Abstract
Glycation as a non-enzymatic protein modification is related to aging and chronic diseases, especially diabetes. Comprehensive analysis of protein glycation will aid in a better understanding of its formation mechanism and biological significance. In this work, we comprehensively investigated protein glycation in human cells (HEK293T, Jurkat, and MCF7 cells). The current results indicated that this non-enzymatic modification was not random, and protein at the extracellular regions and the nucleus were more frequently glycated. Systematic and site-specific analysis of glycated proteins allowed us to study the effect of the primary sequences and secondary structures of proteins on glycation. Furthermore, nearly every enzyme in the glycolytic pathway was found to be glycated and a possible mechanism was proposed. Many glycation sites were also previously reported as acetylation and ubiquitination sites, which strongly suggested that this non-enzymatic modification may disturb protein degradation and gene expression. The current results will facilitate further studies of protein glycation in biomedical and clinical research.
Keywords: Protein glycation, Comprehensive analysis, MS-based proteomics, HEK293T, Jurkat, MCF7 cells
Graphical Abstract
Introduction
Proteins can be modified with different molecules from small to large ones. It is well-known that protein post-translational modifications (PTMs) play extremely important roles in many biological events, including cell signaling, and the regulation of protein interactions, stability and degradation [1]. Many types of modifications, including glycosylation [2–4], phosphorylation [5–7], and methylation [8], are regulated by enzymes. However, some non-enzymatic modifications can take place in cells and their formation is based on the chemical reactivity of certain amino acid residues on proteins such as cysteine, lysine, and arginine. These non-enzymatic PTMs impact protein structures and activities, and are involved in many cellular processes [9, 10]. They are also related to human aging and diseases, especially chronic ones including cardiovascular diseases and diabetes. Therefore, it is of great importance to systematically study non-enzymatic PTMs, which will deepen our understanding of their critical roles in biological systems.
Glycation is one of non-enzymatic modifications, in which reducing sugars or sugar-derived metabolites are covalently attached to primary amine or guanidino groups of proteins through the Maillard reaction [11]. The reaction starts with the condensation between the carbonyl group of reducing sugars and the amine group of proteins to produce an unstable Schiff base, which can undergo an intramolecular rearrangement to form the Amadori product. The relatively stable Amadori on modified proteins can further undergo a series of reactions, including rearrangement and dehydration to form advanced glycation end products (AGEs) [12].
Protein glycation has recently attracted increased attention because it is closely associated with aging and diabetes, and could be a hallmark of other diseases, such as neurodegenerative, cardiovascular, and metabolic diseases [13–15]. Protein glycation is correlated with the concentration of glucose. In normal cells, glucose is tightly regulated in a narrow concentration range, while cancer cells take in more glucose for the glycolysis in order to provide sufficient energy and intermediates for their proliferation [16, 17], which could result in the difference of protein glycation from normal cells.
Despite its importance, systematic analysis of protein glycation is understudied because it is extraordinarily challenging due to the low abundances of many glycated proteins and the typical sub-stoichiometry of glycation. Antibody-based analysis, including immunoassay and Western blot, has provided valuable information on protein glycation [18, 19]. However, the low throughput and the issues related to antibodies, such as the non-specificity and high cost, restrict its applications. With the development of the instrument and computational technology in recent years, mass spectrometry (MS) has become a very powerful tool and provided a unique opportunity to study protein PTMs on a large scale [20–29]. However, due to the complexity of biological samples and the typically low stoichiometry of protein glycation, the enrichment of glycated proteins/peptides is indispensable before MS analysis. Boronate affinity enrichment, which relies on the reversible covalent interactions between the hydroxyl groups of the glycan and boronic acid under basic conditions, has been employed to enrich glycated proteins/peptides [11, 30, 31]. Recently, our group developed a method based on dendrimer-conjugated boronic acid derivative (DBA), which can dramatically enhance the interactions between glycans and boronic acid benefiting from synergistic interactions, to efficiently capture low-abundance N- and O-glycopeptides [32]. The synergistic interactions can also facilitate the enrichment of glycated peptides.
In this work, we systematically and site-specifically analyzed protein glycation in three types of human cells - Jurkat, HEK293T, and MCF7 cells. Several hundreds of glycated proteins were identified and the results indicated that this non-enzymatic modification was not entirely random. Proteins at the extracellular regions and the nucleus were more frequently glycated. The formation of protein glycation is related to protein sequences and secondary structures. Interestingly, almost all enzymes involved in the glycolytic pathway were glycated. In addition, we found that many glycation sites were also reported as the ubiquitination and acetylation sites, which showed that protein stability and the regulation of gene expression may be disturbed by protein glycation. Systematic analysis of protein glycation in human cells helps us have a better understanding of this non-enzymatic modification.
Experimental
Sample Preparation and MS Analysis
As discussed above, benefitting from synergistic interactions with glycans, the DBA method is highly effective to catch glycopeptides in complex biological samples. Using this method, we globally analyzed protein N-glycosylation in human and yeast cells and mouse brain tissues [32]. In addition, we applied the DBA method to analyze O-GlcNAcylated proteins in human cells. Normally there is no cis-diols in GlcNAc and glucose, and therefore the interactions between boronic acid and GlcNAc or glucose are weak. Because of multiple hydroxyl groups of these sugars participating in the synergistic interactions, the DBA method was also effective to enrich O-GlcNAcylated peptides for their global analysis.
In brief, HEK293T, Jurkat, and MCF7 cells were cultured, and after cell lysis, proteins were extracted. Proteins were digested with trypsin, and purified peptides were treated with PNGase F for the removal of N-glycans, and then we enriched glycopeptides with the DBA beads. Enriched glycopeptides were fractionated and analyzed using an online LC-MS/MS system. The resolution was set as 70,000 for full MS and 35,000 for MS2. The detailed experimental procedure is available in the previous paper [32]. The raw files for protein O-GlcNAcylation analysis may also contain much information about protein glycation. Therefore, benefitting from the raw files collected previously [32], here we performed further database search for protein glycation. Glycated proteins were analyzed using different bioinformatics methods, and a possible mechanism for the formation of protein glycation was proposed.
Database Search and Data Filtering
The raw files were first converted to mzXML formats and then were searched using SEQUEST (version 28) [33]. The spectra were searched against a database containing sequences of all human proteins (Homo sapiens) downloaded from UniProt. The following parameters were used for the peptide search: 10 ppm precursor mass tolerance; 0.025 Da fragment ion mass tolerance; fully digested with trypsin; up to three missed cleavages; variable modifications: oxidation of methionine (+15.9949 Da), glycation of lysine or arginine (+162.0528 Da); fixed modifications: carbamidomethylation of cysteine (+57.0214 Da). The raw files are publicly accessible at http://www.peptideatlas.org/PASS/PASS01344.
The target-decoy method was used to evaluate the false discovery rates (FDRs) of glycated peptide identifications [34]. Each sequence from a protein in the protein database (downloaded from Uniprot) was listed in both forward and reverse orders. The quality of glycated peptides was evaluated and controlled by linear discriminant analysis (LDA) integrating several parameters including Xcorr, charge state, and precursor mass accuracy. Peptides with fewer than seven amino acids were removed and the glycated peptide spectral matches were filtered to be less than 1% FDR. The dataset was restricted to only glycated peptides while determining the FDRs.
Glycation Site Localization
The possibility of glycation site localization was calculated based on the fragment ions using a probabilistic algorithm similar to Ascore that considers all possible glycation sites in a peptide and uses the presence of experimental fragment ions unique to each site [35]. The resulting ModScore indicates the possibility of the glycation site, and the sites with a ModScore >13 (P < 0.05) were considered to be well-localized.
Results and Discussion
Identification of Glycation Sites and Glycated Peptides from MCF7 Cells
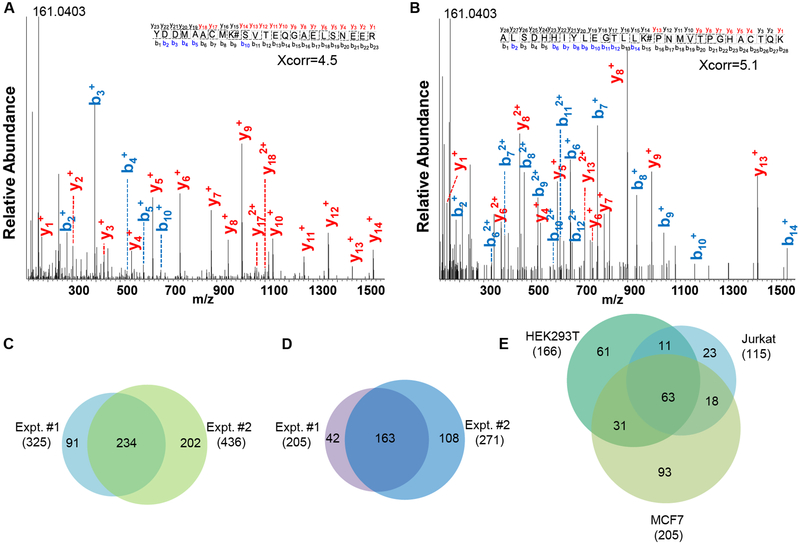
Orbitrap MS has become very powerful for bottom-up proteomics [36, 37] and both MS1 and MS2 were recorded in the Orbitrap cell with high resolution and high mass accuracy for glycated peptide identifications. Two examples of tandem mass spectra for glycated peptide identifications are shown in Figure 1A and 1B. The glycated peptide YDDMAACMK#SVTEQGAELSNEER (# represents the glycation site) from YWHAZ was identified with Xcorr of 4.5 and the mass accuracy of 0.66 ppm. The glycation site was well-localized at K27. Another example is ALSDHHIYLEGTLLK#PNMVTPGHACTQK, which was confidently identified with Xcorr of 5.1 and the mass accuracy of 0.47 ppm, and the modified site was well-localized at K230. The peptide is from fructose-bisphosphate aldolase A, which plays a key role in glycolysis and gluconeogenesis.
Figure 1.
Tandem mass spectra of YDDMAACMK#SVTEQGAELSNEER (# represents the glycation site) (A) and ALSDHHIYLEGTLLK#PNMVTPGHACTQK (B). Identification of glycation sites (C) and glycated proteins (D) from biologically duplicate experiments of MCF7 cells. Identification of glycated proteins from HEK293T, Jurkat, and MCF7 cells in the parallel experiments (E).
Like phosphorylated peptides, the neutral loss could happen for glycated peptides. Previously the pyrylium (loss of three water molecules) and furylium ions (loss of 3×H2O and HCHO) were reported under collision-induced collision (CID) [38]. When manually checking many tandem mass spectra, we barely found these fragments related to the water loss. Instead, some fragments in the low mass range appeared in almost every MS2 spectrum that we checked, such as the fragments at m/z of 161.0403, 136.0752, and 110.0717 Da. The peak at m/z of 161.0403 may be from the glucose residue, which was cleaved from glycated peptides. Other peaks may also be related to the glucose residue, and remain to be investigated. The difference of the neutral loss may be contributed to the different activation methods. In this work, we used higher-energy collision dissociation (HCD) to fragment peptides, and although HCD is also a type of CID, it can produce different fragments. It has been well-documented that the neutral loss from glycopeptides may be different with different activation methods [39, 40]. Different neutral losses were observed for glycopeptides even with varying normalized collision energies (NCE) under HCD [40].
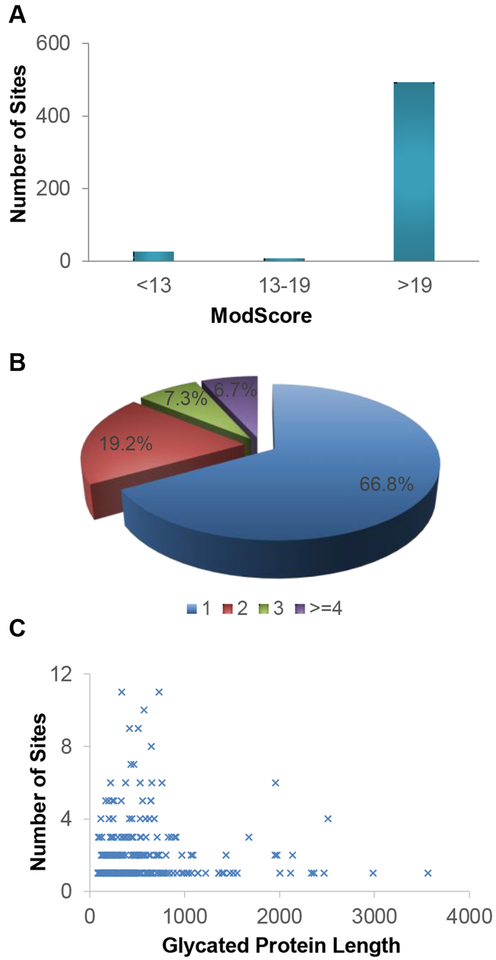
In biological duplicate experiments in MCF7 cells, we identified 325 glycation sites on 205 proteins and 436 sites on 271 glycated proteins, respectively, and all identified sites are listed in Table S1. The overlap of glycation sites and proteins (234 and 163) is reasonably good between the biological duplicate experiments ((Figures 1C and 1D), which indicates that protein glycation, even though it is a non-enzymatic modification, did not occur randomly. Here, ModScore was used to evaluate the possibility of glycation site localization. Of all the identified glycation sites from MCF7 cells, the ModScore values of most sites (94.7%) were larger than 13, which was considered well-localized, and over 93% had a ModScore larger than 19 (corresponding to a P value of less than 0.01) (Figure 2A).
Figure 2.
(A) The ModScore distribution for the identified glycation sites in MCF7 cells. (B) The number of glycation sites identified per glycated protein. (C) The relationship between the number of glycation sites and the length of glycated proteins.
For the identified glycated proteins, most of them (66.8%) carried only one glycation site and 20 proteins had more than three sites (Figure 2B). For example, 10 glycation sites were identified on heat shock protein 60, which is a mitochondrial chaperonin responsible for transporting and refolding of proteins from the cytoplasm into the mitochondria. This protein was reported to be correlated with diabetes, cancer, and immunological disorders [41]. Another heat shock protein, HSP90A, which assists protein folding, transport, maintenance, degradation, and cell signaling [42], was found to possess 8 glycation sites. We also attempted to investigate the relationship between the protein length and the number of glycation sites, and could not find obvious correlation between them (Figure 2C). However, it seemed that proteins with more glycation sites (larger than 3) were likely to be shorter (fewer than 1000 amino acids). Moreover, we evaluated the effect of protein abundance on glycation. The comparison between the abundance distribution of the glycated proteins identified here and all proteins from an online database (PaxDb) [43] is displayed in Figure S1. The result demonstrated that protein glycation was not biased for highly abundant proteins.
Identification of Glycated Peptides in HEK293T, Jurkat, and MCF7 Cells
Protein glycation has been reported to be related to chronic diseases such as diabetes [44]. Glycated proteins in human plasma and erythrocytes were investigated previously because of their association with the glucose concentration in the blood. For example, Zhang et al. analyzed protein glycation in normal and diabetic plasma and erythrocytes, and found that several amino acid residues including alanine (A), valine (V), and glutamic acid (E) appeared more frequently in the vicinity of the glycated lysine residues [31]. Keilhauer et al. analyzed protein glycation in the HeLa lysate and surprisingly found that over 50% protein glycation sites were modified on arginine (R) instead of lysine (K). In addition, they also concluded that HCD fragmentation is well-suited for analyzing glycated peptides [45].
In this work, we systematically studied protein glycation in three different types of human cells. The results about the glycated proteins identified in each cell line are displayed in Figure 1E. We identified 166, 115 and 205 glycated proteins from HEK293T, Jurkat, and MCF7 cells, respectively (Table S2), and 123 proteins were identified in at least two types of cells. We also investigated the distribution of the glycation sites on lysine (K) and arginine (R) in the three cell lines and found that nearly all the identified sites were on lysine and only 4, 1 and 3 glycation sites were identified on arginine (R) from HEK293T, Jurkat, and MCF7 cells, respectively (Figure S2).
The glycated proteins identified in the three cell lines were clustered using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) [46]. Proteins located at extracellular vesicles were the mostly highly enriched with a P value of 1.2×10−37, 1.2×10−40, 1.2×10−60 for Jurkat, HEK293T, and MCF7 cells, respectively (Figure S3A). Some examples of the glycated proteins located at the extracellular vesicles are in Table 1. Since the glucose concentration in the standard culture media (4.5 mg/L in DMEM for HEK293T and MCF7 cells, and 2 mg/L in RPMI 1640 for Jurkat cells) are higher than the physiological concentration in the body (0.75 mg/L) [47], it is reasonable that proteins located at the extracellular region are more likely to react with glucose in the media. Meanwhile, proteins located at membrane-bounded organelle, nucleus, and ribonucleoprotein complex were also highly enriched, which might be due to the fact that proteins in those regions were long-lived and/or were exposed to activated glucose-derived metabolites [48, 49]. All glycated proteins were also categorized according to biological process using DAVID. The highly enriched categories included protein localization to organelle, protein folding, and translation, which indicated that protein glycation might affect these biological processes (Figure S3B).
Table 1.
Examples of identified glycation sites and glycated proteins located at the extracellular vesicles
| UniProt ID | Gene | Site | ModScore | Peptidea | ppm | Xcorr | Annotation |
|---|---|---|---|---|---|---|---|
| P09382 | LEG1 | 64 | 1000 | FNAHGDANTIVCNSK# DGGAWGTEQR | −0.36 | 2.45 | Galectin-1 |
| P06396 | GELS | 277 | 1000 | VHVSEEGTEPEAMLQV LGPK#PALPAGTEDTAK | 1.52 | 4.95 | Gelsolin |
| P19256 | LFA3 | 62 | 1000 | DK#VAELENSEFR | −0.58 | 1.72 | CD58 |
| P25705 | ATPA | 305 | 1000 | DNGK#HALIIYDDLSK | 0.79 | 2.72 | TP synthase subunit alpha |
| 504 | 1000 | ITK#FENAFLSHWSQH QALLGTIR | −1.59 | 3.02 | |||
| P31939 | PUR9 | 66 | 1000 | VK#TLHPAVHAGILAR | −0.01 | 2.46 | Bifunctional purine biosynthesis protein |
| P69905 | HBA | 17 | 1000 | AAWGK#VGAHAGEYG AEALER | −0.40 | 2.71 | Hemoglobin subunit alpha |
| 41 | 1000 | MFLSFPTTK#TYFPHFD LSHGSAQVK | 0.77 | 2.48 |
#-glycation site.
Protein Glycation and Protein Structure
Most studies investigated protein glycation on single or several proteins, while large-scale analysis may provide us more valuable information such as the site preference of glycation. Protein glycation may be attributed to many factors [50]. The type of amino acids near lysine is highly important for glycation. Hydrophobic and acidic amino acids including alanine (A), leucine (L), and glutamic acid (E) frequently appear around the glycated lysine residues. On the contrary, histidine (H) and cysteine (C) seem not to be among the major amino acids in the neighborhood of the glycated lysine [31, 51, 52]. In addition, it has been proposed that pKa values and the microenvironment caused by the 3D structure near the lysine residue may also play important roles in the lysine glycation [53]. However, it should be noted that the results from different reports may contradict with each other [54, 55], which further complicates the understanding of protein glycation.
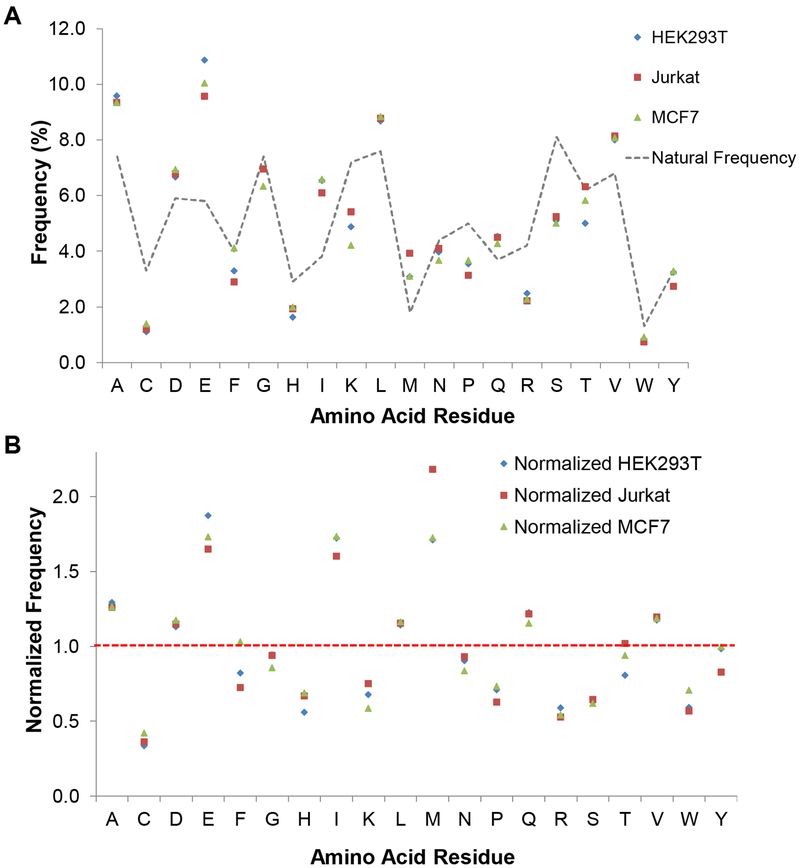
Here we studied the relationship between protein glycation and protein structure based on the results from the three cell lines. First, we calculated the frequency of five amino acids flanking each side of the glycated lysine. The frequency of each amino acid is displayed in Figure 3A and is further normalized by its corresponding natural abundance (Figure 3B). Glutamic acid (E), isoleucine (I), and methionine (M) were found to be the most frequent near the glycated lysine, and compared to their natural abundances, the frequencies of all three amino acids increased by over 60% in the three types of cells. In the current results, acidic and hydrophobic amino acids were found to promote protein glycation, while cysteine (C) and basic amino acids (histidine (H), lysine (K), and arginine (R)) were underrepresented in the proximity of the glycation sites (Figure 3B and Figure S4).
Figure 3.
Frequency (A) and normalized frequency (B) of amino acid residues near the identified lysine glycation sites (five residues flanking the central lysine).
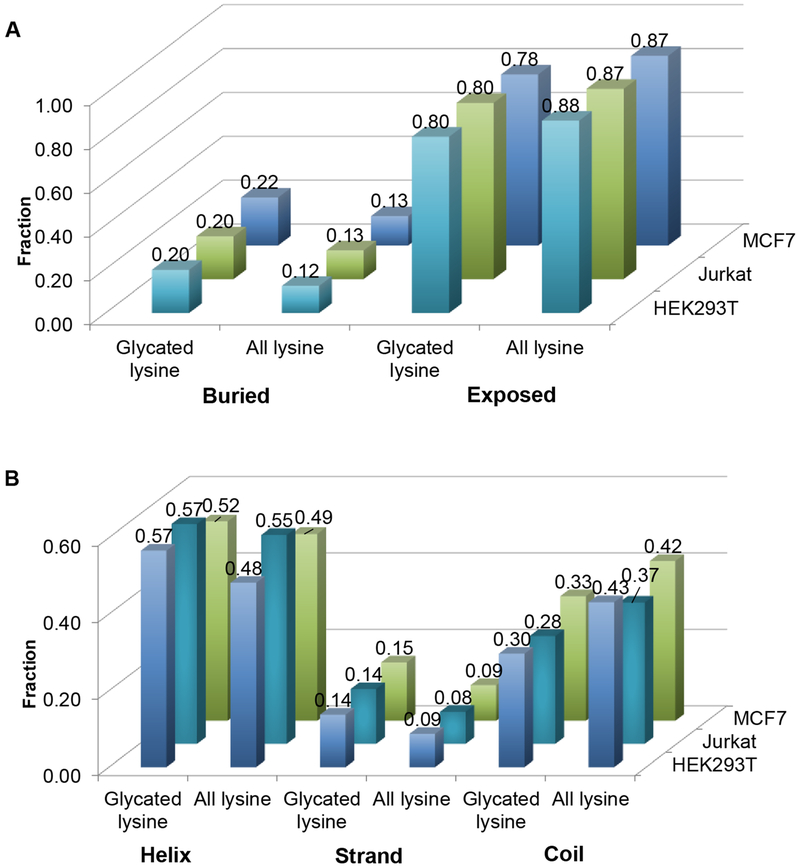
Next, we investigated the effect of protein structure on glycation. NetsurfP [56] was employed to predict the solvent accessibility of the glycated lysine residues and all the lysine residues from the glycated proteins identified here and the results are shown in Figure 4A. It is reasonable that most of the glycated lysine residues (around 80% for all the three cell lines) are exposed to solvent. Interestingly, 20%, 20% and 22% of the glycated lysine residues from HEK293T, Jurkat, and MCF7 cells were buried in the proteins, and these numbers increased by 8%, 7% and 9% compared to the fractions of all the buried lysine residues from the identified glycated proteins. One possibility was that some proteins might not fold properly or be unfolded under certain conditions, which made the buried lysine residues be glycated more easily. Furthermore, the protein structures are dynamic, which makes the buried residues accessible to solvent and glucose under certain conditions.
Figure 4.
Distribution of the solvent accessibility (A) and predicted structure (B) of each glycated lysine site and all lysine residues from the glycated proteins identified in HEK293T, Jurkat, and MCF7 cells.
Glycation may result in local distortion of proteins and change the overall protein structures. For human serum albumin (HSA), glycation leads to a higher propensity for the formation of β-sheet, which causes protein aggregates eventually [57]. Here, we studied the location of the glycated lysine residues in the secondary structures of proteins. NetsurfP was also used to predict the secondary structures of the glycated proteins. Among all the lysine residues, most of them were found at the helix and coil structures while the percentage of lysine at the β-strand structure was the lowest (Figure 4B). This is consistent with the previous result that hydrophilic lysine less frequently occurred in the β-strand [58]. The trends were the same with those of the glycated lysine residues in the three cell lines. The fractions of the glycated lysine residues from HEK293T, Jurkat, and MCF7 in the β-strand were 0.14, 0.14 and 0.15, respectively, which were slightly higher than the fractions of all lysine residues (0.09, 0.08 and 0.09) from the identified glycated proteins (Figure 4B). Actually, the solvent accessibility among the three types of secondary structures is different and the amino acid residues from the β-strand are the least accessible to solvent compared to those in the helix and the coil [59]. Previously, it was believed that glycation may cause protein structure damage [57, 60], but the above results might indicate that proteins with “damaged” structures were easier to be further glycated, which may account for the increased fraction of the glycated lysine residues in the β-strand. These results further demonstrated that protein glycation was complicated.
Protein Glycation and Glycolysis
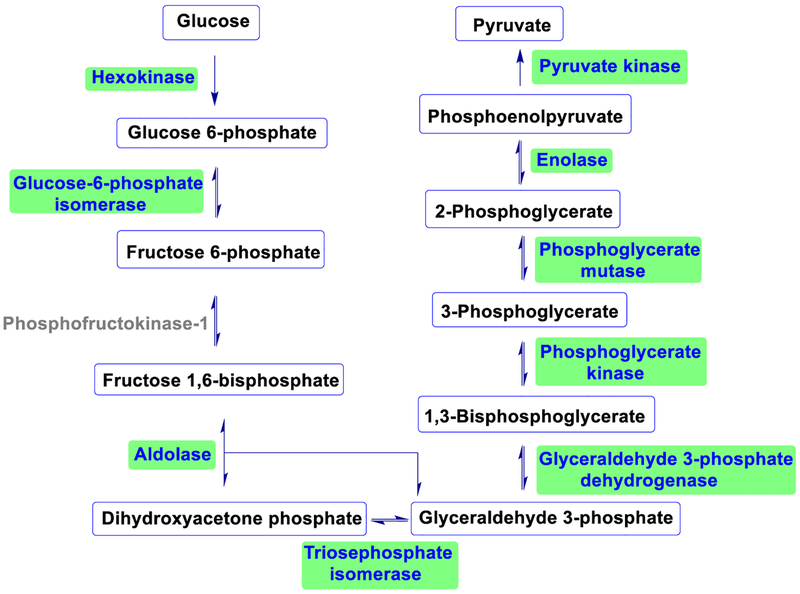
In cells, glucose participates in the glycolysis and can be converted to pyruvate with the energy released through sequential enzymatic transformations [61, 62]. This process generates carbonyl-containing intermediates, including glyceraldehyde-3-phosphate and dihydroxyacetone phosphate. They can be converted to methylglyoxal, which is reactive and may modify proteins. Therefore, an increase in the concentration of glycolytic aldehydes and methylglyoxal promotes protein non-enzymatic modifications. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is a very important enzyme in the glycolytic pathway, and it was reported that glycation on GAPDH would reduce its catalytic activity [63]. Methylglyoxal as a non-enzymatic modification reagent for GAPDH was also reported previously [64]. However, the investigation of glucose on glycating the enzymes in the glycolytic pathway is underrepresented.
We systematically analyzed the glycation of the enzymes involved in the glycolytic pathway using glucose as the glycation reagent, and surprisingly nine of the ten enzymes including GAPDH were glycated except phosphofructokinase-1 (Figure 5 and Table S3). In cells, due to its relatively low concentration, it is generally thought that glucose is not effective to glycate proteins [65]. However, we found that the majority of the glycolytic enzymes were glycated by glucose in cells. As discussed above, the frequencies of acidic amino acids near the glycated lysine residues were relatively high. We checked the glycated peptides from the glycolytic enzymes and also found that aspartic acid (D) and glutamic acid (E) occurred frequently in the proximity of the glycated lysine residues.
Figure 5.
Identified glycated proteins in the glycolytic pathway (highlighted).
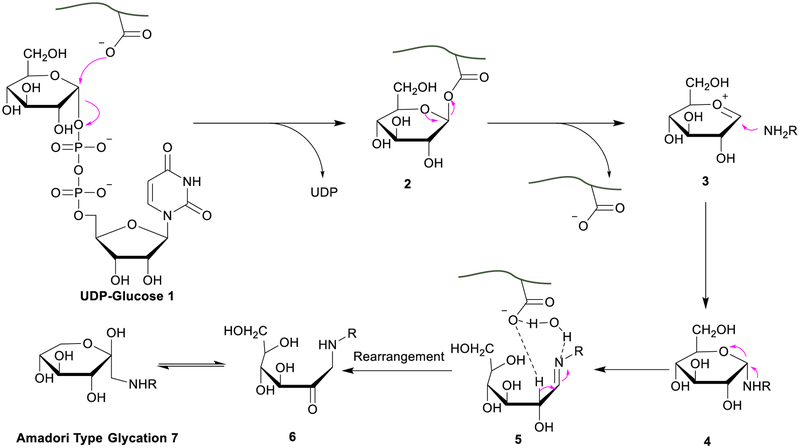
Here we proposed a possible mechanism to explain why nearly every enzyme in the glycolytic pathway was glycated (Figure 6). Uridine diphosphate glucose (UDP-glucose) is the activated form of glucose, participating in the glycolytic pathway, and the concentration of UDP-glucose normally is higher in the region where the glycolysis happens [66]. UDP-glucose may participate in the formation of protein glycation involved in the glycolytic pathway. A double displacement mechanism involving the formation of a covalent intermediate between an acidic residue and the substrate of glycosyltransferase was proposed previously [67, 68]. Similarly, UDP-glucose may be “hijacked” by an acidic amino acid near the lysine residue through an SN2 reaction [67]. Then the amine group of the nearby lysine residue could attack the “trapped” glucose to form a Schiff base. With the help of the proximity effect [69], glucose may become more reactive towards lysine. Another possible way was to generate an oxocarbenium cation-like species as a short-lived intermediate, which can be captured by lysine. The function of the acidic amino acid residues near lysine was to deprotonate and activate lysine [68].
Figure 6.
A proposed mechanism for protein glycation.
Another key step that the acidic amino acids may participate in is the rearrangement of the Schiff base, which is normally considered as the rate-determining step of the Maillard reaction. Theoretical calculation indicated that in aqueous solution, water (H2O) could form a hydrogen bond bridge between the α-hydrogen atom and the imine, which lowers the energy barrier of this step [70]. As shown in Figure 6, carboxylic acid may facilitate the hydrogen bond formation to lower the energy barrier and further promote the rearrangement. Integrating the two factors, the proposed mechanism may be able to account for why the enzymes in the glycolytic pathway were prone to protein glycation. Overall, these current results provide new insights into the glycation on the proteins involved in glycolysis.
Protein Glycation and Other Lysine Modifications (Acetylation and Ubiquitination)
PTMs regulate protein structures, spatial localizations and interactions, and thus control their activities. Over 400 PTMs are listed in UniProt database with acetylation, ubiquitination, and phosphorylation being most studied and common ones [71]. The crosstalk between different modifications has been recognized as an important way for the regulation of cellular events, and numerous studies of well-characterized proteins like p53 and tau have demonstrated that the PTM crosstalk regulates the activities of these proteins [72, 73].
Generally, the crosstalk may be divided into two categories, i.e. positive and negative forms [74]. In the positive crosstalk, one PTM may trigger the addition or removal of another modification, such as enhancing the binding affinity of the protein towards an enzyme catalyzing another modification. For the negative crosstalk, one modified group may compete against another group on a single site or prevent the modification of the protein through masking the binding site. Hart and colleagues studied the competitive crosstalk between O-phosphorylation and O-linked N-acetylglucosamine (O-GlcNAc) [75]. Recently it was reported that O-GlcNAcylation was also involved in the crosstalk with other PTMs such as acetylation and methylation [75, 76]. The PTM crosstalk of histone was reported to have a variety of PTMs involved, including acetylation, methylation, phosphorylation, and ubiquitination [77, 78], which tightly regulated gene expression. However, the correlation between protein glycation and other PTMs has not been systematically studied yet.
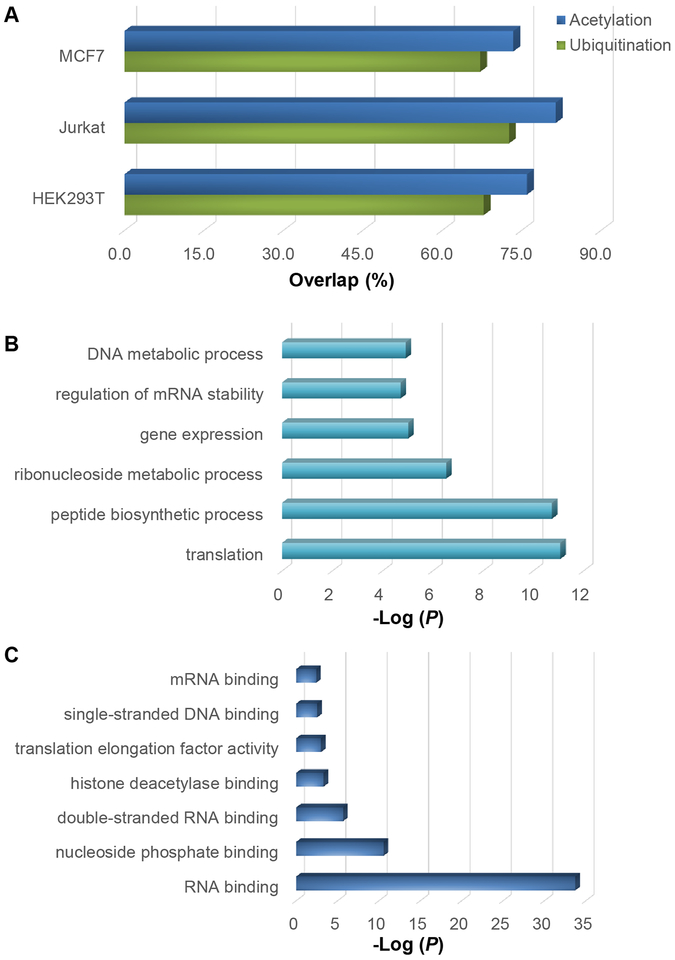
We studied the relationship between protein glycation and two most common Lys modifications, i.e. acetylation and ubiquitination. The ubiquitination sites were from previous reports [79, 80] and the acetylation sites were downloaded from Protein Lysine Modifications Database (PLMD) [81], in which the acetylation sites from the literature are archived. It was found that 67.9%, 72.6% and 67.2% of the identified glycation sites from HEK293T, Jurkat and MCF7 cells were the sites that were reported to be ubiquitinated (Figure 7A). This strongly suggested that glycation may prevent proteins from ubiquitination, and thus affect their degradation by proteasome.
Figure 7.
Overlap between the glycation sites identified here and the reported acetylation or ubiquitination sites (A). Protein clustering based on cellular component (B) and biological process (C) for the proteins with glycation and acetylation.
The overlaps between the identified glycation sites and the acetylation sites were even higher, i.e. 76.0%, 81.5% and 73.5% for HEK293T, Jurkat and MCF7 cells, respectively (Figure 7A), which indicated that glycation may disturb gene expression through the interference of protein acetylation. Gene Ontology (GO) analysis of the overlapped proteins showed a strong enrichment of biological processes associated with translation, gene expression, regulation of mRNA stability, and DNA metabolic process (Figure 7B). For molecular function analysis, proteins involved in RNA binding, histone deacetylase binding, and single-stranded DNA binding were highly enriched (Figure 7C). These results further supported that protein glycation may interfere gene expression and protein translation.
Conclusions
Compared to enzymatic modifications of proteins, non-enzymatic ones have been dramatically understudied. Glycation, as a non-enzymatic modification, is related to the development and progression of chronic diseases, especially diabetes. In this work, we systematically analyzed protein glycation in HEK293T, Jurkat, and MCF7 cells. Proteins at some cellular components such as the extracellular regions and the nucleus were more frequently glycated. Both protein primary sequences and secondary structures affected the formation of protein glycation. Interestingly, nearly every enzyme in the glycolytic pathway was glycated and a possible mechanism was proposed. In addition, we found that many glycation sites were also reported as the ubiquitination and acetylation sites, which strongly suggested that glycation may disturb protein degradation and the regulation of gene expression. The systematic analysis of glycated proteins provides valuable information for further clinical and biomedical investigation of protein glycation.
Supplementary Material
Acknowledgements
This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R01GM118803.
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References
- 1.Walsh CT, Garneau-Tsodikova S, Gatto GJ Jr.: Protein posttranslational modifications: the chemistry of proteome diversifications. Angew. Chem. Int. Ed 44, 734–7372 (2005) [DOI] [PubMed] [Google Scholar]
- 2.Wang XS, Yuan ZF, Fan J, Karch KR, Ball LE, Denu JM, Garcia BA: A Novel Quantitative Mass Spectrometry Platform for Determining Protein O-GlcNAcylation Dynamics. Mol. Cell. Proteomics 15, 2462–2475 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xiao H, Suttapitugsakul S, Sun F, Wu R: Mass Spectrometry-Based Chemical and Enzymatic Methods for Global Analysis of Protein Glycosylation. Acc. Chem. Res 51, 1796–1806 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen W, Smeekens JM, Wu R: Systematic and site-specific analysis of N-sialoglycosylated proteins on the cell surface by integrating click chemistry and MS-based proteomics. Chem. Sci 6, 4681–4689 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi TJ, Gao YQ, Gaffrey MJ, Nicora CD, Fillmore TL, Chrisler WB, Gritsenko MA, Wu CC, He JT, Bloodsworth KJ, Zhao R, Camp DG, Liu T, Rodland KD, Smith RD, Wiley HS, Qian WJ: Sensitive Targeted Quantification of ERK Phosphorylation Dynamics and Stoichiometry in Human Cells without Affinity Enrichment. Anal. Chem 87, 1103–1110 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang M, Wang Y, Zhao M, Wang F, Zhao C: Metabolic profiling analysis of fatty acids from hyperlipidemic rats treated with Gynostemma pentaphyllum and atorvastatin based on GC/MS. Anal. Methods 6, 8660–8667 (2014) [Google Scholar]
- 7.Wu R, Haas W, Dephoure N, Huttlin EL, Zhai B, Sowa ME, Gygi SP: A large-scale method to measure absolute protein phosphorylation stoichiometries. Nat. Methods 8, 677–683 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murn J, Shi Y: The winding path of protein methylation research: milestones and new frontiers. Nat. Rev. Mol. Cell Biol 18, 517–527 (2017) [DOI] [PubMed] [Google Scholar]
- 9.Holmstrom KM, Finkel T: Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat. Rev. Mol. Cell Biol 15, 411–421 (2014) [DOI] [PubMed] [Google Scholar]
- 10.Liu X, Long MJC, Aye Y: Proteomics and Beyond: Cell Decision-Making Shaped by Reactive Electrophiles. Trends Biochem. Sci 44, 75–89 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Q, Ames JM, Smith RD, Baynes JW, Metz TO: A perspective on the Maillard reaction and the analysis of protein glycation by mass spectrometry: probing the pathogenesis of chronic disease. J. Proteome Res 8, 754–769 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thornalley PJ, Battah S, Ahmed N, Karachalias N, Agalou S, Babaei-Jadidi R, Dawnay A: Quantitative screening of advanced glycation endproducts in cellular and extracellular proteins by tandem mass spectrometry. Biochem. J 375, 581–592 (2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Basta G, Schmidt AM, De Caterina R: Advanced glycation end products and vascular inflammation: implications for accelerated atherosclerosis in diabetes. Cardiovasc. Res 63, 582–592 (2004) [DOI] [PubMed] [Google Scholar]
- 14.Wautier JL, Schmidt AM: Protein glycation: a firm link to endothelial cell dysfunction. Circ. Res 95, 233–238 (2004) [DOI] [PubMed] [Google Scholar]
- 15.Yamagishi SI, Nakamura N, Matsui T: Glycation and cardiovascular disease in diabetes: A perspective on the concept of metabolic memory. J. Diabetes 9, 141–148 (2017) [DOI] [PubMed] [Google Scholar]
- 16.Vander Heiden MG, Cantley LC, Thompson CB: Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 324, 1029–1033 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fadaka A, Ajiboye B, Ojo O, Adewale O, Olayide I, Emuowhochere R: Biology of glucose metabolization in cancer cells. J. Oncol. Sci 3, 45–51 (2017) [Google Scholar]
- 18.Ikezaki H, Furusyo N, Ihara T, Hayashi T, Ura K, Hiramine S, Mitsumoto F, Takayama K, Murata M, Kohzuma T, Ai M, Schaefer EJ, Hayashi J: Glycated albumin as a diagnostic tool for diabetes in a general Japanese population. Metabolism. 64, 698–705 (2015) [DOI] [PubMed] [Google Scholar]
- 19.Kim J, Kim CS, Moon MK, Kim JS: Epicatechin breaks preformed glycated serum albumin and reverses the retinal accumulation of advanced glycation end products. Eur. J. Pharmacol 748, 108–114 (2015) [DOI] [PubMed] [Google Scholar]
- 20.Zhu Z, Desaire H: Carbohydrates on Proteins: Site-Specific Glycosylation Analysis by Mass Spectrometry. Annu. Rev. Anal. Chem 8, 463–483 (2015) [DOI] [PubMed] [Google Scholar]
- 21.Chen BF, Hwang L, Ochowicz W, Lin ZQ, Guardado-Alvarez TM, Cai WX, Xiu LC, Dani K, Colah C, Jin S, Ge Y: Coupling functionalized cobalt ferrite nanoparticle enrichment with online LC/MS/MS for top-down phosphoproteomics. Chem. Sci 8, 4306–4311 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simithy J, Sidoli S, Yuan ZF, Coradin M, Bhanu NV, Marchione DM, Klein BJ, Bazilevsky GA, McCullough CE, Magin RS, Kutateladze TG, Snyder NW, Marmorstein R, Garcia BA: Characterization of histone acylations links chromatin modifications with metabolism. Nat. Commun 8, 13 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greer SM, Sidoli S, Coradin M, Jesperser MS, Schwammle V, Jensen ON, Garcia BA, Brodbelt JS: Extensive Characterization of Heavily Modified Histone Tails by 193 nm Ultraviolet Photodissociation Mass Spectrometry via a Middle-Down Strategy. Anal. Chem 90, 10425–10433 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Witze ES, Old WM, Resing KA, Ahn NG: Mapping protein post-translational modifications with mass spectrometry. Nat. Methods 4, 798–806 (2007) [DOI] [PubMed] [Google Scholar]
- 25.Xiao H, Tang GX, Wu R: Site-Specific Quantification of Surface N-Glycoproteins in Statin-Treated Liver Cells. Anal. Chem 88, 3324–3332 (2016) [DOI] [PubMed] [Google Scholar]
- 26.Yue XS, Lukowski JK, Weaver EM, Skube SB, Hummon AB: Quantitative Proteomic and Phosphoproteomic Comparison of 2D and 3D Colon Cancer Cell Culture Models. J. Proteome Res 15, 4265–4276 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao HP, Wu RH: Quantitative investigation of human cell surface N-glycoprotein dynamics. Chem. Sci 8, 268–277 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao HP, Wu RH: Simultaneous Quantitation of Glycoprotein Degradation and Synthesis Rates by Integrating Isotope Labeling, Chemical Enrichment, and Multiplexed Proteomics. Anal. Chem 89, 10361–10367 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu Q, Wang BW, Chen ZW, Urabe G, Glover MS, Shi XD, Guo LW, Kent KC, Li LJ: Electron-transfer/higher-energy collision dissociation (EThcD)-enabled intact glycopeptide/glycoproteome characterization. J. Am. Soc. Mass Spectrom 28, 1751–1764 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Q, Tang N, Schepmoes AA, Phillips LS, Smith RD, Metz TO: Proteomic profiling of nonenzymatically glycated proteins in human plasma and erythrocyte membranes. J. Proteome Res 7, 2025–2032 (2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Q, Monroe ME, Schepmoes AA, Clauss TR, Gritsenko MA, Meng D, Petyuk VA, Smith RD, Metz TO: Comprehensive identification of glycated peptides and their glycation motifs in plasma and erythrocytes of control and diabetic subjects. J. Proteome Res 10, 3076–3088 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao H, Chen W, Smeekens JM, Wu R: An enrichment method based on synergistic and reversible covalent interactions for large-scale analysis of glycoproteins. Nat Commun. 9, 1692 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eng JK, McCormack AL, Yates JR: An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom 5, 976–989 (1994) [DOI] [PubMed] [Google Scholar]
- 34.Elias JE, Gygi SP: Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207 (2007) [DOI] [PubMed] [Google Scholar]
- 35.Beausoleil SA, Villén J, Gerber SA, Rush J, Gygi SP: A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol 24, 1285 (2006) [DOI] [PubMed] [Google Scholar]
- 36.Zubarev RA, Makarov A: Orbitrap mass spectrometry. Anal. Chem 85, 5288–5296 (2013) [DOI] [PubMed] [Google Scholar]
- 37.Chen WX, Smeekens JM, Wu RH: A universal chemical enrichment method for mapping the yeast N-glycoproteome by mass spectrometry (MS). Mol. Cell. Proteomics 13, 1563–1572 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frolov A, Bluher M, Hoffmann R: Glycation sites of human plasma proteins are affected to different extents by hyperglycemic conditions in type 2 diabetes mellitus. Anal. Bioanal. Chem 406, 5755–5763 (2014) [DOI] [PubMed] [Google Scholar]
- 39.Dodds ED: Gas-phase dissociation of glycosylated peptide ions. Mass Spectrom. Rev 31, 666–682 (2012) [DOI] [PubMed] [Google Scholar]
- 40.Cao L, Tolic N, Qu Y, Meng D, Zhao R, Zhang QB, Moore RJ, Zink EM, Lipton MS, Paga-Tolic L, Wu S: Characterization of intact N- and O-linked glycopeptides using higher energy collisional dissociation. Anal. Biochem 452, 96–102 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koll H, Guiard B, Rassow J, Ostermann J, Horwich AL, Neupert W, Hartl FU: Antifolding Activity of Hsp60 Couples Protein Import into the Mitochondrial Matrix with Export to the Intermembrane Space. Cell. 68, 1163–1175 (1992) [DOI] [PubMed] [Google Scholar]
- 42.Pearl LH, Prodromou C: Structure and in vivo function of Hsp90. Curr. Opin. Struct. Biol 10, 46–51 (2000) [DOI] [PubMed] [Google Scholar]
- 43.Wang M, Herrmann CJ, Simonovic M, Szklarczyk D, von Mering C: Version 4.0 of PaxDb: Protein abundance data, integrated across model organisms, tissues, and cell-lines. Proteomics. 15, 3163–3168 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paul RG, Bailey AJ: Glycation of collagen: the basis of its central role in the late complications of ageing and diabetes. Int. J. Biochem. Cell Biol 28, 1297–1310 (1996) [DOI] [PubMed] [Google Scholar]
- 45.Keilhauer EC, Geyer PE, Mann M: HCD Fragmentation of Glycated Peptides. J. Proteome Res 15, 2881–2890 (2016) [DOI] [PubMed] [Google Scholar]
- 46.Huang DW, Sherman BT, Lempicki RA: Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc 4, 44 (2008) [DOI] [PubMed] [Google Scholar]
- 47.Kratz A, Ferraro M, Sluss PM, Lewandrowski KB: Case records of the Massachusetts General Hospital. Weekly clinicopathological exercises. Laboratory reference values. N. Engl. J. Med 351, 1548–1563 (2004) [DOI] [PubMed] [Google Scholar]
- 48.Toyama BH, Savas JN, Park SK, Harris MS, Ingolia NT, Yates JR 3rd, Hetzer MW: Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell. 154, 971–982 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen W, Smeekens JM, Wu R: Systematic study of the dynamics and half-lives of newly synthesized proteins in human cells. Chem. Sci 7, 1393–1400 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johansen MB, Kiemer L, Brunak S: Analysis and prediction of mammalian protein glycation. Glycobiology. 16, 844–853 (2006) [DOI] [PubMed] [Google Scholar]
- 51.Bilova T, Paudel G, Shilyaev N, Schmidt R, Brauch D, Tarakhovskaya E, Milrud S, Smolikova G, Tissier A, Vogt T, Sinz A, Brandt W, Birkemeyer C, Wessjohann LA, Frolov A: Global proteomic analysis of advanced glycation end products in the Arabidopsis proteome provides evidence for age-related glycation hot spots. J. Biol. Chem 292, 15758–15776 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Venkatraman J, Aggarwal K, Balaram P: Helical peptide models for protein glycation: proximity effects in catalysis of the Amadori rearrangement. Chem. Biol 8, 611–625 (2001) [DOI] [PubMed] [Google Scholar]
- 53.Bunn HF, Shapiro R, McManus M, Garrick L, McDonald MJ, Gallop PM, Gabbay KH: Structural heterogeneity of human hemoglobin A due to nonenzymatic glycosylation. J. Biol. Chem 254, 3892–3898 (1979) [PubMed] [Google Scholar]
- 54.Sjoblom NM, Kelsey MMG, Scheck RA: A Systematic Study of Selective Protein Glycation. Angew. Chem. Int. Ed 57, 16077–16082 (2018) [DOI] [PubMed] [Google Scholar]
- 55.Ito S, Nakahari T, Yamamoto D: The structural feature surrounding glycated lysine residues in human hemoglobin. Biomed. Res 32, 217–223 (2011) [DOI] [PubMed] [Google Scholar]
- 56.Petersen B, Petersen TN, Andersen P, Nielsen M, Lundegaard C: A generic method for assignment of reliability scores applied to solvent accessibility predictions. BMC Struct. Biol 9, 51 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.GhoshMoulick R, Bhattacharya J, Roy S, Basak S, Dasgupta AK: Compensatory secondary structure alterations in protein glycation. Biochim. Biophys. Acta 1774, 233–242 (2007) [DOI] [PubMed] [Google Scholar]
- 58.Otaki JM, Tsutsumi M, Gotoh T, Yamamoto H: Secondary structure characterization based on amino acid composition and availability in proteins. J. Chem. Inf. Model 50, 690–700 (2010) [DOI] [PubMed] [Google Scholar]
- 59.Lins L, Thomas A, Brasseur R: Analysis of accessible surface of residues in proteins. Protein Sci 12, 1406–1417 (2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bouma B, Kroon-Batenburg LM, Wu YP, Brunjes B, Posthuma G, Kranenburg O, de Groot PG, Voest EE, Gebbink MF: Glycation induces formation of amyloid cross-beta structure in albumin. J. Biol. Chem 278, 41810–41819 (2003) [DOI] [PubMed] [Google Scholar]
- 61.Lunt SY, Vander Heiden MG: Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell. Dev. Biol 27, 441–464 (2011) [DOI] [PubMed] [Google Scholar]
- 62.Li XB, Gu JD, Zhou QH: Review of aerobic glycolysis and its key enzymes - new targets for lung cancer therapy. Thorac. Cancer 6, 17–24 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.He RQ, Yang MD, Zheng X, Zhou JX: Isolation and Some Properties of Glycated D-Glyceraldehyde-3-Phosphate Dehydrogenase from Rabbit Muscle. Biochem. J 309, 133–139 (1995) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee HJ, Howell SK, Sanford RJ, Beisswenger PJ: Methylglyoxal can modify GAPDH activity and structure. Ann. N. Y. Acad. Sci 1043, 135–145 (2005) [DOI] [PubMed] [Google Scholar]
- 65.Muronetz VI, Melnikova AK, Seferbekova ZN, Barinova KV, Schmalhausen EV: Glycation, Glycolysis, and Neurodegenerative Diseases: Is There Any Connection? Biochemistry (Mosc). 82, 874–886 (2017) [DOI] [PubMed] [Google Scholar]
- 66.Thoden JB, Wohlers TM, Fridovich-Keil JL, Holden HM: Human UDP-galactose 4-epimerase. Accommodation of UDP-N-acetylglucosamine within the active site. J. Biol. Chem 276, 15131–15136 (2001) [DOI] [PubMed] [Google Scholar]
- 67.Sheng F, Jia X, Yep A, Preiss J, Geiger JH: The Crystal Structures of the Open and Catalytically Competent Closed Conformation of Escherichia coli Glycogen Synthase. J. Biol. Chem 284, 17796–17807 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Breton C, Šnajdrová L, Jeanneau C, Koča J, Imberty A: Structures and mechanisms of glycosyltransferases. Glycobiology. 16, 29R–37R (2006) [DOI] [PubMed] [Google Scholar]
- 69.Yang B, Wu H, Schnier PD, Liu Y, Liu J, Wang N, DeGrado WF, Wang L: Proximity-enhanced SuFEx chemical cross-linker for specific and multitargeting cross-linking mass spectrometry. Proc. Natl. Acad. Sci. U. S. A 115, 11162–11167 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ren GR, Zhao LJ, Sun Q, Xie HJ, Lei QF, Fang WJ: Explore the reaction mechanism of the Maillard reaction: a density functional theory study. J. Mol. Model 21, 132 (2015) [DOI] [PubMed] [Google Scholar]
- 71.UniProt C: Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res. 40, D71–75 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bode AM, Dong Z: Post-translational modification of p53 in tumorigenesis. Nat. Rev. Cancer 4, 793–805 (2004) [DOI] [PubMed] [Google Scholar]
- 73.Kontaxi C, Piccardo P, Gill AC: Lysine-Directed Post-translational Modifications of Tau Protein in Alzheimer’s Disease and Related Tauopathies. Front. Mol. Biosci 4, 56 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hunter T: The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol. Cell 28, 730–738 (2007) [DOI] [PubMed] [Google Scholar]
- 75.Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O: Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem 80, 825–858 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sakabe K, Hart GW: O-GlcNAc transferase regulates mitotic chromatin dynamics. J. Biol. Chem 285, 34460–34468 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Latham JA, Dent SY: Cross-regulation of histone modifications. Nat. Struct. Mol. Biol 14, 1017–1024 (2007) [DOI] [PubMed] [Google Scholar]
- 78.Young NL, DiMaggio PA, Plazas-Mayorca MD, Baliban RC, Floudas CA, Garcia BA: High Throughput Characterization of Combinatorial Histone Codes. Mol. Cell. Proteomics 8, 2266–2284 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Akimov V, Barrio-Hernandez I, Hansen SVF, Hallenborg P, Pedersen AK, Bekker-Jensen DB, Puglia M, Christensen SDK, Vanselow JT, Nielsen MM, Kratchmarova I, Kelstrup CD, Olsen JV, Blagoev B: UbiSite approach for comprehensive mapping of lysine and N-terminal ubiquitination sites. Nat. Struct. Mol. Biol 25, 631–640 (2018) [DOI] [PubMed] [Google Scholar]
- 80.Udeshi ND, Svinkina T, Mertins P, Kuhn E, Mani DR, Qiao JW, Carr SA: Refined preparation and use of anti-diglycine remnant (K-epsilon-GG) antibody enables routine quantification of 10,000s of ubiquitination sites in single proteomics experiments. Mol. Cell. Proteomics 12, 825–831 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu Z, Wang Y, Gao T, Pan Z, Cheng H, Yang Q, Cheng Z, Guo A, Ren J, Xue Y: CPLM: a database of protein lysine modifications. Nucleic Acids Res. 42, D531–536 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.