Graphical Abstract
Keywords: HIV, RNase H, NMR, Molecular Docking, Quantum Mechanical calculations
The ribonuclease H (RNH) activity of HIV-1 reverse transcriptase (RT) is essential for viral replication and can be a target for drug development. Yet, no RNH inhibitor to date has substantial antiviral activity to allow advancement into clinical development. Herein, we describe our characterization of the detailed binding mechanisms of RNH active-site inhibitors, YLC2-155 and ZW566, that bind to the RNH domain through divalent metal ions, using NMR, molecular docking and quantum mechanical calculations. In the presence of Mg2+, NMR spectra of RNH exhibited split (two) resonances for some residues upon inhibitor binding, suggesting two binding modes, an observation consistent with the docking results. The relative populations of the two binding conformers were independent of inhibitor or Mg2+ concentration, with one conformation consistently more favored. In our docking study, one distinctive pose of ZW566 showed more interactions with surrounding residues of RNH compared to the analogous binding pose of YLC2-155. Inhibitor titration experiments revealed a lower dissociation constant for ZW566 compared to YLC2-155, in agreement with its higher inhibitory activity. Mg2+ titration data also indicated a stronger dependence on Mg2+ for the RNH interaction with ZW566 compared to YLC2-155. Combined docking and quantum mechanical calculation results suggest that stronger metal coordination as well as more protein-inhibitor interactions may account for the higher binding affinity of ZW566. These findings support the idea that strategies for the development of potent competitive active site RNH inhibitors should not only take into account metal-inhibitor coordination but also protein-inhibitor interaction and conformational selectivity.
Human immunodeficiency virus-1 (HIV-1) reverse transcriptase (RT) is a multi-functional protein, containing DNA polymerase activity and ribonuclease H (RNH) activity.1–5 RT comprises a 66 kDa subunit (p66) and a p66-derived 51 kDa subunit (p51), and both the polymerase and RNH active sites are located in the p66 subunit.6 Although over 10 clinically approved polymerase inhibitors are available, the RNH domain remains the only HIV enzyme not targeted by approved drugs.7–14 Given that RNH activity is crucial for virus replication, RNH inhibitors have the potential to become applicable as an antiviral therapy. Considerable effort has been made to synthesize and test several different chemotypes of RNH inhibitors, including active-site inhibitors that interact with RNH through divalent metal ions.15–17 However, no RNH inhibitor characterized to date has substantial antiviral activity to allow advancement into clinical development. In contrast to the 20-plus years of study on the structures and binding mechanisms of polymerase inhibitors,18–27 structural information for RNH inhibitor interactions is scarce: only recently were crystal structures of RNH active-site inhibitors in complex with RT or isolated RNH domain published.28–33 In all these crystal structures, the inhibitors coordinate the two catalytic metal ions in a similar fashion, while hydrophobic or hydrogen-bonding interactions between the inhibitors and protein differ among the structures. The effects of these varied contacts on binding and inhibition potency are not well characterized.
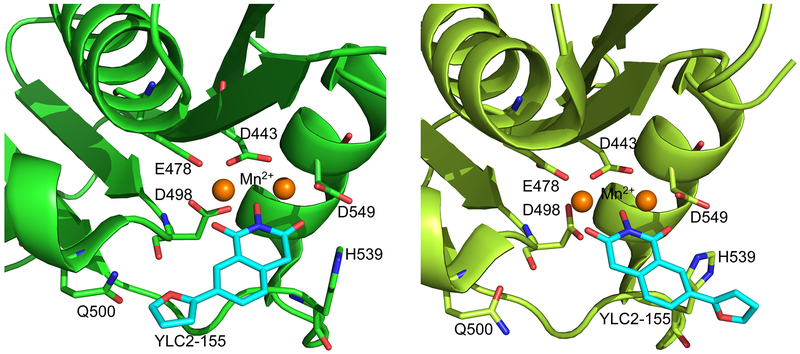
Recently, we published crystal structures of RT in complex with a hydroxyisoquinoline-dione (HID)-type active-site inhibitors, YLC2-155, showing that the inhibitor interacts with the RNH domain in two binding modes (Figure 1).34 Based on both crystal structure and molecular docking, we showed that the inhibitor chelates two Mn2+ ions through its hydroxyl and carbonyl groups, while the remainder of its chemical groups are positioned against RNH in two different configurations, one towards Q500 and the other towards H539 (Figure 1).34 NMR experiments with the isolated RNH domain showed two sets of resonances for some RNH residues when bound to YLC2-155 in the presence of Mg2+.34 However, the detailed mechanism of the molecular interaction was not studied. In addition to the chelating properties of RNH active-site inhibitors, specific interactions of the inhibitors with conserved residues in RNH play an important role in inhibitory activity.13, 35, 36 It is important to investigate these factors to identify inhibitor-interaction mechanisms.
Figure 1.
View of the two binding modes of YLC2-155 to the RNH domain, observed in a crystal structure of the complex (5UV5). Mn2+ was used for crystal structure determination.34
Herein, we report a comprehensive interaction study of the ternary complex comprising the RNH domain bound to Mg2+ and inhibitor, using NMR spectroscopy, differential scanning fluorimetry (DSF), molecular docking and quantum mechanical calculations. For NMR and DSF studies, we used an isolated RNH domain (residues 427 to 560), which is routinely used to investigate active-site inhibitor and metal interactions37, 38 but is inactive due to the absence of the RNH substrate-handle region.39–42 Mg2+ presumably acts as the integral cofactor for HIV-1 RNH activity.43, 44 However, under physiological conditions, RNH in RT is not expected to be fully Mg2+ saturated due to weak binding.45, 46 Thus, an understanding of inhibitor binding and whether it shifts the equilibrium between Mg2+-free and Mg2+-bound states toward the Mg2+-bound state and eventually to the RNH-Mg2+-inhibitor ternary complex is important. To address this, we varied the Mg2+ concentration from 1 to 20 mM, to generate differential populations of Mg2+-free and Mg2+-bound RNH.47–50 We also varied the inhibitor concentration in the presence of 20 mM Mg2+ to generate differential populations of the Mg2+-bound and the inhibitor-bound RNH, and monitored state changes by NMR.
NMR experiments were conducted on two different inhibitors with different inhibition activities, a HID-based active site RNH inhibitor, YLC2-155, and a hydroxypyrimidine-dione (HPD)-type active-site inhibitor, ZW566. The IC50 of ZW566 against RNH activity, as determined with wild-type RT, is 0.011 ± 0.003 μM,51 over 50-fold lower than that of YLC2-155, 0.65 ± 0.25 μM.34, 52, 53 We found that inhibitor binding to RNH is Mg2+-dependent while observation of two inhibitor-bound forms, assessed by the presence of split signals in NMR spectra, is independent of both Mg2+ and inhibitor concentration. Although analysis of inhibitor-titration data showed a 10-fold lower dissociation constant of ZW566 compared to YLC2-155, titration profiles were similar, whereas ZW566 exhibited significantly stronger Mg2+-dependence, showing different Mg2+-titration profiles compared to those of YLC2-155. The thermal stability of RNH upon Mg2+ and inhibitor binding was greater for the ZW566 bound form, compared to the YLC2-155 bound form, as assessed by DSF. We also performed molecular docking and quantum mechanical calculations on both inhibitors to investigate the binding modes, protein-inhibitor interactions, and chelating properties. Both inhibitors bind RNH in two orientations, however, ZW566 exhibits more protein interactions and stronger metal chelating ability than YLC2-155. Overall, our findings provide valuable insights for RNH drug design.
RESULTS
Mg2+ interaction to RNH and Mg2+-dependent inhibitor binding to RNH.
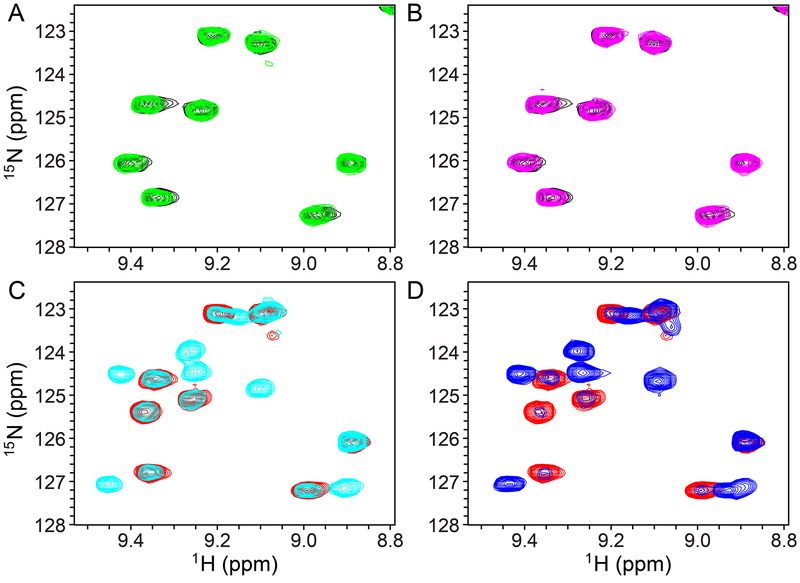
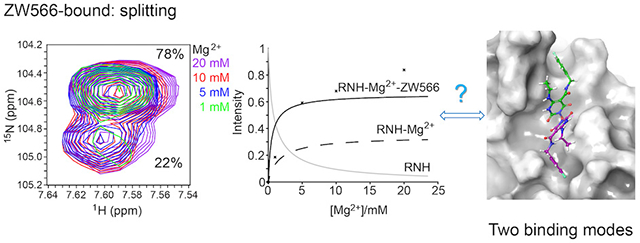
The interaction of RNH with Mg2+ was previously studied using NMR, by us and other groups.47–49 Mg2+ binding produces gradual NMR chemical shift changes, due to fast exchange, and leads to significant chemical shift perturbations of many residues, particularly those near the metal binding site (previously published,47–49 and compare black-colored spectrum in Figure 2A with the red-colored spectrum in Figure 2C). In the present study, 1H-15N HSQC spectra of RNH were recorded to characterize in detail the binding of YLC2-155 and ZW566 to RNH. In the absence of metal ions, no change in the HSQC spectra could be observed, even after adding equimolar inhibitor, (Figures 2A, 2B, and S1), indicating no detectable interaction between RNH and inhibitor. In the presence of 20 mM Mg2+, new signals emerged when the inhibitors were added, and these signals were distinct from those observed in the spectrum of RNH in presence of Mg2+ alone, without inhibitor (Figures 2C, 2D, and S2). These results demonstrate that inhibitor binding is metal ion dependent. The observation of two sets of signals, one arising from the inhibitor-free form and the other from the inhibitor-bound form, is indicative of a slow-exchange phenomenon.
Figure 2.
Selected region of 1H-15N HSQC spectra of 50 μM 15N-labeled RNH in the absence (A and B, black) and presence of 20 mM Mg2+ (C and D, red). 50 μM YLC2-155 (green in A and cyan in C) or ZW566 (magenta in B and blue in D) was added. All spectra were recorded at 293 K. Note, we selected a region that does not include split signals in panel C and D to simply explain spectral changes first. See Figure S1 and S2 for overall spectral changes.
To better understand the binding mechanisms, we assigned the backbone amide resonances for inhibitor-free, YLC2-155-bound, and ZW566-bound RNH in the presence of 20 mM Mg2+, using traditional triple-resonance experiments. Both inhibitor-bound proteins exhibited a greater number of resonances than the inhibitor-free protein (Figure S3 and S4). This is because all protein formed tight ternary complexes in the presence of excess inhibitor, while, in the absence of inhibitor, some signals were broadened due to fast exchange between Mg2+-free and Mg2+-bound protein. Chemical shift analysis showed the largest perturbations on the C-terminal helix (residue 545 - 560) between the YLC2-155 and ZW566 bound proteins (Figure S4C), which may result from more contacts between the C-terminal helix and ZW566 (see below).
Split signals suggest two inhibitor-binding modes.
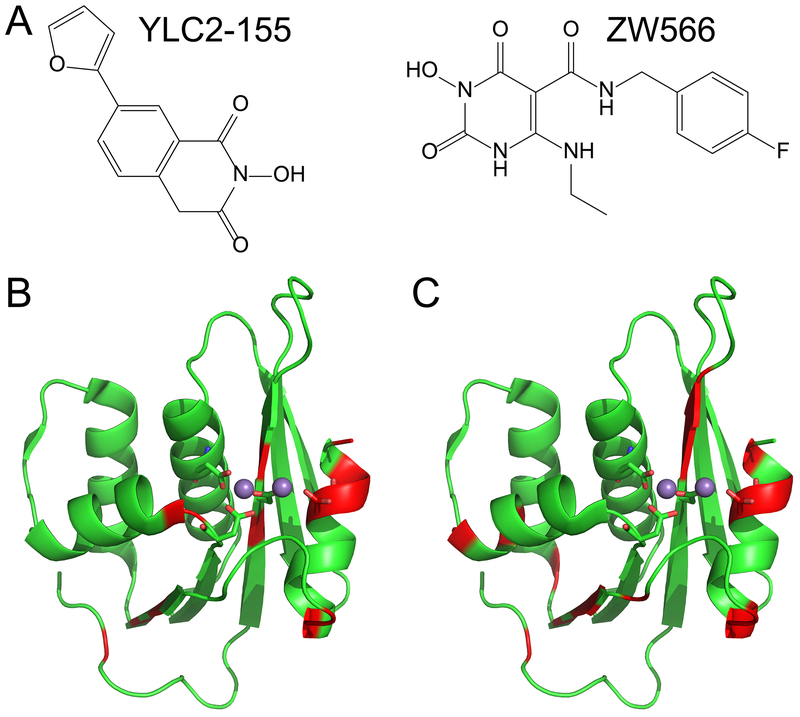
Different from Mg2+-bound protein, two sets of resonances were observed for some residues in the inhibitor-bound proteins (Figure S3). Such splitting of signal indicates that there are two forms of inhibitor-bound protein, consistent with previous observations for YLC2-155 binding.34 These residues were mapped onto the crystal structure of the RNH domain (Table S1 and Figure 3). Similar patterns were observed for the two inhibitors. Since signal splits are not detectable when the chemical shifts overlap (< 40 Hz), residues near the Mg2+ coordinating sites do not necessarily show the split. For the same reason, we could not determine the orientation of the two inhibitor-bound forms separately.
Figure 3.
Structures of the RNH active-site inhibitors used in this study are shown in (A). Residues showing split resonances upon binding of (B) YLC2-155 or (C) ZW566 are mapped onto the crystal structure of the RNH domain (PDB 5UV5), and colored in red. Metal ions are shown as balls, and metal binding residues are displayed in stick representation.
To investigate the reason for our observation of two binding forms for each inhibitor, we carried out two series of NMR titration experiments. Based on our previous crystal structure, we considered it likely that two different orientations of inhibitor resulted in the split signals. To investigate this, 1H-15N HSQC spectra of RNH were recorded to monitor the intensity changes of inhibitor-free and bound signals, when inhibitor was added at different molar ratios, relative to RNH, in the presence of 20 mM Mg2+. The high concentration of Mg2+ was used due to its known weak interaction with RNH (dissociation constant > 1 mM).47–50 In addition, because Mg2+ interaction with RNH in solution can be explained with either a single Mg2+-binding model or a two Mg2+-binding model,45, 47, 48 different numbers of bound Mg2+ possibly influence the two binding modes. For this reason, Mg2+titration was also performed, in which Mg2+ was varied from 1 to 20 mM with a constant 1:1 inhibitor:RNH ratio.
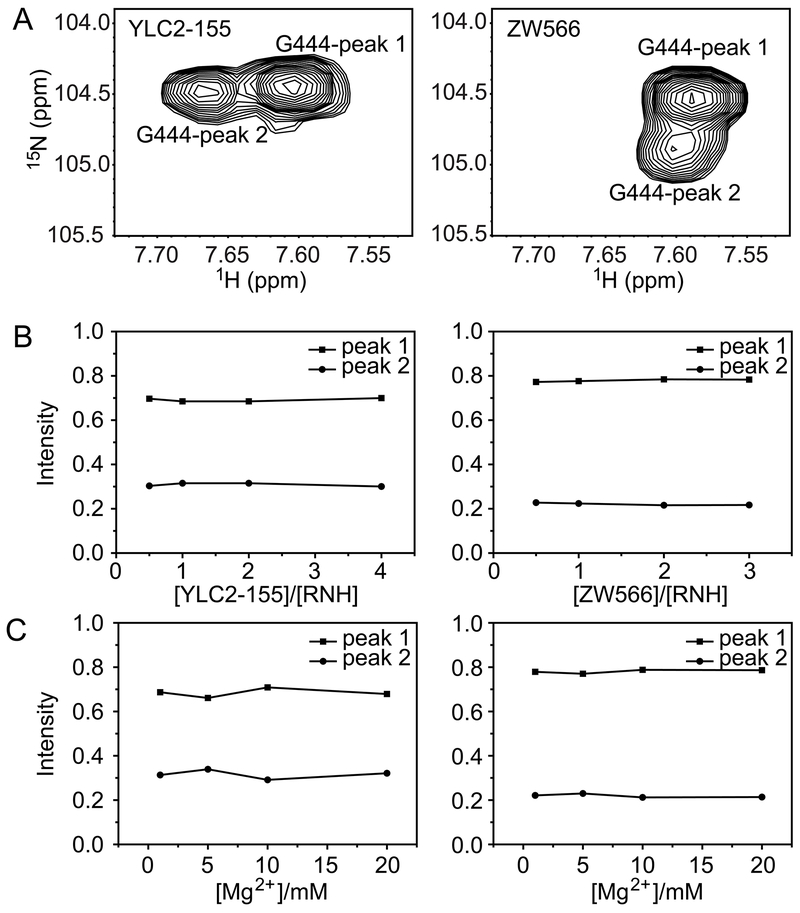
In all NMR titration experiments, peak splitting was always observed (Figures S5 and S6), for those residues shown in Figure 3 and Table S1, indicating that the two binding forms are not dependent on inhibitor or Mg2+ concentration, within the sensitivity of our experiments. These findings demonstrate that split signals are not the result of unbound-bound equilibrium or different numbers of bound Mg2+ or inhibitor, but due to two binding orientations of inhibitor. It is worth noting that there was also no concentration dependence on the proportions of two inhibitor binding forms, which were almost constant for each residue in both inhibitor-bound forms, YLC2-155 or ZW566 (Figure 4, S7 and S8). For residue G444, the proportional difference between the two binding modes of ZW566 is larger than that of YLC2-155 (Figure 4). Unfortunately, due to some split signals overlapping with each other, no data of other residues could be used to directly compare binding of the two inhibitors so that we cannot conform the difference observed on G444. However, for both inhibitors, one binding mode is obviously more favored than the other.
Figure 4.
Analysis of proportions of two binding modes for residue G444 in the (left column) YLC2-155 bound form and (right column) ZW566 bound form. (A) Peak splitting of residue G444. Normalized peak intensities plotted as a function of (B) [inhibitor]/[RNH] molar ratios or (C) Mg2+ concentration.
NMR titration confirms higher ZW566 affinity for RNH than YLC2-155.
Upon addition of inhibitors to the RNH solution in the presence of 20 mM Mg2+, inhibitor-bound signals increased in intensity while the inhibitor-free signals decreased (Figure S5). This observation indicates that inhibitor binding to RNH is in a slow exchange on NMR time scale, which is often observed for a ligand with high affinity. Since no interaction between RNH and inhibitor was detected in the absence of Mg2+ (Figure S1) and the proportion of the two binding forms is not concentration dependent, a two-step binding pathway was assumed for the determination of inhibitor dissociation constants (K2) (Equation 1 and 2). Here, the number of Mg2+ ions bound to RNH is assumed to be two, based on the previous divalent ion geometry of YLC2-155 binding34 and no concentration dependence of the split signals (Figure 4, S7 and S8). Amide resonances that did not exhibit overlap between the inhibitor-free and bound signals were chosen for this analysis (Table S2). Titration curve fits were obtained for 18 or 14 residues of RNH titrated with YLC2-155 or ZW566, respectively (Table S2). Representative titration curves and fits for residues F440 and T470 are shown in Figure 5A–5D. The averaged K2 for residues upon ZW566 titration, 1.9 ± 1.7 μM, is about 10-fold lower than that for YLC2-155 titration, 17.9 ± 7.3 μM. This difference is in agreement with the 50-fold lower IC50 of ZW566 compared to YLC2-155.34, 52, 53
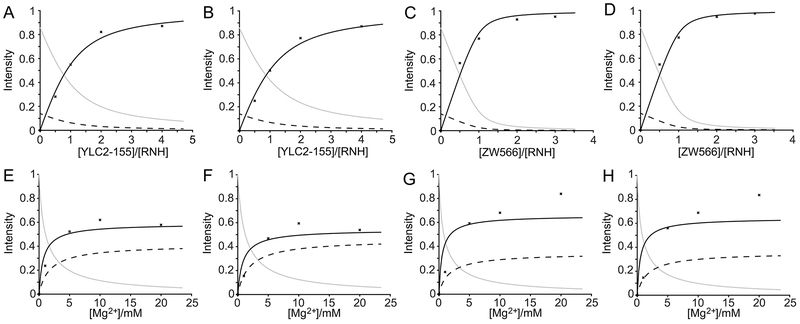
Figure 5.
Inhibitor titration data points and the curve fits for residue F440 (A, C, E and G) and T470 (B, D, F and H) in YLC2-155 (A, B, E and F) or ZW566 (C, D, G and H) titrations. Inhibitor titration was performed in the presence of 50 μM RNH and 20 mM Mg2+. Mg2+ titration was performed in the presence of 50 μM RNH and 50 μM inhibitor. The titration data were analyzed assuming three species: free RNH, RNH-2Mg2+, and RNH-2Mg2+-inhibitor. Normalized peak intensities are plotted as a function of total [inhibitor]/[RNH] molar ratios or Mg2+ concentration. Plots for fitting fractions of RNH, RNH-2Mg2+, and RNH-2Mg2+-Inhibitor are shown as solid gray, dashed black, and solid black line, respectively. Determination of inhibitor dissociation constants (K2) were performed by assuming a two-step mechanism (Equation 1 and 2).
Mg2+ titration elucidates higher Mg2+ sensitivity of ZW566 than YLC2-155.
Similar to inhibitor titration, Mg2+ titration again exhibited a slow exchange regime on the NMR time scale for inhibitor binding, as signals of the RNH-2Mg2+-inhibitor complex increased in intensity (Figure S6). In addition, Mg2+-bound RNH signals shifted with the change of Mg2+ concentration, indicative of a weak interaction of Mg2+ with RNH, which is consistent with the previously reported dissociation constant.45, 47–50 We did a similar analysis with the Mg2+ titration data using the same two-step binding model. Titration curves for residues F440 and T470 are shown as examples (Figure 5E–5H). As illustrated in Figures 5E and 5F, passable fits were obtained for YLC2-155 binding, resulting in an averaged K2 of 15.4 ± 5.4 μM, which is similar to that calculated from inhibitor titration. In contrast, data for ZW566 binding could not be well-fitted (Figure 5G and 5H). Experimental data points showed that a high concentration of Mg2+ promoted binding of both inhibitors, however a plateau was not reached in the case of ZW566. Such observation demonstrates the deviation of the binding model from the two-step system (Equation 1–2): for example, a metal-inhibitor complex formation that is not directly included in the model, may become more significant in the ZW566 interaction with RNH at high Mg2+ concentrations.
Thermal stability experiments indicate higher RNH stability of ZW566 bound form than YLC2-155.
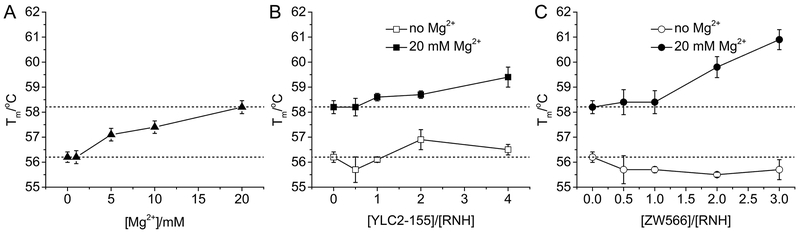
DSF was utilized to determine thermal stability of RNH and the influence of Mg2+ and inhibitor binding. RNH exhibited a melting temperature (Tm) of 56.2 ± 0.2 °C, similar to a previous result determined using DSF but smaller than that determined using circular dichroism (CD), 60.8 ± 0.5 °C.50, 54 This difference in Tm may reflect differences in the melting of structural components: the CD experiments detected the secondary structure change, while DSF can also measure the unfolding of protein tertiary structure. As shown in Figure 6 and Table S3, Tm of RNH slightly increased with the increasing concentration of Mg2+, indicating that Mg2+ binding may promote the thermal stability of RNH (Figure 6A). In the absence of Mg2+, both inhibitors had little effect on RNH stability, in agreement with the NMR result of no interaction between RNH and inhibitor. Both YLC2-155 and ZW566 binding to RNH in the presence of Mg2+ further increased the thermal stability (Figure 6B and 6C). However, ZW566 binding exhibited larger Tm value than YLC2-155 binding, over two standard deviation (i.e., over 95% confident level), suggesting tighter binding of ZW566 (Figure 6B and 6C). Such observation of more significant Mg2+ contribution to ZW566 interaction with RNH, than YLC2-155 interaction, is also consistent with the NMR observations.
Figure 6.
Analysis of RNH thermal stability upon Mg2+ and inhibitor binding. Tm values are plotted against (A) Mg2+ concentration, (B) molar ratio of [YLC2-155]/[RNH], and (C) molar ratio of [ZW566]/[RNH].
Molecular docking calculations show two binding forms for both YLC2-155 and ZW566.
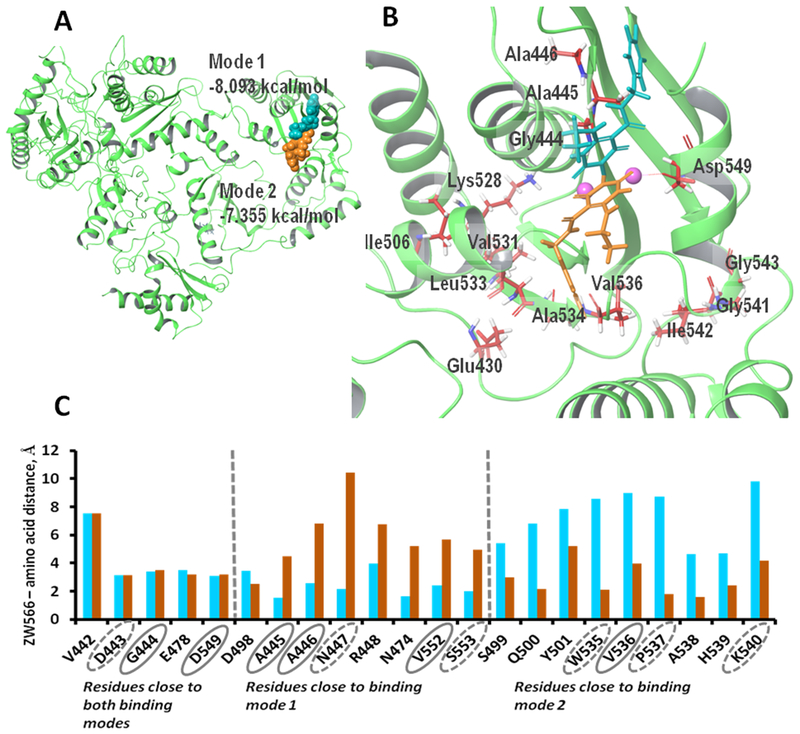
An advanced conformational search predicted 52 conformers for the ZW566 molecule. All conformers were docked using flexible ligand sampling protocol into the active center of the RNH domain, and for each conformer one docking pose was generated. An analysis to obtain the average structure for each orientation, as well as calculated RMSD docking pose-to-average structure (Figure S9) allowed us to conclude that two distinctive binding modes occur for ZW566 (Figure 7). Binding mode 1, obtained from 44 poses (the lowest docking score was −8.09 kcal/mol among the 44 poses), binds to the RNH active site with fluorobenzene ring oriented toward opposite to the p66-p51 interface, whereas binding mode 2, obtained from 8 poses (best docking score was −7.36 kcal/mol among the 8 poses) binds with the flourobenzene ring oriented toward p66-p51 interface (Fig. 7A and 7B). The position of ZW566 relative to several active site residues is different for the two binding modes, including those residues that exhibit split NMR signals and those located adjacent to the split-signal residues (Fig. 7C): in binding mode 1, ZW566 is closer to Ala445, Ala446, and Val552, while binding mode 2 places ZW566 close to Val536.
Figure 7.
Docking simulations show two binding modes for ZW566. (A) Overview of ZW566 binding in the RNH active site. In binding mode 1 (−8.09 kcal/mol, cyan structure), the fluorinated benzene ring of ZW566 points toward the upper part of p66 subunit. In binding mode 2 (−7.36 kcal/mol, orange structure), the fluorinated benzene ring of ZW566 points toward the p66/p51 interface. (B) A close view of the two ZW566 binding modes with surrounding residues that show split NMR signals. (C) Distances from each of the indicated residues to the nearest proximity of ZW566 in mode 1 (cyan columns) and in mode 2 (orange columns). Residues circled with a solid line show split NMR signals; residues circled with a dashed line are adjacent to residues showing split NMR signals.
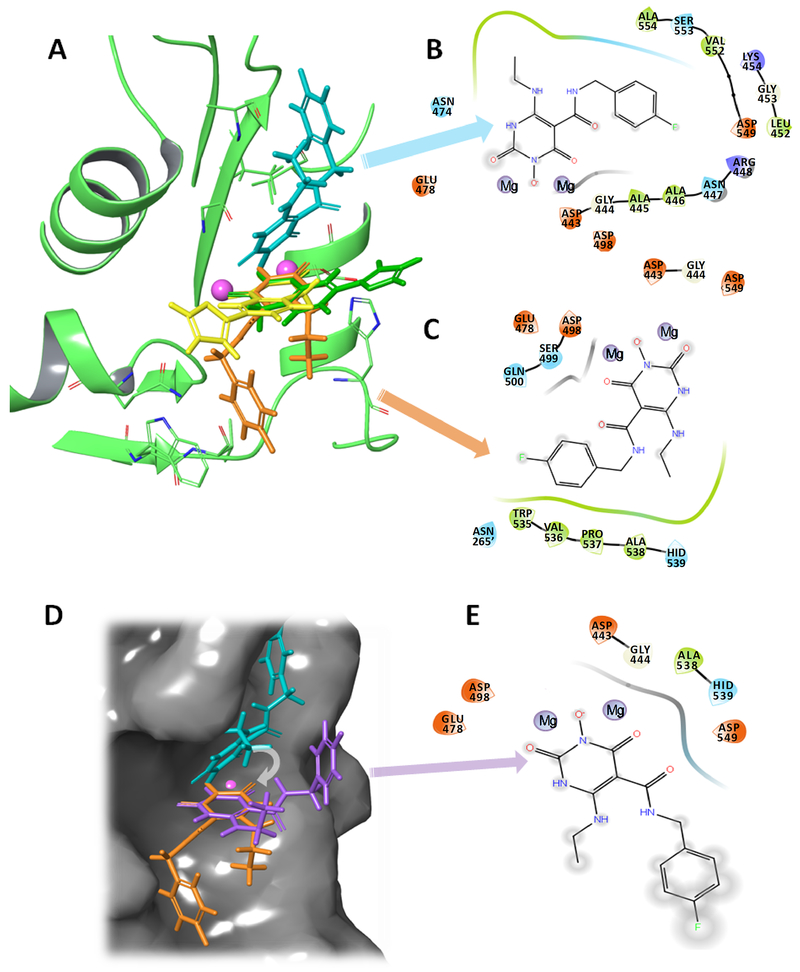
Molecular docking of YLC2-155 to RT was also performed using conditions identical to those applied for ZW566 docking, to directly compare the results for the two inhibitors. Docking poses obtained for YLC2-155 were the same as those published previously, showing two binding poses (Fig. S10).34 ZW566 binding modes 1 and 2 are distinctive from the two binding modes of YLC2-155 (Fig. 8A).34 In both YLC2-155 binding modes, a chelating triad lays in the same plane, approaching the metal cations at a similar angle. ZW566 binding mode 2 is similar to YLC2-155 binding, with the chelating triad overlapping with that of YLC2-155. As ligand interaction diagrams (LID) show, in both binding modes, ZW566 is surrounded by a greater number of residues in close proximity than is YLC2-155 (Fig. 8B and 8C). In contrast, YLC2-155 interaction with RNH is mostly at or around the hydroxyisoquinoline-dione core that chelates the metal ions (Fig. S10A and S10B).34 To understand the importance of ZW566 interactions with these surrounding amino acid residues, we superimposed ZW566 binding mode 1 with the chelating triad of binding mode 2, simulating “YLC2-155-like” binding, when both binding modes chelate metal cations at the same angle (Fig. 8D). LID for this ZW566 superposition shows that binding of ZW566 in a manner similar to YLC2-155 would result in much fewer amino acid contacts and complete exposure of the fluorinated ring and the aminoethyl group to solvent (Fig. 8E).
Figure 8.
The two binding modes of ZW566 are distinctive from the two-mode binding modes of YLC2-155. A) Overlap of ZW566 (cyan and orange structures) and YLC2-155 (yellow and green structures) binding modes. B) Ligand interaction diagram (LID) for ZW566 binding mode 1. C) LID for ZW566 binding mode 2. D) ZW566 binding mode 1 (cyan structure), mode 2 (orange structure) and a simulated “YLC-2155-like binding mode” for ZW566 (purple structure) obtain by superposition of the ZW566 chelating ring in binding mode 1 with the ZW566 chelating ring in binding Mode 2. Structures are shown on the RNH active site surface. E) LID for ZW566 superimposed to simulate YLC2-155 binding mode. Such a binding mode would result in fewer interactions with amino acid residues and greater exposure to solvent. In LID, negatively charged residues are shown in red, positively charged residues are shown in blue, hydrophobic residues are shown in green, polar residues are shown in cyan, and glycine is shown in white. Note, in the LID presentation, residue numbers were embedded into the original graphics. In panel C, dashed residue number indicates that from p51 subunit in RT.
Molecular docking calculations demonstrate stronger ZW566 interaction to the Mg2+ bound RNH than YLC2-155.
The interaction energies contributing to the docking scores were calculated (Table 1). We found that metal binding, hydrophobic interactions, and van der Waals interactions are all noticeably stronger for ZW566 compared to YLC2-155 (Table 1), consistent with the observed higher affinity for ZW566 than YLC2-155 (Figures 5A–5D). The smaller energies for van der Waals and hydrophobic interactions for YLC2-155 could be explained by fewer residue contacts for YLC2-155 compared to ZW566. The energy of Coulombic interactions is comparable for both compounds which is predictable because both ZW566 and YLC2-155 are deprotonated and, despite differences in chemical structures of the two compounds, have very similar surroundings of amino acid residues with partial charges, thus calculated Coulombic energies are similar.
Table 1.
Calculated docking scores and major docking score components (in kcal/mol) calculated for ZW566 and YLC2-155 lowest docking poses
| Compound | Mode | Gscorea | Interaction Energiesb | |||
|---|---|---|---|---|---|---|
| EMB | Ehphob | EvdWc | ECoulc | |||
| ZW566 | 1 | −8.09 | −2.88 | −1.23 | −14.34 | −15.85 |
| 2 | −7.36 | −2.57 | −1.41 | −14.86 | −13.99 | |
| YLC2-155 | 1 | −7.52 | −2.30 | −0.74 | −9.41 | −14.31 |
| 2 | −7.12 | −1.97 | −0.50 | −8.17 | −15.45 | |
Gscore –docking score calculated by Glide.
Energy terms: EMB – energy of metal binding; Ehphob – energy of hydrophobic interactions; EvdW – energy of van der Waals interactions; ECoul – energy of Coulombic interactions calculated by ChemScore empirical scoring function,55 embedded in Glide.
Note that docking scores are significantly smaller in absolute values than individual vdW and Coulombic interaction energies since weighted vdW and Coulombic interaction energies are used in calculation of docking scores.56
Quantum Mechanical Calculations suggest greater negative charges on ZW566 than YLC2-155.
Mg2+ binding to the RNH domain has a dissociation constant greater than 1 mM,45, 50 while the dissociation constants of ZW566 and YLC2-155 binding to RNH are determined as 1.9 ± 1.7 μM and 17.9 ± 7.3 μM, respectively, in the presence of Mg2+. Such observations indicate that the metal-inhibitor interaction could stabilize the ternary complex. In our docking study, we investigated the interaction of each inhibitor with the Mg2+-bound form of RNH. However, metal-ligand interaction energies cannot be correctly estimated by docking algorithms, including the program used in the present study, Glide. An accurate description of metal-ligand coordination involves consideration of the electron density distribution on ligand atoms and of the energy required to move an electron from a ligand molecular orbital to the metal orbitals. This information can only be obtained from quantum mechanical calculations which are not included in docking algorithms. Thus, to better understand metal coordination by ZW566 and YLC2-155, we performed high accuracy quantum mechanical calculations to estimate certain electronic structure properties including partial atomic charges, molecular orbital energies and compositions, and average local ionization energies.
Every charge calculation method has its limitation and care should be taken when interpreting results. Therefore, we calculated the atomic charges of the compounds, using three different methods, to obtain a pattern of charge distribution that is relatively independent of the method of calculation (Table 2). From an electrostatic point of view, when more negative electronic charge is located on the ligand atoms that coordinating the metal, the interaction between metal and ligand will be stronger. All three atomic charge computing methods predicted a greater overall negative charge on the three chelating oxygen atoms of ZW566 compared to the charge on the chelating triad of YLC2-155, suggesting stronger metal interaction of ZW566 than YLC2-155.
Table 2.
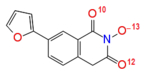
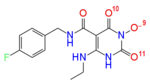
Partial charges (in atomic units) on selected YLC2-155 and ZW566 atoms calculated by different charge estimation methods at B3LYP/6-311G** level of theory
| YLC2-155 | ZW566 | |
|---|---|---|
 |
 |
|
| Natural Charges | O(10) -0.62 | O(11) -0.65 |
| O(13) -0.59 | O(9) -0.61 | |
| O(12) -0.61 | O(10) -0.69 | |
| Total charge on three oxygen atoms | −1.82 | −1.95 |
| Atomic Charges from Electrostatic Potential | O(10) -0.53 | O(11) -0.60 |
| O(13) -0.56 | O(9) -0.57 | |
| O(12) -0.57 | O(10) -0.62 | |
| Total charge on three oxygen atoms | −1.66 | −1.79 |
| Atomic charges from Mulliken population analysis | O(10) -0.38 | O(11) -0.4 |
| O(13) -0.38 | O(9) -0.51 | |
| O(12) -0.49 | O(10) -0.44 | |
| Total charge on three oxygen atoms | −1.25 | −1.35 |
We also carried out molecular orbital analysis for both compounds, to investigate the ease of electron delocalization from ligand orbitals to metal orbitals. Although no significant difference in the orbital energies of the lone oxygen pair orbitals of ZW566 and YLC2-155 is observed (Table S4), the average calculated local ionization energy (ALIE) for ZW566, 66.86 kcal/mol, is about 3.2 kcal/mol lower than that for YLC2-155 (ALIE = 70.09 kcal/mol), suggesting that electrons are more delocalized from ZW566 compared to YLC2-155. The ALIE provides an energetic measure of how easy or difficult it is to remove electrons from regions of a molecule and describes how a molecule would respond to an electrophile, which in our case is a metal cation. The energy difference, 3.2 kcal/mol, profoundly impacts acid/base properties and thus pKa.57 Taken together, all these findings from quantum mechanical calculations suggest that ZW566 makes stronger dative bonds to metal cations than YLC2-155. Such observations may be consistent with the observed higher Mg2+-sensitivity of ZW566 binding than YLC2-155 binding, as determined by NMR (Figure 5E–5H).
DISCUSSION
This study aimed to understand the detailed binding mechanism of HIV-1 RNH active-site inhibitors by comparing two recently developed inhibitors, YLC2-155 and ZW566. We investigated the concentration-dependence of inhibitor and of Mg2+ on interactions with an isolated RNH using NMR spectroscopy. Although some RNH domain signals exhibited split peaks upon inhibitor binding, the relative intensities of the two peaks did not change with inhibitor or Mg2+ concentration. Thus, the split resonances are not due to differences in the number of bound Mg2+ ions or inhibitor but most likely reflect two binding conformations of inhibitor, based on our titration data. Molecular docking also revealed two poses for each inhibitor interaction. Although the docking was performed using full-length RT, i.e., p66/p51, no interaction between inhibitor and other domains of RT was observed. Therefore, although we do not know that the two poses predicted by the two methods are the same, our combined NMR and molecular docking results consistently indicate two binding modes for both inhibitors.
Since the binding energies of the two poses calculated in the docking study are different from each other, our observation in NMR titrations, that the split signals are not sensitive to inhibitor or Mg2+ concentration, is somewhat unexpected. If the two forms observed by these two methods indicate the same forms, this observation may be due to an equilibrium between the two inhibitor binding modes that is established at a slow exchange rate (< 20 ms, assessed from the chemical shift difference). In addition to such observation of two split signals, there are additional limitations in the data analysis: since Mg2+ is a weak binder to RNH, the exact stoichiometry on inhibitor-free RNH is unknown;45, 50 titration with YLC2-155, that has limited solubility, did not exhibit the saturation (Figure 5). Thus, we estimated the dissociation constant (K2 in Equation 4) of inhibitor binding using a ternary RNH-2Mg2+-inhibitor model (Equation 1 and 2), rather than testing multiple inhibitor binding models. Even using the simple model, a lower K2 was observed for ZW566-RNH interaction than for YLC2-155, consistent with the higher inhibition activity of ZW566.34, 51–53
Molecular modeling studies have demonstrated that selective interactions of inhibitors with conserved residues in RNH could promote effective targeting and thus improve the inhibitory potency.13, 35, 36, 58 Our molecular docking results show multiple additional direct contacts between RNH and ZW566 compared to YLC2-155, including those with some highly conserved residues (Figure 8 and S10). These additional interactions could bring about higher thermal stability of RNH and higher binding affinity of inhibitor and make ZW566 a more potent inhibitor than YLC2-155. The relative populations of two binding forms in ZW566 binding are always the same (Figure 4); however, different interaction patterns with RNH residues were observed for the two binding orientations (Figure 7). Selection of one conformation might be a good starting point for further optimization.
Importantly, addition of excess inhibitor mostly shifted the free:bound equilibrium toward the inhibitor-bound form whereas the shift upon addition of excess Mg2+ was less (Figure 5). The titration data also indicate that ZW566 binding is more sensitive to Mg2+ concentration than YLC2-155 binding. These findings motivated us to more thoroughly inspect metal coordination by each inhibitor. We utilized quantum mechanics calculations to accurately assess the charge distribution and molecular orbits of the two inhibitors. Larger overall negative charges on the metal chelating atoms of ZW566 and its lower ALIE suggest a stronger coordination of Mg2+ by ZW566 compared to YLC2-155. Collectively, these results suggest that the metal-inhibitor interaction, in addition to the RNH-inhibitor interaction, plays an important role in known RNH-metal-inhibitor interaction systems.
In summary, the current study, using NMR, molecular docking, and quantum mechanical calculations, investigated factors that affect the inhibitor interaction. Our data consistently indicate stronger binding of ZW566 than YLC2-155 to RNH, probably due to the higher metal chelating ability of ZW566 and its greater number of interactions with the protein, compared to YLC2-155. Both inhibitors bind RNH in two distinct binding modes, with one conformation more favorable than the other. In addition, one of the two binding modes of ZW566, when compared to the analogous binding mode of YLC2-155, shows a greater number of interactions with surrounding residues. A focus on improvements in both metal chelating strength and favorable protein-inhibitor binding modes could be a strategy for the development of more potent RNH active-site inhibitors.
MATERIALS AND METHODS
Protein preparation.
The isolated RNH domain (amino acids 427-560 of HIV-RT) was produced as described previously.54 Briefly, the coding sequence was inserted into pE-SUMO vector that coded for an N-terminal six histidine (His6) tag followed by a SUMO-fusion tag. Protein expression was induced using IPTG in Escherichia coli Rosetta 2 (DE3) cells. The His6-SUMO-fusion protein was purified from the clarified lysate using a HisTrap HP column (GE Healthcare), followed by gel filtration on a HiLoad Superdex 75 26/60 column (GE Healthcare). The fusion protein was then incubated with histidine-tagged ubiquitin-like-protein specific protease (ULP1), and the uncleaved protein, as well as the ULP1, was separated from the cleaved protein with a second passage over the HisTrap HP column. The cleaved RNH domain was collected in flow-through fraction and further purified over the HiLoad Superdex 75 26/60 column. Monomer fractions were collected, concentrated, and stored at −80 °C.
Inhibitors.
YLC2-155 and ZW566 were synthesized as previously described.34, 51, 52 Inhibitors were dissolved in DMSO at a concentration of 50 mM for YLC2-155 and 25 mM for ZW566. The stock solutions were stored at −20 °C.
NMR samples.
All NMR samples were prepared in 20 mM Bis-Tris buffer (pH 7.0) containing 5% D2O. Since we previously found that nonspecific interactions of Mg2+ ions with RNH was affected by NaCl concentration,49 5 M NaCl and 320 mM MgCl2 stock solutions in 20 mM Bis-Tris buffer (pH 7.0) were used to make constant Cl- (50 mM) and various Mg2+ concentrations. All titration samples were made independently. For inhibitor titrations, inhibitor was added to 50 μM 15N-labeled RNH in the presence of 20 mM Mg2+. 1H-15N heteronuclear single quantum coherence (HSQC) spectra were recorded at 1:0, 1:0.5, 1:1, 1:2, 1:4 RNH:YLC2-155 ratios, and 1:0.5, 1:1, 1:2, 1:3 RNH:ZW566 ratios. For Mg2+ titrations, 1H-15N HSQC data were collected for 50 μM 15N-labeled RNH in the presence of 50 μM inhibitor and various concentrations of Mg2+ (0, 1, 5, 10, and 20 mM). 100 μM 13C, 15N-labeled RNH samples in the presence of 20 mM Mg2+ were used for 3D data collection, and an excess of inhibitor was added to saturate the interaction for inhibitor bound samples.
NMR spectroscopy.
All NMR data were recorded at 293 K on Bruker Avance spectrometers, equipped with triple-resonance, z-axis gradient cryo-probes. Data for signal assignments were recorded on 700 and 900 MHz spectrometers. 2D 1H-15N HSQC and 3D HNCA experiments were performed for backbone-assignments of inhibitor-free and bound 13C, 15N-labeled RNH samples in the presence of 20 mM Mg2+. To further confirm some of the assignments, 3D HNCACB data was collected for the ZW566-bound sample in the presence of 20 mM Mg2+, using non-uniform sampling to improve the resolution.59 All titration experiments were done on 800 MHz spectrometer. Spectra were processed using NMRPipe and istHMS, and analyzed with CcpNmr and Sparky.60–63 The chemical shifts of RNH in the YLC2-155 and ZW566 bound forms, in the presence of 20 mM Mg2+, have been deposited in the Biological Magnetic Resonance Data Bank with accession number 27825 and 27826.
Titration data analysis.
Relative intensities were normalized with respect to the inhibitor-free signals, and plotted as a function of Mg2+ concentration or inhibitor:RNH ratios. For both titrations, the decrease in intensity of inhibitor-free signals and the increase of inhibitor-bound signals were used to determine the apparent dissociation constant (K) of inhibitor binding at the RNH active site, using Matlab software (Mathworks). The fit curves for titration data were derived on a two-step pathway:
| (1) |
| (2) |
where P, M, and I stand for RNH, Mg2+, and inhibitor, and PM and PMI refer to Mg2+-bound RNH and RNH-Mg2+-inhibitor ternary complex, respectively. Dissociation constants for the equilibria, K1 and K2, are defined by the following equations:
| (3) |
| (4) |
Here, [M]total and [I]total stand for the total concentration of Mg2+ and inhibitor for each titration, respectively. Total RNH concentration [P]total, equal to [P]+[PM]+[PMI], is constant throughout the titrations. K1 of 3.2 mM was used for fitting, which was previously determined using isothermal titration calorimetry.50 As experimental [PMI] was obtained from the normalized intensity of inhibitor-bound signals, K2 was optimized by minimizing the differences between experimental and calculated [PMI] as well as between [P]total and calculated [P]+[PM]+[PMI].
Differential Scanning Fluorimetry (DSF).
Fluorescence measurements were performed using a microplate reader (FluoDia T70, Photon Technology International, Edison, NJ). Samples were prepared at a concentration of 10 μM for RNH in 20 mM Bis-Tris buffer at pH 7.0, including 400 mM NaCl and 5× SYPRO Orange. Mg2+ was added to 1, 5, 10, and 20 mM to examine the influence of Mg2+ on the thermal stability of RNH. Inhibitors were added at different inhibitor:protein molar ratios, 0.5, 1, 2, and 4 for YLC2-155 and 0.5, 1, 2, 3 for ZW566, in the absence and presence of 20 mM Mg2+ to investigate the effect of inhibitor. 30 μl of sample was loaded to 96-well PCR plate (Bio-Rad, Hercules, CA). The excitation/emission wavelengths of 465 and 590 nm, respectively, were set for SYPRO Orange to measure fluorescence intensity. The temperature was increased from 25 to 75 °C at an increment of 1 °C. All measurements were performed in triple. The melting temperature was calculated using a Boltzmann equation.
Computational Modeling.
Preparation of Structures.
2D structures of YLC2-155 and ZW566 compounds were energy minimized and converted to 3D structures employing LigPrep™ module of Schrodinger Small Molecule Discovery Suite.64 Ionization states populated at pH 7 were also calculated by Epik algorithm embedded in LigPrep™. Possible YLC2-155 and ZW566 conformers were generated by advanced conformational search implemented in ConfiGen™ module. HIV-1 RT structure co-crystallized with YLC2-155 (PDB access code 5UV5) was used as a protein receptor.34 To add hydrogen atoms, minimize energy, and create appropriate protonation states of amino acid side chains to 5UV5 pdb structure, the Protein Preparation Wizard ™ module was used. Manganese ions present in 5UV5 structure were replaced by magnesium ions, and the entire structure was energy minimized.
Docking Experiments.
All obtained conformers were docked into RNH active center using Glide™ algorithm with standard precision. Docking grid was centered on co-crystallized YLC2-155 ligand. Metal coordination constrains were added upon grid preparation to enable metal-mediated ligand binding. Van der Waals radii of non-polar atoms were scaled by 0.8 scaling factor to account for some flexibility of protein backbone and amino acid side chains. Ligand Interaction Diagrams (LIDs) were automatically generated for selected docking poses using Maestro visualizing interface. Other protein graphics were generated using PyMol (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC).
Quantum Mechanical Calculations of the small molecules were carried out using Gaussian 16W quantum chemical suite of programs.65 Density Functional Theory level of approximation was used with correlation-exchange B3LYP functional and 6-311G** basis set.66–70 For each compound, YLC2-155 and ZW566, geometry of one lowest energy conformer was optimized at B3LYP/6-311G** level of theory. Geometry optimization was followed by vibrational analysis to ensure that obtained geometries represent minima on potential energy surface. Then, partial atomic charges and molecular orbital analysis, were calculated at B3LYP/6-311G** theory level. Partial atomic charges were calculated by three different methods: Natural Charge Analysis, Electrostatic Potential, and Mulliken Population Analysis as a part of Natural Bonding Orbital analysis incorporated in Gaussian 16W.71 Molecular orbital analysis were performed by computing orbital energies and populations of the small molecules. Obtained orbitals were used to calculate average local ionization energy (ALIE) which is a sum of orbital energies weighted by the orbital densities and provides an energetic measure of how easy or difficult it is to remove electrons from regions of the molecule.56, 72
Supplementary Material
Figure S1, Overlay of 1H-15N HSQC spectra of 50 μM 15N-labeled RNH in the absence and presence of active-site inhibitors in the absence of Mg2+;
Figure S2, Overlay of 1H-15N HSQC spectra of 50 μM 15N-labeled RNH in the absence and presence of active-site inhibitors in the presence of Mg2+;
Figure S3, 1H-15N HSQC spectra of 100 μM 13C/15N-labeled RNH recorded at 293 K in the presence of 20 mM Mg2+ in the absence of inhibitor, in the presence of YLC2-155 or ZW566, with the resonance assignments;
Figure S4, chemical shift difference between YLC2-155-bound and inhibitor-free RNH, between ZW566-bound and inhibitor-free RNH, and between YLC2-155- and ZW566-bound RNH in the presence of 20 mM Mg2+;
Table S1. Residues showing split signals upon inhibitor binding;
Figure S5, inhibitor titration to RNH in the presence of 20 mM Mg2+;
Figure S6, Mg2+ titration to RNH in the presence of inhibitor;
Figure S7, inhibitor (A) and Mg2+ (B) titration data analysis for proportions of two binding modes in YLC2-155 binding;
Figure S8, inhibitor (A) and Mg2+ (B) titration data analysis for proportions of two binding modes in ZW566 binding;
Table S2, calculated dissociation constants for residues in inhibitor titrations;
Table S3. Melting temperature of RNH upon Mg2+ and/or inhibitor binding;
Figure S9, obtained ZW566 poses by molecular docking study;
Figure S10, LID for YLC2-155 bound in Mode 1 and Mode 2.
Table S4, calculated orbital characters and energies (in eV) of frontier molecular orbitals of YLC2-155 and ZW566.
ACKNOWLEDGMENT
We especially acknowledge Michael A. Parniak who is now deceased, for his long-term support and encouragement of the work. We thank Karen Kirby for discussion, Teresa Brosenitsch for critical reading of the manuscript, and Michael Delk for NMR support.
Funding Sources
This research was supported in part by Chatham University, University of Pittsburgh, and the National Institutes of Health (AI100890 to SGS and ZW, and GM105401 to RI).
ABBREVIATIONS
- HIV
Human immunodeficiency virus
- RT
reverse transcriptase
- RNH
Ribonuclease H
- HID
hydroxyisoquinoline-dione
- HPD
hydroxypyrimidine-dione
- DSF
differential scanning fluorimetry
- CD
circular dichroism
- HSQC
heteronuclear single quantum coherence
- LID
Ligand Interaction Diagram
- ALIE
average local ionization energy
REFERENCES
- [1].Baltimore D (1970) RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature 226, 1209–1211. [DOI] [PubMed] [Google Scholar]
- [2].Temin HM, and Mizutani S (1970) RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature 226, 1211–1213. [DOI] [PubMed] [Google Scholar]
- [3].Temin HM (1993) Retrovirus variation and reverse transcription: abnormal strand transfers result in retrovirus genetic variation. Proc. Natl. Acad. Sci. U. S. A 90, 6900–6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wilhelm M, and Wilhelm FX (2001) Reverse transcription of retroviruses and LTR retrotransposons. Cell. Mol. Life Sci 58, 1246–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Eickbush TH, and Jamburuthugoda VK (2008) The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res. 134, 221–234. DOI: 10.1016/j.virusres.2007.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jacobo-Molina A, and Arnold E (1991) HIV reverse transcriptase structure-function relationships. Biochemistry 30, 6351–6356. [DOI] [PubMed] [Google Scholar]
- [7].Borkow G, Fletcher RS, Barnard J, Arion D, Motakis D, Dmitrienko GI, and Parniak MA (1997) Inhibition of the ribonuclease H and DNA polymerase activities of HIV-1 reverse transcriptase by N-(4-tert-butylbenzoyl)-2-hydroxy-1-naphthaldehyde hydrazone. Biochemistry 36, 3179–3185. DOI: 10.1021/bi9624696 [DOI] [PubMed] [Google Scholar]
- [8].Shaw-Reid CA, Munshi V, Graham P, Wolfe A, Witmer M, Danzeisen R, Olsen DB, Carroll SS, Embrey M, Wai JS, Miller MD, Cole JL, and Hazuda DJ (2003) Inhibition of HIV-1 ribonuclease H by a novel diketo acid, 4-[5-(benzoylamino)thien-2-yl]-2,4-dioxobutanoic acid. J. Biol. Chem 278, 2777–2780. DOI: 10.1074/jbc.C200621200 [DOI] [PubMed] [Google Scholar]
- [9].Budihas SR, Gorshkova I, Gaidamakov S, Wamiru A, Bona MK, Parniak MA, Crouch RJ, McMahon JB, Beutler JA, and Le Grice SF (2005) Selective inhibition of HIV-1 reverse transcriptase-associated ribonuclease H activity by hydroxylated tropolones. Nucleic Acids Res. 33, 1249–1256. DOI: 10.1093/nar/gki268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Takada K, Bermingham A, O’Keefe BR, Wamiru A, Beutler JA, Le Grice SF, Lloyd J, Gustafson KR, and McMahon JB (2007) An HIV RNase H inhibitory 1,3,4,5-tetragalloylapiitol from the African plant Hylodendron gabunensis. J. Nat. Prod 70, 1647–1649. DOI: 10.1021/np0702279 [DOI] [PubMed] [Google Scholar]
- [11].Bokesch HR, Wamiru A, Le Grice SF, Beutler JA, McKee TC, and McMahon JB (2008) HIV-1 ribonuclease H inhibitory phenolic glycosides from Eugenia hyemalis. J. Nat. Prod 71, 1634–1636. DOI: 10.1021/np8002518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chung S, Wendeler M, Rausch JW, Beilhartz G, Gotte M, O’Keefe BR, Bermingham A, Beutler JA, Liu S, Zhuang X, and Le Grice SF (2010) Structure-activity analysis of vinylogous urea inhibitors of human immunodeficiency virus-encoded ribonuclease H. Antimicrob. Agents Chemother 54, 3913–3921. DOI: 10.1128/AAC.00434-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Corona A, Di Leva FS, Thierry S, Pescatori L, Crucitti GC, Subra F, Delelis O, Esposito F, Rigogliuso G, Costi R, Cosconati S, Novellino E, Di Santo R, and Tramontano E (2014) Identification of Highly Conserved Residues Involved in Inhibition of HIV-1 RNase H Function by Diketo Acid Derivatives. Antimicrob. Agents Chemother 58, 6101–6110. DOI: 10.1128/Aac.03605-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Corona A, Desantis J, Massari S, Distinto S, Masaoka T, Sabatini S, Esposito F, Manfroni G, Maccioni E, Cecchetti V, Pannecouque C, Le Grice SFJ, Tramontano E, and Tabarrini O (2016) Studies on cycloheptathiophene-3-carboxamide derivatives as allosteric HIV-1 ribonuclease H inhibitors. ChemMedChem 11, 1709–1720. DOI: 10.1002/cmdc.201600015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ilina T, and Parniak MA (2008) Inhibitors of HIV-1 reverse transcriptase. Adv. Pharmacol 56, 121–167. DOI: 10.1016/S1054-3589(07)56005-9 [DOI] [PubMed] [Google Scholar]
- [16].Tramontano E, and Di Santo R (2010) HIV-1 RT-associated RNase H function inhibitors: recent advances in drug development. Curr. Med. Chem 17, 2837–2853. DOI: 10.2174/092986710792065045 [DOI] [PubMed] [Google Scholar]
- [17].Esposito F, and Tramontano E (2014) Past and future. Current drugs targeting HIV-1 integrase and reverse transcriptase-associated ribonuclease H activity: single and dual active site inhibitors. Antivir. Chem. Chemother 23, 129–144. DOI: 10.3851/IMP2690 [DOI] [PubMed] [Google Scholar]
- [18].Kohlstaedt LA, Wang J, Friedman JM, Rice PA, and Steitz TA (1992) Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science 256, 1783–1790. [DOI] [PubMed] [Google Scholar]
- [19].Clark AD, JacoboMolina A, Clark P, Hughes SH, and Arnold E (1995) Crystallization of human immunodeficiency virus type 1 reverse transcriptase with and without nucleic acid substrates, inhibitors, and an antibody fab fragment. Methods Enzymol. 262, 171–185. [DOI] [PubMed] [Google Scholar]
- [20].Ren J, Esnouf R, Garman E, Somers D, Ross C, Kirby I, Keeling J, Darby G, Jones Y, Stuart D, and Stammers D (1995) High resolution structures of HIV-1 RT from four RT-inhibitor complexes. Nat. Struct. Biol 2, 293–302. DOI: 10.1038/nsb0495-293 [DOI] [PubMed] [Google Scholar]
- [21].Jaeger J, Restle T, and Steitz TA (1998) The structure of HIV-1 reverse transcriptase complexed with an RNA pseudoknot inhibitor. EMBO J. 17, 4535–4542. DOI: 10.1093/emboj/17.15.4535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ren J, Nichols C, Bird LE, Fujiwara T, Sugimoto H, Stuart DI, and Stammers DK (2000) Binding of the second generation non-nucleoside inhibitor S-1153 to HIV-1 reverse transcriptase involves extensive main chain hydrogen bonding. J. Biol. Chem 275, 14316–14320. [DOI] [PubMed] [Google Scholar]
- [23].Pata JD, Stirtan WG, Goldstein SW, and Steitz TA (2004) Structure of HIV-1 reverse transcriptase bound to an inhibitor active against mutant reverse transcriptases resistant to other nonnucleoside inhibitors. Proc. Natl. Acad. Sci. U. S. A 101, 10548–10553. DOI: 10.1073/pnas.0404151101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ren J, Nichols CE, Chamberlain PP, Weaver KL, Short SA, and Stammers DK (2004) Crystal structures of HIV-1 reverse transcriptases mutated at Codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors. J. Mol. Biol 336, 569–578. DOI: 10.1016/j.jmb.2003.12.055 [DOI] [PubMed] [Google Scholar]
- [25].Morningstar ML, Roth T, Farnsworth DW, Smith MK, Watson K, Buckheit RW, Das K, Zhang WY, Arnold E, Julias JG, Hughes SH, and Michejda CJ (2007) Synthesis, biological activity, and crystal structure of potent nonnucleoside inhibitors of HIV-1 reverse transcriptase that retain activity against mutant forms of the enzyme. J. Med. Chem 50, 4003–4015. DOI: 10.1021/jm060103d [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Das K, Bauman JD, Clark AD Jr., Frenkel YV, Lewi PJ, Shatkin AJ, Hughes SH, and Arnold E (2008) High-resolution structures of HIV-1 reverse transcriptase/TMC278 complexes: strategic flexibility explains potency against resistance mutations. Proc. Natl. Acad. Sci. U. S. A 105, 1466–1471. DOI: 10.1073/pnas.0711209105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Tu X, Das K, Han Q, Bauman JD, Clark AD Jr., Hou X, Frenkel YV, Gaffney BL, Jones RA, Boyer PL, Hughes SH, Sarafianos SG, and Arnold E (2010) Structural basis of HIV-1 resistance to AZT by excision. Nat. Struct. Mol. Biol 17, 1202–1209. DOI: 10.1038/nsmb.1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kirschberg TA, Balakrishnan M, Squires NH, Barnes T, Brendza KM, Chen X, Eisenberg EJ, Jin W, Kutty N, Leavitt S, Liclican A, Liu Q, Liu X, Mak J, Perry JK, Wang M, Watkins WJ, and Lansdon EB (2009) RNase H active site inhibitors of human immunodeficiency virus type 1 reverse transcriptase: design, biochemical activity, and structural information. J. Med. Chem 52, 5781–5784. DOI: 10.1021/jm900597q [DOI] [PubMed] [Google Scholar]
- [29].Su HP, Yan Y, Prasad GS, Smith RF, Daniels CL, Abeywickrema PD, Reid JC, Loughran HM, Kornienko M, Sharma S, Grobler JA, Xu B, Sardana V, Allison TJ, Williams PD, Darke PL, Hazuda DJ, and Munshi S (2010) Structural basis for the inhibition of RNase H activity of HIV-1 reverse transcriptase by RNase H active site-directed inhibitors. J. Virol 84, 7625–7633. DOI: 10.1128/JVI.00353-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lansdon EB, Liu Q, Leavitt SA, Balakrishnan M, Perry JK, Lancaster-Moyer C, Kutty N, Liu X, Squires NH, Watkins WJ, and Kirschberg TA (2011) Structural and binding analysis of pyrimidinol carboxylic acid and N-hydroxy quinazolinedione HIV-1 RNase H inhibitors. Antimicrob. Agents Chemother 55, 2905–2915. DOI: 10.1128/AAC.01594-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Chung S, Himmel DM, Jiang JK, Wojtak K, Bauman JD, Rausch JW, Wilson JA, Beutler JA, Thomas CJ, Arnold E, and Le Grice SF (2011) Synthesis, activity, and structural analysis of novel alpha-hydroxytropolone inhibitors of human immunodeficiency virus reverse transcriptase-associated ribonuclease H. J. Med. Chem 54, 4462–4473. DOI: 10.1021/jm2000757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kankanala J, Kirby KA, Liu F, Miller L, Nagy E, Wilson DJ, Parniak MA, Sarafianos SG, and Wang Z (2016) Design, synthesis, and biological evaluations of hydroxypyridonecarboxylic acids as inhibitors of HIV reverse transcriptase associated RNase H. J. Med. Chem 59, 5051–5062. DOI: 10.1021/acs.jmedchem.6b00465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Tang J, Liu F, Nagy E, Miller L, Kirby KA, Wilson DJ, Wu B, Sarafianos SG, Parniak MA, and Wang Z (2016) 3-Hydroxypyrimidine-2,4-diones as selective active site inhibitors of HIV reverse transcriptase-associated RNase H: design, synthesis, and biochemical evaluations. J. Med. Chem 59, 2648–2659. DOI: 10.1021/acs.jmedchem.5b01879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kirby KA, Myshakina NA, Christen MT, Chen YL, Schmidt HA, Huber AD, Xi Z, Kim S, Rao RK, Kramer ST, Yang Q, Singh K, Parniak MA, Wang Z, Ishima R, and Sarafianos SG (2017) A 2-hydroxyisoquinoline-1,3-dione active-site RNase H inhibitor binds in multiple modes to HIV-1 reverse transcriptase. Antimicrob. Agents Chemother 61 DOI: 10.1128/AAC.01351-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Corona A, di Leva FS, Rigogliuso G, Pescatori L, Madia VN, Subra F, Delelis O, Esposito F, Cadeddu M, Costi R, Cosconati S, Novellino E, di Santo R, and Tramontano E (2016) New insights into the interaction between pyrrolyl diketoacids and HIV-1 integrase active site and comparison with RNase H. Antiviral Res. 134, 236–243. DOI: 10.1016/j.antiviral.2016.09.008 [DOI] [PubMed] [Google Scholar]
- [36].Poongavanam V, Corona A, Steinmann C, Scipione L, Grandi N, Pandolfi F, Di Santo R, Costi R, Esposito F, Tramontano E, and Kongsted J (2018) Structure-guided approach identifies a novel class of HIV-1 ribonuclease H inhibitors: binding mode insights through magnesium complexation and site-directed mutagenesis studies. Med. Chem. Commun 9, 562–575. DOI: 10.1039/c7md00600d [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Himmel DM, Maegley KA, Pauly TA, Bauman JD, Das K, Dharia C, Clark AD Jr., Ryan K, Hickey MJ, Love RA, Hughes SH, Bergqvist S, and Arnold E (2009) Structure of HIV-1 reverse transcriptase with the inhibitor beta-Thujaplicinol bound at the RNase H active site. Structure 17, 1625–1635. DOI: 10.1016/j.str.2009.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mueller GA, Pari K, DeRose EF, Kirby TW, and London RE (2004) Backbone dynamics of the RNase H domain of HIV-1 reverse transcriptase. Biochemistry 43, 9332–9342. DOI: 10.1021/bi049555n [DOI] [PubMed] [Google Scholar]
- [39].Keck JL, and Marqusee S (1995) Substitution of a highly basic helix/loop sequence into the RNase H domain of human immunodeficiency virus reverse transcriptase restores its Mn(2+)-dependent RNase H activity. Proc. Natl. Acad. Sci. U. S. A 92, 2740–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kern G, Handel T, and Marqusee S (1998) Characterization of a folding intermediate from HIV-1 ribonuclease H. Protein Sci. 7, 2164–2174. DOI: 10.1002/pro.5560071014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Chattopadhyay D, Finzel BC, Munson SH, Evans DB, Sharma SK, Strakalaitus NA, Brunner DP, Eckenrode FM, Dauter Z, Betzel C, and Einspahr HM (1993) Crystallographic analyses of an active HIV-1 ribonuclease H domain show structural features that distinguish it from the inactive form. Acta Crystallogr., Sect. D: Biol. Crystallogr 49, 423–427. DOI: 10.1107/S0907444993002409 [DOI] [PubMed] [Google Scholar]
- [42].Zheng X, Pedersen LC, Gabel SA, Mueller GA, DeRose EF, and London RE (2016) Unfolding the HIV-1 reverse transcriptase RNase H domain--how to lose a molecular tug-of-war. Nucleic Acids Res. 44, 1776–1788. DOI: 10.1093/nar/gkv1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Cirino NM, Cameron CE, Smith JS, Rausch JW, Roth MJ, Benkovic SJ, and Le Grice SF (1995) Divalent cation modulation of the ribonuclease functions of human immunodeficiency virus reverse transcriptase. Biochemistry 34, 9936–9943. DOI: 10.1021/bi00031a016 [DOI] [PubMed] [Google Scholar]
- [44].Fenstermacher KJ, and DeStefano JJ (2011) Mechanism of HIV reverse transcriptase inhibition by zinc formation of a highly stable enzyme-(primer-template) complex with profoundly diminished catalytic activity. J. Biol. Chem 286, 40433–40442. DOI: 10.1074/jbc.M111.289850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Cowan JA, Ohyama T, Howard K, Rausch JW, Cowan SML, and Le Grice SFJ (2000) Metal-ion stoichiometry of the HIV-1 RT ribonuclease H domain: evidence for two mutually exclusive sites leads to new mechanistic insights on metal-mediated hydrolysis in nucleic acid biochemistry. J. Biol. Inorg. Chem 5, 67–74. DOI: 10.1007/s007750050009 [DOI] [PubMed] [Google Scholar]
- [46].Grubbs RD (2002) Intracellular magnesium and magnesium buffering. Biometals 15, 251–259. DOI: 10.1023/A:1016026831789 [DOI] [PubMed] [Google Scholar]
- [47].Pari K, Mueller GA, DeRose EF, Kirby TW, and London RE (2003) Solution structure of the RNase H domain of the HIV-1 reverse transcriptase in the presence of magnesium. Biochemistry 42, 639–650. DOI: 10.1021/bi0204894 [DOI] [PubMed] [Google Scholar]
- [48].Yan J, Wu H, Tom T, Brodsky O, Maegley K (2011) Targeting divalent metal ions at the active site of the HIV-1 RNase H domain: NMR studies on the interactions of divalent metal ions with RNase H and its inhibitors. Am. J. Anal. Chem 2, 639–649. DOI: 10.4236/ajac.2011.26073 [DOI] [Google Scholar]
- [49].Karki I, Christen MT, Spiriti J, Slack RL, Oda M, Kanaori K, Zuckerman DM, and Ishima R (2016) Entire-dataset analysis of NMR fast-exchange titration spectra: a Mg2+ titration analysis for HIV-1 Ribonuclease H domain J. Phys. Chem. B. 120, 12420–12431. DOI: 10.1021/acs.jpcb.6b08323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Oda M, Xi Z, Inaba S, Slack RL, and Ishima R (2018) Binding thermodynamics of metal ions to HIV-1 ribonuclease H domain. J. Therm. Anal. Calorim DOI: 10.1007/s10973-018-7445-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Wu B, Tang J, Wilson DJ, Huber AD, Casey MC, Ji J, Kankanala J, Xie J, Sarafianos SG, and Wang Z (2016) 3-hydroxypyrimidine-2,4-dione-5-N-benzylcarboxamides potently inhibit HIV-1 integrase and RNase H. J. Med. Chem 59, 6136–6148. DOI: 10.1021/acs.jmedchem.6b00040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Kirby KA, Marchand B, Ong YT, Ndongwe TP, Hachiya A, Michailidis E, Leslie MD, Sietsema DV, Fetterly TL, Dorst CA, Singh K, Wang Z, Parniak MA, and Sarafianos SG (2012) Structural and inhibition studies of the RNase H function of xenotropic murine leukemia virus-related virus reverse transcriptase. Antimicrob. Agents Chemother 56, 2048–2061. DOI: 10.1128/AAC.06000-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Tang J, Vernekar SKV, Chen YL, Miller L, Huber AD, Myshakina N, Sarafianos SG, Parniak MA, and Wang Z (2017) Synthesis, biological evaluation and molecular modeling of 2-Hydroxyisoquinoline-1,3-dione analogues as inhibitors of HIV reverse transcriptase associated ribonuclease H and polymerase. Eur. J. Med. Chem 133, 85–96. DOI: 10.1016/j.ejmech.2017.03.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Slack RL, Spiriti J, Ahn J, Parniak MA, Zuckerman DM, and Ishima R (2015) Structural integrity of the ribonuclease H domain in HIV-1 reverse transcriptase. Proteins 83, 1526–1538. DOI: 10.1002/prot.24843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Eldridge MD, Murray CW, Auton TR, Paolini GV, and Mee RP (1997) Empirical scoring functions .1. The development of a fast empirical scoring function to estimate the binding affinity of ligands in receptor complexes. J. Comput.-Aided Mol. Des 11, 425–445. DOI: 10.1023/A:1007996124545 [DOI] [PubMed] [Google Scholar]
- [56].Schrödinger Release 2018-4: Glide S, LLC, New York, NY, 2018 (2018). [Google Scholar]
- [57].Chamorro E, and Duque-Norena M (2015) Understanding the Highly Varying pKa of Arylamines. A Perspective from the average local ionization condensed-to-atom framework. J. Phys. Chem. A 119, 8156–8162. DOI: 10.1021/acs.jpca.5b03252 [DOI] [PubMed] [Google Scholar]
- [58].Himmel DM, Myshakina NS, Ilina T, Van Ry A, Ho WC, Parniak MA, and Arnold E (2014) Structure of a dihydroxycoumarin active-site inhibitor in complex with the RNase H domain of HIV-1 reverse transcriptase and structure-activity analysis of inhibitor analogs. J. Mol. Biol 426, 2617–2631. DOI: 10.1016/j.jmb.2014.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Hyberts SG, Takeuchi K, and Wagner G (2010) Poisson-gap sampling and forward maximum entropy reconstruction for enhancing the resolution and sensitivity of protein NMR data. J. Am. Chem. Soc 132, 2145–2147. DOI: 10.1021/ja908004w [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, and Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293. [DOI] [PubMed] [Google Scholar]
- [61].Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, and Laue ED (2005) The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59, 687–696. DOI: 10.1002/prot.20449 [DOI] [PubMed] [Google Scholar]
- [62].Hyberts SG, Milbradt AG, Wagner AB, Arthanari H, and Wagner G (2012) Application of iterative soft thresholding for fast reconstruction of NMR data non-uniformly sampled with multidimensional Poisson Gap scheduling. J. Biomol. NMR 52, 315–327. DOI: 10.1007/s10858-012-9611-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Lee W, Tonelli M, and Markley JL (2015) NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31, 1325–1327. DOI: 10.1093/bioinformatics/btu830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].2018-4, R. S (2018) Schrödinger Suite 2018-4 Protein Preparation Wizard; Epik, Schrödinger, LLC, New York, NY, 2018; Impact, Schrödinger, LLC, New York, NY, 2018; Prime, Schrödinger, LLC, New York, NY, 2018; LigPrep, Schrödinger, LLC, New York, NY, 2018; ConfGen, Schrödinger, LLC, New York, NY, 2018; Glide, Schrödinger, LLC, New York, NY, 2018. [Google Scholar]
- [65].Frisch MJT, W. G; Schlegel HB; Scuseria GE; Robb MA; Cheeseman JR; Scalmani G; Barone V; Petersson GA; Nakatsuji H; Li X; Caricato M; Marenich AV; Bloino J; Janesko BG; Gomperts R; Mennucci B; Hratchian HP; Ortiz JV; Izmaylov AF; Sonnenberg JL; Williams-Young D; Ding F; Lipparini F; Egidi F; Goings J; Peng B; Petrone A; Henderson T; Ranasinghe D; Zakrzewski VG; Gao J; Rega N; Zheng G; Liang W; Hada M; Ehara M; Toyota K; Fukuda R; Hasegawa J; Ishida M; Nakajima T; Honda Y; Kitao O; Nakai H; Vreven T; Throssell K; Montgomery JA Jr.; Peralta JE; Ogliaro F; Bearpark MJ; Heyd JJ; Brothers EN; Kudin KN; Staroverov VN; Keith TA; Kobayashi R; Normand J; Raghavachari K; Rendell AP; Burant JC; Iyengar SS; Tomasi J; Cossi M; Millam JM; Klene M; Adamo C; Cammi R; Ochterski JW; Martin RL; Morokuma K; Farkas O; Foresman JB; Fox DJ (2016) Gaussian 16, Revision B.01, Gaussian, Inc., Wallingford CT. [Google Scholar]
- [66].Kohn W, and Sham LJ (1965) Self-consistent equations including exchange and correlation effects. Phys. Rev 140, 1133-&. DOI: 10.1103/PhysRev.140.A1133 [DOI] [Google Scholar]
- [67].Parr RG, and Yang W (1989) Density-functional theory of atoms and molecules, Oxford Univ. Press, Oxford. [Google Scholar]
- [68].Becke AD (1993) Density-functional thermochemistry .3. The role of exact exchange. J. Chem. Phys 98, 5648–5652. DOI: 10.1063/1.464913 [DOI] [Google Scholar]
- [69].Mclean AD, and Chandler GS (1980) Contracted Gaussian-basis sets for molecular calculations .1. 2nd row atoms, Z=11-18. J. Chem. Phys 72, 5639–5648. DOI: 10.1063/1.438980 [DOI] [Google Scholar]
- [70].Krishnan R, Binkley JS, Seeger R, and Pople JA (1980) Self-consistent molecular-orbital methods .20. Basis set for correlated wave-functions. J. Chem. Phys 72, 650–654. DOI: 10.1063/1.438955 [DOI] [Google Scholar]
- [71].Glendening ED, Reed AE, Carpenter JE, and Weinhold F NBO Version 3.1 [Google Scholar]
- [72].Politzer P, Murray JS, and Bulat FA (2010) Average local ionization energy: A review. J. Mol. Model. 16, 1731–1742. DOI: 10.1007/s00894-010-0709-5 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1, Overlay of 1H-15N HSQC spectra of 50 μM 15N-labeled RNH in the absence and presence of active-site inhibitors in the absence of Mg2+;
Figure S2, Overlay of 1H-15N HSQC spectra of 50 μM 15N-labeled RNH in the absence and presence of active-site inhibitors in the presence of Mg2+;
Figure S3, 1H-15N HSQC spectra of 100 μM 13C/15N-labeled RNH recorded at 293 K in the presence of 20 mM Mg2+ in the absence of inhibitor, in the presence of YLC2-155 or ZW566, with the resonance assignments;
Figure S4, chemical shift difference between YLC2-155-bound and inhibitor-free RNH, between ZW566-bound and inhibitor-free RNH, and between YLC2-155- and ZW566-bound RNH in the presence of 20 mM Mg2+;
Table S1. Residues showing split signals upon inhibitor binding;
Figure S5, inhibitor titration to RNH in the presence of 20 mM Mg2+;
Figure S6, Mg2+ titration to RNH in the presence of inhibitor;
Figure S7, inhibitor (A) and Mg2+ (B) titration data analysis for proportions of two binding modes in YLC2-155 binding;
Figure S8, inhibitor (A) and Mg2+ (B) titration data analysis for proportions of two binding modes in ZW566 binding;
Table S2, calculated dissociation constants for residues in inhibitor titrations;
Table S3. Melting temperature of RNH upon Mg2+ and/or inhibitor binding;
Figure S9, obtained ZW566 poses by molecular docking study;
Figure S10, LID for YLC2-155 bound in Mode 1 and Mode 2.
Table S4, calculated orbital characters and energies (in eV) of frontier molecular orbitals of YLC2-155 and ZW566.