8–11. Early efforts preceding the “modern era of radiomics” demonstrated that peritumoral image-based features have diagnostic and predictive utility12–15. Recent efforts have shown the clinical utility of peritumoral radiomic features in studies of lung, breast, and head and neck cancers16–21.
Prior studies have established that some radiomic features are sensitive to tumor segmentation and/or image acquisition hence unstable and non-reproducible22–24, where stability is defined as the consistency of a feature across different segmentations and reproducibility is defined as the consistency of a feature across image acquisition parameters such as patient position and respiration phase. Identifying stable and reproducible features is an important precursor prior to conducting analyses of radiomic data since features with low-fidelity will likely lead to spurious findings and unrepeatable models. Though aforementioned studies have characterized the stability22 and reproducibility23,24 of intratumoral radiomics, no such study to date has been conducted on peritumoral radiomic features.
To address the gap in this domain, we conducted a study to assess the stability and reproducibility of peritumoral radiomic features of lung lesions captured by thoracic computed tomography (CT) scans. This study is also different from prior work conducted on intratumoral radiomics in that the majority of the radiomic features that were evaluated in this study were standardized through algorithms defined by Image Biomarker Standardization Initiative25 (IBSI). To measure stability we utilized the “Moist run” dataset26 from The Cancer Imaging Archive and to measure reproducibility we utilized the Reference Image Database to Evaluate Therapy Response (RIDER) dataset that consists of test-retest data27. Peritumoral ROIs with incremental distances of 3 mm to 12 mm from the tumor boundary were generated by applying morphological image processing operations on tumor segmentation masks. The clinical utility of stable and reproducible peritumoral features was tested on three previously published lung cancer datasets using overall survival (OS) as the endpoint. The stable and reproducible features identified in this study could be applied to a feature selection pipeline for future CT radiomic analyses.
Materials and Methods
Moist run dataset
The “Moist run” dataset was utilized to measure radiomic feature stability. This dataset was constructed by the Quantitative Imaging Network as a lung segmentation challenge26 and consists of 40 CT images of 40 NSCLC patients from five collections of Digital Imaging and Communications Medicine series and one thoracic phantom. Each patient in the dataset had one lesion of interest and the phantom scan had 12 lesions of interest which totals to 52 lesions of interests. The images on this dataset were previously de-identified.
RIDER test-retest dataset
To measure reproducibility of radiomic features, the RIDER test-retest dataset was utilized27. This National Cancer Institute (NCI) dataset was developed to generate an initial consensus on how to harmonize data collection and analysis for quantitative imaging methods. This dataset consisted of 32 NSCLC patients who had two separate non-contrast chest CT scans that were acquired within 15 minutes of each other using the same scanner, acquisition and processing parameters. As such, the only variability between the test and re-test scans would be attributed to patient orientation, respiration, and movement. The images on this dataset were previously de-identified.
Prognostic lung cancer datasets
To test the applied utility of stable and reproducible peritumoral features, three previously published datasets were utilized. One dataset was used for training and two datasets for validation. The training dataset included 62 surgically resected lung adenocarcinoma patients from the H. Lee Moffitt Cancer Center & Research Institute who had CTs two months prior to surgery17,28. The first validation cohort included 47 lung adenocarcinoma patients from the Maastricht Radiation Oncology Clinic (MAASTRO), Maastricht, Netherlands17,28 and the second validation cohort included 103 adenocarcinoma patients who had pre-surgery CTs for radio-genomic analysis29.
Segmentation algorithms
The lesions on the Moist run dataset were previously segmented using 3 different semi-automatic segmentation algorithms. Each segmentation algorithm was implemented using three different initial parameters (i.e., seed point or bounding circle, Supplementary Fig. S1) hence; 9 segmentations per lesion were acquired. Algorithm 1 uses marker-controlled watersheds, geometric active contours and Markov random fields inside a user drawn bounding circle ROI surrounding the lesion. Algorithm 2 requires a single-click inside the lesion as an initial parameter which then automatically generates multiple seed points inside the tumor. Subsequently, a click and grow algorithm was used to generate multiple segmentations that are combined to generate a consensus segmentation. Algorithm 3 uses a “seed circle” as an initial parameter and applies a two-dimensional region growing technique followed by automatic removal of blood vessels and lung parenchyma. Further details of the segmentation algorithms were previously described elsewhere26.
The lesions on the RIDER dataset were previously segmented using a semi-automatic single-click ensemble region growing segmentation algorithm on the Lung Tumor Analysis software program platform (Definiens Developer XD©, Munich, Germany)30. The segmentation workflow contained 4 steps: 1) Pre-processing of automatic organ segmentation; 2) Semi-automated correction of pulmonary boundary; 3) Click and Grow execution; 4) A manual refinement by an expert if needed. Further details of the segmentation algorithms were previously published elsewhere23.
Peritumoral masks
Peritumoral masks were generated as a natural extension of the tumor segmentations by using morphological image processing operations. A disk-shaped structural element with a radius of intended peritumoral distance was used for morphological dilation on tumor segmentations, followed by removal of the tumor region to create “doughnut-shaped” peritumoral masks. Intervals of 3, 6, 9 and 12 mm outside the tumor region created the peritumoral masks. For the first analysis, the peritumoral regions were bounded by the lung parenchyma and for the second analysis the peritumoral regions were not bounded by lung parenchyma (i.e., peritumoral regions were allowed to extend outside of lung parenchyma, Fig. 1). The MATLAB® (version 2018a) scripts to create peritumoral masks from intratumoral masks are available at https://github.com/TunaliIlke/peritumoral_regions/.
Figure 1. Peritumoral masks.
Image on the left shows a CT scan ROI. Image on the middle image shows a peritumoral region that is not bounded by the lung parenchyma mask. Image on the right shows peritumoral region bounded by the lung parenchyma mask. Red region is removed from the peritumoral region as it lies outside the lung parenchyma.
Radiomic features
All images were linearly resampled to a single voxel spacing of 1mm × 1mm × 1mm to standardize spacing across all images. A total of 264 statistical, histogram and texture radiomic features (Supplementary Table 1A) were extracted from the selected peritumoral and intratumoral ROIs using in-house toolboxes created in C++ (https://isocpp.org). Texture features included gray-level co-occurrence matrix (GLCM), gray level run-length matrix (GLRLM), gray level size zone matrix (GLSZM) and neighboring gray tone difference matrix (NGTDM), 3D Laws and wavelet features. All features (except wavelet texture features are defined elsewhere31) were extracted using standardized algorithms defined by the IBSI v525. Histogram, GLCM, GLRLM, GLSZM and NGTDM texture features were extracted using a common bin width of 25 Hounsfield units (HU). Additionally, 41 IBSI standardized shape and size features were extracted from intratumoral masks (Supplementary Table 1B).
Statistical analyses
Statistical analyses were performed using Intercooled Stata/MP 14.2 (StataCorp LP, College Station, TX) and R Project for Statistical Computing version 2.13.1 (http://www.r-project.org). Stability and reproducibility of features were assessed using concordance correlation coefficient (CCC)32. For each feature, CCCs were calculated between different segmentation algorithms, initial parameters and test-retest scans. The CCC values range from 1 to −1, where 1 indicates a perfect correlation between two variables. Similarity between different segmentation approaches were computed using the Jaccard index:
| (1) |
where and are the two segmentation masks being compared. Differences between initial parameters and algorithms by varying distances were tested using Fisher’s exact tests.
Details of the survival analysis are described in the Supplemental Methods. Briefly, survival analyses were performed using Kaplan-Meier survival estimates and the log-rank test33. Overall survival (OS) was the main endpoint for these analyses and an event was defined as date of death. OS was assessed from date of first treatment (e.g., surgery) to the date of death or date of last follow-up. The survival data were right censored at 60-months. All P-values were 2-sided and a P-value less than or equally to 0.05 was deemed statistically significant.
Results
Table 1 presents the similarities between segmentations using Jaccard indices between different initial parameters and algorithms being used. The results demonstrate high similarities (Jaccard index > 0.90) between segmentations that were computed using different initial parameters. On the other hand, moderate similarities (Jaccard index > 0.80) were observed between segmentations that were computed using different segmentation algorithms.
Table 1.
Initial parameter and algorithm comparison by Jaccard index scores.
| Initial parameter comparisons | Jaccard index | ||
|---|---|---|---|
| Algorithm 1-Initial parameter 1 vs Initial parameter 2 | 0.973 | ||
| Algorithm 1-Initial parameter 1 vs Initial parameter 3 | 0.969 | ||
| Algorithm 1-Initial parameter 2 vs Initial parameter 3 | 0.979 | ||
| Algorithm 2-Initial parameter 1 vs Initial parameter 2 | 0.948 | ||
| Algorithm 2-Initial parameter 1 vs Initial parameter 3 | 0.955 | ||
| Algorithm 2-Initial parameter 2 vs Initial parameter 3 | 0.962 | ||
| Algorithm 3-Initial parameter 1 vs Initial parameter 2 | 0.943 | ||
| Algorithm 3-Initial parameter 1 vs Initial parameter 3 | 0.955 | ||
| Algorithm 3-Initial parameter 2 vs Initial parameter 3 | 0.942 | ||
| Algorithm comparisons1 | |||
| Algorithm 1 vs Algorithm 2 | 0.810 | ||
| Algorithm 1 vs Algorithm 3 | 0.827 | ||
| Algorithm 2 vs Algorithm 3 | 0.805 | ||
Algorithms were compared using segmentations created by random selections of initial parameters (1, 2 or 3) for each lesion.
Peritumoral features
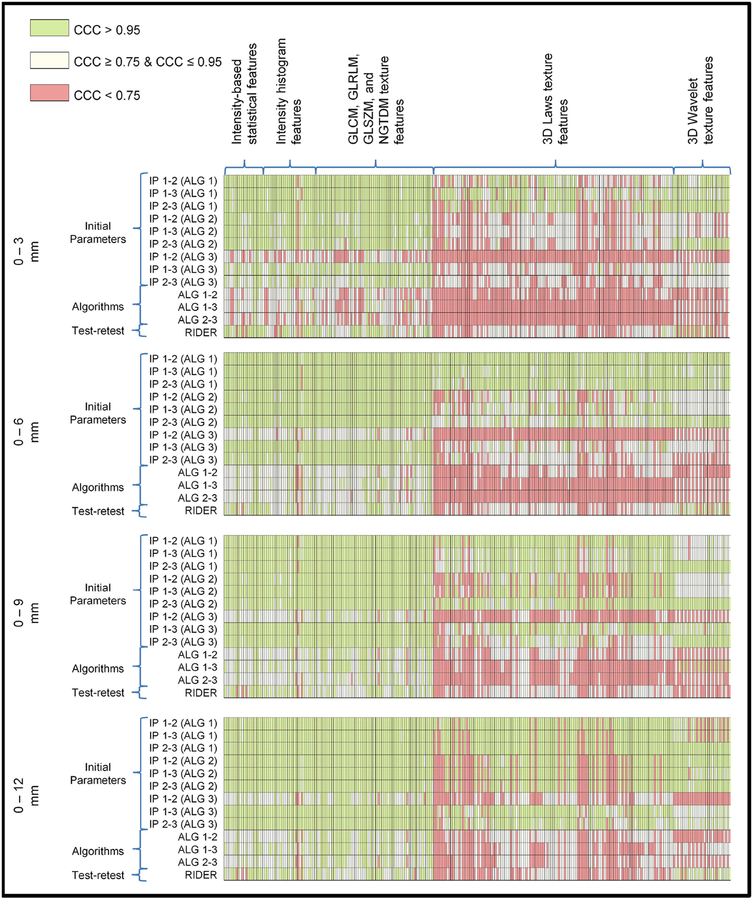
Figure 2 presents CCC groups (high, moderate, low) of peritumoral radiomic features with respect to different algorithms and different initial parameters. The green boxes represent high (CCC > 0.95), yellow boxes represent moderate (CCC ≥ 0.75 & CCC ≤ 0.95) and red boxes represent low (CCC < 0.75) CCCs. A high CCC indicates that the radiomic feature is not sensitive to variation in segmentations, whereas a low CCC indicates that radiomic feature is sensitive to the difference in segmentations. As the peritumoral distance increased, there were significantly higher numbers of moderate or highly stable features (Table 2). The statistical, histogram, and a subset of texture features (GLCM, GLRLM, GLSZM and NGTDM) were found stable (Supplementary Table 2a–c) and reproducible (Table 3) for different initial parameters however, 3D Laws and wavelet texture features (Supplementary Table 2d–e) were found to be significantly less stable and reproducible. Overall, the inter-stability (i.e., stability of features across different segmentation algorithms) was observed to be significantly lower than the intra-stability (i.e., stability of features across different initial parameters using same segmentation algorithms). The overall reproducibility of features were not significantly different as peritumoral distances changed; although a subset of texture features (GLCM, GLRLM, GLSZM and NGTDM) were slightly more reproducible for peritumoral distances above 3 mm (Table 3).
Figure 2. Concordance correlation coefficient groups of peritumoral features bounded by lung parenchyma.
The green boxes represent higher (CCC > 0.95), yellow boxes represent moderate (CCC ≥ 0.75 & CCC ≤ 0.95) and red boxes represent lower (CCC < 0.75) CCCs.
Table 2.
Distribution of stability groups of all peritumoral radiomic features extracted from region-of-interests that are bounded by a lung mask.
| Distancea | |||||||
|---|---|---|---|---|---|---|---|
| 0 – 3 mm | 0 – 6 mm | 0 – 9 mm | 0 – 12 mm | P- Valueb | |||
| Algorithm 1-initial parameter 1 vs initial parameter 2 | |||||||
| CCC < 0.75 (Red) | 30 (11.4) | 0 (0) | 10 (3.8) | 21 (8.0) | |||
| CCC ≥ 0.75 & CCC ≤ 0.95 (Yellow) | 63 (23.9) | 9 (3.4) | 26 (9.9) | 8 (3.0) | |||
| CCC > 0.95 (Green) | 171 (64.7) | 255 (96.6) | 228 (86.3) | 235 (89.0) | <0.001 | ||
| P-value | <0.001 | <0.001 | 0.001 | ||||
| Algorithm 1-initial parameter 1 vs initial parameter 3 | |||||||
| CCC < 0.75 (Red) | 14 (5.3) | 1 (0.4) | 9 (3.4) | 29 (11.0) | |||
| CCC ≥ 0.75 & CCC ≤ 0.95 (Yellow) | 55 (20.8) | 32 (12.1) | 63 (23.9) | 8 (3.0) | |||
| CCC > 0.95 (Green) | 195 (73.9) | 231 (87.5) | 192 (72.7) | 227 (86.0) | <0.001 | ||
| P-value | <0.001 | <0.001 | <0.001 | ||||
| Algorithm 1-initial parameter 2 vs initial parameter 3 | |||||||
| CCC < 0.75 (Red) | 33 (12.5) | 2 (0.8) | 8 (3.0) | 10 (7.6) | |||
| CCC ≥ 0.75 & CCC ≤ 0.95 (Yellow) | 34 (12.9) | 24 (9.1) | 38 (14.4) | 0 (0) | |||
| CCC > 0.95 (Green) | 197 (74.6) | 238 (90.1) | 218 (85.6) | 244 (92.4) | <0.001 | ||
| P-value | <0.001 | 0.022 | <0.001 | ||||
| Algorithm 2-initial parameter 1 vs initial parameter 2 | |||||||
| CCC < 0.75 (Red) | 57 (21.6) | 36 (13.6) | 44 (16.7) | 44 (16.7) | |||
| CCC ≥ 0.75 & CCC ≤ 0.95 (Yellow) | 122 (46.2) | 86 (32.6) | 52 (19.7) | 11 (4.2) | |||
| CCC > 0.95 (Green) | 85 (32.2) | 142 (53.8) | 168 (63.6) | 209 (79.1) | <0.001 | ||
| P-value | <0.001 | 0.004 | <0.001 | ||||
| Algorithm 2-initial parameter 1 vs initial parameter 3 | |||||||
| CCC < 0.75 (Red) | 40 (15.2) | 22 (8.3) | 37 (14.0) | 45 (17.1) | |||
| CCC ≥ 0.75 & CCC ≤ 0.95 (Yellow) | 124 (47.0) | 76 (28.8) | 39 (14.8) | 10 (3.8) | |||
| CCC > 0.95 (Green) | 100 (37.8) | 166 (62.9) | 188 (71.2) | 209 (79.1) | <0.001 | ||
| P-value | <0.001 | <0.001 | <0.001 | ||||
| Algorithm 2-initial parameter 2 vs initial parameter 3 | |||||||
| CCC < 0.75 (Red) | 55 (20.8) | 6 (2.3) | 8 (3.0) | 45 (17.1) | |||
| CCC ≥ 0.75 & CCC ≤ 0.95 (Yellow) | 95 (36.0) | 74 (28.0) | 23 (8.7) | 10 (3.8) | |||
| CCC > 0.95 (Green) | 114 (43.2) | 184 (69.7) | 233 (88.3) | 209 (79.1) | <0.001 | ||
| P-value | <0.001 | <0.001 | <0.001 | ||||
| Algorithm 3-initial parameter 1 vs initial parameter 2 | |||||||
| CCC < 0.75 (Red) | 182 (68.9) | 146 (55.3) | 121 (45.8) | 97 (36.7) | |||
| CCC ≥ 0.75 & CCC ≤ 0.95 (Yellow) | 78 (29.6) | 99 (37.5) | 108 (40.9) | 122 (46.2) | |||
| CCC > 0.95 (Green) | 4 (1.5) | 19 (7.2) | 35 (13.3) | 45 (17.1) | <0.001 | ||
| P-value | <0.001 | 0.023 | 0.096 | ||||
| Algorithm 3-initial parameter 1 vs initial parameter 3 | |||||||
| CCC < 0.75 (Red) | 41 (15.5) | 33 (12.5) | 19 (7.2) | 11 (4.2) | |||
| CCC ≥ 0.75 & CCC ≤ 0.95 (Yellow) | 133 (50.4) | 103 (39.0) | 42 (15.9) | 45 (17.1) | |||
| CCC > 0.95 (Green) | 90 (34.1) | 128 (48.5) | 203 (76.9) | 208 (78.7) | <0.001 | ||
| P-value | 0.004 | <0.001 | 0.335 | ||||
| Algorithm 3-initial parameter 2 vs initial parameter 3 | |||||||
| CCC < 0.75 (Red) | 49 (18.6) | 48 (18.2) | 30 (11.4) | 12 (4.5) | |||
| CCC ≥ 0.75 & CCC ≤ 0.95 (Yellow) | 156 (59.1) | 112 (42.4) | 66 (25.0) | 49 (18.6) | |||
| CCC > 0.95 (Green) | 59 (22.3) | 104 (39.4) | 168 (63.6) | 203 (76.9) | <0.001 | ||
| P-value | <0.001 | <0.001 | 0.001 | ||||
| Algorithm 1 vs Algorithm 2 | |||||||
| CCC < 0.75 (Red) | 148 (56.1) | 102 (38.6) | 69 (26.1) | 73 (27.6) | |||
| CCC ≥ 0.75 & CCC ≤ 0.95 (Yellow) | 108 (40.9) | 140 (53.0) | 151 (57.2) | 142 (53.8) | |||
| CCC > 0.95 (Green) | 8 (3.0) | 22 (8.4) | 44 (16.7) | 49 (18.6) | <0.001 | ||
| P-value | 0.001 | <0.001 | 0.722 | ||||
| Algorithm 1 vs Algorithm 3 | |||||||
| CCC < 0.75 (Red) | 165 (62.5) | 139 (52.6) | 118 (44.7) | 81 (30.7) | |||
| CCC ≥ 0.75 & CCC ≤ 0.95 (Yellow) | 84 (31.8) | 90 (34.1) | 76 (28.8) | 121 (45.8) | |||
| CCC > 0.95 (Green) | 15 (5.7) | 35 (13.3) | 70 (26.5) | 62 (23.5) | <0.001 | ||
| P-value | 0.005 | 0.001 | <0.001 | ||||
| Algorithm 2 vs Algorithm 3 | |||||||
| CCC < 0.75 (Red) | 180 (68.2) | 147 (55.7) | 128 (48.5) | 84 (31.8) | |||
| CCC ≥ 0.75 & CCC ≤ 0.95 (Yellow) | 77 (29.2) | 96 (36.4) | 105 (39.8) | 139 (52.7) | |||
| CCC > 0.95 (Green) | 7 (2.6) | 21 (7.9) | 31 (11.7) | 41 (15.5) | <0.001 | ||
| P-value | 0.002 | 0.158 | <0.001 | ||||
Numbers inside parenthesis are the percentage values.
P-values were generated using Fisher’s Exact test comparing 0–3 mm vs. 0–6 mm, 0–6 mm vs. 0–9 mm, and 0–9 mm vs. 0–12 mm, respectively.
P-value was generated using Fisher’s Exact test for the overall distributions of the four peritumoral distances (3 × 4 contingency table).
Table 3.
Distribution of reproducibility groups of peritumoral radiomic features extracted from region-of-interests that are bounded by a lung mask.
| Distancea | |||||||
|---|---|---|---|---|---|---|---|
| 0 – 3 mm | 0 – 6 mm | 0 – 9 mm | 0 – 12 mm | P- Valueb | |||
| All features | |||||||
| CCC < 0.75 (Red) | 68 (25.8) | 55 (20.8) | 71 (26.9) | 80 (30.3) | |||
| CCC ≥ 0.75 & CCC ≤ 0.95 (Yellow) | 121 (45.8) | 148 (56.1) | 138 (52.3) | 113 (42.8) | |||
| CCC > 0.95 (Green) | 75 (28.4) | 61 (23.1) | 55 (20.8) | 71 (26.9) | <0.001 | ||
| P-Value | 0.068 | 0.263 | 0.081 | ||||
| Statistical features | |||||||
| CCC < 0.75 (Red) | 3 (15.8) | 2 (10.5) | 3 (15.8) | 3 (15.8) | |||
| CCC ≥ 0.75 & CCC ≤ 0.95 (Yellow) | 1 (5.3) | 5 (26.3) | 4 (21.0) | 4 (21.0) | |||
| CCC > 0.95 (Green) | 15 (78.9) | 12 (63.2) | 12 (63.2) | 12 (63.2) | 0.731 | ||
| P-Value | 0.272 | 1.000 | 1.000 | ||||
| Histogram features | |||||||
| CCC < 0.75 (Red) | 5 (17.9) | 2 (7.1) | 4 (14.3) | 4 (14.3) | |||
| CCC ≥ 0.75 & CCC ≤ 0.95 (Yellow) | 7 (25.0) | 15 (53.6) | 10 (35.7) | 10 (35.7) | |||
| CCC > 0.95 (Green) | 16 (57.1) | 11 (39.3) | 14 (50.0) | 14 (50.0) | 0.512 | ||
| P-Value | 0.086 | 0.467 | 1.000 | ||||
| Texturec features | |||||||
| CCC < 0.75 (Red) | 3 (4.8) | 0 (0) | 0 (0) | 1 (1.6) | |||
| CCC ≥ 0.75 & CCC ≤ 0.95 (Yellow) | 28 (45.2) | 39 (62.9) | 33 (53.2) | 31 (50.0) | |||
| CCC > 0.95 (Green) | 31 (50.0) | 23 (37.1) | 29 (46.8) | 30 (48.4) | 0.198 | ||
| P-Value | 0.038 | 0.363 | 0.857 | ||||
| 3D Laws features | |||||||
| CCC < 0.75 (Red) | 44 (35.2) | 44 (35.2) | 44 (35.2) | 68 (54.4) | |||
| CCC ≥ 0.75 & CCC ≤ 0.95 (Yellow) | 79 (63.2) | 81 (64.8) | 81 (64.8) | 57 (45.6) | |||
| CCC > 0.95 (Green) | 2 (1.6) | 0 (0) | 0 (0) | 0 (0) | 0.002 | ||
| P-Value | 0.615 | 1.000 | 0.003 | ||||
| Wavelet features | |||||||
| CCC < 0.75 (Red) | 13 (43.3) | 7 (23.3) | 20 (66.7) | 4 (13.3) | |||
| CCC ≥ 0.75 & CCC ≤ 0.95 (Yellow) | 6 (20.0) | 8 (26.7) | 10 (33.3) | 11 (36.7) | |||
| CCC > 0.95 (Green) | 11 (36.7) | 15 (50.0) | 0 (0) | 15 (50.0) | <0.001 | ||
| P-Value | 0.305 | <0.001 | <0.001 | ||||
Numbers inside parenthesis are the percentage values.
P-values were generated using Fisher’s Exact test comparing 0–3 mm vs. 0–6 mm, 0–6mm vs. 0–9 mm, and 0–9 mm vs. 0–12 mm, respectively.
P-value was generated using Fisher’s Exact test for the overall distributions of the four peritumoral distance (3 × 4 contingency table).
Features consist GLCM, GLRLM, GLSZM and NGTDM texture features.
All aforementioned analyses were performed for features extracted from peritumoral regions that were bounded by a lung parenchyma mask (Supplementary Fig. S2). The stability of features where a lung parenchyma mask was not used to bound the peritumoral region was consistent with the analysis where a lung parenchyma mask was used (Supplementary Table 3a–f). However, peritumoral features were significantly more reproducible with the increasing peritumoral distances when lung parenchyma mask was not used (Supplementary Table 4). The exact CCC values of features are provided in Supplementary Table 1a.
Intratumoral features
Figure 3 presents CCC groups of intratumoral radiomic features with respect to different algorithms and different initial parameters. The majority of the features had low inter-stability (CCC < 0.75) while intra-stability of features were more frequently moderate or high (Supplementary Table 5a). Most size and shape features were found to be highly stable for different initial parameters (Supplementary Table 5g–h). Intensity, size, shape and a subset of a texture features (GLCM, GLRLM, GLSZM and NGTDM), features were at moderately or highly reproducible, while 3D Laws and wavelet texture features were less reproducible (Supplementary Table 6). For all feature categories, intratumoral features had lower median CCC values than their corresponding peritumoral features for both reproducibility (Fig. 4a) and stability (Fig. 4b) assessments. The exact CCC values of features are provided in Supplementary Table 1B.
Figure 3. Concordance correlation coefficient groups of intratumoral features.
The green boxes represent higher (CCC > 0.95), yellow boxes represent moderate (CCC ≥ 0.75 & CCC ≤ 0.95) and red boxes represent lower (CCC < 0.75) CCCs.
Figure 4. Whisker-box plots of CCCs by feature categories. A).
CCCs of features extracted from test-retest dataset (RIDER) from intratumoral ROI and peritumoral ROIs. B) CCCs of features extracted from Moist-run dataset using Algorithm 1 and Algorithm 2 from intratumoral ROI and 3mm peritumoral ROI.
Survival analysis of peritumoral features
Utilizing the training cohort, univariable Cox regression analyses were conducted using only stable and reproducible peritumoral features (0–3 mm, not bounded by a lung mask, n = 63) and the top performing features (n = 5, p < 0.05) were selected for multivariable analysis. These remaining features were included in a stepwise backward elimination Cox regression model and one feature (F300:3D_Wavelet_P2_L2_C11) remained in the final model. The classification and regression tree analysis identified the optimal cut-point (>= 1.18 × 10−4) that discriminated by OS in the training dataset and found that patients categorized in high cut-point had significantly worse OS. Applying the novel cut-off point to two independent cohorts showed that F300:3D_Wavelet_P2_L2_C11 was prognostic in all three cohorts (Supplementary Fig. S3).
Discussion
Radiomics are powerful image-based biomarkers that have been successfully applied for cancer detection, diagnosis, prognosis, prediction of response to treatment, and monitoring of disease status by converting standard-of-care medical images into quantitative data3–7. Because the surrounding peripheral areas of the tumors represent the tumor microenvironment, emerging studies have considered the clinical utility of peritumoral radiomic features16–21. Overall, we found a subset of peritumoral features were stable and reproducible. Features found to be stable regardless of the peritumoral distances included statistical and histogram and a subset of texture features (GLCM, GLRLM, GLSZM and NGTDM). This suggests these features are less affected by changes in the ROIs. We also found that the stability and reproducibility of most 3D Laws and wavelet texture features were inconsistent across the peritumoral regions which have been shown in other studies of intratumoral radiomics23,31. As such, the inclusion of a subset of 3D Laws and wavelet texture features may result in spurious and irreproducible findings. Also, when we assessed the clinical utility of stable and reproducible peritumoral radiomic features in relation to lung cancer survival, we found that stable and reproducible were more likely to be validated than unstable and non-reproducible. Specifically, we found that the top performing stable and reproducible peritumoral feature was prognostic in three previously published datasets17,28,29 (Supplementary Fig. S3).
Although prior studies have been conducted to assess for stability22 and reproduciblity23,24 of intratumoral radiomic features, this is the first study conducted on peritumoral radiomic features and the first study assessing the stability and reproducibility using IBSI radiomic features25. Kalpathy-Cramer et al.22 found that intratumoral size-based CT features were highly stable and shape-based features were less stable. Also, they showed that texture-based features were less stable which is consistent with our findings for peritumoral texture-based features. On the other hand, size and shape-based peritumoral features were not extracted in our study because these feature classes explicitly describe the intratumoral ROI. Balagurunathan et al.23 found that most intratumoral features were reproducible utilizing a semi-automatic segmentation method on test-retest CT imaging which was also consistent with our findings however, we also observed that 3D Laws texture features were less reproducible than the rest of the feature groups. A separate study from Balagurunathan et al.24 assessed lung tumor volumes across different segmentation algorithms and found that larger nodules (≥ 8 mm) were more reproducible. However, volumetric analyses of the peritumoral regions were not conducted in this study.
Because peritumoral masks are natural extensions of the intratumoral masks, the radiomic features extracted from the peritumoral and intratumoral regions could be expected to yield similar stability and reproducibility. Interestingly, the majority of the intratumoral features were unstable, especially when extracted using different segmentation algorithms (Fig. 3). However, peritumoral features were found to be more stable and reproducible than their corresponding intratumoral features. We also showed that peritumoral features further away from the intratumoral region were increasingly more stable. This finding might be related to the existence of homogenous lung parenchyma in distal peritumoral regions compared to intratumoral regions or peritumoral regions proximal to the tumor.
Our analyses also revealed that subsets of features were consistently stable irrespective of the initial parameter (e.g., seed point) for a given segmentation algorithm. These findings are important since there is no ground truth for initial parameters for any segmentation algorithm and it is essential that features are consistent across different users. On the other hand, some of these features were not stable when they were extracted using different segmentation algorithms. These results demonstrate the importance of using the same segmentation algorithm when conducting radiomics research especially when attempting to train, test, and validate findings.
We found no significant differences in stability for features that were extracted from ROIs bounded by a lung parenchyma mask versus ROIs that were not bounded by a lung parenchyma mask. Although peritumoral features of lung tumors near the mediastinum or chest wall may be attenuated, our data suggests that these features were still stable. The clinical utility of including outside of the lung parenchyma to the ROI is currently unknown. Notably, pleural invasion by lung tumors is associated with a poor prognosis34 and peritumoral features extracted from ROIs bounded by lung parenchyma may not accurately capture such a trait. Additionally, the lung parenchyma masks are not always available or are not included in software algorithms.
In 2017 a comprehensive review on the process and developments in radiomics by Lambin et al.35 stated, “…optimal reproducibility and stability enable multicenter studies to maximize the likelihood of a validated radiomic signature being fit-for-purpose in routine clinical use.” To meet this goal, assessing the reproducibility and stability using the framework presented here and by others22,23 provide groundwork to ensure generalizable studies across datasets and institutions. Because the peritumoral region has unique clinical and biological significance, capturing this information using radiomic analyses has tremendous translational utility as demonstrated from previous studies and this study16–21.
Conclusions
In summary, this study identified a subset of stable and reproducible CT radiomic features from the peritumoral region of lung lesions. Because recent studies have shown evidence that peritumoral features have clinical significance16–21, identifying stable and reproducible features is crucial to minimize spurious and non-repeatable results. The stable and reproducible features identified in this study can be used to guide a feature selection pipeline for assessing the clinical utility of peritumoral CT radiomic features.
Supplementary Material
Acknowledgements
The authors thank to Dr. Mahmoud Abdallah and the Image Response Assessment Team (IRAT) Shared Resource Core for their assistance in implementing the IBSI radiomic feature set. The authors would also like to thank to Drs. Jayashree Kalpathy-Cramer, Yoganand Balagurunathan, Dmitry Goldgof and others that contributed to the development of Moist-run dataset that was used in this study.
Funding
Funding support came from a NCI Quantitative Imaging Network Grant (U01-CA143062 to Drs. Gillies and Schabath), from a NCI Early Detection Research Network Grant (U01-CA200464 to Drs. Gillies, and Schabath), and a NCI Grant (U01-CA186145 subcontract to Drs. Schabath and Gillies). This work has also been supported in part by a Cancer Center Support Grant (CCSG) at the H. Lee Moffitt Cancer Center and Research Institute; an NCI designated Comprehensive Cancer Center (P30-CA76292).
Footnotes
Conflict of Interest
Robert J. Gillies is an investor and member of the Advisory Board at HealthMyne, Inc., and has Research support form Helix Biopharma. Sandy Napel is a consultant for Carestream, Inc. and is on the scientific advisory boards of Echo Pixel, Inc., Fovia, Inc., and RadLogics, Inc.elix Biopharma. No other conflicts.
Data Availability
The datasets generated and/or analyzed during the current study are publicly available and can be downloaded from The Cancer Imaging Archive Public Access website however; the corresponding author can also provide datasets on reasonable request.
References
- 1.Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology. 2016;278(2):563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis [published online ahead of print 2012/01/20]. Eur J Cancer. 2012;48(4):441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach [published online ahead of print 2014/06/04]. Nat Commun. 2014;5:4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hawkins S, Wang H, Liu Y, et al. Predicting Malignant Nodules from Screening CT Scans [published online ahead of print 2016/07/17]. J Thorac Oncol. 2016;11(12):2120–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coroller TP, Agrawal V, Narayan V, et al. Radiomic phenotype features predict pathological response in non-small cell lung cancer [published online ahead of print 2016/04/18]. Radiother Oncol. 2016;119(3):480–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fave X, Zhang LF, Yang JZ, et al. Delta-radiomics features for the prediction of patient outcomes in non-small cell lung cancer. Sci Rep-Uk. 2017;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parmar C, Leijenaar RT, Grossmann P, et al. Radiomic feature clusters and prognostic signatures specific for Lung and Head & Neck cancer [published online ahead of print 2015/08/08]. Sci Rep. 2015;5:11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bremnes RM, Camps C, Sirera R. Angiogenesis in non-small cell lung cancer: the prognostic impact of neoangiogenesis and the cytokines VEGF and bFGF in tumours and blood [published online ahead of print 2005/12/20]. Lung Cancer. 2006;51(2):143–158. [DOI] [PubMed] [Google Scholar]
- 9.Christiansen A, Detmar M. Lymphangiogenesis and cancer [published online ahead of print 2012/08/07]. Genes Cancer. 2011;2(12):1146–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer [published online ahead of print 2010/03/23]. Cell. 2010;140(6):883–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pages F, Galon J, Dieu-Nosjean MC, Tartour E, Sautes-Fridman C, Fridman WH. Immune infiltration in human tumors: a prognostic factor that should not be ignored [published online ahead of print 2009/12/01]. Oncogene. 2010;29(8):1093–1102. [DOI] [PubMed] [Google Scholar]
- 12.Way TW, Hadjiiski LM, Sahiner B, et al. Computer-aided diagnosis of pulmonary nodules on CT scans: segmentation and classification using 3D active contours [published online ahead of print 2006/08/11]. Med Phys. 2006;33(7):2323–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Way TW, Sahiner B, Chan HP, et al. Computer-aided diagnosis of pulmonary nodules on CT scans: improvement of classification performance with nodule surface features [published online ahead of print 2009/08/14]. Med Phys. 2009;36(7):3086–3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shah SK, McNitt-Gray MF, Rogers SR, et al. Computer aided characterization of the solitary pulmonary nodule using volumetric and contrast enhancement features. Academic Radiology. 2005;12(10):1310–1319. [DOI] [PubMed] [Google Scholar]
- 15.Hardie RC, Rogers SK, Wilson T, Rogers A. Performance analysis of a new computer aided detection system for identifying lung nodules on chest radiographs. Med Image Anal. 2008;12(3):240–258. [DOI] [PubMed] [Google Scholar]
- 16.Sun R, Limkin EJ, Vakalopoulou M, et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study [published online ahead of print 2018/08/19]. Lancet Oncol. 2018;19(9):1180–1191. [DOI] [PubMed] [Google Scholar]
- 17.Tunali I, Stringfield O, Guvenis A, et al. Radial gradient and radial deviation radiomic features from pre-surgical CT scans are associated with survival among lung adenocarcinoma patients [published online ahead of print 2017/12/10]. Oncotarget. 2017;8(56):96013–96026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braman NM, Etesami M, Prasanna P, et al. Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE-MRI [published online ahead of print 2017/05/20]. Breast Cancer Res. 2017;19(1):57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prasanna P, Patel J, Partovi S, Madabhushi A, Tiwari P. Radiomic features from the peritumoral brain parenchyma on treatment-naive multi-parametric MR imaging predict long versus short-term survival in glioblastoma multiforme: Preliminary findings [published online ahead of print 2016/10/26]. Eur Radiol. 2017;27(10):4188–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dou TH, Coroller TP, van Griethuysen JJM, Mak RH, Aerts H. Peritumoral radiomics features predict distant metastasis in locally advanced NSCLC. PLoS One. 2018;13(11):e0206108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mattonen SA, Davidzon GA, Bakr S, et al. Utility of 18F-FDG Positron Emission Tomography (PET) Tumor and Penumbra Texture Features for Recurrence Prediction in Non-Small Cell Lung Cancer. Tomography. November 2018;IN PRESS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalpathy-Cramer J, Mamomov A, Zhao B, et al. Radiomics of Lung Nodules: A Multi-Institutional Study of Robustness and Agreement of Quantitative Imaging Features [published online ahead of print 2017/02/06]. Tomography. 2016;2(4):430–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balagurunathan Y, Kumar V, Gu Y, et al. Test-retest reproducibility analysis of lung CT image features [published online ahead of print 2014/07/06]. J Digit Imaging. 2014;27(6):805–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balagurunathan Y, Beers A, Kalpathy-Cramer J, et al. Semi-automated pulmonary nodule interval segmentation using the NLST data [published online ahead of print 2018/01/25]. Med Phys. 2018;45(3):1093–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zwanenburg A, Leger S, Vallières M, Löck S. Image biomarker standardisation initiative. https://arxivorg/abs/161207003 2018.
- 26.Kalpathy-Cramer J, Zhao B, Goldgof D, et al. A Comparison of Lung Nodule Segmentation Algorithms: Methods and Results from a Multi-institutional Study [published online ahead of print 2016/02/06]. J Digit Imaging. 2016;29(4):476–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao B, James LP, Moskowitz CS, et al. Evaluating variability in tumor measurements from same-day repeat CT scans of patients with non-small cell lung cancer [published online ahead of print 2009/06/30]. Radiology. 2009;252(1):263–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grove O, Berglund AE, Schabath MB, et al. Quantitative computed tomographic descriptors associate tumor shape complexity and intratumor heterogeneity with prognosis in lung adenocarcinoma [published online ahead of print 2015/03/05]. PLoS One. 2015;10(3):e0118261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schabath MB, Welsh EA, Fulp WJ, et al. Differential association of STK11 and TP53 with KRAS mutation-associated gene expression, proliferation and immune surveillance in lung adenocarcinoma [published online ahead of print 2015/10/20]. Oncogene. 2016;35(24):3209–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gu Y, Kumar V, Hall LO, et al. Automated Delineation of Lung Tumors from CT Images Using a Single Click Ensemble Segmentation Approach. Pattern Recognit. 2013;46(3):692–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balagurunathan Y, Gu Y, Wang H, et al. Reproducibility and Prognosis of Quantitative Features Extracted from CT Images [published online ahead of print 2014/04/29]. Transl Oncol. 2014;7(1):72–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin LI. A concordance correlation coefficient to evaluate reproducibility [published online ahead of print 1989/03/01]. Biometrics. 1989;45(1):255–268. [PubMed] [Google Scholar]
- 33.Klein JP, Moeschberger ML. Survival analysis: techniques for censored and truncated data. 2nd ed New York: Springer; 2003. [Google Scholar]
- 34.Tian D, Pei Y, Zheng Q, et al. Effect of visceral pleural invasion on the prognosis of patients with lymph node negative non-small cell lung cancer [published online ahead of print 2017/03/04]. Thorac Cancer. 2017;8(2):97–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14(12):749–762. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.